Abstract
As in other areas of society, the Internet and the World Wide Web are becoming important topics in medical informatics. This is evident from the recent American Medical Informatics Association's 1996 Annual Fall Symposium, where the theme was “Beyond the Superhighway: Exploiting the Internet with Medical Informatics.” Of the over 330 papers and abstracts published in the Proceedings, one third dealt with the Internet and/or the Web. In some cases, system developers demonstrated how this technology can do old tasks in new ways. In other cases, researchers described new tasks that are now possible with this technology. Still others examined this technology to show how it can be evaluated and improved. This paper summarizes their accomplishments.
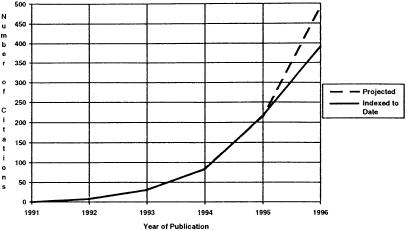
The Internet and the World Wide Web have become household terms. Advertisements in newspapers, in magazines, and on the sides of buses display Uniform Resource Locators (URLs) with the expectation that their potential customers will not only know what they are but have the wherewithal to use them. Even the New York Times relies on the Web to provide its readers with additional material; for example, it may print an excerpt from a speech and point to the full text with a URL.1 Although biomedical researchers, particularly in biotechnology, have been using the Internet for years as a medium for sharing data and conversing, the larger biomedical community has been slow to adopt the technology. The word “Internet” does not appear in the indexed medical literature until 1992.2 Biomedical use of the Internet is now increasing rapidly: although only eight citations with the word “Internet” appear in Medline for 1992, by 1995 there were 217 citations with “Internet”, “World Wide Web”, or “WWW”. With only 80% of the 1996 literature indexed to date, there are already 392 such citations (Fig. 1). Another positive sign is that review papers are starting to appear which describe the workings of the Internet intended for medical informatics researchers3 and the medical community at large.4,5,6
Figure 1.
Medline citations with the words “Internet”, “World Wide Web”, or “WWW” by publication year. The actual count for the 1996 figure is 392, based on approximately 80% of the database currently indexed; the amount projected after 100% of the indexing is completed is 490.
The Proceedings of the Annual Symposium on Computer Applications in and Medical Care (SCAMC) has been indexed in Medline since 1991, and Internet-related research began to appear there in the following year.7,8 The inclusion of the SCAMC Proceedings in Medline has, until now, made only a modest contribution to the numbers shown in Figure 1 (about 10% each year). However, when the 1996 Proceedings is indexed, its contribution will be considerably more (about 30%). The theme in 1996 was “Beyond the Superhighway: Exploiting the Internet with Medical Informatics.”9 The intent of this theme was to highlight ways in which medical informatics researchers are rising above the now-tired metaphors about onramps and roadkill. The scientific program committee planned the meeting to be a forum for showing how developers are actually using this technology and how researchers are applying the techniques of information science to tame the vast chaos of what has been more like an amusement park bumper-car ride than a superhighway. The result was gratifying: the papers, demonstrations, and panels verified that the medical informatics community is indeed taking on the task of exploiting the Internet for health care. This paper reviews the papers and abstracts from the Proceedings and attempts to organize them into a general framework for discussing ways in which the Internet can be “exploited.”
Using the Internet to do Old Things in New Ways
The history of medical informatics is littered with thousands of stand-alone, one-of-a kind applications that demonstrated interesting technologies and possibilities but were unable to go further due to their inability to research potential users. Those applications that were available via networks and telephone lines were often limited to simplistic, awkward, character-based user interfaces, due to technical difficulties with providing distributed graphical user interfaces and the “lowest common denominator” of hardware among users. As with most new technologies, the first use of the Internet has been to attempt to find novel ways to perform existing tasks—in this case, to provide access to applications and information resources in ways that had not been possible previously.
By providing standards for conectivity and information transfer, the Internet solves much of the access problem. At the same time, the Web and Hypertext Markup Language (HTML) address the user interface problem elegantly. The free distribution of Mosaic browsers10 for multiple computing platforms was the final piece to the puzzle; now that software distribution is as simple as sending a URL by e-mail or publishing it in a paper, potential users are running out of excuses for not trying new applications.11 Now that system developers can reach wider audiences, they can get broader support for the development and maintenance of their products and explore new ways to present them.
The most common use of the Internet in health care is, not surprisingly, to provide access to existing online resources. Many of these resources are static databases of reference materials that are being made generally available (pp. 398, 413, 849, 930, 935, 950, 960, 969, 970).* The dream of Integrated Advanced Information Management Systems (IAIMS)12 is now within reach of everyone (p. 859).13 Among these resources are ones sponsored by the federal government, such as Cancer Net (p. 403), CDC Wonder (p. 408), and Medline (pp. 847, 888, 933). Of particular note, Detmer and Shortliffe (p. 933) show how the Internet environment and the Web paradigm can add capabilities to existing resources and, for example, make a better-Medline-than-Medline.14
Many on-line resources fall into the category of research database, including the following: digital images of neuroanatomy (p. 299), cytokine structural and functional data (p. 393), results from clinical trials (p. 433) and brain mapping (p. 892), bone scintigraphy (p. 953), and a variety of epidemiologic registries (pp. 408, 669, 852, 968). Once again, the Web provides new ways to take information previously provided as text and numbers and display it, as in the French communicable disease surveillance database (SentiWeb, p. 669), in graphical and geographical formats.15
Interactive applications are also finding a place on the Internet, including those that perform such tasks as workflow management (p. 577), medical language processing (p. 938), and ventilator simulation (p. 967). Decision support seems to be an especially good fit, as demonstrated by a Medical Logic Module-driven clinical alerting system (p. 189), a health status assessment program (p. 338), a decision analysis tool (p. 925), infectious disease guidelines (p. 942), and a variety of expert systems (pp. 60, 762, 930, 988). One of these, DXplain (p. 988), has been around for over a decade. In 1987, it was described as “evolving,”16 and it has continued to do so in its Web-based incarnation.17
Various institutions are using the Internet to meet their educational mission as well. In some cases, the Web is used for “self-representation,”18 to provide institutional, faculty, and other information of interest to students (pp. 363, 822, 888, 918). Educational materials are also made accessible, as are the programs to test students' knowledge of it (pp. 32, 41, 46, 871, 882, 887, 906, 909, 911, 947, 961). One system, described by Bittorf (p. 46), covers the domain of dermatology with dynamic lectures, case reports, an atlas, and a quiz system.19 Another system, described by Oliver and et al. (p. 887), shows how authors can use Web links in their curricular materials to “capitalize on extra-mural authoritative sources.”20 Physicians can continue their medical education with a radiology self-assessment program (p. 37), and patients can obtain information about breast cancer (p. 861), postpartum depression (p. 915), and general medical questions (p. 976).
A very natural and (when considering security and confidentiality) potentially dangerous use of the Internet is to provide access to clinical information. The Internet itself provides a useful medium for this access (pp. 792, 845, 984), while the Web allows development of new interfaces for reviewing the data from legacy systems (pp. 179, 488, 618, 623, 628, 633, 638, 689, 719, 733, 772, 782, 857, 886, 989). System developers find the Web paradigm a useful place for creating mock-ups and prototyping (pp. 488, 628, 719). Object-oriented techniques in general (pp. 638, 857) and the Java language in particular (pp. 733, 772) are emerging as popular techniques for carrying out this development. Some detailed architecture descriptions can be found in papers by Flanagan et al. (p. 618),21 Klimczak et al. (p. 623),22 and Chueh et al. (p. 638).23 There is also a comprehensive review by Sittig et al. (p. 694) of 31 features found in 9 Web-based systems (with two non-Web systems for comparison).24
Using the Internet to do New Things
Harry Truman once described atomic energy as “a new force too revolutionary to consider in the framework of old ideas.” A half a century later, we are confronted with the Internet—another revolutionary force. Although it can be used to support conventional tasks, it also offers new ways to carry out entirely new tasks for those with sufficient vision.
Two of the areas where new approaches are becoming possible are in electronic publishing (pp. 343, 368, 891, 929, 930, 986) and controlled vocabularies (pp. 164, 940, 945). Publishers have been reticent about embracing electronic distribution of their materials, but with the help of medical informatics researchers, solutions are being found that satisfy the users' desire for access and the publishers' desire for control (for copyright protection and remuneration). Sharek and Greenes (p. 343) are breaking new ground with their development of an electronic version of the New England Journal of Medicine.25 Steffen et al. (p. 891) show that the very definition of the term “publishing” is changing; not only the narrative but also the underlying data can be published in electronic, searchable form.26 In a similar fashion, controlled vocabularies previously available only in printed form or distributed on diskette and tape are now being made available through Internet-based vocabulary servers. McCray et al. (p. 164) show how perhaps the most complex vocabulary of all, the Unified Medical Language System, can be made accessible to system developers (through Application Program Interface calls) and to end users (through Web-based browsers).27
In the clinical area, system developers are exploiting the hypertext and multimedia capabilities of the Web to develop new graphical methods for data display (p. 879) and even real-time data monitoring (p. 729). Distributed display of obstetrical ultrasound images (p. 895) and other radiologic images (pp. 886, 934) is now possible. With technology such as this, it is now possible for the clinical record to evolve into a true multimedia information source, as demonstrated by Lowe et al. (p. 314).28 In addition to extending the clinical data displayed, the medical record can also be made “smarter” through the integration of decision-support tools directly into the record system. For example, Canfield et al. (p. 175) have developed an architecture in which a geriatric clinic record system uses Knowledge Query and Manipulation Language (KQML) agents to retrieve information from outside resources.29 Elhanan et al. (p. 348) integrated a diagnostic decision support system, available at another institution (p. 988),17 into their own clinical record system to provide instant, patient-specific advice.30
The delivery of new types of clinical information will undoubtedly affect our perception of what really constitutes the medical record. Even more exciting is the potential for using the Internet to change the way in which we use the record. These new methods are emerging as the result of Internet-based collaboration. The theme of the 1992 SCAMC was “Supporting Collaboration.” In his Preface in the Proceedings, Program Chair Mark Frisse placed networks at the top of his list of technologies needed to support collaboration.31 As he predicted, the Internet is serving as a medium for collaboration among medical informatics researchers (pp. 125, 826), as exemplified by the description by Shortliffe et al. (p. 125) in which six sites across the United States and Canada are brought together in a “research collaboratory.”32
Many other projects demonstrate that collaboration can be applied to the patient care process as well. Collaboration is being used to influence patient care in indirect ways, such as facilitating physician referrals (p. 742) and educating physicians through international on-line discussions (p. 839). Electronic consultations are being carried out with exchange of reports among physicians (pp. 838, 839) and between patients and physicians (p. 903). Collaboration in direct support of patient care is a novel activity being catalyzed through the Internet. Beuscart et al. (p. 742) have developed a multimedia workstation to support medical codiagnosis,33 while Shank and Lawson (p. 928) describe the coordination of care providers through the use of multimedia e-mail and desktop conferencing.34 Two papers, one by Kohane et al. (p. 608) and the other by van Wingerde et al. (p. 643), describe the difficulties encountered and solutions being applied to pull data together from multiple institution-specific electronic records to form an interinstitutional W3-EMRS.35,36 Forslund et al. (p. 990) add the ability for physicians to edit and annotate data from multiple sources and share these data back with physicians at the original sources.37
Contributions Beyond Medical Informatics
The above analogy of the Internet to Truman's view of atomic energy is apt in another way. Just as the definition of “explosion” changed with the start of the nuclear era, so too has the definition of “information explosion” taken on a new significance in the Internet era. In retrospect, the information explosion we complained about ten years ago seems like so many fireworks compared with the chain reaction one experiences today when searching the Internet or traversing links on the Web. The resulting difficulty in coping with huge amounts of available information is not limited to the health care field but is common to all fields. Given that medical informatics has produced MUMPS (an ANSI-standard computer language used throughout the world), CT scanners (whose mathematical reconstruction formulae are used in fields such as geology and astronomy), and MYCIN (the prototype for rule-based expert systems), it is not surprising, perhaps, that the Proceedings also contains papers describing information solutions applicable to the non-medical Internet community at large.
Many of these contributions result from the authors' efforts to solve problems common to Internet-based and Web-based applications. For example, when Web sites begin to grow and develop, solutions are needed to manage their content (pp. 309, 393). On the other hand, when users retrieve large collections of information from these sites, they need methods for indexing them properly, based on contextual information (p. 846). As the number of Web sites proliferates, keeping track of the sites themselves becomes a management issue, as shown by Williams and Giuse (p. 836), who have developed a database for managing pointers to on-line resources.38
Issues such as security and confidentiality are not limited to patient information. Approaches such as security mediators (p. 120) and encrypted e-mail (p. 850) will be applicable in many domains. A common related problem is the need to scrub databases in order to share information without revealing sensitive information. To this end, Berman et al. (p. 328) show how information in an autopsy database can be scrubbed through generalizable methods.39
Data entry is an important feature of many Web sites. Filling out forms is readily accomplished with HTML, but this is insufficient for more complex data capture tasks. Two papers discuss generalizable methods for advancing the capabilities of Web-based data entry. In one, Kahn and Huynh (p. 478) describe a Data Entry and Report Markup Language (DRML) that can be used for structured reporting.40 In a second paper, Zuckerman and Jacques (p. 906) describe creation of extensions to HTML that support user interaction with Java programs.41
The Web is a phenomenon which is itself worthy of study. For example, with users running independent Web browsers in a stateless architecture, how is an application developer supposed to track individual users' interactions with the application? Feliciano and Altman (p. 757) provide a way: a tool called Lamprey translates all URLs so that they point to Lamprey, which in turn retrieves the desired information and in the process notes the users' request and translates any additional URLs.42 What about the appropriateness of the Web paradigm for various applications? Friedman et al. (p. 2) conducted a study comparing efficiency of retrieval with Boolean versus hypertext retrieval (hypertext won). Finally, lest we become too enamored of this revolutionary technology, Narus and Pryor (p. 767) provide a thoughtful examination of some of the drawbacks of the Web and its visual presentation.43
Summary
This brief review cannot convey all the hard work and creativity behind the papers and abstracts mentioned here. However, there can be no doubt that medical informaticians have indeed begun to exploit the Internet. By stepping back to look at the general categories into which this work falls, we can see some interesting patterns emerge.
The Internet has done for computer communications what the printing press did for the written word. Just by making communications faster and easier, just by extending access to a higher degree, the Internet has brought us to a whole new level. At this level, we will find new things happening with familiar resources. For example, people have always been willing to share their work; now it is available at the touch of a button. This is particularly true of U.S. government resources, which are in the public domain but for which distribution has been a limiting factor. It is also true of application developers who previously had to make a choice: stand-alone systems with good user interfaces versus distributed applications with character-based, command-line interfaces. The National Library of Medicine (NLM) found a middle ground with the Grateful Med user interface to the character-based Medline system, but users still had to obtain and install software to get to Medline. Now all they need is a URL, cut from an e-mail message and pasted into a Web browser. Similarly, the access to clinical information has traditionally been limited to particular sites; now there is the potential for caregivers to obtain information about their patients regardless of the caregiver's location or the location of the information. This kind of access is more than a quantitative improvement; for many, the barrier to access will be lowered below a threshold that for them will mean a qualitative difference. Improved access means improved value, and improved value may mean that the cost/benefit balances that have been preventing the computerization of information (particularly clinical information) may begin to tip favorably.
By nature, it is impossible to predict what new tasks people will solve with the Internet. Some of the examples so far are tantalizing. The emergence of a true multimedia record seems likely. Perhaps clinicians will once again be able to look at all aspects of their patients, including patients' blood smears and x-rays. Perhaps they will be able to see patients for the first time and still know what they looked like a year ago, or how they walked, or what their hearts sounded like. In this way, perhaps the computer, which is blamed for taking us away from our patients, can bring us closer.
The emergence of collaborative patient care is another exciting development. The curbside consult can be replaced by a query to the most appropriate authority who will have the benefit of the patient's record, rather than a brief description. There may be a return of continuity of care, with the computer serving as the stable, ever-present repository of the patient's history and condition. Finally, the emergence of true integrated systems is now limited only by our ability to imagine these integrations. Creators of expert systems are willing to share their applications. Clinical system developers are building Web interfaces. Tying the two together seems a natural next step (and is already happening), making automated decision support available to all.
It is easy to predict, however, that new problems will arise. Intellectual property issues—both copyright and patent—are already making front-page news. Issues of subscription and compensation are not far behind. As with every other aspect of American society (medicine in particular), liability is sure to become an important factor in deciding what really does get shared and how it will be used.
Another prediction that seems sure is that the field of medical informatics has a safe future and that this future will be inextricably linked to the future of telecommunications. Over the course of a few years, the amount of Internet-related work presented at the Symposium has grown from none to one third of the total. My last prediction is that there will be even more and better things to see at the 1997 Symposium (Nashville, October 25-29), where the theme is “The Emergence of `Internetable' Health Care and Systems That Really Work.” See you (and your innovation) there.
Footnotes
This paper refers to over a hundred papers and abstracts in the Proceedings. Inclusion of full citations is reserved for those which are highlighted; the remainder are referred to by page number. Page numbers below 817 refer to full papers, while those above 817 refer to abstracts describing posters and demonstrations.
References
- 1.Anonymous. William Vickery: Letter to the editor. In: The lives they lived. The New York Times Magazine, Dec 29, 1996: 16.
- 2.Cariello NF, Craft TR, Vrieling H, van Zeeland AA, Adams T, Skopek TR. Human HPRT mutant database: software for data entry and retrieval. Environmental and Molecular Mutagenesis. 1992;20: 81-3. [DOI] [PubMed] [Google Scholar]
- 3.Lowe HJ, Lomax EC, Polonkey SE. The World Wide Web: a review of an emerging Internet-based technology for the distribution of biomedical information. J Am Med Inform Assoc. 1996;3: 1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kleeberg P. Medical uses of the Internet. Journal of Medical Systems. 1993;17: 363-6. [DOI] [PubMed] [Google Scholar]
- 5.McKinney WP, Wagner JM, Bunton G, Kirk LM. A guide to Mosaic and the World Wide Web for physicians. MD Comput. 1995;12: 109-14,141. [PubMed] [Google Scholar]
- 6.Metcalf ES, Frisse ME, Hassan SW, Schnase JL. Academic networks: Mosaic and the World Wide Web. Acad Med. 1994;69: 270-273. [DOI] [PubMed] [Google Scholar]
- 7.Fowler J, Barber S, Gilson H, Long KB, Gorry GA. The Medline Retriever. In: Frisse ME (ed). Proc Annu Symp Comput Appl Med Care. New York: McGraw-Hill, 1993: 473-7. [PMC free article] [PubMed]
- 8.Preuss DR, Sequeira EP, Graeff AS. Grateful Med on an institutional local area network. In: Frisse ME, ed. Proc Annu Symp Comput Appl Med Care. New York: McGraw-Hill, 1993: 488-90. [PMC free article] [PubMed]
- 9.Cimino JJ (ed). Proceedings of the 1996 American Medical Informatics Association Annual Fall Symposium, Washington, DC, October 26-30, 1996. Philadelphia: Hanley & Belfus, 1996.
- 10.Schatz BR, Hardin JB. NCSA Mosaic and the World Wide Web: global hypermedia protocols for the Internet. Science. 1994;265: 895-901. [DOI] [PubMed] [Google Scholar]
- 11.Masys DR. Moving ahead on webbed feet. J Am Med Inform Assoc. 1995;2: 332-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindberg DAB. The IAIMS opportunity: the NLM view. Bull Med Libr Assoc. 1988;76: 224-5. [PMC free article] [PubMed] [Google Scholar]
- 13.Blumenthal JL, Seymour AK, Broering NC. IAIMS and the Internet. J Am Med Inform Assoc. 1996;3: 859 (supplement). [Google Scholar]
- 14.Detmer WM, Shortliffe EH. WebMedline: transforming Medline into a hypertext environment with links to full-text documents. J Am Med Inform Assoc. 1996;3: 933 (supplement). [Google Scholar]
- 15.Bioussard E, Flahault A. The SentiWeb method for exploring a database on the net. J Am Med Inform Assoc. 1996;3: 669 (supplement). [PMC free article] [PubMed] [Google Scholar]
- 16.Barnett GO, Cimino JJ, Hupp JA, Hoffer EP. DXplain—An evolving diagnostic decision-support system. J Am Med Inform Assoc 1987;258: 67-74. [DOI] [PubMed] [Google Scholar]
- 17.Barnett GO, Hoffer E, Kim RJ, Famiglietti KT. DXplain on the World Wide Web. J Am Med Inform Assoc. 1996;3: 988 (supplement). [Google Scholar]
- 18.Ganslandt T, Fischer C, Riechman G, Prokosch HU, Köpcke W. Medical Websites in Germany: a status report and critical analysis. J Am Med Inform Assoc. 1996;3: 822 (supplement). [Google Scholar]
- 19.Bittorf A. Teaching resources for dermatology on the WWW—quiz system and dynamic lecture scripts using a HTTP-Database Demon. J Am Med Inform Assoc. 1996;3: 46-50 (supplement). [PMC free article] [PubMed] [Google Scholar]
- 20.Oliver RE, Wingert K, Reid JC, Locatis C. The Virtual Health Care Team: an example of distributed virtual education. J Am Med Inform Assoc. 1996;3: 887 (supplement). [Google Scholar]
- 21.Flanagan JR, Chun J, Wagner JR. Evolution of a legacy system to a Web patient record server: leveraging investment while opening the system. J Am Med Inform Assoc. 1996;3: 618-22 (supplement). [PMC free article] [PubMed] [Google Scholar]
- 22.Klimczak JC, Witten DM, Ruiz M, Mitchell JA, Brilhart JG, Frankenberger ML. Providing location-independent access to patient clinical narratives using Web browsers and a tiered server approach. J Am Med Inform Assoc. 1996;3: 623-7 (supplement). [PMC free article] [PubMed] [Google Scholar]
- 23.Chueh HC, Raila WF, Pappas JJ, Ford M, Zatsman P, Tu J, Barnett GO. A component-based distributed object services architecture for a clinical workstation. J Am Med Inform Assoc. 1996;3: 638-42 (supplement). [PMC free article] [PubMed] [Google Scholar]
- 24.Sittig DF, Kuperman GJ, Teich JM. WWW-based interfaces to clinical information systems: state of the art. J Am Med Inform Assoc. 1996;3: 694-8 (supplement). [PMC free article] [PubMed] [Google Scholar]
- 25.Shareck EP, Greenes RA. Publishing biomedical journals on the World-Wide Web using an open architecture model. J Am Med Inform Assoc. 1996;3: 343-7 (supplement). [PMC free article] [PubMed] [Google Scholar]
- 26.Steffen DL, Yarus S, Burch PE, Wheeler DA. A paradigm for electronic publication. J Am Med Inform Assoc. 1996;3: 891 (supplement). [Google Scholar]
- 27.McCray AT, Razi AM, Bangalore AK, Browne AC, Stavri PZ. The UMLS Knowledge Source Server: a versatile Internet-based research tool. J Am Med Inform Assoc. 1996;3: 164-8 (supplement). [PMC free article] [PubMed] [Google Scholar]
- 28.Lowe HJ, Antipov I, Walker WK, Polonkey SE, Naus GJ. WebReport: a World Wide Web based clinical multimedia reporting system. J Am Med Inform Assoc. 1996;3: 314-8 (supplement). [PMC free article] [PubMed] [Google Scholar]
- 29.Canfield K, Ramesh V, Quirolgico S, Silva M. An intelligent information systems architecture for clinical decision support on the Internet. J Am Med Inform Assoc. 1996;3: 175-8 (supplement). [PMC free article] [PubMed] [Google Scholar]
- 30.Elhanan G, Socratous SA, Cimino JJ. Integrating DXplain into a clinical information system using the World Wide Web. J Am Med Inform Assoc. 1996;3: 348-52 (supplement). [PMC free article] [PubMed] [Google Scholar]
- 31.Frisse ME. Preface. In: Frisse ME (ed). Proc Annu Symp Comput Appl Med Care. New York: McGraw-Hill, 1993: xiii-xiv.
- 32.Shortliffe EH, Patel VL, Greenes RA, Huff SM, Cimino JJ, Barnett GO. Collaborative medical informatics research using the Internet and the World Wide Web. J Am Med Inform Assoc. 1996;3: 125-9 (supplement). [PMC free article] [PubMed] [Google Scholar]
- 33.Beuscart RJ, Molenda S, Souf N, Foucher C, Beuscart-Zephir MC. Assessment of a cooperative workstation. J Am Med Inform Assoc. 1996;3: 742-6 (supplement). [PMC free article] [PubMed] [Google Scholar]
- 34.Shank RR, Lawson RR. ARTEMIS: a WWW-base inranet for clinical information. J Am Med Inform Assoc. 1996;3: 928 (supplement). [Google Scholar]
- 35.Kohane IS, van Wingerde FJ, Fackler JC, et al. Sharing electronic medical records across multiple heterogeneous and competing institutions. J Am Med Inform Assoc. 1996;3: 608-12 (supplement). [PMC free article] [PubMed] [Google Scholar]
- 36.van Wingerde FJ, Schindler J, Kilbridge P, et al. Using HL-7 and the World Wide Web for unifying patient data from remote databases. J Am Med Inform Assoc. 1996;3: 643-7 (supplement). [PMC free article] [PubMed] [Google Scholar]
- 37.Forslund DW, Phillips RL, Kilman DG, Cook JL. TeleMed: a working distributed virtual patient record system. J Am Med Inform Assoc. 1996;3: 990 (supplement). [PMC free article] [PubMed] [Google Scholar]
- 38.Williams TW, Giuse NB. Managing annotated lists of resources on the Web. J Am Med Inform Assoc. 1996;3: 836 (supplement). [Google Scholar]
- 39.Berman JJ, Moore GW, Hutchin GM. Maintaining patient confidentiality in the public domain Internet Autopsy Database (IAD). J Am Med Inform Assoc. 1996;3: 328-32 (supplement). [PMC free article] [PubMed] [Google Scholar]
- 40.Kahn CE, Huynh PN. Knowledge representation for platform-independent structured reporting. J Am Med Inform Assoc. 1996;3: 478-82 (supplement). [PMC free article] [PubMed] [Google Scholar]
- 41.Zuckerman AE, Jacques LB. A Java language program for writing clinical cases using custom extensions to HTML. J Am Med Inform Assoc. 1996;3: 906 (supplement). [Google Scholar]
- 42.Feliciano RM, Altman RB. Lamprey: tracking users on the World Wide Web. J Am Med Inform Assoc. 1996;3: 757-61 (supplement). [PMC free article] [PubMed] [Google Scholar]
- 43.Narus SP, Pryor TA. Beyond the Web: building information systems with clinical context. J Am Med Inform Assoc. 1996;3: 767-71 (supplement). [PMC free article] [PubMed] [Google Scholar]