Reconstruction of relatedness and ancestry from ancient DNA from Medieval burial provides new insights into kinship behavior.
Abstract
From historical and archeological records, it is posited that the European medieval household was a combination of close relatives and recruits. However, this kinship structure has not yet been directly tested at a genomic level on medieval burials. The early 7th century CE burial at Niederstotzingen, discovered in 1962, is the most complete and richest example of Alemannic funerary practice in Germany. Excavations found 13 individuals who were buried with an array of inscribed bridle gear, jewelry, armor, and swords. These artifacts support the view that the individuals had contact with France, northern Italy, and Byzantium. This study analyzed genome-wide sequences recovered from the remains, in tandem with analysis of the archeological context, to reconstruct kinship and the extent of outside contact. Eleven individuals had sufficient DNA preservation to genetically sex them as male and identify nine unique mitochondrial haplotypes and two distinct Y chromosome lineages. Genome-wide analyses were performed on eight individuals to estimate genetic affiliation to modern west Eurasians and genetic kinship at the burial. Five individuals were direct relatives. Three other individuals were not detectably related; two of these showed genomic affinity to southern Europeans. The genetic makeup of the individuals shares no observable pattern with their orientation in the burial or the cultural association of their grave goods, with the five related individuals buried with grave goods associated with three diverse cultural origins. These findings support the idea that not only were kinship and fellowship held in equal regard: Diverse cultural appropriation was practiced among closely related individuals as well.
INTRODUCTION
The Alemanni were a confederation of Germanic tribes that inhabited the eastern Upper Rhine basin and surrounding region (Fig. 1) (1). Roman ethnographers mentioned the Alemanni, but historical records from the 3rd to the 6th century CE contain no regular description of these tribes (2). The upheaval that occurred during the European Migration Period (Völkerwanderung) partly explains the interchangeability of nomenclature with the contemporaneous Suebi people of the same region and periods of geographic discontinuity in the historical record (3). This diverse nomenclature reflects centuries of interactions between Romans and other Germanic groups such as the Franks, Burgundians, Thuringians, Saxons, and Bavarians. With the defeat of the Alemanni by Clovis I of the Franks in 497 CE, Alamannia became a subsumed Duchy of the Merovingian Kingdom. This event solidified the naming of the inhabitants of this region as Alemanni (3). From the 5th to the 8th century CE, integration between the Franks and the Alemanni was reflected by changed burial practices, with households (familia) buried in richly furnished graves (Adelsgrablege) (4). The splendor of these Adelsgräber served to demonstrate the kinship structure, wealth, and status of the familia and also the power of the Franks (Personenverbandstaaten, a system of power based on personal relations rather than fixed territory). Because inclusion in familia during the Merovingian period was not necessarily based on inheritance or provenance, debate continues on the symbolism of these burial rites (5).
Fig. 1. Burial site reconstructions and location.
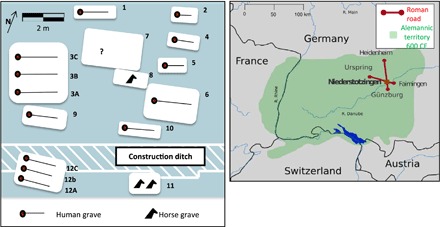
(Left) Burial orientation of human and horse graves at Niederstotzingen. (Right) Location of burial site in southwest Germany.
The 7th century CE Alemannic burial site at Niederstotzingen in southern Germany, used circa 580 to 630 CE, represents the best-preserved example of such an Alemannic Adelsgrablege. Discovered in 1962 (Fig. 1) (6, 7), 13 human skeletal remains (10 adults and 3 infants), along with a varied assemblage of weapons, armor, jewelry, equestrian gear, and remains of three horses, were excavated from 12 graves. The grave goods indicated contacts with Byzantines, Lombards, and Franks (Table 1 and fig. S1). These outside contacts appear to have been facilitated by equestrianism and guardianship of a nearby Roman crossroads. Two of the 12 graves (3 and 12) were multiple burials containing three individuals each, from which it has been inferred that they had close familial relationships (8). Both multiple burials contained Byzantine artifacts, lamella helmet (grave 12, individual 12B), and equestrian gear with Byzantine engravings (grave 3, individual 3A), suggesting eastern Mediterranean contact, whereas those buried with Frankish (individual 9) and Lombard (individual 6) artifacts support contact with eastern France and northern Italy (6, 8, 9).
Table 1. Archeological context, karyotype sex estimates, haplotypes, contamination estimates, and isotopic data from Niederstotzingen.
Strontium values that indicate provenance outside of the region. Full list of mtDNA and NRY haplotypes is given in tables S8 and S9. Complete summary of contamination estimates for mitochondrial and X chromosome on 1240K capture and SE is given in tables S6 and S7. Genetic sex estimates are based on shotgun and 1240K capture. Individuals highlighted in red and green are from two multiple burials, graves 3 and 12 (Fig. 1).
| Individual |
Archeological context |
Age at death estimate (years) |
Genetic sex | MtDNA/NRY haplotype |
Median mtDNA/X (SE) contamination |
Strontium 87Sr/86Sr |
| 1 | — | 40–50 | Male | K1a/R1b1a2a1a1c2b2b1a1 | 0.03/0.002 (0.003) | 0.7089 |
| 2 | — | 9–11 | Male? *† | K1a1b2a1a/- | 0.15/— | 0.7093 |
| 3a | Byzantine | 20–30 | Male | K1a/R1b1a2a1a1c2b2b1a1 | 0.02/0.0145 (0.007) | 0.709 |
| 3b | — | 50–60 | Male | I5a1b/G2a2b1 | 0.03/0.0009 (0.0024) | 0.7104† |
| 3c | — | 20–30 | Male | T2/R1b1a2 | 0.03/0.0404 (0.0203) | 0.7089 |
| 4 | — | 2 | Male | X2b4/- | 0.03/— | 0.7094 |
| 5 | — | 0.5–2 | Male?*† | K1a1b2a1a/- | 0.04/— | 0.7096 |
| 6 | Lombardian | 14–17 | Male | H65a/R1b1a2a1a1c2b2b | 0.02/−0.0007 (0.00117) | 0.7093 |
| 9 | Franconian | 40–50 | Male | X2b4/R1b1a2a1a1c2b2b1a1 | 0.04/−0.0047 (0.00466) | 0.7089 |
| 10 | — | 20–25 | Male* | H1b/R1 | 0.06/— | 0.7105† |
| 12a | — | 25–35 | Male* | H10e1/R1b1 | 0.09/— | 0.7087 |
| 12b | Byzantine | 30–40 | Male | X2b4/R1b1a2a1a1c2b2b1a1 | 0.03/0.0128 (0.0136) | 0.709 |
| 12c | — | 20–30 | Male | U5a1a1/R1b1a2a1a1c2b2b1a1 | 0.03/0.0010 (0.0078) | 0.7093 |
*Statistically insignificant sex estimate but consistent with maleness (table S10 and fig. S4).
†After PMD filtering, individuals 2 and 5 had insufficient reads for accurate sex estimation (table S13).
Since the initial discovery of Niederstotzingen, bioarcheological analyses have provided additional information from the remains (section S2). Strontium and oxygen isotope data from the enamel showed that most individuals are local rather than migrants (Table 1, table S2, and fig. S2), except for individuals 10 and 3B. Polymerase chain reaction (PCR)–based sex estimation and reconstruction of the mitochondrial DNA (mtDNA) hypervariable region offered the first genetic characterization of the individuals (table S1) (8). Despite these findings, there are still questions regarding their genetic sex, kinship, and genetic origin, because of the technical challenges of ancient DNA (aDNA) analysis (10). Typically, DNA extracted from archeological remains contains only trace amounts of endogenous human DNA. Furthermore, multiple waves of migration and recent admixture can make it challenging to genetically distinguish ancient populations inhabiting nearby geographical locations (11, 12). Single-nucleotide polymorphisms (SNP) in recombining regions of autosomes and the X chromosome are especially useful for analysis of population affiliations (13, 14) and kinship (15, 16). These SNPs contain ancestral information from a large number of an individual’s ancestors, unlike the nonrecombining maternally inherited mtDNA and paternally inherited Y chromosome each representing only an ancestral lineage.
To increase the resolution of kinship and population genetic analysis, we analyzed the individuals recovered from Niederstotzingen by using an approach that combined targeted DNA enrichment of more than 1 million SNPs (17) with high-throughput sequencing. Through joint analysis of mtDNA, Y chromosome [the nonrecombining region (NRY)], and autosomal SNPs, we aimed to reconstruct potential familial relationships of the Niederstotzingen individuals and estimate their genetic sex. In combination with archeological and isotopic data, we directly tested historical and archeological hypotheses positing that Alemannic burial practices, through assortment of deceased individuals and their associated artifacts, are a reflection of mobility and kinship structure. Our results provide insight into the medieval household (familia) and the role of multiple burials at Niederstotzingen. Previous studies have shown that the genetic affiliations of ancient people often do not match putative geographic origin of their culture (5, 18). Hence, we investigated the congruence between the biological provenance and the cultural origin of burial rites and goods present at Niederstotzingen. Our results shed new light on the kinship structure in Early Medieval Europe and investigate whether contacts at Niederstotzingen to southern Europe and elsewhere went beyond the exchange of material artifacts. The results show that 11 of the individuals are likely males, suggesting a sex-biased burial practice, 5 of whom are detectably related to at least second degree. These five related individuals had culturally diverse grave goods despite the evidence that all of them showed local isotope signals with northern European genetic affiliations; these data show how diverse cultural appropriation could exist even among close relatives.
RESULTS
Authentication of sequenced data
To assess the quantity and quality of the extracted DNA, we performed shotgun sequencing before capture and mapped the sequences to the human genome. The percentage of endogenous DNA varied considerably between sequenced samples (<1 to 75%), showing different rates of endogenous DNA preservation among individuals and even in subsamplings from the same individual (table S3). Captured libraries showed increased percentage abundance mapping at target loci compared to shotgun libraries: between 273- and 2494-fold target enrichment efficiency for mtDNA and between 22- and 232-fold target enrichment efficiency for the 1240K SNP capture (tables S4 and S5). Deamination patterns at the 5′ and 3′ ends, typical of aDNA damage in non-UDG (uracil-DNA glycosylase)–treated libraries (fig. S3), indicate that there are endogenous sequences present. Low contamination estimates of the uniparental markers obtained with the software Schmutzi on mtDNA and ANGSD (analysis of next-generation sequencing data) on the X chromosome further support the authenticity of our ancient human DNA (Table 1 and tables S6 and S7). Exceptions were individuals 2, 10, and 12A that had a median mtDNA contamination above 5%. For these mitogenomes, low-covered and ambiguous mtDNA positions were further stringently filtered through visual inspection with Integrative Genomics Viewer (19) and compared with PMDtools filtered reads.
Analysis of uniparental markers
mtDNA haplogroups were successfully assigned to all 13 individuals (Table 1). Notably, there are three groups of individuals that share, among the assigned positions, identical haplotypes: individuals 4, 9, and 12B in haplogroup X2b4; individuals 1 and 3A in haplogroup K1a; and individuals 2 and 5 in haplogroup K1a1b2a1a (table S8). Postmortem deamination (PMD) PMDtools filtering (threshold 3) of mtcapture sequences of individuals 2, 10, and 12A (that showed contamination estimates greater than 5%) did not change the main haplogroup; only 12A’s haplogroup becomes less derived (table S14).
Despite low coverage across the Y chromosome in the 1240K, the NRY haplogroups of 10 individuals were successfully recovered (Table 1 and tables S3 and S9). Most individuals belong to the R1b haplogroup (individuals 1, 3A, 3C, 6, 9, 12A, 12B, and 12C), which has the highest frequency (>70%) in modern western European populations (20). Five individuals (1, 3A, 9, 12B, and 12C) share the same marker (Z319) defining haplogroup R1b1a2a1a1c2b2b1a1. Because of incomplete SNP capture and coverage on the Y chromosome, most of the individual’s haplotypes do not overlap across the entire International Society of Genetic Genealogy (ISOGG) Y-haplogroup tree, but multiple positions with the same haplotype and/or consistent root allow estimation of Y chromosome ancestry anyway. For example, individuals 1, 3A, and 6 have R1b lineage and marker Z347 (R1b1a2a1a1c2b2b), which belongs to the same male ancestral lineage as marker Z319. Individual 3B instead carries NRY haplogroup G2a2b1, which is rare in modern north, west, and east European populations (<5%), only reaching common abundance in the Caucasus (>70%), southern Europe, and the Near East (10 to 15%) (21).
Sex determination of individuals
Genetic-based sex estimates, using shotgun and 1240K capture data, show that at least 11 of the individuals were probably male. From shotgun sequences, 1, 3A, 3B, 3C, 4, 6, 9, 12B, and 12C have statistically significant rates of male DNA, while 2, 5, 10, and 12A are low-covered but consistent with the presence of male DNA (Table 1 and table S10). To further validate authenticity of these data, shotgun sequences were filtered for reads with PMD with PMDtools (threshold 3) (22). Sex estimates were largely consistent for filtered data, except 3A, 3C, and 12B, which lost statistical significance, and individuals 2 and 5, which had insufficient reads remaining (table S13). The normalized ratio of X chromosome to autosomal reads provided a robust estimate of sex for shotgun libraries that had extremely low coverage, except for individuals 5 and 12a (table S11) (23). The ratio of captured X and Y chromosome SNPs to autosomal SNPs in the 1240K also shows a similar proportion of X and Y SNPs, further supporting the indication that all the individuals are males (fig. S4 and table S10) (24).
Genotyping and population affinity of genome-wide capture
After 1240K capture, between 29,673 and 374,347 called SNPs overlapped with the Human Origins Database (25, 26), providing enough genomic markers for accurate estimates of genetic affinity (1, 3A, 3B, 3C, 6, 9, 12B, and 12C) (Table 1 and table S5). The projections of the ancient genomic data on principal components analysis (PCA) built with modern west Eurasians show that six individuals from Niederstotzingen (1, 3A, 6, 9, 12B, and 12C, hereafter referred to as Niederstotzingen North) have the greatest affinity with modern northern, eastern, and central Europeans, while individuals 3B and 3C (hereafter referred to as Niederstotzingen South) have the greatest affinity with southern Europeans and individuals from the eastern Mediterranean (Fig. 2). Admixture analysis with five components that had the optimum cross-validation (CV) error (table S12) indicates that Niederstotzingen North individuals have the most similar admixture components to modern-day eastern European populations (fig. S5). Niederstotzingen South individuals have a proportion of components that most resemble modern Mediterranean populations. However, 3B and 3C do not have similar proportions of ancestry to one another unlike Niederstotzingen North individuals.
Fig. 2. PCA plot of Niederstotzingen individuals, modern west Eurasians, and selected ancient Europeans.
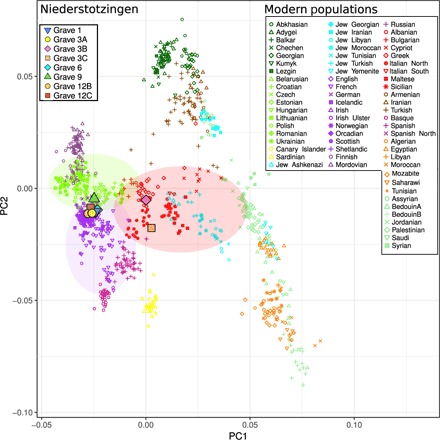
Genome-wide ancient data were projected against modern west Eurasian populations. Colors on PCA indicate more general Eurasian geographic boundaries than countries: dark green, Caucasus; bright green, eastern Europe; yellow, Sardinia and Canary Islands; bright blue, Jewish diaspora; bright purple, western and central Europe; red, southern Europe; dark brown, west Asia; light purple, Spain; dark purple, Russia; pale green, Middle East; orange, North Africa. The transparent circles serve to highlight the genetic overlap between regions of interest.
To formally test the extent of shared genetic drift among individuals, outgroup F3 statistics were applied to the autosomal data using populations guided by PCA projections. F3 statistics (Mbuti; Niederstotzingen individual, modern west Eurasian) concur with PCA and admixture estimates, showing that Niederstotzingen North individuals are closely related to northern and eastern European populations, particularly from Lithuania and Iceland. Niederstotzingen South individuals have the greatest affinity to southern Europeans (figs. S6 to S13), in particular to populations from modern northern Spain, but have a much weaker population affinity to any European population overall, thus suggesting recent admixture between different populations in their ancestry.
Estimation of kinship
To investigate kinship between the individuals, we made pairwise estimates of first- and second-degree kinship based on the proportion of nonmatching autosomal genotypes (P0) in each unique pair from all SNPs that overlapped with the 1240K (Fig. 3 and tables S15 and S16). READ (relationship estimation from ancient DNA) software defines first degree as immediate family (parent-offspring and siblings) and second degree as extended family (cousins, uncles/aunts, grandparent-grandchild, and half-siblings) and uses a normalized P0 value (27). READ software is tailored for aDNA from software designed for modern whole-genome sequencing data (28). To back up READ results, the coefficient of relatedness was estimated from non-normalized P0 without READ software (table S16) (16). Both kinship estimates show first-degree relatedness for pairs 1/3A, 1/6, 1/9, 3A/9, and 9/12B and second-degree relatedness for 1/12B, 3A/6, 3A/12B, and 6/9. Except for 12C, all of the Niederstotzingen North individuals are detectably and closely related. The Niederstotzingen South individuals are not detectably related to each other or any other members of the cohort.
Fig. 3. Reconstruction of first- and second-degree relatedness among all related individuals.
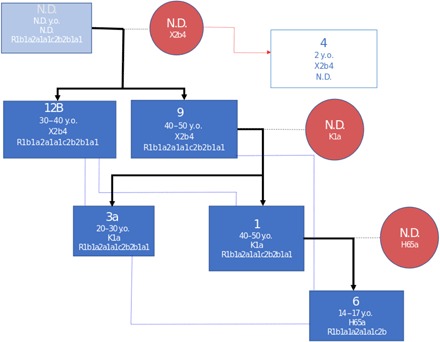
Bold black lines and blue lines indicate first- and second-degree relatedness, respectively. Dark blue squares are identified males with age-at-death estimates years old (y.o.), mtDNA haplotypes, and NRY haplogroups. Red circles represent unidentified females that passed maternal haplotypes to their offspring. The light square represents one male infant that shares its maternal haplotype with individuals 12B and 9. N.D., not determined.
DISCUSSION
The genetic characterization of the individuals at Niederstotzingen offers insights into their kinship structure and origin. We show genome-wide data for all 13 individuals recovered from the site, 8 of which (Fig. 1) had sufficient genomic enrichment to infer genetic affinity to a familial level. These results allow direct testing of historical and archeological hypotheses regarding the symbolism of burial rites from the Early Medieval.
Eleven individuals were probably genetically male. The combined 1240K and shotgun data inform the sex of the three infants (2, 4, and 5) and are consistent with the anthropologically determined sex of all adults, including three adults males with gracile craniofacial features that may be attributable to female osteology (3C, 6, and 12C) (8, 29). The observation that even most of the infants and juveniles were probably male and that grave 7 may have once contained females, which were later exhumed and possibly reburied (supported by the presence of women’s jewelry) (6), suggests that burial rites were sex-biased. Across the Merovingian Kingdom, male-only burials have been observed in this Adelsgräber buried in prominent places such as Roman roads (4). This pattern may reflect the military function of the burial or the social structure of the nobility. Likewise, sex-biased burial patterns have been observed at European sites throughout Late Antiquity (11, 30).
Genomically, the individuals buried at Niederstotzingen can be split into two groups: Niederstotzingen North (1, 3A, 6, 9, 12B, and 12C), who have genomic signals that most resemble modern northern and eastern European populations, and Niederstotzingen South (3B and 3C), who most resemble modern-day Mediterraneans, albeit with recent common ancestry to other Europeans. Niederstotzingen North is composed of those buried with identifiable artifacts: Lombards (individual 6), Franks (individual 9), and Byzantines (individuals 3A and 12B), all of whom have strontium and oxygen isotope signals that support local provenance (fig. S2) (8). Just two individuals, 3B (Niederstotzingen South) and 10 (no sufficient autosomal data, with R1 Y-haplogroup), have nonlocal strontium isotope signals. The δ18O values suggest that individuals 10 and 3B may have originated from a higher-altitude region, possibly the Swiss-German Alpine foothills (8). Combined with the genome affinity of individual 3B to southern Europeans, these data provide direct evidence for incoming mobility at the site and for contact that went beyond exchange of grave goods (4). Familia had holdings across the Merovingian Kingdom and traveled long distances to maintain them; these holdings could have extended from northern Italy to the North Sea. Nobles displayed and accrued power by recruiting outside individuals into the household as part of their traveling retinue. Extravagant burial rites of these familia are symbolic evidence of the Frankish power systems based on people Personenverbandstaaten imposed from the 5th until the 8th century CE (4). The assignment of grave goods and the burial pattern do not follow any apparent pattern with respect to genetic origin or provenance, suggesting that relatedness and fellowship were held in equal regard at this burial.
Further insights into the kinship structure are obtained from the reconstruction of direct relatives at the site. We demonstrated that five of the individuals (1, 3A, 6, 9, and 12B) were kin to at least second degree (Fig. 3 and tables S15 and S16); four of these were buried with distinguishable grave goods (discussed above and in fig. S1). These data show that at Niederstotzingen, at least in death, diverse cultural affiliations could be appropriated even within the same family across just two generations. This finding is somewhat similar to the burial of the Frankish King Childeric in the 5th century CE with a combination of Frankish and Byzantine grave goods that symbolized both his provenance and military service to the Romans (4). The burial of three unrelated individuals (3B, 3C, and 12C) in multiple graves beside the rest of the cohort would imply that this Alemannic group buried their dead based on a combination of familial ties and fellowship. One explanation could be that they were adopted as children from another region to be trained as warriors, which was a common practice at the time; these children were raised with equal regard in the familia (2, 4).
To infer a tentative family tree, we combined autosomal SNPs, NRY haplotypes, mtDNA haplotypes, and age-at-death data. It is possible that pairs 9/1, 9/3A, and 1/6 represent father-son relationships. The results also support that pairs 1/3A and 9/12B are siblings, as they share the same maternal haplotype (K1a and X2B4, respectively). Pairs 3A/6, 1/12B, 6/9, and 3A/12B are second degree–related, that is, they resemble uncles/cousins to sibling pair 1/3A. An infant buried in grave 4 has the same mtDNA haplotype (X2b4) as 9 and 12B, which supports a maternal relation, but there is no further genomic information to confirm this possibility. This family tree is based on the assumption that the individuals died over a relatively short time period. However, archeological evidence suggests that the gravesite may have been used for two generations (circa 580 to 630 CE) (8), which allows the possibility of multiple scenarios when constructing a family tree. Notably, individual 9, who is buried with Frankish grave goods (8), might have represented the highest-ranking individual and head of the household, given his close kinship to the four others and prominence in the burial rites.
The 7th century CE burial in Niederstotzingen represents the best-preserved example of an Alemannic Adelsgrablege. The observation that burial of the remains was close to a Roman crossroads, orientated in a considered way, and associated with rich grave goods points to a noble gravesite of an Alemannic familia with external cultural influences. The high percentage of males in the burial site suggests that this site was intended for a ranked warrior group, meaning that the individuals are not representative of the population existing in 7th century CE Alemannia. The kinship estimates show that kinship structure was organized around the familia, which is defined by close association of related and unrelated individuals united for a common purpose. The apparent kinship structure is consistent with the hypothesized Personenverbandstaaten, which was a system by which Merovingian nobles enforced rule in the Duchies of Alemannia, Thuringia, Burgundy, and elsewhere. Beyond the origin of the grave goods, we show isotopic and genetic evidence for contact with communities external to the region and evidence for shared ancestry between northern and southern Europeans. This finding invites debate on the Alemannic power system that may have been highly influenced by mobility and personal relations.
MATERIALS AND METHODS
Molecular analysis
Teeth were sampled from all 13 individuals previously excavated at Niederstotzingen (Fig. 1 and table S1) (6). aDNA was isolated from dentine with the protocol of Rohland et al. (31). Double-stranded libraries were prepared for sequencing (32, 33), and some libraries were treated with UDG (table S5) (34). To enrich the endogenous content of libraries, mtDNA (35) and autosomal DNA (36) (probe set of 1.24 million autosomal SNPs, hereafter referred to as 1240K) were captured using in-solution probes (17). Libraries were sequenced on Illumina HiSeq 2500, NextSeq, and Illumina HiSeq 4000 platforms using 75–base pair paired-end sequencing kits.
Data analysis
To process raw sequences, default parameters of EAGER (37) were applied. Sequences were mapped to human genome (build Hg19) (38, 39) and the mtDNA reference (rCRS) (40). The minimum mapping and base quality were both 30. MapDamage2.0 quantified deamination at 5′ and 3′ ends in non–UDG-treated libraries to show the presence of aDNA (41). To authenticate the retrieved data, mtDNA and X chromosome contamination estimates were made with Schmutzi (42) and ANGSD (43), respectively. To investigate the impact that modern contamination may have had on the results, non–UDG-treated data (shotgun and mtcapture) were filtered with PMDtools (threshold 3) and analyzed in parallel with non–PMD-filtered reads (22).
To retrieve mtDNA haplotypes, stringent filtering with log2fasta (42) was applied (quality > 20) to create mitogenome sequences. mtDNA haplotypes were assigned with haplofind (phylotree 17) (44, 45). The NRY haplotypes were identified relative to ISOGG database 11.349 (https://isogg.org/tree), and haplotyping was performed with ANGSD haploid caller (43) guided by yhaplo tool (46). Low-coverage (<2X) Y SNPs that were potentially caused by deamination were filtered. To estimate the sex of the individuals, the relative coverage of X and Y chromosomes was calculated from shotgun (23, 47) and 1240K capture (24) sequences.
To investigate genomic data, SAMtools mpileup (38) and PileupCaller (https://github.com/stschiff/sequenceTools/tree/master/src-pileupCaller) called pseudodiploid genotypes for the individuals, at loci that overlapped with the 1240K targeted SNPs, and merged them to a Human Origins Affymetrix (25) modern west Eurasian subset (n = 1063) (26). PCA smartpca (48, 49), admixture (50), and outgroup F3 statistics made estimates of ancestry from genome-wide data (25, 26). Admixture was run from 2 to 12 components (K), cross-validation (--cv) with five replications each, and random seeding (-s 1-1000). The selection of the admixture results to present was based on the lowest CV error, the replicate with the highest log likelihood, and careful interpretation of the results (51). Kinship estimates used the READ tool and the coefficient of relatedness, which were adapted for low-coverage genotypes (16, 27).
Supplementary Material
Acknowledgments
We would like to thank S. Schiffels, S. Clayton, A. Mittnik, T. Lamnidis, C.-C. Wang, C. Jeong, A. Herbig, and A. Immel and all of the colleagues at the Department of Archaeogenetics of the Max Planck Institute for Human History who provided support through advice and access to software and reference data. We are also grateful to our colleagues at the Institute for Mummy Studies in Bolzano for their assistance. Funding: We acknowledge the following funding sources: Provincia Autonoma di Bolzano, grant legge 14 agreement no. 1/40.3, 23.11.2012 (to F.M., N.O., V.C., and A.Z.), and the European Research Council starting grant APGREID (to J.K.). Author contributions: A.Z., J.W., F.M., J.K., and N.O. planned the research. J.K. and A.Z. provided reagents, sequencing, and access to computers. N.O., C.P., V.J.S., and F.M. extracted DNA from samples and prepared it for sequencing. N.O., C.P., V.C., T.D.P., and F.M. analyzed the data. All authors wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. Raw sequencing data are available at the European Nucleotide Archive with the accession number PRJEB26982.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/9/eaao1262/DC1
Supplementary Materials and methods
Fig. S1. Photographs of burial goods with most specific cultural identifying markers.
Fig. S2. Isotope 86Sr/87Sr and δ18O values for enamel from teeth.
Fig. S3. Overlaying shotgun and mtDNA deamination plots from mapDamage quantification against the reference genome.
Fig. S4. Genetic sex estimates from genome-wide capture.
Fig. S5. Admixture estimates for west Eurasians, Niederstotzingen, and selected ancient individuals.
Fig. S6. F3 outgroup statistics for Niederstotzingen 1 using Mbuti as an outgroup.
Fig. S7. F3 outgroup statistics for Niederstotzingen 3A using Mbuti as an outgroup.
Fig. S8. F3 outgroup statistics for Niederstotzingen 3B using Mbuti as an outgroup.
Fig. S9. F3 outgroup statistics for Niederstotzingen 3C using Mbuti as an outgroup.
Fig. S10. F3 outgroup statistics for Niederstotzingen 6 using Mbuti as an outgroup.
Fig. S11. F3 outgroup statistics for Niederstotzingen 9 using Mbuti as an outgroup.
Fig. S12. F3 outgroup statistics for Niederstotzingen 12B using Mbuti as an outgroup.
Fig. S13. F3 outgroup statistics for Niederstotzingen 12C using Mbuti as an outgroup.
Table S1. PCR-based haplotyping of DNA extracts from previous study.
Table S2. Archeological context and isotopes.
Table S3. Shotgun sequencing output and mapping data.
Table S4. mtDNA capture sequencing output data and mapping.
Table S5. 1240K sequencing output data and mapping.
Table S6. Schmutzi estimation of mtDNA contamination.
Table S7. ANGSD X chromosome contamination estimate on 1240K libraries.
Table S8. Haplogroups and private mutations for each mtDNA capture (phylotree 17).
Table S9. Complete list of NRY haplogroups with identifying ISOGG markers for 1240K capture sequences.
Table S10. Sex estimates from shotgun and genome-wide capture data.
Table S11. Sex estimates from shotgun data based on Rx values.
Table S12. Admixture CV error values for each component and individual.
Table S13. Shotgun reads with PMDtools threshold 3 filtered and skoglund sex estimate of filtered reads.
Table S14. Haplogroup calling of selected individuals before and after PMD (threshold 3) filtering.
Table S15. READ pairwise kinship-based estimate.
Table S16. Pairwise estimate of kinship and coefficient of relatedness.
REFERENCES AND NOTES
- 1.D. Geuenich, Geschichte der Alemannen (Kohlhammer, 1997), vol. 575. [Google Scholar]
- 2.P. J. Geary, The Myth of Nations: The Medieval Origins of Europe (Princeton Univ. Press, 2002). [Google Scholar]
- 3.Hummer H. J., The fluidity of barbarian identity: The ethnogenesis of Alemanni and Suebi, AD 200–500. Early Mediev. Eur. 7, 1–27 (1998). [Google Scholar]
- 4.H. Steuer, Archaeology and history: Proposals on the social structure of the Merovingian kingdom, in The Birth of Europe: Archaeology and Social Development in the First Millennium A.D., K. Randsborg, Ed. (L’Erma di Bretschneider, 1989), pp. 100–122. [Google Scholar]
- 5.Eckardt H., Müldner G., Lewis M., People on the move in Roman Britain. World Archaeol. 46, 534–550 (2014). [Google Scholar]
- 6.P. Paulsen, H.-J. Hundt, Alamannische Adelsgräber von Niederstotzingen (Kreis Heidenheim) (Müller & Gräff, 1967). [Google Scholar]
- 7.Die Alamannen, Archäologisches Landesmuseum Baden-Württemberg (Theiss, 1997). [Google Scholar]
- 8.Wahl J., Cipollini G., Coia V., Francken M., Harvati-Papatheodorou K., Kim M.-R., Maixner F., O’Sullivan N., Price T. D., Quast D., Speith N., Zink A., Neue Erkenntnisse zur frühmittelalterlichen Separatgrablege von Niederstotzingen, Kreis Heidenheim. Fundber. Bad. Württ. 34, 341–390 (2014). [Google Scholar]
- 9.I. Stork, Friedhof und Dorf - der exemplarische Fall Lauchheim, in Die Alamannen auf der Ostalb. Frühe Siedler im Raum zwischen Lauchheim und Niederstotzingen, Andreas Gut, Ed. (Regierungspräsidium Stuttgart Landesamt für Denkmalpflege, 2010), vol. 26, pp. 92–105. [Google Scholar]
- 10.Hofreiter M., Paijmans J. L. A., Goodchild H., Speller C. F., Barlow A., Fortes G. G., Thomas J. A., Ludwig A., Collins M. J., The future of ancient DNA: Technical advances and conceptual shifts. Bioessays 37, 284–293 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Martiniano R., Caffell A., Holst M., Hunter-Mann K., Montgomery J., Müldner G., McLaughlin R. L., Teasdale M. D., van Rheenen W., Veldink J. H., van den Berg L. H., Hardiman O., Carroll M., Roskams S., Oxley J., Morgan C., Thomas M. G., Barnes I., McDonnell C., Collins M. J., Bradley D. G., Genomic signals of migration and continuity in Britain before the Anglo-Saxons. Nat. Commun. 7, 10326 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schiffels S., Haak W., Paajanen P., Llamas B., Popescu E., Loe L., Clarke R., Lyons A., Mortimer R., Sayer D., Tyler-Smith C., Cooper A., Durbin R., Iron Age and Anglo-Saxon genomes from East England reveal British migration history. Nat. Commun. 7, 10408 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Günther T., Jakobsson M., Genes mirror migrations and cultures in prehistoric Europe—A population genomic perspective. Curr. Opin. Genet. Dev. 41, 115–123 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Busby G. B. J., Hellenthal G., Montinaro F., Tofanelli S., Bulayeva K., Rudan I., Zemunik T., Hayward C., Toncheva D., Karachanak-Yankova S., Nesheva D., Anagnostou P., Cali F., Brisighelli F., Romano V., Lefranc G., Buresi C., Ben Chibani J., Haj-Khelil A., Denden S., Ploski R., Krajewski P., Hervig T., Moen T., Herrera R. J., Wilson J. F., Myers S., Capelli C., The role of recent admixture in forming the contemporary West Eurasian genomic landscape. Curr. Biol. 25, 2518–2526 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korneliussen T. S., Moltke I., NgsRelate: A software tool for estimating pairwise relatedness from next-generation sequencing data. Bioinformatics 31, 4009–4011 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kennett D. J., Plog S., George R. J., Culleton B. J., Watson A. S., Skoglund P., Rohland N., Mallick S., Stewardson K., Kistler L., LeBlanc S. A., Whiteley P. M., Reich D., Perry G. H., Archaeogenomic evidence reveals prehistoric matrilineal dynasty. Nat. Commun. 8, 14115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathieson I., Lazaridis I., Rohland N., Mallick S., Patterson N., Roodenberg S. A., Harney E., Stewardson K., Fernandes D., Novak M., Sirak K., Gamba C., Jones E. R., Llamas B., Dryomov S., Pickrell J., Arsuaga J. L., de Castro J. M. B., Carbonell E., Gerritsen F., Khokhlov A., Kuznetsov P., Lozano M., Meller H., Mochalov O., Moiseyev V., Guerra M. A. R., Roodenberg J., Vergès J. M., Krause J., Cooper A., Alt K. W., Brown D., Anthony D., Lalueza-Fox C., Haak W., Pinhasi R., Reich D., Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528, 499–503 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones E. R., Zarina G., Moiseyev V., Lightfoot E., Nigst P. R., Manica A., Pinhasi R., Bradley D. G., The Neolithic transition in the Baltic was not driven by admixture with early European farmers. Curr. Biol. 27, 576–582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thorvaldsdóttir H., Robinson J. T., Mesirov J. P., Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myres N. M., Rootsi S., Lin A. A., Järve M., King R. J., Kutuev I., Cabrera V. M., Khusnutdinova E. K., Pshenichnov A., Yunusbayev B., Balanovsky O., Balanovska E., Rudan P., Baldovic M., Herrera R. J., Chiaroni J., Di Cristofaro J., Villems R., Kivisild T., Underhill P. A., A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe. Eur. J. Hum. Genet. 19, 95–101 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rootsi S., Myres N. M., Lin A. A., Järve M., King R. J., Kutuev I., Cabrera V. M., Khusnutdinova E. K., Varendi K., Sahakyan H., Behar D. M., Khusainova R., Balanovsky O., Balanovska E., Rudan P., Yepiskoposyan L., Bahmanimehr A., Farjadian S., Kushniarevich A., Herrera R. J., Grugni V., Battaglia V., Nici C., Crobu F., Karachanak S., Hooshiar Kashani B., Houshmand M., Sanati M. H., Toncheva D., Lisa A., Semino O., Chiaroni J., Di Cristofaro J., Villems R., Kivisild T., Underhill P. A., Distinguishing the co-ancestries of haplogroup G Y-chromosomes in the populations of Europe and the Caucasus. Eur. J. Hum. Genet. 20, 1275–1282 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skoglund P., Northoff B. H., Shunkov M. V., Derevianko A. P., Pääbo S., Krause J., Jakobsson M., Separating endogenous ancient DNA from modern day contamination in a Siberian Neandertal. Proc. Natl. Acad. Sci. U.S.A. 111, 2229–2234 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mittnik A., Wang C.-C., Svoboda J., Krause J., A molecular approach to the sexing of the triple burial at the upper paleolithic site of Dolní Věstonice. PLOS ONE 11, e0163019 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu Q., Posth C., Hajdinjak M., Petr M., Mallick S., Fernandes D., Furtwängler A., Haak W., Meyer M., Mittnik A., Nickel B., Peltzer A., Rohland N., Slon V., Talamo S., Lazaridis I., Lipson M., Mathieson I., Schiffels S., Skoglund P., Derevianko A. P., Drozdov N., Slavinsky V., Tsybankov A., Cremonesi R. G., Mallegni F., Gély B., Vacca E., Morales M. R. G., Straus L. G., Neugebauer-Maresch C., Teschler-Nicola M., Constantin S., Moldovan O. T., Benazzi S., Peresani M., Coppola D., Lari M., Ricci S., Ronchitelli A., Valentin F., Thevenet C., Wehrberger K., Grigorescu D., Rougier H., Crevecoeur I., Flas D., Semal P., Mannino M. A., Cupillard C., Bocherens H., Conard N. J., Harvati K., Moiseyev V., Drucker D. G., Svoboda J., Richards M. P., Caramelli D., Pinhasi R., Kelso J., Patterson N., Krause J., Pääbo S., Reich D., The genetic history of Ice Age Europe. Nature 534, 200–205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patterson N., Moorjani P., Luo Y., Mallick S., Rohland N., Zhan Y., Genschoreck T., Webster T., Reich D., Ancient admixture in human history. Genetics 192, 1065–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lazaridis I., Nadel D., Rollefson G., Merrett D. C., Rohland N., Mallick S., Fernandes D., Novak M., Gamarra B., Sirak K., Connell S., Stewardson K., Harney E., Fu Q., Gonzalez-Fortes G., Jones E. R., Roodenberg S. A., Lengyel G., Bocquentin F., Gasparian B., Monge J. M., Gregg M., Eshed V., Mizrahi A.-S., Meiklejohn C., Gerritsen F., Bejenaru L., Blüher M., Campbell A., Cavalleri G., Comas D., Froguel P., Gilbert E., Kerr S. M., Kovacs P., Krause J., McGettigan D., Merrigan M., Merriwether D. A., O’Reilly S., Richards M. B., Semino O., Shamoon-Pour M., Stefanescu G., Stumvoll M., Tönjes A., Torroni A., Wilson J. F., Yengo L., Hovhannisyan N. A., Patterson N., Pinhasi R., Reich D., Genomic insights into the origin of farming in the ancient Near East. Nature 536, 419–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuhn J. M. M., Jakobsson M., Günther T., Estimating genetic kin relationships in prehistoric populations. PLOS ONE 13, e0195491 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H., Glusman G., Huff C., Caballero J., Roach J. C., Accurate and robust prediction of genetic relationship from whole-genome sequences. PLOS ONE 9, e85437 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.T. Schneider, Die Frauenkrieger von Niederstotzingen, in Amazonen: Geheimnisvolle Kriegerinnen (Edition Minerva GmbH, 2010), pp. 178–181. [Google Scholar]
- 30.Alt K. W., Knipper C., Peters D., Müller W., Maurer A.-F., Kollig I., Nicklisch N., Müller C., Karimnia S., Brandt G., Roth C., Rosner M., Mende B., Schöne B. R., Vida T., von Freeden U., Lombards on the move—An integrative study of the migration period cemetery at Szólád, Hungary. PLOS ONE 9, e110793 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rohland N., Siedel H., Hofreiter M., A rapid column-based ancient DNA extraction method for increased sample throughput. Mol. Ecol. Resour. 10, 677–683 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Meyer M., Kircher M., Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010, pdb.prot5448 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Kircher M., Sawyer S., Meyer M., Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 40, e3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rohland N., Harney E., Mallick S., Nordenfelt S., Reich D., Partial uracil–DNA–glycosylase treatment for screening of ancient DNA. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20130624 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carpenter M. L., Buenrostro J. D., Valdiosera C., Schroeder H., Allentoft M. E., Sikora M., Rasmussen M., Gravel S., Guillén S., Nekhrizov G., Leshtakov K., Dimitrova D., Theodossiev N., Pettener D., Luiselli D., Sandoval K., Moreno-Estrada A., Li Y., Wang J., Gilbert M. T. P., Willerslev E., Greenleaf W. J., Bustamante C. D., Pulling out the 1%: Whole-genome capture for the targeted enrichment of ancient DNA sequencing libraries. Am. J. Hum. Genet. 93, 852–864 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu Q., Meyer M., Gao X., Stenzel U., Burbano H. A., Kelso J., Pääbo S., DNA analysis of an early modern human from Tianyuan Cave, China. Proc. Natl. Acad. Sci. U.S.A. 110, 2223–2227 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peltzer A., Jäger G., Herbig A., Seitz A., Kniep C., Krause J., Nieselt K., EAGER: Efficient ancient genome reconstruction. Genome Biol. 17, 60 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H., Durbin R., Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenbloom K. R., Armstrong J., Barber G. P., Casper J., Clawson H., Diekhans M., Dreszer T. R., Fujita P. A., Guruvadoo L., Haeussler M., Harte R. A., Heitner S., Hickey G., Hinrichs A. S., Hubley R., Karolchik D., Learned K., Lee B. T., Li C. H., Miga K. H., Nguyen N., Paten B., Raney B. J., Smit A. F. A., Speir M. L., Zweig A. S., Haussler D., Kuhn R. M., Kent W. J., The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 43, D670–D681 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andrews R. M., Kubacka I., Chinnery P. F., Lightowlers R. N., Turnbull D. M., Howell N., Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 23, 147 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Jónsson H., Ginolhac A., Schubert M., Johnson P. L. F., Orlando L., mapDamage2.0: Fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29, 1682–1684 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Renaud G., Slon V., Duggan A. T., Kelso J., Schmutzi: Estimation of contamination and endogenous mitochondrial consensus calling for ancient DNA. Genome Biol. 16, 224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korneliussen T. S., Albrechtsen A., Nielsen R., ANGSD: Analysis of next generation sequencing data. BMC Bioinformatics 15, 356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vianello D., Sevini F., Castellani G., Lomartire L., Capri M., Franceschi C., HAPLOFIND: A new method for high-throughput mtDNA haplogroup assignment. Hum. Mutat. 34, 1189–1194 (2013). [DOI] [PubMed] [Google Scholar]
- 45.van Oven M., PhyloTree Build 17: Growing the human mitochondrial DNA tree. Forensic Sci. Int. Genet. Suppl. Ser. 5, e392–e394 (2015). [Google Scholar]
- 46.Poznik G. D., Identifying Y-chromosome haplogroups in arbitrarily large samples of sequenced or genotyped men. bioRxiv 2016, 088716 (2016). [Google Scholar]
- 47.Skoglund P., Storå J., Götherström A., Jakobsson M., Accurate sex identification of ancient human remains using DNA shotgun sequencing. J. Archaeol. Sci. 40, 4477–4482 (2013). [Google Scholar]
- 48.Patterson N., Price A. L., Reich D., Population structure and eigenanalysis. PLOS Genet. 2, e190 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price A. L., Patterson N. J., Plenge R. M., Weinblatt M. E., Shadick N. A., Reich D., Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Alexander D. H., Novembre J., Lange K., Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Falush D., van Dorp L., Lawson D., A tutorial on how (not) to over-interpret STRUCTURE/ADMIXTURE bar plots. bioRxiv, 066431 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maricic T., Whitten M., Pääbo S., Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLOS ONE 5, e14004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schubert M., Ginolhac A., Lindgreen S., Thompson J. F., Al-Rasheid K. A. S., Willerslev E., Krogh A., Orlando L., Improving ancient DNA read mapping against modern reference genomes. BMC Genomics 13, 178 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin; R. 1000 Genome Project Data Processing Subgroup , The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poznik G. D., Identifying Y-chromosome haplogroups in arbitrarily large samples of sequenced or genotyped men. bioRxiv, 088716 (2016). [Google Scholar]
- 56.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A. R., Bender D., Maller J., Sklar P., de Bakker P. I. W., Daly M. J., Sham P. C., PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.R. C. Team, R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2014).
- 58.Francis R. M., pophelper: An R package and web app to analyse and visualize population structure. Mol. Ecol. Resour. 17, 27–32 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/9/eaao1262/DC1
Supplementary Materials and methods
Fig. S1. Photographs of burial goods with most specific cultural identifying markers.
Fig. S2. Isotope 86Sr/87Sr and δ18O values for enamel from teeth.
Fig. S3. Overlaying shotgun and mtDNA deamination plots from mapDamage quantification against the reference genome.
Fig. S4. Genetic sex estimates from genome-wide capture.
Fig. S5. Admixture estimates for west Eurasians, Niederstotzingen, and selected ancient individuals.
Fig. S6. F3 outgroup statistics for Niederstotzingen 1 using Mbuti as an outgroup.
Fig. S7. F3 outgroup statistics for Niederstotzingen 3A using Mbuti as an outgroup.
Fig. S8. F3 outgroup statistics for Niederstotzingen 3B using Mbuti as an outgroup.
Fig. S9. F3 outgroup statistics for Niederstotzingen 3C using Mbuti as an outgroup.
Fig. S10. F3 outgroup statistics for Niederstotzingen 6 using Mbuti as an outgroup.
Fig. S11. F3 outgroup statistics for Niederstotzingen 9 using Mbuti as an outgroup.
Fig. S12. F3 outgroup statistics for Niederstotzingen 12B using Mbuti as an outgroup.
Fig. S13. F3 outgroup statistics for Niederstotzingen 12C using Mbuti as an outgroup.
Table S1. PCR-based haplotyping of DNA extracts from previous study.
Table S2. Archeological context and isotopes.
Table S3. Shotgun sequencing output and mapping data.
Table S4. mtDNA capture sequencing output data and mapping.
Table S5. 1240K sequencing output data and mapping.
Table S6. Schmutzi estimation of mtDNA contamination.
Table S7. ANGSD X chromosome contamination estimate on 1240K libraries.
Table S8. Haplogroups and private mutations for each mtDNA capture (phylotree 17).
Table S9. Complete list of NRY haplogroups with identifying ISOGG markers for 1240K capture sequences.
Table S10. Sex estimates from shotgun and genome-wide capture data.
Table S11. Sex estimates from shotgun data based on Rx values.
Table S12. Admixture CV error values for each component and individual.
Table S13. Shotgun reads with PMDtools threshold 3 filtered and skoglund sex estimate of filtered reads.
Table S14. Haplogroup calling of selected individuals before and after PMD (threshold 3) filtering.
Table S15. READ pairwise kinship-based estimate.
Table S16. Pairwise estimate of kinship and coefficient of relatedness.


