Abstract
The versatility and universality of Ca2+ as intracellular messenger is guaranteed by the compartmentalization of changes in [Ca2+]. In this context, mitochondrial Ca2+ plays a central role, by regulating both specific organelle functions and global cellular events. This versatility is also guaranteed by a cell type-specific Ca2+ signaling toolkit controlling specific cellular functions. Accordingly, mitochondrial Ca2+ uptake is mediated by a multimolecular structure, the MCU complex, which differs among various tissues. Its activity is indeed controlled by different components that cooperate to modulate specific channeling properties. We here investigate the role of MICU3, an EF-hand containing protein expressed at high levels, especially in brain. We show that MICU3 forms a disulfide bond-mediated dimer with MICU1, but not with MICU2, and it acts as enhancer of MCU-dependent mitochondrial Ca2+ uptake. Silencing of MICU3 in primary cortical neurons impairs Ca2+ signals elicited by synaptic activity, thus suggesting a specific role in regulating neuronal function.
Subject terms: Calcium channels, Calcium
Introduction
Calcium (Ca2+) signals widely varies depending on the cell type and the generating stimulus. This diversity is the consequence of a tightly regulated Ca2+ signaling toolkit that characterizes each cell and guarantees the realization of precise cellular events [1–3]. Mitochondria are active players in the regulation of Ca2+ dynamics thanks to dedicated machineries controlling Ca2+ fluxes in and out organelle [4]. Compartmentalization of Ca2+ signals on one hand allows organelle-specific functions and on the other provides a buffer that can shape cytosolic Ca2+ waves [5–7]. Therefore, mitochondrial Ca2+ handling must be intrinsically intertwined with global Ca2+ dynamics and tuned to coordinate the accurate decoding of these signals.
Ca2+ accumulation inside matrix is an electrophoretic process governed by a structure named MCU complex [8], while its efflux occurs by two different mechanisms, i.e a Na+/Ca2+ exchanger (NCX) and a H+/Ca2+ antiporter (HCX). In particular, NCX is mediated by NCLX [9], while the molecular identity of HCX is still controversial [10, 11], although Letm1 can play a role in this pathway [12–14]. The MCU complex includes components of the inner membrane, namely MCU [15, 16], MCUb [17] and EMRE [18], and regulators localized in the intermembrane space, i.e. MICU1 [19] and MICU2 [20]. MCU represents the Ca2+-permeant pore-forming subunit [15], while MCUb acts as dominant-negative by inserting itself in the multimer and inhibiting Ca2+ channeling activity [17]. EMRE is a small protein necessary to form a functional MCU complex in vivo, at least in vertebrates [18, 21, 22]. Finally, MCUR1 was reported to be a regulator of the MCU complex [23, 24], although it was also proposed to function as a COX assembly factor, opening the possibility that the modulation of MCU channel activity can be secondary to the respiratory chain defect [25]. Recently, MCUR1 was shown to act as a scaffold factor also for the MCU complex [26].
MCU is gated by Ca2+ itself and shows low activity at resting extramitochondrial [Ca2+] and it is activated when [Ca2+] is in the micromolar range. Gating depends on a family of EF hands containing proteins, the MICU family. MICU1 is the prototypical member [19] and in several organisms it represents the only component, while in vertebrates two additional isoforms are present, MICU2 and MICU3 [20]. MICU1 was described to act both as inhibitor of MCU [27, 28], and as a cooperative activator [28]. Conversely, MICU2 has been shown to act mainly as a channel inhibitor [29–31]. As to MICU3, its function is still unknown. Importantly, MICU2 requires MICU1 to interact with MCU [29, 31], suggesting that MICU1 represents a common required platform for the physical association of the whole family to the channel. In line with this, we and others previously demonstrated that MICU1 and MICU2 are linked together through a disulfide bond [29, 32]. This complexity allows the precise titration of each component in order to fit the specific Ca2+ signaling profile of each cell to the properties of the mitochondrial Ca2+ transport machinery. However, the tissue-specific characteristics of the MCU complex has been poorly investigated so far. Here we address this point and describe the properties of a novel tissue-specific MCU modulator, MICU3, that acts as a highly potent stimulator of mitochondrial Ca2+ uptake.
Results
MICU3 is a tissue-specific MCU modulator
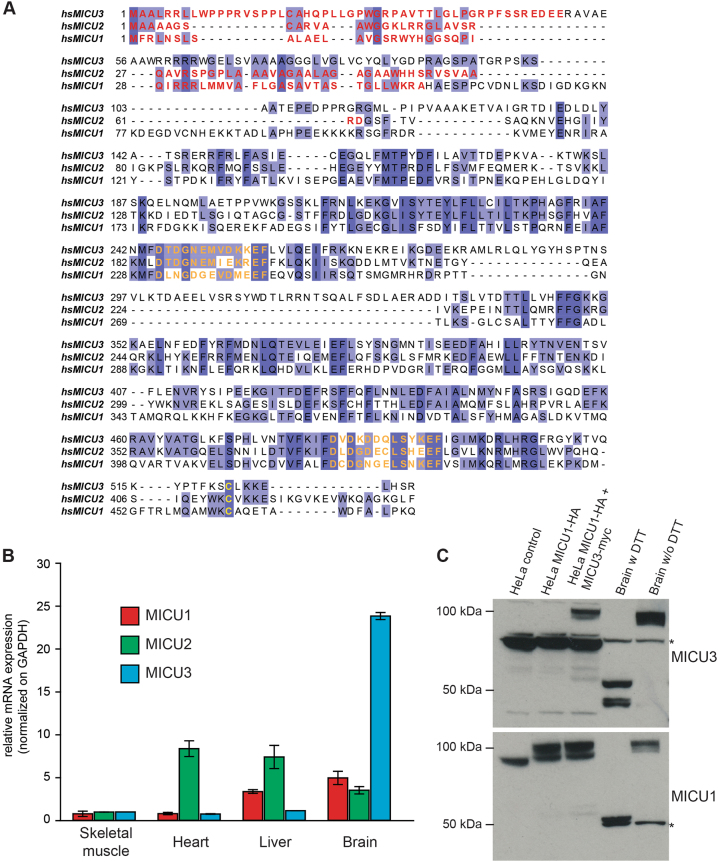
MCU activity widely varies among different tissues [33] to fulfill different metabolic activity. We looked at mRNA expression levels of all components of mitochondrial Ca2+ handling machinery. We used expression data from the GTEx Consortium [34] for human samples and by the ENCODE project [35] for cell lines. As shown in Table 1, human MCU levels are generally low (blue and red indicate low and high expression, respectively), while its associated regulators are more abundant, probably reflecting different stability at the protein level. Conversely, cell lines express MCU at high levels, with the exception of CD20-positive B cells. As to the tissue-specific differences, on one hand MCU is surprisingly abundant in cerebellum and skeletal muscle, despite an unexpected low expression of the Na+/Ca2+ exchanger NCLX (i.e., the main efflux pathway). On the other hand, MCUb, the negative MCU isoform [17], is expressed in adipose tissue, blood and spleen. Most importantly, some MCU complex components show a tissue-specific signature, such as MICU3 that is expressed at high levels in the central nervous system (CNS), which is in line with previous reports [20, 36]. We thus looked at MICU3, an uncharacterized MCU regulator. Its gene is conserved in vertebrates, but it is absent in other organisms where other components of the MCU complex are present, with a conservation profile closely resembling to MICU2. As to protein sequence, MICU3 shows a 34% similarity to MICU1, 47% to MICU2 (while MICU2 is 41% similar to MICU1), and possesses two EF hand domains and a mitochondrial targeting signal. The only noticeable difference compared to the other members of the MICU family is the presence of an additional domain (50 aa) in the central part of the protein (Fig. 1a). We confirmed by qPCR that also in mice MICU3 is expressed at high levels in the brain (Fig. 1b) [20, 36].
Table 1.
Expression of mitochondrial Ca2+ handling genes
| Tissue | MCU | MCUB | EMRE | MICU1 | MICU2 | MICU3 | NCLX |
|---|---|---|---|---|---|---|---|
| CNS - Average | 2.6 | 0.8 | 14.6 | 10.1 | 7.1 | 4.8 | 1.8 |
| Cerebellum | 8.9 | 0.1 | 16.6 | 21.2 | 10.8 | 9.6 | 2.5 |
| Cerebral cortex | 1.2 | 0.5 | 13.6 | 9.9 | 5.1 | 4.1 | 1.5 |
| Hippocampus | 1.7 | 0.7 | 14.6 | 8.1 | 6.1 | 4.7 | 1.7 |
| Hypothalamus | 1.4 | 0.8 | 14.7 | 7.9 | 6.3 | 3.5 | 1.4 |
| Spinal cord | 1.5 | 2.1 | 14.1 | 7.0 | 8.8 | 3.8 | 2.3 |
| Substantia nigra | 1.2 | 0.6 | 14.2 | 6.4 | 5.5 | 2.9 | 1.7 |
| Adipose tissue - average | 2.8 | 3.5 | 11.0 | 6.0 | 5.4 | 2.1 | 4.3 |
| Subcutaneous | 3.1 | 3.7 | 11.1 | 6.2 | 5.3 | 2.5 | 5.1 |
| Visceral | 2.5 | 3.4 | 10.9 | 5.7 | 5.5 | 1.7 | 3.6 |
| Arteries - average | 3.9 | 1.6 | 12.4 | 6.7 | 5.0 | 3.1 | 3.4 |
| Aorta | 4.2 | 1.4 | 12.2 | 6.4 | 4.8 | 2.5 | 3.9 |
| Coronary artery | 3.5 | 2.0 | 12.2 | 5.8 | 5.1 | 2.9 | 3.5 |
| Esophagus - average | 2.6 | 1.4 | 10.9 | 4.3 | 4.0 | 1.1 | 2.6 |
| Mucosa | 3.2 | 1.5 | 9.5 | 3.0 | 4.3 | 0.3 | 2.8 |
| Muscularis mucosa | 2.0 | 1.3 | 12.3 | 5.7 | 3.7 | 1.8 | 2.4 |
| Intestine - average | 2.1 | 1.0 | 15.3 | 6.0 | 6.6 | 1.9 | 5.8 |
| Peyer’s patch | 3.8 | 2.2 | 13.0 | 5.7 | 6.2 | 0.7 | 4.9 |
| Sigmoid colon | 2.1 | 1.2 | 12.2 | 5.3 | 4.1 | 1.4 | 2.3 |
| Transverse colon | 5.0 | 0.9 | 10.4 | 4.8 | 5.2 | 0.4 | 3.5 |
| Glands - average | 2.1 | 1.0 | 15.3 | 6.0 | 6.6 | 1.9 | 5.8 |
| Adrenal gland | 1.3 | 0.3 | 9.6 | 10.1 | 6.2 | 1.0 | 12.6 |
| Pancreas | 2.1 | 0.4 | 18.0 | 3.9 | 6.0 | 1.3 | 4.7 |
| Prostate gland | 1.9 | 1.2 | 16.9 | 5.1 | 6.0 | 2.1 | 5.1 |
| Thyroid gland | 2.1 | 1.2 | 16.2 | 4.7 | 7.5 | 2.1 | 3.8 |
| Blood | 4.3 | 4.3 | 10.6 | 13.4 | 4.3 | 0.1 | 8.6 |
| Breast | 2.6 | 2.8 | 13.6 | 6.7 | 6.3 | 2.2 | 4.5 |
| Heart (left ventricle) | 0.8 | 0.4 | 6.9 | 3.0 | 4.2 | 0.4 | 1.0 |
| Kidney (cortex) | 2.3 | 0.5 | 11.7 | 5.2 | 6.1 | 0.7 | 3.4 |
| Liver | 1.2 | 0.2 | 8.1 | 6.0 | 3.0 | 1.4 | 2.6 |
| Lung | 4.8 | 2.5 | 12.2 | 8.1 | 6.0 | 1.7 | 6.2 |
| Reproductive system - average | 2.9 | 2.4 | 15.8 | 7.7 | 6.5 | 2.2 | 7.1 |
| Skeletal muscle | 4.2 | 0.2 | 10.7 | 5.4 | 4.1 | 0.3 | 0.8 |
| Spleen | 2.5 | 4.0 | 14.3 | 6.9 | 6.5 | 1.5 | 9.2 |
| Tibial nerve | 2.3 | 2.5 | 12.9 | 8.5 | 7.2 | 3.4 | 5.1 |
| ENCODE cell lines | MCU | MCUB | EMRE | MICU1 | MICU2 | MICU3 | NCLX |
| A549 | 18.1 | 2.5 | 2.1 | 6.9 | 3.6 | 0.8 | 0.8 |
| CD20-positive B cell line | 2.4 | 13.9 | 18.5 | 2.6 | 7.9 | 1.0 | 4.2 |
| GM12878 | 3.5 | 7.2 | 4.4 | 2.7 | 5.5 | 0.0 | 0.4 |
| H1-hESC | 8.0 | 1.2 | 2.5 | 2.1 | 1.6 | 0.2 | 1.5 |
| HMEC cell line | 5.1 | 2.6 | 2.7 | 3.6 | 1.7 | 0.1 | 1.0 |
| HUVEC cell line | 6.5 | 2.9 | 4.7 | 3.8 | 1.7 | 0.3 | 4.0 |
| HeLa-S3 | 14.6 | 2.6 | 2.1 | 2.9 | 3.4 | 0.0 | 1.2 |
| HepG2 | 4.8 | 0.4 | 3.4 | 4.6 | 3.0 | 0.6 | 0.8 |
| K562 | 5.0 | 0.4 | 1.8 | 4.6 | 2.5 | 0.0 | 1.1 |
| MCF-7 | 6.1 | 2.6 | 3.5 | 3.6 | 7.7 | 0.0 | 0.7 |
| NHEK cell line | 10.1 | 2.3 | 1.8 | 2.5 | 3.1 | 0.1 | 0.9 |
| NHLF cell line | 12.3 | 4.1 | 3.3 | 12.2 | 5.1 | 2.0 | 1.1 |
| SK-N-SH | 3.6 | 2.1 | 2.0 | 3.3 | 2.8 | 0.5 | 1.3 |
Fig. 1.
MICU3 is a tissue-specific MCU modulator. a Multiple alignment of human MICU1, MICU2 and MICU3 protein sequences. Identical and similar residues are highlighted in dark and light blue respectively. The predicted mitochondrial targeting signal is shown in red. EF hand domains are shown in orange. The conserved cysteine involved in dimerization is shown in yellow. b RNA was extracted from Tibialis Anterior (Skeletal muscle), cardiac ventricles (Heart), liver and whole brain of C57BL/6 mice and real-time PCR of MICU1, MICU2 and MICU3 mRNAs was performed. The data were normalized to the expression levels in skeletal muscle and are shown as the average ± s.d. of two different animals. c HeLa cells were transfected with the indicated constructs and harvested after 48 h. Total proteins were extracted from cells or homogenized brains and subjected to western blotting analysis with MICU1 and MICU3 antibodies. SDS-PAGE was performed in the absence of DTT, except where indicated. *indicates non-specific bands
As to the protein, MICU1 and MICU2 have been shown to form dimers through a disulfide bond [29, 32]. We thus performed immunoblot analysis of brain lysate in both reducing and non-reducing condition. In reduced samples, MICU3 runs at approximately 55 kDa (Fig. 1c), in line with its predicted molecular weight. Conversely, in the absence of reducing agents MICU3 migrates at approximately 100 kDa, in a band recognized also by an antibody against MICU1 (Fig. 1c). In line with mRNA, HeLa cells show no detectable MICU3, thus representing a suitable cell line for its characterization. Indeed, ectopic expression of MICU3 in these cells leads to the appearance of a 100 kDa species detected by both MICU1 and MICU3 antibodies, suggesting a heterodimerization of these two proteins.
MICU3 and MICU1 form a dimer through a disulfide bond
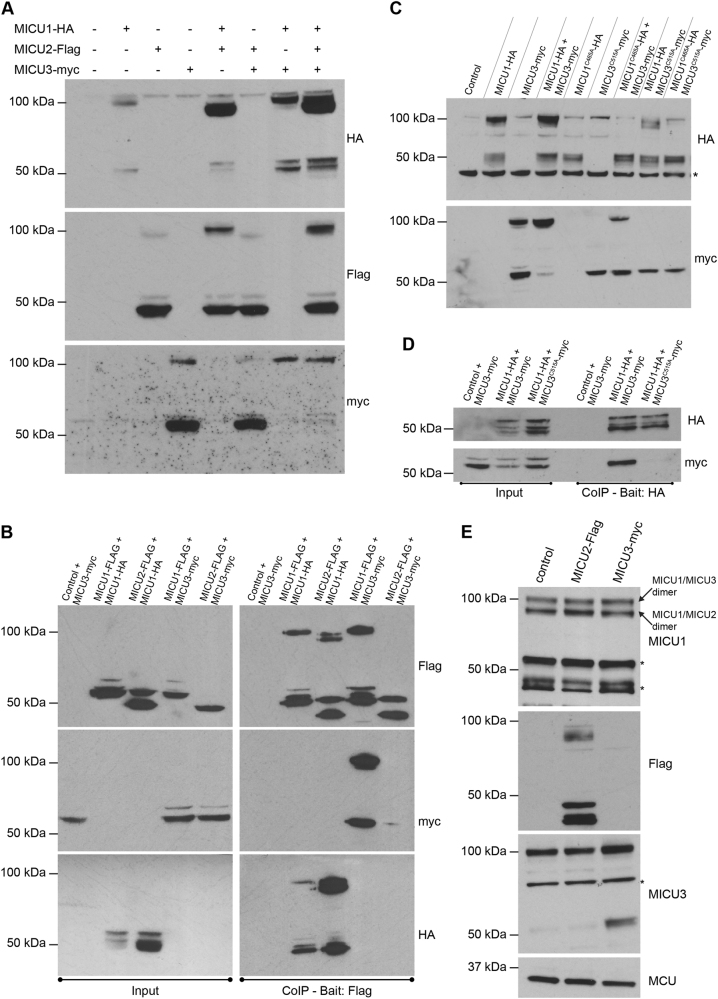
We then expressed different combinations of all MICUs (MICU1-HA, MICU2-Flag and MICU3-myc) in HeLa cells. In non-reducing conditions, overexpression of MICU1 alone leads to the appearance of a predominant 100 kDa band, corresponding to the MICU1 homodimer [29]. Conversely, when MICU2 or MICU3 are expressed alone, the predominant species runs around 50 kDa, indicative of monomers. Co-expression of either MICU2 or MICU3 with MICU1 leads to the formation of the 95 or 100 kDa heterodimer, respectively. Most importantly, the co-expression of MICU2 and MICU3 fails to generate any apparent high molecular weight species, thus suggesting that no MICU2/MICU3 dimer can be formed (Fig. 2a). We then performed also co-immunoprecipitation experiments. Figure 2b demonstrates that MICU1 can interact with itself, MICU2 and MICU3. Conversely, MICU2 can efficiently immunoprecipitate MICU1, but not MICU3 (Fig. 2b), thus confirming that MICU1 acts as a required docking site for the other family members.
Fig. 2.
MICU3 and MICU1 form a dimer through a disulfide bond. a HeLa cells were transfected with the indicated constructs. HeLa cells were harvested after 48 h of transfection, total proteins were extracted and subjected to western blotting analysis with HA, Flag and myc antibodies. SDS-PAGE was performed in the absence of DTT. b HeLa cells were transfected with the indicated constructs. Flag-tagged chimeras were immunoprecipitated from whole cell lysate with a specific agarose resin conjugated to a Flag antibody. The precipitated proteins were immunoblotted with HA, Flag and myc antibodies. SDS-PAGE was performed in the presence of DTT. c HeLa cells were transfected with the indicated constructs. HeLa cells were harvested after 48 h of transfection, total proteins were extracted and subjected to western blotting analysis with HA and myc antibodies. SDS-PAGE was performed in the absence of DTT. d HeLa cells were transfected with the indicated constructs. HA-tagged MICU1 was immunoprecipitated from whole cell lysate with a specific agarose resin conjugated to a HA antibody. The precipitated proteins were immunoblotted with HA and myc antibodies. SDS-PAGE was performed in the presence of DTT. e Primary cortical neurons were transfected with the indicated constructs. Cells were collected after 24 h, total proteins were extracted and subjected to western blotting analysis with MICU1, Flag, MICU3 and MCU antibodies. SDS-PAGE was performed in the absence of DTT. * indicates non-specific bands
As in the case of the MICU1/MICU2, also MICU1/MICU3 interaction is likely to be mediated by a disulfide bridge, since cysteine 515 is conserved among all MICUs (see Fig. 1a). To test this, we generated an epitope-tagged mutant (MICU3C515A-myc) and tested its ability to form dimers. Then we expressed either wild type or the MICU1C465A and MICU3C515A mutants and tested their ability to form dimers. When expressed alone, they gave rise only to the 50 kDa and 55 kDa bands of MICU1 and MICU3, respectively. The same results, however, was obtained when they were co-expressed with wild-type counterparts, indicating that both cysteine residues are necessary for the formation of the MICU1/MICU3 dimers (Fig. 2c). This notion was confirmed by co-immunoprecipitation. HeLa cells were co-transfected with epitope-tagged MICU1 and MICU3 moieties (MICU1-HA, MICU3-myc and MICU3C515A-myc) and immunoprecipitation was performed using MICU1-HA as bait. Cysteine mutation in MICU3 destroyed its interaction with MICU1 (Fig. 2d).
Finally, we looked at the endogenous expression in primary cortical neurons. Figure 2e shows that in non-reducing conditions, MICU1 occurs in two different species, one at 100 kDa and the other at 95 kDa. Due to the antibody limitations, we could not detect endogenous MICU2. However, we overexpressed either MICU2 or MICU3 and saw accumulation of the lower or upper dimer respectively. This demonstrates not only that the 100 kDa specie correspond to the MICU1/MICU3 heterodimer and the 95 kDa specie correspond to the MICU1/MICU2 heterodimer, but also that both dimers are expressed in neurons at similar levels. Overall, these data indicate that the MICU family exists in three different configurations (the MICU1 homodimer, the MICU1/MICU2 and the MICU1/MICU3 heterodimers) and that in neurons the two latter species are present.
MICU3 potently enhances mitochondrial Ca2+ uptake
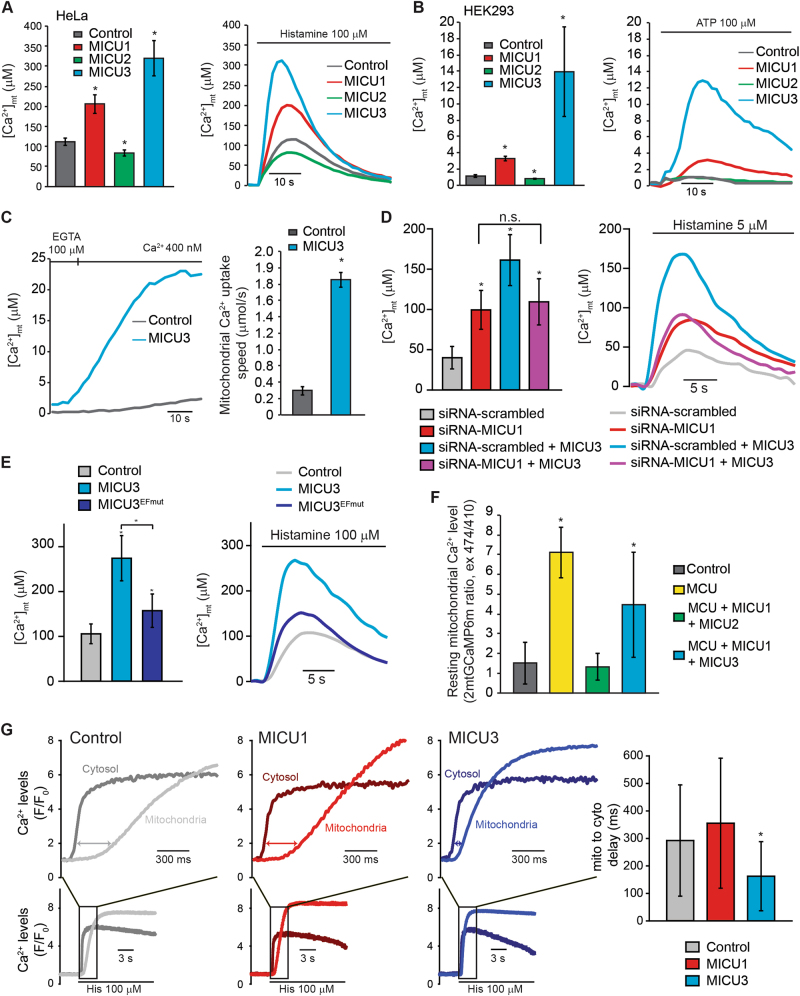
We then investigated the functional role of MICU3. In HeLa cells, MICU3 expression potently stimulated mitochondrial Ca2+ rise evoked by stimulation with the IP3-generating agonist histamine (Fig. 3a). Remarkably, not only MICU3 can easily overtake MICU1 in terms of absolute mitochondrial [Ca2+], but its effect is likely to be underestimated due to probe saturation. We indeed tested MICU3 expression in HEK293 cells, where mitochondrial Ca2+ transients are inherently low. In this experimental model, MICU3 expression causes a 10-fold increase in mitochondrial Ca2+ transients (Fig. 3b), compared to the 3-fold increment provoked by MICU1 overexpression. To better understand the extent of this activation, we performed experiments in permeabilized HeLa cells perfused with a fixed [Ca2+] buffer. After permeabilization, Ca2+ accumulation was initiated by switching the perfusion buffer to IB containing an EGTA-buffered [Ca2+] of 400 nM. In this condition, mitochondrial Ca2+ uptake is only barely detectable in control cells, since this value is near the activation threshold [28]. Conversely, MICU3 expression causes robust matrix Ca2+ accumulation (Fig. 3c).
Fig. 3.
MICU3 potently enhances mitochondrial Ca2+ uptake. a [Ca2+]mt measurements in intact HeLa cells overexpressing MICU1, MICU2 or MICU3 and challenged with maximal (100 μM) histamine stimulation. The bar graph on the left shows the average ± s.d. of [Ca2+]mt peak values, while the right panel displays representative traces. b [Ca2+]mt measurements in intact HEK293 cells overexpressing MICU1, MICU2 or MICU3 and challenged with maximal (100 μM) ATP stimulation. The bar graph on the left shows the average ± s.d. of [Ca2+]mt peak values, while the right panel displays representative traces. c [Ca2+]mt measurements in control and MICU3-expressing permeabilized HeLa cells perfused with 400 nM buffered [Ca2+]. The left panel shows representative traces, while the bar graph on the right shows the average ± s.d. mitochondrial Ca2+ uptake speed expressed in μMol/sec. d [Ca2+]mt measurements in intact HeLa cells co-transfected with pcDNA3.1 or MICU3 together with siRNA-scrambled or siRNA-MICU1 and challenged with low (5 μM) histamine stimulation. The right panel displays representative traces, while the bar graph on the left shows the average ± s.d. of [Ca2+]mt peak values. e [Ca2+]mt measurements in intact HeLa cells transfected with control, MICU3 or MICU3EFmut-expressing plasmids and challenged with maximal (100 μM) histamine stimulation. The right panel displays representative traces, while the bar graph on the left shows the average ± s.d. of [Ca2+]mt peak values. f HeLa cells were transfected with 2mtGCaMP6m together with the indicated constructs. After 24 h, resting mitochondrial Ca2+ levels were evaluated through ratiometric imaging of the mitochondrial targeted GCaMP6m. g Simultaneous high speed imaging of [Ca2+]cyt and [Ca2+]mt in intact HeLa cells overexpressing MICU1 or MICU3 and challenged with maximal (100 μM) histamine stimulation. Representative traces and their relative magnification during Ca2+ increases are shown. The bar graph shows the average time delay ± s.d. between the onset of cytosolic and mitochondrial Ca2+ rises, as indicated by the double-headed arrows. *indicates p < 0.01. n > 30, from at least three independent transfections
Since our biochemical data indicate that MICU3 depends on MICU1 for dimer formation, we wondered whether this is true also in its function. We compared the effect of MICU3 on mitochondrial Ca2+ transients evoked by low histamine stimulation on both control and MICU1-silenced cells. MICU3 has no effect in the absence of MICU1 (Fig. 3d), thus reinforcing the idea of MICU1 as a necessary and common platform for all the members of its family.
Finally, we asked if EF-hand domains in MICU3 are necessary for its function. We compared the effect of wild type and EF-hand mutant MICU3. As expected, MICU3EFmut is not as effective as the wild type in enhancing MCU opening (Fig. 3e). However, MICU3EFmut expressing cells still show higher mitochondrial Ca2+ uptake compared to control, suggesting that MICU3 intrinsically stimulates organelle Ca2+ entry, although the EF-hand domains plays a critical role.
To test if MICU3 can also work as gatekeeper, we investigated MCU gating in unstimulated cells [29]. HeLa cells were transfected with MCU alone or together with the other MICU regulators and the resting mitochondrial [Ca2+] was measured. Control cells show low mitochondrial Ca2+ levels, due to the presence of an endogenous gatekeeper. However, overexpression of MCU alone leads to constitutively elevated organelle Ca2+ levels, due to the imbalanced overexpression of the channel without its gatekeepers. As already demonstrated [29], only the concomitant overexpression of MICU1 and MICU2 can rescue normal mitochondrial Ca2+ levels. Most importantly, the co-expression of MICU1 and MICU3 fails to restore normal organelle Ca2+ handling (Fig. 3f), thus indicating that MICU3 has much lower, if any, gatekeeping functions.
Finally, we performed simultaneous imaging of cytosolic and mitochondrial Ca2+ dynamics with high temporal resolution. We transfected HeLa cells with two distinct Ca2+ indicators, targeted to either cytoplasm (R-GECO1) or mitochondrial matrix (4mtGCaMP6f). In control cells, histamine stimulation triggers a rapid rise in [Ca2+]cyt, followed by an upstroke of [Ca2+]mt that is delayed of approximately 300 ms, in line with previous reports [37] and reflecting MCU activation kinetics (Fig. 3g). Importantly, overexpression of MICU1 does not alter the delay between mitochondrial and cytosolic Ca2+ rises, despite of an overall increase of organelle Ca2+ uptake (see Fig. 3a). Conversely, this delay is shortened (160 ms) in MICU3-expressing cells, indicating an anticipation of MCU opening (Fig. 3g).
MICU3 sustains cytoplasmic Ca2+ rises triggered by synaptic activity
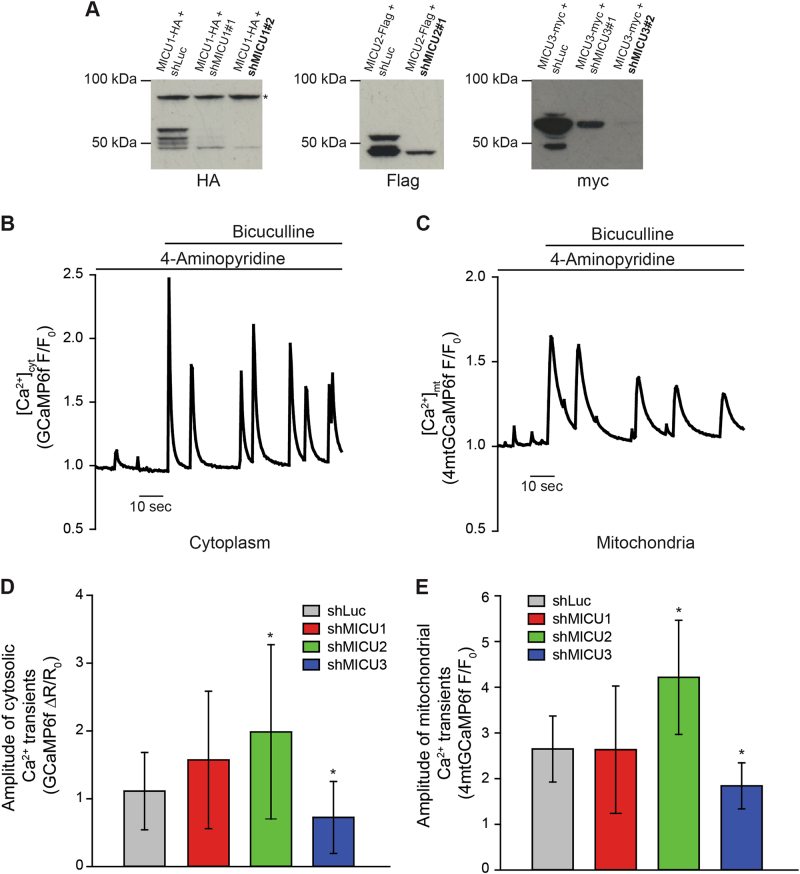
We finally moved to a physiological relevant cellular context. We first developed and validated shRNA targeting the mouse isoforms of the different MICU proteins (Fig. 4a) and expressing mCherry to identify knockdown cells. Then, we cultured primary cortical neurons and monitored Ca2+ dynamics. We treated cells with the 4-aminopyridine (4-AP), a K+ channel blocker, and bicuculline, a GABAA antagonist. This protocol produces transient spikes in cytosolic [Ca2+] triggered by the activation of synaptic NMDA receptors [38, 39]. Here we measured cytosolic Ca2+ dynamics with GCaMP6f and confirmed that treatment with 4-AP/bicuculline triggers cytosolic Ca2+ oscillations (Fig. 4b). In parallel, we monitored mitochondrial Ca2+ dynamics using the same probe targeted to organelle matrix and revealed that also [Ca2+]mt undergoes rapid and transient rises, although with a longer transient (Fig. 4c).
Fig. 4.
MICU3 sustains cytoplasmic Ca2+ rises triggered by synaptic activity. a HeLa cells were transfected with the indicated constructs. HeLa cells were harvested after 48 h of transfection, total proteins were extracted and subjected to western blotting analysis with HA, Flag and myc antibodies. SDS-PAGE was performed in the presence of DTT. b, c Mouse primary cortical neurons (PCN) were transfected with either cytosolic (b) or mitochondrial targeted (c) GCaMP6f. Time-lapse single-cell imaging was performed during the addition of 100 μM 4-Aminopyridine and 10 μM Bicuculline, as indicated. d PCN were transfected with the cytosolic GCaMP6f together with short hairpin RNA targeting MICU1, MICU2 or MICU3. After 24 h, ratiometric imaging of GCaMP6f was performed. The bar graph shows the average ± s.d. amplitude of cytosolic Ca2+ transients expressed as ΔR/R0. e PCN were transfected with the mitochondrial targeted GCaMP6f together with short hairpin RNA targeting MICU1, MICU2 or MICU3. After 24 h, Ca2+ imaging was performed as in b. The bar graph shows the average ± s.d. amplitude of mitochondrial Ca2+ transients expressed as normalized fluorescence (F/F0). *indicates p < 0.05. n > 20 single neurons derived from four independent PCN preparations
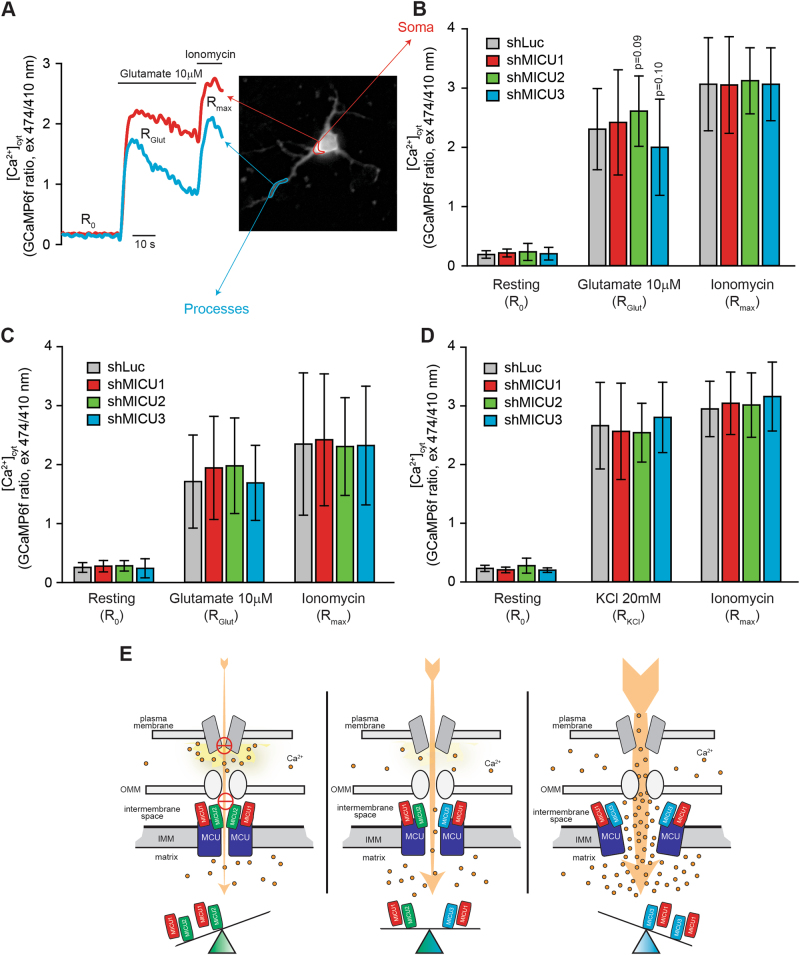
We next investigated how MICU silencing impacts on the amplitude of Ca2+ transients. In the case of mitochondria, MICU2 silencing leads to an increase of organelle Ca2+ uptake (60%). Conversely, MICU3 downregulation causes a substantial decrease (30%) in the amplitude of organelle Ca2+ transients. Finally, silencing of MICU1 has marginal impact on the overall Ca2+ handling, although its absence generates highly heterogeneous organelle Ca2+ responses (Fig. 4e). We next wondered how the altered mitochondrial Ca2+ dynamics are decoded at cytosolic level. Analysis of the amplitude of Ca2+ peaks in the cytoplasm unexpectedly revealed that silencing of MICU2 increase Ca2+ entry (up to 80%). Conversely, downregulation of MICU3 suppresses the amplitude of Ca2+ oscillations by 35% (Fig. 4d). Apparently, this supports the notion of a feedback inhibition of the synaptic NMDA receptor mediated by localized mitochondrial Ca2+ accumulation in a microdomain [40, 41]. Conditions that enhances mitochondrial Ca2+ uptake (e.g. MICU2 silencing) prevents the feedback inhibition of plasma membrane channel and thus sustains Ca2+ entry from the extracellular medium. Conversely, a condition that inhibits the local Ca2+ clearance by mitochondria (e.g. silencing of MICU3) anticipates channel closure thus limiting cytosolic signals. If this hypothesis is correct, this feedback inhibition should be overwhelmed by stimuli provoking large and sustained Ca2+ rises. We thus measured cytosolic Ca2+ dynamics elicited by other stimuli. We first used glutamate (10 μM), i.e. a stimulus causing a large (~10×) and sustained increase of [Ca2+]cyt (Fig. 5a). Using this protocol, silencing of either MICU1, MICU2 or MICU3 does not cause any major difference. However, taking into account only the neuronal soma (a cellular region where mitochondria are more abundant), a tendency still persists. Specifically, downregulation of MICU3 slightly decreases cytosolic transients, while silencing of MICU2 increases the amplitude of glutamate-induced Ca2+ rises (Fig. 5b). Conversely, in neuronal processes no differences could be appreciated (Fig. 5c), thus reinforcing the idea that mitochondria can play a role in the local (but not global) control of cytosolic Ca2+ signals. Finally, we used an additional stimulus causing even larger Ca2+ mobilization (20 mM KCl). As shown in Fig. 5d, silencing of all the members of the MICU family has no effect on the modulation of cytosolic Ca2+ handling.
Fig. 5.
MICU3 has no impact on high amplitude Ca2+ signals. a Mouse primary cortical neurons (PCN) were transfected with the cytosolic GCaMP6f probe. After 24 h, time-lapse imaging was performed and glutamate and ionomycin were added when indicated. b, c PCN were transfected with the mitochondrial targeted GCaMP6f together with short hairpin RNA targeting MICU1, MICU2 or MICU3. After 24 h, Ca2+ imaging was performed as in panel A. Resting, glutamate- and ionomycin-induced Ca2+ rises were calculate in the cell soma (b) or in the neuronal process (c) and shown as bar graphs of the average ± s.d. ratio. d PCN were transfected as in panel B. After 24 h, time-lapse imaging was performed and 20 mM KCl and ionomycin were added. The bar graphs show the average ± s.d. ratio of cytosolic Ca2+ levels. n > 25 single neurons derived from four independent PCN preparations. e Schematic representation of the proposed model. In neurons in normal conditions (middle panel), the balanced expression of MICU1/MICU2 and MICU1/MICU3 dimers ensures the efficient mitochondrial Ca2+ uptake during Ca2+ entry from plasma membrane channels. Downregulation of MICU2 (right panel) causes the loss of normal threshold and anticipates MCU opening thus causing exaggerated organelle Ca2+ accumulation and prolonged opening of plasma membrane Ca2+ channels. Conversely, loss of MICU3 (left panel) leads to inefficient organelle Ca2+ uptake and local Ca2+ clearance, thus accelerating the feedback inhibition of plasma membrane channels
Discussion
The identification of MCU [15, 16] provided the opportunity to interrogate the role of mitochondrial Ca2+ uptake through direct genetic approaches, but also raised new intriguing biological questions. How is the MCU activity controlled at the molecular level? Are there any cell-specific differences in the composition of mitochondrial Ca2+ handling machinery? We here provide novel insights to address these questions, by investigating the role of MICU3, an uncharacterized MCU modulator. Analysis of mRNA expression of all components of mitochondrial Ca2+ handling revealed significant differences among tissues (Table 1), a notion supported by the direct measurements of MCU currents [33, 42].
Overall, if we look at the relative expression levels of the components that positively (i.e. MCU, MICU1 and MICU3) or negatively (i.e. MCUb and MICU2) regulate mitochondrial Ca2+ uptake, we will predict high Ca2+ accumulation in brain and skeletal muscle and low in heart and spleen. Remarkably, these observations are coherent with previous reports [33, 43], although a careful assessment of mitochondrial Ca2+ fluxes in many tissues is still lacking. The MCU complex is notably abundant in the cerebellum, while tissues containing immune cells display an overall composition of the MCU complex that should limit mitochondrial Ca2+ accumulation, due to high MCUb expression. As to the efflux pathway, the Na+/Ca2+ exchanger NCLX [9] is unexpectedly expressed at low level in excitable tissues and abundant in several glands, spleen and blood. This suggests that mitochondrial influx and efflux pathways are independently regulated at the transcriptional level, thus providing an additional mechanism to expand the range of different spatiotemporal Ca2+ signaling patterns that can be generated within mitochondria. However, these conclusions must be interpreted with cautions since mRNA and protein levels can differ significantly. Unfortunately, reliable antibodies for all the components of mitochondrial Ca2+ handling machinery are still lacking.
However, when looking at the tissue-specific composition of the MCU complex, the most relevant trait is the expression of MICU3 within the central nervous system. Previous data [20, 36] and qPCR on mouse tissues (Fig. 1b) confirmed that MICU3 is mainly brain-specific, although recent work showed detectable MICU3 protein also in skeletal muscle [44]. Other members of the MICU family are known to be the key regulators of MCU gating [27–31, 45]. According to our hypothesis [29], MICU2 acts as a genuine MCU inhibitor, thus providing the physiological gatekeeping at low [Ca2+]. Conversely, MICU1 acts predominantly as a cooperative MCU activator at high [Ca2+], although it also actively participates in MCU gatekeeping. Indeed, on one hand MICU1 represents a common platform for the interaction with MCU, and on the other, it still retains partial channel inhibition at low [Ca2+] when MICU2 is absent [28]. We here show that MICU3 exists in a disulfide bond-mediated dimer (Fig. 2), where it acts as a highly potent stimulator of MCU activity, with no gatekeeping function (Fig. 3). Rather, the forced expression of MICU3 can displace the endogenous MICU2, thus causing, at least in part, the loss of the MICU2-dependent threshold.
From the biochemical point of view, MICU3 operates similarly to MICU2. It uniquely forms heterodimers with MICU1 and not with itself or MICU2 (Fig. 2). However, MICU3 also represents the functional opposite of MICU2, since its expression leads to a dramatic increase of mitochondrial Ca2+ uptake (Fig. 3). Interestingly, both MICU2 and MICU3 are conserved in vertebrates, while other organisms possess only MICU1. In organisms with only one component, MICU1 appears to serve the dual role of gatekeeper and cooperative activator. During evolution, two novel isoforms emerged, MICU2 and MICU3 with bona fide role of inhibitor and activator of MCU, respectively. Through this molecular complexity, the MICU family appears perfectly tuned to finely shape the sigmoidal relationship between MCU activity and extramitochondrial [Ca2+].
Accordingly, neuronal Ca2+ signals are generated by the opening of plasma membrane channels, leading to an increase of [Ca2+] that is normally low in amplitude and short. Conversely, for e.g., lymphocytes Ca2+ signals are mainly generated through the prolonged opening of capacitative Ca2+ influx, leading to a sustained increase of [Ca2+]. On the other hand, mitochondrial Ca2+ levels must be carefully controlled in order to both maximize the boost of oxidative metabolism and minimize organelle Ca2+ overload. Even in this case, energy requirements are likely to be higher for the brain compared to other non-excitable tissues, although this potentially endangers neurons to Ca2+-mediated cell death [46]. In line with this view, we found that in neurons MICU3 is the only MICU isoform that upon silencing can effectively decrease mitochondrial Ca2+ transients elicited by synaptic activity (Fig. 4). Remarkably, MCU in neurons still retains an activation threshold determined by MICU2, given that its silencing leads to overwhelming mitochondrial transients. Indeed, neurons simultaneously express both MICU1/MICU2 dimers, that ensure low vicious Ca2+ cycling in resting conditions, and MICU1/MICU3 dimers, that anticipates MCU opening (Fig. 3e), and instantly activate organelle Ca2+ uptake even in the presence of small and rapid cytosolic Ca2+ signals (Fig. 4c).
Overall, how mitochondria can shape global Ca2+ signals in neurons is still controversial and mixed results have been obtained [40, 47]. Mitochondria can in principle act either as passive buffers of global cytoplasmic [Ca2+] rises or as local Ca2+ sponges, thus mediating Ca2+ sequestration in defined cellular subdomains. Indeed, mitochondria positioning to specific subcellular regions is critical for neuronal activity [48, 49]. Consequently, mitochondrial could sustain Ca2+ entry from the extracellular space by locally sequestering Ca2+ and thus delaying the Ca2+-dependent inactivation of plasma membrane Ca2+ channels [50–52], as also suggested by previous reports [41, 53]. In line with this view, silencing of MICU3 causes a decrease in the amplitude of [Ca2+]cyt rises, while MICU2 downregulation exert the opposite effect. Conversely, when high amplitude and sustained [Ca2+] elevation are elicited, all these differences disappeared (Fig. 5). Alternatively, this could be also due to different Ca2+ affinity of the different MICU isoforms. For instance, the EF-hand domains could saturate at diverse Ca2+ levels, thus limiting MICUs activity within specific ranges of [Ca2+]cyt. Altogether, our data indicate that modulation of mitochondrial Ca2+ uptake impacts on the decoding of temporally and spatially restricted Ca2+ signals elicited by synaptic activity (Fig. 5e). However, we cannot exclude other potential indirect mechanisms, such as impaired levels of ATP, metabolites, ROS or expression of channels, pumps and their regulators.
In conclusion, the primary role of MICU3 is to enhance MCU opening in order to guarantee organelle Ca2+ uptake in response also to small and fast increases of cytosolic [Ca2+]. This finding add another level of complexity to the regulation of the MCU complex and further strengthen the potential physiopathological role of the MICU family [54–57].
Materials and methods
Chemicals, cell culture and transfection
All chemicals were purchased from Sigma-Aldrich, unless otherwise specified. Antibodies against MICU1 (WB, 1:1000), MICU2 (WB, 1:1000), MICU3 (WB, 1:1000), MCU (WB, 1:1000) and myc (WB, 1:1000) were purchased from Sigma-Aldrich. Antibodies against Flag (WB, 1:1000) and HA (WB, 1:1000) were purchased from Cell Signaling. Secondary, HRP-conjugated antibodies (WB, 1:5000) were purchased from BioRad.
HeLa (ATCC Number: CCL-2) or HEK293A (ThermoFisher Scientific) cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) (ThermoFisher Scientific), supplemented with 10% fetal bovine serum (FBS) (ThermoFisher Scientific) and transfected with a standard calcium-phosphate procedure. For aequorin measurements, the cells were seeded 24 h before transfection onto 13 mm glass coverslips and allowed to grow to 50% confluence before transfection. For imaging experiments, cells were seeded 24 h before transfection onto 24 mm glass coverslips and allowed to grow to 50% confluence before transfection, unless otherwise specified.
Mouse primary cortical neurons were prepared from p0-p2 newborn C57BL/6J mice. Briefly, the brain cortex of newborn mice was isolated and dissected using a stereomicroscope and digested with trypsin at 37 °C. After two digestion steps, cells were counted and seeded on poly-L-lysine-coated glass coverslips. Neurons were cultured in MEM (ThermoFisher Scientific), supplemented with 10% Horse Serum (ThermoFisher Scientific), N2 supplement (ThermoFisher Scientific), B27 supplement (ThermoFisher Scientific), Sodium Pyruvate (ThermoFisher Scientific), biotin, glucose, L-glutamine, penicillin and streptomycin for 5 DIV before transfection. Neurons were transfected with Lipofectamine 2000 (ThermoFisher Scientific), according to manufacturer’s instruction. Experiments were carried out 24 h after transfection.
Constructs and shRNAs
The following plasmids were used and are described elsewhere: 2mtGCaMP6m [29, 58], 4mtGCaMP6f [59], mtAeq [60], MICU1-HA [29], MICU1-Flag [29], MICU2-Flag [29], MICU1C465A-HA [29], MCU-Flag [15], GCaMP6f, a kind gift from Douglas Kim (Addgene plasmid # 40755) and R-GECO1, a kind gift from Robert Campbell (Addgene plasmid # 32444) [61]. The MICU3-myc chimera was produced by gene synthesis (ThermoFisher Scientific) using the coding sequence derived from NM_030110, and cloned into pcDNA3.1. The MICU3C515A-myc mutant was produced by site-specific mutagenesis using the following primer: 5′-AAGTATCCGACCTTCAAATCCGCCCTGAAAAAGGAACTTCACAGC-3′. MICU3EFmut (MICU3D238A-E249K-D476A-E487K) was produced by site-specific mutagenesis using the following primers:
5′-TTTAGAATAGCTTTCAACATGTTTGCCACTGACGGAAATGAGATGGTGGATAAAAAAGCATTTTTGGTGCTTCAGGAGATAT-3′ and 5′-TGAACACTGTCTTCAAGATTTTTGCTGTTGACAAAGATGACCAATTAAGTTATAAAGCATTTATTGGAATTATGAAAGACAGA-3′). For shRNA plasmids, we started from the pZac2.1-U6-shLuc-ZsGreen backbone described in [62]. ZsGreen has been replaced with mCherry using the NheI and NotI sites. The following sequences were then used to replace the Luc-targeting hairpin: for shMICU1, 5′-GATCGGCTCTGGATTCAGAGACAGAATTCAAGAGATTCTGTCTCTGAATCCAGAGCTTTTTG-3′; for shMICU2, 5′-GATCGGCTGCAGAAGATCATAAGTAAATTCAAGAGATTTACTTATGATCTTCTGCAGTTTTTG-3′; for shMICU3, 5′-GATCGCCCATATTCTCCTGCGGTATATTCAAGAGATATACCGCAGGAGAATATGGGTTTTTG-3′. In all overexpression experiments, pcDNA3.1 was also transfected in control conditions. Similarly, in all knockdown experiments shLuc was used as control. All constructs were verified by Sanger sequencing.
Co-immunoprecipitation
For co-immunoprecipitation experiments, HeLa cells were grown in 10 cm Petri dishes and transfected with the indicated constructs as described above. After 36 h, cells were lysate in an appropriate volume of lysis buffer (125 mM NaCl, 25 mM TRIS-Cl pH7.4, 1 mM EGTA-Tris pH 7.4, 0.5% n-Dodecyl β-D-maltoside, Complete EDTA-free protease inhibitor mixture and PhoSTOP Phosphatase Inhibitor Cocktail (Roche Applied Science)). Whole cell lysate was precleared for 30’ with a control agarose resin (Pierce). 1 mg of precleared proteins from the indicated conditions was incubated with monoclonal αFlag or αHA agarose-conjugated antibody (Sigma-Aldrich) and the co-immunoprecipitation was performed following manufacturer instructions. After three 10’ washes in lysis buffer, the bait was eluted in a non-reducing Laemli sample buffer and denatured for 10’ at 90 °C. The precleared lysate (Input) and the eluted fraction (CoIP) were separated by SDS-PAGE, transferred to Hybond-C Extra membrane (Amersham) and developed according to standard procedures. The same membrane in each panel was stripped for 10’ in StripABlot stripping buffer (Euroclone) and probed with different antibodies as indicated.
SDS-PAGE and western blot
For protein extraction, HeLa cells and mouse primary cortical neurons were grown on 10 cm Petri dishes and transfected with the indicated constructs as described above. After 24 or 48 h, cells were washed twice with PBS, collected and resuspended in the appropriate volume of RIPA buffer (125 mM NaCl, 25 mM TRIS-Cl pH 7.4, 1 mM EGTA-TRIS pH 7.4, 1% Triton-X100, 0.5% sodium deoxycholate, 0.1% SDS and Complete EDTA-free protease inhibitor mixture (Roche Applied Science)) and briefly sonicated. Crude extracts were centrifuged at 15000xg for 10’ to remove debris, and proteins in the supernatant were quantified using the BCA Protein Assay Kit (Pierce). 40 μg of proteins were dissolved in LDS sample buffer (Life Technologies), heated for 10’ at 70 °C and loaded on 4–12% Bis-Tris NuPage gels (ThermoFisher Scientific). Where indicated, proteins were reduced with 100 mM DTT and denatured for 10’ at 90 °C, unless specified. After electrophoretic separation, proteins were transferred onto nitrocellulose membranes and probed with the indicated antibodies. Isotype matched, horseradish peroxidase-conjugated secondary antibodies (BioRad) were used followed by detection by chemiluminescence (SuperSignal Pico, Pierce). Densitometry was performed using the built-in function present in Fiji image processing package based on ImageJ [63, 64].
Aequorin measurements
For measurements of [Ca2+]cyt and [Ca2+]mt, HeLa or HEK293 cells grown on 13 mm round glass coverslips at 50% confluence were transfected with cytosolic (cytAeq), mitochondrial (mtAeq), or low-affinity mitochondrial (mtAeqMut) probes [65, 66] together with the indicated siRNA or constructs. After 24 or 48 h, the coverslip was incubated with 5 μM coelenterazine for 1–2 h in a Krebs-Ringer modified buffer (KRB, in mM: 135 NaCl, 5 KCl, 1 MgSO4, 0.4 K2HPO4, 20 HEPES, 1 CaCl2, pH = 7.4), and then transferred to the perfusion chamber. All aequorin measurements were carried out in KRB. Agonists and other drugs were added to the same medium as specified in the text. The experiments were terminated by lysing the cells with 100 μM digitonin in a hypotonic Ca2+-rich solution (10 mM CaCl2 in H2O), thus discharging the remaining aequorin pool. The light signal was collected and calibrated into [Ca2+] values by an algorithm based on the Ca2+ response curve of aequorin at physiological conditions of pH, [Mg2+], and ionic strength, as previously described [65]. Alternatively, aequorin measurements were carried out on a PerkinElmer EnVision plate reader equipped with a two injector unit. Cells were transfected as described above in 24-well plates and then re-plated into 96-well plates (1:5 dilution) the day before the experiment. After reconstitution with 5 μM coelenterazine, cells were placed in 70 μl of KRB and luminescence from each well was measured for 1 min. During the experiment, agonist was first injected at the desired concentration to activate Ca2+ transients, and then a hypotonic, Ca2+-rich, digitonin-containing solution was added to discharge the remaining aequorin pool. Output data were analyzed and calibrated with a custom-made macro-enabled Excel workbook. In the experiments with permeabilized cells, a buffer mimicking the cytosolic ionic composition (IB) was employed: 130 mM KCl, 10 mM NaCl, 2 mM K2HPO4, 5 mM succinic acid, 5 mM malic acid, 1 mM MgCl, 20 mM HEPES, and 1 mM pyruvate (pH 7) at 37 °C. IB was supplemented with either 100 μM EGTA (IB/EGTA) or a 2 mM EGTA-buffered [Ca2+] of the indicated concentration (IB/Ca2+). Calculated [Ca2+] free was predicted with a custom-made Excel workbook and confirmed fluorimetrically with the Fura2 free acid. HeLa cells were permeabilized by a 1 min perfusion with 100 μM digitonin (added to IB/EGTA) during luminescence measurements. Mitochondrial Ca2+ uptake speed was calculated as the first derivative by using the SLOPE Excel function and smoothed for three time points. The higher value reached during Ca2+ addition represents the maximal Ca2+ uptake speed. All of the results are expressed as means ± SD, and Student’s t test was used for the statistics.
Ca2+ imaging
For the measurements of basal [Ca2+] (Fig. 3f), cells were transfected with 2mtGCamp6m probes [58, 67], together with the indicated constructs. After 24 or 48 h, cells were mounted in an open-bath custom made imaging chamber and maintained in KRB. Imaging was performed on a Zeiss Axiovert 200 microscope equipped with a 40 × /1.3 N.A. Plan-NeoFluar objective. Excitation was performed with a DeltaRAM V high-speed monochromator (Photon Technology International) equipped with a 75 W xenon arc lamp. Images were captured with a high-sensitivity Evolve 512 Delta EMCCD (Photometrics). The system is controlled by MetaMorph 7.5 (Molecular Devices) and was assembled by Crisel Instruments. Cells were alternatively illuminated at 474 and 410 nm and fluorescence was collected through a 515/30 nm band-pass filter (Semrock). Exposure time was set to 200 ms at 474 nm and to 400 ms at 410 nm, in order to account for the low quantum yield at the latter wavelength. At least ten fields were collected per coverslip, and each field was acquired for 10 s (1 frame/s). Analysis was performed with the Fiji distribution of ImageJ. Both images were background corrected frame by frame by subtracting mean pixel values of a cell-free region of interest. Data are presented as the mean of the averaged ratio of all time points.
For the measurements of cytosolic and mitochondrial Ca2+ dynamics after 4AP/bicuculline stimulation (Fig. 4), mouse primary cortical neurons at 5 DIV were transfected with 4mtGCaMP6f together with the indicated constructs. After 24 h, primary neurons were mounted in an open-bath custom made imaging chamber and maintained in KRB. Cells were then stimulated by adding 10 μM bicuculline, an antagonist of GABAA receptors, and 100 μM 4-aminopyridine, a selective blocker of voltage-activated K+ channels (Abcam). Imaging was performed on an Olympus IX71/IX51 inverted microscope equipped with a xenon light source (150 W) for epifluorescence illumination, alternatively illuminated at 474 and 410 nm. Exposure time was set to 200 ms at 474 nm and to 400 ms at 410 nm, in order to account for the low quantum yield at the latter wavelength. Images were collected with a digital camera using a ×40, 1.3 NA oil immersion objective (Olympus). Data were acquired at 1 Hz and analyzed using CellR software (Olympus). Analysis was performed with the Fiji distribution of ImageJ. Both images were background corrected frame by frame by subtracting mean pixel values of a cell-free region of interest. In the case of mitochondrial targeted probe (4mtGCaMP6f), the fluorescence recorded at 410 nm was too dim for a reliable quantification, and thus data are provided as normalized fluorescence at 474 nm only (F/F0, Fig. 4b,c). In the case of cytosolic sensor (GCaMP6f), higher probe expression enabled sufficient signal intensity to perform ratiometric imaging (expressed as R/R0). For the measurements of cytosolic Ca2+ dynamics shown in Fig. 5, imaging was performed on a Zeiss Axiovert 200 microscope equipped with a 40 × /1.3 N.A. Plan-NeoFluar objective. Excitation was performed with a DeltaRAM V high-speed monochromator (Photon Technology International) equipped with a 75 W xenon arc lamp. Images were captured with a high-sensitivity Evolve 512 Delta EMCCD (Photometrics). The system is controlled by MetaMorph 7.5 (Molecular Devices) and was assembled by Crisel Instruments. Cells were alternatively illuminated at 474 and 410 nm and fluorescence was collected through a 515/30 nm band-pass filter (Semrock). Exposure time was set to 150 ms at 474 nm and to 300 ms at 410 nm, in order to account for the low quantum yield at the latter wavelength. Analysis was performed with the Fiji distribution of ImageJ. Both images were background corrected frame by frame by subtracting mean pixel values of a cell-free region of interest. Data are presented as fluorescence ratio (R, 474/410 nm).
For the analysis of the time delay between cytoplasmic and mitochondrial [Ca2+] rises, HeLa cells were co-transfected with 4mtGCaMP6f and R-GECO1, together with control, MICU1 or MICU3 expressing constructs. After 24 h, cells were mounted in an open-bath custom made imaging chamber and maintained in KRB. Imaging was performed on a Zeiss Axiovert 200 microscope equipped with a 40 × /1.3 N.A. Plan-NeoFluar objective. Excitation was performed with a DeltaRAM V high-speed monochromator (Photon Technology International) equipped with a 75 W xenon arc lamp. Images were captured with a high-sensitivity Evolve 512 Delta EMCCD (Photometrics). The system is controlled by MetaMorph 7.5 (Molecular Devices) and was assembled by Crisel Instruments. Cells were alternatively illuminated at 490 and 565 nm and fluorescence was collected through a dual band-pass filter (520/40 and 630/60). Exposure time was set to 10 ms, EM gain at 400, and images were streamed to an SSD using 2 × 2 binning and 20 Mhz readout speed. The final frame rate was 50 images per second. Images were background corrected and linear unmixing was performed to get rid of the residual bleed trough of the two fluorochromes. Data are expressed as F/F0 where F0 is the average intensity of the first 10 frames of the experiment. The delay between the onset of cytosolic and mitochondrial Ca2+ rises was measured by calculating the time between a 50% increase over the baseline of F/F0 in cytosolic and mitochondrial Ca2+ responses.
RNA extraction, reverse transcription and quantitative real-time PCR
For the expression analysis of MICU1, MICU2 and MICU3 were used both different mouse tissues, as indicated. At least 3 samples were prepared for each condition. Total RNA was extracted from 30 mg of each mouse tissue using the SV Total RNA Isolation Kit (Promega) following manufacturer instructions. The RNA was quantified with an Eppendorf Bio Plus photometer. Using an equal amount of RNA from each sample, complementary DNA was generated with a cDNA synthesis kit (SuperScript II, ThermoFisher Scientific) and analyzed by real-time PCR using the SYBR green chemistry (ThermoFisher Scientific). The primers were designed and analyzed with Primer3 [68]. Identity of the amplicons was confirmed by their dissociation profiles and gel analysis. Real-time PCR standard curves were constructed by using serial dilutions of cDNAs of the analyzed samples, using at least five dilution points and the efficiency of all primer sets was between 95 and 105%. The housekeeping gene GAPDH were used as an internal control for cDNA quantification and normalization of the amplified products. Real-time PCR primer sequences (5′–3′) were as follows:
mm-MICU1-fw: GTCGAACTCTCGGACCATGT
mm-MICU1-rv: GTGCTAAGGTGCAGGAGGTG
mm-MICU2-fw: TGGAGCACGACGGAGAGTAT
mm-MICU2-rv: GCCAGCTTCTTGACCAGTGT
mm-MICU3-fw: CGACCTTCAAATCCTGCCTG
mm-MICU3-rv: TCTGCGTGCTCTGACCTTAC
mm-GAPDH-fw: CACCATCTTCCAGGAGCGAG
mm-GAPDH-rv: CCTTCTCCATGGTGGTGAAGAC
RNA-seq and bioinformatics
RNA-seq data were retrieved from ArrayExpress (http://www.ebi.ac.uk/arrayexpress/), expressed as Transcripts Per Kilobase Million (TPM). The E-MTAB-5214 dataset [34] was used for expression analysis of human tissues. The E-GEOD-26284 dataset [35] was used for expression analysis of cell lines. Expression values were then normalized across different tissues or cell lines taking into account the relative abundance of nuclear-encoded mitochondrial transcripts. To do this, we started from the Mitocarta database [69], selected only proteins reported to be expressed in all 14 tissues and excluded all transcripts with TPM < 1. We then calculated the average TPM of this subset of transcripts (approximately 450) and used these values to normalize TPM values of our genes of interest. Multiple alignment was performed starting from human MICU1, MICU2 and MICU3 protein sequences using MUSCLE [70]. Prediction of mitochondrial targeting signals was carried out using Mitoprot [71].
Statistical analysis of data
The data are presented as mean ± S.D. unless specified. Significance was calculated by one-way (or two-way, where needed) ANOVA test, and correlation analysis was performed with the SigmaPlot 12.0 software (Systat Software Inc.) or Excel (Microsoft).
Acknowledgements
This work was supported by grants from the University of Padova (SID 2016 to DDS), the Italian Ministries of Health (Ricerca Finalizzata to R.R.), and of Education, University and Research (FIRB to RR), the European Union (ERC mitoCalcium, no. 294777), NIH (Grant #1P01AG025532-01A1), the Italian Association for Cancer Research (AIRC IG18633 to RR) and Telethon-Italy (GGP16029 to RR). M.P. is supported by an EMBO long-term fellowships (ALTF649-2015; LTFCOFUND2013; and GA-2013-609409). JE holds a post-doctoral fellowship from Junta de Extremadura (jointly financed by the European Regional Development Fund, ERDF, ref. PO14011)
Author Contributions
: MP, VG and JE performed the experiments and analyzed the data. MP, VG, DDS and RR designed the research. DDS and RR wrote the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by-N.Chandel
These authors contributed equally: Maria Patron, Veronica Granatiero.
Contributor Information
Rosario Rizzuto, Phone: +390498273001, Email: rosario.rizzuto@unipd.it.
Diego De Stefani, Phone: +390498276029, Email: diego.destefani@gmail.com.
References
- 1.Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 2.Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4:517–29. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 3.Clapham DE. Calcium signaling. Cell. 2007;131:1047–58. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 4.De Stefani D, Rizzuto R, Pozzan T. Enjoy the trip: calcium in mitochondria back and forth. Annu Rev Biochem. 2016;85:161–92. doi: 10.1146/annurev-biochem-060614-034216. [DOI] [PubMed] [Google Scholar]
- 5.Rizzuto R, Pozzan T. Microdomains of intracellular Ca2 + : molecular determinants and functional consequences. Physiol Rev. 2006;86:369–408. doi: 10.1152/physrev.00004.2005. [DOI] [PubMed] [Google Scholar]
- 6.Rizzuto R, De Stefani D, Raffaello A, Mammucari C. Mitochondria as sensors and regulators of calcium signalling. Nat Rev Mol Cell Biol. 2012;13:566–78. doi: 10.1038/nrm3412. [DOI] [PubMed] [Google Scholar]
- 7.Filadi R, Pozzan T. Generation and functions of second messengers microdomains. Cell Calcium. 2015;58:405–14. doi: 10.1016/j.ceca.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 8.De Stefani D, Patron M, Rizzuto R. Structure and function of the mitochondrial calcium uniporter complex. Biochim Biophys Acta. 2015;1853:2006–11. doi: 10.1016/j.bbamcr.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palty R, Silverman WF, Hershfinkel M, Caporale T, Sensi SL, Parnis J, et al. NCLX is an essential component of mitochondrial Na + /Ca2 + exchange. Proc Natl Acad Sci USA. 2010;107:436–41. doi: 10.1073/pnas.0908099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Marchi U, Santo-Domingo J, Castelbou C, Sekler I, Wiederkehr A, Demaurex N. NCLX protein, but not LETM1, mediates mitochondrial Ca2 + extrusion, thereby limiting Ca2 + -induced NAD(P)H production and modulating matrix redox state. J Biol Chem. 2014;289:20377–85. doi: 10.1074/jbc.M113.540898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nowikovsky K, Pozzan T, Rizzuto R, Scorrano L, Bernardi P. Perspectives on: SGP symposium on mitochondrial physiology and medicine: the pathophysiology of LETM1. J Gen Physiol. 2012;139:445–54. doi: 10.1085/jgp.201110757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang D, Zhao L, Clapham DE. Genome-wide RNAi screen identifies Letm1 as a mitochondrial Ca2 + /H + antiporter. Science (80-) 2009;326:144–7. doi: 10.1126/science.1175145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang D, Zhao L, Clish CB, Clapham DE. Letm1, the mitochondrial Ca2 + /H + antiporter, is essential for normal glucose metabolism and alters brain function in Wolf-Hirschhorn syndrome. Proc Natl Acad Sci USA. 2013;110:E2249–54. doi: 10.1073/pnas.1308558110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai MF, Jiang D, Zhao L, Clapham D, Miller C. Functional reconstitution of the mitochondrial Ca2 + /H + antiporter Letm1. J Gen Physiol. 2014;143:67–73. doi: 10.1085/jgp.201311096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Stefani D, Raffaello A, Teardo E, Szabò I, Rizzuto R, Szabo I, et al. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature. 2011;476:336–40. doi: 10.1038/nature10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baughman JM, Perocchi F, Girgis HS, Plovanich M, Belcher-Timme Ca, Sancak Y, et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;476:341–5. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raffaello A, De Stefani D, Sabbadin D, Teardo E, Merli G, Picard A, et al. The mitochondrial calcium uniporter is a multimer that can include a dominant-negative pore-forming subunit. EMBO J. 2013;32:2362–76. doi: 10.1038/emboj.2013.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sancak Y, Markhard AL, Kitami T, Kovacs-Bogdan E, Kamer KJ, Udeshi ND, et al. EMRE is an essential component of the mitochondrial calcium uniporter complex. Science. 2013;342:1379–82. doi: 10.1126/science.1242993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perocchi F, Gohil VM, Girgis HS, Bao XR, McCombs JE, Palmer AE, et al. MICU1 encodes a mitochondrial EF hand protein required for Ca(2 + ) uptake. Nature. 2010;467:291–6. doi: 10.1038/nature09358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plovanich M, Bogorad RL, Sancak Y, Kamer KJ, Strittmatter L, Li Aa, et al. MICU2, a paralog of MICU1, resides within the mitochondrial uniporter complex to regulate calcium handling. PLoS ONE. 2013;8:e55785. doi: 10.1371/journal.pone.0055785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovács-Bogdán E, Sancak Y, Kamer KJ, Plovanich M, Jambhekar A, Huber RJ, et al. Reconstitution of the mitochondrial calcium uniporter in yeast. Proc Natl Acad Sci USA. 2014;111:8985–90. doi: 10.1073/pnas.1400514111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsai MF, Phillips CB, Ranaghan M, Tsai CW, Wu Y, Williams C, et al. Dual functions of a small regulatory subunit in the mitochondrial calcium uniporter complex. eLife. 2016;5:371–81. doi: 10.7554/eLife.15545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mallilankaraman K, Cárdenas C, Doonan PJ, Chandramoorthy HC, Irrinki KM, Golenár T, et al. MCUR1 is an essential component of mitochondrial Ca(2 + ) uptake that regulates cellular metabolism. Nat Cell Biol. 2012;14:1336–43. doi: 10.1038/ncb2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vais H, Tanis JE, Müller M, Payne R, Mallilankaraman K, Foskett JK. MCUR1, CCDC90A, Is a regulator of the mitochondrial calcium uniporter. Cell Metab. 2015;22:533–5. doi: 10.1016/j.cmet.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paupe V, Prudent J, Dassa EPP, Rendon OZZ, Shoubridge EAA. CCDC90A (MCUR1) is a cytochrome c oxidase assembly factor and not a regulator of the mitochondrial calcium uniporter. Cell Metab. 2015;21:109–16. doi: 10.1016/j.cmet.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 26.Tomar D, Dong Z, Shanmughapriya S, Koch DA, Thomas T, Hoffman NE, et al. MCUR1 is a scaffold factor for the MCU complex function and promotes mitochondrial bioenergetics. Cell Rep. 2016;15:1673–85. doi: 10.1016/j.celrep.2016.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mallilankaraman K, Doonan P, Cardenas C, Chandramoorthy HC, Muller M, Miller R, et al. MICU1 Is an essential gatekeeper for mcu-mediated mitochondrial ca(2 + ) uptake that regulates cell survival. Cell. 2012;151:630–44. doi: 10.1016/j.cell.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Csordas G, Golenar T, Seifert EL, Kamer KJ, Sancak Y, Perocchi F, et al. MICU1 controls both the threshold and cooperative activation of the mitochondrial Ca(2)( + ) uniporter. Cell Metab. 2013;17:976–87. doi: 10.1016/j.cmet.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patron M, Checchetto V, Raffaello A, Teardo E, VecellioReane D, Mantoan M, et al. MICU1 and MICU2 finely tune the mitochondrial Ca2 + uniporter by exerting opposite effects on MCU activity. Mol Cell. 2014;53:726–37. doi: 10.1016/j.molcel.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matesanz-Isabel J, Arias-Del-Val J, Alvarez-Illera P, Fonteriz RI, Montero M, Alvarez J. Functional roles of MICU1 and MICU2 in mitochondrial Ca2 + uptake. Biochim Biophys Acta. 2016;1858:1110–7. doi: 10.1016/j.bbamem.2016.02.022. [DOI] [PubMed] [Google Scholar]
- 31.Kamer KJ, Mootha VK. MICU1 and MICU2 play nonredundant roles in the regulation of the mitochondrial calcium uniporter. EMBO Rep. 2014;15:299–307. doi: 10.1002/embr.201337946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petrungaro C, Zimmermann KM, Küttner V, Fischer M, Dengjel J, Bogeski I, et al. The Ca2 + -dependent release of the Mia40-induced MICU1-MICU2 dimer from MCU regulates mitochondrial Ca2 + uptake. Cell Metab. 2015;22:721–33. doi: 10.1016/j.cmet.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 33.Fieni F, Lee SB, Jan YN, Kirichok Y, Bae Lee S, Jan YN, et al. Activity of the mitochondrial calcium uniporter varies greatly between tissues. Nat Commun. 2012;3:1317. doi: 10.1038/ncomms2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ardlie KG, Deluca DS, Segre AV, Sullivan TJ, Young TR, Gelfand ET, et al. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Márkus NM, Hasel P, Qiu J, Bell KFS, Heron S, Kind PC, et al. Expression of mRNA encoding Mcu and other mitochondrial calcium regulatory genes depends on cell type, neuronal subtype, and ca2 + signaling. PLoS ONE. 2016;11:e0148164. doi: 10.1371/journal.pone.0148164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerencsér AacuteA, Adam-Vizi V. Selective, high-resolution fluorescence imaging of mitochondrial Ca2 + concentration. Cell Calcium. 2001;30:311–21. doi: 10.1054/ceca.2001.0238. [DOI] [PubMed] [Google Scholar]
- 38.Hardingham GE, Arnold FJ, Bading H. Nuclear calcium signaling controls CREB-mediated gene expression triggered by synaptic activity. Nat Neurosci. 2001;4:261–7. doi: 10.1038/85109. [DOI] [PubMed] [Google Scholar]
- 39.Hardingham GE, Fukunaga Y, Bading H. Extrasynaptic NMDARs oppose synaptic NMDARs by triggering CREB shut-off and cell death pathways. Nat Neurosci. 2002;5:405–14. doi: 10.1038/nn835. [DOI] [PubMed] [Google Scholar]
- 40.Budd SL, Nicholls DG. A reevaluation of the role of mitochondria in neuronal Ca2 + homeostasis. J Neurochem. 1996;66:403–11. doi: 10.1046/j.1471-4159.1996.66010403.x. [DOI] [PubMed] [Google Scholar]
- 41.Budd S, Nicholls D. Mitochondria, calcium regulation, and acute glutamate excitotoxicity in cultured cerebellar granule cells. J Neurochem. 1996;67:2282–91. doi: 10.1046/j.1471-4159.1996.67062282.x. [DOI] [PubMed] [Google Scholar]
- 42.Kirichok Y, Krapivinsky G, Clapham DE. The mitochondrial calcium uniporter is a highly selective ion channel. Nature. 2004;427:360–4. doi: 10.1038/nature02246. [DOI] [PubMed] [Google Scholar]
- 43.Williams GS, Boyman L, Lederer WJ. Mitochondrial calcium and the regulation of metabolism in the heart. J Mol Cell Cardiol. 2015;78:35–45. doi: 10.1016/j.yjmcc.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paillard M, Csordás G, Szanda G, Golenár T, Debattisti V, Bartok A, et al. Tissue-specific mitochondrial decoding of cytoplasmic Ca2 + signals is controlled by the stoichiometry of MICU1/2 and MCU. Cell Rep. 2017;18:2291–300. doi: 10.1016/j.celrep.2017.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de la Fuente S, Matesanz-Isabel J, Fonteriz RI, Montero M, Alvarez J. Dynamics of mitochondrial Ca2 + uptake in MICU1-knockdown cells. Biochem J. 2014;458:33–40. doi: 10.1042/BJ20131025. [DOI] [PubMed] [Google Scholar]
- 46.König T, SESE Troumlder, Bakka, Korwitz K, Richter-Dennerlein A, Lampe PAPA R, et al. The m-AAA protease associated with neurodegeneration limits MCU activity in mitochondria. Mol Cell. 2016;64:148–62. doi: 10.1016/j.molcel.2016.08.020. [DOI] [PubMed] [Google Scholar]
- 47.Billups B, Forsythe ID. Presynaptic mitochondrial calcium sequestration influences transmission at mammalian central synapses. J Neurosci. 2002;22:5840–7. doi: 10.1523/JNEUROSCI.22-14-05840.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kang JSS, Tian JHH, Pan PYY, Zald P, Li C, Deng C, et al. Docking of axonal mitochondria by syntaphilin controls their mobility and affects short-term facilitation. Cell. 2008;132:137–48. doi: 10.1016/j.cell.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Macaskill AF, Rinholm JE, Twelvetrees AE, Arancibia-Carcamo IL, Muir J, Fransson A, et al. Miro1 is a calcium sensor for glutamate receptor-dependent localization of mitochondria at synapses. Neuron. 2009;61:541–55. doi: 10.1016/j.neuron.2009.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zong ZQ, Zhou JY, Tanabe T. Molecular determinants of calcium-dependent inactivation in cardiac L-type calcium channels. Biochem Biophys Res Commun. 1994;201:1117–23. doi: 10.1006/bbrc.1994.1821. [DOI] [PubMed] [Google Scholar]
- 51.Peterson BZ, DeMaria CD, Yue DT. Calmodulin is the Ca2 + sensor for Ca2 + -dependent inactivation of L-type calcium channels. Neuron. 1999;22:549–58. doi: 10.1016/S0896-6273(00)80709-6. [DOI] [PubMed] [Google Scholar]
- 52.Imredy JP, Yue DT. Submicroscopic Ca2 + diffusion mediates inhibitory coupling between individual Ca2 + channels. Neuron. 1992;9:197–207. doi: 10.1016/0896-6273(92)90159-B. [DOI] [PubMed] [Google Scholar]
- 53.Nicholls DG. Mitochondria and calcium signaling. Cell Calcium. 2005;38:311–7. doi: 10.1016/j.ceca.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 54.Logan CV, Szabadkai G, Sharpe JA, Parry DA, Torelli S, Childs A-M et al. Loss-of-function mutations in MICU1 cause a brain and muscle disorder linked to primary alterations in mitochondrial calcium signaling. Nat Publ Gr. 2013;46. 10.1038/ng.2851. [DOI] [PubMed]
- 55.Lewis-Smith D, Kamer KJ, Griffin H, Childs AM, Pysden K, Titov D, et al. Homozygous deletion in MICU1 presenting with fatigue and lethargy in childhood. Neurol Genet. 2016;2:e59. doi: 10.1212/NXG.0000000000000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Antony AN, Paillard M, Moffat C, Juskeviciute E, Correnti J, Bolon B, et al. MICU1 regulation of mitochondrial Ca(2 + ) uptake dictates survival and tissue regeneration. Nat Commun. 2016;7:10955. doi: 10.1038/ncomms10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu JC, Liu J, Holmström KM, Menazza S, Parks RJ, Fergusson MM et al. MICU1 serves as a molecular gatekeeper to prevent in vivo mitochondrial calcium overload. 2016. 10.1016/j.celrep.2016.07.011. [DOI] [PMC free article] [PubMed]
- 58.Hill JMM, De Stefani D, Jones AWEW, Ruiz A, Rizzuto R, Szabadkai G. Measuring baseline ca(2 + ) levels in subcellular compartments using genetically engineered fluorescent indicators. Methods Enzym. 2014;543:47–72. doi: 10.1016/B978-0-12-801329-8.00003-9. [DOI] [PubMed] [Google Scholar]
- 59.Tosatto A, Sommaggio R, Kummerow C, Bentham RB, Blacker TS, Berecz T, et al. The mitochondrial calcium uniporter regulates breast cancer progression via HIF‐1α. EMBO Mol Med. 2016;8:569–85. doi: 10.15252/emmm.201606255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rizzuto R, Simpson AW, Brini M, Pozzan T. Rapid changes of mitochondrial Ca2 + revealed by specifically targeted recombinant aequorin. Nature. 1992;358:325–7. doi: 10.1038/358325a0. [DOI] [PubMed] [Google Scholar]
- 61.Zhao Y, Araki S, Wu J, Teramoto T, Chang YF, Nakano M, et al. An expanded palette of genetically encoded Ca2 + indicators. Science (80-) 2011;333:1888–91. doi: 10.1126/science.1208592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mammucari C, Gherardi G, Zamparo I, Raffaello A, Boncompagni S, Chemello F, et al. The mitochondrial calcium uniporter controls skeletal muscle trophism in vivo. Cell Rep. 2015;10:1269–79. doi: 10.1016/j.celrep.2015.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schindelin J, Rueden CT, Hiner MC, Eliceiri KW. The ImageJ ecosystem: An open platform for biomedical image analysis. Mol Reprod Dev. 2015;82:518–29. doi: 10.1002/mrd.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Granatiero V, Patron M, Tosatto A, Merli G, Rizzuto R. Using targeted variants of aequorin to measure Ca2 + levels in intracellular organelles. Cold Spring Harb Protoc. 2014;2014:86–93. doi: 10.1101/pdb.prot072843. [DOI] [PubMed] [Google Scholar]
- 66.Granatiero V, Patron M, Tosatto A, Merli G, Rizzuto R. The use of aequorin and its variants for Ca2 + measurements. Cold Spring Harb Protoc. 2014;2014:9–16. doi: 10.1101/pdb.top066118. [DOI] [PubMed] [Google Scholar]
- 67.Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 69.Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2015;44:D1251–7. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–86. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]