Abstract
Our comparative studies reveal that the two lytic polysaccharide monooxygenases HiLPMO9B and HiLPMO9I from the white-rot conifer pathogen Heterobasidion irregulare display clear difference with respect to their activity against crystalline cellulose and glucomannan. HiLPMO9I produced very little soluble sugar on bacterial microcrystalline cellulose (BMCC). In contrast, HiLPMO9B was much more active against BMCC and even released more soluble sugar than the H. irregulare cellobiohydrolase I, HiCel7A. Furthermore, HiLPMO9B was shown to cooperate with and stimulate the activity of HiCel7A, both when the BMCC was first pretreated with HiLPMO9B, as well as when HiLPMO9B and HiCel7A were added together. No such stimulation was shown by HiLPMO9I. On the other hand, HiLPMO9I was shown to degrade glucomannan, using a C4-oxidizing mechanism, whereas no oxidative cleavage activity of glucomannan was detected for HiLPMO9B. Structural modeling and comparison with other glucomannan-active LPMOs suggest that conserved sugar-interacting residues on the L2, L3 and LC loops may be essential for glucomannan binding, where 4 out of 7 residues are shared by HiLPMO9I, but only one is found in HiLPMO9B. The difference shown between these two H. irregulare LPMOs may reflect distinct biological roles of these enzymes within deconstruction of different plant cell wall polysaccharides during fungal colonization of softwood.
Introduction
Fungal colonization of wood is associated with decomposition of plant biomass to sustain the growing mycelia. The major constituent of woody biomass is secondary cell walls, of which the three major components (cellulose, hemicellulose and lignin) form a complex and rigid structure, in which cellulose is densely packed into microfibrils, which in turn are covered by hemicellulose and lignin [1,2]. The inherent recalcitrance of woody biomass against microbial degradation requires wood-colonizing fungi to produce a diverse array of hydrolytic and oxidative enzymes that act in synergy to degrade individual components of the plant cell walls [3]. The enzymatic degradation of plant cell walls not only removes physical barriers to enable fungal colonization of the host plant but also releases free sugars that can be used as a source of metabolic carbon and energy by the fungus [4]. White-rot fungi is the only subclass among wood-decaying fungi that are able to efficiently degrade both the carbohydrate and lignin fractions of woody biomass [5].
The basidiomycete white-rot fungus Heterobasidion annosum (Fr.) Bref. sensu lato (s.l) is a serious plant pathogenic species complex that causes severe root rot on conifer trees within the boreal and temperate regions [6–8]. The economic losses due to H. annosum s.l. infection for European forest owners due to tree growth reduction and devaluation of timber at harvest reaches at least 790 million euros per year [9]. The H. annosum s.l. complex is a group of five phylogenetically distinct species with partly overlapping geographic distribution and host preferences [9–11]. The species H. irregulare is native within North America where it colonizes and decays mostly softwood trees, such as pine and juniper species [10,12]. H. irregulare infects fresh wood via basidiospores that land on the exposed stump surfaces or wounds on stem, and spread from one tree to another via root-to-root contacts [13]. The mycelium of H. irregulare spreads mainly through the heartwood and sapwood of softwood plants, in which the major carbohydrate components of the secondary plant cell walls are cellulose and glucomannan [14].
The depolymerization of wood polysaccharides by fungal systems is known to involve a complex and well-orchestrated enzymatic cascade [15]. The H. irregulare genome encodes 305 putative carbohydrate-active enzymes [16]. Global gene expression analysis of H. irregulare growing on woody biomaterials revealed that 27 of these 305 genes were up-regulated more than 10-fold compared to when the fungus was cultured in a glucose-containing liquid medium [16]. About half of the up-regulated genes (14 out of 27) were categorized to encode either putative glycoside hydrolases (GHs) or lytic polysaccharide monooxygenases (LPMOs), both of which have been identified as important enzymes responsible for microbial degradation of plant cell wall polysaccharides [16]. In another study, H. irregulare was grown on woody biomass, and the most abundant protein by far in the culture filtrate was identified as cellobiohydrolase HiCel7A [17]. It is the only family 7 glycoside hydrolase in the genome of H. irregulare and it consists of a single catalytic domain without attached linker region and carbohydrate-binding module (CBM). The structure of HiCel7A has been determined and the structure was found to be similar to that of other know GH7 CBHs [17]. It is the only cellulase from H. irregulare that has been biochemically characterized so far.
LPMOs are copper-dependent enzymes that employ an oxidative mechanism to cleave linkages in polysaccharides [18–20], and they have been found to oxidize at either the C1 or the C4 position of pyranose rings of polysaccharides [18,21–23]. The catalytic mechanism of LPMOs require several co-factors including external reducing agent, and reactive oxygen species such as molecular oxygen or hydrogen peroxide [19,21,24,25]. The current categorization divides LPMOs into six auxiliary activity families (AA9, AA10, AA11, AA13, AA14 and AA15) [26–29]. The LPMOs in AA9 family are only found in fungi, including ascomycetes and basidiomycetes [26]. Over the last 10 years, extensive research efforts have been made to understand the biological contribution of LPMOs in plant biomass degradation [30,31]. AA9 LPMOs have been shown to possess different substrate preferences in terms of activities against cellulosic and hemicellulosic substrates, indicating that LPMOs could be recruited for degradation of different polysaccharide components in plant cell walls [18,26,32–37]. They have also been shown to boost the hydrolytic activity of cellulases in terms of saccharification of lignocellulosic substrates [27,38–44], and several AA9 LPMOs show effects of separating fibrils from cellulose fibrillar structures, pointing at important roles in cellulose decrystallization [45–47]. Furthermore, studies with atom force microscopy have revealed that modification of highly resistant microfibril bundles by oxidative cleavage using a C1-oxidizing LPMO could enhance the digestion of crystalline regions by a processive cellobiohydrolase I, providing insights into synergistic cooperation between LPMO and cellulase enzymes [45].
The genome of H. irregulare has been sequenced and annotated, and was found to encode ten putative AA9 LPMOs [16]. More recently, two putative AA14 LPMOs were also discovered in its genome (GenBank ID: XP_009545121.1 and XP_009545122.1) [27]. All of the ten H. irregulare AA9 LPMOs are predicted to have an N-terminal catalytic domain, three of them have an additional C-terminal family 1 cellulose binding module (CBM 1), and one has a C-terminal extension without known function [16]. Among the H. irregulare AA9 LPMOs, HiLPMO9B showed the highest transcription level when the fungus was grown on pine heartwood, and HiLPMO9I when the fungus was grown on the material from the cambial zone of necrotic bark, which suggest that these two LPMOs may play important roles during colonization processes of this fungus on pine trees [16]. Therefore, it is of interest to further understand how these two LPMOs are involved in the depolymerization of plant-cell-wall polysaccharides. We have previously reported the recombinant expression and biochemical characterization of both HiLPMO9B and HiLPMO9I, and the three-dimensional structure and molecular dynamics studies of HiLPMO9B [48]. In brief, HiLPMO9B does not contain a cellulose binding module (CBM) and cleaves cellulose by oxidation at the C1 position of cellulose [48,49], whereas HiLPMO9I is a bi-modular protein with linker-CBM attached to the catalytic domain, and this enzyme oxidizes cellulose at the C4 position, and HiLPMO9I is also found to be active on glucomannan [49].
In this study we present a side-by-side comparison of HiLPMO9B and HiLPMO9I in terms of activity on bacterial microcrystalline cellulose (BMCC) and cooperation with the H. irregulare glycoside hydrolase family 7 cellobiohydrolase HiCel7A, as well as their glucomannan activity and detailed analysis of glucomannan products generated by HiLPMO9I and structural factors of HiLPMO9I important for glucomannan binding.
Materials and methods
Substrates
Bacterial microcrystalline cellulose (BMCC) was prepared by acid hydrolysis of bacterial cellulose from Acetobacter xylinum, extracted from commercially available Nata de Coco as previously described [50]. 20 ml of 2.5 mg/ml BMCC was heated at 95°C in 1 M hydrochloric acid for 1 h. After cooling down to room temperature, the BMCC samples were centrifuged at 4,000 ×g for 5 min and suspended with 20 ml deionized water for 5 cycles. The BMCC was finally resuspended in 20 ml of 50 mM sodium acetate buffer, pH 5.0, and the dry mass content of BMCC was measured using Precisa XM60 moisture analyzer (Precisa Gravimetrics AG, Dietikon, Switzerland). 10 mg/ml konjac glucomannan low viscosity (Megazyme, Wicklow, Ireland) was dissolved in 50 mM sodium acetate buffer, pH 5.0.
Enzyme preparation
The H. irregulare LPMOs, HiLPMO9B (Uniprot ID: W4KMP1) and HiLPMO9I (Uniprot ID: W4K8M0), were heterologously expressed in Pichia pastoris and purified as described previously [48,49]. The wild-type H. irregulare Cel7A (HiCel7A, Uniprot ID: W4KCY5) was produced by growing H. irregulare in liquid cultivation medium containing microcrystalline cellulose substrate Avicel PH-101 as carbon source (Sigma-Aldrich, Steinheim, Germany), and HiCel7A was purified from the culture supernatant as described previously [17]. A GH family 7 endoglucanase (MtEG7) from Myceliophthora thermophila (Uniprot ID: G2QCS4) was heterologously expressed in P. pastoris, and the protein was purified as described previously [51]. Neurospora crassa LPMO9C (NcLPMO9C, Uniprot: Q7SHI8) was produced as described by Borisova, et al [52].
Microcrystalline cellulose activity assay
To investigate the cooperation between the two LPMOs and the H. irregulare cellobiohydrolase on degradation of crystalline cellulose, reactions were carried out by incubating 100 μl of 1 mg/ml BMCC with LPMO-CBH mixtures at different molar ratios (HiCel7A: HiLPMO9B or HiCel7A: HiLPMO9I = 100:0, 75:25, 50:50, 25:75, 0:100) at a constant total enzyme concentration of 0.5 μM, in 50 mM sodium acetate buffer, pH 5.0, in the presence of 1 mM pyrogallol (Sigma-aldrich, Steinheim, Germany), with three biological replicates for each reaction. The reaction mixtures were incubated at 20°C in a thermomixer (Eppendorf, Hamburg, Germany) with orbital shaking at 700 rpm for 48 h. The reactions were stopped by heating the sample at 95°C for 3 min. The insoluble residues of BMCC were collected by centrifugation of the sample at 13,000 ×g for 5 min, and the supernatant was vacuum-filtrated through 0.45 μm polyethersulfone membranes on a filter plate (Pall Corporation, Ann Arbor, MI, USA). The soluble sugars in the liquid fraction were analyzed and quantified using high-performance anion-exchange chromatography (HPAEC) (detailed procedures as described further down). For the reaction with CBH-LPMO ratio at 0:100, the supernatant was subjected to an extra digestion with 0.5 μM HiCel7A at 20°C for 1 h prior to HPAEC analysis.
The insoluble residues of BMCC were recovered from the reactions with CBH-LPMO ratio at 0:100 to investigate the actions of CBH on LPMO-modified crystalline cellulose surface. The BMCC residues were re-suspended to 100 μl in 50 mM sodium acetate buffer, pH 5.0, after 3 cycles of centrifugation and dilution with the same buffer. The LPMO-treated BMCC residues were then incubated with 0.5 μM HiCel7A at 20°C for 6 h, and all the reactions were repeated in triplicate. After heat inactivation and vacuum filtration, the soluble enzyme products were analyzed by HPAEC analysis.
Glucomannan activity assays
Reduction in the dynamic viscosity of glucomannan by the action of purified HiLPMOs was determined using a Lovis 2000 ME microviscometer (Anton Paar, Graz, Austria). The instrument was equipped with a glass capillary tube of 1.59 mm diameter with a 1.5 mm diameter steel ball inside. The instrument was calibrated with deionized water at 30°C prior to the experiments. Viscosity change was determined as a function of difference in “time of fall” [35] of the steel ball while the capillary was positioned at 60° angle. The capillary with reaction sample was inverted when the ball reached the bottom of the capillary. All reactions were carried out at 30°C for 30 min, in a total volume of 500 μl, with 1 mg/ml glucomannan, and 1 μM LPMO in 50 mM sodium acetate buffer, pH 5.0, in the presence of 1 mM ascorbic acid. NcLPMO9C was used as positive control as it had been shown to decrease the viscosity of glucomannan in a previous study [35]. A sample consisting of substrate and ascorbic acid only was used as negative control. At the end of the 30-min viscosity measurement, 200 μl of the sample was drawn from the capillary and subjected to further depolymerization by incubation with 1 μM Myceliophthora thermophila EG7 (MtEG7) at 30°C for 1 h. The depolymerized samples were loaded onto a 1-ml Alltech Carbograph SPE column (Grace Davison Discovery Sciences, Deerfield, IL, USA) and eluted with 30% acetonitrile containing 10 mM ammonium acetate. The eluate was freeze-dried and re-dissolved in deionized water, prior to LC-MS analysis (detailed procedures as described further down) for detection of oxidized products.
Enzyme product analysis
High-performance anion-exchange chromatography (HPAEC)
HPAEC analyses were performed using a CarboPac PA1 2×250 mm analytical column (Dionex Corp., Sunnyvale, CA, USA) in an ICS3000 system equipped with a pulsed amperometric (PAD) detector (Dionex Corp., Sunnyvale, CA, USA) and a 5-μl sample loop. A gradient elution program was applied after a full-loop injection in each analysis. The elution program included the following steps: 0–12 min: 30 mM sodium hydroxide (isocratic), 12–15 min: 30–100 mM sodium hydroxide (linear gradient), 15–40 min: 0–350 mM sodium acetate in 100 mM sodium hydroxide (linear gradient), 40–43 min: 350–1000 mM sodium acetate in 100 mM sodium hydroxide (linear gradient) at a constant flow rate of 0.36 ml/min with a post-column addition of 0.3 M sodium hydroxide at 0.18 ml/min prior to PAD detection.
Liquid chromatography–mass spectrometry (LC-MS)
LC-MS analysis was carried out on a Bruker maXis Impact mass spectrometer (ESI-QTOFMS) (Bruker Daltonics, Bremen, Germany) operated in positive mode scanning m/z from 50 to 1500, with calibration using sodium formate clusters. Sample of 10 μl was injected to an analytical hypercarb porous graphitic carbon (PGC) column (Thermofisher; 3 μm particle size; 2.1 mm diameter) connected to a HP1100 LC system (Hewlett–Packard). The separation was performed using a gradient from 0% to 80% acetonitrile in 0.2% formic acid at 0.4 ml/min (0–15 min: 0% to 27.5% acetonitrile (gradient), 15–17 min: 27.5% to 80% acetonitrile (gradient), 17–26 min: 80% acetonitrile (isocratic)). Precursor ions in the ranges m/z 533–541, 695–705, 857–867 were selected and subjected to collision induced (CID) MS/MS for structure determination (m/z = 0–100: 15 eV; 100–500, 25 eV; 500–1000: 30 eV).
Structural modeling and analysis
A homology model of the HiLPMO9I catalytic domain was built using SWISS-MODEL (http://swissmodel.expasy.org/) using default parameters and the crystal structure (chain A) of NcLPMO9C as template (PDB ID: 4D7U_A; 53% sequence identity). The Ramachandran plots used for validating the HiLPMO9I model was obtained by analysing the model in the PROCHECK analysis server [53] and the quality of the model was assessed by Qualitative Model Energy ANalysis (QMEAN) based on QMEAN score and the Z score obtained [54]. Structure-based alignment of the LPMOs was generated using the program Chimera version 1.11 (http://www.cgl.ucsf.edu/chimera/), using alignment algorithm Needleman-Wunsch and matrix BLOSUM-62 (Gap opening = 12, Gap extension = 1), and the visualization of the protein structures were performed using PyMol molecular graphing software version 1.8.6.0 (DeLano Scientific LLC, San Carlos, USA).
Results
The effects of HiLPMO9B and HiLPMO9I on CBH activity on microcrystalline cellulose
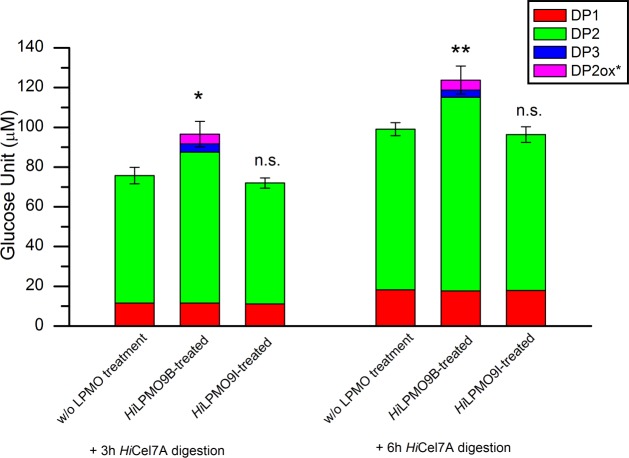
Bacterial microcrystalline cellulose at 1 mg/ml concentration was incubated for 48 h at 20°C, pH 5.0, without and with the addition of 0.5 μM of HiLPMO9B or HiLPMO9I, in the presence of 1mM pyrogallol as reducing agent; thereafter the residual cellulose was washed to remove soluble sugars. The insoluble residue of BMCC was then used to study the release of soluble sugar by 0.5 μM HiCel7A from the LPMO-modified cellulose sample after 3 h and 6 h of digestion at 20°C, pH 5.0, and thereafter quantified using HPAEC. The pretreatment with HiLPMO9B led to approximately 25% increase in total sugar release (including native and oxidized sugar forms) after 3-h and 6-h digestion with HiCel7A, whereas no considerable difference was found between HiLPMO9I-treated BMCC compared to BMCC without prior LPMO treatment (Fig 1). There was no change in the level of glucose released between samples with and without prior LPMO treatment. The pretreatment with HiLPMO9B gave 18% more cellobiose than without LPMO after 3 h HiCel7A digestion, and 20% after 6 h. Also, C1-oxidized cellobiose and cellotriose were detected after incubating HiLPMO9B-treated BMCC with HiCel7A, but not with the other samples (Fig 1).
Fig 1. HiCel7A activity on BMCC substrate residues with or without prior LPMO pretreatment.
Soluble sugar released by HiCel7A from BMCC substrate residues with or without LPMO pretreatment was analyzed by HPAEC. Quantification was based on 3 h and 6 h digestion with 0.5 uM HiCel7A of the BMCC residue after 48 h pretreatment with 0.5 μM HiLPMO9B or HiLPMO9I in presence of 1 mM pyrogallol (LPMO-treated), or with only pyrogallol without LPMO added as negative control (w/o LPMO treatment). Incubations were done at 20°C, pH 5.0. DPn stands for native cello-oligosaccharides, and DP2ox* represents C1-oxidized products (DP1: glucose, DP2: cellobiose, DP3: cellotriose, DP2ox*: C1-oxidized cellobiose). Values are means of triplicates, and the error bars show the standard deviations (error bar = ± standard deviation). Two-tailed t-test indicates that total soluble sugar release of HiCel7A on HiLPMO9B-treated BMCC significantly differ (* p<0.05, ** p<0.01) from the amount on BMCC without LPMO preteatment, but there is no significance difference (n.s. p>0.05) between HiLPMO9I-treated BMCC and BMCC without LPMO pretreatment.
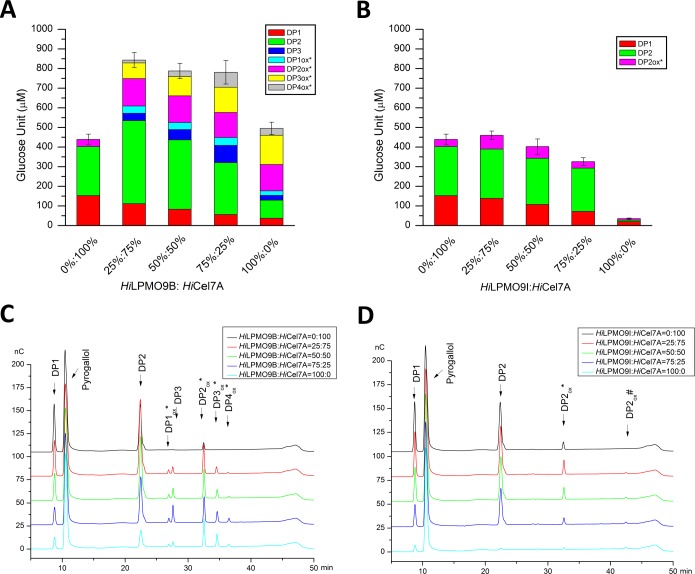
Possible LPMO-cellobiohydrolase cooperation on cellulose was investigated by comparing the saccharification of 1 mg/ml BMCC incubated for 48 h at 20°C, pH 5.0, with 0.5 μM mixtures of HiCel7A+HiLPMO9B, or HiCel7A+HiLPMO9I, and with 1 mM pyrogallol as reducing agent. Released soluble sugars were analyzed and quantified by HPAEC-PAD (Fig 2). The results reveal that the two LPMOs are strikingly different (Fig 2). HiLPMO9I displayed very little activity, whereas HiLPMO9B actually released more soluble sugar than HiCel7A when the enzymes were acting alone on the BMCC substrate. The amount of sugar released by HiLPMO9B is about 15 times higher than for HiLPMO9I. Also, there does not seem to be any cooperative effect when replacing HiCel7A with HiLPMO9I at different ratios; the 0:100, 25:75 and 50:50 replacement experiments are all within standard error (Fig 2B). On the other hand, for HiLPMO9B there is very clear cooperation. The substitution of HiCel7A with HiLPMO9B significantly boosted the overall saccharification level at all three substitution ratios (Fig 2A). When 25% of HiCel7A was replaced with HiLPMO9B there was an increase of the total sugar release by 92% compared to HiCel7A alone, and by 70% compared to using HiLPMO9B alone (Fig 2A). The saccharification levels are similar at 50% and 75% replacement level with HiLPMO9B, giving approximately 80% more soluble sugar than for HiCel7A alone (Fig 2A). As expected, the amount of oxidized oligosaccharides increased with 25% added HiLPMO9B, but there was also an increase in the release of cellobiose. The proportion of oxidized oligosaccharides is further increased and the amount of cellobiose decreased as the HiCel7A is further replaced by HiLPMO9B (Fig 2A).
Fig 2. Saccharification levels after treatment of BMCC with LPMO and HiCel7A at different mixing ratios.
Released soluble sugars were analyzed and quantified using HPAEC analysis after 48-h incubation at 20°C, pH 5.0, of 1 mg/ml BMCC with 0.5 μM of HiCel7A and HiLPMO9B (A) or HiCel7A and HiLPMO9I (B) at indicated ratios, in the presence of 1 mM pyrogallol as reducing agent. DP1-3 indicate native cello-oligosaccharides and DP1-4ox* represent C1-oxidized sugars. The amounts of C4-oxidized sugars were negligible and were not quantified. Values are means of three biological replicates, and error bars correspond to the standard deviation (error bar = ± standard deviation). The quantification is based on HPAEC analysis of soluble sugars released from BMCC by the different combinations of HiCel7A+ HiLPMO9B (C) or HiCel7A+ HiLPMO9I (D). DP 1–3: native cello-oligosaccharides; DPnox* represent C1-oxidized products, and DPnox# stands for C4-oxidized products.
The activity of HiLPMOB and HiLPMO9I activity on glucomannan
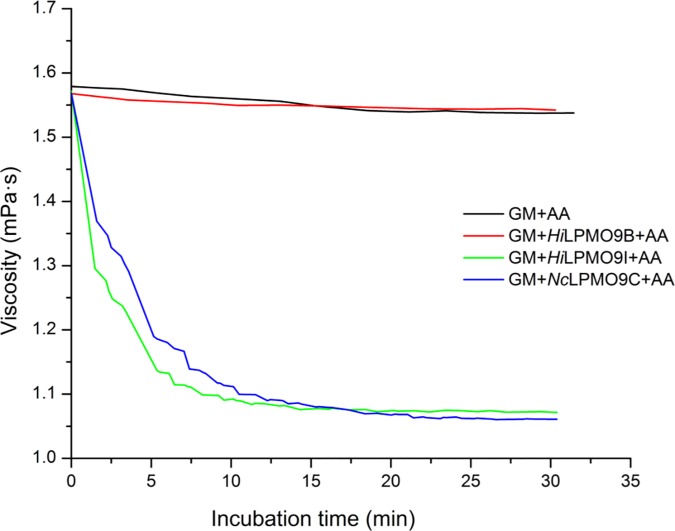
The glucomannan degrading activities of HiLPMO9B and HiLPMO9I were compared in the presence of ascorbic acid as reducing agent using a dynamic viscosity assay (Fig 3). The treatment of glucomannan with HiLPMO9I led to a significant viscosity reduction of glucomannan. The viscosity was reduced by 30% after 15 min incubation with HiLPMO9I, similar to the effect of the positive control NcLPMO9C, which is known to be active on glucomannan [32,35]. Meanwhile, HiLPMO9B did not show any effect on the viscosity of glucomannan and exhibited similar behavior as the negative control without enzyme added.
Fig 3. Viscosity changes of glucommanan during the incubation with two H. irregulare LPMOs using ascorbic acid as reducing agent.
The dynamic viscosity was monitored for 30 min at 30°C, pH 5.0, in a falling-ball capillary viscometer with 1 mg/ml glucomannan, 1 uM LPMO and 1 mM ascorbic acid. NcLPMO9C, a LPMO known to oxidatively cleave glucomannan, was used as a positive control, and a negative control containing only glucomannan (GM) and ascorbic acid (AA) was included.
Product analysis after treating glucomannan with LPMOs
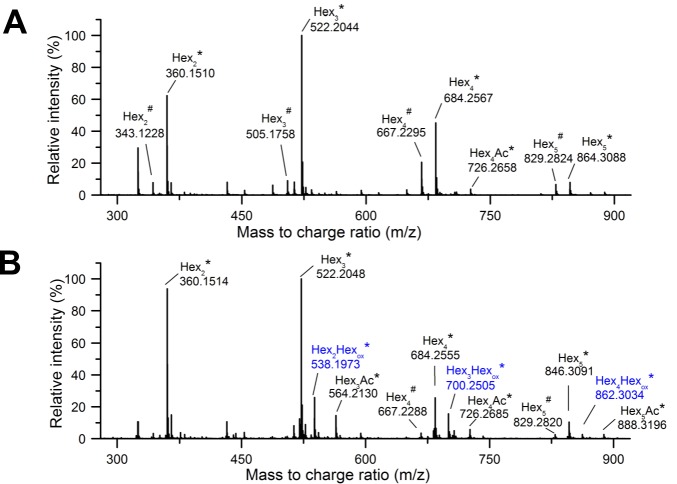
After the 30 min viscosity measurements, samples of the LPMO-treated glucomannan were withdrawn and depolymerized with an endoglucanase enzyme (Myceliophthora thermophila MtEG7) prior to analysis by LC-MS for detection of oxidized saccharide products. For the HiLPMO9B-treated glucomannan, no oxidized sugars could be detected. Ammonium adducts of native non-acetylated oligosaccharides were detected, with mass-to-charge ratios (m/z) = 360, 522, 684, 846; degree of polymerization (DP 2–5), as well as proton adducts at lower levels (m/z = 343, 505, 667, 829; DP 2–5) (Fig 4A). The HiLPMO9I-treated glucomannan gave a similar product pattern, but also displayed signals for oxidized saccharides at m/z 538, 700, 862, i.e. with m/z +16 Da compared to the corresponding native sugars (m/z = 522, 684, 846) ranging from DP 3 to DP 5 (Fig 4B).
Fig 4. LC-MS analysis of oligosaccharides from LPMO-treated products from glucomannan.
Mass spectra are presented as averages over 7.5 to 15 min of the chromatographic analysis of glucomannan treated with HiLPMO9B (A) or HiLPMO9I (B) in presence of ascorbic acid, and subsequent depolymerization with MtEG7. Native (black) and oxidized (blue) glucomannan oligosaccharides were mainly detected as ammonium adducts (*) and to a much less extent as proton adducts (#) in the MS analyses.
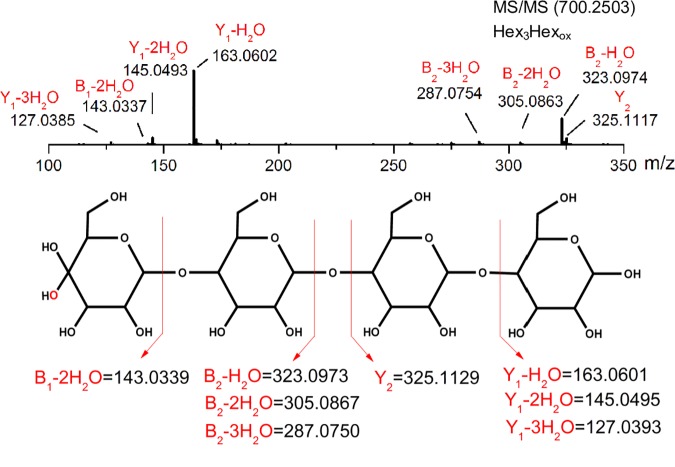
The oxidized oligosaccharides from the reaction of HiLPMO9I with glucomannan were further studied by MS/MS fragmentation to identify the oxidization position on the pyronose ring. All the three oxidized oligosaccharides, the DP3, DP4 and DP5 saccharides with m/z 538, 700, 862, gave very similar fragmentation patterns (Fig 4). The result of the LC-MS/MS analysis of the oxidized DP4 ion (m/z 700) is displayed (Fig 5). These results point towards the sole existence of C4-oxidized sugars products in the MtEG7-depolymerized sample. The MS/MS fragmentation generated three product ions [m/z = 323.09 (B2-H2O), 305.05 (B2-2H2O), 287.07 (B2-3H2O)] from all the three oxidized GM oligosaccharides, which are in good agreement with B2 fragment ions of C4-gemdiols after elimination of water (Fig 5). Additionally, B1 fragment ions after elimination of two water (m/z = 143.03) were also observed for DP3 (m/z = 538.1977), DP4 (m/z = 700.2503) (Fig 5). Fragmentation of the ion with m/z 862.3033 (oxidized Hex5) generated a product ion (m/z = 485.15) in line with B3-2H2O for a C4-gemdiol. Conclusively, C4-oxidized but not C1-oxidized sugars were detected as products from the treatment of glucomannan with HiLPMO9I, and no oxidized sugars at all were detected when the substrate was treated with HiLPMO9B.
Fig 5. LC-MS/MS analysis of the HiLPMO9I-treated glucomannan sample.
MS/MS fragmentation of ion representing oxidized glucomannan tetrasaccharides (upper panel) shows good agreement with the schematic fragmentation pattern of a C4-oxidized gemidiol tetrasaccharides (lower panel).
Structural factors of glucomannan substrate binding
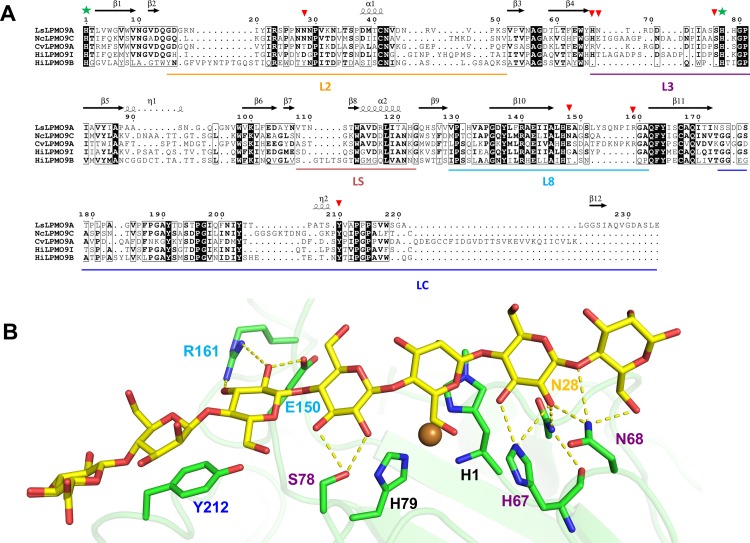
A structure model of the HiLPMO9I catalytic domain, produced using a crystal structure of NcLPMO9C catalytic domain as template (PDB code 4D7U; 53% sequence identity; [55]), was validated with Z score and QMEAN and Ramachandran plot parameters [54]. The QMEAN scoring for model quality estimation indicated a reliable HiLPMO9I model, with Z score of −1.83 and 0.693 as QMEAN score. Ramachandran plot analysis showed 0.6% of amino acid residues in disallowed regions, 97.7% of residues in the allowed region and 1.7% in generously allowed regions [56].The HiLPMO9I structure model was superposed with the crystal structures of HiLPMO9B (PDB code 5NNS; [48]), NcLPMO9C, CvLPMO9A (PDB code 5NLT; [36]) and LsLPMO9A in complex with a GM oligosaccharide (PDB code 5NKW; [36]), to generate a structure-based sequence alignment. The alignment is shown in Fig 6 together with an image of the protein-sugar interactions in the LsLPMO9A/GM-oligosaccharide complex structure. Clearly, HiLPMO9I displays higher similarity with the GM-active NcLPMO9C, CvLPMO9A and LsLPMO9A (53%, 40% and 39% identity, respectively) than does HiLPMO9B (37%, 29% and 28%). HiLPMO9I shares 36% sequence identity with HiLPMO9B. In particular, surface exposed regions of the L2 and L3 loops that interact with GM in the LsLPMO9A structure are more similar in HiLPMO9I than in HiLPMO9B (Fig 6A). One example is the conservative replacement of an asparagine in loop L2 of LsLPMO9A (Asn-28) and NcLMPO9C with an aspartic acid in HiLPMO9I, while there is a hydrophobic tyrosine at the corresponding position in HiLPMO9B. Also, the L3 loop is more similar in length and sequence between these four GM-active LPMOs. The two substrate-interacting residues (His-67 and Ser-78) at either end of the loop are conserved but not Asn-67. Neither of these residues seems to be present in HiLPMO9B structure, which has a much shorter L3 loop. The two GM-interacting residues found on loop L8 of LsLPMO9A, Glu-150 and Arg-161, are not conserved in the GM-active NcLPMO9C and HiLPMO9I (or in HiLPMO9B), but are present in the L8 loop of CvLPMO9A. One interacting residue, Tyr-212 in the LC region, is conserved in all the five aligned sequences, but is also present in most of the known AA9 LPMOs [20], and may thus not necessarily be specific for interaction with glucomannan.
Fig 6. Structural comparison of HiLPMO9B and HiLPMO9I with three other GM-active AA9 LPMOs.
(A) Structure-based sequence alignment of LsLPMO9A (PDB code 5NKW; [36]), CvLPMO9A (PDB code 5NLT; [36]), NcLPMO9C (PDB code 4D7U; [55]), HiLPMO9I and HiLPMO9B (PDB code 5NNS; [48]) catalytic domains, and (B) structural detail of GM-oligosaccharide binding to LsLPMO9A (PDB code 5NKW; [36]). The sequence alignment is based on secondary structure information and superposition of the HiLPMO9B structure with the HiLPMO9I homology model, and three other GM-active LPMOs, NcLPMO9C, CvLPMO9A, and LsLPMO9A in complex with glucomannan oligosaccharides. Fully conserved residues are shown in white letters on a black background. Thin-line boxes indicate residues with more than 60% sequence similarity among the aligned sequences. Secondary structure elements of LsLPMO9A are shown at the top of the sequence alignment, where β stands for beta-sheets, α for alpha-helices, and η for 310-helices, respectively. The surface-exposed loop regions are labeled, and underlined in different colors (Loop L2 in gold; L3 in purple; LS in brown; L8 in cyan; LC in blue). The active-site histidine residues of LsLPMO9A are marked with green stars. Red triangles mark residues of LsLPMO9A that were observed to interact with the GM-oligosaccharide in the structure (PDB code 5NKW).
Discussion
Our results show that the two characterized H. irregulare LPMOs, HiLPMO9B and HiLPMO9I, are quite different in terms of activity on crystalline cellulose and glucomannan, the major polysaccharides of conifer trees. When BMCC was incubated with the two LPMOs, HiLPMO9I produced very little soluble sugar, in contrast to HiLPMO9B, which even exceeded the HiCel7A cellobiohydrolase in soluble sugar yield. The difference between these two LPMOs was more pronounced when comparing the saccharification of BMCC by LPMOs and HiCel7A in conjunction. There was practically no stimulation with HiLPMO9I to HiCel7A, but clear enhancement was seen when using HiLPMO9B. Mixtures of HiLPMO9B and HiCel7A, at different ratios but with the same total protein load, gave higher yields of soluble sugar than either of the enzymes on its own. Apparently, HiLPMO9B is complementary in activity and cooperate with HiCel7A in degradation of cellulose.
The trend was similar when the cellulose sample was treated sequentially, first treated with LPMO and thereafter treated with HiCel7A. No effect was seen by HiLPMO9I, whereas more cellobiose and also C1-oxidized cellobiose was released by HiCel7A from the cellulose pretreated with HiLPMO9B, despite the fact that the total amount of remaining cellulose was reduced. Approximately 8% of the cellulose, and presumably the most accessible regions, should already have been degraded and washed away in form of the soluble sugars generated by HiLPMO9B after 48 h incubation. Nevertheless, more soluble sugars could be released by HiCel7A on the HiLPMO9B-treated BMCC. This indicates that the activity of HiLPMO9B modifies the cellulose substrate to make it more accessible for degradation by HiCel7A.
These results are in good agreement with the findings in previous studies. It has been shown by AFM that the C1-oxidizing Neurospora crassa LPMO9F (NcLPMO9F) caused separation of cellulose fibrils from the crystalline cellulose surface, and the pretreatment of NcLPMO9F could enhance absorption and hydrolytic activity on crystalline cellulose by the cellobiohydrolase TrCel7A from Trichoderma reesei [47]. Similar results were reported in another AFM study wherein the C1/C4-active TrLPMO9A could split larger clusters of crystalline cellulose ribbons into thinner fibrils, and the degradation was accelerated with a combination of TrLPMO9A and TrCel7A [57]. This is in line with molecular simulations to examine the effect of oxidative cleavage on the structure of crystalline cellulose [58]. As expected, the cleavage due to LPMO actions generated new chain ends that are more solvent accessible. Interestingly, the reducing end was the most solvent exposed of the two new ends at the cleavage site, regardless of which end that was oxidized. LPMO action at either C1 or C4 would thus in both cases primarily increase the exposure of new reducing ends on the cellulose, and potentially the accessibility for reducing end-specific cellulases such as TrCel7A and HiCel7A and their productivity.
In contrast to TrCel7A, the corresponding HiCel7A does not contain any linker-CBM, but consists of a single GH7 catalytic domain [17]. HiCel7A is the most secreted enzyme by H. irregulare, and the only reducing-end specific CBH of H. irregulare. It is known from previous studies that GH7 CBHs (and also other cellulases) can access much fewer binding sites on cellulose without than with a CBM [59]. Therefore, to increase the number of accessible binding sites on cellulose for HiCel7A may be even more important in the H. irregulare system, and suggests that this may be the major role for HiLPMO9B. Indeed, HiCel7A appears to be even more stimulated than TrCel7A by the cooperation with an LPMO, although the experiments may not be directly comparable. HiCel7A is apparently able to attack and cleave off C1-oxidized ends (Figs 1 and 2). We also note that HiCel7A did generate small amounts of C1-oxidized cellobiose also when acting alone, i.e. in the absence of any LPMO, indicating that C1-oxidized ends were already present in the cellulose substrate, as shown previously [60].
In previous research, nearly all of AA9 LPMOs were characterized using amorphous cellulose (e.g. PASC) as substrate, to determine the regioselectivity on cellulose. The activity of LPMOs on crystalline cellulose has been much less studied, so information is scarce about the relative efficiency of crystalline cellulose degradation between LPMOs with different regioselectivity. In this study, we found that the C1-oxidizing HiLPMO9B is much more efficient in degrading BMCC cellulose than the C4-oxidizing HiLPMO9I.
When glucomannan was used as the substrate the picture was quite different. No activity was detected for HiLPMO9B, whereas clear activity was displayed by HiLPMO9I. So far only five out of 37 characterized LPMOs are known to have activities against glucomannan, but the regioselectivity and substrate specificity of those GM-active LPMOs vary [61]. LsLPMO9A, CvLPMO9A, NcLPMO9C and Padospora anserina LPMO9H (PaLPMO9H) displayed broad substrate specificity over a range of hemicellulosic substrates other than GM, such as xyloglucan, mixed-linkage β-glucan, and even xylan in some cases [32,36,37], while HiLPMO9I was only active on GM but not on the other hemicellulosic substrates tested [49]. LsLPMO9A, CvLPMO9A, PaLPMO9H oxidizes both C1 and C4 carbons in glucomannan polysaccharides [32,36,37], while NcLPMO9C only oxidizes at C4 [32,35]. Our MS/MS analysis indicates that similar to NcLPMO9C, HiLPMO9I also oxidizes only at the C4 carbon of the pyranose units in glucomannan. The MS/MS data does not, however, show if the C4 oxidation is on glucose or mannose residues in the glucomannan.
Molecular details of an LPMO with glucomannan was originally shown in the LsLPMO9A-GM complex where the substrate is bound to charged/polar residues located on surface-exposed loop regions (loop L2, L3 and L8) via hydrogen bonding, in addition to hydrophobic interaction with the conserved tyrosine residue on loop LC [36]. Structural comparison of four known GM-active LPMOs (NcLPMO9C, LsLPMO9A, CvLPMO9A and HiLPMO9I) highlights similarities in the L2 and L3 loops in terms of proposed sugar-interacting residues, suggesting that those residues may be essential for glucomannan binding. While 4 out of 7 sugar-interacting residues in LsLPMO9A are conserved in HiLPMO9I, only one (Tyr-212) is maintained in HiLPMO9B. All the corresponding charged or polar sugar-interacting residues on the L2 and L3 loops are missing in HiLPMO9B, which could be a possible reason why no activity on glucomanan was detected for this enzyme. Among the GM-active LPMOs, differences in the suface-exposed loop regions and the distribution of sugar-interacting residues may lead to different positioning of the glucomannan chain relative to the active-site copper ion, which may connect to the varied oxidation preference on glucomannan substrates, but this requires further investigation.
The difference of activity between HiLPMO9B and HiLPMO9I on cellulose and glucomannan indicate possible functional diversification within the LPMO system of H. irregulare, presumably connected to the biological behavior of this fungus in colonization on softwood. H. irregulare is known to remove lignin and hemicellulose prior to breaking down cellulose [62], which suggests that deconstruction of glucomannan network may be an initial step that allows hyphal penetration across the plant cell walls. The considerable reduction of glucomannan viscosity shown by HiLPMO9I suggests that this enzyme could be recruited for the disruption of glucomannan network to facilitate the hyphal penetration. HiLPMO9B, on the other hand, is capable of degrading cellulose and increasing substrate accessibility for the HiCel7A cellobiohydrolase, suggesting that HiLPMO9B could play an important role in crystalline cellulose degradation, through cooperation with other cellulases.
In summary, this study shows that the two H. irregulare LPMOs, HiLPMO9B and HiLPMO9I, show different performance in terms of degrading glucomannan and microcrystalline cellulose, which suggests that HiLPMO9B and HiLPMO9I could be involved in the different parts of the enzyme machinery used for decomposition of individual polysaccharide components in the softwood cell wall, with different contribution to the colonization process of this fungus on softwood.
Acknowledgments
We thank Anna Borisova for preparing the HiCel7A sample and Thu Vuong for preparing the C1-oxidized cello-oligosaccharide standards for HPAEC analysis.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was financially supported by the Swedish Energy Agency (http://www.energimyndigheten.se/) through the grant 40144-1 to M.S. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ochoa-Villarreal M, Aispuro-Hernndez E, Vargas-Arispuro I, ngel M. Plant cell wall polymers: function, structure and biological activity of their derivatives In: De Souza Gomes A, editor. Polymerization. InTech; 2012. Available: http://www.intechopen.com/books/polymerization/plant-cell-wall-polymers-function-structure-and-biological-activity-of-their-derivatives [Google Scholar]
- 2.Blanchette RA, Nilsson T, Daniel G, Abad A. Biological Degradation of Wood. Archaeological Wood. American Chemical Society; 1989. pp. 141–174. 10.1021/ba-1990-0225.ch006 [DOI] [Google Scholar]
- 3.Cragg SM, Beckham GT, Bruce NC, Bugg TD, Distel DL, Dupree P, et al. Lignocellulose degradation mechanisms across the Tree of Life. Curr Opin Chem Biol. 2015;29: 108–119. 10.1016/j.cbpa.2015.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lionetti V, Métraux J-P. Plant cell wall in pathogenesis, parasitism and symbiosis. Front Plant Sci. 2014;5 10.3389/fpls.2014.00612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mäkelä MR, Donofrio N, de Vries RP. Plant biomass degradation by fungi. Fungal Genet Biol. 2014;72: 2–9. 10.1016/j.fgb.2014.08.010 [DOI] [PubMed] [Google Scholar]
- 6.Lind M, Stenlid J, Olson Å. Chapter Twelve—Heterobasidion annosum s.l. Genomics In: Martin FM, editor. Advances in Botanical Research. Academic Press; 2014. pp. 371–396. [DOI] [Google Scholar]
- 7.Garbelotto M, Gonthier P. Biology, Epidemiology, and Control of Heterobasidion Species Worldwide. Annu Rev Phytopathol. 2013;51: 39–59. 10.1146/annurev-phyto-082712-102225 [DOI] [PubMed] [Google Scholar]
- 8.Asiegbu FO, Adomas A, Stenlid J. Conifer root and butt rot caused by Heterobasidion annosum (Fr.) Bref. s.l. Mol Plant Pathol. 2005;6: 395–409. 10.1111/j.1364-3703.2005.00295.x [DOI] [PubMed] [Google Scholar]
- 9.Korhonen K. Intersterility groups of Heterobasidion annosum. Commun Instituti For Fenn. 1978;94: 25. [Google Scholar]
- 10.Chase TE, Ullrich RC. Heterobasidion Annosum, root- and butt-rot of trees. Adv Plant Pathol. 1988;6: 501–510. [DOI] [Google Scholar]
- 11.Capretti P, Korhonen K, Mugnai L, Romagnoli C. An intersterility group of Heterobasidion annosum specialized to Abies alba. Eur J For Pathol. 1990;20: 231–240. 10.1111/j.1439-0329.1990.tb01134.x [DOI] [Google Scholar]
- 12.Woodward S, Stenlid J, Karjalainen R, Hüttermann A. Heterobasidion annosum. biology, ecology, impact and control. CAB Int Camb. 1998; Available: http://onlinelibrary.wiley.com/doi/10.1046/j.1365-3059.1999.0366b.x/abstract [Google Scholar]
- 13.Otrosina WJ, Garbelotto M. Heterobasidion occidentale sp. nov. and Heterobasidion irregulare nom. nov.: a disposition of north american Heterobasidion biological species. Fungal Biol. 2010;114: 16–25. 10.1016/j.mycres.2009.09.001 [DOI] [PubMed] [Google Scholar]
- 14.Scheller HV, Ulvskov P. Hemicelluloses. Annu Rev Plant Biol. 2010;61: 263–289. 10.1146/annurev-arplant-042809-112315 [DOI] [PubMed] [Google Scholar]
- 15.van den Brink J, de Vries RP. Fungal enzyme sets for plant polysaccharide degradation. Appl Microbiol Biotechnol. 2011;91: 1477–1492. 10.1007/s00253-011-3473-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olson Å, Aerts A, Asiegbu F, Belbahri L, Bouzid O, Broberg A, et al. Insight into trade-off between wood decay and parasitism from the genome of a fungal forest pathogen. New Phytol. 2012;194: 1001–1013. 10.1111/j.1469-8137.2012.04128.x [DOI] [PubMed] [Google Scholar]
- 17.Momeni MH, Payne CM, Hansson H, Mikkelsen NE, Svedberg J, Engström Å, et al. Structural, Biochemical, and Computational Characterization of the Glycoside Hydrolase Family 7 Cellobiohydrolase of the Tree-killing Fungus Heterobasidion irregulare. J Biol Chem. 2013;288: 5861–5872. 10.1074/jbc.M112.440891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Westereng B, Ishida T, Vaaje-Kolstad G, Wu M, Eijsink VGH, Igarashi K, et al. The Putative Endoglucanase PcGH61D from Phanerochaete chrysosporium is a metal-dependent oxidative enzyme that cleaves cellulose. PLOS ONE. 2011;6: e27807 10.1371/journal.pone.0027807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaaje-Kolstad G, Westereng B, Horn SJ, Liu Z, Zhai H, Sørlie M, et al. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science. 2010;330: 219–222. 10.1126/science.1192231 [DOI] [PubMed] [Google Scholar]
- 20.Leggio LL, Simmons TJ, Poulsen J-CN, Frandsen KEH, Hemsworth GR, Stringer MA, et al. Structure and boosting activity of a starch-degrading lytic polysaccharide monooxygenase. Nat Commun. 2015;6: 5961 10.1038/ncomms6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beeson WT, Phillips CM, Cate JHD, Marletta MA. Oxidative cleavage of cellulose by fungal copper-dependent polysaccharide monooxygenases. J Am Chem Soc. 2012;134: 890–892. 10.1021/ja210657t [DOI] [PubMed] [Google Scholar]
- 22.Isaksen T, Westereng B, Aachmann FL, Agger JW, Kracher D, Kittl R, et al. A C4-oxidizing lytic polysaccharide monooxygenase cleaving both cellulose and cello-oligosaccharides. J Biol Chem. 2014;289: 2632–2642. 10.1074/jbc.M113.530196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bey M, Zhou S, Poidevin L, Henrissat B, Coutinho PM, Berrin J-G, et al. Cello-oligosaccharide oxidation reveals differences between two lytic polysaccharide monooxygenases (family GH61) from Podospora anserina. Appl Environ Microbiol. 2013;79: 488–496. 10.1128/AEM.02942-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bissaro B, Røhr ÅK, Müller G, Chylenski P, Skaugen M, Forsberg Z, et al. Oxidative cleavage of polysaccharides by monocopper enzymes depends on H2O2. Nat Chem Biol. 2017;13: 1123–1128. 10.1038/nchembio.2470 [DOI] [PubMed] [Google Scholar]
- 25.Kracher D, Andlar M, Furtmüller PG, Ludwig R. Active-site copper reduction promotes substrate binding of fungal lytic polysaccharide monooxygenase and reduces stability. J Biol Chem. 2018;293: 1676–1687. 10.1074/jbc.RA117.000109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Busk PK, Lange L. Classification of fungal and bacterial lytic polysaccharide monooxygenases. BMC Genomics. 2015;16: 368 10.1186/s12864-015-1601-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Couturier M, Ladevèze S, Sulzenbacher G, Ciano L, Fanuel M, Moreau C, et al. Lytic xylan oxidases from wood-decay fungi unlock biomass degradation. Nat Chem Biol. 2018;14: 306–310. 10.1038/nchembio.2558 [DOI] [PubMed] [Google Scholar]
- 28.Voshol GP, Vijgenboom E, Punt PJ. The discovery of novel LPMO families with a new Hidden Markov model. BMC Res Notes. 2017;10 10.1186/s13104-017-2429-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sabbadin F, Hemsworth GR, Ciano L, Henrissat B, Dupree P, Tryfona T, et al. An ancient family of lytic polysaccharide monooxygenases with roles in arthropod development and biomass digestion. Nat Commun. 2018;9: 756 10.1038/s41467-018-03142-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johansen KS. Lytic polysaccharide monooxygenases: the microbial power tool for lignocellulose degradation. Trends Plant Sci. 2016;21: 926–936. 10.1016/j.tplants.2016.07.012 [DOI] [PubMed] [Google Scholar]
- 31.Hemsworth GR, Johnston EM, Davies GJ, Walton PH. Lytic polysaccharide monooxygenases in biomass conversion. Trends Biotechnol. 2015;33: 747–761. 10.1016/j.tibtech.2015.09.006 [DOI] [PubMed] [Google Scholar]
- 32.Agger JW, Isaksen T, Várnai A, Vidal-Melgosa S, Willats WGT, Ludwig R, et al. Discovery of LPMO activity on hemicelluloses shows the importance of oxidative processes in plant cell wall degradation. Proc Natl Acad Sci U S A. 2014;111: 6287–6292. 10.1073/pnas.1323629111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frommhagen M, Koetsier MJ, Westphal AH, Visser J, Hinz SWA, Vincken J-P, et al. Lytic polysaccharide monooxygenases from Myceliophthora thermophila C1 differ in substrate preference and reducing agent specificity. Biotechnol Biofuels. 2016;9: 186 10.1186/s13068-016-0594-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennati-Granier C, Garajova S, Champion C, Grisel S, Haon M, Zhou S, et al. Substrate specificity and regioselectivity of fungal AA9 lytic polysaccharide monooxygenases secreted by Podospora anserina. Biotechnol Biofuels. 2015;8: 90 10.1186/s13068-015-0274-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kojima Y, Várnai A, Ishida T, Sunagawa N, Petrovic DM, Igarashi K, et al. A Lytic Polysaccharide Monooxygenase with Broad Xyloglucan Specificity from the Brown-Rot Fungus Gloeophyllum trabeum and Its Action on Cellulose-Xyloglucan Complexes. Appl Environ Microbiol. 2016;82: 6557–6572. 10.1128/AEM.01768-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simmons TJ, Frandsen KEH, Ciano L, Tryfona T, Lenfant N, Poulsen JC, et al. Structural and electronic determinants of lytic polysaccharide monooxygenase reactivity on polysaccharide substrates. Nat Commun. 2017;8 10.1038/s41467-017-01247-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fanuel M, Garajova S, Ropartz D, McGregor N, Brumer H, Rogniaux H, et al. The Podospora anserina lytic polysaccharide monooxygenase PaLPMO9H catalyzes oxidative cleavage of diverse plant cell wall matrix glycans. Biotechnol Biofuels. 2017;10: 63 10.1186/s13068-017-0749-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harris PV, Welner D, McFarland KC, Re E, Navarro Poulsen J-C, Brown K, et al. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family. Biochemistry. 2010;49: 3305–3316. 10.1021/bi100009p [DOI] [PubMed] [Google Scholar]
- 39.Karnaouri A, Muraleedharan MN, Dimarogona M, Topakas E, Rova U, Sandgren M, et al. Recombinant expression of thermostable processive MtEG5 endoglucanase and its synergism with MtLPMO from Myceliophthora thermophila during the hydrolysis of lignocellulosic substrates. Biotechnol Biofuels. 2017;10: 126 10.1186/s13068-017-0813-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim IJ, Youn HJ, Kim KH. Synergism of an auxiliary activity 9 (AA9) from Chaetomium globosum with xylanase on the hydrolysis of xylan and lignocellulose. Process Biochem. 2016;51: 1445–1451. 10.1016/j.procbio.2016.06.017 [DOI] [Google Scholar]
- 41.Langston JA, Shaghasi T, Abbate E, Xu F, Vlasenko E, Sweeney MD. Oxidoreductive cellulose depolymerization by the enzymes cellobiose dehydrogenase and glycoside hydrolase 61. Appl Environ Microbiol. 2011;77: 7007–7015. 10.1128/AEM.05815-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pierce BC, Agger JW, Zhang Z, Wichmann J, Meyer AS. A comparative study on the activity of fungal lytic polysaccharide monooxygenases for the depolymerization of cellulose in soybean spent flakes. Carbohydr Res. 2017;449: 85–94. 10.1016/j.carres.2017.07.004 [DOI] [PubMed] [Google Scholar]
- 43.Jung S, Song Y, Kim HM, Bae H-J. Enhanced lignocellulosic biomass hydrolysis by oxidative lytic polysaccharide monooxygenases (LPMOs) GH61 from Gloeophyllum trabeum. Enzyme Microb Technol. 2015;77: 38–45. 10.1016/j.enzmictec.2015.05.006 [DOI] [PubMed] [Google Scholar]
- 44.Guo Z, Duquesne S, Bozonnet S, Nicaud J-M, Marty A, O’Donohue MJ. Expressing accessory proteins in cellulolytic Yarrowia lipolytica to improve the conversion yield of recalcitrant cellulose. Biotechnol Biofuels. 2017;10: 298 10.1186/s13068-017-0990-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eibinger M, Ganner T, Bubner P, Rošker S, Kracher D, Haltrich D, et al. Cellulose surface degradation by a lytic polysaccharide monooxygenase and its effect on cellulase hydrolytic efficiency. J Biol Chem. 2014;289: 35929–35938. 10.1074/jbc.M114.602227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villares A, Moreau C, Bennati-Granier C, Garajova S, Foucat L, Falourd X, et al. Lytic polysaccharide monooxygenases disrupt the cellulose fibers structure. Sci Rep. 2017;7: srep40262 10.1038/srep40262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eibinger M, Sattelkow J, Ganner T, Plank H, Nidetzky B. Single-molecule study of oxidative enzymatic deconstruction of cellulose. Nat Commun. 2017;8: 894 10.1038/s41467-017-01028-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu B, Kognole AA, Wu M, Westereng B, Crowley MF, Kim S, et al. Structural and molecular dynamics studies of a C1-oxidizing lytic polysaccharide monooxygenase from Heterobasidion irregulare reveal amino acids important for substrate recognition. FEBS J. 2018;285: 2225–2242. 10.1111/febs.14472 [DOI] [PubMed] [Google Scholar]
- 49.Liu B, Olson Å, Wu M, Broberg A, Sandgren M. Biochemical studies of two lytic polysaccharide monooxygenases from the white-rot fungus Heterobasidion irregulare and their roles in lignocellulose degradation. PLOS ONE. 2017;12: e0189479 10.1371/journal.pone.0189479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Väljamäe P, Sild V, Nutt A, Pettersson G, Johansson G. Acid hydrolysis of bacterial cellulose reveals different modes of synergistic action between cellobiohydrolase I and endoglucanase I. Eur J Biochem. 1999;266: 327–334. [DOI] [PubMed] [Google Scholar]
- 51.Karnaouri AC, Topakas E, Christakopoulos P. Cloning, expression, and characterization of a thermostable GH7 endoglucanase from Myceliophthora thermophila capable of high-consistency enzymatic liquefaction. Appl Microbiol Biotechnol. 2014;98: 231–242. 10.1007/s00253-013-4895-9 [DOI] [PubMed] [Google Scholar]
- 52.Borisova AS, Isaksen T, Dimarogona M, Kognole AA, Mathiesen G, Várnai A, et al. Structural and Functional Characterization of a Lytic Polysaccharide Monooxygenase with Broad Substrate Specificity. J Biol Chem. 2015;290: 22955–22969. 10.1074/jbc.M115.660183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benkert P, Tosatto SCE, Schomburg D. QMEAN: A comprehensive scoring function for model quality assessment. Proteins. 2008;71: 261–277. 10.1002/prot.21715 [DOI] [PubMed] [Google Scholar]
- 54.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26: 283–291. 10.1107/S0021889892009944 [DOI] [Google Scholar]
- 55.Borisova AS, Isaksen T, Dimarogona M, Kognole AA, Mathiesen G, Várnai A, et al. Structural and functional characterization of a lytic polysaccharide monooxygenase with broad substrate specificity. J Biol Chem. 2015; jbc.M115.660183. 10.1074/jbc.M115.660183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hollingsworth SA, Karplus PA. A fresh look at the Ramachandran plot and the occurrence of standard structures in proteins. Biomol Concepts. 2010;1: 271–283. 10.1515/BMC.2010.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song B, Li B, Wang X, Shen W, Park S, Collings C, et al. Real-time imaging reveals that lytic polysaccharide monooxygenase promotes cellulase activity by increasing cellulose accessibility. Biotechnol Biofuels. 2018;11: 41 10.1186/s13068-018-1023-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vermaas JV, Crowley MF, Beckham GT, Payne CM. Effects of Lytic Polysaccharide Monooxygenase Oxidation on Cellulose Structure and Binding of Oxidized Cellulose Oligomers to Cellulases. J Phys Chem B. 2015;119: 6129–6143. 10.1021/acs.jpcb.5b00778 [DOI] [PubMed] [Google Scholar]
- 59.Kont R, Kari J, Borch K, Westh P, Väljamäe P. Inter-domain Synergism Is Required for Efficient Feeding of Cellulose Chain into Active Site of Cellobiohydrolase Cel7A. J Biol Chem. 2016;291: 26013–26023. 10.1074/jbc.M116.756007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Szakmary K, Wotawa A, Kubicek CP. Origin of oxidized cellulose degradation products and mechanism of their promotion of cellobiohydrolase I biosynthesis in Trichoderma reesei. Microbiology. 1991;137: 2873–2878. 10.1099/00221287-137-12-2873 [DOI] [Google Scholar]
- 61.Frommhagen M, Westphal AH, Berkel V, H WJ, Kabel MA. Distinct Substrate Specificities and Electron-Donating Systems of Fungal Lytic Polysaccharide Monooxygenases. Front Microbiol. 2018;9 10.3389/fmicb.2018.01080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yakovlev IA, Hietala AM, Courty P-E, Lundell T, Solheim H, Fossdal CG. Genes associated with lignin degradation in the polyphagous white-rot pathogen Heterobasidion irregulare show substrate-specific regulation. Fungal Genet Biol FG B. 2013;56: 17–24. 10.1016/j.fgb.2013.04.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.