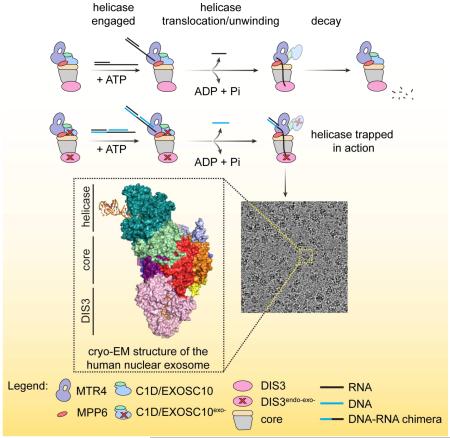
SUMMARY
The ribonucleolytic RNA exosome interacts with RNA helicases to degrade RNA. To understand how the 3′ to 5′ Mtr4 helicase engages RNA and the nuclear exosome, we reconstituted fourteen-subunit Mtr4-containing RNA exosomes from Saccharomyces cerevisiae, Schizosaccharomyces pombe and human and show that they unwind structured substrates to promote degradation. We loaded a human exosome with an optimized DNA-RNA chimera that stalls MTR4 during unwinding and determined its structure to an overall resolution of 3.45 Å by cryo-EM. The structure reveals an RNA-engaged helicase atop the non-catalytic core, with RNA captured within the central channel and DIS3 exoribonuclease active site. MPP6 tethers MTR4 to the exosome through contacts to the RecA domains of MTR4. EXOSC10 remains bound to the core, but its catalytic module and cofactor C1D are displaced by RNA-engaged MTR4. Competition for the exosome core may ensure that RNA is committed to degradation by DIS3 when engaged by MTR4.
Keywords: RNA degradation, ribonuclease, helicase, SF2, RNA-protein complex, translocase, exosome, nuclear
eToc
The cryo-EM structure of the human nuclear exosome-Mtr4 complex loaded with an RNA substrate reveals mechanistic insights into how the helicase unwinds structured RNA substrates to promote degradation
Graphical abstract
INTRODUCTION
Transcripts produced by RNA polymerase I, II or III can encode proteins or become integral components of ribonucleoprotein machineries such as the ribosome, the spliceosome, and telomerase. Pervasive transcription of the genome also generates a plethora of noncoding transcripts (Birney et al., 2007). A fundamental player in processing, quality control, and degradation of a variety of eukaryotic RNA transcripts is the RNA exosome, a multi-subunit 3′ to 5′ ribonucleolytic complex. In the cytoplasm, the RNA exosome is involved in mRNA turnover as well as in co-translational mRNA surveillance during nonsense-mediated decay, non-stop decay and no-go decay. In the nucleus, the RNA exosome functions in maturation and/or quality control of ribosomal RNAs, pre-mRNAs, transfer RNAs, small nuclear and nucleolar RNAs as well as in turnover of unstable or unwanted noncoding transcripts. In addition, the nuclear RNA exosome participates in targeted degradation of meiotic transcripts and assembly of heterochromatin in fission yeast (Chlebowski et al., 2013; Kilchert et al., 2016; Schneider and Tollervey, 2013; Zinder and Lima, 2017). In humans, the exosome is implicated in cellular pathways involved in antiviral defense against HIV-1 and HBV, regulation of developmental signaling cascades in erythroid differentiation, and genome modifications pertinent to immunoglobulin diversification (Kilchert et al., 2016; Lim et al., 2017; McIver et al., 2014; Molleston and Cherry, 2017; Zinder et al., 2016). Given its ubiquitous role in RNA metabolism, it is not surprising that genetic lesions in subunits and co-factors of the exosome are linked to pathological conditions in human including multiple myeloma, pontocerebellar hypoplasia, and retinitis pigmentosa (Morton et al., 2018; Weißbach et al., 2015).
The nuclear RNA exosome includes a nine-subunit core (Exo9) and two associated ribonucleases. Exo9 consists of a pseudohexameric ring of six RNAse PH-like proteins that is capped by three S1/KH domain proteins to form a barrel-like structure with a prominent central channel (Hernández et al., 2006; Liu et al., 2006). Human and S. cerevisiae Exo9 cores are catalytically inert (Dziembowski et al., 2007; Liu et al., 2006) with catalytic activity imparted by the processive exo- and endoribonuclease Dis3 (Rrp44/EXOSC11) (Bonneau et al., 2009; Dziembowski et al., 2007; Lebreton et al., 2008; Liu et al., 2006; Schneider et al., 2009) and the distributive exoribonuclease Rrp6 (EXOSC10/PM/SCL-100) (Allmang et al., 1999; Briggs et al., 1998). As nomenclature differs for S. cerevisiae, S. pombe and human proteins, a list is provided (Figure S1A). In S. cerevisiae, Dis3 binds the bottom of the PH-like ring (Bonneau et al., 2009; Hernández et al., 2006; Wang et al., 2007) in two distinct conformations: one that binds short substrates (<14 nucleotides (nt)) that bypass the Exo9 central channel (open conformation), and one that binds longer substrates (>30 nt) that thread through the Exo9 central channel (closed conformation) (Bonneau et al., 2009; Liu et al., 2014, 2016, Makino et al., 2013, 2015; Zinder et al., 2016). The Rrp6 catalytic module binds the top of the Exo9 core and contacts the S1/KH ring while its C-terminal Exosome Associating Region (EAR) wraps around part of the PH-like ring (Makino et al., 2013; Wasmuth et al., 2014).
Several functions of the nuclear RNA exosome require Mtr4 (SKIV2L2/Dob1), a 3′ to 5′ DExH-box RNA helicase. Mtr4 is involved in exosome-mediated maturation of the large ribosomal subunit 5.8S rRNA (de la Cruz et al., 1998; Tafforeau et al., 2013) and, in human, participates in the removal of internal transcribed spacer 1 (ITS1) for maturation of the small ribosomal subunit 18S rRNA (Sloan et al., 2013; Tafforeau et al., 2013). Mtr4 also works in concert with the nuclear exosome in regulating programmed DNA mutagenesis at DNA/RNA loci during class switch recombination (Lim et al., 2017).
Mtr4 is a central subunit of many protein complexes, namely TRAMP, NEXT, and PAXT/PPC, that aid in RNA quality control by the exosome. TRAMP targets cryptic unstable transcripts (CUTs) and hypomodified tRNAs for destruction by the exosome (Kadaba et al., 2004; Wang et al., 2008; Wyers et al., 2005). NEXT and PAXT/PPC are metazoan-specific Mtr4-containing complexes that were first isolated in human cells (Lubas et al., 2011; Meola et al., 2016; Ogami et al., 2017). NEXT is involved in turnover of noncoding RNAs (PROMPTs and enhancer RNAs) and aberrant 3′ extended transcripts of snRNAs, snoRNAs, and telomerase RNA (Lubas et al., 2015; Tseng et al., 2015). PAXT/PPC targets a more specific class of noncoding RNAs that are typically longer and more extensively polyadenylated than NEXT substrates (Meola et al., 2016; Ogami et al., 2017).
Mtr4 interacts with the exosome via two conserved cofactors, Rrp47 (C1D) and Mpp6. Rrp47 associates with the N-terminus of Rrp6, forming a composite surface that binds an N- terminal helix of Mtr4 (Schuch et al., 2014; Stead et al., 2007). Mpp6 tethers Mtr4 to the exosome core through contacts to Rrp40 (Falk et al., 2017; Lubas et al., 2011; Schilders et al., 2007; Wasmuth et al., 2017). The structural and mechanistic bases underlying the intimate collaboration and interplay between the enzymatic activities of Mtr4 and the RNA exosome remain elusive. Additionally, high resolution structures for Mtr4 bound to nuclear exosomes are lacking, and reported structures of the RNA exosome from human are limited to the nine-subunit core (Liu et al., 2006) and EXOSC10 catalytic module (Januszyk et al., 2011).
Here, we present biochemical characterization of Mtr4-nuclear exosome complexes from the budding yeast Saccharomyces cerevisiae, the fission yeast Schizosaccharomyces pombe and human. Mtr4 helicase activity is enhanced in exosome complexes relative to Mtr4 alone, enabling Dis3-dependent degradation of RNA in RNA or RNA/DNA duplexes. We designed a substrate that engages and then stalls the human MTR4 helicase after it passes RNA through the exosome central channel to the DIS3 exoribonuclease active site and determined a structure of an RNA-engaged human exosome complex by single particle cryo-EM to an overall resolution of 3.45 Å. The structure reveals the architecture of the human RNA exosome and provides atomic details for MTR4 in the process of unwinding and delivering RNA into the exosome, MPP6 contacts to MTR4, and an RNA path from MTR4 through the exosome central channel and into the exoribonuclease active site of DIS3.
RESULTS
Unwinding activities of Mtr4-nuclear RNA exosomes
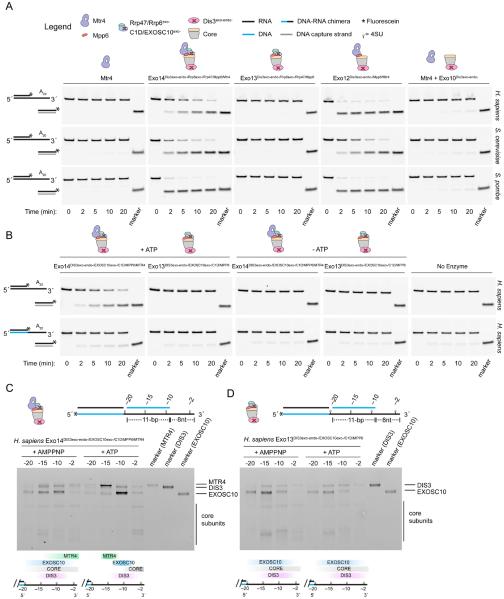
To examine RNA unwinding activity, exosomes from S. cerevisiae, S. pombe and human were reconstituted with catalytically inactive versions of the ribonucleases, or subcomplexes therein, and assayed for ATP-dependent strand displacement in comparison to Mtr4 alone using a substrate consisting of a 16-bp RNA duplex with a 20-nt poly(A) 3′ overhang (Figure 1A). Mtr4 displayed weak helicase activity in isolation, consistent with previous findings using S. cerevisiae Mtr4 (Jia et al., 2012). In contrast, Mtr4 helicase activities were stimulated when incorporated in exosomes containing Mpp6 or Mpp6 and Rrp47/Rrp6.
Figure 1. Mtr4 unwinding and translocation activities in RNA exosomes.
(A) Unwinding time-course assays using a 3′ A20 tailed RNA duplex substrate. Marker indicates position of the fluorescein-labeled (*) displaced strand captured by a DNA trap oligo (gray line). Cartoons represent composition of complexes used. Legend applies to all panels.
(B) Comparative unwinding assays for human complexes using the RNA (top) or DNA-RNA chimera (bottom) translocation strand. Substrate RNA or DNA is indicated by black and blue lines, respectively.
(C and D) UV-induced crosslinking of human exosome complexes to tripartite substrates in the presence of AMPPNP or ATP. Substrates schematically represented (top). Each substrate contains a 5′ fluorescein (*) and a single 4-thiouridine (4SU) at indicated positions in the DNA-RNA chimera translocation strand. Markers shown for indicated subunits (right) with schematics below each gel indicating qualitative changes in crosslinking patterns (bottom). Gel images in panels A-D are representative of three independent experiments.
See also Figure S1.
To capture a “snapshot” of Mtr4-dependent threading of RNA into the exosome, we set out to engineer a substrate that would trap the complex in action. Mtr4 can translocate 3′ to 5′ on RNA (translocation strand) but not on DNA, and it is capable of displacing either DNA or RNA (displacement strand) opposite the translocation strand (Jia et al., 2012; Lim et al., 2017). Utilizing this information, we prepared a substrate containing a DNA-RNA chimera that stalls Mtr4 at the junction after translocation on RNA. Indeed, MTR4- and ATP-dependent duplex unwinding by human exosomes was compromised with a DNA-RNA chimeric translocation strand (Figure 1B).
To confirm that MTR4 translocates to a position proximal to the RNA/DNA duplex, we analyzed UV-induced crosslinking of human complexes to substrates containing 4-thiouridine (4SU) at positions 2, 10, 15 or 20 nt from the 3′ end of the RNA translocation strand (Figure 1C). A DNA strand was annealed to the RNA segment of the chimera to provide an 8 nt 3′ overhang to optimize interactions with MTR4 and minimize direct binding by either EXOSC10 (Rrp6) or DIS3. RNA crosslinks to MTR4 were evident when 4SU is positioned at the 3′ end in the presence of a non-hydrolyzable ATP analog, AMPPNP. Addition of ATP increased RNA crosslinks to MTR4 when 4SU is placed 15 nucleotides from the 3′ end, EXOSC10 when 4SU is placed 10 nucleotides from the 3′ end, and to components of the Exo9 core when 4SU is placed 2 nucleotides from the 3′ end. In contrast, crosslinking patterns did not shift using exosomes lacking MTR4 (Figure 1D). These data are consistent with the helicase stalling at the DNA-RNA junction of the translocation strand with the extruded RNA passing by EXOSC10 and into the Exo9 central channel.
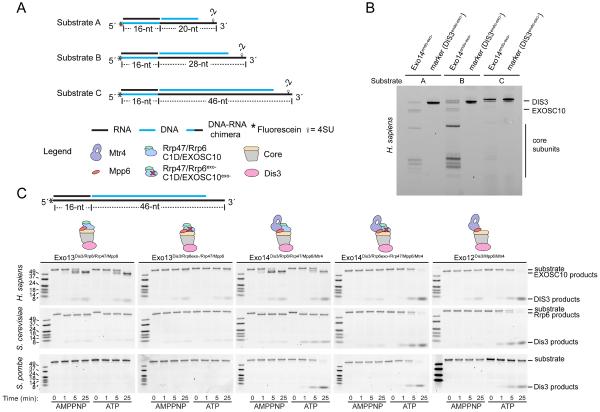
We next determined the ideal length of the translocation strand to reach DIS3 by analyzing UV-induced crosslinking to human RNA exosomes with substrates containing 4SU-labeled DNA-RNA translocation strands with varying RNA lengths (20 nt, 28 nt or 46 nt) (Figure 2A). The 20 nt 3′ RNA substrate generated crosslinking patterns to several Exo9 core subunits and to EXOSC10, with minimal crosslinking to DIS3 (Figure 2B). When extended to 28 nt, crosslinking patterns to Exo9 subunits increased. When extended to 46 nt, crosslinking was largely restricted to DIS3. These data are consistent with MTR4 positioned at the top of the exosome, passing RNA through the central channel and into DIS3.
Figure 2. Crosslinking to human MTR4-exosomes and helicase-dependent degradation.
(A) Schematic representation of tripartite substrates (A-C) used for crosslinking in panel B. Legend applies to all panels. Substrates include a DNA-RNA chimera translocation strand and two complementary strands composed of RNA (black) complementary to the DNA portion of the translocation strand, and DNA (blue) complementary to a portion of the RNA translocation strand. Each substrate includes a 5′ fluorescein and one 4SU at position −2 in the DNA-RNA chimera.
(B) Crosslinking patterns for H. sapiens Exo14DIS3exo-endo-/EXOSC10exo-/C1D/MPP6/MTR4 using substrates in (A) in the presence of ATP. Gel image is representative of three independent experiments. Markers for DIS3 adducts with individual substrates next to each sample.
(C) RNA decay assays using a substrate with RNA for the entire translocation strand. Time-course assays with indicated complexes in the presence of a non-hydrolyzable ATP analogue AMPPNP (left) or ATP (right). Gel images are representative of two independent experiments for S. cerevisiae and three independent experiments for S. pombe and human.
See also Figure S1.
Mtr4-nuclear RNA exosomes promote helicase-dependent decay
To show that Mtr4 unwinds substrates to promote Dis3-dependent decay we performed ribonucleolytic assays with yeast and human exosomes and a tripartite substrate with a translocation strand composed of only RNA (Figure 2C). Distributive RNA decay products of Rrp6 were observed for S. cerevisiae and human complexes independent of ATP and MTR4, products that were lost when an Asp to Asn substitution was introduced into the Rrp6 active site (exo-). Rrp6 activity was not observed for S. pombe exosomes, a point of interest for future studies. In the presence of Mtr4, reactions resulted in ATP-stimulated 4–5 nt Dis3 cleavage products for all three Exo14 complexes. Rrp6 activities appear to compete with Mtr4-dependent Dis3 decay as complexes with Rrp6exo- accumulate more Dis3 products in the presence of ATP. We next confirmed that the chimeric DNA-RNA substrate was a substrate for DIS3 and MTR4. Consistent with data presented in Figure 1, exosomes containing the helicase and Mpp6 or Mpp6 and Rrp47/Rrp6 degraded this substrate in an ATP- and Mtr4-dependent fashion (Figure S1B) and catalyzed ATP-dependent displacement of the DNA strand (Figure S1C).
Combined with our prior observations in the budding yeast system (Wasmuth et al., 2017), these data from three evolutionarily divergent organisms provide evidence that Mpp6- or Mpp6/Rrp47-dependent recruitment of Mtr4 to nuclear exosomes promotes unwinding and Dis3-dependent degradation. Moreover, we show that a DNA-RNA chimera with a 46 nt RNA tail may be well suited to capture a human MTR4-exosome complex with RNA threaded through MTR4 and the Exo9 central channel to DIS3. We loaded a human complex containing inactive ribonucleases with this tripartite substrate and purified the resulting complex by gel filtration. Peak fractions that included all fourteen subunits and the desired ATP- and helicase-dependent nucleic acid reaction products (Figure S1D,E) were used for cryo-EM grid preparation and data collection (see STAR Methods for details).
Cryo-EM structure of a human helicase-containing RNA exosome complex
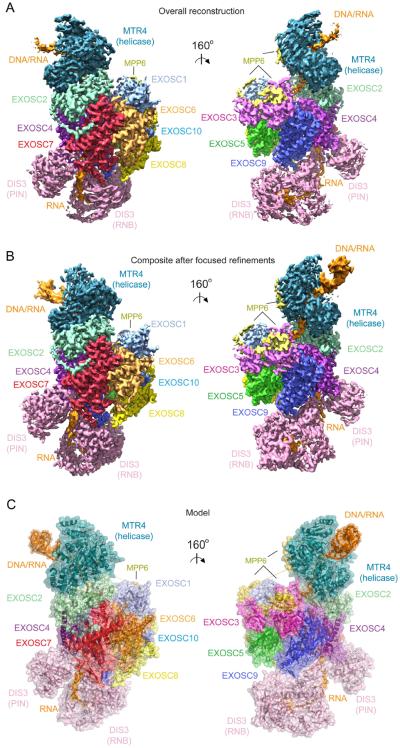
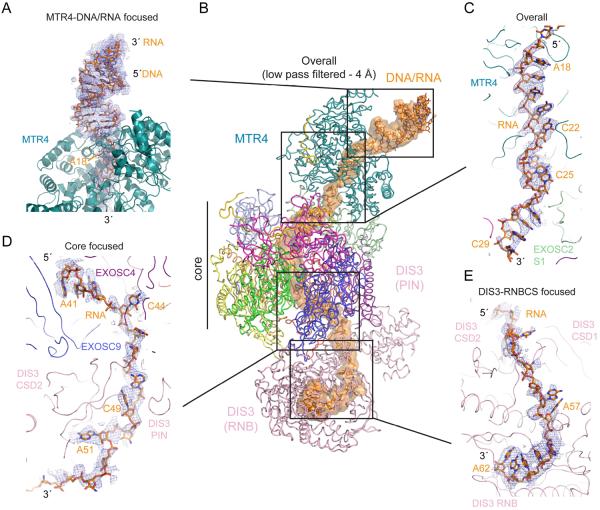
Data processing in cryoSPARC (Punjani et al., 2017) resulted in 278185 particles that classified into four major 3D classes after ab initio 3D reconstructions: the MTR4-exosome complex (45.1%), exosome core with DIS3 (34.0%), and two smaller subcomplexes (10.4% and 10.3%) (Figures S2, S3, S4, S5A and S5B; STAR Methods). Following additional rounds of 2D and 3D classification, the largest class containing the MTR4-exosome complex (122703 particles) was input to 3D refinement in RELION (Scheres, 2012) to reconstruct an overall map with a resolution of 3.45 Å.
Clear electron densities were evident for the core, but electron densities for MTR4 and DIS3 exhibited decreasing order and resolution with increasing distance from the core (Figure 3A and Figure S3). Focused classification and refinement using RELION generated five additional maps with improved continuity and resolution for regions encompassing MTR4 and the DIS3 RNB/CS/S1 (RNBCS) and PIN domains (Figure 3B). These include maps after focused refinement for the core, MPP6 and EXOSC10 (Core; 3.31 Å), after focused refinement for MTR4 (MTR4; 3.54 Å), after 3D classification and focused refinement for MTR4 with more continuous densities for duplex RNA/DNA (MTR4-dsRNA/DNA; 3.91 Å), after 3D classification and focused refinement for the DIS3 RNBCS domains (DIS3-RNBCS; 3.8 Å), and after focused refinement for the PIN domain with a mask encompassing the proximal PH-like proteins (DIS3-PIN; 3.4 Å) (Figure S2). Maps were assessed for estimates of overall and local resolution (Kucukelbir et al., 2014) (Figure S3). Together, these maps were used to build and refine atomic models of the human nuclear RNA exosome engaged in helicase dependent decay (Figures 3C, S4 and S5; Table S1; STAR Methods).
Figure 3. Electron densities and overall structure.
(A) Electron densities from the overall reconstruction.
(B) Composite map generated by combining each map from focused refinement.
(C) Overall structure of the human nuclear exosome. Subunits labeled and uniquely colored.
The structure shows the MTR4 helicase on top of the non-catalytic core with DIS3 located below the core (Figure 3). Subunit arrangement of the non-catalytic core resembles that determined for the human core by x-ray crystallography (Liu et al., 2006). Eleven amino acids of EXOSC10 are observed in the interface between EXOSC8 and EXOSC6 (Figure S6A,B), but its catalytic domain and C1D are not evident in our structure and presumed disordered. DIS3 is positioned at the bottom of the non-catalytic core where it contacts EXOSC4, EXOSC7 and EXOSC9 (Figure S6C-I). Consistent with its high mobility in the reconstructions, the RNB domain is only tethered to the exosome core through minimal contacts to EXOSC9 (Figure S6D) with most core contacts to the CSD and PIN domains (Figure S6E-I). MPP6 tethers MTR4 to the core through interactions with the MTR4 RecA domains, EXOSC1 and EXOSC3.
RNA path from MTR4 to DIS3
Densities for nucleic acid are evident with A-form duplex DNA/RNA and MTR4 at the top of the complex, with the remaining single-stranded RNA threaded through the MTR4 helicase to the central channel and into the DIS3 active site (Figure 4). The human MTR4 helicase core is similar to structures of yeast Mtr4 and includes two RecA-like domains (RecA1 and RecA2), a winged helix domain (WH), and a C-terminal helical bundle (HB) domain (Jackson et al., 2010; Weir et al., 2010). RNA passes through a channel in the helicase between the HB and RecA domains, a hallmark of related RNA and DNA SF2 family helicases such as Ski2 (Halbach et al., 2012) and HEL308 (Büttner et al., 2007), respectively. While largely disordered in overall and MTR4 focused maps, a 4 Å resolution low pass filtered overall map and an additional map generated after focused classification and refinement of an MTR4 subclass revealed continuous densities for the duplex portion of our substrate (Figure 4A,B).
Figure 4. Electron densities and overall path for RNA.
Electron densities (blue mesh) shown superposed on the model for (A) the DNA/RNA duplex and MTR4,
(B) the overall model with electron density for RNA as an orange surface after low pass filtering the overall map to 4.0 A resolution.
(C) densities for single stranded RNA within and below the MTR4 helicase,
(D) the base of the Exo9 core and
(E) entering the DIS3 RNB domain. Densities for the overall path in panel B were obtained by segmenting in Chimera. Electron densities for regions in panels B-E shown from maps as indicated in each panel. Protein chains are labeled and depicted as cartoon or ribbon with RNA as sticks with select residues labeled.
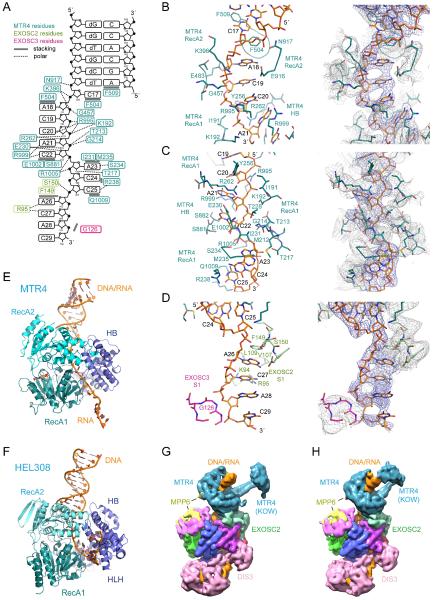
Interactions between MTR4 and the substrate include base stacking interactions and polar contacts to the RNA backbone (Figure 5A). Phe504 and Phe509 reside in the beta hairpin of MTR4 and contact C17 and the base of the duplex, respectively. Phe504 disrupts base stacking interactions between C17 and A18. The nucleobases of A18-C22 are stacked with numerous polar contacts to the RNA backbone in this region (Figure 5B). Base stacking is again disrupted between C22 and A23 via contacts to the C22 nucleobase by Ser881 and Glu1002 and the A23 nucleobase by Ile231 and Met235 with the side chain of Arg1005 projecting toward the phosphate backbone of A23 and C24 (Figure 5C). The nucleobases of A23-C25 stack with the last interactions between MTR4 and substrate involving contacts between the C25 base and Arg238 and Gln1009 (Figure 5C). Interactions between the MTR4 helicase and substrate resemble those observed for the DNA helicase HEL308 insofar as both helicases engage the duplex and disrupt the continuity of base stacking interactions in the single-stranded portion of the substrates before passing the 3′ end to the unique HLH domain in HEL308 or the S1 domain of EXOSC2 in the case of MTR4 (Figure 5D-F). Three MTR4 residues are proximal to the unique 2′ OH moieties in its RNA substrate, but two of these are conserved in HEL308. While DNA and RNA sugar puckers differ, the respective preference for RNA versus DNA for these helicases remains unclear.
Figure 5. Contacts to RNA after duplex unwinding, comparison to HEL308 and conformations of MTR4.
(A) Schematic representation of RNA and protein interactions. MTR4, EXOSC2 and EXOSC3 residues uniquely colored and labeled with polar and stacking interactions indicated by dashed or solid lines. DNA residues lacking the 2′ OH in the DNA-RNA chimera indicated with a lower case d.
(B-D) Stick representation of the structure showing nucleic acid residues and proximal side chains from MTR4, EXOSC2 and EXOSC3 with side chains colored and labeled as in panel A. Electron densities shown to the right overlaying the corresponding areas with blue mesh for RNA and grey mesh for protein.
(E) Ribbon diagram of MTR4 engaged in unwinding RNA in a DNA-RNA chimera
(F) Ribbon diagram of HEL308 engaged in unwinding DNA
(G) Reconstruction shown for one class containing the MTR4 KOW domain in an open conformation.
(H) Reconstruction shown for one class containing the MTR4 KOW domain in a closed conformation.
See also Figure S5.
Additional contacts to the duplex may involve the MTR4 KOW domain, a unique insertion in Mtr4-like SF2 helicases. While disordered in our overall reconstruction, 3D classification uncovered two subclasses with density for the substrate duplex and KOW domain in open and closed conformations (Figure 5G,H and Figure S5D,E). The helical stalk and KOW domain are evident, one in a conformation positioned away from the duplex and the other with the KOW domain apparently engaging the substrate. This latter conformation is consistent with a purported role for the KOW domain in facilitating interactions with structured RNA substrates (Weir et al., 2010).
After extrusion from the helicase, base stacking interactions between C25 and A26 are broken by substrate contacts to EXOSC2 (Figure 5D). The A26 phosphate is within hydrogen bonding distance of the Ser150 backbone amide and side chain hydroxyl while the A26 nucleobase stacks on Phe149 within a hydrophobic cavity that includes Val107. The remaining nucleobases of C27, A28 and C29 are stacked below that of A26 with densities for the RNA backbone proximal to Gly126 in EXOSC3 with Gly126 proximal to the A28 phosphate.
Densities for RNA between positions 29 and 41 are only evident in a low-pass filtered map (Figure 4B), but densities that define positions for the RNA and contacts to the core become clear again after nucleotide A41. A41-A43 nucleobases are stacked with the A41 nucleobase positioned above Phe77 of EXOSC9 with EXOSC9 Arg111, EXOSC4 Arg93 and Lys94 side chains projecting toward the RNA backbone (Figure S7A). The nucleobase of C44 stacks between Thr82-Gly83 and His174 of EXOSC4 with the phosphate of C45 proximal to Arg12 of DIS3. Residues that form the RNA interaction surface between EXOSC4/EXOSC9 are conserved in non-catalytic eukaryotic exosome core structures and are highly similar to residues in phosphorolytic active sites of distantly related PNPase-like exosomes (Liu et al., 2006; Lorentzen and Conti, 2012) (Figure S7B).
Densities for C46-A51 are positioned within a channel created between the DIS3 PIN N-terminal beta hairpin that contacts EXOSC9 and the CSD2 domain of DIS3 (Figure 4D,E; Figure S7A). While densities are less well defined, C52-A56 pass between the DIS3 CSD2 and S1 domains before entering the DIS3 exoribonuclease active site. RNA paths through Dis3 differ in structures of yeast exosomes. In yeast Dis3, RNA does not pass by the CDS2 or S1 domains but instead traverses under CSD1. In our human DIS3 structure, the RNB, CS and S1 domains are oriented similar to budding yeast Dis3 in the open conformation, or to DIS3L2 when bound to RNA (Figure S7C-F). Our data show that human DIS3 can bind RNA in the open conformation after it passes through the central channel. In addition, and despite the high mobility of the DIS3 RNB domain, no class averages were observed with DIS3 adopting a closed conformation (Figure S2). Finally, the RNA path between the exosome core and DIS3 in our structure is ~7 nt longer than observed in yeast through-channel structures, perhaps consistent with the observation that EXOSC10 inactivation results in a 3′ extended 5.8S rRNA species that is 10 nt longer in humans than in yeast (Tafforeau et al., 2013).
The MTR4 helicase is tethered to the exosome via EXOSC2 and MPP6
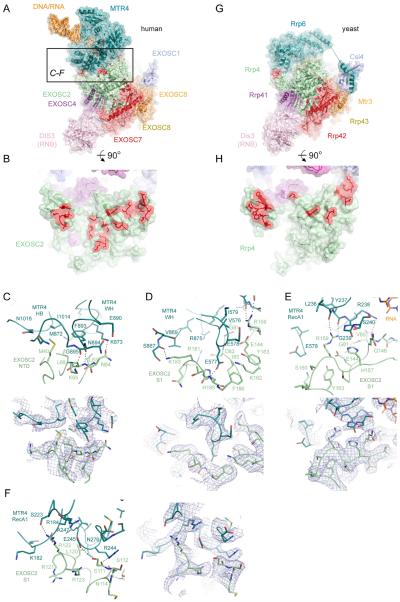
Atomic details for contacts between MTR4 and exosome core have not been reported. Here, we reveal that MTR4 contacts to the core are restricted to interactions between the RecA1, WH, and HB domains of MTR4 and the NTD and S1 domains of EXOSC2 (Figure 6A-F). Contacts include interactions between conserved residues in both proteins in an interface that buries 1440 Å2 of total surface area (Figure S7G). The NTD of EXOSC2 contacts the MTR4 HB and WH domains mainly through an aliphatic surface, centered on EXOSC2 Met40 and Leu66, that engages a hydrophobic pocket on MTR4. The remainder of interactions occur between the S1 domain of EXOSC2 that establishes contacts to residues within the MTR4 RecA1 and WH domains (Figure 6C-F). Several conserved side chain contacts are established in this interface, however several side chain to main chain contacts are of particular note as reciprocal conservation is not readily apparent by analysis of amino acid side chain identities (Figure S7G). These include EXOSC2 Arg159 side chain contacts to backbone carbonyl oxygen atoms of MTR4 Leu236 and Tyr237, MTR4 Glu577 side chain contacts to the backbone amide of EXOSC2 His185, and MTR4 Arg244 side chain contacts to the backbone carbonyl atoms of EXOSC2 Ser111 and Ser112 (Figure 6C-F).
Figure 6. MTR4 contacts to the exosome core and comparison to yeast Rrp6-core interactions.
(A) Overall structure of the human MTR4 RNA exosome complex with a black box indicating the region of interactions with EXOSC2 shown in panels C-F.
(B) Rotated view of panel A to display the EXOSC2 surface that contacts MTR4 with MTR4 removed. Amino acids that engage in direct contact to MTR4 are highlighted in red.
(C-F) Contacts between MTR4 and EXOSC2 N-terminal domain (C) and S1 domain (D-F) with subunits, domains, and side chains labeled and colored as in Figure 3 in ribbon representation with side chains as sticks. Corresponding electron densities (blue mesh) are shown below (D-E) or beside (F) respective panels.
(G) Overall structure of the S. cerevisiae RNA exosome complex (PDBID 5K36).
(H) Rotated view of panel G to display the Rrp4 surface that contacts Rrp6 with Rrp6 removed. Amino acids that engage in direct contact to Rrp6 are highlighted in red.
See also Figure S7G,H.
It is noteworthy that interaction surfaces between EXOSC2 and MTR4 in the human system overlap with Rrp4 surfaces that engage Rrp6 in the yeast system (Wasmuth et al., 2014) (Figure 6G,H and Figure S7H). While the structural basis for contacts between the human EXOSC10 catalytic domain and exosome core remain unknown, it is likely that EXOSC10 occupies a similar position as Rrp6 in yeast exosomes based on similarities in UV-crosslinking patterns and because insertions near the top of the exosome core central channel inhibit both human EXOSC10 and yeast Rrp6 (Wasmuth et al., 2014). These data suggest that EXOSC10 and MTR4 may compete for interactions with the exosome core.
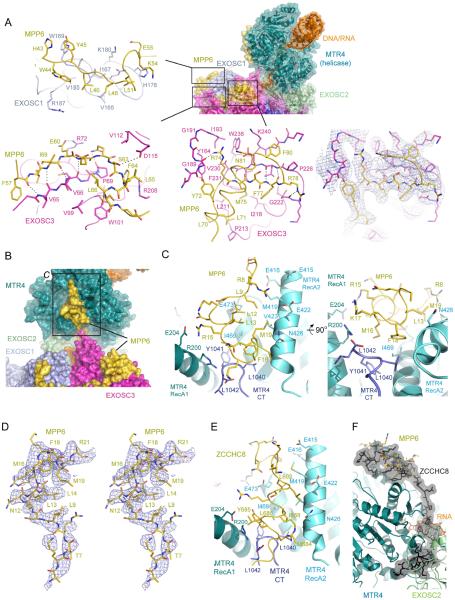
MPP6 tethers MTR4 to the core with MPP6 amino acids 5–21 binding MTR4 and residues 43–93 binding EXOSC3 and EXOSC1 (Figure 7A-C and Figure S7I). MPP6 residues 22–42 and 94–160 are not apparent in our structure. MPP6 contacts to EXOSC3 are similar to yeast Mpp6 fragments bound to a yeast exosome (Wasmuth et al., 2017) or its non-catalytic core (Falk et al., 2017). These studies revealed the importance of an arginine anchor that corresponds to Arg74 in human MPP6 (Figure 7A). As predicted, Arg74 is positioned proximal to EXOSC3 Trp238, a residue mutated in patients with pontocerebellar hypoplasia type I and spinal motor neuron degeneration (Wan et al., 2012) and Gly191, a residue mutated in hereditary spastic paraplegia (Halevy et al., 2014). Human MPP6 forms an additional helix (residues 81–93) that packs above the beta strand that contains the arginine anchor. In addition to contacts to EXOSC3, MPP6 contacts EXOSC1, albeit with weaker density, between MPP6 residues 43–46 and S1 domain of EXOSC1, an interface that is apparently conserved (Figure S7I).
Figure 7. MPP6 interacts with MTR4 and the exosome core and comparison to ZCCHC8 contacts to MTR4.
(A) Contacts between MPP6, EXOSC1 and EXOSC3. Overall structure shown to the right colored and labeled as in Figure 3 with boxes indicating three regions of interaction. Specific interactions are shown in individual panels in stick representation with subunits and amino acids labeled. Electron densities (blue mesh) covering a region that includes MPP6 Arg74 adjacent to the corresponding schematic.
(B) Overall structure of MTR4 highlighting the surface that binds to the N-terminal portion of MPP6 colored and labeled as in Figure 3.
(C) Orthogonal views of interactions between MPP6 and MTR4 with MTR4 depicted in cartoon with different shades of blue to indicate the labeled domains. MPP6 is shown in ribbon representation. Side chains are shown in stick representation with select amino acids labeled.
(D) Stereo view of electron density (blue mesh) for MPP6 amino acids that contact MTR4.
(E) Close-up of interactions between a ZCCHC8 C-terminal domain and MTR4 (PDBID: 6C90) in the same orientation for MTR4/MPP6 as in panel C.
(F) The ZCCHC8 C-terminal domain binds to MTR4 through interfaces that are mutually exclusive to those used by MTR4 to interact with MPP6 and EXOSC2.
See also Figure S7.
Most relevant to MTR4 recruitment is that MPP6 residues 5–21 contact MTR4 in an interface between its two RecA domains (Figure 7B-F). Contacts primarily involve hydrophobic interactions between side chains in MPP6 and the MTR4 RecA2 domain, regions that are highly conserved in both proteins (Figure S7I). Consistent with our structure, Mtr4 recruitment is disrupted in the yeast system by N-terminal deletions of Mpp6 (Wasmuth et al., 2017). Furthermore, mutagenesis experiments designed to disrupt the structurally validated interaction between yeast Rrp47-Rrp6 NTD and Mtr4Nterm did not alter yeast viability (Schuch et al., 2014), however synthetic lethality was observed when these mutants were combined with an Mtr4 C-terminal GFP tag. Based on our structure, a C-terminal fusion to GFP could interfere with MPP6 interactions as the MTR4 surface that engages MPP6 is just above the Mtr4 C-terminus (Figure 7C).
Finally, MTR4 surfaces between RecA1 and RecA2 that engage MPP6 overlap with those used by ZCCHC8 to interact with MTR4 in the context of the NEXT complex (Puno and Lima, 2018; Figure 7E,F). In the case of ZCCHC8, an amphipathic helix is situated in the opposite orientation between the RecA1 and RecA2 domains. Furthermore, this fragment of ZCCHC8 extends interactions over MTR4 surfaces that engage EXOSC2 in our structure. Together, these data suggest that interactions between this element of ZCCHC8 and MTR4 are mutually exclusive with MTR4 contacts to both the exosome core and to MPP6. It remains unclear how Mpp6-exosomes from human and yeast systems stimulate Mtr4 helicase activity (Figure 1), but it is intriguing that the C-terminal ZCCHC8 amphipathic helix occupies the same position as MPP6 and is required for stimulation of MTR4 ATPase and helicase activities in the NEXT complex (Puno and Lima, 2018).
DISCUSSION
Experiments presented here illustrate that the human, budding yeast and fission yeast Mtr4 helicases can promote unwinding and degradation of structured RNA substrates when associated with the nuclear exosome and its cofactors. Previous in vivo studies have shown a role for the nuclear exosome and MTR4 in RNA/DNA hybrid resolution and have suggested that this function may play a broader role in maintaining genomic stability and in controlling targeted strand-specific DNA mutagenesis at loci relevant to antibody diversification (Lim et al., 2017; Pefanis et al., 2015). Here we show that Mtr4-containing exosomes can displace a DNA strand while translocating on RNA to ultimately degrade the RNA within a DNA/RNA heteroduplex.
Our structure of the human nuclear exosome complex shows that MTR4 engages the non-catalytic core via interactions with MPP6, the RNA substrate, and contacts to EXOSC2. After RNA is extruded from the helicase, two regions within the exosome core central channel may assist in capturing single-stranded RNA. Immediately after extrusion from the helicase, residues on the S1 domain surface of EXOSC2 interact with single-stranded RNA before entering the PH-like ring. Single-stranded RNA is again captured at the base of the central channel within a conserved pocket between EXOSC4 and EXOSC9. We showed previously that amino acids in this surface can bind tungstate, a phosphate mimetic, and that residues within this pocket share the highest degree of similarity to residues in bona fide active sites of phosphorolytic enzymes such as PNPase and archael exosomes (Liu et al., 2006). Indeed, comparison of our structure to the RNA-binding pocket of a phosphorolytic archaeal exosome reveals further similarities (Lorentzen and Conti, 2012) (Figure S7A,B). These observations suggest that non-catalytic eukaryotic exosome cores may have conserved elements within this site to enable capture, but not cleavage, of single-stranded RNA.
After passing through the exosome core, the RNA path to DIS3 follows a path reminiscent of DIS3L2 and related bacterial RNAses via contacts to the S1 and CSD2 domains (Faehnle et al., 2014; Frazão et al., 2006), a path that differs from budding yeast Dis3 where RNA traverses below CSD1 (Lorentzen et al., 2008; Makino et al., 2013; Zinder et al., 2016). Moreover, human DIS3 remains in an open conformation when bound to RNA via the central channel, a conformation that is distinct from the closed through-channel conformation observed in S. cerevisiae (Liu et al., 2014; Makino et al., 2013). While the physiological relevance of yeast Dis3 conformations appears clear (Delan-Forino et al., 2017; Han and van Hoof, 2016), the features of the yeast exosome core that stabilize different conformations of Dis3 in budding yeast are not conserved in human (Zinder et al., 2016). It is tempting to speculate that higher eukaryotes may have evolved other mechanisms for distinguishing between substrates. One way this could be achieved is through specialization of catalytic subunits within subcellular compartments. In the nucleus of human cells, the exosome core is associated with DIS3 while in the cytoplasm it is associated with DIS3L, a paralog of DIS3 (Tomecki et al., 2010). Moreover, nucleoli accumulate EXOSC10 but exclude DIS3, suggesting the existence of an EXOSC10 nucleolar exosome.
Our structure provides an atomic model and insights for a helicase engaged human nuclear exosome after a substrate is fully committed to decay, i.e. after the RNA-bound helicase has engaged the core, displaced C1D/EXOSC10, and after RNA has passed through the helicase and central channel and into the active site of DIS3. Our biochemical data in budding yeast and human systems show that EXOSC10-mediated 3′ trimming of RNA competes with helicase-dependent decay insofar as helicase-dependent decay is enhanced when a catalytically inactivated form of EXOSC10/Rrp6 is used. It will remain a challenge to capture exosome complexes in conformations associated with these earlier steps. It is noteworthy that binding sites for MPP6 and ZCCHC8 of NEXT overlap on the surface of MTR4. Moreover, MTR4 surfaces used to bind EXOSC2 overlap with those used by yeast Rrp6 to bind Rrp4. While this process is unidirectional for substrates targeted for decay, it is intriguing that the process can be reversed during rRNA processing where Dis3 acts first, followed by Rrp6 trimming to generate the mature rRNA (Fromm et al., 2017). A reconstruction was recently reported for the yeast nuclear exosome engaged in processing the pre-60S ribosome (Schuller et al., 2018). While the reconstruction of the yeast nuclear exosome component of the structure lacked side chain densities, the overall arrangement of subunits is consistent with our structure of the human nuclear exosome-MTR4 complex suggesting that contacts between the helicase and the exosome core may be conserved independent of whether exosomes are in the process of degrading or processing their substrates.
Together, these observations suggest a sequential exchange of protein-protein and protein-RNA interactions prior to decay and that the fate of any particular substrate will depend on the dynamic and competitive exchange of factors within nuclear exosomes, cofactors and the substrates themselves. Moreover, several exosome subunits and cofactors are known to be post-translationally modified in response to environmental cues (Knight et al., 2016; Matsumoto-Taniura et al., 1996; Stehmeier and Muller, 2009). This additional layer of regulation has been largely unexplored but is likely to add further complexity to this highly regulated system. Further studies will be needed to fully appreciate how different co-factors regulate exosome function, particularly in the more complex mammalian system where additional complexes such as NEXT and PAXT provide further specificity.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents may be directed to and will be fulfilled by the Lead Contact Christopher D. Lima (limac@mskcc.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
E. cloni 10G competent bacterial cells were used for molecular cloning of exosome expression plasmids. Protein subunits were expressed in E. coli BL21CodonPlus (DE3)-RIL competent cells, E. coli One Shot BL21 (DE3) STAR chemically competent cells, or E. coli SoluBL21 (DE3) chemically competent cells. Standard culture was performed as follows: Cells were grown with appropriate antibiotics to OD600 1.0 to 1.8 in Superbroth medium at 37°C, supplemented with 100 % ethanol to a final concentration of 2 % and transferred to an ice bath for 30 minutes. Protein expression was induced with 0.25 mM IPTG overnight at 18°C. Protocol variations for individual exosome subunits can be found in method details.
METHOD DETAILS
Exosome Purification and Complex Reconstitution
S. cerevisiae RNA exosomes.
Subunits were expressed, purified, and reconstituted into complexes as described previously (Greimann and Lima, 2008; Wasmuth et al., 2017). Where indicated, Rrp6 contains a D238N mutation and Dis3 contains both D171N and D551N mutations to render them catalytically inactive.
S. pombe RNA exosomes.
All DNA sequences for Schizosaccharomyces pombe were obtained from PomBase S. pombe (Database issue: D695–9) genome data (Wood et al., 2012). cDNA sequences for S. pombe genes rrp41, rrp42, rrp43, rrp45, rrp46, mtr3, rrp4, rrp40, csl4, rrp6, and dis3 were codon-optimized, synthesized for expression in E. coli, and inserted into sub-cloning vectors (DNA 2.0/ATUM, Newark, CA). Additional codon-optimized cDNA sequences rrp47/C1D, mpp6, and mtr4 were obtained from IDT DNA and cloned using pGEM vectors.
Expression of S. pombe proteins was achieved by sub-cloning DNA sequences into the MCS1 and MCS2 of plasmid pRSF-SMT3-Duet1 (Novagen) vector by use of restriction sites engineered into positions flanking the coding sequence. S. pombe rrp41 and rrp45 cDNA were inserted into the MCS1 and MCS2, respectively. S. pombe rrp42 and mtr3 cDNA were inserted into the MCS1 and MCS2, respectively. S. pombe rrp43 and rrp46 cDNA were inserted into the MCS1 and MCS2, respectively. S. pombe rrp6 and rrp47 cDNA were inserted into the MCS1 and MCS2, respectively. S. pombe csl4, rrp4, rrp40, dis3, mpp6, and mtr4 were each cloned separately solely into MCS1. Additional point mutations were produced by QuikChange mutagenesis (Agilent Technologies) and sequenced for fidelity to render Rrp6 and Dis3 catalytically inert: the exoribonucleolytic mutation for S. pombe Rrp6 was D243N and the endo- and exoribonucleolytic mutations for S. pombe Dis3 were D166N and D516N, respectively. All wild-type and mutant protein expression plasmids were transformed into One Shot BL21 Star (DE3) (Thermo Fisher Scientific) for protein expression. For expression, cells were grown in shaker flasks to 1.0 OD600, and induced overnight at 18°C with 0.25 mM IPTG in the presence of 2% ethanol. After induction, cells were harvested and flash-frozen in liquid nitrogen.
Individual S. pombe protein subunits (spCsl4, spRrp4, spRrp40, spDis3, spMtr4, and spMpp6) and heterodimers (spRrp41/Rrp45, spRrp42/Mtr3, spRrp43/Rrp46, spRrp6/Rrp47) were each purified separately. To prepare cell lysate for protein purification, sonication was performed using a cell disrupter in 50 mM Tris-HCl pH 8.0, 20% (w/v) sucrose, 350 mM NaCl, 20 mM imidazole, 0.1% IGEPAL, 1 mM PMSF, 1 mM β-mercaptoethanol (BME), and 10 ug/mL DNase on the thawed cells. Lysate was then centrifuged using a Beckman JA-20 rotor at 44,000×g. Supernatant was applied to Ni-NTA resin (QIAGEN) for purification, and each of the His-tagged Smt3 fusion proteins and Smt3-fusion protein heterodimer pairs were eluted in 20 mM Tris-HCl pH 8.0, 350 mM NaCl, 250 mM imidazole, and 1 mM BME. For further purification, Smt3 fusion proteins were cleaved by Ulp1 overnight at 4°C and fractionated by size exclusion chromatography for spCsl4, spRrp4, spRrp40 with a HiLoad Superdex 75 PG 26/60 (GE Healthcare) and for spRrp41/Rrp45, spRrp42/Mtr3, spRrp43/Rrp46, spRrp6/Rrp47, spDis3, spMtr4, and spMpp6 on a HiLoad Superdex 200 PG 26/60 (GE Healthcare) in Sizing Buffer: 20 mM Tris-HCl pH 8.0, 350 mM NaCl, and 1 mM BME. For each protein subunit or heterodimer sample, peak fractions were pooled, concentrated to 5–10 mg/mL, and flash-frozen prior to reconstitution.
S. pombe complexes were reconstituted in three steps: reconstitution of the spExo9 core, reconstitution of the core with the exoribonucleolytic subunits and co-factors (spRrp6/Rrp47, spDis3, spMpp6) to form spExo13, and reconstitution of the spExo14 by addition of the helicase Mtr4 to spExo13. To form the spExo9 core, each of S. pombe core subunits (spCsl4, spRrp4, spRrp40, spRrp41/Rrp45, spRrp42/Mtr3, spRrp43/Rrp46) were added together at equimolar concentrations. The mixture was then dialyzed overnight at 4°C to 20 mM Tris-HCl pH 8.0, 100 mM NaCl, 1 mM BME. The dialysate was separated from free subunits using a HiLoad Superdex 200 PG 26/60 (GE Healthcare) in 20 mM Tris-HCl pH 8.0, 100 mM NaCl, and 1 mM BME, and then peak fractions were further purified with a linear salt gradient via a Mono Q 10/100 GL column (GE Healthcare) with Buffer A: 20 mM Tris-HCl pH 8.0, 100mM NaCl, and 1 mM BME and Buffer B: 20 mM Tris-HCl pH 8.0, 1 M NaCl, and 1 mM BME. To reconstitute spExo13, the resulting spExo9 core was mixed with 1.5-fold molar excess of purified spRrp6/Rrp47 and spDis3 and 3-fold molar excess of spMpp6. Mixtures were then dialyzed overnight at 4°C to 20 mM Tris-HCl pH 8.0, 100 mM NaCl, 1 mM BME. The dialysate was then applied to a HiLoad Superdex 200 PG 26/60 (GE Healthcare) in 20 mM Tris-HCl pH 8.0, 100mM NaCl, and 1 mM BME to separate away free enzymes from the reconstituted complex. Peak fractions containing stoichiometric quantities of all subunits of spExo13 were pooled and added to a 1.5-fold molar excess of spMtr4 to reconstitute spExo14. Mixtures were then dialyzed in two steps overnight at 4°C to 20 mM Tris-HCl pH 8.0, 50 mM NaCl, 1 mM BME. The dialysate was then applied to a Superdex 200 Increase 10/300 GL column (GE Healthcare) in 20 mM Tris-HCl pH 8.0, 50 mM NaCl, 0.5 mM MgCl2, 0.5 mM TCEP to remove excess spMtr4. All reconstitutions were concentrated to 7–10 mg/mL by centrifugation in an Amicon Ultra-15 30,000 MWCO filtration unit (Millipore-Sigma), flash frozen in liquid nitrogen, and stored at −80°C.
H. sapiens RNA exosomes.
RNA exosome core subunits were expressed, purified and reconstituted as described previously (Greimann and Lima, 2008; Wasmuth et al., 2014). Full-length human EXOSC10 was codon-optimized for expression in E. coli using the GeneArt platform (Thermo Fisher Scientific) and inserted into a pRSF-His6Smt3-Duet vector. To improve expression, constructs were generated to remove aa 805–885 and replace aa 649–704 with the linker sequence SRGSGSGSGSGS to yield EXOSC10 1–804Δ649–704. Catalytically inert EXOSC10 was generated by introducing a D313N mutation. hRrp47 (C1D) was generated from a cDNA library (Ambion) and cloned into MCS2 of the pRSF-His6Smt3-EXOSC10 vector. Cloning of different pRSF-Duet-Smt3-EXOSC10/C1D constructs was performed using custom primers and Gibson assembly cloning (New England Biolabs). BL21 (DE3) Codon Plus RIL STAR cells were transformed with the EXOSC10/C1D heterodimer construct and induction, lysis and Ni-affinity purification were performed as described for human exosome core components, however, Ulp1 cleavage of the Smt3 tag was not performed, instead Smt3- EXOSC10/C1D was purified by size exclusion using a HiLoad Superdex 200 PG 26/60 (GE Healthcare) column equilibrated with gel filtration buffer (20 mM Tris-HCl pH 8.0, 350 mM NaCl, 1 mM BME). Peak heterodimer-containing fractions were dialyzed overnight against buffer containing 20 mM Tris-HCl pH 8.0, 250 mM NaCI, and 2 mM TCEP and purified using Heparin HiTrap HP 5 ml affinity columns (GE Healthcare, Buffer A: 20 mM Tris-HCl pH 8.0, 250 mM NaCl, 2 mM TCEP; Buffer B: 20 mM Tris-HCl pH 8.0, 1 M NaCl, 2 mM TCEP, 0–40% buffer B over 20 CV).
hDIS3 was generated from a cDNA library and cloned into pGEX-4T2 using SmaI and NotI restriction sites to generate a GST fusion. Inactivating mutations to the endonucleolytic and exonucleolytic sites (D146N and D487N, respectively) were introduced by QuikChange mutagenesis (Agilent Technologies). Expression and induction was performed following the established protocol for the human Exo9 core. For lysis, cells were supplemented to a volume of 250 ml with hDIS3 lysis buffer (20 mM Tris pH 8.6, 350 mM NaCl, 1 mM BME, 0.1% v/v IGEPAL, 10 μg/ml DNAse I, 1 mM PMSF) and disrupted by sonication over three times 2 min at 65% intensity with 1 s on, 3 s off intervals using a Branson Digital Sonifier. Cell lysates were cleared by centrifugation at 44,000×g (Beckman JA-20) for 45 minutes at 4°C. For pull-down of GST-hDIS3, 5 ml of Glutathione Sepharose™ 4B (GE Healthcare) were equilibrated with binding buffer (20 mM Tris pH 8.6, 350 mM NaCl, 1 mM BME), added to the supernatant and incubated for 1 h at 4°C. Protein-bound resin was washed with 40 volumes of binding buffer and GST-DIS3 was eluted after 15 min of incubation in 7.5 ml elution buffer (20 mM Tris pH 8.6, 350 mM NaCl, 15 mM reduced L-glutathione). Thrombin was added at a concentration of 1.5 U/mg hDIS3 followed by overnight incubation at 4°C. Cleavage products were separated by gel filtration using a HiLoad Superdex 200 PG 26/60 (GE Healthcare) column equilibrated with gel filtration buffer (20 mM Tris-HCl pH 8.0, 350 mM NaCl, 1 mM BME). Peak fractions were dialyzed overnight against buffer containing 20 mM Tris-HCl pH 8.0, 50 mM NaCl, and 2 mM TCEP. Sample was then purified using a MonoQ 10/100 GL column (GE healthcare; buffer A: 20 mM Tris-HCl pH 50 mM NaCl, 2 mM TCEP; buffer B: 20 mM Tris-HCl pH 8.0, 1 M NaCl, 2 mM TCEP, 0–40% buffer B over 20 CV).
Human M-Phase-Phosphoprotein 6 (MPP6) gene was PCR-amplified from placenta cDNA pool (Ambion) using Pfu Turbo DNA polymerase (Stratagene) and digested PCR products were inserted into pSMT3-TOPO. Expression and purification was performed analogous to what has been described previously for the human core subunits Csl4, Rrp4 and Rrp46 (Greimann and Lima, 2008; Wasmuth et al., 2014).
Human MTR4 (SKIV2L2, DOB1) was amplified from human kidney cDNA (Invitrogen) and subcloned into pET-28a with an N-terminal His6Smt3 tag. Culture and purification were performed as described for the H. sapiens core subunits using a HiLoad SD200 PG 26/60 size exclusion column. Peak fractions were pooled, dialyzed overnight at 4°C against 100 mM NaCl, 20 mM Tris-HCl pH8.0, 2 mM TCEP and purified over a Heparin HiTrap HP 5ml affinity column (GE Healthcare, Buffer A: 20 mM Tris-HCl pH 8.0, 100 mM NaCl, 2 mM TCEP; Buffer B: 20 mM Tris-HCl pH 8.0, 1 M NaCl, 2 mM TCEP; 0–100% buffer B over 20 CV).
All individual subunits were concentrated to ~10 mg/ml using appropriately sized Amicon® centrifugal filter tubes (Merck Millipore Ltd.), flash-frozen and stored at −80°C. For reconstitution of H. sapiens 12-, 13-, and 14-component RNA exosomes, the Exo9 core was mixed with 4-fold molar excess of EXOSC10/C1D, 2-fold molar excess of DIS3, 7.5-fold molar excess of MPP6 and 2-fold molar excess of MTR4 in 20 mM Tris-HCl 8.0, 350 mM NaCl, 2 mM TCEP supplemented with Ulp1 at a mass ratio of 1:1000 relative to EXOSC10. The mixture was dialyzed for 4 h at 4°C in buffer containing 20 mM Tris-HCl 8.0, 100 mM NaCl, 2 mM TCEP, followed by overnight dialysis in 20 mM Tris-HCl 8.0, 50 mM NaCl, 2 mM TCEP, 0.5 mM MgCl2. Reconstituted complexes were purified by size-exclusion chromatography using a Superdex 200 Increase 10/300 GL column (GE Healthcare) and peak fractions containing stoichiometric amounts of all subunits were concentrated to 10 mg/ml using Amicon® UltraCel® 30K MWCO centrifugal filters (Merck Millipore Ltd.), flash-frozen in liquid nitrogen, and stored at −80°C. Reconstitution of 12- and 13-component exosomes was performed analogously.
Biochemical Assays
Substrate Preparation.
Oligonucleotides were synthesized and HPLC-purified by Integrated DNA Technologies (Coralville, IA) and Dharmacon Inc (Lafayette, CO). Lyophilized RNA was suspended in annealing buffer (20 mM Tris-HCl pH 7.0, 100 mM potassium acetate) and stored at −80°C. Duplex substrates were prepared by heating an equimolar mixture of each strand to 95°C followed by cooling to 16°C for at least an hour. Annealed substrates were purified by size exclusion chromatography using Superdex 200 increase 10/300 GL column (GE Healthcare) pre-equilibrated with annealing buffer. Oligonucleotides used to prepare helicase and decay substrates include the following 5′ to 3′ sequences (DNA sequences are underlined): Fluorescein-AGC ACC GUA AAG ACG C (RNA displacement strand, Figure 1A,B), GCG UCU UUA CGG UGC UAA AAA AAA AAA AAA AAA AAA (RNA translocation strand, Figure 1A), GCG UCU UUA CGG UGC UAA AAA AAA AAA AAA AAA AAA (DNA-RNA chimera translocation strand, Figure 1B), AGC ACC GUA AAG ACG C (RNA displacement strand, Figure 2C, Figure S1B,C), GTG TGG TGT GGT GTG GTG TGG TGT GGT GTG GTG TGG T (DNA displacement strand, Figure 2C) Fluorescein-GCG UCU UUA CGG UGC U CA CCA CAC CAC ACC ACA CCA CAC CAC ACC ACA CCA CAC AAA AAA AA (RNA translocation strand, Figure 2C), Fluorescein-GCG TCT TTA CGG TGC T CA CCA CAC CAC ACC ACA CCA CAC CAC ACC ACA CCA CAC AAA AAA AA (RNA/DNA translocation strand, Figure S1B), GCG TCT TTA CGG TGC T CA CCA CAC CAC ACC ACA CCA CAC CAC ACC ACA CCA CAC AAA AAA AA (DNA-RNA translocation strand, Figure S1C), Fluorescein-GTG TGG TGT GGT GTG GTG TGG TGT GGT GTG GTG TGG T (DNA displacement strand, Figure S1C). See also Table S2.
Unwinding assays.
Assays in main Figure 1A,B were carried out at 22°C in a buffer containing 20 mM Tris-HCl pH 7.0, 50 mM NaCl, 0.5 mM MgCl2, 5 mM BME, and 1 U/μl RNAse inhibitor, human placenta (New England Biolabs). Substrates (10 nM) were pre-incubated with 200 nM protein for at least 5 minutes. Reactions were initiated by addition of 2 mM ATP, 2 mM MgCl2, and 100 nM DNA trap (5′ GCGTCTTTACGGTGCT 3′). Aliquots were taken at indicated times and quenched using 0.25% v/v SDS, 5 mM EDTA, 10% v/v glycerol and 80 mU/μl Proteinase K (New England Biolabs). Samples were resolved in Novex 20% TBE gels (Thermo Fisher Scientific), imaged using Typhoon FLA 9500 laser scanner (GE Healthcare) and analyzed using ImageJ (Schneider et al., 2012).
For helicase time course assays in Figure S1C.
S. cerevisiae. Final concentrations were 20 nM exosome, 22 nM Mtr4, 1 mM ATP or AMPPNP, 0.5 UμL RNAse inhibitor (New England Biolabs), and 10 nM RNA substrate in RNA decay buffer. Exosome complexes or buffer was incubated on ice with Mtr4 at 2 μM and 2.2 μM, respectively, in RNA decay buffer for 1 hour prior to initiating the reaction. A mix containing RNA and RNAse inhibitor (all at 1.25× final concentration) was incubated at 20°C for 5 min prior to addition of 1/10 volume of 200 nM enzyme. The reaction was then initiated with 1/10 volume of an initiation mix containing 10 mM ATP or AMPPNP and 300 nM DNA trap oligo (sequence = 5′ ACACCACACCACACCAC 3′). Reactions were quenched after the indicated incubation times by adding 10 μL of reaction to 5 μL of stop mix (0.3 % w/v SDS, 30 mM EDTA pH 8.0, 3 U/mL proteinase K (New England Biolabs)) followed by proteinase K digestion at 37°C for 1 hour and flash freezing in liquid nitrogen for storage at −80°C. 3.5 μL sample was loaded per lane and run for 45 minutes on 4–20% polyacrylamide-TBE gels (Life Technologies) using 0.5× TBE as running buffer. Gels were imaged on Typhoon FLA 9500 laser scanner (GE Healthcare) using the FAM setting at 600V and analyzed using ImageJ (Schneider et al., 2012). H. sapiens. Assays were performed as described for S. cerevisiae using reconstituted complexes. Quenching buffer was modified to contain 80 U/mL Proteinase K with overnight digestion at 30 °C. S. pombe. Assays were performed as described for S. cerevisiae.
Site-directed 4-thiouridine UV crosslinking assay.
Protein samples (200 nM) were incubated with 100 nM 5′ fluorescein- and internal 4SU-labeled RNA in a buffer containing 20 mM Tris-HCl pH 7.0, 100 mM NaCl, 2.5 mM MgCl, 5 mM BME, and 2 mM AMPPNP or ATP. To induce crosslinking, samples were exposed to long-range UV (365 nm) for 20 minutes at 4°C using a 4W handheld UV lamp (UVP). RNA-protein adducts were separated using NuPage 4–12% Bis-Tris protein gels (Thermo Fisher Scientific), visualized using Typhoon FLA 9500 laser scanner (GE Healthcare) and analyzed using ImageJ (Schneider et al., 2012). Oligonucleotides used to prepare substrates include the following 5′ to 3′ sequences of the translocation strands: Fluorescein-GCG TCT TTA CGG TGC TAA AAA AAA AAA AAA AAA A(4SU)A, Fluorescein-GCG TCT TTA CGG TGC TAA AAA AAA AA(4SU) AAA AAA AAA, Fluorescein-GCG TCT TTA CGG TGC TAA AAA (4SU)AA AAA AAA AAA AAA, Fluorescein-GCG TCT TTA CGG TGCT(4SU)A AAA AAA AAA AAA AAA AAA, Fluorescein-GCG TCT TTA CGG TGC T CAC ACC ACA CCA CAC CAC ACA AAA AA(4SU) A, Fluorescein-GCG TCT TTA CGG TGC T CAC CAC ACC ACA CCA CAC CAC ACC ACA CCA CAC CAC ACA AAA AA(4-S-U) A; RNA top strand: AGC ACC GUA AAG ACG C; and DNA displacement strands: TTT TTT TTT TT, TTA TTT TTT TT, TTT TTT ATT TT, GTG TGG TGT GGT GTG GT GT, GTG TGG TGT GGT GTG GTG TGG TGT GGT GTG GTG TGG T; DNA sequences are underlined. See also Table S2.
RNA decay assays.
S. cerevisiae.
For the decay time courses in Figure 2C and Figure S1B, final concentrations were 20 nM exosome, 22 nM or 0 nM Mtr4, 1 mM ATP or AMPPNP, 0.5 U/μL RNAse inhibitor (New England Biolabs), and 10 nM RNA substrate in RNA decay buffer (20 mM HEPES-KOH pH 7.5, 50 mM potassium acetate, 1.1 mM magnesium acetate, 2.5 mM DTT, 0.01 % IGEPAL). 50 mM ATP and AMPPNP stock solutions were adjusted to pH 7.0 with KOH. Exosome complexes were incubated on ice with Mtr4 (or buffer) at 2 μM and 2.2 μM, respectively, in RNA decay buffer for 1 hour prior to diluting 1:10 in RNA decay buffer and initiating the reaction. A mix containing RNA, ATP or AMP-PNP, and RNAse inhibitor (all at 1.1× final concentration) was incubated at 20°C for 5 min prior to initiation with 1/10 volume of 200 nM enzyme mix. Reactions were quenched after the indicated incubation times by adding 10 μL of reaction to 5 μL of stop mix (0.3 % w/v SDS, 30 mM EDTA pH 8.0, 3 U/mL proteinase K (New England Biolabs) followed by proteinase K digestion at 37°C for 1 hour and flash freezing in liquid nitrogen for storage at −80°C. For native gels, 3.5 μl sample was loaded per lane and run for 45 minutes on 4–20% acrylamide-TBE gels using 0.5× TBE as running buffer. Gels were imaged on a typhoon FLA 9500 instrument (GE Healthcare) using the FAM setting at 600V and analyzed using ImageJ (Schneider et al., 2012).
H. sapiens.
Assays were performed as described for S. cerevisiae but using reconstituted complexes at a final concentration of 100 nM. Quenching buffer was modified to contain 80 U/ml Proteinase K (New England Biolabs) and digestion was carried out at 30°C over night.
S. pombe.
Assays were performed as described for S. cerevisiae but using reconstituted complexes at a final concentration of 100 nM for Exo13 and Exo14. For exosomes lacking Rrp6/Rrp47, Exo10Dis3 was added to Mpp6 and Mtr4.
Electron Microscopy Sample Preparation and Imaging
Substrate-loading hExo14 for cryo-electron microscopy.
Large-scale loading reactions of human Exo14 were performed in a total volume of 100 μl (20 mM Tris pH 8.0, 50 mM NaCl, 0.5 mM MgCl2, 2.5 mM BME) at 4.5 μM hExo14 with stoichiometric amounts of substrate. DNA-RNA/DNA substrate and complex were mixed and incubated on ice for 15 min and loading was initiated by adding 10 μl of 20 mM Mg·ATP, 45 μM trap and incubated at 22°C for 2 h. 50 μl of 20 mM Tris pH 8.0, 50 mM NaCl, 6 mM AMPPNP, 2.5 mM MgCl2, 1 mM BME was added to quench the reaction, cleared by high-speed centrifugation and purified using a Superdex 200 Increase 10/300 GL column (GE Healthcare) in 20 mM Tris pH 8.0, 100 mM NaCl, 0.5 mM MgCl2, 1 mM BME. Peak fractions were supplemented with quenching buffer to achieve a final concentration of 20 mM Tris-HCl pH 8.0, 100 mM NaCl, 2.5 mM MgCl2, 2 mM AMPPNP, 1 mM BME. Sample was concentrated to >1 mg/ml using Amicon® UltraCel® 30K MWCO centrifugal filters (Merck Millipore Ltd.), supplemented with 0.05% CHAPSO and immediately used for grid preparation. Aliquots for analysis of nucleic acid were added to loading buffer (20 mM Tris pH 8.0, 100 mM NaCl, 2.5 mM BME, 0.5 mM MgCl2, 20% v/v glycerol, 10 mM EDTA, 0.2 % v/v SDS, 50 U/ml Proteinase K) and incubated at 37°C for 3 h. Aliquots for analysis of protein were added to 4× LDS sample buffer supplemented with 5% v/v BME and separated by SDS-PAGE followed by Coomassie or SYPRO Ruby staining.
Cryo electron microscopy data collection.
Quantifoil® R 2/2 gold grids were glow-discharged for 60 sec at 0.37 mBar, 15 mA in a Pelco easiGlow™ air system (TED PELLA, Inc.). Four μl of a suspension of RNA-loaded hExo14 complexes at a concentration of 1.75 μM protein were applied per grid and plunge-frozen in liquid ethane using a Vitrobot Mark IV (FEI) at 20°C, 100% humidity (30 sec wait time, 2.5 sec blot time). Grids were screened for suitable particle density using an MSKCC in-house Titan Krios 300KV (FEI) microscope and Serial EM data collection software (Mastronarde, 2005). Data collection was performed on a Titan Krios 300KV (FEI) instrument using a K2 Summit direct detector (Gatan, Inc.) and Leginon data acquisition interface (Suloway et al., 2005). A total of 1604 movies (50 frames/movie, 10 sec exposure time) were collected in super-resolution mode with a defocus range from −1.0 to −3.0 μm at a dose rate of 10.0 e-/px/sec and a total dose of 85.23 e-/Å2/movie. The exposure of the movies was filtered in a dose-dependent manner to weight the amplitudes of each movie frame to preserve low resolution contrast across the exposure while favoring high resolution information from early frames that sustained less radiation damage, as described (Grant and Grigorieff, 2015). The calibrated pixel size was 1.07 Å/px.
Image Processing and Map Reconstruction
Structure determination.
Movies were processed with MotionCor2 to generate dose-weighted aligned and averaged micrographs alongside aligned movies (Zheng et al., 2017). 1439 micrographs were selected for further processing based on their appearance, CTF estimation, and number of particles. In RELION (Scheres, 2012), a few hundred particles were selected to generate a small set of 2D classes that were then used as templates for autopicking the remaining particles. This particle stack was input to cryoSPARC (Punjani et al., 2017) to obtain an initial set of 2D class averages and to remove junk particles or particles that could not be classified. This procedure resulted in 278185 particles whose coordinates were used in RELION for movie processing and particle polishing. After another round of 2D classification in RELION, the resulting stack containing 271036 particles was then used in cryoSPARC to generate four ab initio 3D reconstructions, one for the helicase exosome complex (122703 particles), one for the core and DIS3 (64191 particles), and one for the core (49326 particles). The latter two include diffuse densities at the top of the core that may be EXOSC10 or nucleic acid. The remaining particles contributed to a fourth reconstruction that appears to encompass partial complexes of the Exo9 core. Particles associated with the first three models were combined and used as input to cryoSPARC for heterogeneous refinement followed by homogeneous refinement. Further inspection of the helicase complex revealed well-resolved densities for the nine-subunit exosome core, most of MPP6, a peptide of EXOSC10 bound to the exosome core, and lower resolution densities for the helicase, nucleic acid and DIS3.
The 122703 particles for the helicase complex were then transferred to RELION for an independent round of 2D classification that resulted in 122284 particles that were input to 3D refinement, generating a map that closely resembled the map obtained from cryoSPARC (correlation coefficient = 0.985). The resolution of this map was assessed to be 3.45 Å (or 3.55 Å) depending on whether the half-maps were masked to exclude (or include) the disordered KOW domain of MTR4. All map resolutions throughout were assessed using FSC of 0.143. To better resolve densities for the core, MTR4 and DIS3, aligned particles were used as input for focused 3D classification or focused 3D refinement. For the core, 3D classification did not reveal any major subclasses, however focused 3D refinement improved densities for the core (RELION 3.31 Å; phenix.mtriage 3.23 Å) at the expense of densities for MTR4 and DIS3. Focused refinement also resulted in an improved map for MTR4 (RELION 3.54 Å; phenix.mtriage 3.45 Å), although densities for duplex DNA/RNA portion of the substrate or the KOW domain remained poorly resolved. Densities for the DIS3 RNB and CSD domains could not be improved through focused refinement using all of the particles, but 3D classification revealed a major class (91483 particles) that was used in focused 3D refinement encompassing all of DIS3, followed by focused refinement with local searches with each of the three individual segments, the DIS3 PIN, cold-shock domains (CSDs), and the ribonuclease domain (RNB). While focused refinement on the RNB improved densities, focused refinement on the CSDs or PIN did not, presumably due to limited signal in these regions. Densities for the CSDs and RNB were improved by combining both regions in focused refinement (RELION 3.90 Å; phenix.mtriage 3.58 Å) while densities for the PIN were improved by focused refinement using a mask that encompassed the PIN domain and three proximal PH-ring subunits of the core (RELION 3.40 Å; phenix.mtriage 3.57 Å). To better resolve densities for the duplex region of the DNA/RNA substrate and KOW domain, focused 3D classification of MTR4 was conducted in two rounds, one without resolution limits, and a second round that limited resolution to 6 Å that resulted in several subclasses, some of which were refined. In the first round, two small subclasses (3643 and 2973 particles) revealed densities for the KOW domain after refinement at estimated resolutions 7.75 Å and 8.22 Å, respectively (RELION). The second round of 3D classification resulted in a class with resolved densities for the duplex region of our substrate. This class was subjected to focused refinement as described above for MTR4 and resulted in an improved map (RELION 3.90 Å; phenix.mtriage 3.82 Å).
Model Building and Refinement
The model for the human Exo9 nine-subunit core (PDB: 2NN6) was used alongside models for the yeast eleven and twelve-subunit exosome bound to Rrp6 and Mpp6 (PDB: 5K36 and 5VZJ) to manually inspect and rebuild the human exosome core using Coot (Emsley et al., 2010). MPP6, EXOSC10 and DIS3 were manually rebuilt based on electron densities using models based on similarities to available yeast counterparts or mouse DIS3L2 (PDB: 4PMW). The MTR4 core was rebuilt after docking a high-resolution x-ray structure of human MTR4 (PDBID: 6C90) into density. A portion of the two helical stalks that bridge the helicase and KOW domains were included based on homology to the yeast enzyme, and densities from the two lower-resolution maps with visible KOW domains. Models were fit into density and refined in real space using Coot (Emsley et al., 2010) (RRID:SCR_014222). Resulting models were refined in Phenix using real-space with ADP refinement (Adams et al., 2010) (RRID:SCR_014224) against individual maps obtained from focused 3D refinement. A final composite model was generated (PDB: 6D6R). This model was refined against the overall map using the starting model as a restraint with ADP refinement to generate a refined model in the overall reconstruction (PDB: 6D6Q). Figures depicting structures and maps were prepared using PyMol (Schrödinger, 2015) (RRID:SCR_000305) or Chimera (Pettersen et al., 2004). Structure quality was assessed using MolProbity (Chen et al., 2010) (RRID:SCR_014226).
QUANTIFICATION AND STATISTICAL ANALYSIS
Number of replicates of biochemical assays are indicated in corresponding figure legends. Cryo-EM data collection and refinement statistics for the overall data set and data sets used for focused refinement can be found in Table S1. Details on cryo-EM data analysis are provided in method details.
DATA AND SOFTWARE AVAILABILITY
Accession numbers (www.rcsb.org) to access atomic coordinates reported in Figure S5A are PDB: 6D6Q and 6D6R after refinement in the overall reconstruction and the focused reconstructions, respectively. Six electron density maps correspond to deposited PDBs, the overall map (EMDB 7808), a composite map from combining focused reconstructions (EMDB 7809), and each map resulting from focused reconstructions for the core (EMDB 7810), DIS3 PIN (EMDB 7812), MTR4 (EMDB 7813), MTR4 with duplex DNA/RNA (EMDB 7815), the DIS3 RNBCS domains (EMDB 7814) after alignment to the overall map. Two additional maps are deposited with resolved densities for the MTR4 KOW domain in open or closed conformations, respectively (EMDB 7818, EMDB 7819).
Supplementary Material
(A) Nomenclature of exosome subunits and co-factors from Homo sapiens, Saccharomyces cerevisiae, and Schizosaccharomyces pombe. (B) Time-course assays for degradation of the DNA-RNA chimera in the presence of AMPPNP (left) or ATP (right) (similar to Figure 2C). The substrate used is schematically represented at the top; cartoons are labeled to represent subunit compositions. (C) ATP-dependent unwinding of the DNA-RNA chimera by 12-, 13- and 14-component exosomes as well as Mtr4. Marker (M) for single stranded species with sizes indicated to the right and marker (M2) for the displaced fluorescein-labelled strand with trap oligo. Gel images in panels B and C are representative of two independent experiment for S. cerevisiae and three independent experiments for S. pombe and human. (D) Sybr Gold-stained gels of proteinase K-digested fractions after RNA-unwinding and size exclusion chromatography. Top: Fractions 17–22 with corresponding RNA markers (MS=substrate; MP=loaded product) at indicated concentrations. Bottom: Fractions 24–29 with corresponding RNA markers. Band intensity does not scale between individual gels. * denotes peak fractions containing unwound reaction product. (E) Coomassie stained SDS-PAGE protein fractions corresponding to panel D. Gel images in panels D and E are representative of two independent experiments. Load = RNA unwinding reactions prior to gel filtration; M = molecular weight marker.
Boxed maps (green) with labels represent endpoints for those maps available for download in EMDB.
(A) Schematic representing the angular distribution of particles contributing to the overall reconstruction as reported by RELION and displayed using Chimera with the overall reconstruction rendered as isosurface inside the sphere in four orientations. (B,C) Overall reconstruction and gold-standard FSC. (D,E) Reconstruction after focused refinement on the core and gold-standard FSC. (F,G) Reconstruction after focused refinement on the DIS3-PIN and gold-standard FSC. (H,I) Reconstruction after focused refinement on the DIS3-RNBCS and gold-standard FSC. (J,K) Reconstruction after focused refinement on MTR4 and gold-standard FSC. (L,M) Reconstruction after focused refinement on a subclass of MTR4 particles elicited more continuous densities for the duplex DNA/RNA segment of the substrate.The six reconstructions analyzed in this study (and boxed in Figure S2) are isosurface rendered and colored according to local resolution as calculated by RESMAP (Kucukelbir et al., 2014). Separate from this calculation, the maps were low-pass filtered at 4.5 Å resolution to enable visualization of the most disordered regions at low contour threshold (left), at higher contour threshold (middle), and as a central slice (right) in panels B, D, F, H, J, and L. Color code for resolution shown adjacent to each map. Gold-standard FSC curves corresponding to each reconstruction with FSC=0.143 represented as a dashed line and resolutions indicated are shown at far right.
Labels indicated for each map as labeled in Figure S2 and S3. Panels showing regions around: (A) MTR4 in the overall map. (B) MTR4 in the MTR4-focused map. (C) MTR4 the MTR4-dsDNA/RNA focused map. (D) Regions within the core for the overall map (E) Regions within the core for the core focused map. (F) Densities surrounding the PIN domain of DIS3 for the overall map (G) Densities surrounding the PIN domain of DIS3 for the DIS3 PIN focused map. (H) Densities surrounding the DIS3 RNB and cold-shock domains for the overall map. (I) Densities surrounding the DIS3 RNB and cold-shock domains for the DIS3 RNBCS focused map.
(A) 2D class averages obtained from cryoSPARC calculated using all particles selected for the final 3D reconstruction. (B) A representative micrograph from the dataset used to determine the structure showing particles (left) and with green circles depicted around selected particles (right). Scale bar at lower right (30 nm). (C) Gold-standard FSC between the respective coordinate models for the overall refinement and local regions of the complex as indicated as calculated by phenix.mtriage. FSC values at 0.5 and 0.143 are reported in panel A. The DIS3 PIN focused refinement utilized a mask covering the PIN and most of the three adjacent PH-like proteins, however the gold-standard FSC between the DIS3 PIN model and the corresponding focused refinement map overestimated the resolution of the DIS3 PIN if core subunits were included. Therefore, the DIS3 PIN was compared to a modified version of the focused refinement map whereby densities attributed to the core subunits were removed. (D) Gold-standard FSC curve (FSC=0.143, dotted line) from RELION reconstruction shown in Figure 5G. (E) Gold-standard FSC curve (FSC=0.143, dotted line) from RELION reconstruction shown in Figure 5H.
(A) Structural representation of contacts between EXOSC10 and EXOSC6 as well as EXOSC8 colored as in Figure 3. (B) Stereo view of electron densities overlaying the corresponding areas shown in panel A with blue mesh for EXOSC10 and grey mesh for EXOSC6 and EXOSC8. (C) View of DIS3 contacts to the core with boxes representing regions indicated in detail in panels D-I. Subunits, domains and RNA are labeled and colored as in Figure 3. (D) Interactions between EXOSC9 and the DIS3 RNB domain. (E) Interactions between EXOSC9 and the DIS3 CSD2 domain. (F) Interactions between the DIS3 CSD1 domain and EXOSC7 and EXOSC8. (G) Interactions between the DIS3 CSD2 domain and EXOSC9 and EXOSC4. (H) Interactions between EXOSC4 and the DIS3 PIN domain. (I) Interactions between the DIS3 PIN domain and EXOSC4 and EXOSC7.Side chains shown in stick representation, labeled by residue number (colored by subunit), polar interactions indicated by dashed lines between atoms, and domains indicated by labels.
(A) RNA contacts at a conserved interface between EXOSC9 and EXOSC4. (B) RNA contacts within the active site between Rrp41/Rrp42 in the phosphorolytic archaeal S. solfataricus exosome (PDBID: 2C37). Structures were aligned with protein and RNA shown in stick representation with amino acids colored and labeled and bases in the human complex labeled. RNA bound Dis3 structures shown to depict different RNA paths for (C) Human DIS3. (D) Mouse DIS3L2 (PDBID: 4PMW). (E) S. cerevisiae Dis3 from a core bound structure in the ‘open’ conformation (PDBID: 5K36). (F) S. cerevisiae Dis3 from a core bound structure in the ‘closed’ conformation (PDBID: 4IFD). RNB domains were used to align the respective Dis3 structures with Dis3 domains colored as CSD1 (orange), CSD2 (limon), RNB (pink), and S1 (purple). RNA in respective structures is depicted as a blue tube to highlight the different binding modes. The PIN domains for human DIS3 and budding yeast structures are omitted for clarity. (G) Amino acid sequence alignment of Mtr4 from various species. Alignment generated using Muscle alignment tool in Jalview (Waterhouse et al., 2009) with conservation based on BLOSUM62 scoring matrix. EXOSC2 residues that interact with MTR4 in this structure are indicated by green bars above the sequence. (H) Alignment of EXOSC2 and Rrp4 proteins generated using MUSCLE multiple sequence alignment tool. Conservation based on BLOSUM62 scoring in Jalview. Colored bars indicate surfaces involved in protein-protein interactions based on assessment of the human structure (this work) and a previously published S. cerevisiae structure (Zinder et al., 2016)(PDBID: 5K36). (I) Sequence alignment of MPP6 from indicated organisms. Alignment generated using MUSCLE alignment tool and manually corrected for S. cerevisiae and H. sapiens based on alignment of the human structure to a published structure from yeast (PDBID: 5VZJ)(Wasmuth et al., 2017). Conservation indicated by shading and based on BLOSUM62 scoring in Jalview (Waterhouse et al., 2009). Tick marks correspond to 10 amino acid intervals for the human MPP6 sequence. Colored bars indicate residues involved in protein-protein interactions with MTR4, EXOSC1 and EXOSC3 (or Rrp40) with dark shading indicating direct contacts based on PDBe PISA (cutoff for buried area 50%) and further visual assessment of the corresponding density for the human structure. Dark gray lines above or below the sequence alignment indicate residues modeled in this work or previous work in the yeast system, respectively. C-terminal amino acids from all species were omitted for clarity.
HIGHLIGHTS.
-
-
Structure of a human nuclear exosome-MTR4-RNA complex at 3.45 Å overall resolution
-
-
DNA-RNA chimera translocation strand can stall the MTR4 helicase
-
-
Mtr4 helicase activities are stimulated in yeast and human exosomes
-
-
Mutual exclusivity among exosome cofactors implies a sequential process for RNA decay
ACKNOWLEDGEMENTS
We thank Rich Hite, Nikola Pavletich, Steve Long, and Lima Lab members for advice, and we thank Fangyu Liu for her contributions to reconstituting S. pombe exosomes. Work was conducted in part at the cryo-EM facility at MSKCC and Simons Electron Microscopy Center and National Resource for Automated Molecular Microscopy located at the New York Structural Biology Center, the latter supported by grants from the Simons Foundation (SF349247), NYSTAR, NIH NIGMS (GM103310), Agouron Institute (F00316), and NIH (S10 OD019994-01). This research was supported the National Institute of General Medical Sciences of the NIH (R01GM079196 and R35GM118080 to C.D.L. and NIH-NCI-Cancer Center Support Grant (P30 CA008748). The content is solely the responsibility of the authors and does not represent the official views of the NIH. E.-M.W. is supported by the Human Frontier Science Program. C.D.L. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
E.-M.W., M.R.P, K.J., J.C.Z, and C.D.L. designed the experiments. M.R.P. and C.D.L. designed and optimized substrates for structural work. E.-M.W., M.A.D. and C.D.L. determined the structures. E.-M.W., M.R.P, K.J., and J.C.Z. performed the enzyme assays and biochemical experiments. E.-M.W., M.R.P, K.J., J.C.Z, M.A.D. and C.D.L. wrote the manuscript.
DECLARARATION OF INTERESTS
The authors declare no competing financial interests.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, and Tollervey D (1999). Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 18, 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Stamatoyannopoulos JA, et al. (2007). Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau F, Basquin J, Ebert J, Lorentzen E, and Conti E (2009). The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell 139, 547–559. [DOI] [PubMed] [Google Scholar]
- Briggs MW, Burkard KTD, and Butler JS (1998). Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J. Biol. Chem 273, 13255–13263. [DOI] [PubMed] [Google Scholar]
- Büttner K, Nehring S, and Hopfner K-P (2007). Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat. Struct. Mol. Biol 14, 647–652. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray W, Richardson JS, Richardson DC, and IUCr (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlebowski A, Lubas M, Jensen TH, and Dziembowski A (2013). RNA decay machines: The exosome. Biochim. Biophys. Acta - Gene Regul. Mech 1829, 552–560. [DOI] [PubMed] [Google Scholar]
- Delan-Forino C, Schneider C, and Tollervey D (2017). Transcriptome-wide analysis of alternative routes for RNA substrates into the exosome complex. PLoS Genet. 13, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowski A, Lorentzen E, Conti E, and Séraphin B (2007). A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol 14, 15–22. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K, and lUCr (2010). Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faehnle CR, Walleshauser J, and Joshua-Tor L (2014). Mechanism of Dis3l2 substrate recognition in the Lin28-let-7 pathway. Nature 514, 252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk S, Bonneau F, Ebert J, Kögel A, and Conti E (2017). Mpp6 incorporation in the nuclear exosome contributes to RNA channeling through the Mtr4 helicase. Cell Rep. 20, 2279–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazão C, McVey CE, Amblar M, Barbas A, Vonrhein C, Arraiano CM, and Carrondo MA (2006). Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA- bound complex. Nature 443, 110–114. [DOI] [PubMed] [Google Scholar]
- Fromm L, Falk S, Flemming D, Schuller JM, Thoms M, Conti E, and Hurt E (2017). Reconstitution of the complete pathway of ITS2 processing at the pre-ribosome. Nat. Commun 8, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant T, and Grigorieff N (2015). Measuring the optimal exposure for single particle cryo-EM using a 2.6 A reconstruction of rotavirus VP6. eLife 4, e06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greimann JC, and Lima CD (2008). Reconstitution of RNA Exosomes from Human and Saccharomyces cerevisiae. Cloning, Expression, Purification, and Activity Assays. Methods Enzymol. 448, 185–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbach F, Rode M, and Conti E (2012). The crystal structure of S. cerevisiae Ski2, a DExH helicase associated with the cytoplasmic functions of the exosome. RNA 18, 124–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halevy A, Lerer I, Cohen R, Kornreich L, Shuper A, Gamliel M, Zimerman B. El, Korabi I, Meiner V, Straussberg R, et al. (2014). Novel EXOSC3 mutation causes complicated hereditary spastic paraplegia. J. Neurol 261, 2165–2169. [DOI] [PubMed] [Google Scholar]
- Han J, and van Hoof A (2016). The RNA exosome channeling and direct access conformations have distinct in vivo functions. Cell Rep. 16, 3348–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández H, Dziembowski A, Taverner T, Séraphin B, and Robinson CV (2006). Subunit architecture of multimeric complexes isolated directly from cells. EMBO Rep. 7, 605–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RN, Klauer AA, Hintze BJ, Robinson H, Van Hoof A, and Johnson SJ (2010). The crystal structure of Mtr4 reveals a novel arch domain required for rRNA processing. EMBO J. 29, 2205–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januszyk K, Liu Q, and Lima CD (2011). Activities of human RRP6 and structure of the human RRP6 catalytic domain. RNA 17, 1566–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Wang X, Anderson JT, and Jankowsky E (2012). RNA unwinding by the Trf4/Air2/Mtr4 polyadenylation (TRAMP) complex. Proc. Natl. Acad. Sci. U. S. A 109, 7292–7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, and Anderson J (2004). Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 18, 1227–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilchert C, Wittmann S, and Vasiljeva L (2016). The regulation and functions of the nuclear RNA exosome complex. Nat. Rev. Mol. Cell Biol 17, 227–239. [DOI] [PubMed] [Google Scholar]
- Knight JRP, Bastide A, Peretti D, Roobol A, Roobol J, Mallucci GR, Smales CM, and Willis AE (2016). Cooling-induced SUMOylation of EXOSC10 down-regulates ribosome biogenesis. RNA 22, 623–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A, Sigworth FJ, and Tagare HD (2014). Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J, Kressler D, Tollervey D, and Linder P (1998). Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 17, 1128–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebreton A, Tomecki R, Dziembowski A, and Séraphin B (2008). Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature 456, 993–996. [DOI] [PubMed] [Google Scholar]
- Lim J, Giri PK, Kazadi D, Laffleur B, Zhang W, Grinstein V, Pefanis E, Brown LM, Ladewig E, Martin O, et al. (2017). Nuclear proximity of Mtr4 to RNA exosome restricts DNA mutational asymmetry. Cell 169, 523–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JJ, Bratkowski MA, Liu X, Niu CY, Ke A, and Wang HW (2014). Visualization of distinct substrate-recruitment pathways in the yeast exosome by em. Nat. Struct. Mol. Biol 21, 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JJ, Niu CY, Wu Y, Tan D, Wang Y, Ye M. Da, Liu Y, Zhao W, Zhou K, Liu QS, et al. (2016). CryoEM structure of yeast cytoplasmic exosome complex. Cell Res. 26, 822–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, and Lima CD (2006). Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 127, 1223–1237. [DOI] [PubMed] [Google Scholar]
- Lorentzen E, and Conti E (2012). Crystal structure of a 9-subunit archaeal exosome in pre- catalytic states of the phosphorolytic reaction. Archaea 2012, 721869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentzen E, Basquin J, Tomecki R, Dziembowski A, and Conti E (2008). Structure of the active subunit of the yeast exosome core, Rrp44: diverse modes of substrate recruitment in the RNase II nuclease family. Mol. Cell 29, 717–728. [DOI] [PubMed] [Google Scholar]
- Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, Andersen JS, Dziembowski A, and Jensen TH (2011). Interaction profiling identifies the human nuclear exosome targeting complex. Mol. Cell 43, 624–637. [DOI] [PubMed] [Google Scholar]
- Lubas M, Andersen PR, Schein A, Dziembowski A, Kudla G, and Jensen TH (2015). The human nuclear exosome targeting complex is loaded onto newly synthesized RNA to direct early ribonucleolysis. Cell Rep. 10, 178–192. [DOI] [PubMed] [Google Scholar]
- Makino DL, Baumgartner M, and Conti E (2013). Crystal structure of an rna-bound 11-subunit eukaryotic exosome complex. Nature 495, 70–75. [DOI] [PubMed] [Google Scholar]
- Makino DL, Schuch B, Stegmann E, Baumgartner M, Basquin C, and Conti E (2015). RNA degradation paths in a 12-subunit nuclear exosome complex. Nature 524, 54–58. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol 152, 36–51. [DOI] [PubMed] [Google Scholar]
- Matsumoto-Taniura N, Pirollet F, Monroe R, Gerace L, and Westendorf JM (1996). Identification of novel M phase phosphoproteins by expression cloning. Mol. Biol. Cell 7, 1455–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIver SC, Kang Y-A, DeVilbiss AW, O’Driscoll CA, Ouellette JN, Pope NJ, Camprecios G, Chang C-J, Yang D, Bouhassira EE, et al. (2014). The exosome complex establishes a barricade to erythroid maturation. Blood 124, 2285–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meola N, Domanski M, Karadoulama E, Chen Y, Gentil C, Pultz D, Vitting-Seerup K, Lykke-Andersen S, Andersen JS, Sandelin A, et al. (2016). Identification of a nuclear exosome decay pathway for processed transcripts. Mol. Cell 64, 520–533. [DOI] [PubMed] [Google Scholar]
- Molleston J, and Cherry S (2017). Attacked from All Sides: RNA Decay in Antiviral Defense. Viruses 9, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton DJ, Kuiper EG, Jones SK, Leung SW, Corbett AH, and Fasken MB (2018). The RNA exosome and RNA exosome-linked disease. RNA 24, 127–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogami K, Richard P, Chen Y, Hoque M, Li W, Moresco JJ, Yates JR, Tian B, and Manley JL (2017). An Mtr4/ZFC3H1 complex facilitates turnover of unstable nuclear RNAs to prevent their cytoplasmic transport and global translational repression. Genes Dev. 31, 1257–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pefanis E, Wang J, Rothschild G, Lim J, Kazadi D, Sun J, Federation A, Chao J, Elliott O, Liu Z-P, et al. (2015). RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 161, 774–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera - A visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Puno MR and Lima CD (2018). A structural basis for MTR4-ZCCHC8 interactions that stimulate the MTR4 helicase in the nuclear exosome targeting complex. Proc. Natl. Acad. Sci. USA Published online May 29, 2018 10.1073/pnas.1803530115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). CryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296. [DOI] [PubMed] [Google Scholar]
- Scheres SHW (2012). RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilders G, van Dijk E, and Pruijn GJM (2007). C1D and hMtr4p associate with the human exosome subunit PM/Scl-100 and are involved in pre-rRNA processing. Nucleic Acids Res. 35, 2564–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, and Tollervey D (2013). Threading the barrel of the RNA exosome. Trends Biochem. Sci 38, 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Leung E, Brown J, and Tollervey D (2009). The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res. 37, 1127–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger L (2015). The PyMol Molecular Graphics System, Version 1.8.
- Schuch B, Feigenbutz M, Makino DL, Falk S, Basquin C, Mitchell P, and Conti E (2014). The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase. EMBO J. 33, 2829–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller JM, Falk S, Fromm L, Hurt E, and Conti E (2018). Structure of the nuclear exosome captured on a maturing preribosome. Science 5428, eaar5428. [DOI] [PubMed] [Google Scholar]
- Sloan KE, Mattijssen S, Lebaron S, Tollervey D, Pruijn GJM, and Watkins NJ (2013). Both endonucleolytic and exonucleolytic cleavage mediate ITS1 removal during human ribosomal RNA processing. J. Cell Biol 200, 577–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead JA, Costello JL, Livingstone MJ, and Mitchell P (2007). The PMC2NT domain of the catalytic exosome subunit Rrp6p provides the interface for binding with its cofactor Rrp47p, a nucleic acid-binding protein. Nucleic Acids Res. 35, 5556–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehmeier P, and Muller S (2009). Phospho-Regulated SUMO Interaction Modules Connect the SUMO System to CK2 Signaling. Mol. Cell 33, 400–409. [DOI] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter S, and Carragher B (2005). Automated molecular microscopy: The new Leginon system. J. Struct. Biol 151, 41–60. [DOI] [PubMed] [Google Scholar]
- Tafforeau L, Zorbas C, Langhendries J-L, Mullineux S-T, Stamatopoulou V, Mullier R, Wacheul L, and Lafontaine DLJ (2013). The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of pre-rRNA processing factors. Mol. Cell 51, 539–551. [DOI] [PubMed] [Google Scholar]
- Tomecki R, Kristiansen MS, Lykke-Andersen S, Chlebowski A, Larsen KM, Szczesny RJ, Drazkowska K, Pastula A, Andersen JS, Stepien PP, et al. (2010). The human core exosome interacts with differentially localized processive RNases: HDIS3 and hDIS3L. EMBO J. 29, 2342–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C-K, Wang H-F, Burns AM, Schroeder MR, Gaspari M, and Baumann P (2015). Human telomerase RNA processing and quality Control. Cell Rep. 13, 2232–2243. [DOI] [PubMed] [Google Scholar]
- Wan J, Yourshaw M, Mamsa H, Rudnik-Schöneborn S, Menezes MP, Hong JE, Leong DW, Senderek J, Salman MS, Chitayat D, et al. (2012). Mutations in the RNA exosome component gene EXOSC3 cause pontocerebellar hypoplasia and spinal motor neuron degeneration. Nat. Genet 44, 704–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H-W, Wang J, Ding F, Callahan K, Bratkowski MA, Butler JS, Nogales E, and Ke A (2007). Architecture of the yeast Rrp44 exosome complex suggests routes of RNA recruitment for 3′ end processing. Proc. Natl. Acad. Sci 104, 16844–16849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Jia H, Jankowsky E, and Anderson JT (2008). Degradation of hypomodified tRNA(iMet) in vivo involves RNA-dependent ATPase activity of the DExH helicase Mtr4p. RNA 14, 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth EV, Zinder JC, Zattas D, Das M, and Lima CD (2017). Structure and reconstitution of yeast Mpp6-nuclear exosome complexes reveals that Mpp6 stimulates RNA decay and recruits the Mtr4 helicase. Elife 6, e29062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth EV, Januszyk K, and Lima CD (2014). Structure of an Rrp6-RNA exosome complex bound to poly(A) RNA. Nature 511, 435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, and Barton GJ (2009). Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir JR, Bonneau F, Hentschel J, and Conti E (2010). Structural analysis reveals the characteristic features of Mtr4, a DExH helicase involved in nuclear RNA processing and surveillance. Proc. Natl. Acad. Sci. U. S. A 107, 12139–12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weißbach S, Langer C, Puppe B, Nedeva T, Bach E, Kull M, Bargou R, Einsele H, Rosenwald A, Knop S, et al. (2015). The molecular spectrum and clinical impact of DIS3 mutations in multiple myeloma. Br. J. Haematol 169, 57–70. [DOI] [PubMed] [Google Scholar]
- Wood V, Harris MA, McDowall MD, Rutherford K, Vaughan BW, Staines DM, Aslett M, Lock A, Bahler J, Kersey PJ, et al. (2012). PomBase: A comprehensive online resource for fission yeast. Nucleic Acids Res. 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle J-C, Dufour M-E, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. (2005). Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121, 725–737. [DOI] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder JC, and Lima CD (2017). Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors. Genes Dev. 31, 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder JC, Wasmuth EV, and Lima CD (2016). Nuclear RNA exosome at 3.1 Å reveals substrate specificities, RNA paths, and allosteric inhibition of Rrp44/Dis3. Mol. Cell 64, 734–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Nomenclature of exosome subunits and co-factors from Homo sapiens, Saccharomyces cerevisiae, and Schizosaccharomyces pombe. (B) Time-course assays for degradation of the DNA-RNA chimera in the presence of AMPPNP (left) or ATP (right) (similar to Figure 2C). The substrate used is schematically represented at the top; cartoons are labeled to represent subunit compositions. (C) ATP-dependent unwinding of the DNA-RNA chimera by 12-, 13- and 14-component exosomes as well as Mtr4. Marker (M) for single stranded species with sizes indicated to the right and marker (M2) for the displaced fluorescein-labelled strand with trap oligo. Gel images in panels B and C are representative of two independent experiment for S. cerevisiae and three independent experiments for S. pombe and human. (D) Sybr Gold-stained gels of proteinase K-digested fractions after RNA-unwinding and size exclusion chromatography. Top: Fractions 17–22 with corresponding RNA markers (MS=substrate; MP=loaded product) at indicated concentrations. Bottom: Fractions 24–29 with corresponding RNA markers. Band intensity does not scale between individual gels. * denotes peak fractions containing unwound reaction product. (E) Coomassie stained SDS-PAGE protein fractions corresponding to panel D. Gel images in panels D and E are representative of two independent experiments. Load = RNA unwinding reactions prior to gel filtration; M = molecular weight marker.
Boxed maps (green) with labels represent endpoints for those maps available for download in EMDB.
(A) Schematic representing the angular distribution of particles contributing to the overall reconstruction as reported by RELION and displayed using Chimera with the overall reconstruction rendered as isosurface inside the sphere in four orientations. (B,C) Overall reconstruction and gold-standard FSC. (D,E) Reconstruction after focused refinement on the core and gold-standard FSC. (F,G) Reconstruction after focused refinement on the DIS3-PIN and gold-standard FSC. (H,I) Reconstruction after focused refinement on the DIS3-RNBCS and gold-standard FSC. (J,K) Reconstruction after focused refinement on MTR4 and gold-standard FSC. (L,M) Reconstruction after focused refinement on a subclass of MTR4 particles elicited more continuous densities for the duplex DNA/RNA segment of the substrate.The six reconstructions analyzed in this study (and boxed in Figure S2) are isosurface rendered and colored according to local resolution as calculated by RESMAP (Kucukelbir et al., 2014). Separate from this calculation, the maps were low-pass filtered at 4.5 Å resolution to enable visualization of the most disordered regions at low contour threshold (left), at higher contour threshold (middle), and as a central slice (right) in panels B, D, F, H, J, and L. Color code for resolution shown adjacent to each map. Gold-standard FSC curves corresponding to each reconstruction with FSC=0.143 represented as a dashed line and resolutions indicated are shown at far right.
Labels indicated for each map as labeled in Figure S2 and S3. Panels showing regions around: (A) MTR4 in the overall map. (B) MTR4 in the MTR4-focused map. (C) MTR4 the MTR4-dsDNA/RNA focused map. (D) Regions within the core for the overall map (E) Regions within the core for the core focused map. (F) Densities surrounding the PIN domain of DIS3 for the overall map (G) Densities surrounding the PIN domain of DIS3 for the DIS3 PIN focused map. (H) Densities surrounding the DIS3 RNB and cold-shock domains for the overall map. (I) Densities surrounding the DIS3 RNB and cold-shock domains for the DIS3 RNBCS focused map.
(A) 2D class averages obtained from cryoSPARC calculated using all particles selected for the final 3D reconstruction. (B) A representative micrograph from the dataset used to determine the structure showing particles (left) and with green circles depicted around selected particles (right). Scale bar at lower right (30 nm). (C) Gold-standard FSC between the respective coordinate models for the overall refinement and local regions of the complex as indicated as calculated by phenix.mtriage. FSC values at 0.5 and 0.143 are reported in panel A. The DIS3 PIN focused refinement utilized a mask covering the PIN and most of the three adjacent PH-like proteins, however the gold-standard FSC between the DIS3 PIN model and the corresponding focused refinement map overestimated the resolution of the DIS3 PIN if core subunits were included. Therefore, the DIS3 PIN was compared to a modified version of the focused refinement map whereby densities attributed to the core subunits were removed. (D) Gold-standard FSC curve (FSC=0.143, dotted line) from RELION reconstruction shown in Figure 5G. (E) Gold-standard FSC curve (FSC=0.143, dotted line) from RELION reconstruction shown in Figure 5H.
(A) Structural representation of contacts between EXOSC10 and EXOSC6 as well as EXOSC8 colored as in Figure 3. (B) Stereo view of electron densities overlaying the corresponding areas shown in panel A with blue mesh for EXOSC10 and grey mesh for EXOSC6 and EXOSC8. (C) View of DIS3 contacts to the core with boxes representing regions indicated in detail in panels D-I. Subunits, domains and RNA are labeled and colored as in Figure 3. (D) Interactions between EXOSC9 and the DIS3 RNB domain. (E) Interactions between EXOSC9 and the DIS3 CSD2 domain. (F) Interactions between the DIS3 CSD1 domain and EXOSC7 and EXOSC8. (G) Interactions between the DIS3 CSD2 domain and EXOSC9 and EXOSC4. (H) Interactions between EXOSC4 and the DIS3 PIN domain. (I) Interactions between the DIS3 PIN domain and EXOSC4 and EXOSC7.Side chains shown in stick representation, labeled by residue number (colored by subunit), polar interactions indicated by dashed lines between atoms, and domains indicated by labels.
(A) RNA contacts at a conserved interface between EXOSC9 and EXOSC4. (B) RNA contacts within the active site between Rrp41/Rrp42 in the phosphorolytic archaeal S. solfataricus exosome (PDBID: 2C37). Structures were aligned with protein and RNA shown in stick representation with amino acids colored and labeled and bases in the human complex labeled. RNA bound Dis3 structures shown to depict different RNA paths for (C) Human DIS3. (D) Mouse DIS3L2 (PDBID: 4PMW). (E) S. cerevisiae Dis3 from a core bound structure in the ‘open’ conformation (PDBID: 5K36). (F) S. cerevisiae Dis3 from a core bound structure in the ‘closed’ conformation (PDBID: 4IFD). RNB domains were used to align the respective Dis3 structures with Dis3 domains colored as CSD1 (orange), CSD2 (limon), RNB (pink), and S1 (purple). RNA in respective structures is depicted as a blue tube to highlight the different binding modes. The PIN domains for human DIS3 and budding yeast structures are omitted for clarity. (G) Amino acid sequence alignment of Mtr4 from various species. Alignment generated using Muscle alignment tool in Jalview (Waterhouse et al., 2009) with conservation based on BLOSUM62 scoring matrix. EXOSC2 residues that interact with MTR4 in this structure are indicated by green bars above the sequence. (H) Alignment of EXOSC2 and Rrp4 proteins generated using MUSCLE multiple sequence alignment tool. Conservation based on BLOSUM62 scoring in Jalview. Colored bars indicate surfaces involved in protein-protein interactions based on assessment of the human structure (this work) and a previously published S. cerevisiae structure (Zinder et al., 2016)(PDBID: 5K36). (I) Sequence alignment of MPP6 from indicated organisms. Alignment generated using MUSCLE alignment tool and manually corrected for S. cerevisiae and H. sapiens based on alignment of the human structure to a published structure from yeast (PDBID: 5VZJ)(Wasmuth et al., 2017). Conservation indicated by shading and based on BLOSUM62 scoring in Jalview (Waterhouse et al., 2009). Tick marks correspond to 10 amino acid intervals for the human MPP6 sequence. Colored bars indicate residues involved in protein-protein interactions with MTR4, EXOSC1 and EXOSC3 (or Rrp40) with dark shading indicating direct contacts based on PDBe PISA (cutoff for buried area 50%) and further visual assessment of the corresponding density for the human structure. Dark gray lines above or below the sequence alignment indicate residues modeled in this work or previous work in the yeast system, respectively. C-terminal amino acids from all species were omitted for clarity.