Abstract
Neoadjuvant treatment (NAT) of breast cancer (BCa) is an option for patients with the locally advanced disease. It has been compared with standard adjuvant therapy with the aim of improving prognosis and surgical outcome. Moreover, the response of the tumor to the therapy provides useful information for patient management. The pathological examination of the tissue sections after surgery is the gold-standard to estimate the residual tumor and the assessment of cellularity is an important component of tumor burden assessment. In the current clinical practice, tumor cellularity is manually estimated by pathologists on hematoxylin and eosin (H&E) stained slides, the quality, and reliability of which might be impaired by inter-observer variability which potentially affects prognostic power assessment in NAT trials. This procedure is also qualitative and time-consuming. In this paper, we describe a method of automatically assessing cellularity. A pipeline to automatically segment nuclei figures and estimate residual cancer cellularity from within patches and whole slide images (WSIs) of BCa was developed. We have compared the performance of our proposed pipeline in estimating residual cancer cellularity with that of two expert pathologists. We found an intra-class agreement coefficient (ICC) of 0.89 (95% CI of [0.70, 0.95]) between pathologists, 0.74 (95% CI of [0.70, 0.77]) between pathologist #1 and proposed method, and 0.75 (95% CI of [0.71, 0.79]) between pathologist #2 and proposed method. We have also successfully applied our proposed technique on a WSI to locate areas with high concentration of residual cancer. The main advantage of our approach is that it is fully automatic and can be used to find areas with high cellularity in WSIs. This provides a first step in developing an automatic technique for post-NAT tumor response assessment from pathology slides.
Keywords: pathology image analysis, breast cancer, machine learning, neoadjuvant therapy
Breast cancer (BCa) is one of the most common occurring cancers in women (1). There are several treatment options available to treat BCa depending on its type or stage which include surgery, radiation therapy, chemotherapy, or targeted therapy. Neoadjuvant (preoperative) therapy (NAT) is a treatment approach for selected high-risk and/or locally advanced BCa patients in which chemotherapy or targeted therapy is administered before the surgery (2). There have been several advantages reported for the neoadjuvant treatment including: (a) tumor size reduction for patients with large tumors which makes them eligible for surgical resection or candidates for breast-conserving surgery instead of mastectomy; (b) efficacy of the therapy can be assessed in vivo which helps in the modification or change of treatment for patients who show little to no respond to the therapy; and (c) association of tumor response to the therapy is facilitated by comparing tissue samples before, during, and after treatment to identify gene expression profiles associated with tumor response and/or development of novel therapeutic agents (2–5). It is desirable to achieve a pathologic complete response (pCR) after the course of NAT as this is directly correlated to prognosis and efficacy of the treatment (6–8). pCR is defined as the absence of residual cancer in the breast tissue and associated axillary lymph nodes at the completion of NAT (9). Tumor response during NAT is monitored by physical examination (palpation), ultrasonography, and mammography (10,11). While accurate prediction of the residual cancer is crucial after NAT in assessing the efficacy of the therapy, clinical and radiologic assessment of the response often under-or over-estimates the amount of residual cancer (5,10,12).
Although reporting treatment response for post-NAT is not part of the synoptic reporting system suggested by American Joint Committee on Cancer (AJCC) manual, a descriptive assessment of response is commonly reported qualitatively in health care centers. Currently, the pathologic examination of the tissue sections after surgery is the gold-standard to estimate the residual tumor. Quantitative measurement of the residual cancer burden after NAT is even more beneficial for drug trials to assess the success of newly developed drugs. In (13,14), different proposed systems for evaluating the pathologic response to NAT are summarized. Currently, tumor burden is manually estimated by pathologists on hematoxylin and eosin (H&E) stained slides, the quality and reliability of which might be impaired by inter-observer variability which potentially affects pCR and prognostic power assessment in NAT trials (9,15). This procedure is also qualitative and time-consuming in the current practice (16–19).
Although there is no standardized definition of the factors considered to assess post-treatment tissue sections for pCR (20,21), the cellularity fraction of cancer within tumor bed (TB) seems to be a better prognostic indicator of the efficacy of the treatment compared to other factors (12,20,22). The objective of this article is to present a pipeline to automatically assess the cellularity of residual tumor from post-treatment pathology image patches and whole slides of breast cancer. An automated method could provide pathologists a second opinion assessment on the tissue regions and provides bases to gain more information from post-NAT tissue micro-environment (23).
Background
Many features used in the analysis of pathology slides are inspired by visual markers characterized by pathologists. A large subset of these features is derived from nuclei objects present within the tissue micro-environment. Therefore, before attempting to assess the tumor cellularity, it is an important first step to detect and segment nuclei objects from regions of interest (ROIs). While some studies focus on producing high quality nuclei segmentation for subsequent automated processing of pathology images (24,25), others claim perfect nuclei segmentation does not necessarily lead to a better classification performance (26). Most of the currently present nuclei segmentation techniques in the literature utilize one or a combination of watershed segmentation (27–30), graph-cuts (31), matching-based (32), Laplacian of Gaussian (LoG) (31), active contours (24,30,33), or pixel classification (33–35) methods combined with pre-or post-processing phases (36).
Once the nuclei figures are segmented, features defining their morphology, texture, and spatial relationship with one another are calculated to represent different nuclei classes in a multi-dimensional feature space (37,38). Morphological features characterize size and shape of nuclei figures such as area, perimeter, major axis, minor axis, shape factor, etc. (39), texture features are used to describe gray level pixels inter-relations with each other or provide cell interpretations in a multi-scale fashion such as Daubechies wavelet features (40) or Gabor wavelet features (41), and spatial relationship features are derived to summarize the way nuclei figures are located with respect to each other to form disease micro-environment such as graph-based features (41–44) or spin-context (45,46).
After deriving appropriate features characterizing nuclei figures, statistical models such as support vector machines (SVM) (47), Naive Bayes (48), neural networks (47), and decision trees (49) are employed to learn and distinguish the differences between patterns of nuclei figures. Nuclei classification approaches have been used as a processing step toward various pathology automated analysis projects in the literature including identification of intraductal proliferative lesions (50), correcting for copy number profiles between samples of BCa (51), nuclei grading (52,53), and other (54).
Recently, deep learning based methods, particularly those based on convolutional neural networks (CNNs) have gained much attention in the medical image analysis community due to their better performance in some applications compared to the traditional machine learning approaches (55–57). They work by iteratively improving learned data representations by deriving features that are originally calculated from the data itself (usually by applying some texture filters on the training images) with the aim of achieving maximum class separability. Compared to the traditional machine learning approaches, deep learning methods do not require the design of hand-crafted features, scale well to large datasets, and can easily be adopted to other applications. However, deep learning methods need large datasets to outperform traditional learning methods, and it is not easy to correlate their findings with theoretical medical foundations.
In this study, we chose to use traditional machine learning approaches to assess residual cancer cellularity due to our limited dataset size. First, we divide whole slide images into smaller patches, and from within the smaller patches or regions of interest (ROI) detect and segment nuclei figures. Segmented malignant nuclei figures are then distinguished from lymphocyte and normal epithelial figures using an SVM classifier. The cellularity estimation is then performed using distinguished malignant epithelial figures for every ROI.
The combination of methods presented in this article creates a novel pipeline to tackle the problem of assessing tumor cellularity from post treated tissue sections, a problem that has not been addressed before. For this purpose, a unique set of features was designed to better represent the differences among different classes of nuclei figures. Furthermore, to calibrate between computer assessed cellularity and that of pathologists from malignant nuclei figures, a novel calibration approach was considered. Our approach has the advantage that it mirrors the work flow of the pathologists and it is possible to interpret what the pipeline is doing at each step.
Dataset
Pathology Whole Slide Images
The dataset used to train and validate the tumor cellularity assessment consists of n = 121 H&E stained post neoadjuvant whole slide images (WSIs) from 64 patients. They were prepared and scanned at 20X magnification (0.5 μm/pixel) in the Department of Anatomic Pathology at Sunnybrook Health Sciences Centre (SBHSC), Toronto, Canada after receiving appropriate ethical approval from the hospital and were annotated by expert pathologists using the Sedeen Viewer (Pathcore, Toronto, Canada). Both collaborating pathologists in this study are specialist in anatomical pathology with breast pathology fellowship training at the University of Toronto with >10 years of experience in practice. 67 slides (corresponding to 31 patients) were used to train and 25 slides (corresponding to 15 patients) were used to validate the nuclei classification statistical models (training part—also referred to as sets A and B, respectively in the text). Furthermore, 29 slides (corresponding to 18 patients) were used to validate the cellularity assessment pipeline (validation part—also referred to as set C in the text). Table 1 illustrates the histopathologic characterization of patients included in the training and validation sets used in this study. The training and validation sets were gathered from consecutive cases with residual disease between years 2014–2015 for the training set and 2016 for the validation set. As shown in Table 1, patient data used for this study include different hormonal status, grade, and clinical response categories. Although we were focused on cases with the residual disease, eight patient cases were specifically added to the training set only to include the complete pathologic response category. Some of the eligibility criteria for selecting patients for NAT trials include having a clinical stage of II or III, presence of objective measurable lesion(s), and previously untreated disease. Here, we briefly explain the dataset preparations used for nuclei object classification and cellularity assessment pipelines.
Table 1.
Histopathologic characterization of the 64 patients in training and validation sets
| CHARACTERISTIC | TRAINING SET (N = 92) | VALIDATION SET (N = 29) |
|---|---|---|
| Age | 50 (± 12.6) | 51 (± 12.7) |
| Tumor bed size (mm) | 51.3 (± 35.7) | 84.7 (± 82.2) |
| Hormonal status | No. of patients (%) | |
| ER | 63 (68%) | 19 (65%) |
| PR | 46 (50%) | 17 (57%) |
| HER2 | 58 (63%) | 19 (65%) |
| Grade | No. of patients (%) | |
| 1 | 12 (13%) | 4 (14%) |
| 2 | 49 (53%) | 14 (48%) |
| 3 | 18 (19%) | 11 (38%) |
| N/A | 13 (14%) | 0 |
| Histology | No. of patients (%) | |
| IDC | 65 (71%) | 27 (93%) |
| ILC | 10 (11%) | 2 (7%) |
| Other | 16 (17%) | 0 |
| Clinical response | No. of patients (%) | |
| CPR | 8 (9%) | 0 |
| PDR | 77 (84%) | 27 (93%) |
| NDR | 5 (5%) | 2 (7%) |
| Other | 1 (1%) | 0 |
IDC: intra-ductal carcinoma, ILC: intra-lobular carcinoma, CPR: complete pathological response, PDR: probable or definite response, NDR: no definite response.
Nuclei figure annotation.
Nuclei figures were marked by an expert pathologist by clicking on the centroid of individual nuclei from within a rectangular ROI on the tissue slides of the training part (sets A and B). Size and location of ROIs were arbitrarily chosen from within the tumor bed by the expert pathologist in such a way that they contained a mixture of nuclei figure types (benign or malignant epithelial, and lymphocyte). Figure 1 depicts an example of a rectangular ROI from within which nuclei centroids were marked by the expert pathologist. The pathologist only marked nuclei figures if she could be certain about their type; nuclei that were out of plane or out of focus were not marked. >30,000 nuclei (n = 3,868 lymphocyte, n = 10,407 benign epithelial, and n = 16,419 malignant epithelial figures) were marked by the pathologist from 166 ROIs in sets A and B which included a mixture of lymphocyte, benign, and malignant epithelial nuclei figures.
Figure 1.
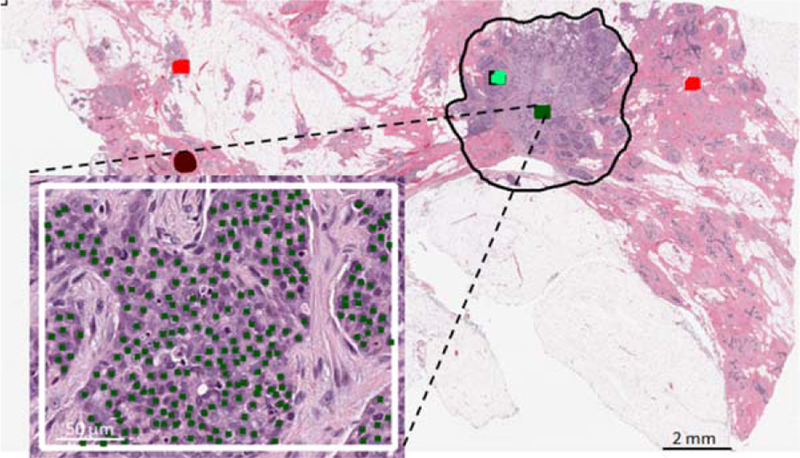
Subset of nuclei annotations by expert pathologist. Centroids of different nuclei classes (malignant epithelial in this demonstration) were marked by the pathologist. Regions of interest were selected from within the tumor bed regions (black contour). [Color figure can be viewed at wileyonlinelibrary.com]
Cellularity assessment within patches.
In order to evaluate the proposed cellularity assessment pipeline, ROI patches from image slides in the validation part (set C) were extracted by an expert pathologist (pathologist #1) and their individual cellularity was reported as a scalar value between 0 and 100%. All image patches were selected from within tumor bed regions and were 512×512 pixels (corresponding to an area of about 1 mm2). To ensure a balanced selection between various patches with different cellularity portions, the pathologist was asked to select representative ROIs in four roughly estimated cellularity categories of 0 (normal), 1–30 (low cellularity), 31–70 (medium cellularity), and 71–100% (high cellularity). N = 1121 ROI patches were reviewed by the pathologist from 29 WSIs. Figure 2 shows a WSI from which a subset of cellularity assessment dataset used for this study were chosen with a representative high cellularity patch. A second pathologist (pathologist #2) was also asked to score the same patches marked by pathologist #1 and their results were stored to obtain the inter-rater variability among pathologists and compare with that of the performance of the presented automated method.
Figure 2.
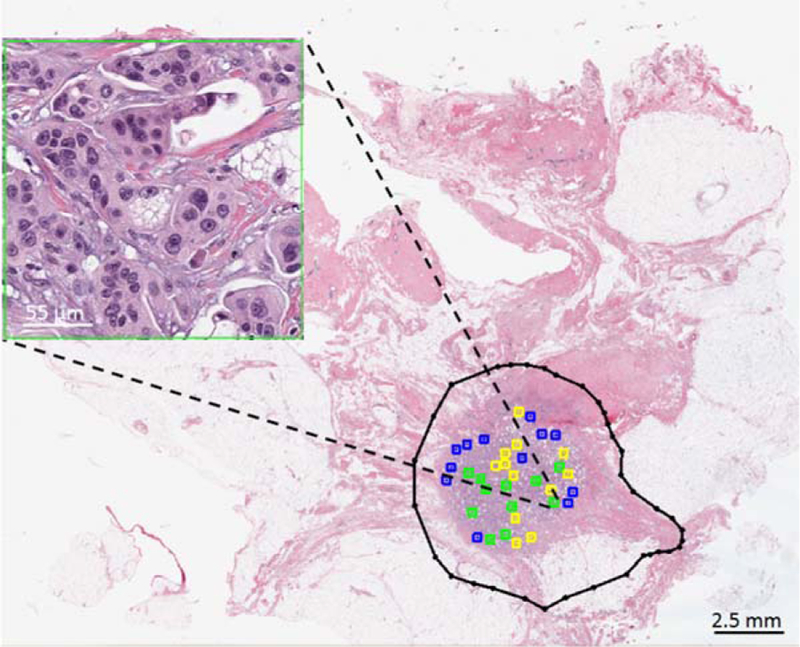
Subset of cellularity assessment dataset from image patches annotated by expert pathologist. 512×512 pixels patches were selected by the pathologist from within the tumor bed (black contour) and were given a score to depict the cellularity of the malignant components within the given patch. Four categories of patches were chosen by the pathologist: 0% (pink), 1–30% (blue), 31–70% (yellow), and 71–100% (green). The image patch shown in this figure was scored as 80% by pathologist #1. [Color figure can be viewed at wileyonlinelibrary.com]
Methodology and Experimental Setup
Segmentation and classification of nuclei is an important step to analyze pathology slides. In this study, in order to assess the cellularity of tumor bed after NAT, we particularly need to be able to distinguish between lymphocyte, benign and malignant epithelial nuclei. Therefore the following multi-step process was considered to estimate the cellularity of residual tumor from pathology images: (a) segment nuclei figures from regions within tumor bed, (b) learn a cascaded statistical model to distinguish between nuclei classes (malignant epithelial, benign epithelial, and lymphocytes); use set A to train and tune the statistical models and set B to validate their performances, and (c) estimate the cellularity of residual cancer from patches using classification results of malignant epithelial figures in step 2; use set C to validate cellularity assessment pipeline.
Nuclei Figure Segmentation
In (35), we proposed a nuclei segmentation method to extract nuclei figures from pathology slides. Briefly, the segmentation technique works by decorrelating and stretching the original RGB colorspace in image slides. Next, the now decorrelated and stretched RGB colorspace was converted into Lab colorspace to reduce the dependency of the pixel values to intensity information and used multilevel thresholding and marker controlled watershed algorithms to segment foreground and separate overlapping nuclei figures. The F1-score of the nuclei segmentation technique was 0.9 when evaluated on 7931 nuclei from 36 images from a publicly available benchmark dataset in (24).
For this study, the effect of color variation within slides was minimized before segmenting nuclei figures by standardizing their color to a reference image. The reference image was chosen from an image slide not used in this study and was selected due to its good color contrast between different image components corresponding to various tissue regions (fat, stroma, and nuclei). The reference and selected images were both converted to Lab color space and selected (source) image color channels were standardized to those of reference image using the following equations:
where lt, at, and bt are the l, a, and b channels of the target image respectively; ls, as, and bs are the l, a, and b channels of the reference image respectively; , , and are the means of l, a, and b channels of the source image respectively; , , and are the means of the reference image, , , and are standard deviations of the l, a, and b channels of the source image respectively; and , , and are standard deviations of the l, a, and b channels of the reference image respectively. The target Lab image was then converted back to the RGB colorspace
Nuclei Figure Labeling
In order to train a classifier to differentiate between normal, malignant, and lymphocyte nuclei, it was necessary to label each segmented figure using the manually labeled data. Therefore, to ensure only one label is assigned to every segmented figure, centroids of segmented nuclei figures were matched with at least one entry in the groundtruth sets A and B. To match the centroids in any given patch, the Euclidean distance between the segmented figure and all nuclei centroids in the groundtruth training set were compared. Given the Euclidean distance of any two nuclei centroids in segmented group and groundtruth training set being lesser than 10 pixels (5 μm), the label of the nucleus in the groundtruth training set was assigned to the closest segmented figure. The assigned label was then removed from the pool of available groundtruth nuclei to avoid assigning label of one nucleus to multiple segmented figures. >72% of nuclei figures (n = 21779) in the groundtruth training set were matched with one segmented nuclei.
Nuclei Figure Classification
After nuclei were segmented from color standardized pathology images, they were converted to gray scale and their intensity level was normalized to have zero mean and standard deviation of one. For every nuclei figure, 125-dimensional feature vectors describing shape (mean and standard deviation of area, perimeter, eccentricity of all nuclei within a square with edges of size S = 0.125 mm and with the nuclei figure’s centroid as its center, and shape factor ), intensity (mean, mode, median, standard deviation, skewness, and kurtosis of the gray scale histogram of pixels within the segmented nuclei figure), texture (10 dimensional normalized histogram of uniform rotation-invariant local binary patterns (58) from 8-neighborhood pixels from within the segmented nuclei figure, and 88 dimensional Haralick textures (59) at 0, 45, 90, and 135 degrees from within the segmented nuclei figure), and 14 dimensional spatial features (comprised of 12 for number of nuclei present around the considered nuclei in spiral growing circles with diameters starting from 0.04 mm to 0.125 mm with a step size of 0.04 mm, including mean and standard deviation of the values from all 12 circles) were calculated from each individual segmented nuclei figures.
The nuclei features extracted from 92 slides (sets A and B) were divided into two parts: set A (n = 13821 nuclei objects from 67 slides) to train statistical models to distinguish between three nuclei classes (lymphocyte, malignant epithelial, and benign epithelial) and set B (n = 7958 nuclei objects from 25 slides) to check generalization performances of statistical models learned from set A. We trained a cascaded classifier from set A to first distinguish lymphocyte from epithelial figures (L vs. BM, n = 2260 lymphocyte, and n = 11561 epithelial figures) and then to separate epithelial figures into benign versus malignant figures (B vs. M, n = 3157 benign, and n = 8404 malignant figures). A five-fold cross-validation scheme was used for both cascades to train SVM classifiers with non-linear radial basis function (RBF) kernels. We used the SVM implementation present in the libsvm (60) library. In each fold, the optimum hyperplane parameters were found by testing different possible combinations of the classifier’s trade-off parameter (C) and kernel width (γ) in order to maximize the accuracy of classification. A final model for each classifier cascade was then generated by considering the whole set A (for each cascade) and combining the hyperplane parameters corresponding to the best classification performance by taking the median of the five parameters (corresponding to fivefolds). The overall trained model generated for each cascade was then used to validate the classification performance on nuclei objects present in the independent and unseen set B. Stromal nuclei were eliminated from the segmented figures both for tumor cellularity assessment and nuclei figure classification by removing the objects with the ratio of major to minor axes equal or >3.
Residual Cancer Cellularity Assessment
Once statistical models were trained to classify nuclei figures to any of lymphocyte, normal, and malignant epithelial classes, they were used to assess the cellularity of image patches in set C. In order to reduce the false-positive and false-negative detection rates on images in set C, a k-nearest neighbor approach (with k = 8) was used to correct nuclei labels that were mostly surrounded by one class of nuclei. Using this post-processing approach, about 2% improvement in the performance accuracies was observed from the images in set B.
Once the individual nuclei figures were classified to the three known classes and the malignant epithelial candidates were identified, a morphological dilation with a structuring element of size s = 11 was applied on the malignant figures to expand the malignant figures to account for the presence of cytoplasm around each nucleus. The cellularity of any given image patch was then calculated as the area covered by malignant figures divided by the area of the image patch. In order to correct for variation between the area calculated from morphologically expanded figures and the actual scores given by the pathologist in set C, the information on a portion of patches (n = 758) in set B that was previously annotated by pathologist #1 was used to find correction parameters using a linear regression.
Results
Nuclei Object Classification
Table 2 summarizes the mean performance of the 5 fold cross-validation on set A consisting of n = 13821 nuclei objects from 67 slides and Table 3 summarizes the performance of applying the model generated from set A on the unseen and independent testing set B. Comparing the performances reported in Tables 2 and 3 indicates that the developed classification pipeline is generalizable on independent sets to classify nuclei objects with AUCLvsBM = 0.87 and AUCBvsM = 0.86. The same model was used in subsequent analysis to assess residual cancer cellularity. Figure 3 shows the performance of nuclei classification on two representative patches from the validation dataset (set C) one covered by normal epithelial and another by lymphocyte and malignant epithelial components.
Table 2.
Mean performance of 5-fold cross-validation on set A (n = 13821 nuclei objects from 67 slides)
| AUC | ACCURACY (SD) % | SENSITIVITY % | SPECIFICITY % | |
|---|---|---|---|---|
| L vs BM | 0.97 | 95 (± 0.1) | 79 | 99 |
| B vs M | 0.86 | 83 (± 0.5) | 57 | 92 |
Table 3.
Performance of applying the generated model from data used in Table 2 on n = 7958 nuclei objects from the independent testing set B from 25 slides.
| CLASS | ACCURACY % | SENSITIVITY % | SPECIFICITY % |
|---|---|---|---|
| Lymphocyte (L) | 92 | 80 | 94 |
| Benign epithelial (B) | 75 | 50 | 92 |
| Malignant epithelial (M) | 77 | 91 | 63 |
Figure 3.
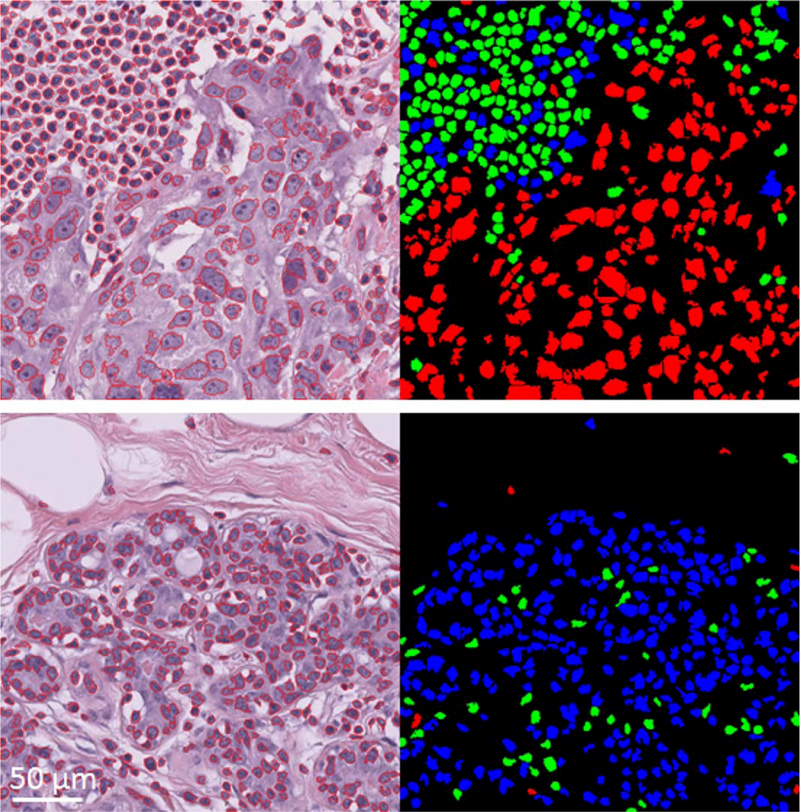
Examples of nuclei figure segmentation and classification on two representative patches of size 512×512 pixels from set C. On top is an image patch covered by lymphocyte and malignant epithelial components and on bottom is an image patch covered by benign epithelial nuclei. Their classified mask images are shown to their right. Green represents lymphocyte, blue represents benign epithelial, and red represents malignant epithelial classified objects. [Color figure can be viewed at wileyonlinelibrary.com]
Residual Cancer Cellularity Assessment
In order to compare the performance of the proposed cellularity assessment pipeline with that of the expert pathologists, they were reported both in terms of categorical and correlational agreements. Table 4 summarizes the confusion matrices of different cellularity categories comparing pathologists and the automated method. Considering the first confusion matrix in Table 4 (pathologist #2 vs. pathologist #1), it can be seen that the two pathologists were mostly in agreement when they were looking at normal or low cellularity patches, while their level of agreement decreased when annotating medium and high cellularity patches with pathologist #2 tending to assign a higher value of cellularity. The Cohen’s Kappa agreement between pathologist #1 vs. pathologist #2 is κ = 0.66. On the other hand, comparing the automated method with pathologists #1 and #2, we see that while accuracy for the medium cellularity patches is very good, the automated method seems to perform less well for normal and low cellularity patches. The Cohen’s Kappa agreements between pathologist #1 vs. automated method and pathologist #2 vs. automated method are κ = 0.42 and κ = 0.43 respectively. In some cases, cellularity fraction of pathology regions may be reported in four groups of 0–25%, 26–50%, 51–75%, and 76–100%. To compare the performance of the pathologists and automated method when residual cellularity is to be categorized in the aforementioned four groups, the Cohen’s Kappa agreement was calculated. The agreement coefficients were found to be κ = 0.55 for pathologist #1 vs. pathologist #2, κ = 0.38 for pathologist #1 and automated method, and κ = 0.42 for pathologist #2 and automated method respectively.
Table 4.
Categorical agreements between pathologists #1, pathologist #2 and the proposed pipeline (automated) for n = 1121 patches of size 512×512 pixels (set C).
| PATHOLOGIST 1 |
|||||
|---|---|---|---|---|---|
| NORMAL | LOW | MEDIUM | HIGH | ||
| Pathologist 2 | Normal | 237 | 5 | 0 | 0 |
| Low | 0 | 202 | 23 | 0 | |
| Medium | 0 | 102 | 197 | 2 | |
| High | 0 | 3 | 155 | 195 | |
| AUTOMATED |
|||||
| NORMAL | LOW | MEDIUM | HIGH | ||
| Pathologist 1 | Normal | 102 | 97 | 34 | 4 |
| Low | 13 | 143 | 152 | 4 | |
| Medium | 4 | 19 | 291 | 61 | |
| High | 5 | 10 | 68 | 114 | |
| AUTOMATED |
|||||
| NORMAL | LOW | MEDIUM | HIGH | ||
| Pathologist 2 | Normal | 105 | 99 | 34 | 4 |
| Low | 10 | 129 | 85 | 1 | |
| Medium | 4 | 28 | 246 | 23 | |
| High | 5 | 13 | 180 | 155 | |
Table 5 shows the degree of absolute intraclass agreements (also known as intraclass correlation—ICC) among the expert pathologists and the proposed automated method in this article.
Table 5.
Intra-class correlation coefficients (ICC) comparing the agreement levels between pathologist #1, pathologist #2 and the proposed pipeline (automated) for n = 1121 patches of size 512×512 pixels (set C)
| ICC COEFFICIENT (95% CI) | |
|---|---|
| Pathologist1 vs. Pathologist2 | 0.89 [0.70, 0.95] |
| Pathologist1 vs. Automated | 0.74 [0.70, 0.77] |
| Pathologist2 vs. Automated | 0.75 [0.71, 0.79] |
Figure 4 shows the heat scatter plot of the agreement between pathologists and the automated method. It is clear from this figure that the cellularity of most of the patches was assessed closely to those of the pathologists as most of the patches laid close to the correlation (black) line. There are some other patches for which the assessment pipeline fails to estimate their true cellularity assigned by the pathologists (located at the top-left or bottom-right of the graphs in Fig. 4) due to failure in nuclei segmentation or classification, and presence of large difference between the cellularity reported by one of the pathologists and that of estimated by the automated pipeline. Figure 5 shows some examples where the automated method fails to correctly estimate the cancer cellularity compared to the pathologists due to the aforementioned reasons. From the 124 patches (11% of patches) where the difference between the automated cellularity measurement and at least one of the pathologists were >30%, 52 (42%) were caused by misclassification of nuclei objects in an entire patch (Fig. 5a, b), 18 (14%) were caused by failure in the nuclei segmentation procedure (Fig. 5a), and 54 (43%) were due to errors in calibration (Fig. 5c).
Figure 4.
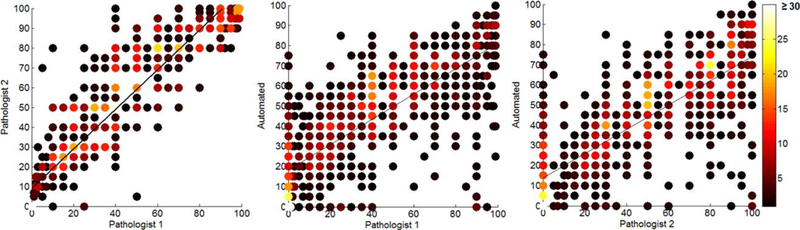
Heat scatter plot of the agreement between the pathologists and the automated method. The color bar indicates the number of patches for each point/circle. As can be seen, slopes of correlation lines for automated method versus pathologist #1 and #2 plots are close to each other and are different from that of the two pathologist’s plot. [Color figure can be viewed at wileyonlinelibrary.com]
Figure 5.
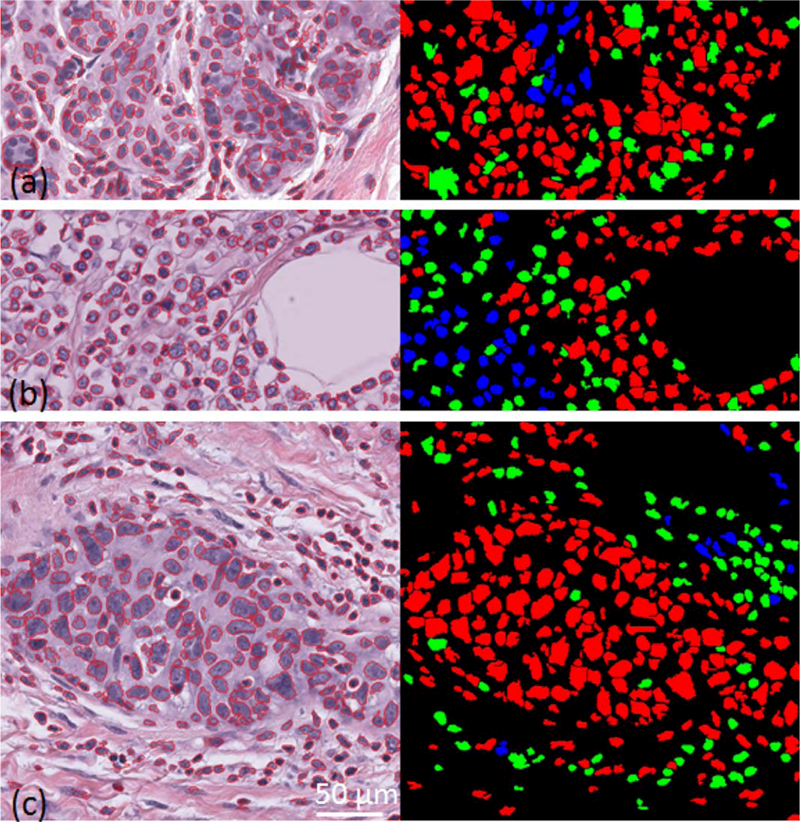
Image examples where the automated pipeline fails to correctly assess the cellularity of patches with reference to that reported by pathologists: (a) part of a patch with normal epithelial tissue structure where segmentation fails to correctly identify true nuclei boundaries due to them being out-of-focus, (b) part of a patch with cancer cells are within fatty tissue; this type of tissue structures were not present in the training set and so was new to the classification pipeline, (c) an example of a patch where the difference between the assessed cellularity by automated method was large compared to at least one pathologist. [Color figure can be viewed at wileyonlinelibrary.com]
To test the performance of our pipeline with top ranked features that are most effective for the nuclei classification task, we used various feature selection methods that are compared in (60). We found that the classification performance was not improved when feature selection methods were used and that our original 125-dimensional feature vector, which was designed using domain knowledge, gave the best results. We have reported the top 10 features chosen by the Fisher’s method (62) in Table 6. As shown, top ranking features correspond to Haralick texture features. This finding also matches with other research article findings in the literature (50).
Table 6.
Top 10 features chosen by Fisher’s feature selection method (62) for the nuclei classification task
| TOP 10 FEATURE RANKS FOR NUCLEI CLASSIFICATION TASK | |
|---|---|
| LVS. BM | B VS. M |
| Cluster Shade at 135° | SD histogram of gray scale pixel intensity |
| Cluster Shade at 0° | Mean histogram of gray scale pixel intensity |
| Cluster Shade at 90° | Cluster Shade at 45° |
| Cluster Shade at 45° | Cluster Shade at 135° |
| Autocorrelation at 135° | Cluster Shade at 90° |
| Sum variance at 135° | Cluster Shade at 0° |
| Autocorrelation at 45° | Correlation at 45° |
| Sum variance at 135° | Correlation at 135° |
| Sum variance at 90° | Energy at 45° |
| Sum variance at 0° | Energy at 135° |
As shown top features correspond to Haralick texture features.
We have also applied the proposed automated method on a whole slide image to find regions of high residual cancer (Fig. 6). As seen in Fig. 6, our proposed method was able to find regions with high residual cancer cellularity while neglecting regions that contained normal components.
Figure 6.
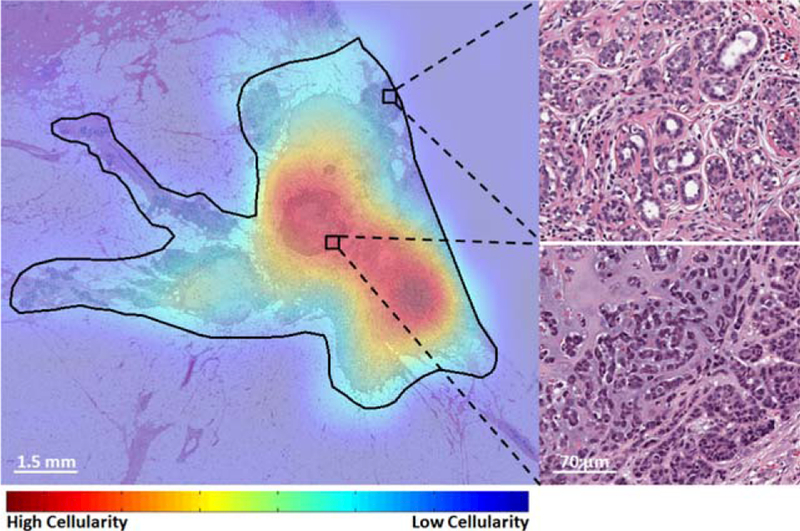
Cellularity assessment heat map from within a tumor bed region (black contour) on a whole slide image with two representing benign (top-right) and malignant (bottom-right) regions. [Color figure can be viewed at wileyonlinelibrary.com]
Discussion and Conclusion
In this study, we developed a framework to automatically estimate residual cancer cellularity from patches within whole slide images of BCa. We extensively validated the framework using a dataset consisting of 64 patients (n = 121 WSIs) and we demonstrated that the automated method correlated well with the pathologist’s assessment of cellularity. This allows cellularity to be automatically assessed over a WSI and can be used as part of a pipeline to assess tumor burden post NAT.
It is possible that improving the accuracy of the nuclei segmentation would also improve results of classification performance, however, in (26) authors used both perfectly and imperfectly segmented nuclei figures to train a classifier to detect malignant objects from breast cancer tissues and found that perfect segmentation does not ensure better classification performance. Asking pathologists to generate accurate segmentation ground truth labels for additional training and testing would have been prohibitively time consuming and, although more sophisticated cell segmentation methods exist, they tend to be too slow to be practical for processing WSIs. For these reasons we decided to not to spend additional time optimizing this step.
The percentage area occupied by malignant nuclei is not the same as the percentage cellularity estimated by pathologists as it does not take into account the other cellular components present in the image. We, therefore, had to convert the percentage of nuclei to cellularity using a calibration curve based on a training dataset. We did not take into account local variations in cell density within each patch which may invalidate our assumption that there is a linear relationship between nuclei area and cellularity and this is a potential source of error.
We chose to use an SVM classifier as we found that it performed well on related tasks (63,64). We did explore the use of a random forest classifier on a small subset of our nuclei figure classification dataset and found that the performance of the SVM was better. There is a slight imbalance in the datasets for training nuclei figure classification but we found that the performance of the SVM classifier did not change when we applied weighting to correct for this, suggesting that the imbalance did not affect accuracy.
While the aim of this study was to develop an automated method to assess the residual tumor cellularity in image patches of BCa, it is worth mentioning that it is clinically desirable to be able to identify patients who have an estimated residual disease of lesser than 20% after the completion of NAT (near complete or complete pathologic response). Using our proposed automated pipeline, we were able to correctly identify 68%, and 75% of the patches with residual tumor cellularity of lesser than 20% identified by pathologist #1 and pathologist #2 respectively.
In order to have a completely automated system to assess residual tumor after NAT, areas of tumor bed and ductal carcinoma in-situ (DCIS) also need to be identified. Tumor bed regions are where tumor cells are currently present or might have been present when the neoadjuvant treatment started. To identify tumor bed regions, pathologists look at features indicating the process of tissue healing after it was affected by the therapy. Regions with features like the high concentration of fibroblast cells within fibrosis, foamy macrophages with pigments, calcifications, aggregates of lymphocytes, areas of tumor necrosis, thin collagen bundle strands, and small capillaries are indications for the tissue that is healing and hence are considered as a part of tumor bed. It is technically a difficult task to identify each of the mentioned features in the pathology slides using image analysis methods with hand-crafted features due to their high variability and subjectivity in their identification. However, CNN learning based methods could be employed to tackle this problem due to their ability to derive suitable features from the data itself.
Acknowledgments
This research is funded by the Canadian Cancer Society (grant number 703006) and with added support from the National Institutes of Health (grant number U24CA199374–01).
Grant sponsor: Canadian Cancer Society, Grant number: grant number 703006
Grant sponsor: National Institutes of Health, Grant number: U24CA199374-01
Literature Cited
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin 2016;66:7–30. [DOI] [PubMed] [Google Scholar]
- 2.Thompson AM, Moulder-Thompson SL. Neoadjuvant treatment of breast cancer. Ann Oncol 2012;23:231–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faneyte IF, Schrama JG, Peterse JL, Remijnse PL, Rodenhuis S, van de Vijver MJ. Breast cancer response to neoadjuvant chemotherapy: Predictive markers and relation with outcome. Br J Cancer 2003;88:406–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaufmann M Recommendations from an international expert panel on the use of neoadjuvant (primary) systemic treatment of operable breast cancer: An update. J Clin Oncol 2006;24:1940–1949. [DOI] [PubMed] [Google Scholar]
- 5.Schott AF, Hayes DF. Defining the benefits of neoadjuvant chemotherapy for breast cancer. J Clin Oncol 2012;30:1747–1749. [DOI] [PubMed] [Google Scholar]
- 6.Hermanek P, Wittekind C. Residual tumor (R) classification and prognosis. Semin Surg Oncol 1994;10:12–20. [DOI] [PubMed] [Google Scholar]
- 7.Nahleh Z, Sivasubramaniam D, Dhaliwal S, Sundarajan V, Komrokji R. Residual cancer burden in locally advanced breast cancer–a superior tool. Pdf Med Oncol 2008; 15:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mougalian SS, Hernandez M, Lei X, Lynch S, Kuerer HM, Symmans WF, Theriault RL, Fornage BD, Hsu L, Buchholz TA, et al. Ten-year outcomes of patients with breast cancer with cytologically confirmed axillary lymph node metastases and pathologic complete response after primary systemic chemotherapy. JAMA Oncol 2015; 1439:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraser Symmans W, Peintinger F, Hatzis C, Rajan R, Kuerer H, Valero V, Assad L, Poniecka A, Hennessy B, Green M, et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J Clin Oncol 2007;25:4414– 4422. [DOI] [PubMed] [Google Scholar]
- 10.Chagpar AB, Middleton LP, Sahin AA, Dempsey P, Buzdar AU, Mirza AN, Ames FC, Babiera GV, Feig BW, Hunt KK, et al. Accuracy of physical examination, ultrasonography, and mammography in predicting residual pathologic tumor size in patients treated with neoadjuvant chemotherapy. Ann Surg 2006;243:257–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu Jing L, Hong Z, Bing O, Bao Ming L, Xiao Yun X, Wen Jing Z, Xin Bao Z, Zi Zhuo Z, Hai Yun Y, Hui Z. Ultrasonic elastography features of phyllodes tumors of the breast: A clinical research. PLoS One 2014;9:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar S, Ashok Badhe B, Krishnan KM, Sagili H. Study of tumour cellularity in locally advanced breast carcinoma on neo-adjuvant chemotherapy. J Clin Diagn Res 2014;8:9–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sahoo S, Lester SC. Pathology of breast carcinomas after neoadjuvant chemotherapy an overview with recommendations on specimen processing and reporting. Arch Pathol Lab Med 2009;133:633–642. [DOI] [PubMed] [Google Scholar]
- 14.Von Minckwitz G, Untch M, Uwe Blohmer J, Costa SD, Eidtmann H, Fasching PA, Gerber B, Eiermann W, Hilfrich J, Huober J, et al. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J Clin Oncol 2012;30:1796–1804. [DOI] [PubMed] [Google Scholar]
- 15.Smits AJJ, Alain Kummer J, de Bruin PC, Bol M, van den Tweel JG, Seldenrijk KA, Willems SM, Johan A. Offerhaus G, de Weger RA, van Diest PJ, et al. The estimation of tumor cell percentage for molecular testing by pathologists is not accurate. Mod Pathol 2014;27:168–174. [DOI] [PubMed] [Google Scholar]
- 16.Al-Adhadh AN, Cavill I. Assessment of cellularity in bone marrow fragments. J Clin Pathol 1983;36:176–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abrial C, Thivat E, Tacca O, Durando X, Mouret-Reynier MA, Gimbergues P, Penault-Llorca F, Chollet P. Measurement of residual disease after neoadjuvant chemotherapy. J Clin Oncol 2008;26:3093–3094. [DOI] [PubMed] [Google Scholar]
- 18.Peintinger F, Sinn B, Hatzis C, Albarracin C, Downs-Kelly E, Morkowski J, Gould R, Fraser Symmans W. Reproducibility of residual cancer burden for prognostic assessment of breast cancer after neoadjuvant chemotherapy. Mod Pathol 2015;28: 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naidoo K, Parham DM, Pinder SE. An audit of residual cancer burden reproducibility in a UK context. Histopathology 2017;70:217–222. [DOI] [PubMed] [Google Scholar]
- 20.Park CK, Jung W-H, Koo JS. Pathologic evaluation of breast cancers after neoadjuvant therapy. J Pathol Transl Med 2016;50:173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Provenzano E, Bossuyt V, Viale G, Cameron D, Badve S, Denkert C, MacGrogan G, Penault-Llorca F, Boughey J, Curigliano G, et al. Standardization of pathologic evaluation and reporting of postneoadjuvant specimens in clinical trials of breast cancer: Recommendations from an international working group. Mod Pathol 2015;28:1185–1201. [DOI] [PubMed] [Google Scholar]
- 22.Rajan R, Poniecka A, Smith TL, Yang Y, Frye D, Pusztai L, Fiterman DJ, Gal-Gombos E, Whitman G, Rouzier R, et al. Change in tumor cellularity of breast carcinoma after neoadjuvant chemotherapy as a variable in the pathologic assessment of response. Am Cancer Soc 2004;100:1365–1373. [DOI] [PubMed] [Google Scholar]
- 23.Sta˚lhammar G, Fuentes Martinez N, Lippert M, Tobin NP, Mølholm I, Kis L, Rosin G, Rantalainen M, Pedersen L, Bergh J, et al. Digital image analysis outperforms manual biomarker assessment in breast cancer. Mod Pathol 2016;2:1–12. [DOI] [PubMed] [Google Scholar]
- 24.Wienert S, Heim D, Saeger K, Stenzinger A, Beil M, Hufnagl P, Dietel M, Denkert C, Klauschen F. Detection and segmentation of cell nuclei in virtual microscopy images: A minimum-model approach. Sci Rep 2012;2:503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irshad H, Roux L, Racoceanu D. Methods for nuclei detection, segmentation, and classification in digital histopathology: A review current status and future potential. IEEE Rev Biomed Eng 2014;7:97–114. [DOI] [PubMed] [Google Scholar]
- 26.Boucheron LE, Manjunath BS, Harvey NR. Use of Imperfectly Segmented Nuclei in the Classification of Histopathology Images of Breast Cancer, Vol. 1 In IEEE International Conference on Acoustics Speech and Signal Processing (ICASSP); 2010. pp 666–669. [Google Scholar]
- 27.Yang X, Li H, Zhou X. Nuclei segmentation using marker-controlled watershed, tracking using mean-shift, and Kalman filter in time-lapse microscopy. IEEE Trans Circ Syst I 2006;53:2405–2414. [Google Scholar]
- 28.Cheng J, Rajapakse JC. Segmentation of clustered nuclei with shape markers and marking function. IEEE Trans Biomed Eng 2009;56:741–748. [DOI] [PubMed] [Google Scholar]
- 29.Veta M, van Diest PJ, Kornegoor R, Huisman A, Viergever MA, Pluim JPW. Automatic nuclei segmentation in H&E stained breast cancer histopathology images. PLoS One 2013;8:e70221–e70jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mouelhi A, Sayadi M, Fnaiech F, Mrad K, Romdhane KB. Automatic image segmentation of nuclear stained breast tissue sections using color active contour model and an improved watershed method. Biomed Signal Process Control 2013;8:421–436. [Google Scholar]
- 31.Al-Kofahi Y, Lassoued W, Lee W, Roysam B. Improved automatic detection and segmentation of cell nuclei in histopathology images. IEEE Trans Bio-Med Eng 2010;57: 841–852. [DOI] [PubMed] [Google Scholar]
- 32.Liu C, Shang F, Ozolek JA, Rohde GK. Detecting and segmenting cell nuclei in two-dimensional microscopy images. J Pathol Inform 2016;7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vink JP, Van Leeuwen MB, Van Deurzen CHM, De Haan G. Efficient nucleus detector in histopathology images. J Microsc 2013;249:124–135. [DOI] [PubMed] [Google Scholar]
- 34.Jung C, Kim C, Wan Chae S, Oh S. Unsupervised segmentation of overlapped nuclei using Bayesian classification. IEEE Trans Biomed Eng 2010;57:2825–2832. [DOI] [PubMed] [Google Scholar]
- 35.Sirinukunwattana K, Ahmed Raza SE, Wah Tsang Y, Snead DRJ, Cree IA, Rajpoot NM. Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images. IEEE Trans Med Imag 2016;35:1196–1206. [DOI] [PubMed] [Google Scholar]
- 36.Peikari M, Martel AL. Automatic cell detection and segmentation from H and E stained pathology slides using colorspace decorrelation stretching. SPIE Med Imaging 2016. [Google Scholar]
- 37.Kothari S, Phan JH, Stokes TH, Wang MD. Pathology imaging informatics for quantitative analysis of whole-slide images. J Am Med Inform Assoc 2013;20:1099–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gandomkar Z, Brennan PC, Mello Thoms C. Computer-based image analysis in breast pathology. J Pathol Inform 2016;43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang M, Zhou X, Li F, Huckins J, King RW, Wong STC. Novel cell segmentation and online SVM for cell cycle phase identification in automated microscopy. Bioinformatics 2008;24:94–101. [DOI] [PubMed] [Google Scholar]
- 40.Huang K, Murphy RF. From quantitative microscopy to automated image understanding. J Biomed Opt 2004;9:893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doyle S, Agner S, Madabushi A, Feldman M, Tomaszewski J. Automated grading of breast cancer histopathology using spectral clustering with textural and architectural image features;. In IEEE International Symposium on Biomedical Imaging: From Nano to Macro. Paris, France; 2008. pp 496–499. [Google Scholar]
- 42.Carraro DM, Elias EV, Andrade VP. Ductal carcinoma in situ of the breast: Morphological and molecular features implicated in progression. Biosci Rep 2013;34:19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fukuma K, Surya Prasath VB, Kawanaka H, Aronow BJ, Takase H. A Study on feature extraction and disease stage classification for glioma pathology images. Procedia-Procedia Comput Sci 2016;96:2150–2156. [Google Scholar]
- 44.Lomenie N, Racoceanu D. Point set morphological filtering and semantic spatial configuration modeling: Application to microscopic image and bio-structure analysis. Pattern Recogn 2012;45:2894–2911. [Google Scholar]
- 45.Bamford P, Lovell B. Unsupervised cell nucleus segmentation with active contours. Signal Process 1998;71:203–213. [Google Scholar]
- 46.Tu Z, Bai X. Auto-context and its application to high-level vision tasks and 3D brain image segmentation. IEEE Trans Pattern Anal Mach Intel 2010;32:1744–1757. [DOI] [PubMed] [Google Scholar]
- 47.Mourice George Y, Helmy Zayed H, Ismail Roushdy M, Mohamed Elbagoury B. Remote computer-aided breast cancer detection and diagnosis system based on cytological images. IEEE Syst J 2014;8:949–964. [Google Scholar]
- 48.Filipczuk P, Fevens T, Krzyzak A, Monczak R. Computer-aided breast cancer diagnosis based on the analysis of cytological images of fine needle biopsies. IEEE Trans Med Imag 2013;32:2169–2178. [DOI] [PubMed] [Google Scholar]
- 49.XiongKim XY, Baek Y. Analysis of breast cancer using data mining & statistical techniques Towson, Maryland: Softw Eng Artif Intell Netw Parallel/Distrib Comput A; 2005;82–87. [Google Scholar]
- 50.Yamada M, Saito A, Yamamoto Y, Cosatto E, Kurata A, Nagao T, Tateishi A, Kuroda M. Quantitative nucleic features are effective for discrimination of intraductal proliferative lesions of the breast. J Pathol Inform 2016;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan Y, Failmezger H, Rueda OM, Raza Ali H, Gr€af S, Chin S-F, Schwarz RF, Curtis C, Dunning MJ, Bardwell H, et al. Quantitative image analysis of cellular heterogeneity in breast tumors complements genomic profiling. Sci Transl Med 2012;4: 157ra143. [DOI] [PubMed] [Google Scholar]
- 52.Weyn B, Van De Wouwer G, Van Daele A, Scheunders P, Van Dyck D, Van Marck E, Jacob W. Automated breast tumor diagnosis and grading based on wavelet chromatin texture description. Cytometry 1998;33:32–40. [DOI] [PubMed] [Google Scholar]
- 53.Petushi S, Garcia FU, Haber MM, Katsinis C, Tozeren A. Large-scale computations on histology images reveal grade-differentiating parameters for breast cancer. BMC Med Imag 2006;6:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang P, Hu X, Li Y, Liu Q, Zhu X. Automatic cell nuclei segmentation and classification of breast cancer histopathology images. Signal Process 2016;122:1–13. [Google Scholar]
- 55.Janowczyk A, Madabhushi A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J Pathol Inform 2016;7:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Romo-Bucheli D, Janowczyk A, Gilmore H, Romero E, Madabhushi A. A deep learning based strategy for identifying and associating mitotic activity with gene expression derived risk categories in estrogen receptor positive breast cancers. Cytometry Part A 2017;91A:566–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sirinukunwattana K, Ahmed Raza SE, Tsang Y-W, Snead D, Cree I, Nasir RB. A spatially constrained deep learning framework for detection of epithelial tumor nuclei in cancer histology images. International Workshop on Patch-based Techniques in Medical Imaging, Vol 9467 Lecture Notes in Computer Science. Munich, Germany: Springer International Publishing; 2015. pp. 154–162. [Google Scholar]
- 58.Wang L, He D-C. Texture classification using texture spectrum. Pattern Recogn 1990;23:905–910. [Google Scholar]
- 59.Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans Syst, Man Cybernet 1973;3:610–621. [Google Scholar]
- 60.Chang C-C, Lin C-J. LIBSVM: A library for support vector machines. ACM Trans Intel Syst Technol 2011;2:27 1–27:27 [Google Scholar]
- 61.Giorgio R Report: Feature selection techniques for classification, Vol 1; 2016; arXiv preprint arXiv:1607.01327. [Google Scholar]
- 62.Gu Q, Li Z, Han J. Generalized Fisher Score for Feature Selection 2012. [Google Scholar]
- 63.Peikari M, Gangeh MJ, Zubovits M, Clarke JG, Martel A. Triaging diagnostically relevant regions from pathology whole slides of breast cancer: A texture based approach. IEEE Trans Med Imag 2015; pp. 307–315. [DOI] [PubMed] [Google Scholar]
- 64.Peikari M, Zubovits JT, Clarke GM, Martel AL. Clustering Analysis for Semi-supervised Learning Improves Classification Performance of Digital Pathology. In Machine Learning in Medical Imaging −6th International Workshop {MLMI} 2015, Held in Conjunction with {MICCAI} 2015, Munich, Germany, October 5, 2015, Proceedings; 2015. pp 263–270. [Google Scholar]


