Abstract
Nonneuronal cell types in the CNS are increasingly implicated as critical players in brain health and disease. While gene expression profiling of bulk brain tissue is routinely used to examine alterations in the brain under various conditions, it does not capture changes that occur within single cell types or allow interrogation of crosstalk among cell types. To this end, we have developed a concurrent brain cell type acquisition (CoBrA) methodology, enabling the isolation and profiling of microglia, astrocytes, endothelia, and oligodendrocytes from a single adult mouse forebrain. By identifying and validating anti-ACSA-2 and anti-CD49a antibodies as cell surface markers for astrocytes and vascular endothelial cells, respectively, and using established antibodies to isolate microglia and oligodendrocytes, we document that these 4 major cell types are isolated with high purity and RNA quality. We validated our procedure by performing acute peripheral LPS challenge, while highlighting the underappreciated changes occurring in astrocytes and vascular endothelia in addition to microglia. Furthermore, we assessed cell type–specific gene expression changes in response to amyloid pathology in a mouse model of Alzheimer’s disease. Our CoBrA methodology can be readily implemented to interrogate multiple CNS cell types in any mouse model at any age.
Keywords: Inflammation, Neuroscience
Keywords: Alzheimer’s disease, Expression profiling, Neurodegeneration
A streamlined methodology that allows investigation of cell type-specific gene expression and crosstalk between cell types in a single mouse forebrain is described.
Introduction
One of the principle underlying challenges of transcriptional analysis in the brain is the cellular complexity of the CNS, which comprises a conglomeration of cell types, the majority being astrocytes, microglia, neurons, vascular endothelia, and oligodendrocytes (1–3). This cellular complexity restricts the utility of traditional transcriptomic analyses, as results from the whole brain, or even specific substructures, do not reveal the cellular specificity of gene expression or changes thereof. Analyzing transcriptional changes as an average across all types of constituent cells comprising the bulk tissue leads to a dramatic loss of power to detect changes that may occur in single cell types or in response to specific insults (4, 5). Most problematic, different cell types may display distinct or even opposing differential gene expression changes compared with bulk brain transcriptomics under specific pathophysiological conditions. Thus, profiles obtained using the whole brain may be misleading or even erroneous.
In order to gain a greater understanding into the complexity of intracellular signaling in the brain, methodologies have been developed to dissociate and isolate distinct cell types from the mouse brain using reporter lines, such as ALDH1l1-EGFP, to label astrocytes and/or antibodies, such as CD45 and CD11b, to mark microglia. Utilizing analogous antibodies and reporter lines and varied strategies of dissociation, neurons (6–10), astrocytes (6, 7, 10–13), microglia (6, 7, 12, 14–20), endothelia (21, 22), and oligodendrocytes (6, 7, 23, 24) have been collected separately. In addition, multiple cell types have been isolated and profiled concurrently by using a combinatorial approach of reporter lines and/or multiple antibodies (1, 5, 6). RiboTagging has also been used in single cell types to investigate gene expression in actively translated mRNAs in neurons (25), microglia (26), and astrocytes (27). However, genetic reporter lines and RiboTagging are often intractable to disease models because they require complicated multiallele crosses. Attempts using antibody-based strategies have required pooling of brain samples and/or fixation to preserve the transcriptomic profile. These strategies raise further technical and biologic limitations related to mRNA integrity and individual animal variance (5). Low mRNA yield requires further RNA amplification, potentially creating artifacts and sample bias (28). Further studies have noted additional artifacts in the form of preparation-induced transcriptomic changes (26), such as artificial inflammatory signals due to endotoxin-contaminated reagents like Percoll. Single-cell transcriptional analysis has been increasingly used to profile specific cell types, providing a great deal of information related to cell type subpopulations (29–34). However, single-cell RNA sequencing does not result in high coverage of the transcriptome, and the results obtained are often unable to be integrated for systems biology applications designed to analyze cellular crosstalk in the CNS in health and disease (29, 35).
To this end, we developed what we believe to be a novel FACS-based method to isolate the major nonneuronal cell types, namely microglia, astrocytes, vascular endothelia, and oligodendrocytes, from (minimally) half of a single adult mouse forebrain, while eschewing the pitfalls mentioned above. The resulting isolated cell types are viable, can be cultured in vitro, and yield high-quality RNA with sufficient quantity for RNA sequencing. Using this methodology, we tested how multiple brain cell types respond to inflammatory conditions following LPS stimulation and to amyloid pathology in a mouse model of Alzheimer’s disease (AD). Our results reveal complex interplay of multiple cell types and highlight the importance of analyzing multiple cell types concurrently from the same mouse brain. Our methodology does not require genetic reporter lines and thus can be rapidly implemented to interrogate any mouse model. We believe this is the first report of a concurrent cell type isolation protocol with these capabilities.
Results
Concurrent brain cell type acquisition from a single adult mouse forebrain.
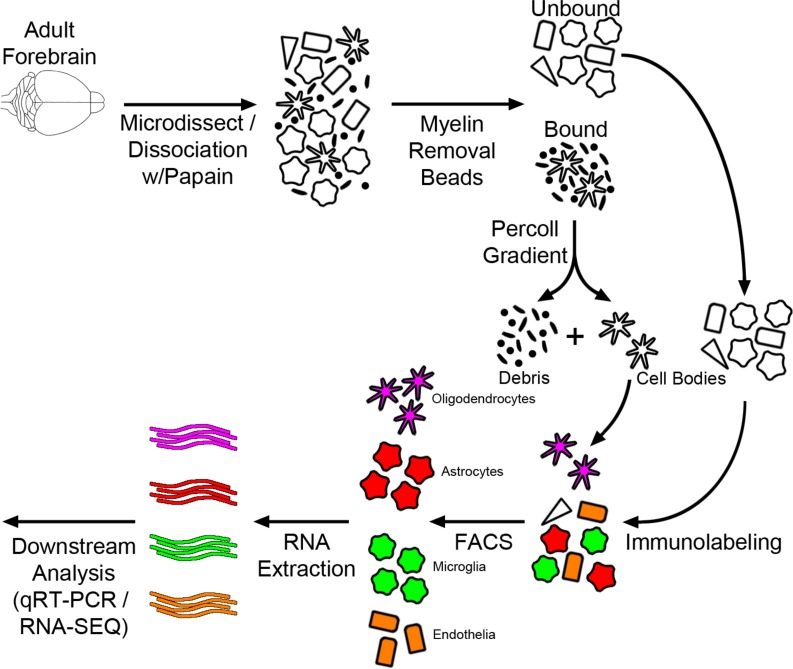
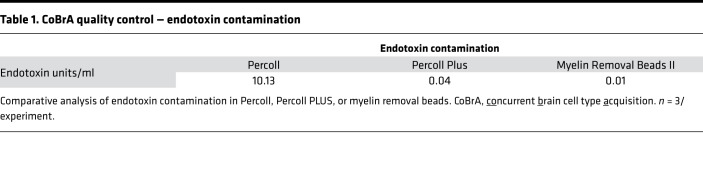
In order to evaluate the interactions and functional contributions of individual cell types in the brain under different conditions, we set out to develop and refine an approach to purify and isolate all major nonneuronal cell types residing in the adult brain. To this end we built upon and refined previous endeavors to dissociate mouse brain using a combination of mechanical and enzymatic digestion techniques and developed a methodology we term concurrent brain cell type acquisition (CoBrA) for isolation of astrocytes, vascular endothelia, microglia, and oligodendrocytes (Figure 1A). Microdissection followed by papain digestion and trituration resulted in high cell yield and optimal conservation of antibody epitopes for cell type–specific immunolabeling. Once the brain is dissociated, we found that the most effective method to remove myelin and cellular debris without significantly altering cellular transcription profiles is by using Miltenyi Biotec’s Myelin Removal Beads II filtration system. Unlike the commonly used compound Percoll, the myelin removal beads are devoid of endotoxin (Table 1), minimizing the risk of inducing artificial inflammatory responses during the procedure (26). The use of Miltenyi Biotec Myelin Removal Beads II has proven to be an effective means of maintaining homeostatic signaling within microglia, as the transcriptional results obtained mirror those of RiboTag-purified microglia (26). This procedure is not without its downside, as it removed most cells closely associated with myelin in vivo, including approximately 10% of astrocytes and microglia and most, if not all, oligodendrocytes (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.121109DS1). Because of the intimate association between myelin and oligodendrocytes in particular, it was necessary to utilize low-endotoxin Percoll PLUS to isolate oligodendrocytes from the myelin-bound fraction and then add these cells back to the main cell pool immediately prior to immunolabeling (Figure 1A).
Figure 1. Concurrent brain cell type acquisition.
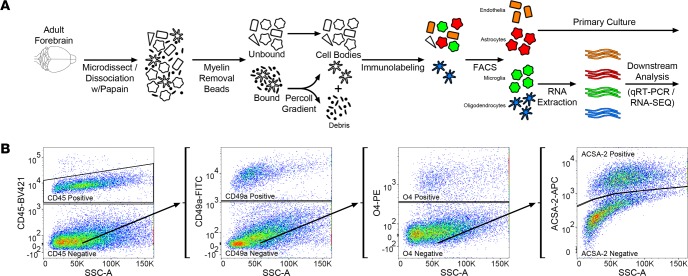
(A) Schematic representation of concurrent brain cell type acquisition (CoBrA) from mouse forebrain. (B) Representative flow cytometry work flow, gating strategy, and antibody separation using the CD45-BV421 (microglia), CD49a-FITC (vascular endothelia), O4-PE (oligodendrocytes), and ACSA-2-APC (astrocyte) antibodies.
Table 1. CoBrA quality control — endotoxin contamination.
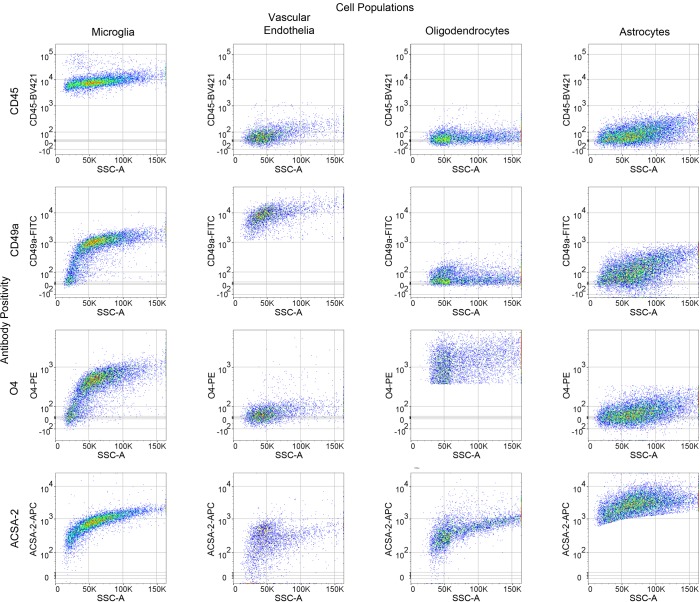
Following debris removal, single cells were labeled using a panel of antibodies to detect all major nonneuronal cell types and sorted using a BD FACSAria II cell sorter. These include the well-established antibodies against CD45 (36) and O4 (37, 38), the astrocyte-specific anti-ACSA-2 antibody that has been recently validated by ourselves and independent groups (27, 39, 40), (Supplemental Figure 2), and the anti-CD49a antibody (Figure 1B, Figure 2, and Supplemental Figure 2). Employing the commonly used ALDH1l1-EGFP reporter (41) believed to mark the majority of astrocytes, we found that the anti-ACSA-2 antibody labeled all GFP+ astrocytes in addition to a sizable GFP– astrocyte population (Supplemental Figure 3A), suggesting that this reporter line does not mark all astrocytes in adult brains. This observation has recently been reported elsewhere, supporting the validity of the anti-ACSA-2 antibody and the idea that this antibody may label astrocytes missed by the ALDH1l1 reporter line, although the extent of additional astrocytes labeled may depend upon age of the mouse or isolation method (39). We discovered that the CD49a antibody is a robust antibody and is specific for vascular endothelia in the brain (Figure 1B, Figure 2, and Supplemental Figure 2).
Figure 2. Representative antibody cross-reactivity based upon gating strategy for each cell population.
n = 3.
In order to perform CoBrA from a single mouse forebrain, the CD45 antibody, commonly used for the detection and isolation of microglia and immune cell lineages, was first used to label CD45mid microglial cells, excluding CD45+ lymphocytes and monocytes from isolation and removing microglia from the CD45– nonmicroglial populations (vascular endothelia, astrocytes, and oligodendrocytes) (Figure 1B). This gating strategy was followed by gates to isolate CD49a+ vascular endothelia cells from CD45–CD49a– cells (astrocytes and oligodendrocytes) and from O4+ oligodendrocytes, leaving CD45–CD49a–O4– ACSA-2+ astrocytes (Figure 1B and Figure 2). The remaining quadruple-negative events appear to constitute one-third of the cellular identity (as verified by FSC and SSC parameters), suggesting that most intact cells were captured using these 4 antibodies. However, not all cells of the brain were labeled by our panel of antibodies, suggesting an opportunity for other cell types to be isolated (Supplemental Figure 3B).
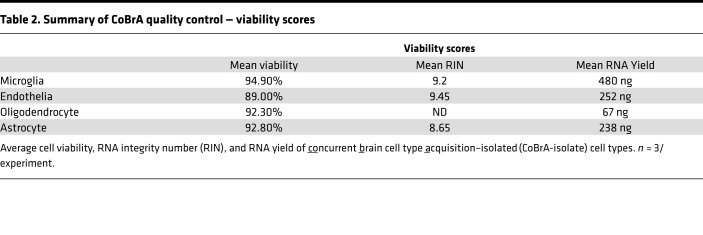
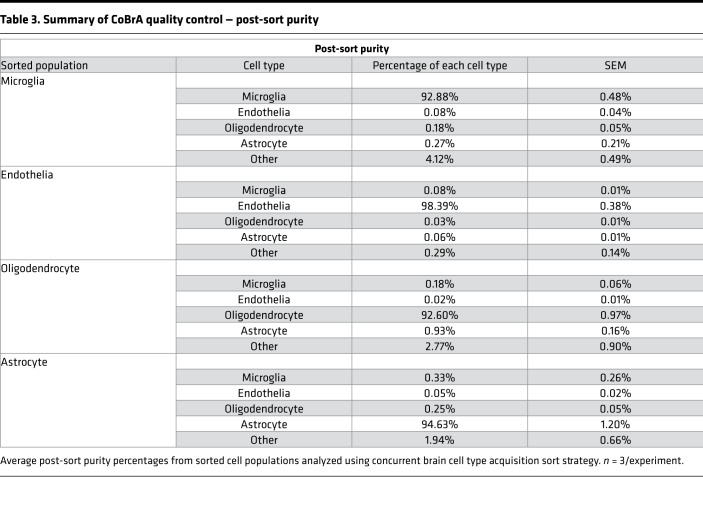
Following CoBrA isolation, the separated cell populations were immediately pelleted and lysed in Qiagen RLT buffer containing 1% β-mercaptoethanol or were pelleted and plated for in vitro culture and purity analyses. Cells isolated in this manner were more than 90% viable, as indicated by the LIVE/DEAD Blue viability dye (Supplemental Figure 3C), and the isolated RNA was of high quality, with an average RNA integrity number greater than or equal to 8.65 for all cell types with the exception of oligodendrocytes, whose score was not detectable due to low RNA yield (Table 2). We also found that sorted cell populations remained highly pure following post-sort FACS analysis (Table 3 and Supplemental Figure 4).
Table 2. Summary of CoBrA quality control — viability scores.
Table 3. Summary of CoBrA quality control — post-sort purity.
It is worth noting that the FACS-based cell type isolation may be confounded by autofluorescence, which is known to increase with age (Supplemental Figure 5). To circumvent this issue, we designed our gating strategy (Figure 1B) to remove the cells with the greatest autofluorescence (microglia) first, making this protocol amenable to analysis of any disease model at any age.
CoBrA cells isolated from adult brain are pure and can be cultured in vitro.
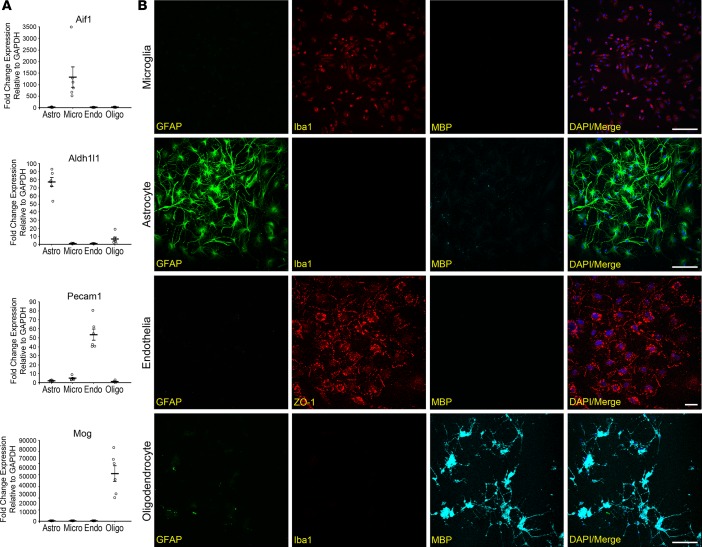
Quantitative real-time PCR (qRT-PCR) analysis was used to determine the purity of the dissociated cell types. Using the cell-type markers Aif1 and Cd68 (microglia), Aldh1l1 and Gfap (astrocytes), Pecam1 and Cdh5 (vascular endothelia), and Mog (oligodendrocytes), we demonstrated that the isolated cells predominantly expressed the markers specific for the respective cell types (Figure 3A and Supplemental Figure 6). Furthermore, these FACS-sorted cells can be cultured in vitro, and their identities were verified by staining with cellular protein markers: IBA1 (microglia), GFAP (astrocytes), ZO-1 (vascular endothelia), and MBP (oligodendrocytes) (Figure 3B). These combined data displayed the potential for CoBrA in allowing for both transcriptional analysis and mechanistic studies of individual cell types.
Figure 3. Purity and viability analysis of concurrent brain cell type acquisition cell types.
Concurrent brain cell type acquisition–isolated (CoBrA-isolated) cell types are highly pure and viable in vitro. (A) qRT-PCR analysis of CoBrA-isolated cell populations using the purity markers Aif1 (microglia), Aldh1l1 (astrocytes), Pecam1 (vascular endothelia), and Mog (oligodendrocytes) across all 4 populations. n = 6. (B) Representative immunofluorescent images of CoBrA-isolated primary cultures triple stained with cell type–specific antibodies as marked. Green, CD45+; red, ACSA-2+; orange, CD49a+; and blue, O4+. Scale bar: 50 microns.
Inflammatory profile of CoBrA-isolated cells following peripheral LPS challenge reveals contribution of vascular endothelia.
In order to validate our CoBrA methodology, we performed peripheral i.p. injection of LPS (4 μg/g) in 2-month-old C57BL/6J mice (42). After 24 hours of LPS stimulation, half of the forebrain was used for bulk brain qRT-PCR analysis, while the other half was used to perform CoBrA and cell type–specific qRT-PCR.
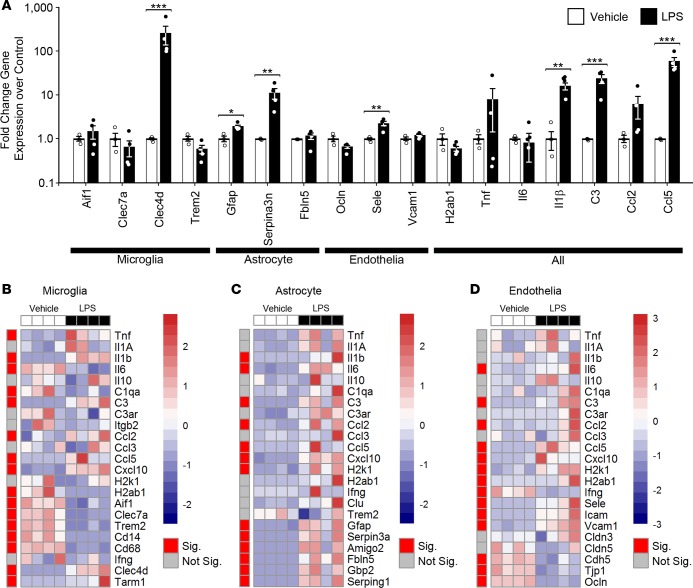
Bulk brain qRT-PCR analysis of LPS-injected mice revealed washed out signals when compared with cell type–specific gene expression (Figure 4). For instance, when comparing gene expression changes primarily thought to occur in microglia to changes in the bulk forebrain tissue, the only conserved detectable change was in Clec4d, with the observed downregulation of Aif1, Clec7a, and Trem2 in isolated microglia being lost in the bulk forebrain tissue (Figure 4, A and B, and Supplemental Table 2). This same observation generally held true for genes primarily expressed in astrocytes and in vascular endothelia, with significant expression changes in Fbln5, Ocln, and Vcam1 being lost in the bulk forebrain tissue analysis (Figure 4, A, C, and D, red, and Supplemental Table 2). Gene expression changes known to arise in multiple cell types were generally conserved in both sample types, including Il1b, C3, and Ccl5. However, significant changes in some genes were confounded or absent due to their differential regulation in separate cell types, including Il6 and H2ab1, which were either significantly or trending upregulated in astrocytes and vascular endothelia but downregulated in microglia (Figure 4 and Supplemental Table 2). Similar results were obtained by others when comparing the differential expressed genes of bulk brain (43) challenged with peripheral LPS to those of microglia (5, 26, 44) or astrocytes (5, 13, 44). These findings validate our system and emphasize the importance of cell type–specific analysis, as significant expression changes are masked when using whole brain.
Figure 4. Comparison of transcriptional profiles in bulk brain and concurrently isolated cell types following peripheral LPS administration.
(A) qRT-PCR data from bulk brain of selected genes in response to LPS. *P < 0.05; **P < 0.01; ***P < 0.001, t test. n = 3, vehicle; n = 4, LPS. (B–D) Heatmaps of qRT-PCR data from CoBrA-isolated microglia (B), astrocytes (C), and vascular endothelia (D) in response to LPS. Sig., P < 0.05 (red); Not Sig., P ≥ 0.05 (gray), t test. The red-and-blue scale bar represents the Z-score, the deviation from the mean by standard deviation units.
Deeper analysis into the CoBrA-isolated cell types revealed that microglia possess the expected inflammatory profile, including significant upregulation in the proinflammatory cytokines Tnf and Il1b; in the chemokines Ccl2, Ccl5, and Cxcl10; and in genes known to be upregulated in an LPS-specific manner, Clec4d and Tarm1, in particular (Figure 4B and Supplemental Table 2) (5, 45). Multiple gene sets were also significantly downregulated, as anticipated, including genes that typify disease-associated microglia (DAM), such as Clec7a and Trem2; genes associated with MHC presentation, such as H2ab1 and H2k1 (trending as downregulated); and genes associated with phagocytosis, such as Cd68 (Figure 4B) (31, 33, 46, 47). Interestingly, the microgliosis marker Aif1 was significantly downregulated in microglia following LPS treatment but was unchanged in the whole brain. This suggests that an increase in cell number may be responsible for the maintenance or slight increase of Aif1 transcript levels often reported in whole brain transcriptomic analyses (Figure 4, A and B, and Supplemental Table 2). Purified astrocytes similarly confirm an upregulated inflammatory response, with all significant gene expression changes, including proinflammatory cytokines (Il1β and Il6), chemokines (Ccl2, Ccl5, and Cxcl10), genes involved in MHC presentation (H2k1 and H2ab1 [trending as upregulated]), and genes identifying pan-reactive (Gfap and Serpin3a) and A1-reactive (Amigo2, Fbln5, Gbp2, and Serping1) astrocytes (Figure 4C and Supplemental Table 2) (5, 11, 48).
Interestingly, vascular endothelia, a cell type underrepresented in the field of brain transcriptomic studies, appeared to undergo significant changes in response to LPS, evidenced by the upregulation of the proinflammatory cytokine, Il6; chemokines (Ccl2, Ccl5, and Cxcl10); MHC presentation (H2k1 and H2ab1); cell adhesion molecules (Sele, Vcam1, and Icam1); and downregulation of tight-junction proteins (Tjp1, Ocln, and Cdh5) (Figure 4D and Supplemental Table 2) (49). These changes indicate that vascular endothelia may be central to the initiation and transmission of the LPS response from the periphery to the CNS via cytokines, chemokines, and extracellular remodeling.
Amyloid pathology primarily and differentially alters gene expression within microglia and astrocytes.
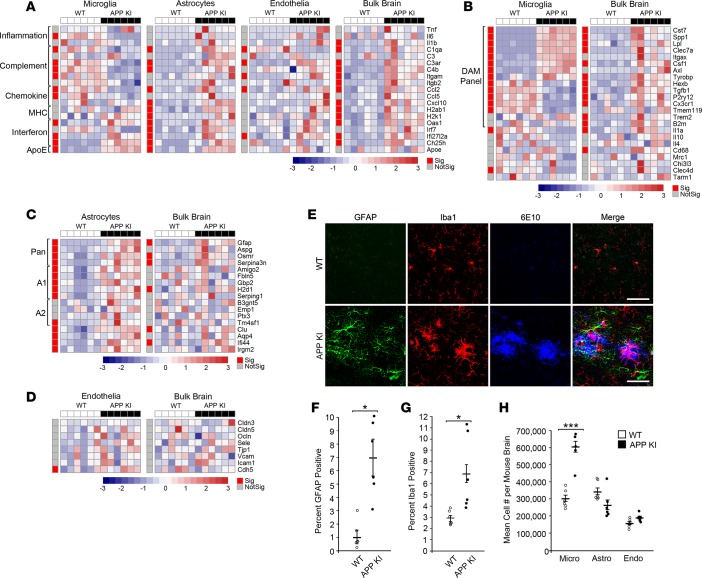
To explore the value of this methodology in the framework of neurodegenerative disease, we compared the bulk brain and CoBrA-isolated cell populations of the APP NL-G-F knockin (KI) mouse model of AD and littermate wild-type controls at 9 months of age (Figure 5) (50). Analysis of genes expressed across all individual cell types revealed that general inflammatory genes, Tnf, Il6, and Il1b, were not significantly altered, with the exception of a 3-fold decrease in Il6 in microglia and a 3-fold increase in Il1b in the bulk forebrain (Figure 5A). Consistent with reports using other AD models (51), Apoe is upregulated in both microglia and astrocytes of APP KI mice and such an increase has been proposed to serve as a clearance mechanism (52). Our bulk brain data are also consistent with recently published bulk transcriptomic data in the APP KI model showing increases in genes generally thought to be microglial (including Cst7, Clec7a, Lpl, Hexb, Spp1, Trem2, Tyrobp, and C3ar1) and astrocytic (including Gfap, Serpina3, and Osmr) (Figure 5, A–C) (53). In agreement with previous bulk brain data from AD mouse models (53–55), we found that genes of the complement pathway were also significantly upregulated (C1qa, C3ar, C4b, Itgam, and Itgb2) at the bulk tissue level. However, analysis of CoBrA-isolated cell populations revealed a more intricate pattern of complement gene expression changes. Some complement genes were upregulated in astrocytes (C1qa, C3ar, and C4b) and oligodendrocytes (C3ar), were mixed in vascular endothelia (C1qa downregulated; C3ar and Itgb2 upregulated) and, surprisingly, mostly downregulated in microglia (C1qa, C3ar, C4b, and Itgam) (Figure 5A, Supplemental Figure 7, and Supplemental Table 3). This same pattern was detected with regard to the significant changes in various chemokine genes, with expression being upregulated in astrocytes (Ccl2 and Cxcl10), vascular endothelia (Ccl2), and the whole brain (Ccl2 and Cxcl10), while being downregulated in microglia (Ccl2 and Ccl5) (Figure 5A and Supplemental Table 3). These results suggest that astrocytes are the main initiators of complement- and chemokine-mediated signaling in response to amyloid and that microglia are potentially dysfunctional, having a reduced phagocytic ability due to decreases in complement signaling (55, 56). Interestingly, MHC molecules were upregulated in the whole brain, with this upregulation appearing to be strongly influenced by astrocytes (Figure 5A and Supplemental Table 3). Correlation analysis of the data from the CoBrA cell populations in relation to bulk brain suggests that astrocytes are the primary drivers of differential gene expression in the APP KI mice at 9 months (Supplemental Figure 8). In contrast, it appeared that the interferon response was upregulated in bulk forebrain tissue and was globally upregulated in all cell types, suggesting amyloid pathology may trigger broad interferon signaling (Figure 5A, Supplemental Figure 7, and Supplemental Table 3). Overall, the highly differential expression of specific inflammatory mediators and receptors across the analyzed cell types hints at complex cellular crosstalk rather than a simple, generalized proinflammatory phenomenon.
Figure 5. Cell type–specific response to amyloid pathology in APP NL-G-F knockin mice.
(A–D) Heatmap of qRT-PCR data from concurrent brain cell type acquisition–isolated (CoBrA-isolated) microglia, astrocytes, and vascular endothelia, with comparison to that of bulk brain. (A) Gene expression profiles of microglia, astrocytes, vascular endothelia, and bulk brain against selected genes involved in inflammation, complement, chemokine, MHC, interferon pathways, and ApoE. (B–D) Comparison of genes specific to microglia (B), astrocyte (C), and vascular endothelia (D) between CoBrA-isolated cell types and bulk brain. Sig., P < 0.05 (red); Not Sig., P ≥ 0.05 (gray), t test. The red-and-blue scale bar represents the Z-score, the deviation from the mean by standard deviation units. (E) Representative confocal microscopy images of brain tissue from APP NL-G-F knockin (APP KI) mice and wild-type littermates at 9 months. Green, GFAP; red, Iba1; and blue, 6E10. Scale bar: 50 microns. (F) Quantitation of GFAP+ signal. n = 6/group. (G) Quantitation of Iba1 + signal. n = 6/group. (H) Mean cell number isolated per mouse forebrain by FACS from wild-type and APP KI mice. n = 6/group. Micro, microglia; Astro, astrocytes; Endo, vascular endothelia. ***P < 0.001, t test.
Analysis of microglia-specific genes strongly indicates that the DAM response detected in the 5XFAD mouse model (31, 33) is also observed in the APP KI mice. This microglial state was demonstrated by the upregulation of Cst7, Spp1, Lpl, Clec7a, Itgax, Csf1, Axl, and Tyrobp and the concomitant downregulation of Hexb, Tgfb1, P2ry12, Cx3cr1, and Tmem119 in our CoBrA microglia (Figure 5B and Supplemental Table 3) (31, 33). Interestingly, the profile was lost in the bulk brain, where the genes downregulated as part of the DAM profile were instead upregulated, further demonstrating the importance of cell type isolation and characterization (Figure 5B and Supplemental Table 3). Critically, the genes found to be upregulated in microglia in response to LPS, Clec4d and Tarm1 (5, 26), were downregulated in APP KI microglia either significantly or with a trend toward downregulation, respectively, after cell dissociation, signifying that CoBrA did not artificially activate LPS-specific response genes (Figure 5B and Supplemental Table 3).
Astrocytes appeared to be highly activated in the APP KI mouse model, with all pan-reactive (Gfap, Aspg, Osrm, and Serpina3n) and A1-reactive (Amigo2, Fbln5, Gbp2, H2d1, and Serping1) genes being significantly upregulated (11, 48). A2-reactive astrocyte genes were upregulated either significantly (Tm4sf1) or trending toward upregulation (B3gnt5, Emp1, and Ptx3) (Figure 5C and Supplemental Table 3) (11, 48). In bulk forebrain analysis, the pan-reactive profile was mostly maintained while the A1 and A2 panels become inconclusive (Figure 5C and Supplemental Table 3). These data indicate that astrocytes are activated by amyloid pathology and that A1-like activation is most prevalent within subpopulations of reactive astrocytes. Finally, with the exception of Cdh5, no vascular endothelia and oligodendrocyte genes were significantly affected in APP KI mice (Figure 5D and Supplemental Figure 8, and Supplemental Table 3) (57).
The APP KI mice used in these experiments display prominent microgliosis and astrogliosis by histology (Figure 5E–G). Our CoBrA isolation of these two cell types provided confirmation of this, with nearly twice the yield of microglia cell numbers from the APP KI brains and increased reactive transcripts in the purified astrocytes (Figure 5C), without increased numbers (Figure 5H). With this in mind, we can deduce that the reason DAM-downregulated genes (Hexb, Tgfb1, P2ry12, Cx3cr1, and Tmem119) are accurately measured in isolated cells but masked in bulk tissue (Figure 5B and Supplemental Table 2) is due to increased abundance of microglia in the tissue.
These combined results imply that astrocytes and microglia are the two primary cell types that respond to amyloid pathology in the APP KI mouse model and further support the idea that microglia cell number likely plays a large role in changes observed in bulk brain transcriptomic data from AD mouse models (5).
Discussion
Our streamlined CoBrA methodology allows for the investigation of cell type–specific gene expression and crosstalk among the major nonneuronal cell types in a single mouse forebrain. Furthermore, it does so in a manner that bypasses most of the downsides of previously published procedures, all of which incorporate one or more of the following pitfalls. (a) Insufficient cell number or RNA yield can necessitate the pooling of mouse brains or RNA amplification (1, 5); here, we demonstrated that sufficient RNA for qRT-PCR and RNA seq was obtained from as little as one hemisphere of a mouse forebrain using CoBrA, leaving the remaining hemisphere for correlative analysis. (b) Isolation of only a single cell type precludes comparison between coexisting cell types (10, 12, 13, 15, 24, 29, 31); using CoBrA methodology we were able to isolate all major CNS cell types, with the exception of neurons, permitting in-depth, cross-cell type analysis and integration of the brain in any state. (c) Use of reporter lines to label specific cell types (13, 33, 48, 55), complicates disease model studies requiring crosses of multiple alleles; by relying solely on antibodies, our approach can be immediately applied to any existing mouse model. (d) The requirement that mice be harvested at embryonic or preadult states to perform dissociation severely limits the application of age-related analyses; our technique and gating strategy affords isolation and interrogation of adult brains of any age. (e) The use of fixation for internal cell type epitope labeling leads to a decrease in RNA integrity and restricts multicell-type applications (38); by using live cells throughout the dissociation and isolation process, we were able to obtain high-quality RNA from highly pure populations of nonneuronal cell types. (f) Use of Percoll to perform debris removal potentially causes an artificial inflammatory response due to endotoxin contamination; we avoided using Percoll and instead used Myelin Removal Beads II, which are endotoxin free.
Using peripheral LPS challenge, we were able to not only validate our methodology, but also explore the underlying consequences of peripheral LPS to the brain. On a technical level, this experiment confirms the power of this technique where it corroborates previous work analyzing the LPS response in specific cell types (5, 11, 31, 33, 48, 49). On a biological level, our technique allows us to detect the response of the vascular endothelia to this stimulus that would otherwise be obstructed by other cell types in bulk brain analysis. Given the observed changes, it is plausible that vascular endothelia mediate peripheral inflammation via response to circulating inflammatory cytokines and transmission of this response via cytokine signaling (Il6) or through remodeling of tight junction proteins (Tjp1, Ocln, Cdh5), weakening the blood-brain barrier and facilitating passive diffusion of peripheral inflammatory cytokines into the brain (49).
In order to investigate how amyloid pathology alters brain biology on a cell type level, we set out to assess the APP NL-G-F KI model of AD, which shares many transcriptional changes seen in human AD (53). Our bulk brain analysis of microglia and astrocyte genes is consistent with the reported APP KI transcriptomic data (53). Upon examination of cell type–specific changes, we noted that most of the observed cellular changes occur in microglia and astrocytes, with little to none occurring in vascular endothelia or oligodendrocytes. Upon closer inspection, we determined that cell type analysis was sufficient to detect changes occurring in subcell types, specifically shown by the differences in the DAM-downregulated markers between microglia and whole brain (Figure 5B). This difference further corroborates previous hypotheses and suggests that there may be an increase in microglia cell number, skewing bulk brain analyses toward a microglia profile due to microgliosis (48). Though single-cell resolution reveals that transcriptional changes do not apply broadly to the whole microglia population, our data suggest that single-cell analysis is not required to observe the total DAM profile and that cell type analysis of microglia is sufficient (31, 33). Additionally, the fact that the inflammatory, complement, and chemokine pathways are downregulated in sorted microglia but upregulated in astrocytes supports the notion that astrocytes may play a larger role in the inflammatory response to amyloid pathology than previously recognized. To our surprise astrocyte number was not significantly higher than microglia number in the wild-type or APP KI preparations. It is possible that some astrocytes are retained in the negative population. Given the nature of the preparation it is also possible that astrocytes are more sensitive to mechanical dissociation. However, assuming uniform loss across all preparations, we would expect to see increases in astrocyte number, similar to those observed in the microglia population, if amyloid were to induce astrocyte proliferation. Thus, we can deduce that the astrogliosis seen in the APP KI brains is not due to increased cell number but instead to the increased presence of GFAP+ reactive astrocytes, an observation corroborated by transcript analysis.
Overall, our analyses of CoBrA-isolated cell populations from these two disease models highlight the importance of examining purified populations to (a) greatly magnify changes within individual cell types and (b) add robustness against cell number differences between animals.
This methodology is not without considerations. First and foremost is the reality that, like all other techniques to attain single cells, forceful dissociation is required. Of note, neurons do not appear to be recovered by this process, as none of the available extracellular neuronal antibodies were able to positively identify and sort neurons (data not shown). Furthermore, this procedure likely strips away most cell processes, leaving only the cell body and RNA contained within it. The dissociation process also takes time, most of it on ice, but relies upon enzymatic digestion using papain at elevated temperatures before cells are ultimately lysed or plated in media. An additional caveat is the use of a single antibody for CD45mid gating of microglia. This strategy inadvertently allows the inclusion of other nonlymphocytic or nonmonocytic myeloid lineage contaminants in the microglia population. However, with proper saline perfusions, these cell types are removed from the tissue preparation, and little evidence suggests that these cells actively reside within the brain parenchyma in substantial numbers. TMEM119 antibodies may serve as a viable substitute (20). However, given the downregulation of Tmem119 in DAM cells, using the TMEM119 antibody may not suffice in isolating all microglial subtypes of certain disease models, resulting in unwanted misrepresentation of transcriptomic analyses. Thus, using traditional CD45mid marker identification allows for the identification of all potential microglial subtypes and allows for the exclusion of nearly all peripheral immune cells, which express higher levels of CD45.
The CoBrA method detailed here is the first to our knowledge to describe the use of CD49a to isolate vascular endothelia as well as to use the anti-ACSA-2 antibody to purify astrocytes in a combinatorial approach. Our method is readily and easily applicable to any mouse model and at any age and affords the ability to delve deep into physiological and disease-related transcriptomic alterations in a cell type–specific manner. Excitingly, this approach also provides proof of principle that isolated cells can be cultured, potentially unlocking the ability to address a myriad of molecular and cellular questions. We envision that this approach will have a significant effect in elucidating the underpinnings of the inner workings of the brain, beginning with basic biology and culminating with pathogenesis and therapeutic testing.
Methods
Mice.
Mice were housed 2–5 per cage in a specific pathogen–free mouse facility on a 12-hour-light/dark cycle, with ad libitum access to food and water. Wild-type, APP NL-G-F KI mice (42), and Aldh1l1-EGFP astrocyte reporter mice (41) were used in this study, and they were maintained on the C57BL/6J background. Homozygous APP KI mice were derived by intercrossing with the heterozygous APP NL-G-F KI mice, and wild-type littermates were used as controls. Both female and male mice were used.
Eight- to ten-week-old C57BL/6 male and female mice were injected i.p. with LPS (MilliporeSigma, 0111:B4 L3024) at 4 μg/g. Control animals were injected with an equal volume of PBS. Animals were monitored for 24 hours and perfused with saline before dissection.
CoBrA.
Adult mice were perfused with ice-cold PBS, forebrains were microdissected, and either half or whole forebrains were gently minced with sterile razor blades and resuspended in 2.5 ml HBSS without Ca2+ and without Mg2+-containing activated papain (half vial [>50 units], Worthington Biochemical, LK003172) and DNase (quarter vial [>250 units], Worthington Biochemical, LK003178). The resuspended forebrains were incubated with gently rocking for 30 minutes at 37°C. Halfway through the incubation, the minced forebrains were triturated 4 times using a fire-polished glass Pasteur pipette. After incubation, 2.5 ml ice-cold HBSS without Ca2+ and Mg2+ and with 2 mM EDTA and 0.5% BSA (HBSS+) was added to the suspension, and the cells were pelleted at 310 g at 4°C for 5 minutes. The supernatant was removed, and pellet was resuspended in 1,000 μl of HBSS+, transferred to an ice-cold 1.7-ml Eppendorf tube, and triturated gently 3 times with a fire-polished glass Pasteur pipette. Cells and tissue were centrifuged for 15 seconds at 100 g, and the supernatant (containing dissociated cells) was carefully transferred to a prechilled 15-ml conical tube. This step was repeated until all/most cells were dissociated. Completing this step correctly and gently is critical to obtain a high yield of healthy cells. The contents of the 15-ml conical tube was filtered through a prewetted 40-μm cell strainer (BD, 352340) into a prechilled 50-ml conical tube and centrifuged at 310 g at 4°C for 5 minutes. Myelin Removal Beads II (Miltenyi Biotec, 130-096-433) and LS columns (Miltenyi Biotec, 130-042-401) were used according to the manufacturer’s protocol to remove myelin debris. The resulting flow-through containing single cells was centrifuged at 310 g at 4°C for 5 minutes. The resulting pellet from myelin removal bead flow-through contained a single-cell suspension of microglia, astrocytes, and vascular endothelia. In order to isolate oligodendrocytes, the LS column was further loaded with 3 ml HBSS+, removed from magnet, plunged into a clean 15-ml conical tube, and centrifuged at 310 G at 4°C for 5 minutes. The resulting pellet was resuspended in 5 ml per half forebrain in 20% isotonic 4°C Percoll PLUS (MilliporeSigma, E0414-250ML) in 1× PBS and centrifuged with low break at 310 g at 4°C for 20 minutes. The resulting pellet containing oligodendrocytes and some astrocytes and microglia were washed with HBSS+ to remove any traces of Percoll PLUS and then added back to the main single cell suspension.
The total dissociated single cells were incubated 200 μl HBSS+ containing 1:100 Mouse BD Fc Block (BD Biosciences, 553141) for 5 minutes. For cell type labeling, the following antibodies were used: 1:200 rat anti-CD45-BV421 (BD Biosciences, catalog 563890); 1:200 anti-CD49a-VioBright FITC (Miltenyi Biotec, 130-107-636); 1:200 anti-O4-PE (Miltenyi Biotec, 130-109-199); 1:100 anti-ACSA-2-APC (Miltenyi Biotec, 130-116-245), and LIVE/DEAD Fixable Blue Dead Cell (Thermo Fisher, catalog L23105). Prior to FACS sorting, the cells were washed twice and resuspended in 500 μl in HBSS+. FACS sorting was performed using a BD Aria II on the 100-micron nozzle. Cells were sorted into 1.7-ml Eppendorf tubes containing 200 μl HBSS+ before subsequent centrifugation and resuspension of pellets for downstream applications (see the Quantitative RT-PCR and Immunofluorescent staining of cultured cells sections for details). It is important to coat the 1.7-ml Eppendorf tubes with HBSS+ by simple vortex prior to sorting to optimize pelleting of sorted cells.
Quantitative RT-PCR.
Single-cell-type pellets (see the CoBrA section) were lysed in Qiagen RLT buffer containing 1% β-mercaptoethanol immediately following sorting, and RNA was extracted using the RNeasy Micro kit (Qiagen, 74004). Brain tissue samples were lysed in Qiagen RLT buffer containing 1% β-mercaptoethanol, but RNA was extracted using the RNeasy Mini Kit (Qiagen, 74106). Reverse transcription was performed using the iScript Reverse Transcription Supermix (Bio-Rad, 1708840) according to the manufacturer’s protocol. All RNA isolated from cell pellets was converted into cDNA. Quantitative RT-PCR was performed using iTaq Universal SYBR Green Supermix (Bio-Rad, 172-5120) on a CFX384 Touch Real-Time PCR Detection System. Primer sequences used are listed in Supplemental Table 1.
In vitro culture of sorted cell populations for immunofluorescence.
Coverslips (Electron Microcopy Sciences, 72196-12) were coated with 1× PBS containing a mixture of 25 μg/ml human collagen IV (MilliporeSigma, C5533) and 25 μg/ml human fibronectin (Corning, 356008) for astrocytes, endothelia, and oligodendrocytes or 10 μg/ml human fibronectin for microglia. All coating was performed using 0.4 ml coating mixture for 2–4 hours at 37°C. Following sorting, cells were seeded at approximately 8.25 × 104/cm2, 4 × 105/cm2, 2.75 × 105/cm2, and 1.5 × 105 cells/cm2 for oligodendrocytes, microglia, astrocytes, and endothelia, respectively. Astrocytes and oligodendrocytes were seeded and maintained in DMEM containing 5% FBS, B27 supplement (Thermo Fisher, 17504044), 20 ng/ml rhFGF (R&D Systems, 233-FB-025/CF), 10 ng/ml rhBDNF (Miltenyi Biotec, 130-096-285), and 100 ng/ml insulin (MilliporeSigma, I2643). Endothelial cells were seeded and maintained in EGM-2 MV Bulletkit medium (Lonza, CC-3202). Microglia were seeded and maintained in DMEM containing 10% FBS, 10 ng/ml rmGMCSF (R&D Systems, 415-ML/CF), and 10 ng/ml rmMCSF (R&D Systems, 416-ML/CF). All media were changed every third day. Cultures were maintained for 1 week prior to immunofluorescent staining.
Immunofluorescent staining of cultured cells.
Cells were washed once with 1× PBS prior to fixing. Astrocytes, oligodendrocytes, and microglia were fixed in 4% PFA for 30 minutes. Endothelial cells were fixed on ice in an ice-cold mixture of 50% acetone and 50% methanol for 20 minutes. Cells were then blocked and permeabilized in PBS containing 0.4% Triton X-100, 1% BSA, and 5% donkey serum at room temperature for 1 hour. Cells were stained using primary antibodies: chicken anti-GFAP (1:2,000, Abcam, 4674); mouse anti-MBP (1:500, Covance, SMI-99P); rabbit anti-Iba1 (1:1,000, Wako, 019-19741); and rabbit anti-ZO1 (1:250, Cell Signaling Technologies, 5406S). Then, they were diluted in blocking buffer and lightly rocked overnight at 4°C. Cells were washed 3 times with PBS, and secondary antibodies were diluted 1:1,000 and incubated at room temperature in blocking buffer. Cells were washed 3 times in PBS, and coverslips were mounted onto slides using Vectashield mounting medium (Vector Labs, H-1200). Confocal microscopy was performed using the Leica DM-2500.
Immunostaining of brain sections and image quantification.
Mice were perfused with 4% PFA, and brains were post-fixed in 4% PFA overnight at 4°C followed by cryoprotection in 40% sucrose. Thirty-micrometer coronal sections were cut with a microtome and stored in cryoprotectant at −20°C. For each experiment, sections were collected randomly from at least 3 animals. Sections were extensively washed in PBS; blocked with PBS containing 0.4% Triton X-100, 1% BSA, and 5% donkey serum for 30 minutes; and then incubated in primary antibody diluted in blocking solution overnight at 4°C (mouse anti-GFAP [Millipore, MAB3402]; rabbit anti-Iba1 [Wako, 019-19741]; mouse anti-6E10 [MilliporeSigma, A1474]). After washing, sections were incubated in secondary antibody for 1 hour at room temperature and mounted in DAPI solution after final washing. The brain sections were imaged using a Leica laser confocal microscope. For the quantification of reactive glial cells in the mouse cortex and hippocampus, the sections were scanned using an EVOS FL Auto system. Images were then processed by ImageJ (NIH), and background was subtracted by the software for fluorescence images before quantification.
Statistics.
All data are presented as mean ± SEM. Pairwise comparisons were analyzed using a 2-tailed Student’s t test for all analyses. P values of less than or equal to 0.05 were considered statistically significant. P values higher than 0.05 were considered nonsignificant.
Study approval.
The present studies in animals were reviewed and approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine and are in accordance with the NIH guidelines.
Author contributions
DBS, NEP, and HZ conceived of the project and designed the experiments. BW performed Aβ immunostaining, and ERR generated a subset of the qPCR results. All other experiments and data analysis were performed by DBS and NEP. T. Saito and T. Saido provided the APP NL-G-F KI mice and made comments on the manuscript. DBS wrote the manuscript, with extensive input and revision from NEP, ERR, and HZ. All authors read and approved the final manuscript.
Supplementary Material
Acknowledgments
We thank B. Deneen (Baylor College of Medicine) for providing the Aldh1l1-EGFP reporter line and C. Beeton, J. Sederstrom, and the Baylor College of Medicine Cytometry and Cell Sorting Core, supported by a National Cancer Institute grant (CA125123), for FACS analysis. We are grateful to N. Aithmitti, A. Cole, and B. Contreras for expert technical support and members of the Zheng laboratory for stimulating discussions. This study was supported by grants from the NIH (R01 NS093652, R01 AG020670, R01 AG032051, and RF1 AG054111 to HZ).
Version 1. 07/12/2018
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(13): e121109. https://doi.org/10.1172/jci.insight.121109.
Contributor Information
Takashi Saito, Email: takasai@brain.riken.jp.
Baiping Wang, Email: baipingw@bcm.edu.
Hui Zheng, Email: huiz@bcm.tmc.edu.
References
- 1.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34(36):11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen NJ, Barres BA. Neuroscience: Glia - more than just brain glue. Nature. 2009;457(7230):675–677. doi: 10.1038/457675a. [DOI] [PubMed] [Google Scholar]
- 3.Sharma K, et al. Cell type- and brain region-resolved mouse brain proteome. Nat Neurosci. 2015;18(12):1819–1831. doi: 10.1038/nn.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okaty BW, Sugino K, Nelson SB. Cell type-specific transcriptomics in the brain. J Neurosci. 2011;31(19):6939–6943. doi: 10.1523/JNEUROSCI.0626-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Srinivasan K, et al. Untangling the brain’s neuroinflammatory and neurodegenerative transcriptional responses. Nat Commun. 2016;7:11295. doi: 10.1038/ncomms11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28(1):264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doyle JP, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135(4):749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heiman M, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135(4):738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lovatt D, et al. The transcriptome and metabolic gene signature of protoplasmic astrocytes in the adult murine cortex. J Neurosci. 2007;27(45):12255–12266. doi: 10.1523/JNEUROSCI.3404-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rossner MJ, et al. Global transcriptome analysis of genetically identified neurons in the adult cortex. J Neurosci. 2006;26(39):9956–9966. doi: 10.1523/JNEUROSCI.0468-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liddelow SA, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541(7638):481–487. doi: 10.1038/nature21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orre M, et al. Acute isolation and transcriptome characterization of cortical astrocytes and microglia from young and aged mice. Neurobiol Aging. 2014;35(1):1–14. doi: 10.1016/j.neurobiolaging.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 13.Zamanian JL, et al. Genomic analysis of reactive astrogliosis. J Neurosci. 2012;32(18):6391–6410. doi: 10.1523/JNEUROSCI.6221-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beutner C, et al. Unique transcriptome signature of mouse microglia. Glia. 2013;61(9):1429–1442. doi: 10.1002/glia.22524. [DOI] [PubMed] [Google Scholar]
- 15.Cardona AE, Huang D, Sasse ME, Ransohoff RM. Isolation of murine microglial cells for RNA analysis or flow cytometry. Nat Protoc. 2006;1(4):1947–1951. doi: 10.1038/nprot.2006.327. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, et al. TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell. 2015;160(6):1061–1071. doi: 10.1016/j.cell.2015.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harms AS, Tansey MG. Isolation of murine postnatal brain microglia for phenotypic characterization using magnetic cell separation technology. Methods Mol Biol. 2013;1041:33–39. doi: 10.1007/978-1-62703-520-0_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JK, Tansey MG. Microglia isolation from adult mouse brain. Methods Mol Biol. 2013;1041:17–23. doi: 10.1007/978-1-62703-520-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jay TR, et al. Disease progression-dependent effects of TREM2 deficiency in a mouse model of Alzheimer’s disease. J Neurosci. 2017;37(3):637–647. doi: 10.1523/JNEUROSCI.2110-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bennett ML, et al. New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci USA. 2016;113(12):E1738–E1746. doi: 10.1073/pnas.1525528113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daneman R, Zhou L, Agalliu D, Cahoy JD, Kaushal A, Barres BA. The mouse blood-brain barrier transcriptome: a new resource for understanding the development and function of brain endothelial cells. PLoS ONE. 2010;5(10):e13741. doi: 10.1371/journal.pone.0013741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huntley MA, Bien-Ly N, Daneman R, Watts RJ. Dissecting gene expression at the blood-brain barrier. Front Neurosci. 2014;8:355. doi: 10.3389/fnins.2014.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dugas JC, Tai YC, Speed TP, Ngai J, Barres BA. Functional genomic analysis of oligodendrocyte differentiation. J Neurosci. 2006;26(43):10967–10983. doi: 10.1523/JNEUROSCI.2572-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson AP, Rodgers JM, Goings GE, Miller SD. Characterization of oligodendroglial populations in mouse demyelinating disease using flow cytometry: clues for MS pathogenesis. PLoS One. 2014;9(9):e107649. doi: 10.1371/journal.pone.0107649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci USA. 2009;106(33):13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kang SS, Baker KE, Wang X, Kocher JP, Fryer JD. Translational Profiling Of Microglia Reveals Artifacts Of Cell Sorting. bioRxiv. https://www.biorxiv.org/content/early/2017/05/09/135566 Published May 7, 2017. Accessed June 18, 2018.
- 27.Boisvert MM, Erikson GA, Shokhirev MN, Allen NJ. The aging astrocyte transcriptome from multiple regions of the mouse brain. Cell Rep. 2018;22(1):269–285. doi: 10.1016/j.celrep.2017.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parekh S, Ziegenhain C, Vieth B, Enard W, Hellmann I. The impact of amplification on differential expression analyses by RNA-seq. Sci Rep. 2016;6:25533. doi: 10.1038/srep25533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeisel A, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347(6226):1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 30.Dulken BW, Leeman DS, Boutet SC, Hebestreit K, Brunet A. Single-cell transcriptomic analysis defines heterogeneity and transcriptional dynamics in the adult neural stem cell lineage. Cell Rep. 2017;18(3):777–790. doi: 10.1016/j.celrep.2016.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keren-Shaul H, et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell. 2017;169(7):1276–1290.e17. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 32.Friedman BA, et al. Diverse brain myeloid expression profiles reveal distinct microglial activation states and aspects of Alzheimer’s disease not evident in mouse models. Cell Rep. 2018;22(3):832–847. doi: 10.1016/j.celrep.2017.12.066. [DOI] [PubMed] [Google Scholar]
- 33.Mathys H, et al. Temporal tracking of microglia activation in neurodegeneration at single-cell resolution. Cell Rep. 2017;21(2):366–380. doi: 10.1016/j.celrep.2017.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cuevas-Diaz Duran R, Wei H, Wu JQ. Single-cell RNA-sequencing of the brain. Clin Transl Med. 2017;6(1):20. doi: 10.1186/s40169-017-0150-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ofengeim D, Giagtzoglou N, Huh D, Zou C, Yuan J. Single-cell RNA sequencing: unraveling the brain one cell at a time. Trends Mol Med. 2017;23(6):563–576. doi: 10.1016/j.molmed.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Penfold PL, Provis JM, Liew SC. Human retinal microglia express phenotypic characteristics in common with dendritic antigen-presenting cells. J Neuroimmunol. 1993;45(1-2):183–191. doi: 10.1016/0165-5728(93)90179-3. [DOI] [PubMed] [Google Scholar]
- 37.Schnitzer J, Schachner M. Developmental expression of cell type-specific markers in mouse cerebellar cells in vitro. J Neuroimmunol. 1981;1(4):471–487. doi: 10.1016/0165-5728(81)90024-2. [DOI] [PubMed] [Google Scholar]
- 38.Schnitzer J, Schachner M. Expression of Thy-1, H-2, and NS-4 cell surface antigens and tetanus toxin receptors in early postnatal and adult mouse cerebellum. J Neuroimmunol. 1981;1(4):429–456. doi: 10.1016/0165-5728(81)90022-9. [DOI] [PubMed] [Google Scholar]
- 39.Batiuk MY, et al. An immunoaffinity-based method for isolating ultrapure adult astrocytes based on ATP1B2 targeting by the ACSA-2 antibody. J Biol Chem. 2017;292(21):8874–8891. doi: 10.1074/jbc.M116.765313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kantzer CG, et al. Anti-ACSA-2 defines a novel monoclonal antibody for prospective isolation of living neonatal and adult astrocytes. Glia. 2017;65(6):990–1004. doi: 10.1002/glia.23140. [DOI] [PubMed] [Google Scholar]
- 41.Tsai HH, et al. Regional astrocyte allocation regulates CNS synaptogenesis and repair. Science. 2012;337(6092):358–362. doi: 10.1126/science.1222381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Erickson MA, Banks WA. Cytokine and chemokine responses in serum and brain after single and repeated injections of lipopolysaccharide: multiplex quantification with path analysis. Brain Behav Immun. 2011;25(8):1637–1648. doi: 10.1016/j.bbi.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kang SS, et al. Lipocalin-2 protects the brain during inflammatory conditions. Mol Psychiatry. 2018;23(2):344–350. doi: 10.1038/mp.2016.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hasegawa-Ishii S, Inaba M, Umegaki H, Unno K, Wakabayashi K, Shimada A. Endotoxemia-induced cytokine-mediated responses of hippocampal astrocytes transmitted by cells of the brain-immune interface. Sci Rep. 2016;6:25457. doi: 10.1038/srep25457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Das A, et al. Transcriptome sequencing of microglial cells stimulated with TLR3 and TLR4 ligands. BMC Genomics. 2015;16:517. doi: 10.1186/s12864-015-1728-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu R, Shen Q, Xu P, Luo JJ, Tang Y. Phagocytosis of microglia in the central nervous system diseases. Mol Neurobiol. 2014;49(3):1422–1434. doi: 10.1007/s12035-013-8620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walker DG, Lue LF. Immune phenotypes of microglia in human neurodegenerative disease: challenges to detecting microglial polarization in human brains. Alzheimers Res Ther. 2015;7(1):56. doi: 10.1186/s13195-015-0139-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clarke LE, Liddelow SA, Chakraborty C, Münch AE, Heiman M, Barres BA. Normal aging induces A1-like astrocyte reactivity. Proc Natl Acad Sci USA. 2018;115(8):E1896–E1905. doi: 10.1073/pnas.1800165115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vasilache AM, Qian H, Blomqvist A. Immune challenge by intraperitoneal administration of lipopolysaccharide directs gene expression in distinct blood-brain barrier cells toward enhanced prostaglandin E(2) signaling. Brain Behav Immun. 2015;48:31–41. doi: 10.1016/j.bbi.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 50.Saito T, et al. Single App knock-in mouse models of Alzheimer’s disease. Nat Neurosci. 2014;17(5):661–663. doi: 10.1038/nn.3697. [DOI] [PubMed] [Google Scholar]
- 51.Krasemann S, et al. The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity. 2017;47(3):566–581.e9. doi: 10.1016/j.immuni.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang Q, et al. ApoE promotes the proteolytic degradation of Abeta. Neuron. 2008;58(5):681–693. doi: 10.1016/j.neuron.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Castillo E, et al. Comparative profiling of cortical gene expression in Alzheimer’s disease patients and mouse models demonstrates a link between amyloidosis and neuroinflammation. Sci Rep. 2017;7(1):17762. doi: 10.1038/s41598-017-17999-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rogers J, et al. Complement activation by beta-amyloid in Alzheimer disease. Proc Natl Acad Sci USA. 1992;89(21):10016–10020. doi: 10.1073/pnas.89.21.10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hong S, et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science. 2016;352(6286):712–716. doi: 10.1126/science.aad8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fu H, et al. Complement component C3 and complement receptor type 3 contribute to the phagocytosis and clearance of fibrillar Aβ by microglia. Glia. 2012;60(6):993–1003. doi: 10.1002/glia.22331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Domingues HS, Portugal CC, Socodato R, Relvas JB. Oligodendrocyte, astrocyte, and microglia crosstalk in myelin development, damage, and repair. Front Cell Dev Biol. 2016;4:71. doi: 10.3389/fcell.2016.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.