Abstract
Background: Urticaria is a common allergic disease that affects all age groups. Allergic disorders are diagnosed at allergy testing centers using skin tests. Though skin tests are the gold standard tests for allergy diagnosis, specialists are required to interpret the observations and test results. Hence, a computer-assisted medical decision-making (CMD) system can be used as an aid for decision support, by junior clinicians, in order to diagnose the presence of urticaria. Methods: The data from intradermal skin test results of 778 patients, who exhibited allergic symptoms, are considered for this study. Based on food habits and the history of a patient, 40 relevant allergens are tested. Allergen extracts are used for skin test. Ten independent runs of 10-fold cross-validation are used to train the system. The performance of the CMD system is evaluated using a set of test samples. The test samples were also presented to the junior clinicians at the allergy testing center to diagnose the presence or absence of urticaria. Results: From a set of 91 features, a subset of 41 relevant features is chosen based on the relevance score of the feature selection algorithm. The Bayes classification approach achieves a classification accuracy of 96.92% over the test samples. The junior clinicians were able to classify the test samples with an average accuracy of 75.68%. Conclusion: A probabilistic classification approach is used for identifying the presence or absence of urticaria based on intradermal skin test results. In the absence of an allergy specialist, the CDM system assists junior clinicians in clinical decision making.
Keywords: allergy and immunology, decision aids, computer-assisted diagnosis, Bayesian statistical methods
Introduction
Allergy is considered to be an abnormal reaction of the body to a previously encountered allergen or trigger introduced by inhalation, ingestion, injection, or skin contact among atopic people. The symptoms of an allergic disorder are often manifested by itchy eyes, running nose, nasal discharge, coughing, shortness of breath, wheezing, itching, and rashes.1 Around 300 million people worldwide have allergic disorders, and approximately 50% of them live in developing countries.2 According to the World Allergy Organization, in India, more than 30% of the population is known to suffer from an allergic ailment. In a study by Kumar and others, out of 1860 patients screened, 1097 (58.9%) gave history of food allergy.3 Allergy symptoms and their manifestations have a profound impact on the quality of life. Allergic diseases with explicit symptoms often hold back the daily activities of people and affect their personal and professional tasks.4 Allergic diseases are rising all over the world, and some commonly known allergies include asthma, rhinitis, anaphylaxis, nasobronchial allergy, eczema, urticaria, and angioedema.
Urticaria is a heterogeneous group of diseases. It is characterized by the appearance of a wheal, which may consist of the following three features: a central swelling that varies in size; an associated itching or burning sensation; and discomforts over a fleeting duration of usually 1 to 24 hours.5 Urticaria is the fourth most prevalent allergic disease after rhinitis, asthma, and drug allergy.6 Urticaria has a strong impact on school performance and is also the cause of the highest number of absence from work. Though urticaria seems to be an allergic reaction, the disease is autoimmune and idiopathic. Around 15% to 20% of people have urticaria at least once in their lifetime.4 Due to the heterogeneity of the disease, the etiology remains unexplained. Hence, the diagnosis and treatment of urticaria is still a challenge to physicians and allergists.
Urticaria is classified based on the duration of its physical manifestations. Broadly, spontaneous urticaria and physical urticaria are the two well-defined classes of urticaria. A short description about classification of urticaria is presented in Table 1.7
Table 1.
Urticaria Classifications by Group and Subgroup
| Urticaria Group and Subgroups | Characteristics and Eliciting Factors |
|---|---|
| Spontaneous | |
| Acute | Spontaneous wheals, <6 weeks |
| Chronic | Spontaneous wheals, >6 weeks |
| Physical | |
| Acquired cold | Cold air, wind, food, objects |
| Delayed pressure | Vertical pressure |
| Heat | Localized heat |
| Solar | Ultraviolet and/or visible light |
| Dermographic | Mechanical shearing force |
| Vibratory | Vibratory forces (e.g., pneumatic hammer) |
| Other disorders | |
| Aquagenic | Water |
| Cholinergic | Increase of body temperature |
| Contact | Contact with urticariogenic substance |
| Exercise induced | Physical exercise |
Acute urticaria resolves within 6 weeks, whereas chronic urticaria lasts longer. Acute urticaria is more common in young adults and children. Acute allergic symptoms may be due to release of mediators from mast cells, whereas chronic symptoms may be due to eosinophil-mediated tissue damage.1,8 Generally, in patients suffering from urticaria, a trigger causes the skin cells to release chemicals such as histamine. These chemicals cause fluids to leak from tiny blood vessels under the skin surface. The fluid accumulates and manifests in the form of wheals. Chemicals also cause the blood vessels to dilate, which causes the flare around the wheals. When the trigger induces allergic symptoms, an allergy evaluation may be sought to identify the potential trigger for the allergic symptoms. Other causes for acute urticaria include sea food, allergy to insects, environment, and transfusion reaction. If the symptoms prolong for more than 6 weeks, the condition is classified as chronic urticaria; 20% to 30% of acute urticaria turns out to be chronic urticaria.9
Intradermal skin tests are used to find the allergens that trigger allergic symptoms. Skin test helps diagnose immunoglobulin E–mediated hypersensitivity specifically. In patients with symptoms of urticaria and who are doubtful of being allergic to particular food items or aero allergens, skin prick tests can be used to identify the potential allergens.
There are several factors and issues that have to be considered while conducting a skin test. While measuring and recording the response of a skin test, the following issues need to be considered: time to measure response, making a permanent record of the response, and measurement and grading of the response. For interpretation of the response, the following issues need to be considered: proficiency of the test, analytical performance, reactivity versus sensitivity, and criteria for a positive response. Furthermore, there are internal and external variables that influence the skin test results: site of injection, distance between injection sites, time (season) of testing, age, race, gender, socioeconomic status, tobacco smoke exposure, and medication.8
Hence, skin testing of a specific immunoglobulin E is not easily accessible and cannot be interpreted precisely by junior physicians and immunologists. But quality control measures and proper performance of skin testing are very important to produce correct results. Timely identification of allergens is important as it may reduce the impact and manifestation of symptoms.
Though intradermal skin test is an effective and efficient way to identify allergic triggers, allergy specialists or experienced immunologists are required to suggest remedial measures based on the observations of the test. Clinicians and immunologists at allergy clinics and specialty centers have to make decisions on whether a diagnosed disease is due to allergic triggers or other factors. Furthermore, they have to provide recommendations on what kind of ingestants, inhalants, and contactants to avoid and other treatment options if necessary. These clinical decisions depend on the patient’s history, food habits, environment, and the results of the skin tests. The general patterns and knowledge models devised by clinical experts should be made available to medical trainees, immunologists, and junior clinicians through computer-aided clinical decision support systems. This raises the performance and confidence of physicians in dealing with more difficult and ambiguous cases.
Over the past decade, due to the availability of vast medical data, computer-assisted medical decision-making (CMD) systems are widely used at clinics and health centers to provide decisions and solutions. In most situations, a CMD system cannot be considered to be a gold standard, but it can be used by junior clinicians in the absence of experts to verify and assert their decisions. Computer-assisted systems are used for diagnosis, decision making, and decision support in various medical applications such as cancer care,10,11 heart disease diagnosis,12 thrombosis diagnosis,13 diagnosis and treatment of lung disorders,14,15 drug reaction analysis,16 and allergy diagnosis.17
A CMD system gets medical data (e.g., patient description) as input, processes the data, extracts useful knowledge from the data, and finally makes decisions or predictions.18 The core tasks of CMD systems are often based on typical data mining tasks such as data cleaning, normalization, data reduction, association analysis, classification, and clustering.
The CMD system assists junior clinicians at allergy centers to diagnose patients with urticaria. The diagnostic result obtained from the system indicates weather a patient shows positive or negative symptoms of acute urticaria. The system supports the clinician to decide whether the reported disease is acute (triggered by allergens) or not based on the results of the intradermal skin tests, in the absence of an expert immunologist.
Methods
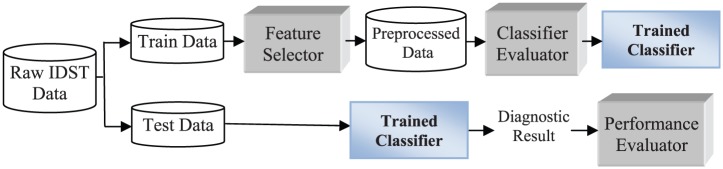
The proposed CMD system framework consists of a feature selector, a classifier evaluator, a Bayes classifier, and a performance evaluator (see Figure 1).
Figure 1.
CMD system framework (IDST = intradermal skin test).
Feature Selector
Feature selection (attribute reduction) is a data preprocessing technique whereby the dimensionality of the data is reduced. Removal of irrelevant and redundant features enhances the efficiency of classifier. In this work, the feature selector uses an instance-based learning approach19 for selecting relevant features. For a given dataset S, let Strain be the training set and Stest the testing set. Let |Strain| denote the sample size of Strain and τ be the relevance threshold ranging from 0 to 1. Consider X and Y to be two instances (samples), whose corresponding nominal values for the kth attribute are xk and yk. Then, the difference between the nominal values of xk and yk is given by
The CMD system uses the RELIEF algorithm,20 presented in Figure 2, to select a set of relevant features for training the classifier.
Figure 2.
RELIEF algorithm.
The algorithm chooses an instance X, a near-hit instance of X and a near-miss instance of X. A near-hit instance is an instance that is in the neighborhood of X and belongs to the same class of X. A near-miss instance also belongs to the same neighborhood but belongs to a different class. The feature weight vector W is updated for each feature. The algorithm chooses the features whose weight (relevance) satisfies the relevance threshold (τ).
Classifier Evaluator
Classification is a typical data mining task and also the core of a decision-making or decision-support system. The inducer (learning algorithm) constructs a classifier model (knowledge model) from a set of class-labelled training samples. The classifier assigns a class label to an unknown instance (test sample) based on the classifier model. Classification approaches differ by the algorithm used for induction and also the knowledge representation model. For example, an associative classifier uses the a priori approach for rule induction and an IF-THEN rule format for representing the classifier model. The multi-layer perceptron classifier uses the gradient descent–based backpropagation algorithm for induction (training), and the trained network constitutes the knowledge model. Each classifier has its own pros and cons; hence, no classifier can be considered as the “universal best” for all applications and domains.
The classifier evaluator is used to choose a suitable classifier for this CDM system for the diagnosis of allergic disorders. The evaluator uses k-fold cross-validation, which is an appropriate method to be used for an unbiased evaluation of classifiers.21 Cross-validation with k folds is a technique whereby the preprocessed Strain data are randomly split into k folds of approximately equal size. The classifier (model) is trained and tested k times. Each time (k− 1) folds are used for training and the remaining one fold is used for testing.
Naïve Bayes Classifier
The CMD system developed in this work uses a probabilistic approach for classification.22 Consider an instance X = (x1, x2, x3, . . . , xp, xc), where x1, x2, x3, . . . , xp are the values for features f1, f2, f3, . . . , fp, respectively, and xc is the class label that can either be positive or negative. The probability of an instance X being in class c is
X is classified as positive class if
The features (allergens) are independent of each other for a given class. Hence,
where is called the naïve Bayesian classifier.21
Performance Evaluator
The performance of the CMD system primarily depends on the classification efficiency of the classifier. The performance evaluator assesses the classification efficiency using four evaluation measures presented in Equations (2) to (5). The four measures, namely, Precision, Sensitivity, Specificity, and Accuracy, differ in their criterion of evaluation. Precision evaluates the agreement of the class label with the positive labels predicted by the classifier. Sensitivity is used to evaluate the effectiveness of a classifier to identify positive labels, whereas Specificity evaluates how effectively a classifier identifies negative labels. Accuracy evaluates the overall classification efficiency of the classifier. Table 2 presents the confusion matrix. True positives (tp) refer to those samples that are positive and correctly diagnosed as positive (patient has urticaria and is allergic). Likewise, true negatives (tn) refer to those samples that are negative and correctly diagnosed as negative (patient does not have urticaria). False positives (fp) are those samples that are diagnosed as positive by the system/clinician but are actually negative as per the expert’s diagnosis (gold standard). False negatives (fn) are those samples that are affected with urticaria but diagnosed as negative by the system/clinician.
Table 2.
Confusion Matrix
| Condition |
|||
|---|---|---|---|
| Positive | Negative | ||
| Test | Positive | tp | fp |
| Negative | fn | tn | |
Data Set Description
Intradermal skin test data were collected from 778 patients who visited the Good Samaritan Lab and Allergy Centre, Chennai, Tamilnadu, India, between 1 March and 20 June 2013. The patients were referred by ENT surgeons and general physicians because of skin diseases, itching, or other plausible allergic symptoms. A total of 365 males and 413 females, of all age groups, were included in the study.
Intradermal Test Method
After analyzing the medical history of a patient, the allergist determines whether skin testing is appropriate for the patient. The allergist also determines the list of selected allergens to be tested. Allergen extracts, negative controls (saline), and positive controls (histamine) were used for performing the skin tests. The upper half of the volar surface of the forearm was selected for the test. It was cleansed with alcohol and a pen is used to label the area in a grid-like pattern to depict where the extract (allergen) is to be applied. About 0.01 mL of the allergen is injected into the epidermis using a sterile, disposable, plastic 1-mL tuberculin syringe. Patients were asked to stop taking antihistamines and anti-allergic drugs and medications. Table 3 lists the medication to be avoided before allergy testing.
Table 3.
Medication to Be Avoided Before Allergy Testing
| Medication | Duration (Days) |
|---|---|
| First-generation antihistamines | 2–3 |
| Nonsedating antihistamines | 7 |
| Tricyclic antidepressants | 7–14 |
| Benzodiazepines | 7–14 |
| Topical corticosteroids | 14–21 |
Consecutive observations, on an hourly basis, were taken. A positive reaction is depicted by a wheal and a flare reaction.8,23 A negative response to a skin test usually indicates that the patient is not sensitive to that allergen. For patients who reported a delayed response to the test, the reactions were incorporated in the results.
Results
The raw data obtained from the intradermal skin test results were split into training data (Strain) and testing data (Stest) using a holdout approach.24 Out of 778 samples, 518 samples were used for training (|Strain| = 518) and the rest were used for testing. The Strain has 92 attributes (features), which includes the class attribute. The next section presents a worked-out example, and then experimental results are presented.
Worked-Out Example
Set of Sample Instances
| Cotton Dust | Wheat | Chicken | Prawn | Brinjal | Carrot | Dhal | Sneezing | Itching | Swelling | Class | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | R | NR | NR | NR | R | NR | NR | No | Yes | Yes | Positive |
| 2 | R | 0 | 0 | 0 | NR | NR | NR | No | Yes | Yes | Negative |
| 3 | NR | R | R | R | NR | NR | NR | Yes | No | No | Negative |
| 4 | R | R | R | NR | NR | NR | R | No | Yes | Yes | Positive |
| 5 | R | 0 | NR | R | R | NR | NR | No | Yes | Yes | Positive |
| 6 | R | NR | R | R | R | NR | 0 | Yes | Yes | Yes | Positive |
| 7 | R | NR | NR | 0 | NR | R | 0 | No | No | No | Negative |
| 8 | R | NR | 0 | 0 | R | NR | NR | Yes | No | No | Negative |
| 9 | R | R | 0 | 0 | NR | NR | R | Yes | No | No | Negative |
| 10 | NR | NR | NR | NR | NR | 0 | R | No | No | No | Negative |
| 11 | R | NR | R | R | R | NR | NR | No | Yes | Yes | Positive |
| 12 | R | N | NR | 0 | NR | NR | R | Yes | No | No | Negative |
Note: R = reactive; NR = not reactive; 0 = not tested/not associated.
Let us consider the last two instances as test samples and the rest as training samples.
The number of samples in each class, corresponding to each attribute-value is presented below.
| Attribute | CLASS | |
|---|---|---|
| POSITIVE | NEGATIVE | |
| (4) | (6) | |
| Cotton Dust | ||
| R | 4.0 | 4.0 |
| NR | 0.0 | 2.0 |
| [total] 4.0 | 6.0 | |
| Wheat | ||
| NR | 2.0 | 3.0 |
| 0 | 1.0 | 1.0 |
| R | 1.0 | 2.0 |
| [total] 4.0 | 6.0 | |
| Chicken | ||
| NR | 2.0 | 2.0 |
| 0 | 0.0 | 3.0 |
| R | 2.0 | 1.0 |
| [total] 4.0 | 6.0 | |
| Prawn | ||
| NR | 2.0 | 1.0 |
| 0 | 0.0 | 4.0 |
| R | 2.0 | 1.0 |
| [total] | 4.0 | 6.0 |
| Attribute | CLASS | |
|---|---|---|
| POSITIVE | NEGATIVE | |
| (4) | (6) | |
| Brinjal | ||
| R | 3.0 | 1.0 |
| NR | 0.0 | 2.0 |
| [total] | 5.0 | 6.0 |
| Dhal | ||
| NR | 2.0 | 3.0 |
| 0 | 1.0 | 2.0 |
| R | 1.0 | 1.0 |
| [total] | 4.0 | 6.0 |
| Swelling | ||
| YES | 4.0 | 1.0 |
| NO | 0.0 | 5.0 |
| [total] | 4.0 | 6.0 |
| Sneezing | ||
| NO | 3.0 | 3.0 |
| YES | 1.0 | 3.0 |
| [total] | 4.0 | 6.0 |
| Itching | ||
| YES | 4.0 | 1.0 |
| 0 | 0.0 | 5.0 |
| [total] | 4.0 | 6.0 |
The Prior probabilities for each class from the training samples are computed as follows:
P(CLASS=POSITIVE) = 4/10 = 0.4
P(CLASS=NEGATIVE) = 6/10 = 0.6
Consider the test instance (X1):
| Cotton Dust | Wheat | Chicken | Prawn | Brinjal | Carrot | Dhal | Sneezing | Itching | Swelling | |
|---|---|---|---|---|---|---|---|---|---|---|
| 11 | R | NR | R | R | R | NR | NR | NO | YES | YES |
The Conditional Probabilities are computed as follows:
P(Cotton dust = R|CLASS = POSITIVE) = 4/4 = 1.00
P(Cotton dust = R|CLASS = NEGATIVE) = 4/6 = 0.66
P(Wheat = NR|CLASS = POSITIVE) = 2/4 = 0.50
P(Wheat = NR|CLASS = NEGATIVE) = 3/6 = 0.50
P(Chicken = R|CLASS = POSITIVE) = 2/4 = 0.50
P(Chicken = R|CLASS = NEGATIVE) = 1/6 = 0.16
P(Prawn = R|CLASS = POSITIVE) = 2/4 = 0.50
P(Prawn = R|CLASS = NEGATIVE) = 1/6 = 0.16
P(Brinjal = R|CLASS = POSITIVE) = 3/4 = 0.75
P(Brinjal = R|CLASS = NEGATIVE) = 1/6 = 0.16
P(Carrot = NR|CLASS = POSITIVE) = 4/4 = 1.00
P(Carrot = NR|CLASS = NEGATIVE) = 4/6 = 0.66
P(Dhal = NR|CLASS = POSITIVE) = 2/4 = 0.50
P(Dhal = NR|CLASS = NEGATIVE) = 3/6 = 0.50
P(Sneezing = NO|CLASS = POSITIVE) = 3/4 = 0.75
P(Sneezing = NO|CLASS = NEGATIVE) = 3/6 = 0.50
P(Itching = YES|CLASS = POSITIVE) = 4/4 = 1.00
P(Itching = YES|CLASS = NEGATIVE) = 1/6 = 0.16
P(Swelling = YES|CLASS = POSITIVE) = 4/4 = 1.00
P(Swelling = YES|CLASS = NEGATIVE) = 1/6 = 0.16
P(X1|CLASS=POSITIVE) = P(Cotton dust = R |CLASS = POSITIVE) ×
P(Wheat = NR |CLASS = POSITIVE) ×
P(Chicken = R |CLASS = POSITIVE) ×
P(Prawn = R |CLASS = POSITIVE) ×
P(Brinjal = R |CLASS = POSITIVE) ×
P(Carrot = NR |CLASS = POSITIVE) ×
P(Dhal = NR |CLASS = POSITIVE) ×
P(Sneezing = NO |CLASS = POSITIVE) ×
P(Itching = YES |CLASS = POSITIVE) ×
P(Swelling = YES |CLASS = POSITIVE)
= 1.0 × 0.5 × 0.5 × 0.5 × 0.75 × 0.1 × 0.5 × .75 × 1.0 × 1.0
= 0.0351
P(X1|CLASS=NEGATIVE) = P(Cotton dust = R |CLASS = NEGATIVE) ×
P(Wheat = NR |CLASS = NEGATIVE) ×
P(Chicken = R |CLASS = NEGATIVE) ×
P(Prawn = R |CLASS = NEGATIVE) ×
P(Brinjal = R |CLASS = NEGATIVE) ×
P(Carrot = NR |CLASS = NEGATIVE) ×
P(Dhal = NR |CLASS = NEGATIVE) ×
P(Sneezing = NO |CLASS = NEGATIVE) ×
P(Itching = YES |CLASS = NEGATIVE) ×
P(Swelling = YES |CLASS = NEGATIVE)
= 0.66 × 0.5 × 0.16 × 0.16 × 0.16 × 0.66 × 0.50 × .50 × 0.16 × 0.16
= 0.00000057
To find the class that maximizes P(X1|CLASS)×P(CLASS), the following is computed:
P(X1|CLASS=POSITIVE) × P(CLASS=POSITIVE) = 0.0351 × 0.4 = 0.0104
P(X1|CLASS=NEGATIVE) × P(CLASS=NEGATIVE) = 0.00000057 × 0.6 = 0.00000034
Therefore, the naïve Bayesian classifier classifies instance X1 as a CLASS = POSITIVE. Hence, the test instance X1 is diagnosed as positive to Acute/Allergic Urticaria.
Experimental Results
Table 4 shows the complete list of features that consists of a list of attributes that include the allergens, allergic symptoms, physical attributes, and the class label.
Table 4.
List of Allergens, Allergic Symptoms, and Patient Details
| Inhalants, Contactants, and Ingestants (Allergens) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | House dust | 21 | Fish 1a | 41 | Avaraikai (Broad beans) | 61 | Grama | 81 | Running nose |
| 2 | Cotton dust | 22 | Fish 2a | 42 | Kovaikai (Coccinia grandis) | 62 | Channa | 82 | Sneeze |
| 3 | Aspergillus | 23 | Crab | 43 | Kothavarai (Cluster beans) | 63 | Dhal | 83 | Cough |
| 4 | Pollen | 24 | Prawns | 44 | Lady’s finger | 64 | Maida | 84 | Wheezing |
| 5 | Parthenium | 25 | Shark | 45 | Malli (Coriander) | 65 | Oats | 85 | Nasal blocks |
| 6 | Cockroach | 26 | Gourdsa | 46 | Mango | 66 | Ragi | 86 | Headache |
| 7 | Cat dander | 27 | Bananaa | 47 | Mushroom | 67 | Rice | 87 | Itching |
| 8 | Dog fur | 28 | Beans | 48 | Nuckol (Brassica oleracea) | 68 | Wheat | 88 | Rashes |
| 9 | Road dust | 29 | Beet root | 49 | Onion | 69 | Coconut | 89 | Age |
| 10 | Old paper Dust | 30 | Brinjal | 50 | Peas | 70 | Oila | 90 | Gender |
| 11 | PS dust | 31 | Cabbage | 51 | Potroot | 71 | Garlic | 91 | Family history |
| 12 | Milk (P) | 32 | Capsicum | 52 | Paneer (“Farmer’s cheese”) | 72 | Ginger | 92 | Class |
| 13 | Milk (B) | 33 | Chillie | 53 | Potato | 73 | Pepper | ||
| 14 | Curd | 34 | Cauliflower | 54 | Pumpkin | 74 | Tamarind | ||
| 15 | Coffee | 35 | Carrot | 55 | Pudina (Mentha spicata) | 75 | Aginomoto | ||
| 16 | Tea | 36 | Radish | 56 | Chow chow (Chayota edulis) | 76 | Spicesa | ||
| 17 | Beef | 37 | Corn | 57 | Tomato | 77 | Coco | ||
| 18 | Chicken | 38 | Cucumber | 58 | Tondaikai (Trichosanthes dioica) | 78 | Horlicks | ||
| 19 | Mutton | 39 | Drumstick | 59 | Plantain stem | 79 | Boost | ||
| 20 | Egg | 40 | Greensa | 60 | Yams | 80 | Nutsa | ||
Customized based on patient history.
The feature evaluator ranks the features of Strain based on their relevance value. The relevance threshold (τ) was set to 0.01. From among the 91 features (excluding class), 41 features were selected. The selected features with the same 518 samples constitute the preprocessed data. The complete list of features, ranked by their relevance value, is presented in Table 5.
Table 5.
Relief Relevance Values
| Allergen (Feature) | Relevance Value | Allergen (Feature) | Relevance Value | Allergen (Feature) | Relevance Value |
|---|---|---|---|---|---|
| Red rashes | 0.63514 | Channa | 0.02317 | Malli | 0 |
| Swelling | 0.62355 | Coffee | 0.02124 | Road dust | −0.00193 |
| Itching | 0.60425 | Pumpkin | 0.02124 | Fish 1 | −0.00193 |
| Cough | 0.22201 | Chicken | 0.02124 | Cat dander | −0.00193 |
| Running nose | 0.17954 | Headache | 0.01931 | Cockroach | −0.00193 |
| Wheeze/blocks | 0.14865 | Garlic | 0.01931 | Nuckol | −0.00386 |
| Sneeze | 0.14093 | F_history | 0.01737 | Chillie | −0.00386 |
| Coconut | 0.09653 | Wheat | 0.01544 | PS dust | −0.00386 |
| Lady’s finger | 0.07915 | Pepper | 0.01351 | Cucumber | −0.00386 |
| Carrot | 0.07915 | Peas | 0.01351 | Spices | −0.00386 |
| Tamarind | 0.07336 | Prawns | 0.01158 | Pudina | −0.00579 |
| Greens | 0.05985 | Beef | 0.00965 | Mutton | −0.00579 |
| Curd | 0.05598 | Mushroom | 0.00772 | Milk (P) | −0.00579 |
| Tea | 0.04826 | Capsicum | 0.00772 | House dust | −0.00772 |
| Egg | 0.04826 | Kovaikai | 0.00579 | Parthenium | −0.00965 |
| Brinjal | 0.04633 | Chow chow | 0.00386 | Pollen | −0.00965 |
| Oats | 0.03861 | Paneer | 0.00386 | Onion | −0.01351 |
| Radish | 0.03861 | Oil | 0.00386 | Beans | −0.01351 |
| Dhal | 0.03668 | Cotton dust | 0.00386 | Maida | −0.01544 |
| Yams | 0.03282 | Cabbage | 0.00193 | Potato | −0.01544 |
| Drumstick | 0.03282 | Gram | 0.00193 | Dog fur | −0.01931 |
| Aginomoto | 0.03282 | Corn | 0.00193 | Kothavarai | −0.01931 |
| Banana | 0.03282 | Tondaikai | 0 | Potroot | −0.01931 |
| Aspergilus | 0.03089 | Shark | 0 | Vazpoo/thandu | −0.0251 |
| Ragi | 0.02703 | Nuts | 0 | Fish2 | −0.0251 |
| Avaraikai | 0.02703 | Horlicks | 0 | Age | −0.02736 |
| Crab | 0.02703 | Boost | 0 | Milk(B) | −0.03282 |
| Ginger | 0.02703 | Coco | 0 | Gourds | −0.03475 |
| Tomato | 0.02703 | Rice | 0 | Beet root | −0.03475 |
| Cauliflower | 0.0251 | Paper dust | 0 | Mango | −0.03861 |
The classifier evaluator accesses the performance of class-based associative classifier (CBA), decision tree classifier (C4.5), support vector machine (SVM), multi-layer perceptron (MLP), naïve Bayes classifier (NB), and k-nearest neighbor classifier (kNN).24 In order to make the evaluation unbiased, cross-validation is applied over the same features and same partitions of the preprocessed data. The samples in each partition remain the same when each fold is iteratively tested. However, different runs had different samples in the folds in order to avoid the variations and perturbations that may exist due to cross-validation. The evaluator carries out 10 independent runs of 10-fold cross-validation. The complete results of cross-validations are presented in Online Appendix 1. Figure 3 presents the classification accuracy of six classifiers.
Figure 3.
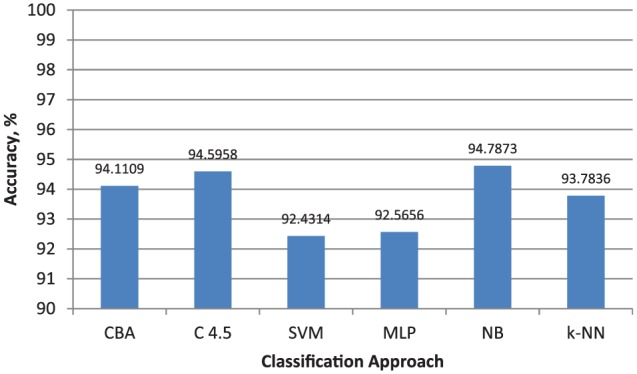
Classification accuracy of six classifiers (C4.5 = decision tree classifier; CBA = class-based associative classifier; kNN = k-nearest neighbor classifier; MLP = multilayer perceptron; NB = naïve Bayes classifier; SVM = support vector machine).
The naïve Bayes classifier was tested with the test data (Stest). A set of sample test instances were also presented to three junior clinicians working at the Good Samaritan Lab and Allergy Centre, Chennai. The clinicians diagnosed the test instances in the absence of the expert. The performance of the clinicians was evaluated using the same performance evaluation measures used by the performance evaluator. The classification performance of the clinicians and the CMD system over the test instances is presented in Table 6.
Table 6.
Performance Evaluation on IDST Test Data
| Clinician |
||||
|---|---|---|---|---|
| 1 | 2 | 3 | CMD System | |
| Sensitivity | 0.5000 | 0.9782 | 0.4782 | 0.969 |
| Specificity | 0.8103 | 0.8275 | 0.6206 | 0.969 |
| Precision | 0.6764 | 0.8181 | 0.5000 | 0.964 |
| Accuracy, % | 67.30 | 89.42 | 70.33 | 96.9231 |
Note: IDST = intradermal skin test.
The significance of the classifier evaluation results was evaluated using Student’s two-tailed paired t test.25 The significance level of the test was set to 0.05 (5%). From the observations, it was inferred that there is a significant improvement in the classification accuracy of the NB Classifier. The run numbers of the 10-fold cross-validation, accuracies obtained, and the corresponding P values for the classifiers are shown in Table 7.
Table 7.
Statistical Significance of Classifier Evaluator
| Run Number | NB | CBA | SVM | C4.5 | MLP | kNN |
|---|---|---|---|---|---|---|
| 1 | 94.59276 | 94.2081446 | 91.1161386 | 94.7813 | 91.8816 | 93.23906 |
| 2 | 94.80015 | 94.2232276 | 92.4773753 | 94.80015 | 91.90045 | 93.82353 |
| 3 | 94.9736 | 93.8159877 | 92.85822 | 94.79638 | 93.05053 | 94.20814 |
| 4 | 94.77376 | 93.9969833 | 92.0701355 | 94.0083 | 92.07768 | 94.01584 |
| 5 | 94.78884 | 94.0233783 | 92.6621416 | 94.21569 | 93.43514 | 93.43891 |
| 6 | 94.79261 | 94.2081446 | 92.6621415 | 94.21569 | 93.24661 | 93.43891 |
| 7 | 94.79638 | 94.2043739 | 92.4698339 | 94.78884 | 92.26998 | 93.43514 |
| 8 | 94.78507 | 94.2006031 | 92.0814477 | 93.23529 | 91.87783 | 93.81599 |
| 9 | 94.58899 | 94.0196077 | 93.8235292 | 96.13876 | 93.43891 | 94.20814 |
| 10 | 94.98115 | 94.2081446 | 92.0927599 | 94.97738 | 92.47738 | 94.21192 |
| P | 1.4164e−06 | 2.2574e−06 | 0.4679 | 4.9900e−05 | 0.025153 |
Note: NB = naïve Bayes classifier; CBA = class-based associative classifier; SVM = support vector machine; C4.5 = decision tree classifier; MLP = multilayer perceptron; kNN = k-nearest neighbor classifier.
Discussion
The inhalants, contactants, and ingestants of an individual are influenced by food habits, biocoenosis, elements of the biosphere, and social environment of an individual. The interactions and adaptations of an individual are prone to be based on socioeconomic status, cultures, traditions, religious beliefs, people groups, and physical environmental factors such as seasons, weather conditions, heat, and humidity. The list of allergens enumerated in Table 4 is neither exhaustive nor generic. In Chennai, a place of diverse people groups, it is not feasible to either capture or generalize all the characteristics of food, behavior, and lifestyle of a population. However, even in the midst of all these limitations, an attempt has been made by a panel of experienced immunologists and medical experts at the allergy center for framing a list of allergens (inhalants, contactants, and ingestants) that is used for analyzing the history of a patient. It can be observed from the list (Table 4) that some of the allergens are customized after a thorough analysis of the history and background of the patient. Tests are continually performed at the center for more than four decades and the list of allergens are annually revised based on the people’s present food habits and environmental conditions.
A patient may be associated with many inhalants, contactants, and ingestants; however, it is not possible to test all possible allergic triggers. According to international allergic testing standards, it is suggested that an upper limit on the number of pricks is up to 40 for intradermal skin tests.26 Though this is a strict limitation for skin tests, it is followed in most allergy testing centers. The feature evaluator with a relevance threshold (τ) of 0.01 selects 41 features that include allergens and allergic symptoms. This ensures that the number of features (allergens) selected by the feature evaluator is in accordance to the allergic testing standards.
All the features selected by the feature evaluator are nominal and are independent of the values of the other attributes. Based on the results of the classifier evaluator the Bayes classifier is used to validate the CMD system using the test data. Bayesian classification approach is well suited for data that are nominal and satisfy the class conditional independence assumption.27 Laplacian correction is used for probability estimation when zero probability values are encountered.24
The evaluation measures used by the performance evaluator to assess the performance of the CMD system are used to evaluate the performance of the clinicians too. From the classification performance of the clinicians, it can be observed that there is a high deviation from one clinician to another. Hence, in the absence of the allergist, clinicians may use the CMD system as a secondary consideration for decision making in the diagnosis of urticaria based on skin test results.
The framework presented in this work is purely developed and validated by using trivial mathematical and statistical models. The data mining process is fully automated and does not require the intervention of an expert to tune or adjust the system. However, the CMD system is intended to replicate expert judgement. Therefore, its purpose in clinical utility is to prompt the clinician to reconsider and confirm his or her decision in the absence of an expert. Clinical judgement is far more comprehensive than pure mathematics.28 There may exist additional subconscious factors that are overlooked by the model.
Conclusion
Medical decision-making systems are widely used for diagnosis. They are also used by junior clinicians and medical students to confirm their decisions. In the diagnosis of allergic disorders, it is not desirable to use a CMD system for complete analysis and diagnosis but as an aid for decision making. The framework of the CMD system used in this work is generic and can be used for a different location. However, the efficiency and efficacy of the system depend on the data distribution, skin test results, and other biological and clinical factors. The medical data that are given as input to the system are important. The testing methods at the allergy center are well established, and the list of allergens is chosen and revised based on the changing food habits of the people and environmental conditions. Hence, the decisions suggested by the system are meaningful and reliable. A focused study on the population and the environmental factors would enable system designers to develop more customized CMD systems. There are allergic disorders and triggers whose causes are ill-defined or unknown. A better biological insight of these disorders may allure the interest of knowledge engineers to develop appropriate CMD systems to enhance and support medical decision making.
Supplementary Material
Acknowledgments
We sincerely thank Mr. Baktha Singh Lazarus, Microbiologist, Joyce Clinical Lab and Allergy Testing Centre, Marthandam 629165, Tamilnadu, India, for providing valuable suggestions and guidelines for this work. The authors also thank the clinicians at the Good Samaritan Lab Services and Allergy Testing Centre, Kilpauk, Chennai 600010, India, for their collaboration and support during the evaluation and testing phases.
Footnotes
The appendix for this article is available on the Medical Decision Making Policy & Practice Web site at http://journals.sagepub.com/doi/suppl/10.1177/2381468316677752.
References
- 1. Blumenthal MN, Rosenberg A. Definition of an allergen (immunobiology). In: Lockey RF, Samuel BC, eds. Allergens and Immunotherapy. 2nd ed.New York: Marcel Dekker; 1999. p 39–51. [Google Scholar]
- 2. Pawankar R, Baena-Cagnani CE, Bousquet J, et al. State of world allergy report 2008: allergy and chronic respiratory diseases. World Allergy Organ J. 2008;1(Suppl. 1):S4–S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kumar R, Kumari D, Srivastava P, et al. Identification of IgE-mediated food allergy and allergens in older children and adults with asthma and allergic rhinitis. Indian J Chest Dis Allied Sci. 2010;52(4):217–24. [PubMed] [Google Scholar]
- 4. Godse KV, Zawar V, Krupashankar D, et al. Consensus statement on the management of urticaria. Indian J Dermatol. 2011;56(5):485–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sachdeva S, Gupta V, Amin SS, Tahseen M. Chronic urticaria. Indian J Dermatol. 2011;56(6):622–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ferrer M. Epidemiology, healthcare, resources, use and clinical features of different types of urticaria. Alergologica 2005. J Investig Allergol Clin Immunol. 2009;19(Suppl. 2):21–6. [PubMed] [Google Scholar]
- 7. Zuberbier T, Bindslev-Jensen C, Canonica W, et al. EAACI/GA2LEN/EDF guideline: definition, classification and diagnosis of urticaria. Allergy. 2006;61(3):316–20. [DOI] [PubMed] [Google Scholar]
- 8. Kemp SF, Lockey RF, eds. Diagnostic Testing of Allergic Disease. New York: Marcel Dekker; 2000. [Google Scholar]
- 9. Kathuria PC. Urticaria and its management. Indian J Allergy Asthma Immunol. 2011;25(1):33–7. [Google Scholar]
- 10. Chang PL, Li YC, Wang TM, Huang ST, Hsieh ML, Tsui KH. Evaluation of a decision-support system for preoperative staging of prostate cancer. Med Decis Making. 1999;19(4):419–27. [DOI] [PubMed] [Google Scholar]
- 11. Barnato AE, Llewellyn-Thomas HA, Peters EM, Siminoff L, Collins ED, Barry MJ. Communication and decision making in cancer care: setting research priorities for decision support/patients’ decision aids. Med Decis Making. 2007;27(5):626–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lutfey KE, Link CL, Marceau LD, et al. Diagnostic certainty as a source of medical practice variation in coronary heart disease: results from a cross-national experiment of clinical decision making. Med Decis Making. 2009;29(5):606–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jane NY, Nehemiah KH, Arputharaj K. A temporal mining framework for classifying un-evenly spaced clinical data: an approach for building effective clinical decision-making system. Appl Clin Inform. 2016;7(1):1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Elizabeth DS, Nehemiah HK, Retmin Raj CS, Kannan A. Computer-aided diagnosis of lung cancer based on analysis of the significant slice of chest computed tomography image. IET Image Process. 2012;6(6):697–705. [Google Scholar]
- 15. Khanna D, Furst DE, Clements PJ, Tashkin DP, Eckman MH. Oral cyclophosphamide for active scleroderma lung disease: a decision analysis. Med Decis Making. 2008;28(6):926–37. [DOI] [PubMed] [Google Scholar]
- 16. Nehemiah HK, Kannan A. A diagnostic decision support system for adverse drug reaction using temporal reasoning. Int J Artif Intell Machine Learn. 2006;6(2):79–86. [Google Scholar]
- 17. Jabez Christopher J, Khanna Nehemiah H, Kannan A. A clinical decision support system for diagnosis of allergic rhinitis based on intradermal skin tests. Comput Biol Med. 2015;65(10):76–84. [DOI] [PubMed] [Google Scholar]
- 18. Lele RD. Computers in Medicine: Progress in Medical Informatics. New Delhi, India: Tata McGraw-Hill;2005. [Google Scholar]
- 19. Aha DW, Kibler D, Albert MK. Instance-based learning algorithms. Mach Learn. 1991;6(1):37–66. [Google Scholar]
- 20. Kira K, Rendell LA. A practical approach to feature selection. In: Proceedings of the Ninth International Workshop on Machine Learning San Francisco: Morgan Kaufmann; 1992. p 249–56. [Google Scholar]
- 21. Kohavi R. A study of cross-validation and bootstrap for accuracy estimation and model selection. In: IJCAI’95: Proceedings of the 14th International Joint Conference on Artificial Intelligence San Francisco: Morgan Kaufmann; 1995. p 1137–45. [Google Scholar]
- 22. Duda RO, Hart PE, Strok DG. Pattern Classification (2nd ed.). New York: John Wiley; 2001. [Google Scholar]
- 23. Smith PH, Condemi JJ, Baum J, Rosier RN. Computer-assisted measurement of skin test wheals. J Allergy Clin Immunol. 1996;97(1):208. [Google Scholar]
- 24. Han J, Kamber M. Data Mining: Concepts and Techniques. San Francisco: Morgan Kaufmann;2006. [Google Scholar]
- 25. Box GEP, Stuart Hunter J, Hunter WG. Statistics for Experimenters: Design, Innovation, and Discovery. New York: Wiley;2005. [Google Scholar]
- 26. Turkeltaub PC. Percutaneous and intracutaneous diagnostic tests of IgE-mediated diseases (immediate hypersensitivity). Clin Allergy Immunol. 1999;15:53–87. [PubMed] [Google Scholar]
- 27. Domingos P. Beyond independence: conditions for optimality of the simple Bayesian classifier. In: Proceedings of the 13th International Conference on Machine Learning; Bari, Italy 1996. p 105–12. [Google Scholar]
- 28. de Vries M, Witteman CL, Holland RW, Dijksterhuis A. The unconscious thought effect in clinical decision making: an example in diagnosis. Med Decis Making. 2010;30(5):578–81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.