Abstract
BACKGROUND. Human papillomavirus–related (HPV-related) oropharyngeal squamous cell carcinomas (OPSCCs) have an excellent response rate to platinum-based chemoradiotherapy. Genomic differences between primary HPV-related OPSCCs that do or do not recur are unknown. Furthermore, it is unclear if HPV-related OPSCCs that recur share a genomic landscape with HPV-negative head and neck cancers (HNCs).
METHODS. We utilized whole exome sequencing to analyze somatic nucleotide (SNVs) and copy number variants (CNVs) among a unique set of 51 primary HPV-related OPSCCs, including 35 that did not recur and 16 that recurred. We evaluated 12 metachronous recurrent OPSCCs (7 with paired primary OPSCCs) and 33 primary HPV-unrelated oral cavity and OPSCCs.
RESULTS.KMT2D was the most frequently mutated gene among primary HPV-related OPSCCs (n = 51; 14%) and among metachronous recurrent OPSCCs (n = 12; 42%). Primary HPV-related OPSCCs that recurred shared a genomic landscape with primary HPV-related OPSCCs that did not recur. However, TSC2, BRIP1, NBN, and NFE2L2 mutations occurred in primary OPSCCs that recurred but not in those that did not recur. Moreover, primary HPV-related OPSCCs that recur harbor features of HPV-unrelated HNCs, notably including MAPK, JAK/STAT, and differentiation signaling pathway aberrations. Metachronous recurrent OPSCCs shared a genomic landscape with HPV-unrelated HNCs, including a high frequency of TP53, CASP8, FAT1, HLA-A, AJUBA, and NSD1 genomic alterations.
CONCLUSION. Overall, primary HPV-related OPSCCs that recur share a genomic landscape with nonrecurrent OPSCCs. Metachronous recurrent OPSCCs share genomic features with HPV-negative HNCs. These data aim to guide future deescalation endeavors and functional experiments.
FUNDING. This study is supported by the American Cancer Society (RSG TBG-123653), funding support for RAH (T32DC00018, Research Training in Otolaryngology, University of Washington), funds to EM from Seattle Translational Tumor Research (Fred Hutchinson Cancer Research Center), and center funds from the Fred Hutchinson Cancer Research Center to EM. UD is supported by the Department of Veterans Affairs, Biomedical Laboratory Research and Development (BLR&D), grant IO1-oo23456, and funds from the Pittsburgh Foundation and PNC Foundation.
Keywords: Oncology
Keywords: Cancer, Head & neck cancer, Molecular diagnosis
Whole genomic sequencing identifies genomic alterations associated with recurrence in HPV-related oropharyngeal squamous cell carcinomas.
Introduction
As human papillomavirus (HPV) infects more than half of US adults (1), and HPV-related oropharyngeal squamous cell carcinoma (OPSCC) incidence is rising, there is a public health need to both prevent these tumors and optimize their treatment. HPV-related OPSCC is expected to surpass the incidence of cervical cancer by 2020 while HPV-unrelated (i.e., HPV-negative) head and neck cancers (HNCs) are decreasing in incidence (2). HPV-related OPSCCs respond favorably to concurrent platinum-based chemoradiation (3), and disease progression is rare (4, 5). Patients with recurrence fare poorly, with 40% mortality at 2-years after disease progression (5). Ongoing clinical investigation in this population is focused on deintensified curative-intent strategies (6). Illuminating genomic aberrations associated with recurrence may help us tailor treatment strategies.
Compared with HPV-unrelated HNC, HPV-related OPSCCs are often, but not always, characterized by a relative paucity of mutations (7). As opposed to carcinogen-related HNC, mutations in HPV-related OPSCCs are, in part, a function of impaired DNA damage response machinery (8). Furthermore, HPV16/18 oncoproteins E6 and E7 mediate inactivation (9–12) of p53 and pRb, contributing to genomic instability. HPV-oncoprotein–mediated genomic instability (13) further accelerates tumorigenesis. While HPV genomic integration is not necessary for tumorigenesis, preliminary data suggest viral integration into cancer-associated genes in some HPV-related OPSCCs (14, 15). Despite our current understanding of this pathogen-mediated cancer, the non-HPV genomic drivers associated with recurrence in this population have yet to be elucidated.
Existing genomic data from primary HPV-related OPSCCs that recur is sparse, including only 7 patients from The Cancer Genome Atlas (TCGA) (16) and 2 from a precision oncology study (17). In the latter study, data from 1 metachronous recurrent HPV-related tumor was analyzed. Given the stark survival differences between HPV-related and the more aggressive HPV-unrelated squamous cell carcinomas (3), we hypothesized that primary HPV-related OPSCC tumors from recurrent cases share genomic features with HPV-unrelated HNC. This includes a greater mutational burden, genomic instability, and a propensity for tumor suppressor inactivation (16, 18). Furthermore, we hypothesized that the mutational landscape of tumors in the recurrent group would harbor a preponderance of mutations in mitogenic signaling (e.g., PI3K, JAK/STAT, MAPK, receptor tyrosine kinase [RTK]), cell cycle, cell death, survival, and differentiation pathways, as observed in HPV-unrelated HNC in TCGA (16).
In this study, we assembled the largest-to-date set of HPV-related OPSCCs, to our knowledge, and a set of rare HPV-related OPSCCs that recurred, including both the primary and metachronous recurrent tumors. This is also the largest sampling of paired primary and matched metachronous recurrent HPV-related OPSCCs, to our knowledge. We sought insight to the following questions: (a) Do primary HPV-associated OPSCCs that do or do not recur differ in their genomic landscape? (b) Do primary OPSCCs that recur harbor features of HPV-negative tumors? (c) Do metachronous recurrent OPSCCs differ genomically from their matched index tumor? (d) Do metachronous recurrent OPSCCs share a genomic landscape with HPV-negative tumors?
Results
Patient baseline characteristics.
Clinical data from 84 patients from the University of Washington (UW), the University of Pittsburgh (UPitt), and TCGA were evaluated: 51 primary HPV-related OPSCCs (35 did not recur and16 recurred) and 33 primary HPV-unrelated (i.e., HPV-negative) oral cavity squamous cell carcinomas (OCSCCs) and OPSCCs (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.99327DS1). A total of 12 metachronous recurrent tumors were analyzed. Of the 16 primary HPV-related OPSCCs that recurred, we analyzed 7 paired metachronous recurrent HPV-related OPSCCs (5 from UPitt, 2 from UW). We analyzed 5 additional metachronous recurrent HPV-related OPSCCs for which we did not have a paired primary sample available (4 from UPitt, 1 from UW). Patients ranged from 19–83 years old. Patients tended to be over 55 years of age, especially those with HPV-unrelated tumors, and were predominantly male (Table 1). Based on the recently revised AJCC 8th edition staging manual (19), only 17% (n = 6 of 35) of primary HPV-related tumors without recurrence and 19% (n = 3 of 16) of the primary HPV-related tumors that recurred were advanced stage (stage 3). Notably, nearly one-half (49%, n = 17) of patients with HPV-related OPSCCs that did not recur had a history of tobacco use greater than 10 years compared with 31% (n = 5) of patients with HPV-related OPSCCs that recurred. There were no deaths among the HPV-related OPSCCs that did not recur, vs. 6 deaths among the primary HPV-related OPSCCs that recurred. Median overall survival (OS) was 20.4 months for the primary HPV-related OPSCCs that did not recur vs. 36.4 months for the primary HPV-related cases that recurred. HPV-unrelated HNCs had a median OS of 30 months.
Table 1. Baseline characteristics of patients with oropharyngeal squamous cell carcinoma according to recurrence.
Somatic mutations.
Tumors were included in genomic analyses based on whole exome sequencing (WES) quality control, tumor identity, and contamination analyses (Figure 1A and Supplemental Table 1). Variant calling and curation were performed as described in the Methods and Supplemental Figure 2. Mean fold tumor target coverage for WES data for our tumors was 98×, with 96% of target bases over 20× coverage (Supplemental Table 2 and Supplemental Figure 3A). Mean fold normal sample target coverage was 91×, with 96% of target bases above 20× coverage (Supplemental Table 2).
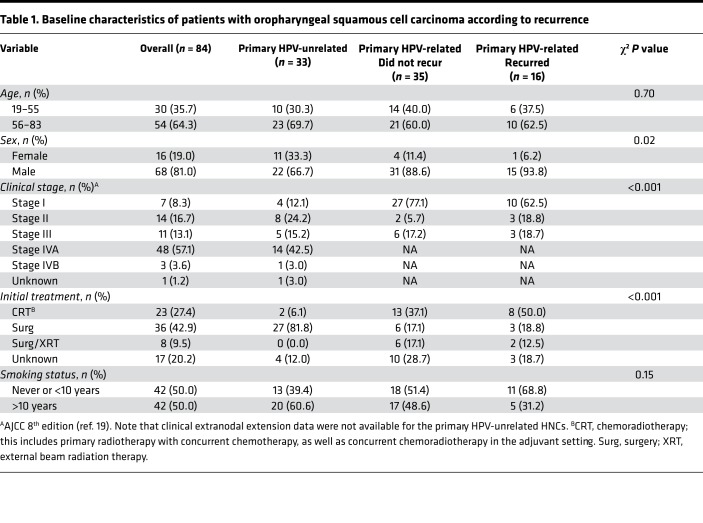
Figure 1. Mutational landscape of primary HPV-related OPSCCs (n = 51).
(A) Conceptual diagram illustrating study aims and sample sizes. (B) Genes mutated in at least 5% of primary OPSCC tumors are shown in order of descending frequency. Each column represents a patient. Rows represent genes. q values from the MutSigCV and VEST algorithms are listed in the right 2 columns. Colored bars represent mutations as described in the legend. Gray bars represent a nonmutated gene of a given tumor. (C) Amino acid changes associated with respective KMT2D mutations in this study population are illustrated. Colored bars represent functional domains of the amino acid sequence. Gray areas between colored bars represent nonfunctional domains of the amino acid sequence.
The median (interquartile range; IQR) mutations per megabase (Mb) per tumor was 1.52 (2.65) and 1.91 (6.94) among the primary HPV-related OPSCCs that did not recur (n = 35) or did recur (n = 16), respectively. Mutations per Mb was 4.59 (8.29) for metachronous recurrent OPSCCs (n = 12) and 2.03 (1.64) for primary HPV-unrelated OCSCCs and OPSCCs (n = 33). The mutational burden was marginally greater among metachronous recurrent tumors than HPV-unrelated tumors (FDR q = 0.10, Dunn’s test) or primary HPV-related OPSCCs that did not recur (FDR q = 0.13, Dunn’s test; Supplemental Figure 4). Among 51 primary HPV-related OPSCCs, we observed 6,063 nonsynonymous and 6,363 synonymous somatic nucleotide variants (SNVs). Thirty-five primary HPV-related OPSCC tumors that did not recur had 3,689 nonsynonymous and 3,180 synonymous SNVs. Sixteen primary HPV-related OPSCC tumors that did recur had 2,374 nonsynonymous and 3,183 synonymous SNVs. Twelve metachronous recurrent HPV-related OPSCC tumors had 7,385 nonsynonymous and 3,102 synonymous SNVs. The most frequently mutated genes across all primary HPV-related OPSCCs were those involved in epigenetic regulation (KMT2D), Notch signaling regulation (SPEN), cell motility (ANK3), mitogenic signaling (FGFR3), and differentiation (zinc finger protein 750, ZNF750). MutSigCV and Variant Effect Scoring Tool (VEST) analyses identified significant mutations in ZNF750 and Myosin heavy chain 9(MYH9) across all primary HPV-related OPSCCs (FDR q < 0.1; Supplemental Table 3, left columns and Figure 1B). Mutations in genes involved in mitogenic and/or NF-κB signaling, including FGFR3, PTEN, PIK3R1, and CYLD, were also among the top gene mutations by MutSigCV and VEST analyses across tumors. Notably, primary HPV-related OPSCCs that did or did not recur were not enriched in TP53 mutations, in contrast with HPV-unrelated tumors. The HPV-unrelated OCSCC/OPSCC mutational landscape was consistent with previously observed findings (16) (i.e., CDKN2A, TP53, NOTCH1, FAT1, CASP8, PIK3CA, KMT2D, NSD1, HLA-A mutations; Supplemental Table 3, right columns).
We also analyzed our samples with the OncoPlex, 262-gene, deep sequencing assay (mean 312× coverage) to test the feasibility of identifying clinically relevant, actionable genomic targets using a clinically implemented sequencing platform (Supplemental Table 2 and Supplemental Figure 3B) (20). A substantial fraction of OncoPlex mutation calls were also detected by WES (66% overall), although OncoPlex detected additional cancer drivers, including PIK3CA mutations in 2 primary HPV-related OPSCCs that did not recur and 4 primary HPV-related OPSCCs that recurred (Supplemental Tables 4 and 5). Moreover, the OncoPlex assay affirmed that 15 (63%) of our OPSCCs had at least 1 mutation in currently actionable genes described by Pritchard et al. (20). Ten (42%) of our samples had more than 1 mutation in currently actionable genes. Eighteen (75%) of our samples had at least 1 mutation in genes expected to be actionable in the near future. Thus, basket trials in HNC may benefit from targeted sequencing using the OncoPlex assay.
To prioritize genes critical to tumorigenesis and/or potentially targetable mutations, we generated a composite list of 467 driver genes (Supplemental Table 6). Driver gene mutations affecting at least 5% of primary HPV-related OPSCCs are illustrated in Figure 1B. MutSigCV and VEST analyses of the primary HPV-related OPSCCs illustrated significant mutations in many of the same genes as in the above analysis, as well as additional driver genes: KMT2D, PIK3CA, FGFR3, CYLD, EP300, RB1, FBXW7, and PTEN (Figure 1B, Supplemental Table 3, left columns). KMT2D, a lysine methyltransferase that regulates the chromatin state at Notch and other genes (21), was the most frequently mutated gene across all primary HPV-related OPSCC tumors, affecting multiple regions of the amino acid sequence (Figure 1C). Truncating mutations, including KMT2DQ56*, KMT2DS1632*, and KMT2DQ4687*, were observed in several samples. KMT2DQ4347*, found in 1 sample, has also been observed in nonmelanoma skin cancer (22).
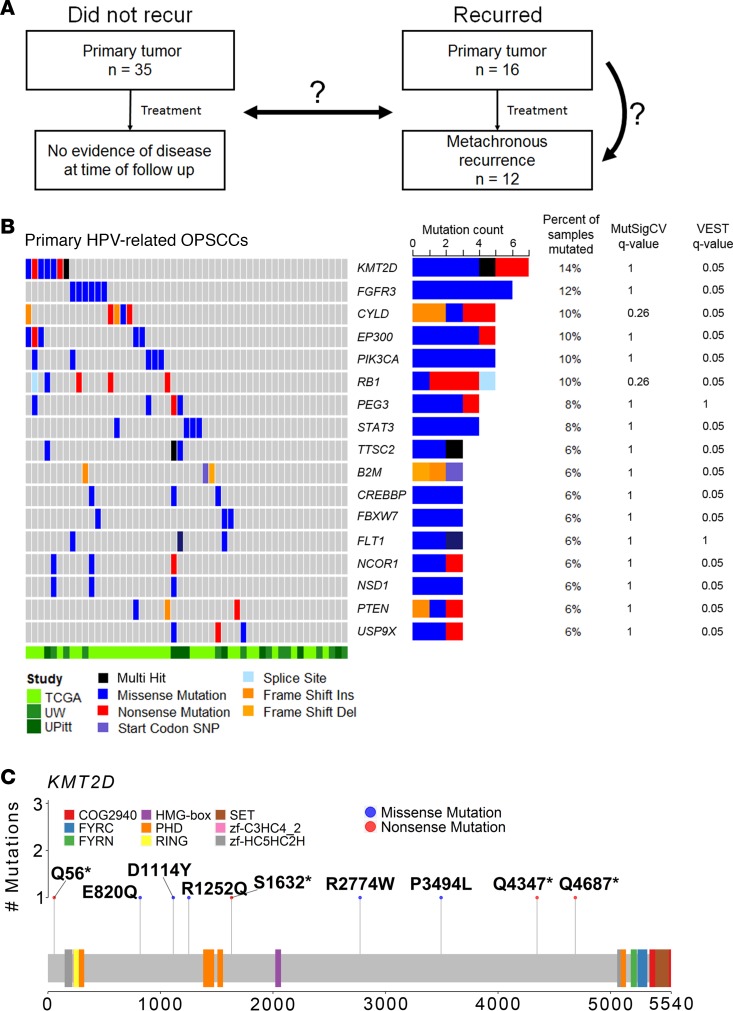
To differentiate mutational patterns among driver genes between primary HPV-related OPSCCs that did or did not recur, we evaluated SNV frequency between groups (Figure 2A). No significant differences in the frequency of mutations were observed between tumors that did or did not recur. However, we note that mutations of genes involved in mitogenic signaling (i.e., TSC2), DNA damage response (i.e., BRIP1 and NBN), and oxidative stress response (i.e., NFE2L2) were identified only among the primary OPSCCs that recurred (Figure 2A). Using MutSigCV and VEST analyses to identify putative mutational drivers of primary HPV-related OPSCCs that recurred, we observed statistically significant mutations in PIK3CA and FAT1 (FDR q < 0.1, Supplemental Table 3). PIK3CA was also significantly mutated among primary HPV-related OPSCCs that did not recur. In contrast, mutations affecting immune signaling genes B2M and TRAF3 were observed exclusively in primary HPV-related OPSCCs that did not recur (Figure 2A), although the frequency of these gene mutations was not statistically significantly than the frequency of mutations for the primary OPSCCs that recurred. Among metachronous recurrent OPSCCs, KMT2D, MTOR, ALK, NFE2L2, and FGFR3 were significantly mutated based on VEST analysis (Supplemental Table 3). Signaling pathway genes that activate DNA damage repair (DDR; ATM and FANCA) were commonly mutated de novo among metachronous recurrences (Figure 2B). Gene mutations occurring at both tumor onset and recurrence tended to involve mitogenic signaling pathways (i.e., FLT1, PIK3R6, and TSC2; Figure 2B). Moreover, not only was KMT2D significantly mutated among 42% of the metachronous recurrent tumors, but it was also mutated both at tumor onset and at recurrence in one-third of our 7 matched primary and subsequent recurrent pairs (Figure 2B). Strikingly, 1 sample had a KMT2D missense mutation at tumor onset (KMT2DR1252Q), which was lost at recurrence, while 2 new KMT2D missense mutations were gained at subsequent recurrence (KMT2DE5425K and KMT2DI3435M). Another sample had a truncating KMT2D mutation at both onset and recurrence (KMT2DQ4347*), in addition to gaining a de novo missense mutation at recurrence (KMT2DE820Q).
Figure 2. Mutational differences between primary HPV-related OPSCC tumors that did not (n = 35) or did recur (n = 16) and mutational characteristics of metachronous recurrences.
(A) Genes mutated in at least 2 samples across primary HPV-related OPSSC that did or did not recur were analyzed. Columns represent individual patients. Genes (rows) are sorted by descending mutation frequency across all samples. While there were no statistically different gene mutation frequencies between groups, between-group comparisons with a Fisher exact q < 1.0 are denoted by an asterisk. Colored bars represent mutations as annotated in the legend, and gray bars represent nonmutated genes for a respective tumor. (B) Mutation patterns in 7 paired index primary and subsequent metachronous recurrent HPV-related OPSCCs. Gene mutations (rows) affecting at least 2 patients are illustrated. Genes mutated both in the index primary and paired metachronous recurrence are annotated in dark blue. Genes mutated only in the index primary tumor are annotated in light blue (i.e., lost at recurrence). Genes mutated only in a metachronous recurrence but not the index primary are annotated in red (i.e., gained at recurrence).
Copy number variants.
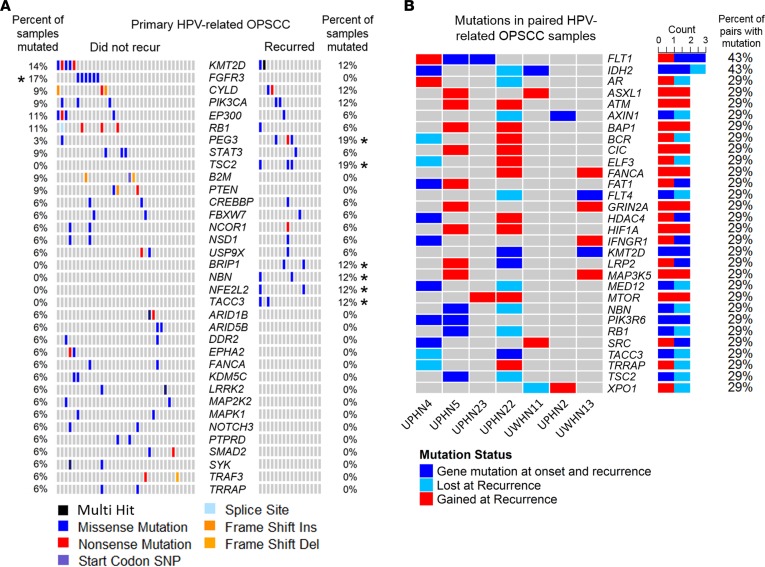
Copy number variants (CNVs) were inferred using WES tumor and matched normal coverage results from TCGA and our data. Figure 3A depicts raw copy number estimates for primary HPV-related OPSCCs that did (n = 16) or did not recur (n = 35), metachronous recurrent tumors (n = 12), and primary HPV-unrelated OCSCCs and OPSCCs (n = 33). Primary HPV-related OPSCCs demonstrated gains of 3q, 8q, 14q, and 20q with copy number losses of 2q, 3p, 10q, 11p, 11q, 13q, and 14q among other regions (Figure 3A, Supplemental Figure 5, and Supplemental Tables 7 and 8). The fraction of exome harboring copy number amplifications did not differ between primary HPV-related OPSCCs that did or did not recur (median [IQR]: 7.8% (14%) vs. 8% (12%), P = 0.80; Wilcoxon rank sum test) nor did copy number deletions (median [IQR], 11% [8%] vs. 10% [10%], P = 0.46; Supplemental Table 9). Metachronous recurrent OPSCCs harbored a greater fraction of amplifications (median [IQR]: 25% [32%]) and deletions (median [IQR]: 19% [10%]) than primary HPV-related OPSCCs that recurred (P = 0.023 and P = 0.029, respectively).
Figure 3. DNA copy number alterations.
(A) Log2 copy number alteration data for primary HPV-related OPSCC tumors that did not (n = 35, blue box) or did recur (n = 16, red box). Metachronous recurrent HPV-related OPSCCs (n = 12, green box) and primary HPV-unrelated OCSCCs and OPSCCs (n = 33, black box) are also illustrated. Rows represent cytobands. Columns represent samples. Copy number gains are represented in shades of red. Log2 copy number ratio equal to 1 is represented in darker red, with smaller gains in lighter shades of red. Copy number losses are represented by shades of blue. Log2 copy number ratio equal to –1 is represented by dark shades of blue, with smaller losses in lighter shades of blue. (B) Percent of samples with a copy gain or loss greater than log2 of 0.2 or less than log2 of –0.2. Rows represent cytobands. Fraction of tumors with copy loss or gain greater is on the x axis. Left panel: primary HPV-related OPSCCs that did vs. did not recur. Right panel: primary HPV-related OPSCCs that recurred vs. metachronous recurrent HPV-related OPSCCs. (C) GISTIC2.0 significant amplifications and deletions across primary HPV-related OPSCCs that did not recur (left panels) or recurred (middle panels) and metachronous recurrent HPV-related OPSCCs (right panels). Selected genes are annotated on respective cytobands. Green line, significance threshold (FDR q = 0.25).
HPV-related primary OPSCC tumors that did not recur featured frequent amplifications of 3q (PIK3CA and SOX2) and 8q, in addition to focal amplifications of CCND1, ANO1, FADD, and E2F1 (Figures 3, B and C, and Supplemental Table 10). Copy number deletions in the tumors that did not recur were prevalent at multiple sites on 1p, 2q, 10q, 11q, 13q, 14q, and 20p, among which key tumor suppressors (PTEN, TP73, and RB1), DDR (ATM), epigenetic regulatory (MLL), and genes involved in cell death regulation (CASP1/4/5/12) are located (Figure 3, B and C, and Supplemental Table 11).
Primary HPV-related OPSCC tumors that recurred featured frequent amplification of 3q, 8q, and 20q (Figure 3, B and C, and Supplemental Table 12). Focal deletions were noted on several chromosomes including 14q, which encodes JAG2, a ligand for Notch signaling (Figure 3, B and C, and Supplemental Table 13). KMT2A (MLL) — or lysine methyltransferase 2A — involved in chromosomal translocations in the development of acute leukemia, was frequently deleted. Other deletions on 11q were also featured among the tumors that recurred, implicating DDR response and cell-death constituents (CASP1/4/5/12, ATM, BIRC2/3). Amplifications among metachronous recurrent HPV-related OPSCCs included 3q, 8q, and 20q (Figure 3, B and C, and Supplemental Table 14). There were several regions with significant deletions involving 2q, 9q, 10q, 11q, 14q, 16q, and 20q, among others (Figure 3C and Supplemental Table 15). Focal deletions included NOTCH1, as well as apoptotic cascade (CASP1/4/5/8/10/12), DDR (ATM), and PI3K signaling constituents (AKT1 and TSC2).
Comparison with HPV-unrelated HNC.
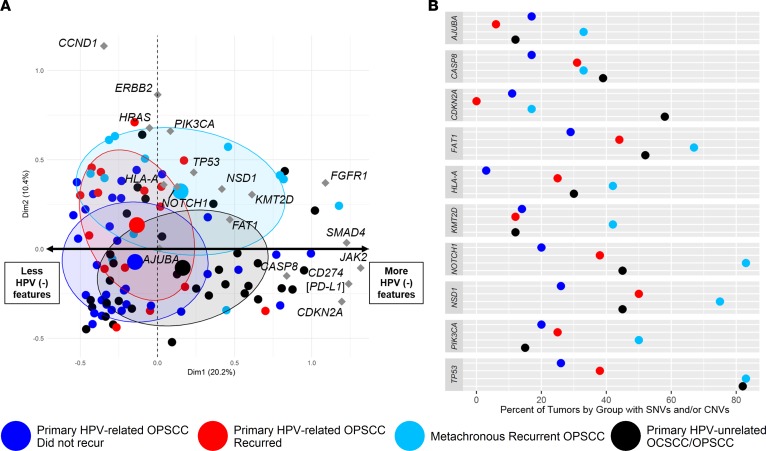
Next, we compared HPV-related OPSCCs with HPV-unrelated OCSCCs/OPSCCs. We generated a list of 41 HPV-negative–like genes based on TCGA mutational and copy number data and prior investigations (e.g., CDKN2A, TP53, NOTCH1, FAT1, AJUBA, CASP8, PIK3CA, KMT2D, NSD1, HLA-A; Supplemental Table 16) (7, 16, 18). We considered both the mutational and copy number status for the following analyses. Multiple correspondence analysis (MCA) — similar to principal component analysis but used for categorical data — was performed as a method to identify variation and patterns in the genomic aberrations between the primary and metachronous recurrent HPV-related OPSCCs relative to HPV-unrelated HNCs (23). MCA analysis suggested that the metachronous recurrent OPSCCs share features with HPV-unrelated tumors, while retaining some similarities with HPV-related tumors (Figure 4A). Primary HPV-related OPSCCs that did or did not recur had more genomic features in common with each other than with HPV-unrelated tumors. Genomic aberrations with the greatest contribution to the first dimension of variance were PSIP1, JAK2, CD274 (i.e., PD-L1), PTPRD, PDE4D, and CDKN2A, among others (Figure 4A and Supplemental Figure 6A). In other words, genomic alterations in these genes accounted for the primary differences between groups. Genomic aberrations with the greatest contribution to the second dimension were CTTN, CCND1, and FADD (Supplemental Figure 6B).
Figure 4. Integrated analysis of genomic aberrations.
(A) Multiple correspondence analysis (MCA) was performed to compare patterns of genomic alterations (i.e., somatic mutation and copy number variants) between primary HPV-related OPSCCs that did (n = 16) or did not recur (n = 35), metachronous recurrent OPSCCs (n = 12), and primary HPV-unrelated OCSCCs and OPSCCs (n = 33). Associations between genomic features are represented graphically in the MCA plot illustrating the statistical relationships among distinct features. The analysis considers 41 HPV-negative–like genes. A sample that has a mutation and/or copy number aberration is defined as having a genomic aberration for a given gene. Small colored dots represent individual tumors. Large dots represent the average position on the first 2 principal dimensions of variance among all tumors within a group. There is one large point for each of the 4 groups. Diamonds represent the contribution of the selected genes to each of the first 2 principal dimensions. Groups that plot close to each other on the ordinate and/or abscissa are related statistically. MCA coordinates are encompassed by 95% confidence ellipses. Samples clustering on the positive pole of the x axis harbor more HPV-negative–like genomic features, while those on the negative pole of the x axis harbor fewer HPN-negative–like features. (B) Dot plot illustrating the percent of tumors in each of the 4 groups with a somatic mutation and/or copy number variant in a set of 10 genes that are frequently altered in HPV-negative head and neck cancer.
NOTCH1 was deleted and/or mutated in 31% and 6%, respectively, of primary HPV-related OPSCCs that recurred compared with 6% and 3% of primary HPV-related OPSCCs that did not recur (Figure 4B and Supplemental Table 17). TP53 was deleted and/or mutated in 38% and 0%, respectively, of primary HPV-related OPSCCs that recurred vs. 26% and 3% of primary OPSCCs that did not recur (Figure 4B and Supplemental Table 17). In comparison, up to 75% and 58% of metachronous recurrent OPSCCs had TP53 and NOTCH1 deletions, respectively, although no mutations were found in these genes among the metachronous recurrent OPSCCs (Supplemental Table 17). Statistically significant differences in deletion frequencies between the primary HPV-related OPSCCs that did or did not recur were noted among NSD1, HLA-A, EGFR, CTTN, and HRAS, with these genes have a higher deletion frequency among the tumors that recurred (Supplemental Table 17). In comparison, we observed a statistically greater proportion of TP53 mutations among the metachronous recurrent OPSCCs than among the primary HPV-related OPSCCs that recurred (Supplemental Table 17). While not statistically different, CDKN2A, NOTCH1, and CD274 were deleted in 17%, 58%, and 25%, respectively, of metachronous recurrent HPV-related OPSCCs compared with 0%, 31%, and 12%, respectively, of primary HPV-related OPSCCs that recurred.
Next, we evaluated the fraction of tumors among each group with a somatic mutation and/or CNV in a set of 10 genes frequently altered in HPV-unrelated HNC (Figure 4B). We performed Fisher’s exact tests comparing the frequency of variants among the 3 HPV-related OPSCC groups with the HPV-unrelated HNC group. Compared with the HPV-unrelated HNCs, we observed a significantly greater proportion of metachronous recurrent HPV-related OPSCCs with genomic aberrations in NOTCH1 (83% vs. 45%, respectively), KMT2D (42% vs. 12%), and PIK3CA (50% vs. 15%). Interestingly, the frequency of genomic alterations among TP53, CASP8, FAT1, HLA-A, AJUBA, and NSD1 was not statistically different between metachronous recurrent OPSCCs and HPV-unrelated HNCs. Additionally, compared with primary HPV-related OPSCCs that did not recur, HPV-unrelated HNCs had a significantly higher frequency of TP53 (82% vs. 26%, respectively), CDKN2A (58% vs. 11%), HLA-A (30% vs. 3%), NOTCH1 (45% vs. 20%), CASP8 (39% vs. 17%), and FAT1 (52% vs. 29%) genomic aberrations. HPV-unrelated HNCs had a significantly higher proportion of CDKN2A (58% vs. 0%) and TP53 (82% vs. 38%) genomic aberrations than primary HPV-related OPSCCs that recurred. Remarkably, primary HPV-related OPSCCs that recurred shared several genomic features with HPV-unrelated HNCs, illustrating a lack of statistically different proportions in PIK3CA, CASP8, NOTCH1, FAT1, KMT2D, HLA-A, AJUBA, and NSD1 gene alterations. Among the 10 HPV-negative–like genes of interest, the only significant difference in the proportion of genomic aberrations between primary HPV-related OPSCCs that did or did not recur was in HLA-A (25% among those that recurred vs. 3% among those that did not recur).
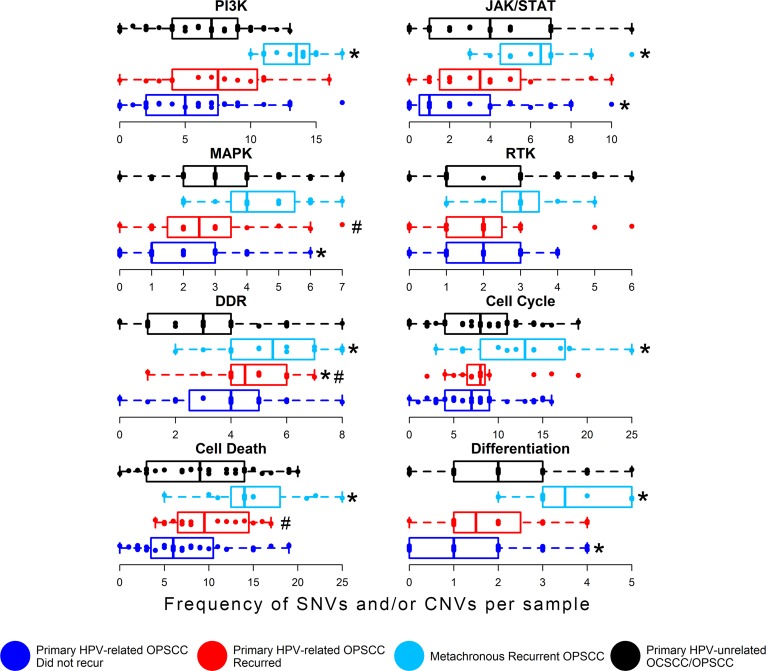
We were also interested in investigating genomic aberrations among a set of biological pathways central to OPSCC tumorigenesis and with potentially targetable constituents, including mitogenic signaling, DDR, cell cycle and death, and differentiation pathways. Pathway constituents are defined in the Methods and in Supplemental Table 18. Figure 5 compares genomic aberrations relative to HPV-unrelated tumors. We observed statistically greater frequencies of genomic aberrations in multiple pathways, including PI3K, DDR, cell death, JAK/STAT, cell cycle, and differentiation among the metachronous recurrent OPSCCs compared with HPV-unrelated HNCs (Figure 5 and Supplemental Table 19). We observed a statistically higher frequency of aberrations in the DDR pathway among the primary HPV-related OPSCCs that recurred compared with HPV-unrelated HNCs. We did not observe any statistically significant differences in biological pathway genomic alterations between the primary HPV-related OPSCCs that did or did not recur. However, primary HPV-related OPSCCs that recurred had a non–statistically significantly greater frequency of genomic aberrations in MAPK, DDR, and cell death pathways compared with primary HPV-related OPSCCs that did not recur (Figure 5). In contrast, compared with HPV-unrelated HNCs, the primary HPV-related OPSCCs that did not recur did not differ statistically in the frequency of genomic alterations in PI3K, DDR, cell death, RTK, or cell cycle pathways. However, the primary HPV-related OPSCCs that did not recur had statistically fewer genomic aberrations in MAPK, JAK/STAT, and differentiation pathways (Figure 5). Potentially targetable genes (http://archive.broadinstitute.org/cancer/cga/target) contributing to these differences are illustrated in Supplemental Figure 7.
Figure 5. Analysis of key gene alterations grouped by biological function.
Somatic mutation and/or copy number alterations per tumor. Pathway constituents are described in the text. Dark blue boxes and points represent primary HPV-related OPSCCs that did not recur, and red boxes and points represent primary HPV-related OPSCCs that did recur. Metachronous recurrent OPSCCs are illustrated in light blue. TCGA HPV-unrelated OCSCCs and OPSCCs are represented in black. Boxes represent median and quartiles. Whiskers represent minimum and maximum. RTK, receptor tyrosine kinase; DDR, DNA damage repair; *, statistically significant differences in genomic aberration between the HPV-related OPSCCs vs. the HPV-unrelated control group (FDR q < 0.1, Dunn’s test). #, non–statistically significant differences between HPV-related OPSCCs that did vs. did not recur (FDR q < 0.15, Dunn’s test).
Discussion
The US is experiencing epidemic levels of HPV infection (1), while the incidence of HPV-related HNC is rapidly rising (2). While treatment outcomes with current therapies are favorable and recurrence is infrequent, the non–viral-related molecular underpinnings of recurrence remain undefined. Moreover, HPV-related HNC affects a younger age group, translating to long-lasting impact on productivity, longevity, and quality of life (24). Short-term mortality among HPV-related HNCs that recur is upwards of 40% (5), warranting investigation into targeted therapies for mitigating recurrence from the outset of treatment and investigation into how to treat recurrent tumors when they arise.
In this report, we identified critical findings regarding genomic aberrations associated with HPV-related OPSCC tumor recurrence. First, we observed that primary HPV-related OPSCCs that did or did not recur shared a genomic landscape trending toward differences in DDR, MAP kinase, and cell death repair pathways, though not statistically significant. In contrast, metachronous recurrent OPSCCs grouped with HPV-unrelated HNCs. Additionally, we observed that KMT2D was the most frequently mutated gene across HPV-related OPSCC tumors with a propensity for truncating mutations, suggesting a tumor suppressor role. Moreover, we observed that PIK3CA and KMT2D mutations cooccurred in only 1 sample, suggesting the intriguing possibility that these genes act in the same pathway. KMT2D mutations occurred in the majority of metachronous recurrent HPV-related OPSCCs, as well as paired primary and metachronous recurrences. Metachronous recurrent HPV-related OPSCCs were remarkable for a high mutational burden. Primary HPV-related OPSCCs that did not recur were notable for mutations involving PI3K and immune signaling pathway aberrations. Primary HPV-related OPSCCs that recurred exclusively featured mutations in DDR (BRIP1 and NBN) as well as PI3K (TSC2) and oxidative stress (NFE2L2) genes. Metachronous recurrent OPSCCs featured de novo mutations impacting DDR machinery. The copy number landscape was similar between primary HPV-related OPSCCs. Primary HPV-related OPSCCs that recurred and metachronous recurrent OPSCCs shared deletions in Notch and PI3K family members. Overall, these data offer appealing opportunities for further work aiming to mitigate HPV-related OPSCC recurrence.
The most frequent genomic aberrations across primary HPV-related OPSCCs included KMT2D, FGFR3, CYLD, EP300, and PIK3CA mutations, as well as 3q amplification, E2F1 amplification, and intact 9p21 (containing CDKN2A), plus a paucity of TP53 mutations. Among primary HPV-related OPSCCs that recurred, we observed a predilection for genomic aberrations in mitogenic signaling (i.e., PIK3CA, AKT1, IGF1R, and TSC2) and DDR constituents (i.e., ATM, PARP1, MLH1, and BRCA2). Analogously, Lui et al. illustrated a relationship between PI3K pathway mutations and advanced stage HNC (25). Genomic aberrations in genes associated with DDR mechanisms were also gained at recurrence (e.g., ATM, FANCA, and TRRAP), whereas gene mutations in mitogenic signaling constituents tended to be shared at onset and recurrence (e.g., FLT1, PIK3R6, and TSC2). Primary HPV-related OPSCCs that did not recur were also notable for gene mutations in mitogenic signaling constituents (e.g., PIK3CA), although they tended toward a predilection for mutations in immune signaling genes (e.g., B2M and TRAF3). On the other hand, metachronous recurrent OPSCCs were markedly enriched in CD274 (PDL1) deletions.
Additionally, we observed frequent mutations and copy number aberrations affecting Notch family members. Differentiation pathways, predominantly involving Notch mutations, are frequently implicated in head and neck carcinogenesis (7, 16, 18), cutaneous malignancies, and lung cancers (26). Interestingly, our analysis considering all mutations across primary HPV-related OPSCCs revealed that ZNF750 was significantly mutated. ZNF750 plays a role in terminal epidermal differentiation as a target of TP63 (27).
A surprising finding was the prevalence of KMT2D mutations. To our knowledge, this is the first study to identify a predilection for KMT2D in metachronous recurrent HPV-related OPSCCs. KMT2D, also known as MLL2, is infrequently involved in pathogen-mediated cancers. Prior investigations illustrated compelling evidence of a founder role for KMT2D in follicular lymphoma (28). Some describe KMT2D as an oncogene (29), while others describe KMT2D function as a tumor suppressor (30). Target genes of KMT2D include Notch family members (21), CD40, JAK/STAT, TLRs, B cell receptor signaling pathways, and other tumor suppressor genes such as TNFAIP3, SOCS3, and TNFRSF14 (30). KMT2D also converges on MAPK signaling pathways in Kabuki syndrome, a disorder with developmental delay and congenital anomalies, resulting in attenuated MEK/ERK activation (31). In Kabuki syndrome, KMT2D is often inactivated (31). In the current study, MAPK, cell death, and DDR pathway aberrations had the strongest association with recurrence. Another mitogenic signaling network implicated in KMT2D chromatin regulatory function is PI3K signaling. Toska et al. elegantly illustrated the role of PI3Kα inhibition on enhancing KMT2D activity in estrogen receptor–positive breast cancer as a mechanism of resistance to PI3Kα inhibition (32). Strikingly, of 5 overall PIK3CA mutations among our primary HPV-related OPSCCs, 4 were in non–KMT2D-altered samples, suggesting a trend toward mutual exclusivity between PIK3CA and KMT2D. These data suggest the intriguing possibility that PI3Kα and KMT2D act in the same pathway, therefore serving as potentially novel targets in OPSCCs.
In our study, we observed a KMT2DQ4347* truncating mutation in the primary HPV-related OPSCC tumor of a patient initially treated with surgery and radiotherapy. Another primary tumor in our study treated with surgery upfront and with adjuvant concurrent chemoradiation had a KMT2DR1252Q missense mutation. Further investigation into the interaction between downstream KMT2D targets with mitogenic signaling pathways as they relate to chemoradiotherapy resistance could lead to the identification of synthetic lethal treatment combinations.
Work using a targeted sequencing panel by Morris et al. (17) found that recurrent and metastatic HPV-related HNCs were genomically similar to HPV-unrelated HNC. Notably, our data suggest similarities between metachronous recurrent OPSCCs and primary HPV-unrelated OCSCCs/OPSCCs. Interestingly, we found that the mutational burden was similar between primary HPV-unrelated and -related OPSCCs, while metachronous recurrent tumors had a predilection for a greater mutational burden. Given that mutation-associated neoantigens are potential determinants of response to immune checkpoint inhibitors (33), our data suggest a potential role for checkpoint inhibitors in metachronous recurrent OPSCCs. In contrast, Stransky et al. (7) found that the mutation rate of HPV-related tumors was approximately half that of HPV-unrelated HNC. One plausible reason for this difference is the prevalence of smoking among patients in this study population, although there was a smaller percentage of patients with greater than 10-year smoking histories among the primary OPSCCs that recurred vs. those that did not recur. Lastly, the mutation rate may be related to DDR machinery alterations, with some HPV-related OPSCCs having a greater overall mutation rate in association with DDR genomic alterations.
Analysis of the combined somatic mutation and copy number landscape between HPV-unrelated tumors and primary HPV-related OPSCC tumors suggested greater overlap between the primary HPV-related OPSCC tumors that did or did not recur than with the HPV-unrelated tumors. However, there were notable similarities between the primary HPV-related OPSCC tumors that recurred and HPV-unrelated tumors. For example, NOTCH1 and TP53 deletions occurred more frequently among the primary HPV-related OPSCCs that recurred than the primary HPV-related OPSCCs that did not recur. Also, AJUBA, FAT1, HLA-A, and NFE2L2 were mutated among primary HPV-related OPSCCs that recurred but not in the primary HPV-related OPSCCs that did not recur. Lastly, mitogenic signaling constituents (PI3K, JAK/STAT, and MAPK) had a similar burden of genomic derangements between primary HPV-related OPSCCs that recurred and HPV-unrelated tumors. In contrast, primary HPV-related OPSCCs that did not recur had a lower rate of genomic alterations in JAK/STAT and MAPK pathways but not in the PI3K pathway relative to HPN-unrelated HNCs. These data suggest that primary HPV-related OPSCC tumors that recur share an overall genomic landscape with those that do not recur, although tumors that recur acquire HPV-negative–like aberrations that could guide treatment stratification in clinical trials and future studies.
When comparing the mutational profile between primary HPV-related OPSCCs that did or did not recur, we observed several genes that were only mutated among tumors that did not recur, such as B2M, a gene encoding β2-microglobulun, which plays a role in antigen presentation. Similarly, a recent study evaluating the effectiveness of anti-PD1 antibody therapy across 12 tumor types noted that B2M was not mutated in any of the primary tumors from treatment-resistant cases (33). Furthermore, with respect to immune therapy, we also observed a significant role of CD274 (PDL1) alterations in distinguishing HPV-unrelated tumors and metachronous recurrent OPSCCs from primary HPV-related OPSCCs. Specifically, CD274 was deleted in 25% of metachronous recurrent OPSCCs compared with 11% and 12% of primary HPV-related OPSCCs that did not and did recur, respectively.
Interestingly, there were several genes mutated only among primary HPV-related OPSCCs that recurred. These involved diverse signaling pathways, including oxidative stress response (NFE2L2), mitogenic signaling (TSC2), and DNA damage response genes (BRIP1 and NBN). DDR (34) and mitogenic signaling constituents (35–38) have previously been implicated in chemoradiation resistance. Not only were DNA damage and mitogenic signaling pathway mutations common in the primary HPV-related OPSCCs that recurred, metachronous recurrent OPSCCs had a predilection for de novo mutations in DNA damage response genes. Moreover, mutations in mitogenic signaling constituents were frequently shared between paired index primary and metachronous recurrent tumors.
Per our review, this is the first study to our knowledge to implement both WES and a clinically implemented targeted sequencing assay to examine the genomic profile of metachronous recurrent HPV-related OPSCCs. This builds on a study by Morris et al. (17) that assessed the relationship between metastatic and recurrent HPV-related HNCs with HPV-negative HNC using the clinically implemented Memorial Sloan Kettering–Integrated Mutation Profiling of Actionable Cancer Target targeted sequencing assay (22). The HPV-related tumors from Morris et al. (17) included 12 distant metastases, 5 index primaries, and 3 locoregional metachronous recurrences from 3 subsites and an unknown primary site. In contrast, we compared primary HPV-related OPSCCs that did or did not recur with metachronous recurrences to identify genomic aberrations associated with recurrence that may be present from the outset, persist, or develop de novo.
Our study implemented WES, in addition to the OncoPlex targeted sequencing assay, to interrogate a wide array of recurrence-promoting genetic alterations while testing the utility of a clinically implemented sequencing assay in identifying potentially actionable targets from the outset of treatment and at recurrence. We did find that the agreement between WES and OncoPlex was lower than described in other studies (39, 40). However, this was the same general trend we would expect — higher depth of coverage sequencing of a targeted panel of cancer-related genes will yield a higher detection rate than WES, especially for variants that are present at lower variant allele frequencies. Differences in library preparation, off-target reads, tumor heterogeneity, sequencing instruments, and tumor-only sequencing may contribute to differences in variant calls between sequencing techniques. Furthermore, the relative advantages and disadvantages of each sequencing technology should be considered. For example, targeted sequencing may be invaluable for guiding treatment of tumors with actionable genes (e.g., ALK mutations). On the other hand, WES may have utility in guiding treatments that depend on markers such as mutational burden, which is associated with response to immunotherapies in some cases (41–43). Overall, future work seeks to address the utility and role of targeted sequencing compared with whole exome or whole genome sequencing.
An additional factor impacting the consideration of targeted vs. whole exome or whole genome sequencing is the issue of tumor clonality and heterogeneity. Tumors are both spatially and temporally heterogenous and often composed of a single founding clone and several subclones, and there is evidence to suggest that intratumor heterogeneity and clonality impact response to therapy and treatment resistance (44–46). This presents an issue for tumor sampling and sequencing, as subclones may be missed that go on to form a recurrence. This also presents an intriguing avenue for future research. Understanding clonal architecture and evolutionary pressures that drive the formation of subclones could help us predict which subclones are likely to recur. Future research efforts may further implement functional genomics approaches to target major clones and subclones. In summary, targeted and whole exome or whole genome sequencing may have unique roles in these endeavors.
Our sample size, coupled with substantial heterogeneity in terms of staging and therapy, limits our ability to detect weak or moderate genomic differences between recurrent and nonrecurrent HPV-related OPSCCs. However, identifying these genomic differences for which our study is appropriately powered provides insight into potential candidate drivers to target in basket trials, especially given the limited targeted therapies available. Another limitation is that we used a mixture of formalin-fixed paraffin-embedded (FFPE) and frozen samples. However, investigators have noted high concordance (>90%) in mutational data comparing clinically derived FFPE and fresh frozen samples (47). There is also potential for misclassification of HPV status, given that we used p16 status as a marker. Our study nonetheless reflects clinical practice, as p16 status is most often used to infer HPV status in HNC prognosis and treatment. Lastly, our study is less prone to confounding by smoking, given the relatively low smoking prevalence among the primary HPV-related OPSCCs that recurred compared with the OPSCCs that did not recur. Finally, this study provides a unique glimpse into the molecular landscape of paired primary and metachronous recurrent HPV-related OPSCCs. These findings underscore the critical need for appropriate correlative studies in the conduct of clinical trials in both curative and palliative settings.
In summary, we observed that primary OPSCCs that recur share a mutational landscape in common with primary HPV-related OPSCCs that do not recur with differences in DDR, MAP kinase, and cell death constituents. In comparison, the molecular landscape of metachronous recurrent OPSCCs shared many features with HPV-unrelated HNC. Further, we noted a potentially novel key role for KMT2D in metachronous recurrent HPV-related OPSCCs, coupled with the intriguing possibility that KMT2D and PIK3CA act in the same pathway. Collectively, these data provide intriguing avenues for future research evaluating mechanisms of treatment resistance while aiming to mitigate HPV-related OPSCC recurrence from the outset of therapy.
Methods
Study design and subjects
A cross-sectional study design was implemented. WES data from TCGA (16) were downloaded, and our WES data were derived from archival and fresh frozen tissue samples. Tissue from our patients who consented to participate in genomic analysis studies were collected between 2010 and 2015. p16 immunohistochemical staining was used as a surrogate for HPV status in the TCGA and our data (3). TCGA clinical data from HPV-related and -unrelated cases were downloaded from the cBioPortal database (http://www.cbioportal.org/) for a total of 527 subjects. Subjects were included if they had either a known HPV in situ hybridization result or if they had a p16 result. This narrowed the dataset to 128 possible subjects (80 HPV-unrelated and 48 HPV-related). Subjects with HPV-related OPSCC primary tumors were included if their recurrence status was known, thus excluding another 5 possible participants. We further limited the TCGA dataset to subjects with HPV-unrelated HNC of the oral cavity or oropharyngeal subsites, resulting in a total of 59 possible subjects. TCGA HPV-related cases were limited to oropharyngeal subsites, resulting in a total of 36 possible subjects. TCGA primary HPV-unrelated OCSCC and OPSCC SNV data were downloaded for analysis in addition to the UW and UPitt samples. Binary alignment/map (BAM) files for the TCGA tumor HPV-unrelated and HPV-related samples were downloaded for inferring CNVs, given that this was the method by which we analyzed CNVs for the UW and UPitt samples. We procured available TCGA BAM files for 33 HPV-unrelated HNCs and 32 HPV-related OPSCCs (Supplemental Figure 1). The final dataset consisted of the UW and UPitt samples, as well as TCGA samples for which we had both somatic nucleotide and copy number data (derived from WES BAM files). Additional details pertaining to TCGA study variables used for subject inclusion are found in the Supplemental Methods.
Data collection
Clinical data.
TCGA clinical data were downloaded from cBioPortal (48, 49). Clinical data were abstracted by study investigators. OPSCCs include subsites as defined in the AJCC Cancer Staging Manual, 7th Edition (50).
Molecular data.
Mutation annotation format (MAF) files containing SNVs from TCGA were obtained through FireBrowse (http://firebrowse.org/) (Supplemental Methods). Copy number data for TCGA subjects were inferred through secondary analyses of controlled-access GRCh37/hg19 tumor and normal BAM files obtained from the National Cancer Institute Genomic Data Commons legacy portal (https://portal.gdc.cancer.gov/legacy-archive/search/f) (Supplemental Methods).
The following pertains to laboratory processing, sequencing, and analyses of our tumor and normal (tissue or blood) samples. Our frozen tissue specimens were stored in the Seattle Translational Tumor Research HNC biorepository at the Fred Hutchinson Cancer Research Center. H&E slides were prepared for each specimen and reviewed by a study pathologist (E.Q. Konnick). Samples with at least 40% tumor based on histology content were included. Pieces of frozen tissue up to 5 mg were used for DNA extraction.
Archival tissue specimens from our samples were processed via fixation in 10% neutral buffered formalin (Thermo Fisher Scientific), dehydrated in ethanol (Thermo Fisher Scientific), and embedded with paraffin wax (Thermo Fisher Scientific) (FFPE). H&E slides were prepared, reviewed by staff pathologists at the Universities of Washington and Pittsburgh, and areas with high tumor density (>75% tumor cells) were marked. Punch biopsies (2-mm) were taken from the FFPE tumor dense regions. FFPE tissue was deparaffinized with xylenes, washed in consecutive ethanol rinses (100% and 70%), and heated to remove formalin cross-linking (51).
Tumor DNA was extracted using an AllPrep DNA/RNA Micro Kit (Qiagen). Double-stranded DNA was quantified using an Invitrogen Quant-iT PicoGreen dsDNA assay kit (Thermo Fisher Scientific). Sample integrity was evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies).
Key covariates
Key covariates include age, stage, sex, and tobacco-use history. Age was categorized. Stage was defined as per the AJCC Cancer Staging Manual, 8th Edition (19). Tobacco use history was defined as either greater than or less than 10 years due to the absence of information on more detailed measures (e.g., pack-years) in the medical records.
Sequencing and alignment
Sequencing and alignment steps for TCGA data were described previously (16, 17). Our samples were prepared for sequencing as follows. Libraries were prepared via standard protocols using the Agilent SureSelect Human All Exon v6 kit on a PerkinElmer Sciclone G3 NGSx workstation and sequenced on an Illumina HiSeq 2500 sequencing system in high output mode (100 bp, paired-end). Quality control was performed on the raw reads using FastQC (https://goo.gl/cyJgXF), and adapters were removed. Preprocessed reads were aligned to the human genome reference sequence assembly (GRCh37/hg19) with the Burrows-Wheeler Aligner (52). Picard software was used to mark and remove PCR duplicates. Base quality score recalibration was performed with the Genome Analysis Toolkit (GATK) base quality score recalibration pipeline (53). Variant discovery analysis was performed as suggested by the Broad Institute’s GATK Best Practices (53). Tumor genomic DNA samples were also sent to UW Department of Laboratory Medicine for targeted sequencing with the UW-OncoPlex assay. The library construction and sequencing of tumor genomic DNA were performed as previously described (20). UW-OncoPlex variant calls were curated based on their in-house analytical pipeline. Furthermore, we evaluated coverage at UW-OncoPlex variant call sites on the corresponding whole exome sequencing germline data. We further excluded UW-OncoPlex variants where there was greater than or equal to 3% variant calls or more than 1 variant base read on the corresponding WES germline data. While this technique aimed to minimize germline variants called by UW-OncoPlex, there is a possibility that there were some germline variants detected by this targeted sequencing method. Of note, we used the same tumor genomic DNA for both OncoPlex and exome sequencing.
SNV/indel analysis
Analysis of tumor contamination was performed with CONPAIR (54). Samples with >20% contamination were excluded from analyses (Supplemental Table 1). SNV and small insertions or deletions (indel) detection was performed with GATK MuTect2 using default filters (53, 55). All detected variants were functionally annotated with ANNOVAR (56). Variant call format (VCF) files were converted to MAF files for downstream analyses using vcf2maf (https://github.com/mskcc/vcf2maf). Further molecular filtering parameters as described in the Supplemental Methods and Supplemental Figure 2 were applied to all somatic variants to further minimize potential false positives, germline variants, and problematic variants (e.g., TTN). Manual review of sequencing alignments using Integrative Genome Viewer (57) was used to confirm mutations in 5 random nonsynonymous variants and 1 or more of PIK3CA, FGFR3, TP53, CDKN2A, RB1, TSC2, ZNF750, and/or DDR2 variants. Filtered MAF files were merged into 1 MAF file for downstream analyses. An additional MAF file was generated, limited to variants affecting a set of 467 driver genes as described by Vogelstein et al. (58), Hedberg et al. (59), and the UW OncoPlex assay (20) (Supplemental Table 6).
MAF files were processed with MutSigCV v1.2 (60) to identify significantly mutated genes, given patient-specific somatic mutation frequency, mutation spectrum, and genomic position-based factors (e.g., DNA replication timing and chromatin state estimation). VEST was used to detect significant pathogenic mutations (61) and was implemented with the Cancer-Related Analysis of Variants Toolkit (62). MAF files, limited to the driver gene set, were used to analyze the mutational profile across samples and between groups using functionalities of the “maftools” (63) R/bioconductoR package.
CNV analysis
Somatic CNV detection was performed using Aberration Detection in Tumor Exome (ADTEx) (64). ADTEx incorporates a functionality to normalize sequencing coverage data for tumor and normal samples. Coverage files for input to ADTEx were prepared from BAM files using bedtools 2.21.0 (65). Analyses were performed using in-house shell scripts. Segmented copy number data generated by the R/bioconductoR package, “DNAcopy”, incorporated into ADTEx were log2 transformed using R statistical programming software. Segmentation and marker files were prepared and processed with GISTIC2.0 v6.10 (66) to test for significant CNVs across tumors where an FDR of 0.25 is used to test for significance. Input parameters are described in the Supplemental Methods. We did not observe a difference in copy number alterations based on derivation from WES or SNP array (Supplemental Figure 8). We also assessed for batch effect among the TCGA, UW, and UPitt WES data. We did not observe a difference in CNV data by origin, although there appeared to be outliers for each dataset (Supplemental Figure 9).
Combined somatic mutation and copy number genomic analyses
SNV and CNV were assessed concomitantly to test differences between groups. Using GISTIC-thresholded CNV calls by gene, we categorized genomic aberrations labeled by GISTIC as “2” as an amplification and GISTIC category “–1” or “–2” as a copy number deletion. For each sample and each gene, genomic aberrations were categorized as “0” or “1” (i.e., absent or present, respectively). An aberration was defined as the presence of either a nonsynonymous mutation, high-level amplification (as categorized by GISTIC), or copy number loss (as categorized by GISTIC). The merged SNV and CNV data were used for MCA, which was performed using the R package “FactoMineR”. MCA analysis was performed as a method to identify variation and patterns in the genomic aberrations among the groups of interest (i.e., primary HPV-related OPSCCs that did not or did recur, metachronous recurrent OPSCCs, and HPV-unrelated HNC). For each sample, every gene covered by WES was categorized as either having a genomic aberration (i.e., SNV or CNV) or not. Associations between genomic features are represented graphically in the MCA plot, illustrating the statistical relationships among distinct features. The average position for each group is plotted to aid in distinguishing genomic similarities and differences between groups and identifying the dimensions with the most data variability. Thus, groups that plot close to each other on the ordinate and/or abscissa are related statistically. The first 2 MCA dimensions are plotted accounting for the greatest source of variation among the samples. The genomic aberrations included in the analysis were those occurring in a set of 41 HPV-negative–like genes (Supplemental Table 16) that were significantly affected by SNVs or CNVs in TCGA (16). The integrated SNV and CNV data were also categorized for pathway analyses limited to variants affecting genes within pathways of interest, including mitogenic pathways (PI3K, JAK/STAT, MAPK, RTK), DNA damage response, cell cycle, cell death, and differentiation pathways. Pathway constituents were defined as follows: PI3K (PIK3CA/D/G, PIK3AP1, PIK3C2A/B/G, PIK3IP1, PIK3R1/2/3/4/5/6, AKT1/2/3, MTOR, PTEN, PDK1, TSC1/2, RICTOR, RPTOR); JAK/STAT (JAK1/2/3, STAT1/2/3/4/5A/5B/6, SOCS3, SHP2, IL6, IL6R, IL6ST); MAPK (ERK1/2, MEK1/2, RAF1, ARAF, BRAF, HRAS, KRAS, NRAS, SHC1/2/3, GRB2); and RTK (EGFR, FGFR1/2/3, ERBB2, EGFR1, EPHA2, DDR2, MET). DDR, cell cycle, cell death, and differentiation pathway components are outlined in Supplemental Table 18.
Statistics
R programming software was used to perform statistical analyses. To test differences in proportions between comparison groups, we utilized Fisher’s exact test. To test for stochastic dominance between multiple groups, we utilized a Kruskal-Wallis test followed by Dunn’s test to assess multiple pairwise comparisons between groups. To account for multiple hypothesis testing, the FDR was controlled using the method of Benjamini and Hochberg. q values less than 0.1 were used to determine if findings were statistically significant. q < 0.25 are used by the GISTIC algorithm to determine if findings are of significance.
Study approval
This study protocol was review and approved by the Fred Hutchinson Cancer Research Center Institutional Review Office. Written consent was obtained for genomic characterization of tumor tissues with matched normal tissue or blood for all participants prior to inclusion in this study. This study abided by the Declaration of Helsinki principles.
Author contributions
EM, UD, and RAH conceived the experiment. RAH, MK, HP, JZ, and EQK collected the data. RAH, MK, EQK, and QZ analyzed the data. RAH, MK, HP, EQK, QZ, EM, UD, CC, CSC, SMS, SGL, and CPR interpreted the data. RAH, EQK, UD, and EM drafted the article. All authors participated in critical revision of the article. RAH, UD, and EM were involved in final approval of the article.
Supplementary Material
Acknowledgments
The authors would like to thank the patients for their participation in this project. We thank Navonil De Sarkar for advice and guidance on selecting and implementing copy number analyses. We thank David Coffey and Daniel Faden for advice and guidance with genomic analyses. We thank the members of our genomics shared resource at the Fred Hutchinson Cancer Research Center, including director Jeffrey Delrow, Ryan Basom for his guidance on bioinformatics and programming, and Andy Marty and Alyssa Dawson for their support with multiplexed sequencing. We thank David Gold and Paul Sampson of the UW Department of Biostatistics for biostatistical guidance and support in choosing appropriate regression models to test the association between genomic aberrations and tumor recurrence. We also thank Yuzheng Zhang for biostatistical and computational guidance. We thank Colin Pritchard for analysis of samples that underwent the OncoPlex assay. We thank Michael Kao and Chang Xu for facilitating sample collection and processing. This study is supported by the American Cancer Society (RSG TBG-123653), funding support for RAH (T32DC00018, Research Training in Otolaryngology, UW), funds to EM from Seattle Translational Tumor Research (Fred Hutchinson Cancer Research Center), and center funds from the Fred Hutchinson Cancer Research Center to EM. UD is supported by the Department of Veterans Affairs, BLR&D, IO1-oo23456, and funds from the Pittsburgh Foundation and PNC Foundation.
Version 1. 07/25/2018
Electronic publication
Version 2. 08/03/2018
The affiliations section was updated.
Funding Statement
Research Training in Otolaryngology
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists. This work does not represent the views of the US Government nor the Department of Veterans Affairs.
Reference information: JCI Insight. 2018;3(14):e99327. https://doi.org/10.1172/jci.insight.99327.
Contributor Information
Mark Kubik, Email: kubikmw@upmc.edu.
Qing Zhang, Email: qing@fredhutch.org.
Seok-Geun Lee, Email: seokgeun@gmail.com.
Heuijoon Park, Email: hpark2@fredhutch.org.
Jianan Zhang, Email: jzhang23@fredhutch.org.
Chu Chen, Email: cchen@fhcrc.org.
Cristina P. Rodriguez, Email: rodrigcr@seattlecca.org.
Umamaheswar Duvvuri, Email: duvvuriu@upmc.edu.
References
- 1.McQuillan G, Kruszon-Moran D, Markowitz LE, Unger ER, Paulose-Ram R. Prevalence of HPV in Adults Aged 18-69: United States, 2011-2014. NCHS Data Brief. 2017;(280):1–8. [PubMed] [Google Scholar]
- 2.Chaturvedi AK, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29(32):4294–4301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ang KK, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363(1):24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin BM, et al. Long-term prognosis and risk factors among patients with HPV-associated oropharyngeal squamous cell carcinoma. Cancer. 2013;119(19):3462–3471. doi: 10.1002/cncr.28250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fakhry C, et al. Human papillomavirus and overall survival after progression of oropharyngeal squamous cell carcinoma. J Clin Oncol. 2014;32(30):3365–3373. doi: 10.1200/JCO.2014.55.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelly JR, Husain ZA, Burtness B. Treatment de-intensification strategies for head and neck cancer. Eur J Cancer. 2016;68:125–133. doi: 10.1016/j.ejca.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wallace NA, Galloway DA. Manipulation of cellular DNA damage repair machinery facilitates propagation of human papillomaviruses. Semin Cancer Biol. 2014;26:30–42. doi: 10.1016/j.semcancer.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63(6):1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 10.Werness BA, Levine AJ, Howley PM. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248(4951):76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 11.Boyer SN, Wazer DE, Band V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996;56(20):4620–4624. [PubMed] [Google Scholar]
- 12.Wazer DE, Liu XL, Chu Q, Gao Q, Band V. Immortalization of distinct human mammary epithelial cell types by human papilloma virus 16 E6 or E7. Proc Natl Acad Sci USA. 1995;92(9):3687–3691. doi: 10.1073/pnas.92.9.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen JJ. Genomic Instability Induced By Human Papillomavirus Oncogenes. N Am J Med Sci (Boston) 2010;3(2):43–47. doi: 10.7156/v3i2p043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mesri EA, Feitelson MA, Munger K. Human viral oncogenesis: a cancer hallmarks analysis. Cell Host Microbe. 2014;15(3):266–282. doi: 10.1016/j.chom.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walline HM, et al. Genomic Integration of High-Risk HPV Alters Gene Expression in Oropharyngeal Squamous Cell Carcinoma. Mol Cancer Res. 2016;14(10):941–952. doi: 10.1158/1541-7786.MCR-16-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas Network Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris LG, et al. The molecular landscape of recurrent metastatic head neck cancers: insights from a precision oncology sequencing platform. JAMA Oncol. 2017;3(2):244–255. doi: 10.1001/jamaoncol.2016.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agrawal N, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333(6046):1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Amin N, et al. AJCC Cancer Staging Manual. 8th Ed. New York, NY: Springer International Publishing; 2017. [Google Scholar]
- 20.Pritchard CC, et al. Validation and implementation of targeted capture and sequencing for the detection of actionable mutation, copy number variation, and gene rearrangement in clinical cancer specimens. J Mol Diagn. 2014;16(1):56–67. doi: 10.1016/j.jmoldx.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oswald F, et al. A phospho-dependent mechanism involving NCoR and KMT2D controls a permissive chromatin state at Notch target genes. Nucleic Acids Res. 2016;44(10):4703–4720. doi: 10.1093/nar/gkw105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zehir A, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mancini R, et al. Tumor Regression Grade After Neoadjuvant Chemoradiation and Surgery for Low Rectal Cancer Evaluated by Multiple Correspondence Analysis: Ten Years as Minimum Follow-up. Clin Colorectal Cancer. 2018;17(1):e13–e19. doi: 10.1016/j.clcc.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Sharma A, et al. Human papillomavirus-positive oral cavity and oropharyngeal cancer patients do not have better quality-of-life trajectories. Otolaryngol Head Neck Surg. 2012;146(5):739–745. doi: 10.1177/0194599811434707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lui VW, et al. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discov. 2013;3(7):761–769. doi: 10.1158/2159-8290.CD-13-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang NJ, et al. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc Natl Acad Sci USA. 2011;108(43):17761–17766. doi: 10.1073/pnas.1114669108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sen GL, et al. ZNF750 is a p63 target gene that induces KLF4 to drive terminal epidermal differentiation. Dev Cell. 2012;22(3):669–677. doi: 10.1016/j.devcel.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Green MR, et al. Hierarchy in somatic mutations arising during genomic evolution and progression of follicular lymphoma. Blood. 2013;121(9):1604–1611. doi: 10.1182/blood-2012-09-457283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo C, et al. KMT2D maintains neoplastic cell proliferation and global histone H3 lysine 4 monomethylation. Oncotarget. 2013;4(11):2144–2153. doi: 10.18632/oncotarget.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ortega-Molina A, et al. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat Med. 2015;21(10):1199–1208. doi: 10.1038/nm.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bögershausen N, et al. RAP1-mediated MEK/ERK pathway defects in Kabuki syndrome. J Clin Invest. 2015;125(9):3585–3599. doi: 10.1172/JCI80102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toska E, et al. PI3K pathway regulates ER-dependent transcription in breast cancer through the epigenetic regulator KMT2D. Science. 2017;355(6331):1324–1330. doi: 10.1126/science.aah6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le DT, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bouwman P, Jonkers J. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat Rev Cancer. 2012;12(9):587–598. doi: 10.1038/nrc3342. [DOI] [PubMed] [Google Scholar]
- 35.Xia S, Zhao Y, Yu S, Zhang M. Activated PI3K/Akt/COX-2 pathway induces resistance to radiation in human cervical cancer HeLa cells. Cancer Biother Radiopharm. 2010;25(3):317–323. doi: 10.1089/cbr.2009.0707. [DOI] [PubMed] [Google Scholar]
- 36.Yu HG, et al. Phosphoinositide 3-kinase/Akt pathway plays an important role in chemoresistance of gastric cancer cells against etoposide and doxorubicin induced cell death. Int J Cancer. 2008;122(2):433–443. doi: 10.1002/ijc.23049. [DOI] [PubMed] [Google Scholar]
- 37.Jin W, Wu L, Liang K, Liu B, Lu Y, Fan Z. Roles of the PI-3K and MEK pathways in Ras-mediated chemoresistance in breast cancer cells. Br J Cancer. 2003;89(1):185–191. doi: 10.1038/sj.bjc.6601048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menendez JA, Vellon L, Mehmi I, Teng PK, Griggs DW, Lupu R. A novel CYR61-triggered ‘CYR61-alphavbeta3 integrin loop’ regulates breast cancer cell survival and chemosensitivity through activation of ERK1/ERK2 MAPK signaling pathway. Oncogene. 2005;24(5):761–779. doi: 10.1038/sj.onc.1208238. [DOI] [PubMed] [Google Scholar]
- 39.Chang YS, Huang HD, Yeh KT, Chang JG. Evaluation of whole exome sequencing by targeted gene sequencing and Sanger sequencing. Clin Chim Acta. 2017;471:222–232. doi: 10.1016/j.cca.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 40.Miller EM, et al. Development and validation of a targeted next generation DNA sequencing panel outperforming whole exome sequencing for the identification of clinically relevant genetic variants. Oncotarget. 2017;8(60):102033–102045. doi: 10.18632/oncotarget.22116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goodman AM, et al. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol Cancer Ther. 2017;16(11):2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanna GJ, et al. Frameshift events predict anti-PD-1/L1 response in head and neck cancer. JCI Insight. 2018;3(4):98811. doi: 10.1172/jci.insight.98811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pai SG, et al. Correlation of tumor mutational burden and treatment outcomes in patients with colorectal cancer. J Gastrointest Oncol. 2017;8(5):858–866. doi: 10.21037/jgo.2017.06.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller CA, et al. SciClone: inferring clonal architecture and tracking the spatial and temporal patterns of tumor evolution. PLoS Comput Biol. 2014;10(8):e1003665. doi: 10.1371/journal.pcbi.1003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yap TA, Gerlinger M, Futreal PA, Pusztai L, Swanton C. Intratumor heterogeneity: seeing the wood for the trees. Sci Transl Med. 2012;4(127):127ps10. doi: 10.1126/scitranslmed.3003854. [DOI] [PubMed] [Google Scholar]
- 46.Mroz EA, Tward AD, Tward AM, Hammon RJ, Ren Y, Rocco JW. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from the Cancer Genome Atlas. PLoS Med. 2015;12(2):e1001786. doi: 10.1371/journal.pmed.1001786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Allen EM, et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med. 2014;20(6):682–688. doi: 10.1038/nm.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Edge S, Byrd D, Compton C, Fritz A, Greene F, Trotti A. AJCC Cancer Staging Manual, 7th Edition. New York, NY: Springer-Verlag New York; 2010. [Google Scholar]
- 51.Shi SR, et al. DNA extraction from archival formalin-fixed, paraffin-embedded tissues: heat-induced retrieval in alkaline solution. Histochem Cell Biol. 2004;122(3):211–218. doi: 10.1007/s00418-004-0693-x. [DOI] [PubMed] [Google Scholar]
- 52.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bergmann EA, Chen BJ, Arora K, Vacic V, Zody MC. Conpair: concordance and contamination estimator for matched tumor-normal pairs. Bioinformatics. 2016;32(20):3196–3198. doi: 10.1093/bioinformatics/btw389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hedberg ML, et al. Genetic landscape of metastatic and recurrent head and neck squamous cell carcinoma. J Clin Invest. 2016;126(1):169–180. doi: 10.1172/JCI82066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Douville C, et al. CRAVAT: cancer-related analysis of variants toolkit. Bioinformatics. 2013;29(5):647–648. doi: 10.1093/bioinformatics/btt017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carter H, Douville C, Stenson PD, Cooper DN, Karchin R. Identifying Mendelian disease genes with the variant effect scoring tool. BMC Genomics. 2013;14 Suppl 3:S3. doi: 10.1186/1471-2164-14-S3-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mayakonda A, Koeffler HP. Maftools: Efficient analysis, visualization and summarization of MAF files from large-scale cohort based cancer studies. bioRxiv. https://www.biorxiv.org/content/early/2016/05/11/052662 Published May 11, 2016. Accessed June 26, 2018.
- 64.Amarasinghe KC, et al. Inferring copy number and genotype in tumour exome data. BMC Genomics. 2014;15:732. doi: 10.1186/1471-2164-15-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Quinlan AR. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr Protoc Bioinformatics. 2014;47:11.12.1–11.1234. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.