Abstract
The tough, hydrogel glue produced by the slug Arion subfuscus achieves impressive performance through metal-based, protein cross-links. The primary sequence of these proteins was determined through transcriptome sequencing and proteome analysis by tandem mass spectrometry. The main proteins that correlate with adhesive function are a group of eleven small, highly abundant lectin-like proteins. These proteins matched the ligand-binding C-lectin, C1q or H-lectin domains. The variety of different lectin-like proteins and their potential for oligomerization suggests that they function as versatile and potent cross-linkers. In addition, the glue contains five matrilin-like proteins that are rich in von Willebrand Factor A (VWA) and EGF domains. Both C-lectins and VWA domains are known to bind to ligands using divalent cations. These findings are consistent with the double network mechanism proposed for slug glue, with divalent ions serving as sacrificial bonds to dissipate energy.
Keywords: glue, hydrogel, double-network, RNA-Seq, lectin, von Willebrand Factor
Introduction
The slug Arion subfuscus (Draparnaud 1805) produces a defensive secretion from its dorsal surface that is a remarkably tough, adhesive hydrogel (Smith 2016). This material is dilute, containing roughly 97% water (Smith 2016). Unlike most dilute mucous gels, it is stiff, with an average elastic modulus of 30 kPa (Wilks et al. 2015). This is stiffer than 7% gelatin and approaches the stiffness of 20% gelatin (Eysturskard et al. 2009; Czerner et al. 2015). Unlike gelatin and other stiff gels, however, slug glue can be stretched over ten times its initial length (Wilks et al. 2015). The combination of stiffness and deformability makes it an unusually tough gel.
Previous work has demonstrated that proteins play a central role in the mechanics of the glue. They stiffen the glue, providing cohesive strength (Pawlicki et al. 2004, Braun et al. 2013), and they create an interpenetrating polymer network along with polysaccharides to toughen the glue through a double network mechanism (Wilks et al. 2015). Such double networks can be orders of magnitude tougher than simple gels (Gong, 2010). The main polysaccharide in the glue has been tentatively identified as heparan sulfate (Wilks et al. 2015). In contrast, little is known of the structure of the proteins.
Current structural information about selected slug glue proteins consists of general characterizations such as isoelectric point, apparent mass on SDS-PAGE, and amino acid composition (Smith 2016). Detailed structural information would complement the available information on the composition and mechanics of the glue and lead to much deeper insight. Thus, it would guide biomimetic syntheses of gels with similar properties.
More than ten different proteins are common in the glue and these proteins appear to have different functions (Smith 2016). Many are equally abundant in the glue and in the much weaker mucus used for locomotion, but several are characteristic of the glue (Pawlicki et al. 2004). The proteins that are characteristic of the glue can stiffen gels, and their presence correlates with a dramatic rise in toughness and adhesiveness (Pawlicki et al. 2004). They are metal-binding proteins, and the glue’s mechanics depend on metals such as iron, calcium and zinc, which may bind to different proteins and have different effects (Werneke et al. 2007). Interactions involving metal ions directly link polymers, while metal-catalyzed oxidation leads to other cross-links (Bradshaw et al. 2011; Braun et al. 2013). Finally, some of the proteins likely contribute to adhesive interactions with the substrate. The structural basis for all these actions is unknown. Thus, RNA-Seq was used to deduce the full amino acid sequence of the main proteins in slug glue.
Materials and Methods
The proteins in the glue of the slug Arion subfuscus were identified from a de novo generated transcriptome using tandem mass spectrometry. The proteins were identified as “Asmp” (Arion subfuscus mucus protein) followed by their apparent molecular mass (in thousands) on SDS-PAGE. Since proteins were only separated by one dimension, rather than two-dimensions, there were situations where multiple proteins were found in the same band due to their similar mass. In these cases, the different proteins were indicated by appended letters.
RNA-Seq
Total RNA was purified from the dorsal body wall of the slugs. Slugs were collected from forests near Ithaca, NY. Their anterior half was cut off with a scalpel and the viscera removed. The ventral surface of the foot was then removed leaving only the dorsal body wall. This portion was immediately placed in 10 volumes of cold RNAlater RNA Stabilization Reagent and stored overnight at 4°C. Total RNA was purified using a PureLink RNA mini kit (ThermoFisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions. The initial lysis was performed with a rotor/stator homogenizer using a ratio of 100 mg body wall per 2 ml of lysis buffer. After isolation, RNA concentration and purity was analyzed on a NanoDrop 2000 spectrophotometer (ThermoFisher Scientific). Since the ratio of 260:230nm absorbance indicated likely contamination from mucus polysaccharides, the RNA collected in the first round was purified a second time.
Purified slug total RNA was treated with DNAse I (Ambion, ThermoFisher Scientific) and column purified using an RNeasy mini kit (Qiagen, Germantown, MD, USA). First and second strand cDNA synthesis was performed using the Ovation RNA-Seq system V2 (NuGEN, San Carlos, CA, USA) with 100 ng total RNA following the manufacturer’s instructions. cDNA was fragmented by sonication and concentrated using Agencourt RNA-Clean XP beads (Beckman Coulter, Indianapolis, IN, USA) following the manufacturer’s instructions. Fragmentation analysis showed effective fragmentation with an average size of 323 bp. Adapters were ligated in preparation for RNA-seq using the Encore Rapid DR Multiplex System (NuGen) following the manufacturer’s instructions. The library was purified using Agencourt RNA-Clean XP beads and subjected to multiplex Illumina sequencing on a HiSeq2500 platform (Illumina, San Diego, CA, USA) using 100 bp, paired end reads.
Transcriptome assembly
De novo assembly of the reads was performed using Trinity software (Haas et al. 2013). Read quality was first analyzed and quality control was performed using tools based on the FASTX-toolkit available on the Galaxy web site (usegalaxy.org). FASTQ Quality filter was performed, eliminating all reads that did not have at least 90% of the cycles with quality scores of at least 30. Artifacts were removed using FASTX artifacts filter. Assembly was performed on the edited data using the Trinity software package on the supercomputing facilities of the University of Pittsburgh (PSC Blacklight). The resulting output file was analyzed on PSC Blacklight with Transdecoder software to identify and translate the longest open reading frames (ORFs) for each gene.
Protein identification
Proteins in the glue were separated by SDS-PAGE, digested by trypsin and analyzed by tandem mass spectrometry. Discontinuous polyacrylamide gels were used, with 7.5%, 10% or 15% acrylamide concentrations to resolve different proteins following the detailed protocols of Hames (1990). Samples of slug glue were collected following the methods of Werneke et al. (2007). This involved gently rubbing the dorsal surface of the slug with a metal spatula, which led the slug to secrete a large volume of adhesive mucus as a defense mechanism. Adhesive mucus samples were then homogenized in SDS-PAGE sample buffer (0.0625 M Tris-Cl, pH 6.8, 2% SDS, 5% 2-mercaptoethanol, 10% glycerol) to a concentration of 20 mg ml−1. Since most molluscan adhesive gels contain ~ 1–3% protein (Smith 2016), this concentration would amount to roughly 0.2–0.6 mg ml−1of protein. Bands were excised on a cleaned plastic surface and sent to the Taplin mass spectrometry facility (Harvard University). There, in-gel tryptic digests and microcapillary liquid chromatography followed by tandem mass spectrometry were performed. The identified peptides were used to search the database of translated polypeptides generated from the transcriptome. This procedure was replicated for each band.
Since one-dimensional gels were used, there were often minor proteins present in the same band as the major proteins from the glue. The major proteins in each band were distinguished from low abundance proteins using spectral counts. Spectral counts provide a reliable way to estimate protein abundance in an LC-MS/MS experiment (Liu et al. 2004), especially when adjusting for the fact that larger proteins have more observable peptides (Kito & Ito 2008). An abundance index was calculated by dividing the total number of MS/MS spectra that mapped to a specific transcript by the number of unique peptides observed for that transcript. Only proteins that had a score greater than 1.5 were considered, thus eliminating proteins where most of its peptides were only detected once. Additionally, only transcripts identified by more than one peptide were considered.
The identification of the primary glue proteins based on spectral counts was further verified by several different methods. The proteins coded by the transcripts were analyzed using the ProtParam program (ExPASy Bioinformatics Resource Portal, Expasy.org) (Gasteiger et al. 2005) to determine the predicted molecular weight and pI. The relative abundance of transcripts coding for the protein was calculated using RSEM software. Finally, proteins coded by the transcripts were analyzed with the signal peptide prediction software PrediSi (predisi.de). Protein identity was considered to be reliable if it was based on a substantially larger number of spectral counts than contaminating proteins, the transcript coded for roughly the correct mass, a signal peptide indicating secretion was present, and the transcripts were among the most abundant in the dorsal body wall. Relative transcript abundance is an effective criterion in this case, because the adhesive secretion can amount to over 5% of the animal’s body mass (Martin & Deyrup-Olsen 1986).
In the case of Asmp44, Asmp57a, Asmp114 and Asmp203, the protein matched a transcript that was not complete due to the absence of a start or stop codon. For the first three of these, 5’ and 3’ rapid amplification of cDNA ends (RACE) was performed. This was not done for Asmp203, since its sequence was deemed sufficient for analysis. Invitrogen 3’ RACE (ThermoFisher Scientific) and Clontech Takara SMARTer RACE (for 5’ sequencing) (Mountain View, CA, USA) kits were used following the manufacturer’s instructions. RNA was prepared as described previously, followed by cDNA synthesis and Polymerase Chain Reaction (PCR) amplification using the known sequence of the partial transcript to design a gene specific primer, and an anchor primer supplied with the kit. All PCR reactions were performed using New England BioLabs Taq DNA polymerase (Ipswich, MA, USA), following the manufacturer’s suggested protocol, but with a gradient program to determine the ideal annealing temperatures for each primer pair. PCR products were extracted from agarose gels using the Omega Bio-Tek E.Z.N.A. Gel Extraction Kit (Norcross, GA, USA) following the recommended protocol. The extracted DNA was sequenced by GenScript (Piscataway, NJ, USA).
The identified ORFs minus signal peptides were analyzed with Basic Local Alignment Search Tool (BLAST) (Altschul et al. 1990) through the National Center for Biotechnology Information (NCBI). Pairwise and grouped comparisons between the sequences were also performed using multiple sequence alignment (Clustal Omega, EMBL-EBI) (Sievers et al. 2011). The amino acid composition was calculated using ProtParam software. Amino acids that were substantially more common or rare relative to typical proteins were identified by comparison to the average abundance of each amino acid among all proteins in the current UniProtKB/SwissProt database (UniProtKB/Swiss-Prot UniProt release 2017_03). In addition, secondary structure predictions were performed using the Gor IV secondary structure prediction method (Garnier et al. 1996; Combet et al. 2000). Coiled-coil predictions were made for some proteins using the Coils program (Lupas et al. 1991; Combet et al. 2000). Overall hydrophobicity was predicted using the GRAVY (Grand Average Hydropathicity) scale available through the ProtParam program. The ProtScale tool available through ExPASy was used to identify whether there were regions that were unusually hydrophilic or hydrophobic, using the Kyte-Doolittle scale (Kyte & Doolittle 1982). Tandem repeats in selected proteins were identified using the Tandem Repeats Finder (Benson 1999). Potential N-, O-, and C-glycosylation sites were identified with NetNGlyc 1.0 (Gupta and Brunak, 2002), NetOGlyc 4.0 (Steentoft et al. 2013), and NetCGlyc 1.0 (Julenius 2007) based on human or mammalian sequence data (Center for Biological Sequence Analysis, cbs.dtu.dk).
Results
Protein identification
The sequences of nineteen proteins representing the most abundant components of slug glue were determined (Supplemental material). Ten bands or regions on SDS-PAGE were analyzed by LC-MS/MS (Fig. 1). The proteins with relative mobility indicating a size equal to or greater than 40 kDa were sharp, single bands except Asmp57, which could be distinguished as two closely adjacent bands on lower percentage gels, and Asmp203, which was a sharp band on some gels, but fainter on others (Fig.1). The proteins with a relative mobility indicating a molecular mass less than 20 kDa were found in multiple closely spaced bands. In previous work, these proteins were collectively identified as Asmp15 because the bands varied in number and relative intensity, and it was not clear if they were separate proteins or different size variants of the same protein (Pawlicki et al. 2004; Werneke et al. 2007; Smith et al. 2009; Bradshaw et al. 2011; Wilks et al. 2015). Most commonly, there were two dark bands with a region below containing several less intense bands (Fig.1). These three regions were analyzed by LC-MS/MS.
Figure 1.
SDS-PAGE of a sample of A. subfuscus glue. Bands that were selected for LC-MS/MS analysis are labeled on the left. Proteins are identified as A. subfuscus mucus protein (Asmp) followed by the estimated mass in kDa based on molecular weight markers run on an adjacent lane. Asmp15 typically consists of multiple bands across a broad region. In previously published work, this group of proteins has appeared as one prominent band with several less intense bands or two to four similarly dark bands (Pawlicki et al., 2004; Werneke et al., 2007; Smith et al. 2009; Bradshaw et al. 2011; Wilks et al. 2015). Because of this variation, previous work had not distinguished them individually.
The bands for the higher molecular weight proteins in the glue (≥40 kDa) each contained one highly abundant protein with minimal contaminants (Table 1). While MS/MS was able to detect other proteins in each band, in all cases only one protein met the criteria of being identified by more than one peptide, having an average of more than 1.5 spectral counts of each peptide, and not being the main component of an adjacent band. The identified proteins greatly exceeded these minimal criteria. The proteins shown in Table 1 were identified by an average of 27 peptides each. The average number of spectral counts per unique peptide was 10. All of the identified transcripts were ranked in the top few hundred transcripts out of more than 100,000 assembled transcripts. For Asmp40, Asmp44, Asmp57b, and Asmp61, the molecular mass predicted by the identified transcript was on average only 3.6% different from the mass estimated by SDS-PAGE. The predicted masses for Asmp114 and Asmp165 were significantly shorter than their mobility on SDS-PAGE suggested. Both of these proteins were predicted to have numerous potential O-glycosylation sites and several potential N-glycosylation sites. Asmp203 was also predicted to have numerous potential glycosylation sites, while the other proteins in the glue typically had much fewer potential sites. Only one potential C-glycosylation site was detected; this was on Asmp15h, but it only barely met the prediction threshold. Asmp57a and Asmp203 were only partially sequenced. In summary, the transcripts identified as coding for the proteins in each band were among the most abundant of all transcripts in the dorsal body wall, and were identified by far more spectral counts than the next most common transcripts identified in the band. All of the full-length transcripts except for Asmp57b included signal peptides at the N-terminus, indicating that they were secreted.
Table 1:
Higher molecular weight proteins identified from SDS-PAGE by LC-MS/MS.
| Name | Predicted mass1 (kDa) | Predicted pI | Transcript abundance rank | Conserved domains |
|---|---|---|---|---|
| Asmp40 | 40.3 | 5.1 | 27 | None |
| Asmp44 | 45.8 | 5.5 | 102 | EGF, VWA |
| Asmp57a | >38.61 | 5.1 | 55 | EGF, VWA |
| Asmp57b | 55.3 | 6.6 | 310 | Catalase |
| Asmp61 | 56.9 | 5.5 | 61 | EGF, VWA |
| Asmp114 | 86.0 | 5.2 | 163 | EGF, VWA |
| Asmp165 | 120.5 | 5.0 | 57 | EGF, VWA |
| Asmp203 | >163.51 | 5.1 | 53 | Tyrosinase, hemocyanin |
Mass predicted from the open reading frame, with the predicted signal peptide removed. Asmp57a and Asmp203 were not complete, lacking a start or stop codon respectively.
All proteins were identified using MS/MS on digested SDS-PAGE bands. Only transcripts identified by more than one peptide, with an abundance index >1.5 were considered. Abundance rank was determined by RSEM and indicates the rank out of the entire transcriptome. Conserved domains were determined by BLAST. EGF = Epidermal Growth Factor, VWA = von Willebrand Factor A.
The bands for the lower molecular weight proteins in the glue contained at least 11 abundant proteins, all with similarity to lectins (Table 2). On average, each protein was identified by 6 unique peptides, and there were on average 17 spectral counts per unique peptide. The identified transcripts were all among the most abundant in the dorsal body wall, with an average rank of 45 out of the entire transcriptome. Six of the proteins were among the top thirty in the transcriptome. For perspective, the most common of these transcripts (Asmp15a) was more abundant than transcripts for paramyosin, tropomyosin and myosin light chain, and 27% as abundant as actin in the muscular dorsal body wall, based on fragments per kilobase million (FPKM). All of the transcripts included signal peptides, and the predicted molecular masses after signal peptide cleavage matched the mass estimated from SDS-PAGE well.
Table 2.
Proteins with an apparent mass < 20 kDa, identified as Asmp15 from the top, middle and low parts of the region. Table 2
| Name | Predicted mass (kDa) | pI | Transcript abundance rank | Conserved domains |
|---|---|---|---|---|
| Top | ||||
| Asmp15a | 15.7 | 8.6 | 14 | C-lectin |
| Asmp15b | 15.8 | 6.1 | 20 | C-lectin |
| Asmp15c | 16.3 | 4.9 | 25 | C-lectin |
| Middle | ||||
| Asmp15d | 14.7 | 8.9 | 29 | C1q |
| Asmp15e | 15.4 | 8.8 | 15 | C1q |
| Asmp15f | 14.9 | 6.9 | 17 | C-lectin |
| Low | ||||
| Asmp15g | 11.1 | 9.4 | 36 | H-lectin |
| Asmp15h | 12.1 | 9.2 | 63 | H-lectin |
| Asmp15i | 14.7 | 8.1 | 37 | C1q |
| Asmp15j | 12.1 | 8.0 | 175 | H-lectin |
| Asmp15k | 14.6 | 6.4 | 58 | C1q |
All proteins identified using MS/MS on digested SDS-PAGE bands. Only proteins identified by more than one peptide, with an abundance index >1.5 were considered. Mass was predicted from the open reading frame, with the predicted signal peptide removed. Abundance rank was determined by RSEM and indicates the rank out of the entire transcriptome. Conserved domains were determined by BLAST.
Conserved domains and protein families
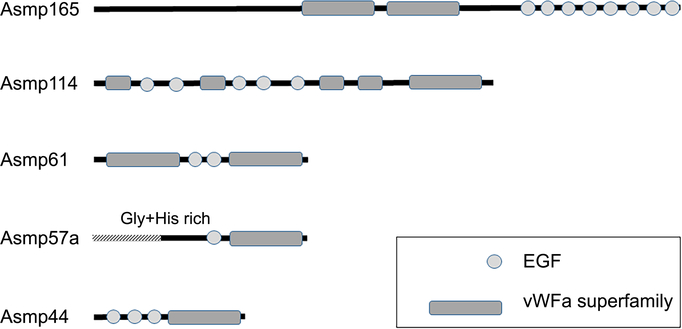
Analysis by BLAST showed that most of the proteins fit into groups defined by the presence of specific conserved domains (Tables 1 + 2). Five of the proteins larger than 40 kDa contained one or more von Willebrand Factor A (VWA) domains and multiple epidermal growth factor (EGF) repeats (Fig. 2). These proteins had closest sequence similarity to a number of matrilins, especially matrilin 2. Thus, they were named matrilin-like proteins. Nevertheless, while they had significant identity with regions of known matrilins, they typically only matched along 50–80% of the sequence, and the highest identity within that region was between 27–39%. The slug glue proteins also had sequence similarity to two matrilin-like proteins from the mucus of the slug Ambigolimax valentianus (Li & Graham 2007), with slightly higher identity in the covered regions. The VWA domains of the matrilin-like proteins all contained metal ion dependent adhesion sites (MIDAS). Asmp61 contained one VWA domain on each end of the protein. In addition, most of the EGF domains were predicted to bind calcium. All of the matrilin-like proteins were predicted to have between 13–24% alpha helix, 23–32% extended strand (beta), roughly 10% beta turn and 34–46% random coil. Because matrilins are characterized by an alpha-helical, coiled-coil forming region at the C-terminus (Wagener et al. 2005), this region was analyzed further in the slug proteins. The slug proteins asmp44, asmp57a, asmp61 and asmp114 all had a short, roughly 20 amino acid sequence at the C-terminal end after the C-terminal VWA domain. These were all predicted to form alpha helices, but they lacked the heptad repeat structure characteristic of the coiled-coil regions in matrilins.
Figure 2.
Notable domains found in the matrilin-like proteins. Proteins are drawn to scale based on their apparent mass on SDS-PAGE. The relative position and size of the conserved domains is proportional to the number of amino acids residues in each, relative to the whole protein. The shaded region of Asmp57a is unsequenced.
Asmp57a was not fully sequenced; the transcript that matched this protein lacked an open reading frame due to the absence of a start codon, and 5’ RACE added some sequence, but was not able to complete the sequence. The 113 residues at the N-terminal part of the protein coded by this transcript consisted primarily of glycine- and histidine-rich repeats with a total of 45% glycine and 21% histidine. There were two repeats of RGDGHPHGEDHKHGEGQG, three repeats of GGGXXFFL with a Y substituting for an F in one case and A substituting for L in another and four repeats of GGGHGHHT with Q substituting for an H in one case. Following this there was a sequence of seven consecutive glycine residues. 5’RACE beyond this region identified similar glycine- and histidine-rich repeats, but the repetitive nature of this region was associated with inconsistencies which complicated the attempt to integrate the sequence data.
In addition to the matrilin-like proteins, there were three other proteins ≥40 kDa. Asmp57b appeared to be the enzyme catalase; it was the same size as catalase and shared 69–70% identity with catalase from scallops (Mizuhopecten) and water fleas (Daphnia). Asmp203 appeared to be hemocyanin, since the whole transcript had 70% identity with a region of hemocyanin beta subunit from the snail Helix. Asmp203 had a high RSEM rank, but was only identified by 2.2 spectra per peptide, and it was not always a clear band on SDS-PAGE of glue samples. Because it was not consistently present, and had high identity with a possible contaminant, RACE was not used to complete the sequence. Finally, BLAST identified no similar proteins for asmp40, and no conserved domains.
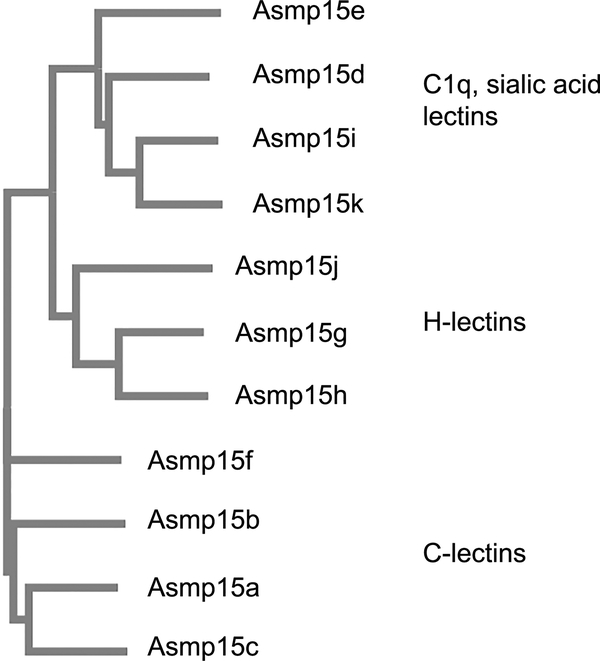
The proteins identified in the group that migrated on SDS-PAGE with apparent masses near 15 kDa all had similarity to known lectins. While the percent identity ranged from only 31–54%, they matched the known lectins in size, with the transcripts covering 89–100% of the matched protein. The proteins near 15 kDa were grouped in three primary categories by Clustal (Fig. 3). One group consisted of the proteins that were most commonly found in the top band in this region, and these were identified as C-lectins. Another group consisted of C1q domain containing proteins, and these were found most commonly in the lower of the two dark bands in this region of the gel. In both cases, the conserved domain encompassed 85–90% of the sequence. The C1q domain-containing proteins had similarity to sialic acid lectins from the garden snail Cepaea or the abalone Haliotis (35–42% identity). The last group found in the region of fainter bands below the two intense bands consisted of proteins with conserved H-lectin domains, and similarity to discoidin II and Helix agglutinins (31–46% identity).
Figure 3.
Relatedness among proteins identified as Asmp15. Relationships were determined using Clustal Omega software.
The proteins containing C-lectin domains consisted of three abundant proteins present in roughly similar quantities, and one less abundant protein. The transcripts were similarly abundant based on RSEM values (Table 2), and the ratio of total spectral counts was 1 : 1 : 0.8 : 0.5. These proteins had conserved pairs of cysteine residues, with the 3rd and 6th being present in all, and the 1st and 2nd or 4th and 5th absent only in Asmp15f and Asmp15c respectively (Fig. 4a). The cysteines occurred in highly conserved positions characteristic of C-lectins, where they typically form disulfide bonds (Drickamer 1988). Additionally, the slug C-lectins contained W—GEPN and WND sequences before and after the 4th cysteine respectively, as well as a glutamate residue just upstream of that cysteine; these are conserved glycan and calcium-binding sequences in C-lectins (Drickamer 1988; Zelensky & Gready 2005; Cummings & McEver 2009). The secondary structure predictions suggested a composition of roughly half random coil with the rest consisting of a mix of alpha helix and beta sheet. The locations of the coil, helix and beta sheet were consistent with the general structure of the C-lectin carbohydrate recognition domain (Weis et al. 1992). Despite their similarity, there was significant sequence variation among the C-lectins. The most closely related were Asmp15a and Asmp15c, which shared 47% identity. Asmp15c and Asmp15f only shared 33% identity. Their predicted iso-electric points of 4.9, 6.1, 6.9 and 8.6 differed widely.
Figure 4.
(a) Clustal alignment of the four C-lectin domain containing proteins. Cysteine residues are boxed. All tryptophan and phenylalanine residues are highlighted in bold, as are tyrosines found at locations where the presence of an aromatic amino acid is conserved. Shaded asterisks indicate matches with consensus calcium and carbohydrate-binding sites. (b) Clustal alignment of C1q-containing proteins, with all tyrosine and phenylalanine residues highlighted in bold. There are no conserved cysteine residues. Asterisks indicate conserved sites, and dots indicate sites that contain amino acids with similar properties. Letters on the left are the letter identifiers appended to Asmp15 to distinguish the different proteins.
The C-lectins exhibited notable differences in the relative frequency of specific amino acids relative to the global average on UniProtKB/SwissProt. They had high levels of asparagine (6.2–10.6% vs. 4.0% global average), and were unusually rich in aromatic amino acids (12.3–15.7% vs. 7.7% global average), especially tryptophan (4.4–5.8% vs. 1.0% global average). The position of the aromatic amino acid residues was highly conserved, with sites holding aromatic amino acids making up a disproportionate amount of the conserved residues (Fig. 4a). Notably, almost all the tryptophan and phenylalanine occurred where the presence of an aromatic amino acid was conserved (Fig. 4a).
The slug glue C-lectins were most similar to incilarin (47–54% identity), a C-lectin from the mucus of the slug Incilaria fruhstorferi (Yuasa et al. 1998). The most common other matches (30–40% identity) were perlucin, ladderlectin, aggrecan core protein, and mannose-binding protein. Overall, these proteins were generally hydrophilic, with a Grand Average Hydropathicity (GRAVY) score of −0.44 to −0.81 (scale ranges from −2 to +2 with negative numbers being hydrophilic and positive numbers being hydrophobic). The hydrophobic and hydrophilic residues were spread throughout the protein, with no clear regionalization.
The proteins containing C1q domains consisted of four main proteins. They aligned with the globular C1q domain identified by Ressl et al. (2015), matching the length and conserved residues of this domain. They did not contain the collagen domain of other C1q-like proteins. Of the four C1q-containing slug glue proteins, Asmp15d and Asmp15e were more abundant based on RSEM, and had significantly more spectral counts. The ratio of spectral counts of these two was almost exactly 1:1 (366 : 365). The other two were markedly less abundant (< 1/10 of the spectral counts). Another similar protein was also identified, but not included in Table 2 because it was only identified by one peptide, though this peptide had a similar number of spectral counts to Asmp15i and Asmp15k, had a similar RSEM rank, a high pI (9.5), and was predicted to be in the same size range.
The four main C1q proteins were similar in many ways, but as with the C-lectins, they did not share strong sequence identity. The two most closely related, Asmp15i and Asmp15k, shared 55% identity but the others shared only 30–40% identity. Their isoelectric points were typically basic, ranging from 8.1 to 8.9, with only Asmp15k being more neutral (6.4). The amino acid compositions of these proteins were unusual in their relatively high concentrations of the aromatic amino acids tyrosine and phenylalanine (11.8 – 14% combined vs 6.7% global average), though with only 0–0.7% tryptophan. As with the C-lectins, the positions of these aromatic amino acids was typically conserved (Fig. 4b). These proteins were most similar to sialic acid lectins from other molluscs. Overall, these proteins were neither hydrophilic nor hydrophobic, with GRAVY scores between −0.19 and 0.21.
The final group of lectin-like proteins were the proteins with H-lectin domains. These were somewhat less abundant than the others based on RSEM ranks, spectral counts and staining intensity on SDS-PAGE. Asmp15g and Asmp15h were somewhat more abundant, but overall comparable to the less abundant of the C1q-containing proteins. The two most common H-lectins were found in nearly equal amounts (the ratio of spectral counts was 1:0.9). Like the other proteins in this size range, they did not share a high percentage of amino acid identity with each other. The most closely related of the group, Asmp15g and Asmp15h, shared 50% identity. This group of proteins was more strongly basic, with predicted pI values ranging from 8 to 9.4. They were unusually rich in glutamine (6.1–6.7% vs. 3.9% global average) and the hydroxylated amino acids serine and threonine (average 20.1% combined vs 11.9% global average). The H-lectins were slightly hydrophilic overall, with GRAVY scores between −0.22 and −0.56. There was one other transcript identified that did not meet the criteria for inclusion in Table 2 because it was only identified by one peptide, but it was notable for a high spectral count (comparable to the other proteins in this group), high RSEM value (19), and high pI (9.2). It did not match any known proteins, and it was characterized by a charged, glutamate-rich domain near the N-terminus (-EEENER-), closely followed by a 39 amino acid sequence containing 31 glycine and leucine residues (79%).
Discussion
RNA sequencing and mass spectrometry identified all the major proteins in the glue, most of which fall within two classes, lectin-like proteins and matrilin-like proteins (VWA and EGF-rich). These were among the most abundant proteins produced by the dorsal body wall of the slug. The lectin-like proteins were notable in that, in each case, nearly their entire sequence consisted of a single, conserved binding domain. Despite fitting into broad ligand-binding categories, they differed significantly among each other in sequence. The number and diversity of lectin-like proteins suggests the potential for binding to a wide variety of ligands, thus possibly serving roles in adhesion and cross-linking. This is consistent with previous evidence showing that these proteins were correlated with adhesion and caused gel-stiffening (Pawlicki et al. 2004). The matrilin-like proteins contained multiple conserved domains that are known to contribute to intermolecular cross-linking, especially via divalent metal ions such as calcium (Whittaker and Hynes 2002). In the glue, these proteins form large, non-covalently linked complexes that appear to contribute substantially to cohesive strength (Wilks et al. 2015). Their similar structure is consistent with a role as bulk adhesive polymers. In summary, the abundance of two types of protein that are characteristically involved in intermolecular interactions fits well with the function of the glue. Furthermore, these results suggest a model for glue function.
Lectin-like proteins
The lectin-like proteins formed a group of bands with an apparent mass near 15 kDa on SDS-PAGE. These proteins fit into three distinct categories: C-lectins, C1q-containing proteins and H-lectins. The C1q-containing proteins had significant sequence similarity to sialic acid lectins. The C-lectins are particularly interesting, since C-lectin domains typically depend on calcium for ligand binding (Drickamer 1988). Previous work has demonstrated that the lectin-like proteins in slug glue bind iron tightly, and iron and calcium share similar ligands (Werneke et al. 2007). Calcium also plays a central role in stiffening the glue (Braun et al. 2013). C1q-like proteins may also use calcium in ligand binding (Ressl et al. 2015), though unlike the C-lectins, the C1q-containing proteins in slug glue do not have the conserved residues implicated in calcium binding.
Within each of the three categories of lectin-like proteins in slug glue, there were three or four abundant proteins. Each shared the same general size and conserved domains, yet they typically shared only 30–50% amino acid identity with each other. The C-lectins in particular had widely different predicted isoelectric points. Thus, the group of all lectin-like proteins consisted of at least eleven proteins with significant diversity among them.
The sequence variation among these proteins suggests that each may have a different binding activity, thus creating many possible interaction sites. Different proteins bearing C1q domains can have very different ligand specificities (Kishore et al. 2004; Gaboriaud et al. 2011). Similarly, C-lectins include many different proteins that interact with markedly different ligands. Like immunoglobins, C-lectins have a common structure with a stable scaffold that allows them to present a highly variable binding domain (McMahon et al. 2005). This is due in part to highly conserved cysteines that form disulfide bonds, creating a characteristic C-lectin fold (Drickamer 1988; Cummings & McEver 2009). Because of the diversity of lectins, with each forming specific interactions, lectins often serve in the innate immune system of invertebrates (Loker 2010). This function could be co-opted to serve adhesive and cross-linking functions.
The range of potential ligands for lectins has been studied extensively for C-lectins. In an analysis of the C. elegans genome, roughly 180 proteins with C-type lectin domains were identified, but only 10% were predicted to bind to carbohydrates (Drickamer & Dodd 1999). Different C-lectins have been shown to bind to proteins, nucleic acids, lipoproteins and inorganic surfaces (Drickamer 1999; Zelensky & Gready 2005). Thus, they represent a versatile class of ligand-binding proteins. Notably, one of the common matches for the C-lectins in slug glue was perlucin. Perlucin is the primary organic component of abalone shells, and can trigger calcium carbonate precipitation from solution (Weiss et al. 2000). Mineral binding is not unusual among C-lectins; proteins such as tetranectin, ovocleidin and lithostathine are C-lectins that are the major components of the matrix of bone, egg shells and pancreatic stones respectively (Patard et al. 2003). Ladderlectin was also identified as having significant sequence similarity to some C-lectin proteins in slug glue. Ladderlectin is a chitin-binding lectin used in the trout immune system (Russell et al. 2008). In the case of slug glue, it is plausible that some of the lectin-like proteins interact with carbohydrates in the glue, some interact with proteins in the glue, and some interact with different ligands on the surfaces to which the glue may adhere.
Another important feature of lectins is that they typically form oligomers. If each lectin-like protein in slug glue only bound to one other molecule, that would not lead to cross-links. In contrast, a lectin oligomer could bring several polymers together. C-lectins typically form homo and heterotrimers, as well as dimers (Zelensky & Gready 2005; Cummings & McEver 2009), H-lectins such as Helix agglutinin can form hexamers by non-covalent trimerization of disulfide-linked dimers (Sanchez et al. 2006), and C1q domains also typically form homo or heterotrimers (Kishore & Reid 2000; Ressl et al. 2015). Such oligomers create a “bouquet” of binding sites. A heterotrimer, for instance, would present three different binding faces. Such a molecule could bring together three different surfaces. This is the case for human C1q (Gaboriaud et al. 2011). Given this, it is striking that the three most abundant C-lectins in slug glue were present in nearly equal amounts (1:1:0.8 ratio), as were the two most abundant C1q proteins (1:1), and the two most abundant H-lectins (1:0.9). This is consistent with hetero-oligomerization. The possibility of using lectin oligomers presenting similar or different binding regions is a novel and exciting design approach that could guide biomimetic glue development.
It is worth noting that many of the lectins in slug glue were predicted to be basic, especially the H-lectins. This feature would also be important in the glue. The remainder of the glue is strongly polyanionic, especially the polysaccharides, which have been tentatively identified as heparan sulfate (Wilks et al. 2015). Thus, a basic protein would likely interact electrostatically with the polyanions, and also serve to balance their charge (Smith 2013). It would also interact well with most environmental surfaces, which tend to be negatively charged due to the exopolysaccharides of bacterial biofilms (Stewart et al. 2011). C1q-domains are noted for being good ligands for polyanionic molecules (Kishore et al. 2004).
Another interesting feature of the lectin-like proteins was the prevalence of conserved aromatic amino acid residues. Aromatic side chains are ubiquitous in the carbohydrate binding modules of glycoside hydrolases, where they play a primary role in determining specificity and affinity (Boraston et al. 2004). In many cases it is the hydrophobicity of aromatic side chains that is essential (Boraston et al. 2004), but aromatic side chains can also serve as good cross-linking sites because the electron-rich pi orbitals interact well with positively charged side chains, creating unusually high binding energies for non-covalent interactions (Dougherty 2007; Gebbie et al. 2017). Such cation pi interactions may play an important role in the cohesive strength of the mussel byssus (Gebbie et al. 2017). It is particularly noteworthy that the position of the aromatic residues was strongly conserved in the C-lectins and C1q-containing proteins. Furthermore, aromatic amino acids made up a disproportionate amount of the conserved residues. This high conservation combined with the known role of aromatic side chains in carbohydrate binding and cation-pi interactions suggests that interactions involving these amino acids play a significant role in the function of this glue.
Matrilin-like proteins
The other major group of proteins in the glue were the matrilin-like proteins. These included the proteins identified as Asmp44, Asmp57a, Asmp61, Asmp114 and Asmp165. They were characterized by EGF repeats and VWA domains. These proteins appeared to be related to two matrilin-like proteins identified by Li and Graham (2007) in the mucus of the slug Lehmannia (Ambigolimax) valentiana. Li and Graham (2007) hypothesized that these proteins were part of an interrelated family of proteins that use VWA domains to cross-link components of the mucus. The findings for A. subfuscus glue supported that hypothesis. The matrilin-like proteins appear to play a role in both non-adhesive and adhesive mucus. They are found in both types of mucus in A. subfuscus (Pawlicki et al. 2004). In addition, the mucus studied by Li and Graham (2007) was from a slug in a different family that is not known for producing a defensive secretion, and Li and Graham reported that the dorsal mucus was not notably different from the mucus used in locomotion. Thus, the matrilin-like proteins may represent a common way to create a general-purpose mucus in slugs.
Matrilin-like proteins are likely to play an important role in mucus mechanics because they commonly serve to cross-link extracellular matrix proteins, and they are typically involved in multiprotein complex formation (Whittaker & Hynes 2002). Matrilins are best known for their role in creating fibrillary structure in cartilage, and binding to aggrecan core protein in cartilage (Wagener et al. 2005). As with slug glue, cartilage is a tough hydrogel consisting of protein and polysaccharide networks. Notably, some of the C-lectins had significant similarity to aggrecan core protein.
An important aspect of matrilins is that they contain Metal Ion Dependent Adhesion Sites (MIDAS), which use calcium or magnesium in cross-linking interactions (Whittaker & Hynes 2002). Similarly, some EGF domains bind to calcium and can be involved in interactions with other proteins (Rao et al. 1995). Thus, this group of proteins likely binds substantial amounts of calcium and uses it in intermolecular cross-links. The ability to cross-link polymers using calcium is consistent with the unusually high concentration of calcium in the glue, and the known importance of metal ions such as calcium in the glue’s stiffness (Braun et al. 2013).
A calcium cross-linked network fits well with the double network mechanism for gel toughening. Unlike typical molluscan mucus, which is highly deformable because it consists of a single network of unusually large polysaccharides (Smith 2016), slug glue is highly extensible but also stiff. This property is attributable to its double network of polysaccharides and proteins. The combination of a deformable network and a stiff network can give values of toughness that are 2–3 orders of magnitude greater than either network separately (Gong 2010; Haque et al. 2012). Wilks et al. (2015) demonstrated such an increase in toughness in slug glue, and hypothesized that the polysaccharides provide hidden length, allowing the glue to deform extensively before fracture. They also hypothesized that the proteins stiffen the gel, and the cross-links between them act as sacrificial bonds to dissipate energy. These would rupture continuously as the glue extends, leading to large energy dissipations and thus toughness.
Calcium-based interactions between matrilins and lectins are likely to form the sacrificial bonds of the double network. They would provide relatively stable but reversible links that, by virtue of their large numbers, could account for the large amount of energy required to fracture the glue. Calcium is a “hard” ligand that is coordinated well by carboxyl groups and other hard ligands (Lippard & Berg 1994). In this respect it is similar to iron, and it could contribute despite its lower affinity, due to its greater abundance (Hwang et al. 2010). These properties would work well for a double network. If the sacrificial bonds were too strong, they might not fail, leading to the glue fracturing by simple crack propagation along the fracture plane, rather than dissipation through a large volume. Thus, the glue would be brittle. For the mechanism to work efficiently, it is best to have a large number of relatively stable bonds that can fracture at the stresses typically seen during deformation of the polysaccharide network. The presence of a large number of calcium-binding, fiber cross-linking proteins is consistent with this model.
As with the lectin-like proteins, most matrilins form trimers or tetramers (Wagener et al. 2005; Klatt et al. 2011). They oligomerize through C-terminal alpha helices that form coiled coils (Wagener et al. 2005; Klatt et al. 2011). Most of the matrilin-like proteins in slug glue were predicted to have alpha helical regions at the C-terminus. Nevertheless, these did not match the expected heptad repeat for coiled-coil formation. Thus, it is unclear if the matrilin-like proteins of slug glue oligomerize.
Another interesting aspect of matrilins is that they can achieve versatility in ligand-binding through variation created by alternative splicing (Klatt et al. 2011). Although the different proteins in slug glue did not appear to be splice variants based on the transcriptome assembly, there was considerable variation in the arrangement of VWA and EGF domains in the matrilin-like proteins of slug glue (Fig. 2). Asmp61, which unlike the other matrilin-like proteins is characteristic of the glue, had VWA domains on either end of the protein. This configuration is similar to typical matrilins (Wagener et al. 2005), and it would seem to render the protein better able to form cross-links. In contrast, Asmp114, had four smaller VWA domains and one normal sized one at the C-terminus, while Asmp44 and Asmp57a had only one VWA domain. In addition, Bradshaw et al. (2011) found variation in the extent of oxidation among these proteins, and this oxidation appeared to contribute to their ability to cross-link (Bradshaw et al. 2011; Braun et al. 2013).
Asmp57a was notable in that it had an unusually glycine- and histidine-rich region. The full sequence of this protein was difficult to determine, presumably because of the repeats. Highly repetitive regions and homopolymeric strings of bases can be difficult to assemble in a de novo transcriptome. They can also complicate the design of unique primers for RACE applications. In any case, the sequence of this protein suggests an important role. Histidine is a common zinc and copper binding motif, and zinc is abundant in slug glue (Werneke et al. 2007). Furthermore, the amino acid composition recalls the structure of the zinc-binding protein Nvjp-1a, which hardens the jaws of nereid worms (Broomell et al. 2008). Nvjp-1a has 36% glycine and 27% histidine (Broomell et al. 2008), which is similar to the 5’ end of the transcript for Asmp57a (45% gly and 21% his). A similar proportion of glycine and histidine was found in spinalin, a protein implicated in strengthening the spines of cnidarian nematocysts (Koch et al. 1998). Histidine-rich regions of the protein mcfp-4 from the mussel byssus may also have a role in cross-linking (Zhao and Waite 2006). Interestingly, Braun et al. (2013) found that zinc removal did not reduce the stiffness of slug glue, so it may have some other mechanical function.
It is also worth noting that Asmp165 and Asmp114 were the only fully sequenced proteins whose predicted masses did not closely match their apparent masses on SDS-PAGE. Bioinformatics analysis found that these proteins contained numerous potential glycosylation sites, especially for O-glycosylation. Such glycosylation could explain the discrepancy in mass. Asmp165 co-purifies with glycosaminoglycans in the glue (Wilks et al. 2015), suggesting a possible connection.
The use of lectins, von Willebrand domains, and EGF domains in biological adhesives is not unique to slug glue. C-lectin domains have been shown to serve as interfacial linkers, joining soft tissues to the byssus of the fan shell Atrina pectinata (Yoo et al. 2016). Galactose-binding lectin domains have been detected in sea star and sea urchin adhesive proteins (Hennebert et al. 2014; Lebesgue et al. 2016), as well as proteins from the adhesive disk of the freshwater cnidarian Hydra (Rodrigues et al. 2016). These proteins may be discoidin-like (Hennebert et al. 2015; Lebesgue et al. 2016), as was found for one of the H-lectins in slug glue. In the echinoderm glues, multiple lectin domains were present in larger proteins, as opposed to the situation in slug glue where each protein consisted of a single lectin domain, though the lectin-like proteins of slug glue may oligomerize to provide multiple binding sites in one larger complex. In addition to lectin domains, the primary sea star adhesive protein contained von Willebrand Factor D domains, while a matrix protein from mussel byssus contained two VWA domains joined by a short link region (Suhre et al. 2014). EGF domains have also been found in several other adhesive proteins. The protein Mfp-2 from mussel byssal plaques consists of eleven tandem EGF repeats. These can bind to calcium, and calcium binding leads to significant interactions between the proteins. These supplement the stronger DOPA-iron interactions (Hwang et al. 2010). The major adhesive protein of the sea star Asterias rubens contains one EGF domain, which is predicted to bind to calcium (Hennebert et al. 2014). Such calcium-binding EGF domains are common in extracellular proteins and can mediate protein-protein interactions (Rao et al. 1995).
Other slug glue proteins
The protein identified as Asmp40 was not identified as similar to any known protein, and it had no conserved domains. This protein is interesting because it forms large complexes that may be dissociated by treatments that would prevent or disrupt cross-linking due to metal catalyzed oxidation (Smith et al. 2009; Wilks et al. 2015). Since it did not have any domains that matched known calcium-binding domains, it is possible that this protein is oxidatively cross-linked instead. Because of the size of the complexes that it forms, it is likely that it plays a mechanical role. During purification of the large polysaccharides, substantial amounts of this protein were found in the same fractions as the polysaccharides, suggesting a possible association (Wilks et al. 2015).
There were two other proteins that were moderately abundant in the glue, and these were common proteins that might be present as contaminants, though it is possible that they play a role in adhesion. Asmp203 was identified as having 70% sequence identity with a subunit of hemocyanin. Hemocyanin is a major constituent of molluscan blood. Since the glue itself contains a large amount of fluid that presumably derives from the hemolymph, and Asmp203 was present irregularly in the glue, it is parsimonious to assume that it was a contaminant. Nevertheless, it is a metal-binding protein with conserved tyrosinase domains, though these are not catalytic in hemocyanin.
The other protein that may or may not be a contaminant was catalase. Catalase plays an important role in protecting many tissues from reactive oxygen species by converting hydrogen peroxide into water and oxygen (Zamocky et al. 2008). Since metal-catalyzed oxidation seems to play an important role in glue cross-linking and it involves reactive oxygen species, the presence of catalase might be a means of regulating that reaction. Nevertheless, catalase is an abundant and ubiquitous protein (Zamocky et al. 2008), and the transcript coding for it lacked a signal peptide, suggesting that it was not secreted. Thus, its presence is suggestive, but it could be a contaminant.
Summary and model
The structure of the sequenced proteins suggests a mechanism that is consistent with previous hypotheses of glue function. In this model, the normal mucus consists of heparan sulfate, which due to its polyanionic character would extend and form a deformable, physically entangled network. The non-adhesive mucus also contains several of the proteins that were identified as matrilin-like, and these may link together to increase the viscosity of the material.
The difference between the glue and the non-adhesive mucus is the marked increase in the concentration of Asmp61 and Asmp15a-k (Pawlicki et al. 2004). Based on their structure, these appear to be better cross-linking proteins. Asmp61 has VWA domains on either end and may thus introduce cross-links into the protein matrix. Asmp15a-k may function in cohesion and/or adhesion. The interaction between lectins and their ligands is potent (Yoo et al. 2016), and the addition of these proteins to the mucus could transform the material. The lectin-like proteins may also provide a range of binding motifs, with some binding to carbohydrates in the glue or on the surface, some binding to proteins, and some binding to inorganic surfaces. If, like many lectins, they can oligomerize, they would form multivalent complexes that could bring together different components of the glue and surface. The cross-links involving matrilins and C-lectins likely involve divalent metal ions. These would provide a large number of sacrificial bonds to enhance energy dissipation in the double network. Failure would require full deformation of the polysaccharide network and in the process that would rupture a large volume of bonds in the protein network (Wilks et al. 2015).
This study focused on the abundant proteins in slug glue, which are likely to play a structural role as fibers or cross-linkers. Presumably there are also enzymes that play important roles in the glue. Further work is necessary to identify these enzymes, which are likely to be found in much smaller quantities, and might not be visible as sharp bands on SDS-PAGE.
Supplementary Material
Acknowledgments
The authors are grateful for the assistance and expertise of Christoph Schorl, PhD, director of the Genomics Core Facility at Brown University.
Funding
This work was supported by the National Science Foundation under an XSEDE start-up grant to AMS [number MCB130091] and by a Rhode Island Research Alliance Collaborative Grant Award to JMB. JMB was also supported by grants from the National Institute of General Medical Sciences (P20GM103537 and P20GM104317) of the National Institutes of Health. CWR was supported by a grant from the National Institute of General Medical Sciences of the National Institutes of Health (P20GM103430).
Footnotes
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Benson G 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J. 382:769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw A, Salt M, Bell A, Zeitler M, Litra N, Smith AM. 2011. Cross-linking by protein oxidation in the rapidly setting gel-based glues of slugs. J Exp Biol. 214:1699–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun M, Menges M, Opoku F, Smith AM. 2013. The relative contribution of calcium, zinc and oxidation-based cross-links to the stiffness of Arion subfuscus glue. J Exp Biol. 216:1475–1483. [DOI] [PubMed] [Google Scholar]
- Broomell CC, Chase SF, Laue T, Waite JH. 2008. Cutting edge structural protein from the jaws of Nereis virens. Biomacromolecules. 9:1669–1677. [DOI] [PubMed] [Google Scholar]
- Combet C, Blanchet C, Geourjon C, Deleage G. 2000. NPS@: Network protein sequence analysis. Trends Biochem Sci. 25:147–150. [DOI] [PubMed] [Google Scholar]
- Cummings RD, McEver RP. 2009. C-type lectins In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of glycobiology, 2nd edition. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; Chapter 31. [PubMed] [Google Scholar]
- Czerner M, Fellay LS, Suárez MP, Frontini PM, Fasce LA. 2015. Determination of elastic modulus of gelatin gels by indentation experiments. Procedia Mater Sci. 8:287–296. [Google Scholar]
- Dougherty DA. 2007. Cation-π interactions involving aromatic amino acids. J Nutr. 137:1504S–1508S. [DOI] [PubMed] [Google Scholar]
- Drickamer K 1988. Two distinct classes of carbohydrate-recognition domains in animal lectins. J Biol Chem. 263:9557–9560. [PubMed] [Google Scholar]
- Drickamer K 1999. C-type lectin-like domains. Curr Opin Struct Biol. 9:585–590. [DOI] [PubMed] [Google Scholar]
- Drickamer K, Dodd RB. 1999. C-type lectin-like domains in Caenorhabditis elegans: predictions from the complete genome sequence. Glycobiology. 9:1357–1369. [DOI] [PubMed] [Google Scholar]
- Eysturskard J, Haug IJ, Ulset A-S, Draget KI. 2009. Mechanical properties of mammalian and fish gelatins based on their average molecular weight and molecular weight distribution. Food Hydrocolloids. 23:2315–2321. [Google Scholar]
- Gaboriaud C, Frachet P, Thielens NM, Arlaud GJ. 2011. The human C1q globular domain: structure and recognition of non-immune self ligands. Front Immunol. 2:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier J, Gibrat JF, Robson B. 1996. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 266:540–553. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. 2005. Protein identification and analysis tools on the ExPASy server In: Walker JM, editor. The proteomics protocols handbook. New York City: Humana Press; p. 571–607. [Google Scholar]
- Gebbie MA, Wei W, Schrader AM, Cristiani TR, Dobbs HA, Idso M, Chmelka BF, Waite JH, Israelachvilli JN. 2017. Tuning underwater adhesion with cation-π interactions. Nat Chem. 9:473–479. [DOI] [PubMed] [Google Scholar]
- Gong JP. 2010. Why are double network hydrogels so tough? Soft Matter. 6:2583–2590. [Google Scholar]
- Gupta R, Brunak S. 2002. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac Symp Biocomput. 7:310–322. [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M., Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat protoc. 8:1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hames BD. 1990. One dimensional polyacrylamide gel electrophoresis In: Hames BD, Rickwood D, editors. Gel electrophoresis of proteins: a practical approach. Oxford: IRL Press; p. 1–147. [Google Scholar]
- Haque MA, Kurokawa T, Gong JP. 2012. Super tough double network hydrogels and their application as biomaterials. Polymer. 53:1805–1822. [Google Scholar]
- Hennebert E, Leroy B, Wattiez R, Ladurner P. 2015. An integrated transcriptomic and proteomic analysis of sea star epidermal secretions identifies proteins involved in defense and adhesion. J Proteomics. 128:83–91. [DOI] [PubMed] [Google Scholar]
- Hennebert E, Wattiez R, Demeuldre M, Ladurner P, Hwang DS, Waite JH, Flammang P. 2014. Sea star tenacity mediated by a protein that fragments, then aggregates. Proc Natl Acad Sci USA. 111:6317–6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang DS,Zeng H, Masic A, Harrington MJ, Israelachvili JN, Waite JH. 2010. Protein- and metal-dependent interactions of a prominent protein in mussel adhesive plaques. J Biol Chem 285:25850–25858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julenius K 2007. NetCGlyc 1.0: prediction of mammalian C-mannosylation sites. Glycobiology. 17:868–876.the [DOI] [PubMed] [Google Scholar]
- Kishore U, Ghai R, Greenhough TJ, Shrive AK, Bonifati DM, Gadjeva MG, Waters P, Kojouharova MS, Chakraborty T, Agrawal A. 2004. Structural and functional anatomy of the globular domain of complement protein C1q. Immunol Lett. 95:113–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore U, Reid KBM. 2000. C1q: structure, function and receptors. Immunopharmacology. 49:159–170. [DOI] [PubMed] [Google Scholar]
- Kito K, Ito T. 2008. Mass spectrometry-based approaches toward absolute quantitative proteomics. Curr Genomics. 9:263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatt AR, Becker AKA, Neacsu CD, Paulsson M, Wagener R. 2011. The matrilins: modulators of extracellular matrix assembly. Int J Biochem Cell Biol. 43:320–330. [DOI] [PubMed] [Google Scholar]
- Koch AW, Holstein TW, Mala C, Kurz E, Engel J, David CN. 1998. Spinalin, a new glycine- and histidine-rich protein in spines of Hydra nematocysts. J Cell Sci. 111:1545–1554. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. 1982. A simple method for displaying the hydropathic character of a protein. 157:105–132. [DOI] [PubMed] [Google Scholar]
- Lebesgue N, da Costa G, Ribeiro RM, Ribeiro-Silva C, Martins GG, Matranga V, Scholten A, Cordeiro C, Heck AJR, Santos R. 2016. Deciphering the molecular mechanisms underlying sea urchin reversible adhesion: a quantitative proteomics approach. J Proteomics. 138:61–71. [DOI] [PubMed] [Google Scholar]
- Li D, Graham LD. 2007. Epidermal secretions of terrestrial flatworms and slugs: Lehmannia valentiana mucus contains matrilin-like proteins. Comp Biochem Physiol B. 148:231–244. [DOI] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR. 2004. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 76:4193–4201. [DOI] [PubMed] [Google Scholar]
- Lippard SL, Berg JM. 1994. Principles of Bioinorganic Chemistry. Mill Valley (CA): University Science Books. [Google Scholar]
- Loker ES. 2010. Gastropod immunobiology. Adv Exp Med Biol. 708:17–43. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. 1991. Predicting coiled coils from protein sequences. Science. 252:1162–1164. [DOI] [PubMed] [Google Scholar]
- Martin AW, Deyrup-Olsen I. 1986. Function of the epithelial channel cells of the body wall of the terrestrial slug Ariolimax columbianus. J Exp Biol. 121:301–314. [Google Scholar]
- McMahon SA, Miller JL, Lawton JA, Kerkow DE, Hodes A, Marti-Renom MA, Doulatov S, Narayanan E, Sali A, Miller JF, Ghosh P. 2005. The C-type lectin fold as an evolutionary solution for massive sequence variation. Nat Struct Mol Biol. 12:886–892. [DOI] [PubMed] [Google Scholar]
- Patard L, Lallemand J-Y, Stoven V. 2003. An insight into the role of human pancreatic lithostathine. J Pancreas. 4:92–103. [PubMed] [Google Scholar]
- Pawlicki JM, Pease LB, Pierce CM, Startz TP, Zhang Y, Smith AM. 2004. The effect of molluscan glue proteins on gel mechanics. J Exp Biol. 207:1127–1135. [DOI] [PubMed] [Google Scholar]
- Rao Z, Handford P, Mayhew M, Knott V, Brownlee GG, Stuart D. 1995. The structure of a Ca(2+)-binding epidermal growth factor-like domain: its role in protein-protein interactions. Cell. 82:131–141. [DOI] [PubMed] [Google Scholar]
- Ressl S, Vu BK, Vivona S, Martinelli DC, Südhof TC, Brunger AT. 2015. Structures of C1q-like proteins reveal unique features among the C1q/TNF superfamily. Structure. 23:688–699. [DOI] [PubMed] [Google Scholar]
- Rodrigues M, Ostermann T, Kremeser L, Lindner H, Beisel C, Berezikov E, Hobmayer B, Ladurner P. 2016. Profiling of adhesive-related genes in the freshwater cnidarian Hydra magnipapillata by transcriptomics and proteomics. Biofouling. 32:1115–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell S, Young KM, Smith M, Hayes MA, Lumsden JS. 2008. Cloning, binding properties, and tissue localization of rainbow trout (Oncorhynchus mykiss) ladderlectin. Fish Shellfish Immunol. 24:669–683. [DOI] [PubMed] [Google Scholar]
- Sanchez J-F, Lescar J, Chazalet V, Audfray A, Gagnon J, Alvarez R, Breton C, Imberty A, Mitchell EP. 2006. Biochemical and structural analysis of Helix pomatia agglutinin: a hexameric lectin with a novel fold. J Biol Chem. 281:20171–20180. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM. 2013. Multiple metal-based cross-links: protein oxidation and metal coordination in a biological glue In: Santos R, Aldred N, Gorb S, Flammang P, editors. Biological and biomimetic adhesives: challenges and opportunities. Cambridge (UK): Royal Society of Chemistry; p. 3–15. [Google Scholar]
- Smith AM. 2016. The biochemistry and mechanics of gastropod adhesive gels. In: Smith AM, editor. Biological adhesives. Cham: Springer; p. 177–192. [Google Scholar]
- Smith AM, Robinson TM, Salt MD, Hamilton KS, Silvia BE, Blasiak R. 2009. Robust cross-links in molluscan adhesive gels: testing for contributions from hydrophobic and electrostatic interactions. Comp Biochem Physiol B. 152:110–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT-BG, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L, et al. 2013. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 32:1478–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart RJ, Ransom TC, Hlady V. 2011. Natural underwater adhesives. J Polym Sci B Polym Phys. 49:757–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre MH, Gertz M, Steegborn C, Scheibel T. 2014. Structural and functional features of a collagen-binding matrix protein from the mussel byssus. Nat Commun. 5:3392. [DOI] [PubMed] [Google Scholar]
- Wagener R, Ehlen HWA, Ko Y-P, Kobbe B, Mann HH, Sengle G, Paulsson M. 2005. The matrilins - adaptor proteins in the extracellular matrix. FEBS Lett. 579:3323–3329. [DOI] [PubMed] [Google Scholar]
- Waite JH. 2017. Mussel adhesion – essential footwork. J Exp Biol. 220:517–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis WI, Drickamer K, Hendrickson WA. 1992. Structure of a C-type mannose-binding protein complexed with an oligosaccharide. Nature. 360:127–134. [DOI] [PubMed] [Google Scholar]
- Weiss IM, Kaufmann S, Mann K, Fritz M. 2000. Purification and characterization of perlucin and perlustrin, two new proteins from the shell of the mollusk Haliotis laevigata. Bioch Biophys Res Comm. 267:17–21. [DOI] [PubMed] [Google Scholar]
- Werneke SW, Swann C, Farquharson L, Hamilton KS, Smith AM. 2007. The role of metals in molluscan adhesive gels. J Exp Biol. 210:2137–2145. [DOI] [PubMed] [Google Scholar]
- Whittaker CA, Hynes RO. 2002. Distribution and evolution of von Willebrand/Integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol Biol Cell. 13:3369–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilks AM, Rabice SR, Garbacz HS, Harro CC, Smith AM. 2015. Double-network gels and the toughness of terrestrial slug glue. J Exp Biol. 218:3128–3137. [DOI] [PubMed] [Google Scholar]
- Yoo HY, Iordachescu M, Huang J, Hennebert E, Kim S, Rho S, Foo M, Flammang P, Zeng H, Hwang D, Waite JH, Hwang DS. 2016. Sugary interfaces mitigate contact damage where stiff meets soft. Nat Commun. 7:11923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa HJ, Furuta E, Nakamura A, Takagi T. 1998. Cloning and sequencing of three C-type lectins from body surface mucus of the land slug, Incilaria fruhstorferi. Comp Biochem Physiol B. 119:479–484. [DOI] [PubMed] [Google Scholar]
- Zamocky M, Furtmuller PG, Obinger C. 2008. Evolution of catalases from bacteria to humans. Antioxid Redox Signaling. 10:1527–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelensky AN, Gready JE. 2005. The C-type lectin-like domain superfamily. FEBS J. 272:6179–6217. [DOI] [PubMed] [Google Scholar]
- Zhao H, Waite JH. 2006. Proteins in load-bearing junctions: the histidine-rich metal-binding protein of mussel byssus. Biochemistry. 45:14223–14231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.