Abstract
Introduction
Melanoma is a malignant tumor that seriously affects patients. The pathogenesis of malignant melanoma is complex, and the cell cycle is closely related to tumor progression. Based on the catalog of cancer somatic mutations, we found that overexpression of the E2F3 gene ranked first in percentage increase in not only melanoma but also in all human cancer tissues. However, there are few studies on the high expression of E2F3 and its carcinogenic mechanism in melanoma.
Methods and results
We found that E2F3 showed extensive copy number amplification that was positively correlated with the expression level. Patients with high copy number had a significantly poorer prognosis. We also found that E2F3 levels were significantly negatively correlated with promoter methylation. However, we showed that the E2F3 promoter region is hypomethylated, and in normal cells or tumor cells, the methylation level did not correlate with expression. Finally, we knocked down the E2F3 gene in melanoma cells by shRNA. Colony formation, anchorage-dependent growth, and EdU cell proliferation experiments showed a significant decrease in proliferation. Flow cytometry showed a significant increase in the G0/G1 ratio.
Conclusion
It can be speculated that copy number amplification and other mechanisms result in the high expression of E2F3 in melanoma, which promotes tumor progression by involving the cell cycle. E2F3 is a good target for the treatment of melanoma.
Keywords: E2F3, melanoma, mechanism, overexpression, copy number, methylation
Video abstract
Introduction
Malignant melanoma is the most aggressive malignant tumor in the skin, and there is a clear trend toward familial clustering.1 Of all patients who die of skin tumors, approximately 80% have malignant melanoma.2 In 2018, the number of new cases of malignant melanoma in the US was estimated to be 91,270, and the estimated death toll was 9,320.3
The cell cycle includes four consecutive phases, from the stationary phase (G0 phase) to the proliferative phases (G1 phase, S phase, G2 phase, and M phase) and back to the stationary phase.4 The key to developing malignant cells is the destruction of normal cell cycle progression, especially in G1 phase. Subsequently, cell cycle-associated regulatory proteins and signal transduction pathways are altered to allow tumor cells to successfully undergo cell cycle progression, lose their ability to differentiate, and undergo malignant proliferation.5
The mechanistic mechanism of melanoma involves the abnormal expression of multiple genes associated with cell cycle. The most important of these genes are those that play an important role in normal cell cycle activity but are overexpressed in cancer, interfering with the cell cycle and promoting the development of tumors. For example, the USP39 gene is highly expressed in melanoma. Silencing USP39 can inhibit the proliferation of melanoma cells in vitro and in vivo, induce G0/G1 arrest, and promote apoptosis. The STAT3 gene is highly expressed in melanoma, and STAT3 promotes tumor cell growth by regulating the expression of genes involved in cell survival and proliferation.6 The cyclin E gene is also highly expressed in melanoma. Cyclin E interacts with CDK2 to form the cyclin E-CDK2 complex, which is highly expressed in both local and metastatic melanoma.7
With the release of relevant cancer data in The Cancer Genome Atlas (TCGA) database, cancer-related research has advanced. In this study, we found in the catalog of cancer somatic mutations (TCGA database) that the E2F3 gene ranked first with a 28.12% increase in expression in melanoma. However, few studies have examined the role of E2F3 in melanoma. We will explore the mechanisms that lead to elevated E2F3 in melanoma and evaluate its associated carcinogenic properties.
Methods
Bioinformatics
The catalog of cancer somatic mutations8 is the world’s greatest source of manual mapping of somatic mutations related to human cancer and is used to assess methylation mutations in human cancer tissues. TCGA melanoma expression level data, copy number data, and methylation data were downloaded from the TCGA website (https://cancergenome.nih.gov/). The Oncomine website (https://www.oncomine.org/resource/login.html) provided E2F3 expression analysis in the TCGA and gene expression omnibus (GEO) (accession number GSE3189 and GSE7553) databases. In addition, we have provided two supplemental tables on the data used in this article. More details about the raw data can be found in Tables S1 and S2.
Cell culture
The human malignant melanoma cell lines SK-MEL-28, MuM-2C, A-375, and HaCaT were purchased from American Type Culture Collection, Manassas, VA, USA, and the cells were maintained according to the manufacturer’s protocol. The cells were cultured with Dulbecco’s Modified Eagle’s Medium (DMEM; Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal bovine serum and antibiotics (100 µg/mL streptomycin and 100 U/mL penicillin). All cells were cultured in a humidified environment at 37°C and 5% CO2. The use of the cell lines was approved by the ethics board. of Xiangya Hospital of Central South University.
Lentiviral production and transduction
The shRNA targeting E2F3 (5′-TTGCGTTACTTTAA GTACTAA-3′; shR-E2F3; Thermo Fisher Scientific) was designed, and the scramble sequence (5′-TTCTCCGAACG TGTCACGT-3′) was used as a lentivirus negative control (NC). The human malignant melanoma cell line A375 was transfected with E2F3-shRNA and NC-shRNA viruses. The transfection efficiency of the recombinant lentivirus was tested by fluorescence microscopy (Olympus Corporation, Tokyo, Japan), and RT-PCR was used to assess the efficiency of knockdown.
RNA purification and reverse transcription PCR
Total RNA was extracted using TRIZOL reagent (Thermo Fisher Scientific). The reverse transcription reaction was performed using a ReverTra Ace qPCR RT Master Mix with a gDNA remover (Toyobo Co., Ltd., Osaka, Japan). The cDNA was amplified by the KODSYBR® qPCR Mix (Toyobo Co., Ltd.) on a LightCycler® 480II System (Hoffmann-La Roche, Basel, Switzerland) according to the manufacturer’s instructions. The primer sequences for the target gene were as follows: forward primer 5′-TATCCCTAAACCCGCTTCC-3′, reverse primer 5′-TTCACAAACGGTCCTTCTA-3′.
DNA purification
DNA was isolated using the Easypure Genomic DNA Kit (TransGen Biotech, Beijing, People’s Republic of China) according to the manufacturer’s instructions. The concentration of DNA was assessed spectrophotometrically and confirmed by gel electrophoresis, and the DNA was stored at −20°C. The results were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene expression.
Analysis of DNA methylation
For bisulfite-polymerase chain reaction, the BSP primer was designed in MethPrimer 2.0.9 Then, the bisulfate-modified DNA was amplified with forward and reverse primers for target genes (bisulfite sequencing or restriction PCR, BSP). The DNA fragments were purified using the Gene JET Gel Extraction Kit (Thermo Fisher Scientific) and mixed at the appropriate proportions. According to the kit instructions, the second-generation sequencing library was prepared with the VAHTSTM Turbo DNA Library Prep Kit (Vazyme Biotech Co., Ltd., Nanjing, China) for Illumina Kits (Illumina, Inc., San Diego, CA, USA), and sequencing was performed with the Illumina MiSeq high-throughput sequencing platform (Illumina, Inc.). The primers for the bisulfite sequencing were as follows: forward primer 5′-GGATYGTTTTAGGTTAGGGAGT-3′, reverse primer 5′-AAAARGAAAACAACTAAATTCC-3′.
Plate colony formation assay
Melanoma cells transfected with E2F3-shRNA lentivirus were used to prepare cell suspensions and counted. Two hundred cells were added to a six-well plate. The cells were further cultured in a cell incubator for 14 days, the cell culture medium was changed every 3 days, and the cell state was observed. Photographs were taken with a fluorescence microscope before the end of the experiment, and the cells were washed twice with PBS. Then, 500 µL of Giemsa dye was added to each well, and the cells were stained for 10–20 minutes. The cells were washed three times with ddH2O and photographed with a digital camera.
Anchorage-dependent growth
The cells were suspended in 0.3% agar medium (DMEM containing 10% FBS) and plated on a 0.6% agar matrix layer at a concentration of 3.0×104 cells/60 mm dish. The cells were cultured in a humidified environment (5% CO2) at 37°C. The number of colonies that were 50 µm or larger were counted after 2 weeks.
EdU cell proliferation assay
Lentivirus-transfected melanoma cells in the logarithmic growth phase were seeded in 96-well plates. Then, 100 µL of 50 µmol/L EdU medium was added per well, and the plates were washed with PBS after 2 hours and fixed. Each well was sequentially supplemented with 100 µL of 1× Apollo staining reaction solution and 100 µL of 1× Hoechst 33,342 nuclear staining reaction solution, evaluated by EdU detection and photographed by inverted fluorescence microscopy.
Cell cycle analysis
Melanoma cells transfected with E2F3-shRNA lentivirus were incubated in six-well plates. When the cells reached 80% confluence, the medium was removed, the cells were suspended, and the supernatant was discarded by centrifugation. The cell pellet was washed with cold PBS. Cells were collected by centrifugation and fixed in 75% cold ethanol for 1 hour. Prior to analysis, the cells were washed with PBS again and suspended in a solution containing 10 µg/mL RNase A and 50 µg/mL propidium iodide. The cell suspension was filtered at a rate of 300 cells/second, and the cell cycle was analyzed by a FACSCalibur™ flow cytometer (BD Biosciences, San Jose, CA, USA).
Statistical analysis
GraphPad Prism 7 software was used to analyze the data. Methylation, copy number variation (CNV), and expression levels between normal and cancer tissues were assessed by a two-tailed unpaired Student’s t-test, and the error bars in the figure represent the SD or SEM. Spearman’s correlation coefficient (r) was used to determine the correlation. Receiver operator characteristic curves were constructed based on the level of E2F3 gene expression. The data were considered significant at P<0.05.
Results
High expression of the E2F3 gene in melanoma
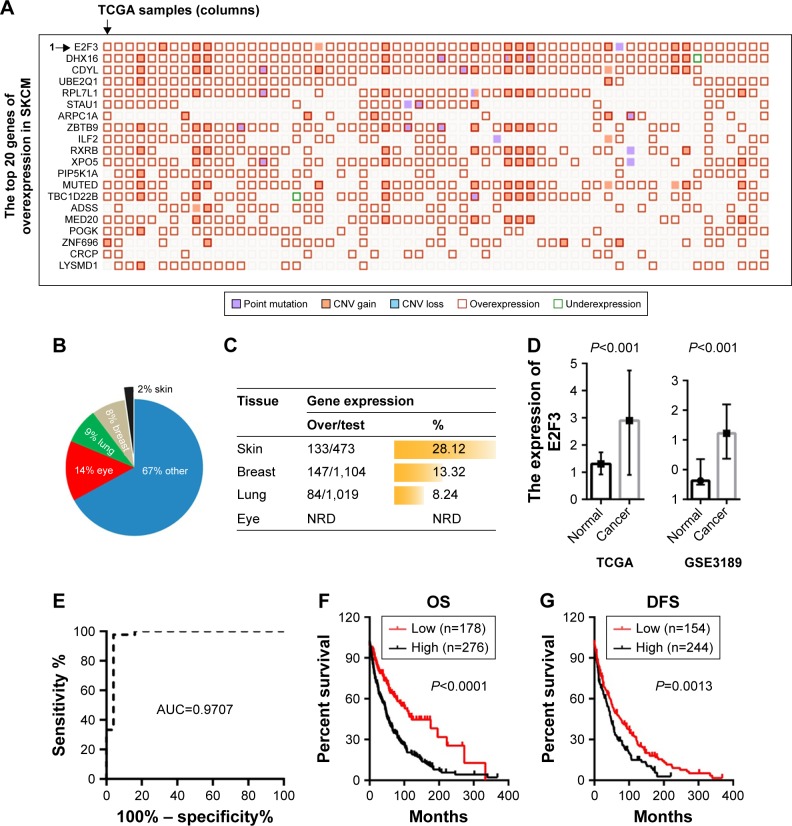
Based on the TCGA data, we found that among the genes with high expression in melanoma, the E2F3 gene ranked first with an increase of 28.12% (Figure 1A and C). The E2F3 gene may promote the development of melanoma. It is worth noting that E2F3-related research in the skin is extremely rare, while studies in the eye, lung, and breast are more common (Figure 1B). We found that the lung and breast ranked second and third, respectively, while melanoma ranked first (Figure 1C). Therefore, it is necessary to study the relationship between E2F3 and melanoma. Further study found that E2F3 gene expression was increased in the TCGA and GEO databases (Figure 1D), and the ROC curve of GSE3189 revealed that E2F3 can identify melanoma tissue (AUC=0.9709) (Figure 1E). Survival analysis showed that high E2F3 expression results in decreases in OS and disease-free survival (DFS) of patients (Figure 1F and G).
Figure 1.
High expression of the E2F3 gene in melanoma and its influence on prognosis.
Notes: (A) A “Mutation Matrix” plot between genes and samples of melanoma tissue that contains 20 top ranked genes (rows) and TCGA samples (columns) with each box representing a Gene-Sample combination. The E2F3 gene ranked as the top melanoma-related gene. (B) E2F3 has been studied in many tissues, including eye, lung, and breast, but studies of the skin are extremely rare. (C) The overexpression percentage (%) is represented as a histogram across different primary tissue types, and the overexpression percentage of the E2F3 gene in melanoma is the highest. (D) The E2F3 gene shows an increased expression level in melanoma tissues compared with normal tissues. The left was derived from the TCGA database, and the right was from the GSE3189 (more details can be found at https://www.oncomine.org/). (E) The ROC curve of GSE3189. (F) The overexpression of the E2F3 gene affected the OS of 454 melanoma patients. (G) The overexpression of the E2F3 gene affected the DFS (disease free since initial treatment) of 398 melanoma patients.
Abbreviations: CNV, copy number variation; DFS, disease-free survival; NRD, no related data; OS, overall survival; SKCM, skin cutaneous melanoma; TCGA, The Cancer Genome Atlas.
The relationship between CNV and E2F3 expression level
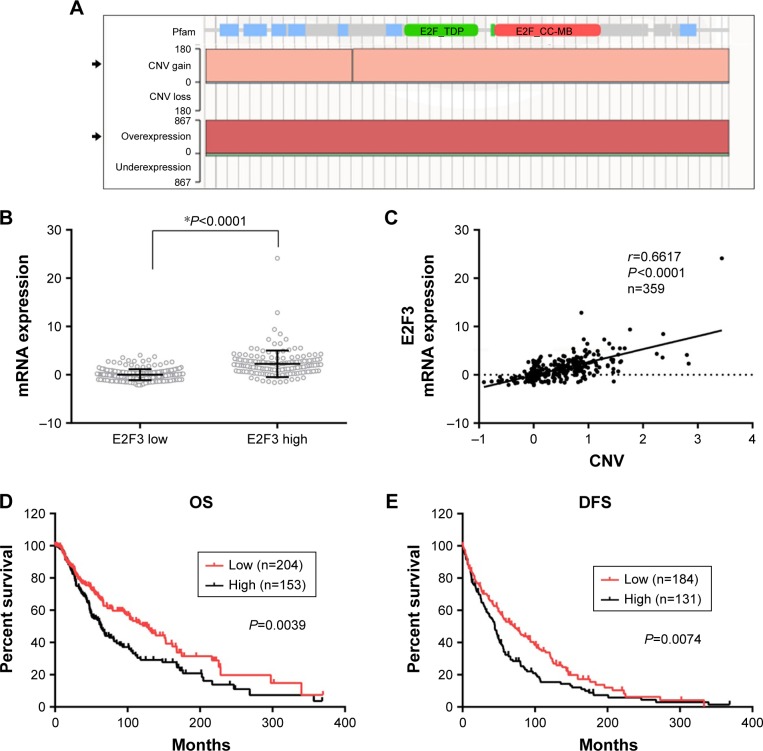
We sought to explore the cause of the high expression of E2F3. First, we found that E2F3 showed extensive copy number amplification (CNA) and high DNA expression in solid cancers (Figure 2A), and amplification was the main cause of high gene expression. Therefore, we hypothesized that the high expression of E2F3 in melanoma was caused by CNA. The TCGA database showed that the expression E2F3 level significantly increased in tissues with high copy number compared with tissues with low copy number (Figure 2B). The expression of the E2F3 gene was significantly positively correlated with CNV level (Figure 2C), showing that CNA is one of the causes of the increase in E2F3. More importantly, we found that CNV was significantly associated with clinical prognosis, with significantly lower OS and DFS in patients with high copy number (Figure 2D and E).
Figure 2.
Copy number variation results in the high expression of the E2F3 gene and affects prognosis.
Notes: (A) COSMIC shows that E2F3 variations in solid cancer are dominated by CNV gains and overexpression. (B) The E2F3 gene shows an increased expression level in patients with high E2F3 copy number (unpaired t-test, *P<0.0001). (C) The expression of the E2F3 gene is significantly positively correlated with the CNV level. (D) The copy number of the E2F3 gene affects the OS of 357 melanoma patients. (E) The copy number of the E2F3 gene affects the DFS of 315 melanoma patients.
Abbreviations: CNV, copy number variation; COSMIC, catalog of cancer somatic mutations; DFS, disease-free survival; OS, overall survival.
The relationship between DNA methylation and E2F3 gene expression in the promoter region
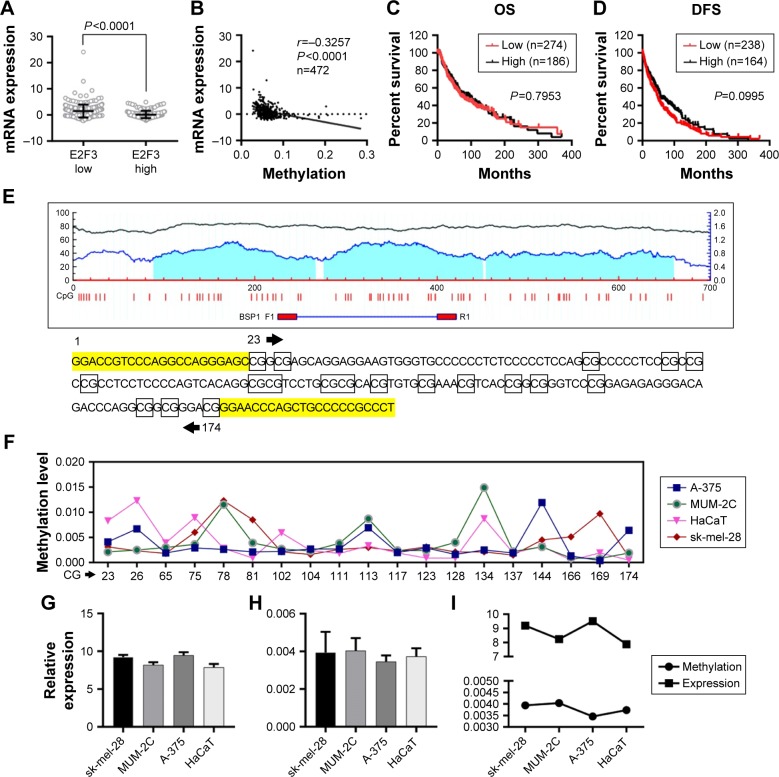
In addition to CNA, promoter methylation is another mechanism that controls expression levels that should not be overlooked. In investigating whether high E2F3 expression is regulated by methylation, we found that E2F3 expression in hypermethylated tissues was obviously lower than that in tissues with low methylation by analyzing the TCGA database (Figure 3A), and the methylation level was significantly negatively correlated with the E2F3 expression level (Figure 3B). However, we showed that methylation level did not affect OS or DFS (Figure 3C and D). The promoter region of E2F3 was heavily enriched with CpG islands (Figure 3E), further confirming this trend. The methylation status of 19 CpG sites in the promoter region in four cell lines was analyzed by bisulfite sequencing PCR. The results showed that the promoter region was hypom-ethylated in both normal cells and malignant tumor cells and that there was no correlation between methylation and expression levels (Figure 3F–I; Tables S3 and S4). These results suggest that methylation may not be the cause of the elevated E2F3 expression.
Figure 3.
DNA methylation does not result in the high expression of the E2F3 gene or affect prognosis.
Notes: (A) The E2F3 gene shows an increased expression level in patients with hypomethylation. (B) The expression of the E2F3 gene is significantly inversely correlated with the methylation level. (C and D) The methylation of the E2F3 gene does not affect OS and DFS. (E) The promoter region of E2F3 is enriched with CpG islands (more details can be found in MethPrimer at http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi). The BSP1 synthesis sequence F1R1 contains 19 CG sites. (F) Methylation of 19 CG sites in four cell lines. (G) Relative expression by PCR. (H) Methylation levels in cells. (I) Methylation and expression levels are inconsistent in the four cell lines.
Abbreviations: DFS, disease-free survival; OS, overall survival.
Carcinogenic properties of the E2F3 gene
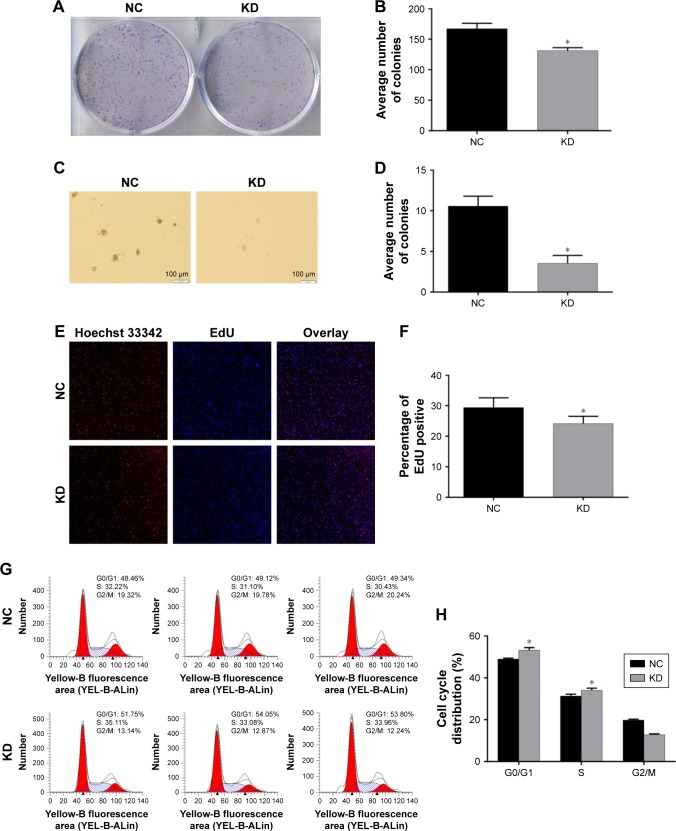
Research about whether high E2F3 expression can promote the progression of melanoma is still lacking. To solve this problem, we successfully knocked down the E2F3 gene in melanoma cells. Melanoma cells that lacked the E2F3 gene showed reduced proliferation in colony formation (Figure 4A and B), anchorage-dependent growth (Figure 4C and D), and EdU cell proliferation experiments (Figure 4E and F). These experiments show that E2F3 can affect cell proliferation. At the same time, we found that the G0/G1 population increased significantly after E2F3 gene knockdown (Figure 4G and H), which indicates that E2F3 regulates the cell cycle and promotes melanoma.
Figure 4.
Carcinogenic properties of the E2F3 gene.
Notes: (A) Colony formation of the cells after transient transfection with E2F3-shRNA or NC-shRNA. (B) Histogram showing colony formation in the groups. (C) Images of anchorage-dependent growth. (D) The average number of colonies decreased following E2F3 gene knockdown. (E) The E2F3 gene affects cell proliferation. Proliferation was assessed using an EdU cell proliferation assay kit (magnification, 100×). (F) The cell proliferation rate decreased following E2F3 gene knockdown. (G) The cell cycle distribution in melanoma cells transfected with E2F3-shRNA or NC-shRNA. (H) Histogram showing the cell cycle distribution (%). All the results are shown as the mean ± SEM, *P<0.05.
Abbreviations: KD, knockdown; NC, negative control.
Discussion
The E2F3 gene is located on chromosome 6p22 and is 91.5 kb in length. E2F3 is a major member of the E2F family and plays an important role in regulating the cell cycle, proliferation, and apoptosis.10
Feber et al found E2F3 gene amplification on chromosome 6p22 in the bladder cancer cell lines TCCSUP and HT1376. Analysis showed that higher clinical stage and pathological grade of bladder cancer corresponded with a higher positive rate of E2F3 expression.11 Silven Bilke et al found that approximately 2/3 of prostate cancers showed high expression of E2F3. E2F3 can be used as an independent factor to predict the OS of prostate cancer patients through multivariate analysis.12 Ren et al found that miR-449a inhibits the expression of E2F3, which decreased when miR-449a was overexpressed in the lung cancer cell lines A549 and 95D. Another study found cell cycle arrest in G1 phase and inhibition of cell proliferation.13 Libertini et al found that the expression of E2F3 in laryngeal squamous cell carcinoma was significantly higher than that in normal nasopharyngeal mucosa (P<0.001), and less differentiation correlated with a higher expression level (P<0.05).14 However, it is worth noting that we found that the expression level of the E2F3 gene was the highest in melanoma, ranking first among all tissues, but few studies have explored this relationship. Therefore, it is necessary to conduct in-depth studies. We also confirmed that E2F3 can interfere with the cell cycle to promote the development of melanoma.
Many studies have found that CNV is an important factor affecting the prognosis of cancer patients. Specific CNVs can be used to assess prognosis.15 A meta-analysis showed that MET gene amplification is inversely related to the OS of non-small-cell lung cancer patients and increases the risk of death.16 Liu et al reported that lung cancer patients with four copies of the mitogen-activated protein kinase-activated protein kinase 2 (MAPKAPK2) gene promoter have a shorter median survival and a 47% increased risk of death compared to those with two or three copies, which shows that the MAPKAPK2 gene can be used as a genetic marker to predict the prognosis of lung cancer.17 We confirmed that E2F3 was also regulated by this mechanism.
In addition to CNV, promoter methylation has been shown to affect gene expression in numerous studies. For example, Kim et al found that hypomethylation of the NAT1 gene may increase the transcriptional activation of breast cancer genes. Therefore, the hypomethylated NAT1 gene plays an important role in the development of breast cancer.18 Other related studies include those on FGFR (sarcoma),19 PRAME (leukemia),20 and IGF2 (colorectal cancer).21 The promoter hypermethylation of these genes is closely related to the occurrence of the corresponding tumors. In our study, we found that the expression level of the E2F3 gene was negatively correlated with methylation but did not affect the prognosis of patients. Moreover, there was no correlation between the methylation and expression levels in four cell lines. It can be inferred that methylation may be one of the reasons for the high expression of E2F3, but more research needs to be done.
In summary, we discussed the mechanism of the high expression of E2F3 in melanoma. Finally, we knocked down the E2F3 gene with lentivirus, which significantly reduced the proliferation of melanoma cells and increased the proportion of cells in the G0 phase. Therefore, it can be speculated that E2F3 can promote cancer. However, considering the extensive and high expression of E2F3, more in-depth research is needed.
Supplementary materials
Table S1.
| Samples | Name column | E2F3 |
|---|---|---|
| GSM71671 | Normal | 215.25 |
| GSM71672 | Normal | 153.95 |
| GSM71673 | Normal | 265.3 |
| GSM71674 | Normal | 199.4 |
| GSM71675 | Normal | 327.95 |
| GSM71676 | Normal | 163.7 |
| GSM71677 | Normal | 203.3 |
| GSM71678 | Nevus | 319.5 |
| GSM71679 | Nevus | 283.85 |
| GSM71680 | Nevus | 369.7 |
| GSM71681 | Nevus | 225.4 |
| GSM71682 | Nevus | 199.2 |
| GSM71683 | Nevus | 270.55 |
| GSM71684 | Nevus | 295.1 |
| GSM71685 | Nevus | 304.3 |
| GSM71686 | Nevus | 326.3 |
| GSM71687 | Nevus | 290.7 |
| GSM71688 | Nevus | 306.95 |
| GSM71689 | Nevus | 372.55 |
| GSM71690 | Nevus | 184.05 |
| GSM71691 | Nevus | 365.15 |
| GSM71692 | Nevus | 369.8 |
| GSM71693 | Nevus | 294.65 |
| GSM71694 | Nevus | 367.75 |
| GSM71695 | Nevus | 642 |
| GSM71696 | Melanoma | 584.9 |
| GSM71697 | Melanoma | 669.35 |
| GSM71698 | Melanoma | 439.55 |
| GSM71699 | Melanoma | 493.05 |
| GSM71700 | Melanoma | 417.3 |
| GSM71701 | Melanoma | 549.8 |
| GSM71702 | Melanoma | 649.4 |
| GSM71703 | Melanoma | 558.9 |
| GSM71704 | Melanoma | 449.25 |
| GSM71705 | Melanoma | 689.95 |
| GSM71706 | Melanoma | 713.9 |
| GSM71707 | Melanoma | 786.1 |
| GSM71708 | Melanoma | 1,071.75 |
| GSM71709 | Melanoma | 405.55 |
| GSM71710 | Melanoma | 686.95 |
| GSM71711 | Melanoma | 634.85 |
| GSM71712 | Melanoma | 464.4 |
| GSM71713 | Melanoma | 624.65 |
| GSM71714 | Melanoma | 610.55 |
| GSM71715 | Melanoma | 676.65 |
| GSM71716 | Melanoma | 810.25 |
| GSM71717 | Melanoma | 659.05 |
| GSM71718 | Melanoma | 498.05 |
| GSM71719 | Melanoma | 604.75 |
| GSM71720 | Melanoma | 368.95 |
| GSM71721 | Melanoma | 526.55 |
| GSM71722 | Melanoma | 511.15 |
| GSM71723 | Melanoma | 582.1 |
| GSM71724 | Melanoma | 519.85 |
| GSM71725 | Melanoma | 740.1 |
| GSM71726 | Melanoma | 602.05 |
| GSM71727 | Melanoma | 399.4 |
| GSM71728 | Melanoma | 435.4 |
| GSM71729 | Melanoma | 423.1 |
| GSM71730 | Melanoma | 423 |
| GSM71731 | Melanoma | 557.65 |
| GSM71732 | Melanoma | 537.55 |
| GSM71733 | Melanoma | 536.95 |
| GSM71734 | Melanoma | 539.95 |
| GSM71735 | Melanoma | 733.9 |
| GSM71736 | Melanoma | 780.4 |
| GSM71737 | Melanoma | 999.3 |
| GSM71738 | Melanoma | 669.75 |
| GSM71739 | Melanoma | 563.9 |
| GSM71740 | Melanoma | 490.6 |
Table S2.
| Samples | Name column | E2F3 |
|---|---|---|
| GSM183222 | Melanoma in situ | 596.8995 |
| GSM183223 | Melanoma in situ | 485.8525 |
| GSM183226 | Metastatic melanoma | 1,043.514 |
| GSM183227 | Metastatic melanoma | 1,065.36 |
| GSM183228 | Metastatic melanoma | 1,097.4305 |
| GSM183229 | Metastatic melanoma | 1,367.071 |
| GSM183230 | Metastatic melanoma | 904.909 |
| GSM183231 | Metastatic melanoma | 966.979 |
| GSM183232 | Metastatic melanoma | 795.534 |
| GSM183233 | Metastatic melanoma | 1,049.4855 |
| GSM183252 | Metastatic melanoma | 1,675.07 |
| GSM183253 | Metastatic melanoma | 1,161.0115 |
| GSM183254 | Metastatic melanoma | 676.6665 |
| GSM183255 | Metastatic melanoma | 1,252.875 |
| GSM183256 | Metastatic melanoma | 1,091.715 |
| GSM183257 | Metastatic melanoma | 1,446.735 |
| GSM183273 | Metastatic melanoma | 763.7105 |
| GSM183274 | Metastatic melanoma | 1,067.1765 |
| GSM183275 | Metastatic melanoma | 972.223 |
| GSM183276 | Metastatic melanoma | 984.8085 |
| GSM183277 | Metastatic melanoma | 935.2895 |
| GSM183278 | Metastatic melanoma | 1,323.74 |
| GSM183279 | Metastatic melanoma | 757.3935 |
| GSM183280 | Metastatic melanoma | 1,052.9955 |
| GSM183281 | Metastatic melanoma | 970.099 |
| GSM183282 | Metastatic melanoma | 1,097.2135 |
| GSM183283 | Metastatic melanoma | 1,349.745 |
| GSM183284 | Metastatic melanoma | 973.492 |
| GSM183285 | Metastatic melanoma | 959.895 |
| GSM183286 | Metastatic melanoma | 967.631 |
| GSM183287 | Metastatic melanoma | 1,336.92 |
| GSM183288 | Metastatic melanoma | 918.0085 |
| GSM183289 | Metastatic melanoma | 1,649.06 |
| GSM183290 | Metastatic melanoma | 1,127.6965 |
| GSM183291 | Metastatic melanoma | 1,256.515 |
| GSM183292 | Metastatic melanoma | 1,410.8 |
| GSM183293 | Metastatic melanoma | 1,364.66 |
| GSM183294 | Metastatic melanoma | 3,495.27 |
| GSM183295 | Metastatic melanoma | 921.3215 |
| GSM183296 | Metastatic melanoma | 1,392.965 |
| GSM183297 | Metastatic melanoma | 582.005 |
| GSM183298 | Metastatic melanoma | 1,088.088 |
| GSM183299 | Normal human epidermal melanocytes | 821.5565 |
| GSM183234 | Normal skin | 289.57 |
| GSM183300 | Normal skin | 554.652 |
| GSM183301 | Normal skin | 360.1345 |
| GSM183302 | Normal skin | 359.8905 |
| GSM183224 | Primary melanoma | 454.667 |
| GSM183225 | Primary melanoma | 472.3105 |
| GSM183235 | Primary melanoma | 2,032.71 |
| GSM183258 | Primary melanoma | 3,043.785 |
| GSM183259 | Primary melanoma | 1,404.6755 |
| GSM183260 | Primary melanoma | 1,513.8725 |
| GSM183261 | Primary melanoma | 1,540.7 |
| GSM183262 | Primary melanoma | 797.319 |
| GSM183263 | Primary melanoma | 1,008.3395 |
| GSM183264 | Primary melanoma | 733.7925 |
| GSM183265 | Primary melanoma | 1,466.6135 |
| GSM183266 | Primary melanoma | 838.3575 |
| GSM183303 | Primary melanoma | 1,035.0555 |
| GSM183304 | Primary melanoma | 612.168 |
Table S3.
E2F3 gene relative expression of by PCR
| Cell name | GAPDH | E2F3 | ΔCt |
|---|---|---|---|
| SK-MEL-28 | 13.07 | 22.12 | 9.05 |
| 12.8 | 22.11 | 9.31 | |
| 12.93 | 22.15 | 9.22 | |
| MUM-2C | 12.81 | 20.85 | 8.04 |
| 12.76 | 20.97 | 8.21 | |
| 12.47 | 20.91 | 8.44 | |
| A375 | 12.43 | 21.7 | 9.27 |
| 12.23 | 21.75 | 9.52 | |
| 12.16 | 21.91 | 9.75 | |
| HaCaT | 13.35 | 20.99 | 7.64 |
| 13.31 | 21.25 | 7.94 | |
| 13.27 | 21.32 | 8.05 |
Table S4.
E2F3 pair all sample site methylation information table
| E2F3 pair methylation position | SK-MEL-28 | MUM-2C | A-375 | HaCaT |
|---|---|---|---|---|
| 23 | 0.0031 | 0.0041 | 0.0041 | 0.0033 |
| 26 | 0.0039 | 0.0045 | 0.0037 | 0.0043 |
| 65 | 0.0038 | 0.003 | 0.0029 | 0.0039 |
| 75 | 0.0041 | 0.0036 | 0.0029 | 0.0049 |
| 78 | 0.0033 | 0.0045 | 0.0026 | 0.0038 |
| 81 | 0.0035 | 0.0039 | 0.0021 | 0.0039 |
| 102 | 0.0021 | 0.0027 | 0.0022 | 0.0039 |
| 104 | 0.0036 | 0.0043 | 0.0027 | 0.0024 |
| 111 | 0.0036 | 0.0038 | 0.0027 | 0.0038 |
| 113 | 0.003 | 0.0047 | 0.0035 | 0.0033 |
| 117 | 0.0033 | 0.0039 | 0.003 | 0.0039 |
| 123 | 0.0031 | 0.0024 | 0.0032 | 0.0039 |
| 128 | 0.0051 | 0.004 | 0.0036 | 0.0039 |
| 134 | 0.0021 | 0.0049 | 0.0025 | 0.0037 |
| 137 | 0.0035 | 0.0042 | 0.0039 | 0.0039 |
| 144 | 0.0045 | 0.0041 | 0.0042 | 0.0032 |
| 166 | 0.0051 | 0.0049 | 0.0033 | 0.0035 |
| 169 | 0.0077 | 0.0042 | 0.0034 | 0.0029 |
| 174 | 0.0031 | 0.0043 | 0.0034 | 0.0035 |
Acknowledgments
This work was supported by the Zensure Leader Plan of the Third Xiangya Hospital of Central South University (grant number 20170302).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Rastrelli M, Tropea S, Rossi CR, Alaibac M. Melanoma: epidemiology, risk factors, pathogenesis, diagnosis and classification. In Vivo. 2014;28(6):1005–1011. [PubMed] [Google Scholar]
- 2.Trotter SC, Sroa N, Winkelmann RR, Olencki T, Bechtel M. A Global Review of Melanoma Follow-up Guidelines. J Clin Aesthet Dermatol. 2013;6(9):18–26. [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 4.Norbury C, Nurse P. Animal cell cycles and their control. Annu Rev Biochem. 1992;61:441–470. doi: 10.1146/annurev.bi.61.070192.002301. [DOI] [PubMed] [Google Scholar]
- 5.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411(6835):366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 6.Kortylewski M, Jove R, Yu H. Targeting STAT3 affects melanoma on multiple fronts. Cancer Metastasis Rev. 2005;24(2):315–327. doi: 10.1007/s10555-005-1580-1. [DOI] [PubMed] [Google Scholar]
- 7.Georgieva J, Sinha P, Schadendorf D. Expression of cyclins and cyclin dependent kinases in human benign and malignant melanocytic lesions. J Clin Pathol. 2001;54(3):229–235. doi: 10.1136/jcp.54.3.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forbes SA, Beare D, Boutselakis H, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45(D1):D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18(11):1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 10.Miles WO, Tschöp K, Herr A, Ji JY, Dyson NJ. Pumilio facilitates miRNA regulation of the E2F3 oncogene. Genes Dev. 2012;26(4):356–368. doi: 10.1101/gad.182568.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feber A, Clark J, Goodwin G, et al. Amplification and overexpression of E2F3 in human bladder cancer. Oncogene. 2004;23(8):1627–1630. doi: 10.1038/sj.onc.1207274. [DOI] [PubMed] [Google Scholar]
- 12.Bilke S, Schwentner R, Yang F, et al. Oncogenic ETS fusions deregulate E2F3 target genes in Ewing sarcoma and prostate cancer. Genome Res. 2013;23(11):1797–1809. doi: 10.1101/gr.151340.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren XS, Yin MH, Zhang X, et al. Tumor-suppressive microRNA-449a induces growth arrest and senescence by targeting E2F3 in human lung cancer cells. Cancer Lett. 2014;344(2):195–203. doi: 10.1016/j.canlet.2013.10.031. [DOI] [PubMed] [Google Scholar]
- 14.Libertini SJ, Chen H, Al-Bataina B, et al. The interleukin 6 receptor is a direct transcriptional target of E2F3 in prostate tumor derived cells. Prostate. 2012;72(6):649–660. doi: 10.1002/pros.21468. [DOI] [PubMed] [Google Scholar]
- 15.Kumaran M, Cass CE, Graham K, et al. Germline copy number variations are associated with breast cancer risk and prognosis. Sci Rep. 2017;7(1):14621. doi: 10.1038/s41598-017-14799-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye S, Li J, Hao K, Yan J, Zhou H. The Efficacy and Risk Profile of c-Met inhibitors in Non-small Cell Lung Cancer: a Meta-analysis. Sci Rep. 2016;6:35770. doi: 10.1038/srep35770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu B, Yang L, Huang B, et al. A functional copy-number variation in MAPKAPK2 predicts risk and prognosis of lung cancer. Am J Hum Genet. 2012;91(2):384–390. doi: 10.1016/j.ajhg.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim SJ, Kang HS, Chang HL, et al. Promoter hypomethylation of the N-acetyltransferase 1 gene in breast cancer. Oncol Rep. 2008;19(3):663–668. [PubMed] [Google Scholar]
- 19.Goldstein M, Meller I, Orr-Urtreger A. FGFR1 overexpression in primary rhabdomyosarcoma tumors is associated with hypomethylation of a 5′ CpG island and abnormal expression of the AKT1, NOG, and BMP4 genes. Genes Chromosomes Cancer. 2007;46(11):1028–1038. doi: 10.1002/gcc.20489. [DOI] [PubMed] [Google Scholar]
- 20.Roman-Gomez J, Jimenez-Velasco A, Castillejo JA, et al. Promoter hypermethylation of cancer-related genes: a strong independent prognostic factor in acute lymphoblastic leukemia. Blood. 2004;104(8):2492–2498. doi: 10.1182/blood-2004-03-0954. [DOI] [PubMed] [Google Scholar]
- 21.Cui H, Onyango P, Brandenburg S, Wu Y, Hsieh CL, Feinberg AP. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res. 2002;62(22):6442–6446. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
| Samples | Name column | E2F3 |
|---|---|---|
| GSM71671 | Normal | 215.25 |
| GSM71672 | Normal | 153.95 |
| GSM71673 | Normal | 265.3 |
| GSM71674 | Normal | 199.4 |
| GSM71675 | Normal | 327.95 |
| GSM71676 | Normal | 163.7 |
| GSM71677 | Normal | 203.3 |
| GSM71678 | Nevus | 319.5 |
| GSM71679 | Nevus | 283.85 |
| GSM71680 | Nevus | 369.7 |
| GSM71681 | Nevus | 225.4 |
| GSM71682 | Nevus | 199.2 |
| GSM71683 | Nevus | 270.55 |
| GSM71684 | Nevus | 295.1 |
| GSM71685 | Nevus | 304.3 |
| GSM71686 | Nevus | 326.3 |
| GSM71687 | Nevus | 290.7 |
| GSM71688 | Nevus | 306.95 |
| GSM71689 | Nevus | 372.55 |
| GSM71690 | Nevus | 184.05 |
| GSM71691 | Nevus | 365.15 |
| GSM71692 | Nevus | 369.8 |
| GSM71693 | Nevus | 294.65 |
| GSM71694 | Nevus | 367.75 |
| GSM71695 | Nevus | 642 |
| GSM71696 | Melanoma | 584.9 |
| GSM71697 | Melanoma | 669.35 |
| GSM71698 | Melanoma | 439.55 |
| GSM71699 | Melanoma | 493.05 |
| GSM71700 | Melanoma | 417.3 |
| GSM71701 | Melanoma | 549.8 |
| GSM71702 | Melanoma | 649.4 |
| GSM71703 | Melanoma | 558.9 |
| GSM71704 | Melanoma | 449.25 |
| GSM71705 | Melanoma | 689.95 |
| GSM71706 | Melanoma | 713.9 |
| GSM71707 | Melanoma | 786.1 |
| GSM71708 | Melanoma | 1,071.75 |
| GSM71709 | Melanoma | 405.55 |
| GSM71710 | Melanoma | 686.95 |
| GSM71711 | Melanoma | 634.85 |
| GSM71712 | Melanoma | 464.4 |
| GSM71713 | Melanoma | 624.65 |
| GSM71714 | Melanoma | 610.55 |
| GSM71715 | Melanoma | 676.65 |
| GSM71716 | Melanoma | 810.25 |
| GSM71717 | Melanoma | 659.05 |
| GSM71718 | Melanoma | 498.05 |
| GSM71719 | Melanoma | 604.75 |
| GSM71720 | Melanoma | 368.95 |
| GSM71721 | Melanoma | 526.55 |
| GSM71722 | Melanoma | 511.15 |
| GSM71723 | Melanoma | 582.1 |
| GSM71724 | Melanoma | 519.85 |
| GSM71725 | Melanoma | 740.1 |
| GSM71726 | Melanoma | 602.05 |
| GSM71727 | Melanoma | 399.4 |
| GSM71728 | Melanoma | 435.4 |
| GSM71729 | Melanoma | 423.1 |
| GSM71730 | Melanoma | 423 |
| GSM71731 | Melanoma | 557.65 |
| GSM71732 | Melanoma | 537.55 |
| GSM71733 | Melanoma | 536.95 |
| GSM71734 | Melanoma | 539.95 |
| GSM71735 | Melanoma | 733.9 |
| GSM71736 | Melanoma | 780.4 |
| GSM71737 | Melanoma | 999.3 |
| GSM71738 | Melanoma | 669.75 |
| GSM71739 | Melanoma | 563.9 |
| GSM71740 | Melanoma | 490.6 |
Table S2.
| Samples | Name column | E2F3 |
|---|---|---|
| GSM183222 | Melanoma in situ | 596.8995 |
| GSM183223 | Melanoma in situ | 485.8525 |
| GSM183226 | Metastatic melanoma | 1,043.514 |
| GSM183227 | Metastatic melanoma | 1,065.36 |
| GSM183228 | Metastatic melanoma | 1,097.4305 |
| GSM183229 | Metastatic melanoma | 1,367.071 |
| GSM183230 | Metastatic melanoma | 904.909 |
| GSM183231 | Metastatic melanoma | 966.979 |
| GSM183232 | Metastatic melanoma | 795.534 |
| GSM183233 | Metastatic melanoma | 1,049.4855 |
| GSM183252 | Metastatic melanoma | 1,675.07 |
| GSM183253 | Metastatic melanoma | 1,161.0115 |
| GSM183254 | Metastatic melanoma | 676.6665 |
| GSM183255 | Metastatic melanoma | 1,252.875 |
| GSM183256 | Metastatic melanoma | 1,091.715 |
| GSM183257 | Metastatic melanoma | 1,446.735 |
| GSM183273 | Metastatic melanoma | 763.7105 |
| GSM183274 | Metastatic melanoma | 1,067.1765 |
| GSM183275 | Metastatic melanoma | 972.223 |
| GSM183276 | Metastatic melanoma | 984.8085 |
| GSM183277 | Metastatic melanoma | 935.2895 |
| GSM183278 | Metastatic melanoma | 1,323.74 |
| GSM183279 | Metastatic melanoma | 757.3935 |
| GSM183280 | Metastatic melanoma | 1,052.9955 |
| GSM183281 | Metastatic melanoma | 970.099 |
| GSM183282 | Metastatic melanoma | 1,097.2135 |
| GSM183283 | Metastatic melanoma | 1,349.745 |
| GSM183284 | Metastatic melanoma | 973.492 |
| GSM183285 | Metastatic melanoma | 959.895 |
| GSM183286 | Metastatic melanoma | 967.631 |
| GSM183287 | Metastatic melanoma | 1,336.92 |
| GSM183288 | Metastatic melanoma | 918.0085 |
| GSM183289 | Metastatic melanoma | 1,649.06 |
| GSM183290 | Metastatic melanoma | 1,127.6965 |
| GSM183291 | Metastatic melanoma | 1,256.515 |
| GSM183292 | Metastatic melanoma | 1,410.8 |
| GSM183293 | Metastatic melanoma | 1,364.66 |
| GSM183294 | Metastatic melanoma | 3,495.27 |
| GSM183295 | Metastatic melanoma | 921.3215 |
| GSM183296 | Metastatic melanoma | 1,392.965 |
| GSM183297 | Metastatic melanoma | 582.005 |
| GSM183298 | Metastatic melanoma | 1,088.088 |
| GSM183299 | Normal human epidermal melanocytes | 821.5565 |
| GSM183234 | Normal skin | 289.57 |
| GSM183300 | Normal skin | 554.652 |
| GSM183301 | Normal skin | 360.1345 |
| GSM183302 | Normal skin | 359.8905 |
| GSM183224 | Primary melanoma | 454.667 |
| GSM183225 | Primary melanoma | 472.3105 |
| GSM183235 | Primary melanoma | 2,032.71 |
| GSM183258 | Primary melanoma | 3,043.785 |
| GSM183259 | Primary melanoma | 1,404.6755 |
| GSM183260 | Primary melanoma | 1,513.8725 |
| GSM183261 | Primary melanoma | 1,540.7 |
| GSM183262 | Primary melanoma | 797.319 |
| GSM183263 | Primary melanoma | 1,008.3395 |
| GSM183264 | Primary melanoma | 733.7925 |
| GSM183265 | Primary melanoma | 1,466.6135 |
| GSM183266 | Primary melanoma | 838.3575 |
| GSM183303 | Primary melanoma | 1,035.0555 |
| GSM183304 | Primary melanoma | 612.168 |
Table S3.
E2F3 gene relative expression of by PCR
| Cell name | GAPDH | E2F3 | ΔCt |
|---|---|---|---|
| SK-MEL-28 | 13.07 | 22.12 | 9.05 |
| 12.8 | 22.11 | 9.31 | |
| 12.93 | 22.15 | 9.22 | |
| MUM-2C | 12.81 | 20.85 | 8.04 |
| 12.76 | 20.97 | 8.21 | |
| 12.47 | 20.91 | 8.44 | |
| A375 | 12.43 | 21.7 | 9.27 |
| 12.23 | 21.75 | 9.52 | |
| 12.16 | 21.91 | 9.75 | |
| HaCaT | 13.35 | 20.99 | 7.64 |
| 13.31 | 21.25 | 7.94 | |
| 13.27 | 21.32 | 8.05 |
Table S4.
E2F3 pair all sample site methylation information table
| E2F3 pair methylation position | SK-MEL-28 | MUM-2C | A-375 | HaCaT |
|---|---|---|---|---|
| 23 | 0.0031 | 0.0041 | 0.0041 | 0.0033 |
| 26 | 0.0039 | 0.0045 | 0.0037 | 0.0043 |
| 65 | 0.0038 | 0.003 | 0.0029 | 0.0039 |
| 75 | 0.0041 | 0.0036 | 0.0029 | 0.0049 |
| 78 | 0.0033 | 0.0045 | 0.0026 | 0.0038 |
| 81 | 0.0035 | 0.0039 | 0.0021 | 0.0039 |
| 102 | 0.0021 | 0.0027 | 0.0022 | 0.0039 |
| 104 | 0.0036 | 0.0043 | 0.0027 | 0.0024 |
| 111 | 0.0036 | 0.0038 | 0.0027 | 0.0038 |
| 113 | 0.003 | 0.0047 | 0.0035 | 0.0033 |
| 117 | 0.0033 | 0.0039 | 0.003 | 0.0039 |
| 123 | 0.0031 | 0.0024 | 0.0032 | 0.0039 |
| 128 | 0.0051 | 0.004 | 0.0036 | 0.0039 |
| 134 | 0.0021 | 0.0049 | 0.0025 | 0.0037 |
| 137 | 0.0035 | 0.0042 | 0.0039 | 0.0039 |
| 144 | 0.0045 | 0.0041 | 0.0042 | 0.0032 |
| 166 | 0.0051 | 0.0049 | 0.0033 | 0.0035 |
| 169 | 0.0077 | 0.0042 | 0.0034 | 0.0029 |
| 174 | 0.0031 | 0.0043 | 0.0034 | 0.0035 |