Abstract
Head and neck cancers consist of a heterogeneous group of cancers that are difficult to treat successfully. Limited screening options lead to advanced stages at presentation, and difficulty with treatment options and post-treatment surveillance can lead to poor outcomes. In this setting, tools for early and precise detection of disease will be highly valuable. Liquid biopsies, or use of analytes in blood, saliva, and other body fluid samples, provide new avenues for cancer screening with the potential for early detection, treatment modification, and surveillance of head and neck cancers. Early studies of liquid biopsies in specific head and neck cancers have had encouraging results. Nevertheless, various challenges remain before routine adoption into clinical use is feasible. With continued advancement in the field of liquid biopsies, there is great promise for clinical implementation and significant improvement in head and neck cancer care.
Introduction
Head and neck cancers, particularly advanced head and neck squamous cell carcinomas (HNSCC), remain a challenging disease to treat successfully. Existing screening options are primarily limited to clinical exams and radiological tests, which are imprecise and may result in delayed detection. Subsequently, many head and neck cancer patients thus present with advanced stage disease, leading to poorer outcomes. As a result, little progress has been made in improving survival from head and neck cancer. In this setting, early detection and surveillance of head and neck cancers has long been a goal for optimizing patient outcomes. Additionally, rather than invasive diagnostic testing, the concept of a quick liquid sample to diagnose disease has long been appealing.
In response to these difficulties in cancer detection, development of new screening modalities has garnered significant interest in recent years. Key amongst these are “liquid biopsies,” or the collection and examination of analytes in body fluid samples, such as blood and saliva specimens. Liquid biopsies are proposed to offer the potential for early identification of initial cancers or early detection of tumor recurrences or metastases. Importantly, they offer a less invasive and potentially less expensive screening option than their traditional screening and diagnostic techniques (e.g. invasive biopsies or frequent radiologic exams). Currently, such biopsy techniques are being investigated for utilization in multiple paradigms, including early cancer detection or screening, considerations for treatment planning and therapies, and surveillance for disease persistence and recurrence.
Various types of liquid biopsies have been proposed, but the ones most rigorously being investigated and implemented are circulating tumor DNA (ctDNA) and circulating tumor cells (CTCs). Additional analytes under study, but with less current data, include tumor antigens, cell free coding and noncoding RNAs, as well as extracellular vesicles and exosomes. Briefly, ctDNA has been identified as a subset of total circulating cell free DNA (cfDNA) in the bloodstream. While total cfDNA is raised in conditions ranging from infection to trauma (Wan et al., 2017), ctDNA is believed to be shed by tumor cells undergoing apoptosis or necrosis (Schwarzenbach et al., 2011). Circulating tumor cells are thought to be free tumor cells in the bloodstream arising from primary tumors in low numbers (Alix-Panabieres et al., 2012). Each has their distinct advantages and disadvantages, and advancements in both recent years has been dramatic. In cancers of the head and neck, each has significant potential for improving cancer detection and treatment.
Current Liquid Biopsy Efforts in Head and Neck Cancers
Nasopharyngeal Carcinoma
While there certainly has been extensive research in liquid biopsies across various cancer types, liquid biopsy in head and neck cancers have played a seminal role in their advancement. The earliest and most robust clinical application of liquid biopsy in head and neck cancer was with the detection of Epstein-Barr virus (EBV) cfDNA in nasopharyngeal cancers. In the late 1990s, EBV cfDNA was identified in the serum of patients with nasopharyngeal carcinoma (NPC) (Lo et al., 1999a; Lo et al., 1999b; Mutirangura et al., 1998), and subsequently was found to correlate with NPC prognosis and recurrence (Chan et al., 2002; Le et al., 2005; Wang et al., 2013). In a recent landmark study, Chan et al. examined over 20,000 Southeast Asian male subjects and showed that EBV cfDNA in plasma samples could be useful in screening for early asymptomatic NPC in populations with a high prevalence of the disease (Chan et al., 2017). Impressively, the investigators were able to identify a large number of patients with early-stage NPC. As this cancer often presents at late stages with nodal metastases, this liquid biopsy has the potential for significant improvement in early treatment and a resultant survival benefit. Furthermore, recent modifications of their screening and analysis techniques have improved sensitivity and specificity of this method, improving clinical feasibility. Thus, EBV cfDNA is arguably the gold standard for clinical use of a liquid biopsy in head and neck cancers.
HPV-Positive Head and Neck Squamous Cell Carcinoma
An additional etiologic virus in head and neck squamous cell carcinomas, human papillomavirus (HPV), is being actively investigated as a potential target for liquid biomarkers. HPV-positive HNSCCs represent a clinically and genetically distinct subset of HNSCCs with increasing prevalence and currently comprising the majority of oropharyngeal SCCs (Fakhry et al., 2008; Kim et al., 2015; Vokes et al., 2015). Predictable genetic alterations underlie HPV positive HNSCCs, such as the presence of HPV-16 viral DNA, p16INK4A overexpression, increased mutation rates in PIK3CA, and FGFR3 dysregulation (Cancer Genome Atlas, 2015; Chung et al., 2015; Seiwert et al., 2015; Slebos et al., 2006; Stransky et al., 2011). Similar to the studies done for EBV in NPC, researchers have found HPV DNA present in both blood (Ahn et al., 2014; Capone et al., 2000; Mazurek et al., 2016) and saliva (Ahn et al., 2014; Chai et al., 2016) samples of HPV-positive HNSCC patients, offering the possibility of a simple blood draw or saliva swab to detect cancer. Notably, HPV detection sensitivity varies by the tested body fluid (Ahn et al., 2014; Spector et al., 2017; Wang et al., 2015), and correlations with clinical outcomes have been shown to differ by which type of sample was used (Ahn et al., 2014). On the other hand, pretreatment serum HPV DNA, while correlated with OPSCC staging, has not been found to predict disease recurrence (Dahlstrom et al., 2015). While encouraging, much remains to be optimized in terms of sensitivity and specificity of liquid biopsies for HPV DNA, with reported sensitivities ranging from 47-100% and most reports less than 90% (Wang et al., 2015). Of note, given low recurrence rates for OPSCC and myriad of tools for detecting HPV through liquid biopsies, future studies will likely require large sample sizes and standardized combinatory HPV detection methodology.
HPV-Negative Head and Neck Squamous Cell Carcinoma
For HPV-negative head and neck cancers, unlike other cancers, there is no ubiquitous activating mutation or biomarker. However, there are a number of promising uses for liquid biopsies for these HNSCCs. There is also evidence that CTCs are an independent prognostic marker for disease-free and overall survival in HNSCC in particular (Grobe et al., 2014; Hristozova et al., 2011; Kulasinghe et al., 2015; Tinhofer et al., 2014), although many of these studies are small, and include a number of head and neck tumor sites (Buglione et al., 2012; Grisanti et al., 2014; Jatana et al., 2010). Interestingly, a recent study has correlated PD-L1 CTCs levels in HNSCC with poorer survival outcomes (Strati et al., 2017). In this era of immunotherapeutics (with anti-PD-1 agents approved in HNSCC) and immune surveillance in cancers, identification of PD-L1 positive circulating cells may prove to have significant prognostic potential, and may inform use of adjuvant immunotherapeutics.
Initial proof of principle analyses of the ability to detect ctDNA in saliva or plasma specimens in HNSCCs have been encouraging. Wang et al. (Wang et al., 2015) were able to identify up to 100% of ctDNA from saliva and plasma from various HNSCCs. Of note their sensitivity range across tumor stages and subsites was quite broad, suggesting further optimization is necessarily for such detection methods. Moreover, for HPV-negative tumors, they utilized genomic sequencing of the primary tumor to subsequently identify the mutated ctDNA, which may be of limited practicality for a cost-effective screening modality.
The Cancer Genome Atlas demonstrated that specific genes and pathways are commonly altered in head and neck cancer, namely TP53, NOTCH1, PIK3CA, among other genes (Cancer Genome Atlas, 2015). There have been efforts, especially in other cancers, to develop sets of probes to screen for commonly mutated genes in ctDNA. In one study assessing patients with colorectal, breast, lung or ovarian cancer with a 58 gene panel, mutation detection rate was between 59-71% in early stage disease (Phallen et al., 2017). While a suboptimal sensitivity rate, this is an exciting initial step towards developing gene panels of commonly mutated genes to screen for disease. In a similar fashion, panels against commonly mutated HNSCC genes may have analogous applicability. As assays expand from assessing individual “hotspot” mutations to cover entire genes or multiple genes, however, the cost of the assay increases exponentially. Thus, in order to control cost, the selection of what genomic loci to monitor is highly dependent on the purpose of the assay: early detection assays should monitor the alterations that are most common across all tumors, and disease monitoring assays should focus more on the alterations most likely associated with an individual’s tumor.
Other Head and Neck Cancers
There is limited data but significant potential for liquid biopsies in other head and neck cancers. Gene fusions are common and tumor-type specific in salivary cancer (Andersson & Stenman, 2016; Stenman et al., 2014), which affords the potential for a molecular target for liquid biopsies. In particular, MYB-NFIB gene fusions (Ho et al., 2013; Stephens et al., 2013) are common in adenoid cystic carcinomas, while CRTC1/3-MAML2 gene fusions have been identified in high frequency in mucoepidermoid carcinomas (Birkeland et al., 2017). These salivary cancers are frequently high-grade malignancies that have a propensity for recurrence and metastasis, often many years after initial treatment (Bhayani et al., 2012). Thus, these fusion genes have potential as targeted options for detection with liquid biopsies.
Thyroid cancers, while overall carrying a good prognosis, have subsets of cancer with higher rates of recurrence or may degenerate into anaplastic thyroid cancer. Researchers are studying whether blood tests for the common BRAF V600E mutation found in the more common differentiated thyroid carcinoma tumor types could predict which subset of well-differentiated thyroid cancer carry a worse prognosis (Lubitz et al., 2016; Nikiforova et al., 2003). Building on understanding of the genetics of anaplastic thyroid carcinoma (Kunstman et al., 2015; Landa et al., 2016; Sykorova et al., 2015), recent work has identified ctDNA with mutations in commonly altered genes: BRAF, PIK3CA, NRAS, PTEN, and TP53 (Sandulache et al., 2017), suggesting a role for liquid biopsies in detection and treatment stratification of aggressive thyroid cancers.
Future Clinical implementation of Liquid Biopsies in Head and Neck Cancer
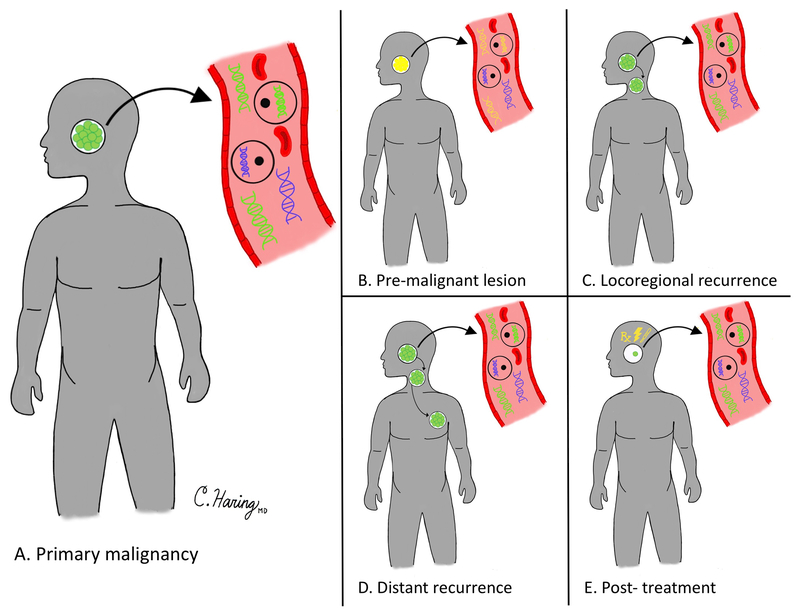
Building on these initial data, one can imagine numerous potential modalities in which to employ liquid biopsies for head and neck cancer detection. This can include initial disease screening or detection, evaluation of disease persistence or recurrence after initial treatment regimens, and screening for recurrence in patients successfully treated from their cancer (Figure). Moreover, unique to head and neck cancers, saliva may be able to be used as a source for liquid biopsy. While none of these liquid biomarkers have been fully adopted for standard use in head and neck cancers, many are increasingly being considered for clinical care.
Figure. Time points for the Potential Utilization of Liquid Biopsies.
Cancer ctDNA or CTCs (green) can be differentiated from non-cancer cell free DNA and cells (blue). Liquid biopsies can be utilized to screen for primary cancers or premalignant lesions (A, B), or to detect recurrence or post-treatment persistence (C-E).
Screening for Initial Disease
Cost-effective and sensitive liquid biopsy screening of at-risk patients remains an enticing prospect for head and neck cancers. Importantly, a critical decision point is how to optimize patient selection to improve the value of these tests (i.e. maximizing positive and negative predictive values for the tests). For instance, with a carefully selected NPC cohort (an age-appropriate Southeast Asian male population) and with a sensitive and specific EBV cfDNA analysis, only 34 out of over 20,000 patients were ultimately diagnosed with NPC (Chan et al., 2017). Certainly for other cancers, there is a need to optimize screening for patient populations at highest risk for development of disease or recurrence who may not be detected at early time points with our current methodologies. For instance, EBV screening for NPC would not have the same level of clinical value in other cohorts with low incidence of NPC. Similarly, there may be limited value in screening all smokers for head and neck cancer liquid biopsies, but rather those with especially high risk based on risk-stratification or those with premalignant lesions. For HPV-positive HNSCCs, lack of traditional risk factors makes it difficult to identify an at-risk population. As rates of high-risk HPV infection are better defined epidemiologically, considerations into screening paradigms may develop. Liquid biopsies could also play a role in early tumorigenesis screening for patients with hereditary cancer syndromes including Fanconi’s anemia and Li-Fraumeni syndrome. Such an application of liquid biopsies could reduce the frequency of radiologic imaging and perhaps provide a higher sensitivity than current clinical screening parameters. Future work remains to explore distinguishing germline and somatic genetic changes in liquid biopsies for such patients.
Response to Initial Therapies/Treatment Opportunities
For existing cancers, liquid biopsies can be tailored to evaluate response to treatment, as well as to screen for disease persistence, recurrence and metastases based on their unique genetic signatures. Preliminary work has explored the dynamics of cfDNA during treatment (Mazurek et al., 2016), which opens the possibility that liquid biopsies could select patients for whom treatment should either be de-escalated or intensified. This has great implications for induction therapy regimens, which are commonly employed in certain head and neck cancers, such as laryngeal squamous cell carcinoma (Urba et al., 2006). Similarly, assessment of cfDNA or CTCs in window of opportunity trials, where short neoadjuvant courses of new therapies (e.g. anti-PD-1 antibodies) are delivered prior to definitive therapy. Given the recent investigations on PD-L1 expression on CTCs in head and neck cancer, assessment of changes in responsive to nivolumab or pembrolizumab may have interesting prognostic results (Strati et al., 2017).
Detection of Recurrent and Metastatic Disease
In consideration for screening criteria for cancer persistence or recurrence, liquid biopsies could focus on patients who are at highest risk for recurrent head and neck cancer after treatment. For instance, high-cost screening protocols may not be prioritized for most HPV-associated HNSCC, where the majority of patients respond well to treatment and do not develop recurrences (Ang et al., 2010). As we further identify clinical and biologic predictors of poor outcomes in this cohort of patients, we may begin to consider utilization of liquid biopsies for these patients. Similarly, for other head and neck cancer patients, careful patient selection for liquid biopsy surveillance will be critical.
Liquid biopsies offer an additional benefit in apart for identification of recurrent and metastatic disease in previously treated patients. Initial studies have identified mutations and other genetic alterations in CTCs and ctDNA, distinct from the primary tumor, that may correlate with clinical resistance mechanisms. For instance, patients who initially responded to anti-EGFR therapy but subsequently relapsed demonstrated consistent mutations in downstream pathways in their ctDNA (Bettegowda et al., 2014). Further investigation into unique gained mutations in ctDNA and CTCs may have significant value in determining subsequent treatment options. There is a need to consider the cost and the comprehensiveness of sequencing both the primary tumor and CTCs/ctDNA in such scenarios. Alternatively, given the current limitations of individualized direct primary tumor sequencing and biomarker detection protocols, some groups have begun working on overall oncogenic signature patterns in liquid biomarkers, thus not relying on individual genetic mutations, but rather a common “cancer signature” for screening purposes (Zviran et al., 2018).
Conclusion
Overall, liquid biopsies are an exciting new field of study with enormous potential for improving early cancer detection, treatment modification, and surveillance for disease recurrence. Initial efforts for liquid biopsies in head and neck cancer have had encouraging results, particularly with NPC and HPV-positive HNSCCs. Much remains to be done in regards to optimization of liquid biopsies and incorporating them into routine clinical use. Current challenges, particularly in HPV-negative HNSCC, where there is not a ubiquitous mutation, are to identify appropriate liquid biopsy screening parameters. With further refinement of methodology, and implementation into clinical protocols, there exists considerable potential for liquid biopsies to play a significant role in the improved care of head and neck cancer patients.
Acknowledgments
Grant Support: J.C.B. received funding from NIH Grants U01-DE025184, P30-CA046592 and R01-CA194536. J.C.B and M.E.S. also received funding from the American Head and Neck Society.
Footnotes
Disclosure
The authors have no relevant disclosures.
References
- Ahn SM, Chan JY, Zhang Z, Wang H, Khan Z, Bishop JA, Westra W, Koch WM, Califano JA. Saliva and plasma quantitative polymerase chain reaction-based detection and surveillance of human papillomavirus-related head and neck cancer. JAMA Otolaryngol Head Neck Surg 140(9):846–854, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alix-Panabieres C, Schwarzenbach H, Pantel K. Circulating tumor cells and circulating tumor DNA. Annu Rev Med 63:199–215, 2012. [DOI] [PubMed] [Google Scholar]
- Andersson MK, Stenman G. The landscape of gene fusions and somatic mutations in salivary gland neoplasms - Implications for diagnosis and therapy. Oral Oncol 57:63–69, 2016. [DOI] [PubMed] [Google Scholar]
- Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, Westra WH, Chung CH, Jordan RC, Lu C, Kim H, Axelrod R, Silverman CC, Redmond KP, Gillison ML. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med 363(1):24–35, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, Antonarakis ES, Azad NS, Bardelli A, Brem H, Cameron JL, Lee CC, Fecher LA, Gallia GL, Gibbs P, Le D, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 6(224):224ra224, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhayani MK, Yener M, El-Naggar A, Garden A, Hanna EY, Weber RS, Kupferman ME. Prognosis and risk factors for early-stage adenoid cystic carcinoma of the major salivary glands. Cancer 118(11):2872–2878, 2012. [DOI] [PubMed] [Google Scholar]
- Birkeland AC, Foltin SK, Michmerhuizen NL, Hoesli RC, Rosko AJ, Byrd S, Yanik M, Nor JE, Bradford CR, Prince ME, Carey TE, Mchugh JB, Spector ME, Brenner JC. Correlation of Crtc1/3-Maml2 fusion status, grade and survival in mucoepidermoid carcinoma. Oral Oncol 68:5–8, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buglione M, Grisanti S, Almici C, Mangoni M, Polli C, Consoli F, Verardi R, Costa L, Paiar F, Pasinetti N, Bolzoni A, Marini M, Simoncini E, Nicolai P, Biti G, Magrini SM. Circulating tumour cells in locally advanced head and neck cancer: preliminary report about their possible role in predicting response to non-surgical treatment and survival. Eur J Cancer 48(16):3019–3026, 2012. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517(7536):576–582, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capone RB, Pai SI, Koch WM, Gillison ML, Danish HN, Westra WH, Daniel R, Shah KV, Sidransky D. Detection and quantitation of human papillomavirus (HPV) DNA in the sera of patients with HPV-associated head and neck squamous cell carcinoma. Clin Cancer Res 6(11):4171–4175, 2000. [PubMed] [Google Scholar]
- Chai RC, Lim Y, Frazer IH, Wan Y, Perry C, Jones L, Lambie D, Punyadeera C. A pilot study to compare the detection of HPV-16 biomarkers in salivary oral rinses with tumour p16(INK4a) expression in head and neck squamous cell carcinoma patients. BMC Cancer 16:178, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AT, Lo YM, Zee B, Chan LY, Ma BB, Leung SF, Mo F, Lai M, Ho S, Huang DP, Johnson PJ. Plasma Epstein-Barr virus DNA and residual disease after radiotherapy for undifferentiated nasopharyngeal carcinoma. J Natl Cancer Inst 94(21):1614–1619, 2002. [DOI] [PubMed] [Google Scholar]
- Chan KCA, Woo JKS, King A, Zee BCY, Lam WKJ, Chan SL, Chu SWI, Mak C, Tse IOL, Leung SYM, Chan G, Hui EP, Ma BBY, Chiu RWK, Leung SF, Van Hasselt AC, Chan ATC, Lo YMD. Analysis of Plasma Epstein-Barr Virus DNA to Screen for Nasopharyngeal Cancer. N Engl J Med 377(6):513–522, 2017. [DOI] [PubMed] [Google Scholar]
- Chung CH, Guthrie VB, Masica DL, Tokheim C, Kang H, Richmon J, Agrawal N, Fakhry C, Quon H, Subramaniam RM, Zuo Z, Seiwert T, Chalmers ZR, Frampton GM, Ali SM, Yelensky R, Stephens PJ, Miller VA, Karchin R, Bishop JA. Genomic alterations in head and neck squamous cell carcinoma determined by cancer gene-targeted sequencing. Ann Oncol 26(6):1216–1223, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlstrom KR, Li G, Hussey CS, Vo JT, Wei Q, Zhao C, Sturgis EM. Circulating human papillomavirus DNA as a marker for disease extent and recurrence among patients with oropharyngeal cancer. Cancer 121(19):3455–3464, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhry C, Westra WH, Li S, Cmelak A, Ridge JA, Pinto H, Forastiere A, Gillison ML. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst 100(4):261–269, 2008. [DOI] [PubMed] [Google Scholar]
- Grisanti S, Almici C, Consoli F, Buglione M, Verardi R, Bolzoni-Villaret A, Bianchetti A, Ciccarese C, Mangoni M, Ferrari L, Biti G, Marini M, Ferrari VD, Nicolai P, Magrini SM, Berruti A. Circulating tumor cells in patients with recurrent or metastatic head and neck carcinoma: prognostic and predictive significance. PLoS One 9(8):e103918, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobe A, Blessmann M, Hanken H, Friedrich RE, Schon G, Wikner J, Effenberger KE, Kluwe L, Heiland M, Pantel K, Riethdorf S. Prognostic relevance of circulating tumor cells in blood and disseminated tumor cells in bone marrow of patients with squamous cell carcinoma of the oral cavity. Clin Cancer Res 20(2):425–433, 2014. [DOI] [PubMed] [Google Scholar]
- Ho AS, Kannan K, Roy DM, Morris LG, Ganly I, Katabi N, Ramaswami D, Walsh LA, Eng S, Huse JT, Zhang J, Dolgalev I, Huberman K, Heguy A, Viale A, Drobnjak M, Leversha MA, Rice CE, Singh B, Iyer NG, et al. The mutational landscape of adenoid cystic carcinoma. Nat Genet 45(7):791–798, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hristozova T, Konschak R, Stromberger C, Fusi A, Liu Z, Weichert W, Stenzinger A, Budach V, Keilholz U, Tinhofer I. The presence of circulating tumor cells (CTCs) correlates with lymph node metastasis in nonresectable squamous cell carcinoma of the head and neck region (SCCHN). Ann Oncol 22(8):1878–1885, 2011. [DOI] [PubMed] [Google Scholar]
- Jatana KR, Balasubramanian P, Lang JC, Yang L, Jatana CA, White E, Agrawal A, Ozer E, Schuller DE, Teknos TN, Chalmers JJ. Significance of circulating tumor cells in patients with squamous cell carcinoma of the head and neck: initial results. Arch Otolaryngol Head Neck Surg 136(12):1274–1279, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KY, Zhang X, Cha IH. Identification of human papillomavirus status specific biomarker in head and neck cancer. Head Neck 37(9):1310–1318, 2015. [DOI] [PubMed] [Google Scholar]
- Kulasinghe A, Perry C, Jovanovic L, Nelson C, Punyadeera C. Circulating tumour cells in metastatic head and neck cancers. Int J Cancer 136(11):2515–2523, 2015. [DOI] [PubMed] [Google Scholar]
- Kunstman JW, Juhlin CC, Goh G, Brown TC, Stenman A, Healy JM, Rubinstein JC, Choi M, Kiss N, Nelson-Williams C, Mane S, Rimm DL, Prasad ML, Hoog A, Zedenius J, Larsson C, Korah R, Lifton RP, Carling T. Characterization of the mutational landscape of anaplastic thyroid cancer via whole-exome sequencing. Hum Mol Genet 24(8):2318–2329, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landa I, Ibrahimpasic T, Boucai L, Sinha R, Knauf JA, Shah RH, Dogan S, Ricarte-Filho JC, Krishnamoorthy GP, Xu B, Schultz N, Berger MF, Sander C, Taylor BS, Ghossein R, Ganly I, Fagin JA. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J Clin Invest 126(3):1052–1066, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le QT, Jones CD, Yau TK, Shirazi HA, Wong PH, Thomas EN, Patterson BK, Lee AW, Zehnder JL. A comparison study of different PCR assays in measuring circulating plasma epstein-barr virus DNA levels in patients with nasopharyngeal carcinoma. Clin Cancer Res 11(16):5700–5707, 2005. [DOI] [PubMed] [Google Scholar]
- Lo YM, Chan LY, Chan AT, Leung SF, Lo KW, Zhang J, Lee JC, Hjelm NM, Johnson PJ, Huang DP. Quantitative and temporal correlation between circulating cell-free Epstein-Barr virus DNA and tumor recurrence in nasopharyngeal carcinoma. Cancer Res 59(21):5452–5455, 1999a. [PubMed] [Google Scholar]
- Lo YM, Chan LY, Lo KW, Leung SF, Zhang J, Chan AT, Lee JC, Hjelm NM, Johnson PJ, Huang DP. Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res 59(6):1188–1191, 1999b. [PubMed] [Google Scholar]
- Lubitz CC, Parangi S, Holm TM, Bernasconi MJ, Schalck AP, Suh H, Economopoulos KP, Gunda V, Donovan SE, Sadow PM, Wirth LJ, Sullivan RJ, Panka DJ. Detection of Circulating BRAF(V600E) in Patients with Papillary Thyroid Carcinoma. J Mol Diagn 18(1):100–108, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurek AM, Rutkowski T, Fiszer-Kierzkowska A, Malusecka E, Skladowski K. Assessment of the total cfDNA and HPV16/18 detection in plasma samples of head and neck squamous cell carcinoma patients. Oral Oncol 54:36–41, 2016. [DOI] [PubMed] [Google Scholar]
- Mutirangura A, Pornthanakasem W, Theamboonlers A, Sriuranpong V, Lertsanguansinchi P, Yenrudi S, Voravud N, Supiyaphun P, Poovorawan Y. Epstein-Barr viral DNA in serum of patients with nasopharyngeal carcinoma. Clin Cancer Res 4(3):665–669, 1998. [PubMed] [Google Scholar]
- Nikiforova MN, Kimura ET, Gandhi M, Biddinger PW, Knauf JA, Basolo F, Zhu Z, Giannini R, Salvatore G, Fusco A, Santoro M, Fagin JA, Nikiforov YE. BRAF mutations in thyroid tumors are restricted to papillary carcinomas and anaplastic or poorly differentiated carcinomas arising from papillary carcinomas. J Clin Endocrinol Metab 88(11):5399–5404, 2003. [DOI] [PubMed] [Google Scholar]
- Phallen J, Sausen M, Adleff V, Leal A, Hruban C, White J, Anagnostou V, Fiksel J, Cristiano S, Papp E, Speir S, Reinert T, Orntoft MW, Woodward BD, Murphy D, Parpart-Li S, Riley D, Nesselbush M, Sengamalay N, Georgiadis A, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med 9(403), 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandulache VC, Williams MD, Lai SY, Lu C, William WN, Busaidy NL, Cote GJ, Singh RR, Luthra R, Cabanillas ME. Real-Time Genomic Characterization Utilizing Circulating Cell-Free DNA in Patients with Anaplastic Thyroid Carcinoma. Thyroid 27(1):81–87, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 11(6):426–437, 2011. [DOI] [PubMed] [Google Scholar]
- Seiwert TY, Zuo Z, Keck MK, Khattri A, Pedamallu CS, Stricker T, Brown C, Pugh TJ, Stojanov P, Cho J, Lawrence MS, Getz G, Bragelmann J, Deboer R, Weichselbaum RR, Langerman A, Portugal L, Blair E, Stenson K, Lingen MW, et al. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin Cancer Res 21(3):632–641, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slebos RJ, Yi Y, Ely K, Carter J, Evjen A, Zhang X, Shyr Y, Murphy BM, Cmelak AJ, Burkey BB, Netterville JL, Levy S, Yarbrough WG, Chung CH. Gene expression differences associated with human papillomavirus status in head and neck squamous cell carcinoma. Clin Cancer Res 12(3 Pt 1):701–709, 2006. [DOI] [PubMed] [Google Scholar]
- Spector ME, Sacco AG, Bellile E, Taylor JMG, Jones T, Sun K, Brown WC, Birkeland AC, Bradford CR, Wolf GT, Prince ME, Moyer JS, Malloy K, Swiecicki P, Eisbruch A, Mchugh JB, Chepeha DB, Rozek L, Worden FP. E6 and E7 Antibody Levels Are Potential Biomarkers of Recurrence in Patients with Advanced-Stage Human Papillomavirus-Positive Oropharyngeal Squamous Cell Carcinoma. Clin Cancer Res 23(11):2723–2729, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenman G, Persson F, Andersson MK. Diagnostic and therapeutic implications of new molecular biomarkers in salivary gland cancers. Oral Oncol 50(8):683–690, 2014. [DOI] [PubMed] [Google Scholar]
- Stephens PJ, Davies HR, Mitani Y, Van Loo P, Shlien A, Tarpey PS, Papaemmanuil E, Cheverton A, Bignell GR, Butler AP, Gamble J, Gamble S, Hardy C, Hinton J, Jia M, Jayakumar A, Jones D, Latimer C, Mclaren S, Mcbride DJ, et al. Whole exome sequencing of adenoid cystic carcinoma. J Clin Invest 123(7):2965–2968, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, Mckenna A, Shefler E, Ramos AH, Stojanov P, Carter SL, Voet D, Cortes ML, Auclair D, Berger MF, Saksena G, Guiducci C, et al. The mutational landscape of head and neck squamous cell carcinoma. Science 333(6046):1157–1160, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strati A, Koutsodontis G, Papaxoinis G, Angelidis I, Zavridou M, Economopoulou P, Kotsantis I, Avgeris M, Mazel M, Perisanidis C, Sasaki C, Alix-Panabieres C, Lianidou E, Psyrri A. Prognostic significance of PD-L1 expression on circulating tumor cells in patients with head and neck squamous cell carcinoma. Ann Oncol 28(8):1923–1933, 2017. [DOI] [PubMed] [Google Scholar]
- Sykorova V, Dvorakova S, Vcelak J, Vaclavikova E, Halkova T, Kodetova D, Lastuvka P, Betka J, Vlcek P, Reboun M, Katra R, Bendlova B. Search for new genetic biomarkers in poorly differentiated and anaplastic thyroid carcinomas using next generation sequencing. Anticancer Res 35(4):2029–2036, 2015. [PubMed] [Google Scholar]
- Tinhofer I, Konschak R, Stromberger C, Raguse JD, Dreyer JH, Johrens K, Keilholz U, Budach V. Detection of circulating tumor cells for prediction of recurrence after adjuvant chemoradiation in locally advanced squamous cell carcinoma of the head and neck. Ann Oncol 25(10):2042–2047, 2014. [DOI] [PubMed] [Google Scholar]
- Urba S, Wolf G, Eisbruch A, Worden F, Lee J, Bradford C, Teknos T, Chepeha D, Prince M, Hogikyan N, Taylor J. Single-cycle induction chemotherapy selects patients with advanced laryngeal cancer for combined chemoradiation: a new treatment paradigm. J Clin Oncol 24(4):593–598, 2006. [DOI] [PubMed] [Google Scholar]
- Vokes EE, Agrawal N, Seiwert TY. HPV-Associated Head and Neck Cancer. J Natl Cancer Inst 107(12):djv344, 2015. [DOI] [PubMed] [Google Scholar]
- Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, Pacey S, Baird R, Rosenfeld N. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 17(4):223–238, 2017. [DOI] [PubMed] [Google Scholar]
- Wang WY, Twu CW, Chen HH, Jiang RS, Wu CT, Liang KL, Shih YT, Chen CC, Lin PJ, Liu YC, Lin JC. Long-term survival analysis of nasopharyngeal carcinoma by plasma Epstein-Barr virus DNA levels. Cancer 119(5):963–970, 2013. [DOI] [PubMed] [Google Scholar]
- Wang Y, Springer S, Mulvey CL, Silliman N, Schaefer J, Sausen M, James N, Rettig EM, Guo T, Pickering CR, Bishop JA, Chung CH, Califano JA, Eisele DW, Fakhry C, Gourin CG, Ha PK, Kang H, Kiess A, Koch WM, et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci Transl Med 7(293):293ra104, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zviran A, Hill STK, Schulman R, Shah M, Deochand S, Ha G, Reed S, Rotem D, Gydush G, Rhoades J, Huang K, Liao W, Maloney D, Ormans N, Malbari M, Spinelli CF, Kazancioglu S, Robine N, Adalsteinsson V, Houck-Loomis B, et al. (2018) Genome-wide cell-free DNA mutation integration for sensitive detection. American Association for Cancer Research; Chicago, IL. [Google Scholar]