Abstract
The treatment and management of early stage estrogen receptor positive (ER1) breast cancer is hindered by the difficulty in identifying patients who require adjuvant chemotherapy in contrast to those that will respond to hormonal therapy. To distinguish between the more and less aggressive breast tumors, which is a fundamental criterion for the selection of an appropriate treatment plan, Oncotype DX (ODX) and other gene expression tests are typically employed. While informative, these gene expression tests are expensive, tissue destructive, and require specialized facilities. Bloom-Richardson (BR) grade, the common scheme employed in breast cancer grading, has been shown to be correlated with the Oncotype DX risk score. Unfortunately, studies have also shown that the BR grade determined experiences notable inter-observer variability. One of the constituent categories in BR grading is the mitotic index. The goal of this study was to develop a deep learning (DL) classifier to identify mitotic figures from whole slides images of ER+ breast cancer, the hypothesis being that the number of mitoses identified by the DL classifier would correlate with the corresponding Oncotype DX risk categories. The mitosis detector yielded an average F-score of 0.556 in the AMIDA mitosis dataset using a 6-fold validation setup. For a cohort of 174 whole slide images with early stage ER+ breast cancer for which the corresponding Oncotype DX score was available, the distributions of the number of mitoses identified by the DL classifier was found to be significantly different between the high vs low Oncotype DX risk groups (P < 0.01). Comparisons of other risk groups, using both ODX score and histological grade, were also found to present significantly different automated mitoses distributions. Additionally, a support vector machine classifier trained to separate low/ high Oncotype DX risk categories using the mitotic count determined by the DL classifier yielded a 83.19% classification accuracy.
Keywords: breast cancer risk, mitosis detection, whole slide images, digital pathology
MODERN treatment of early stage estrogen receptor positive (ER+) breast cancer requires a precise identification of which cases will benefit from additional adjuvant chemotherapy versus those indicating solely hormonal therapy. A distinction between the more and less aggressive breast tumors, required for planning the treatment, is usually performed with Oncotype DX (ODX) and other gene expression tests. In general, the more aggressive cancers require adjuvant chemotherapy while the more benign respond well to hormonal therapy alone. However these gene expression tests tend to be expensive, tissue destructive, and require specialized facilities for processing.
The Oncotype DX risk score has been demonstrated to be highly correlated with breast cancer grade (1–3). Unfortunately, the grades yielded by the standard Bloom-Richardson system (BR) have been found to be highly variable as a result of both observer experience and tumor presentation (4). The BR consists of assigning a value to stratify three properties: (a) the nuclear pleomorphism score aims to characterize the variance in nuclear size and appearance, (b) the tubule density score is intended to reflect the percentage of cancer tissue that contains normal tubules, and (c) the mitotic index aims to quantify the number of mitoses in a specific number of high power fields (HPFs; typically 10) (5). Although the number of mitoses is regarded as an important prognostic indicator, the overall predictive power is limited by significant inter-reader variability due to potentially differently selected fields (6). In spite of these challenges, studies have shown that Oncotype DX is highly correlated with the mitotic grade for ER+ breast cancers (7).
The recent addition of whole slide imaging capabilities to pathology has sparked notable interest in the use of automated computerized histologic predictors of tumor grade and outcome (8–11). In the context of breast cancer, Basavanhally et al. (12) showed that nuclear graphs built using Delaunay Triangulation and Minimum Spanning Trees can be used for distinguishing low and high recurrence score breast cancer cases.
Of late, there has been substantial interest in automating the process of identifying mitotic figures in whole slide pathology images (13). Larsen et al. (14) used color intensity histograms, gradient orientation histogram and shape index histograms to identify mitoses. Wang et al. (15) proposed to use a convolution neural network (CNN) and a set of handcrafted features combined with a random forest classifier to identify mitotic figures in Whole Slide Images (WSI). Janowczyk et al. (16) used deep learning (DL) approaches to perform several histologic image analysis tasks, including mitotic identification. Veta et al. (17) evaluated the performance of several state-of-the-art mitotic detection methods and found that a Deep Neural Network (DNN) yielded the best overall performance with an overall F-score of 0.61 (18) in the test set of the AMIDA2013 challenge (17).
A neural network architecture consisting of more than two layers is commonly referred as a deep neural network. In supervised classification settings, a DNN uses the back propagation algorithm to update its internal weights according to the label of input exemplars (19). The DNN for mitotic detection is usually trained using a set of image patches that are within a defined d pixel radius to a mitotic nuclei (18).
The contributions of the work presented in this article are twofold. Firstly, we aim to evaluate a customized DNN for automatic quantification of mitosis in WSI. Secondly, we seek to evaluate whether the automated mitotic index identified by the DNN correlates with the risk categories determined by ODX.
Experimental Methods
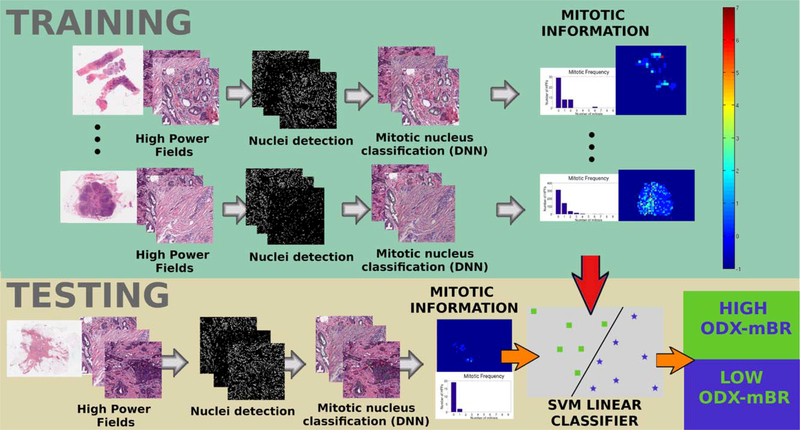
The methodology used to automate mitotic nuclei identification and classify WSI into risk BCa groups as defined by both ODX score and BR grading is illustrated in Figure 1.
Figure 1.
Overall workflow showing the steps to analyze the correlation of the automated mitotic count with ODX and to use mitotic Information In BCa risk stratification. In the training stage, HPFs from several WSI are extracted and a nuclei detection method is applied on each HPF. Each of the candidate nuclei is classied as mitotic or not using a DNN classifier. Subsequently, the mitotic information is used to train a linear support vector machine classifier. During testing, the nuclei identification algorithm and the DNN classifier are used again in the cancerous regions of the WSI. Finally, the resulting mitotic information is used by the support vector machine classifier to predict the WSI risk either as high or low. [Color figure can be viewed at wileyonlinelibrary.com]
DNN for Mitotic Identification
The mitosis detection module involved the training of a DNN model with Hematoxylin and Eosin stained images. Candidate nuclei were found by an automated nuclei detection algorithm at 40 × magnification. Candidate nuclei, separated by less than 20 pixels from the expert annotations, were labeled as mitotic. The approach starts by mapping each RGB point to a grayscale intensity value (20) using: (blue ratio transform). The resulting image highlights the chromatin information (observable in mitotic nuclei) and an adaptive threshold converts the blue ratio image into a binary map, using the gray-level histogram information. The optimal threshold is supposed to maximize the separability of two resulting pixel clusters (21). Subsequently, a set of morphological operations (closing and opening) further improve the selection of the candidate nuclei by removing noise. The connected components are finally selected as candidate nuclei. The centroid of each candidate nucleus corresponds to the center of the associated RGB color patch (size 64 × 64 at 40× magnification), which is then fed into the DL classifier to determine whether or not it is a mitotic nucleus.
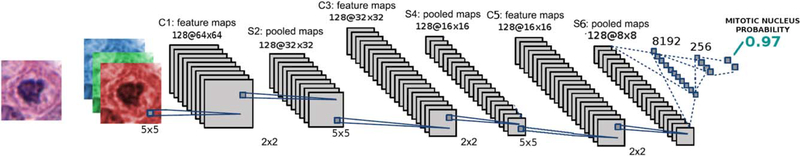
The DNN architecture in Figure 2 consists of three identical blocks, each composed of: a CNN, a Batch normalization layer, a Rectifier Linear Unit (ReLU), and a maximum pool (max pool) operator. Each CNN layer corresponds to a set of convolutional filters learned from the mitotic and non-mitotic classes. The Batch normalization layer imposes a transformation of the neural weights to solve the internal covariate shift problem (22). In practice, this normalization layer allows the method to achieve improved learning rates during the DNN training stage. The proposed network architecture reduces the input patch to an output signal which coincides with the probability that the input contains a mitotic event. Stochastic gradient descent is used to minimize the negative log likelihood loss criterion, which is a measure that quantifies the classification error in the neural network.
Figure 2.
DL architecture for mitosis detection. A 64 × 64 size patch is extracted at 40× magnification. The patch contains a candidate nucleus, which is fed to the DNN. The DNN is composed of several convolutional neural networks, each followed by nonlinear and pooling operations. At the end, two fully connected layers are used to estimate the probability of a patch containing a mitotic figure. The probability that the image patch contains a mitotic nucleus is then assigned by the classifier. [Color figure can be viewed at wileyonlinelibrary.com]
The light design of the DNN, along with the nuclei sampling strategy, reduces the computational burden at test time as the cancerous regions in WSI can cover over 500 HPFs at 40 × magnification. As such, an appropriate strategy combined with a suitable DNN architecture is necessary to meet the time constraints and requirements of a typical clinical workflow.
The DNN classifier was trained using image patches extracted from two open access datasets: 225 [from MITOS2012 (23)] and 516 mitotic events [from AMIDA2013 (17)]. Non-mitotic candidate nuclei were extracted using an automatic nuclei detector. The performance of the DNN was evaluated with the AMIDA2013 training dataset under a 6fold cross-validation setup and each fold was split at the patient level. The Fscore, Precision and Recall measures (24) for the mitotic nuclei class were computed across the 6-folds. The average and the standard deviation for the Fscore, precision and recall measures of the 6 folds were: 0.556 ± 0.21, 0.47 ± 0.24 and 0.78 ± 0.11, respectively. The mitotic classifier was also evaluated on the MITOS2012 testing set (using the AMIDA2013 and MITOS2012 training sets to train the DNN) obtaining a 0.78 Fscore.
Support Vector Machine for BCa Risk Stratification
The DNN mitotic detector can be used on several HPFs from a WSI to assess the mitotic frequency in the respective cancerous regions. The obtained mitotic frequency information is then used to build a feature vector of 10 dimensions, each bin (dimension) representing the number of HPFs with {k ϵ 0,1…8, and k ≥ 9} mitotic figures. Since the cancerous regions in each BCa case have a different number of HPFs, the feature vectors are normalized. Each feature vector is centered and scaled by a unitary standard deviation. The collected feature vectors were used to train a support vector machine with a linear kernel to distinguish BCa cases labeled either as low or high risk.
Experimental Design
Whole Slide Data Description
A cohort of 174 WSI were diagnosed with the BR grading system and with the ODX test. The dataset was split into (a) High (24 WSI, ODX > 30), (b) Intermediate (55 cases and 18 ≤ ODX ≤ 30), and (c) Low Risk categories (95 WSI and ODX < 18). Additionally, BR score was used to determine another two risk subgroups: (d) 15 cases with both high ODX and high BR grade (BR > 7) and (e) 42 cases with both low ODX and low BR grade (BR < 6).
From the 174 WSI, a total of 20082 HPFs were extracted. Each HPF corresponds to an image of 2000 × 2000 pixels at 40× magnification (a pixel is ≈ 0.0625 μm2). The performance of the DNN mitosis detector described in subsection “DNN for mitotic identification” was also evaluated on a subset of the images from these 174 WSI. A total of 40 HPFs were randomly selected from BCa cases that have either high ODX-high grade or low ODX-low grade. Mitoses in these images were annotated by an expert pathologist collaborator. Evaluation of the DNN performance yielded an overall 0.56 F-score measure in the aforementioned set of images. This measure is within the range previously reported (0.556 ± 0.21) in subsection “DNN for mitotic identification.” A detailed presentation of the results is included in the Supporting Information File B.
Experiments and Statistical Analysis
The correlation of the automated mitosis with the ODX and BR groups was assessed in two different ways. First, a t-test evaluated the differences of the obtained mitotic counting for the different risk groups defined in subsection “Whole Slide Data Description.” Second, to evaluate the hypothesis that the automatically estimated mitotic count enables discrimination of the high/low ODX groups, a Support Vector Machine classifier was trained and used to classify WSI into high and low risk BCa.
A mean “mitotic count per ten HPFs” is computed for each WSI. The distribution is then compared for the two risk groups using a two tail t-test. The null hypothesis is that both distributions are sampled from the same normal distribution with equal mean. The t-test is applied using an equal variance for the different risk groups, specifically: (a) The high ODX group against the low ODX group, (b) The high ODX group against both the intermediate and low ODX group, (c) The high ODX-high grade (HH Group) against the low ODX-low grade score (LL group), (d) The high ODX-high grade (HH Group) against the rest of cases (HHc group), and (e) The low ODX-low grade (LL group) against the rest of cases (LLc group). The main hypothesis to test in each of the aforementioned experiments is that the BCa groups with higher ODX score and BR grading have a higher number of mitotic figures.
The discriminability of the number of mitotic figures automatically identified by the DL classifier was assessed in a BCa risk stratification task. The support vector machine classifier, described in subsection “Support Vector Machine for BCa risk stratification” was evaluated using a leave-one-out cross-validation scheme for each of the BCa cases (WSI) labeled either as high risk (BCa cases with high ODX score - 24 cases) or low risk (BCa cases with low ODX score - 95 cases). The SVM classifier was trained with a regularization hyperparameter C = 1.
Experimental Results and Discussion
Statistical Analysis
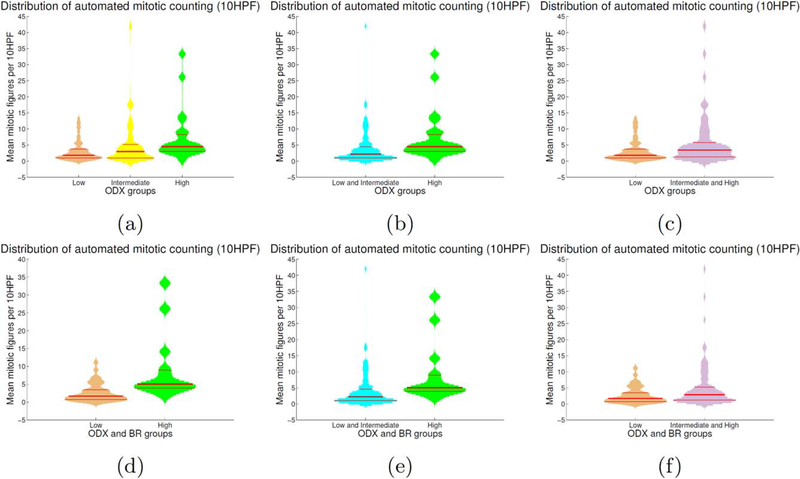
The DNN classifier was applied to the 174 WSI previously described in subsection “Whole Slide Data Description.” Qualitative results for high, intermediate, and low ODX cases can be seen in Figure 3. Distributions of the average mitotic count for the high, intermediate, and low ODX groups are depicted as violin plots in Figure 4 (see caption for violin plot description). Additionally, the intermediate group is paired either with the high or the low group to compare the resulting distributions. The mean mitotic distribution in 10 HPF was also computed for high ODX and BR scores (HH) and for low ODX and BR scores (LL), as illustrated in the bottom row of Figure 4. The distributions of both groups was compared against the rest of BCa cases. The t-test results are presented in Table 1.
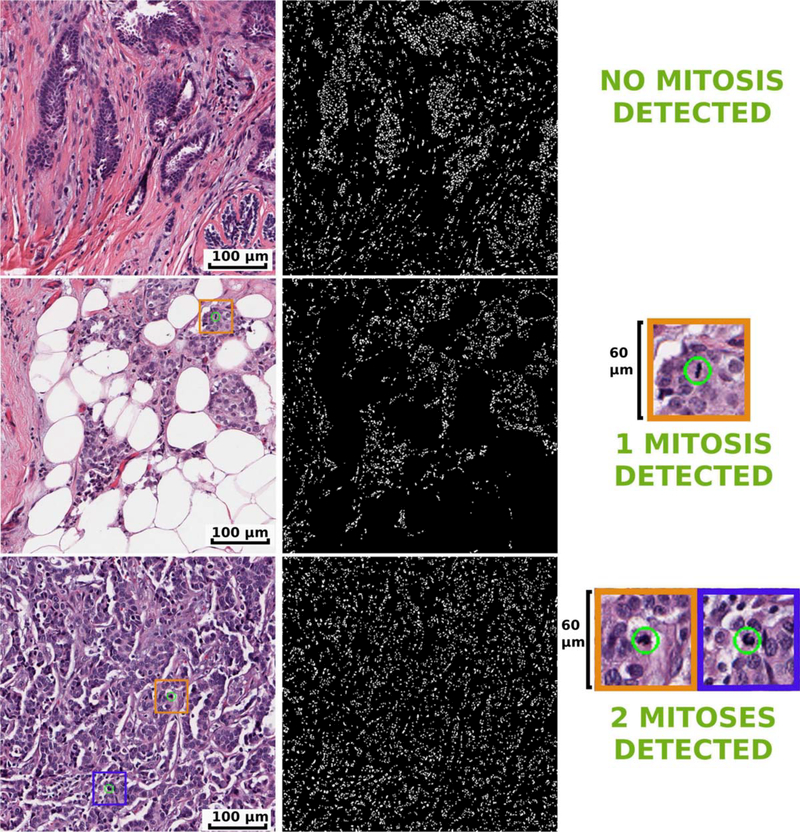
Figure 3.
Mitosis identification process in HPFs extracted from low ODX BCa (top row), intermediate ODX BCa (middle row) and high ODX BCa (bottom row). In the first column, results of the mitosis detection for the original HPF at 40× magnification is depicted. In the second column, the mask after the nuclei detection process is presented. The third column shows a close up for the DNN detection of the mitoses (green circles) depicted in the left most column. Mitosis are rare events: at the selected HPF size the low ODX BCa cases usually does not have mitosis. Meanwhile, intermediate and high ODX HPFs have a slightly larger mitosis count. [Color figure can be viewed at wileyonlinelibrary.com]
Figure 4.
Violin plots depicting the mean mitotic activity in 10 HPFs for the different risk ODX risk groups. The histogram associated with each violin plot is smoothed using a normal kernel. Red lines in the violin plot show the location of the lower quartile (q1), the median, and the upper quartile (q3). Low (orange), intermediate (yellow) and high (green) ODX groups are shown in the top row (a). The mitotic distribution of the low and intermediate groups (cyan) against the high ODX group is presented in (b). The mitotic distribution of the low group against the intermediate and high ODX groups (purple) are presented in (c). The mitotic distribution for the groups with low ODX-low grade and high ODX-high grade are depicted in (d). The mitotic distribution forthe high ODX-high grade against the rest of BCa cases, and low ODX-low grade against the rest of BCa cases are presented in (e, f), respectively. [Color figure can be viewed at wileyonlinelibrary.com]
Table 1.
Statistical evaluation of the DNN mitotic classifier in distinguishinging different breast cancer ODX risk groups: high ODX risk group (H), low ODX risk group (L), and intermediate ODX risk group (I)
| RISK GROUP COMPARISON | P-VALUES (EQUALVARIANCE) |
|---|---|
| H vs. L | 0.1×10−3 |
| H vs. L and I | 3.6×10−3 |
| H and I vs. L | 2.3×10−3 |
| HH vs. LL | 0.2×10−3 |
| LL vs. rest of BCa cases (LLc) | 0.032 |
| HH vs. rest of BCa cases (HHc) | 0.6×10−3 |
Additionally, the Bloom Richardson grading scheme was also used to define the high ODX-high grade group (HH) and the low ODX-low grade group (LL). Note that statistically significant differences were observed for all the six comparative experiments performed.
Results in Figure 4 and Table 1 reveal that the average number of mitoses for the low ODX group is significantly different than those obtained for the high risk group. The low ODX group obtained a mean number of 3.1 mitoses per 10 HPFs while the high ODX group obtained a mean number of 7.2 mitoses per 10 HPFs (p = 0.1×10−3 with 95% CI [ − 6.2, −2.1]). When grouping the intermediate ODX group with the low ODX group, the statistical test shows that the resultant mitoses distribution is different with respect to the high ODX group distribution, with a mean number of 3.8 mitoses per 10 HPFs for the low/intermediate ODX group (p=3.6×10−3 with 95% CI [ − 5.8, −1.15]). Besides, if the intermediate ODX group is combined with the high ODX group, the mixed distribution results different to the low ODX group distribution, with a mean number of 5.6 mitoses per 10 HPFs for the intermediate/high ODX group (p=2.3×10−3 with 95% CI [ − 4.1, −0.9]).
The analysis of the groups defined by both ODX score and BR grade also show a different number of mitotic figures. Cases with a low ODX-low grade yielded a mean number of 2.7 mitoses per 10 HPFs while those with high ODX-high grade showed a mean number of 8.8 mitoses per 10 HPFs (p=0.2×10−3 with 95% CI [ − 9.2, −3.1]). The low ODX-low grade cases continue to show a different number of mitoses when compared with all the remaining cases in the dataset (those that did not have both low ODX and low grade). In this case, the average number of mitoses per 10 HPFs for the 132 (LLc) cases was 4.74. The two distributions are closer but still significantly different (p=0.03 with 95% CI [ − 3.98, −0.18]). Finally, when comparing the high ODX-high grade cases with the remaining cases, a significant difference was found. For those cases that did not have both high ODX and high grade, the mean number of mitoses per 10 HPFs was 3.8 (p=0.6X10−3 with 95% CI [ − 7.8, −2.2).
Support Vector Machine Classification Results
The distribution of mitoses per HPF was used to train a linear support vector machine classifier to discriminate the high/low ODX BCa cases, yielding an 83.19% average accuracy. Now, if one considers that the outcome of the leave one out evaluation in this case is binary, an error of the classification gives a null accuracy for that classifier. From the 119 different experiments, 20 resulted in a null accuracy. The confusion matrix, shown in Table 2, was obtained by computing the aggregation of the confusion matrices over each fold of the leave-one-out experiment (119 experiments in this case). The results reveal that most of the WSIs with low ODX score are correctly classified as Low Risk. Leveraging a SVM classifier resulted in a mean increase of ~15% in the observed accuracy (83.19%) versus using a simple thresholding method (68.07%). The details on how to compute the optimal threshold are described in the Supporting Information File A.
Table 2.
Confusion matrix forthe risk stratification task using mitotic information
| LOW ODX | HIGH ODX | |
|---|---|---|
| Classified as LR | 83 | 8 |
| Classified as HR | 12 | 16 |
This confusion matrix corresponds to the aggregation of 119 confusion matrices obtained over leave-one-out cross-validation run from the 119 test sets. The BCa cases are classified either as low risk (LR) or high risk (HR).
Concluding Remarks
In this article, we rigorously investigated the problem of automated computation of the mitotic activity in WSI, a potential histologic image biomarker of the disease risk and aggressiveness in ER+ breast cancers. A DNN classifier was developed to identify mitoses and to evaluate the correlation of the automatically determined mitotic index with the risk category determined by the Oncotype DX test. Moreover, the mitotic count was evaluated in terms of its ability to distinguish the low and high ODX risk categories as well as low and high grades. Likewise, the strategy was able to discriminate cases with different permutations of ODX risk and grade. On a cohort of 174 WSI, the automated mitotic count was significantly different for the low ODX group when compared against the high ODX risk, thereby demonstrating that this automated quantification captures important prognostic information. This result also extends to the BCa cases group with both low ODX and low grade. However, the mitotic detector failed to show differences between high and intermediate ODX groups.
The mitotic information extracted using the DNN achieves a sufficient level of precision as to deal with patient tissue variability and to preserve enough prognostic information to allow a support vector machine classifier to distinguish between BCa cases with high ODX and low ODX score.
The lightweight DNN approach herein presented shows a similar performance to other DNN approaches, yet the particular sampling strategy herein integrated analyzes a HPF in around 8 seconds using a conventional GPU (Nvidia Quadro K4000). Other DNN approaches in the literature report a full HPF processing in about 8 min (18). In a typical WSI, some of the cancerous regions may have >500 HPFs. Consequently, the lightweight DNN approach herein introduced is more appropriate for a diagnostic routine workflow.
The results of this study have significant clinical applicability even though the high and intermediate risk ODX groups cannot be separated via the DNN. Patients with low ODX have demonstrated low risk, and those patients are unlikely to derive a significant benefit from adjuvant chemotherapy. It is less clear that it is safe to omit chemotherapy in patients with intermediate and high risk scores, and oncologists routinely have to discuss the risks and benefits of chemotherapy in these circumstances.
Mitotic activity of a tumor, whether measured as number of mitoses per high power field (as in traditional histopathologic grading), proliferation index as measured by Ki67 count by immunohistochemistry, or as measured by gene expression by predictive/prognostic assays such as ODX cannot be used alone to separate patients into low, intermediate, or high risk categories. Mitotic activity is an important component of these analyses but it is not the lone determinant of the score. The fact that the mitotic activity figures from the DNN can alone separate low versus intermediate/high is quite remarkable and has direct clinical utility. It is possible that if this model were coupled with additional algorithms aimed at other pathologic features in the future, the risk category could be defined further.
Future work on improving the robustness of the mitotic detector to stain variation and including image information in the Z-axis (i.e., pathologist frequently change the Z-axis of optical microscopes to determine if a confounding nuclei is mitotic) might improve the mitosis detector and allow for even more accurate estimation of mitotic rate. In addition, future work will involve independent validation of these findings using a separate test cohort.
Supplementary Material
Acknowledgments
Part of the experiments presented in this article were carried out using the GridUIS-2 experimental testbed, being developed under the Universidad Industrial de Santander (SC3UIS) High Performance and Scientific Computing Centre, development action with support from UIS Vicerrectoria de Investigación y Extension (VIE-UIS) and several UIS research groups as well as other funding bodies. The authors would also like to acknowledge NVIDIA for their gift of a Titan X GPU to support DL research in our group. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Grant sponsor: National Cancer Institute of the National Institutes of Health, Grant numbers: R01CA202752–01A1, R21CA179327–01, R21CA195152–01, and 1U24CA199374–01
Grant sponsor: National Institute of Diabetes and Digestive and Kidney Diseases, Grant number: R01DK098503–02
Grant sponsor: DOD Prostate Cancer Synergistic Idea Development Award, Grant number: PC120857
Grant sponsor: DOD Lung Cancer Idea Development New Investigator Award, Grant number: LC130463
Grant sponsor: the DOD Prostate Cancer Idea Development Award
Grant sponsor: Case Comprehensive Cancer Center Pilot Grant
Grant sponsor: VelaSano Grant from the Cleveland Clinic
Grant sponsor: Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering at Case Western Reserve University
Grant sponsor: Universidad Nacional de Colombia by means of “Convocatoria del programa nacional de proyectos para el fortalecimiento de la investigación, la creación y la innovación en posgrados de la Universidad Nacional de Colombia”
Grant sponsor: Colciencias by means of Convocatoria 711–2015 para proyectos de ciencia, tecnología e innovacón en salud
Footnotes
Anant Madabhushi is scientific advisory board member, scientific consultant and equity holder in Inspirata Inc., and equity holder in Elucid Bioimaging. David Romo-Bucheli, Andrew Janowczyk, Hannah Gilmore and Eduardo Romero do not have any potential conflict of interest.
Literature Cited
- 1.Khoury T, Huang X, Chen X, Wang D, Liu S, Opyrchal M. Comprehensive Histologic Scoring to Maximize the Predictability of Pathology-generated Equation of Breast Cancer Oncotype DX Recurrence Score. Appl Immunohistochem Mol Morphol 2016;24:703–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thaker NG, Hoffman KE, Stauder MC, Shaitelman SF, Strom EA, Tereffe W, Smith BD, Perkins GH, Huo L, Munsell MF, et al. The 21-gene recurrence score complements IBTR! Estimates in early-stage, hormone receptor-positive, HER2-normal, lymph node-negative breast cancer. Springerplus 2015;4:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Acs G, Kiluk J, Loftus L, Laronga C. Comparison of Oncotype DX and Mammostrat risk estimations and correlations with histologic tumor features in low-grade, estrogen receptor-positive invasive breast carcinomas. Mod Pathol 2013;26:1451–1460. [DOI] [PubMed] [Google Scholar]
- 4.Dalton LW, Page DL, Dupont WD. Histologic grading of breast carcinoma. A reproducibility study. Cancer 1994;73:2765–2770. [DOI] [PubMed] [Google Scholar]
- 5.Fan F, Thomas P Tumors of the Breast In: Damjanov I, Fan F, editors. Cancer Grading Manual. New York: Springer; 2007. pp 75–81. [Google Scholar]
- 6.Meyer JS, Alvarez C, Milikowski C, Olson N, Russo I, Russo J, Glass A, Zehnbauer BA, Lister K, Parwaresch R. Breast carcinoma malignancy grading by Bloom-Richardson system vs proliferation index: Reproducibility of grade and advantages of proliferation index. Mod Pathol 2005;18:1067–1078. [DOI] [PubMed] [Google Scholar]
- 7.Flanagan MB, Dabbs DJ, Brufsky AM, Beriwal S, Bhargava R. Histopathologic variables predict Oncotype DX recurrence score. Mod Pathol 2008;21:1255–1261. [DOI] [PubMed] [Google Scholar]
- 8.Lewis JS Jr, Ali S, Luo J, Thorstad WL, Madabhushi A. A quantitative histomorphometric classifier (QuHbIC) identifies aggressive versus indolent p16-positive oropharyngeal squamous cell carcinoma. Am J Surg Pathol 2014;38:128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee G, Sparks R, Ali S, Shih NN, Feldman MD, Spangler E, Rebbeck T, Tomaszewski JE, Madabhushi A. Co-Occurring Gland Angularity in Localized Subgraphs: Predicting Biochemical Recurrence in Intermediate-Risk Prostate Cancer Patients. PloS One 2014;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee G, Ali S, Veltri R, Epstein JI, Christudass C, Madabhushi A. Cell orientation entropy (COrE): Predicting biochemical recurrence from prostate cancer tissue microarrays Medical Image Computing and Computer-Assisted Intervention MIC-CAI 2013. Springer; 2013. pp 396–403. [DOI] [PubMed] [Google Scholar]
- 11.Chang H, Fontenay GV, Han J, Cong G, Baehner FL, Gray JW, Spellman PT, Parvin B. Morphometic analysis of TCGA glioblastoma multiforme. BMC Bioinformatics 2011;12:484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basavanhally A, Xu J, Madabhushi A, Ganesan S. Computer-aided prognosis of ER1 breast cancer histopathology and correlating survival outcome with oncotype DX assay. IEEE Int Symp Biomed Imaging 2009;851–854. [Google Scholar]
- 13.Veta M, Pluim JP, Diest PJ, van Viergever M, et al. Breast cancer histopathology image analysis: A review. IEEE Trans Biomed Eng 2014;61:1400–1411. [DOI] [PubMed] [Google Scholar]
- 14.Larsen ABL, Vestergaard JS, Larsen R. HEp-2 cell classification using shape index histograms with donut-shaped spatial pooling. IEEE Trans Med Imaging 2014;33:1573–1580. [DOI] [PubMed] [Google Scholar]
- 15.Wang H, Cruz-Roa A, Basavanhally A, Gilmore H, Shih N, Feldman M, Tomaszewski J, Gonzalez F, Madabhushi A. Mitosis detection in breast cancer pathology images by combining handcrafted and convolutional neural network features. J Med Imaging 2014;1:034003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janowczyk A, Madabhushi A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J Pathol Inform 2016;7: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Veta M, Diest PJ, van Willems SM, Wang H, Madabhushi A, Cruz-Roa A, Gonzalez F, Larsen AB, Vestergaard JS, Dahl AB, et al. Assessment of algorithms for mitosis detection in breast cancer histopathology images. Med Image Anal 2015;20: 237–248. [DOI] [PubMed] [Google Scholar]
- 18.Ciresan DC, Giusti A, Gambardella LM, Schmidhuber J. Mitosis Detection in Breast Cancer Histology Images with Deep Neural Networks In: Mori K, Sakuma I, Sato Y, Barillot C, Navab N, editors Medical Image Computing and Computer-Assisted Intervention MICCAI 2013. Vol 8150 Lecture Notes in Computer Science. Berlin Heidelberg: Springer; 2013. pp 411–418. [DOI] [PubMed] [Google Scholar]
- 19.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Adv Neur In 2012;1097–1105. [Google Scholar]
- 20.Chang H, Loss LA, Spellman PT, Borowsky A, Parvin B. Batch-invariant nuclear segmentation in whole mount histology sections. IEEE Int Symp Biomed Imaging 2012; 856–859. [Google Scholar]
- 21.Otsu N A Threshold Selection Method from Gray-Level Histograms. IEEE Trans Syst Man Cyb 1979;9:62–66. [Google Scholar]
- 22.Ioffe S, Szegedy C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. arXiv 1502.03167 2015. [Google Scholar]
- 23.Roux L, Racoceanu D, Loménie N, Kulikova M, Irshad H, Klossa J, Capron F, Genestie C, Le Naour G, Gurcan MN. Mitosis detection in breast cancer histological images An ICPR 2012 contest. J Pathol Inform 2013;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Powers DM. Evaluation: from precision, recall and F-measure to ROC, informedness, markedness and correlation. J Mach Learn Tech 2011;2:37–63. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.