Abstract
Holoprosencephaly (HPE) is a structural brain anomaly characterized by failure of the forebrain to separate during early embryogenesis. Both genetic and environmental etiologies of HPE have been discovered over the last three decades. Traditionally, the genetic work-up for HPE has been a karyotype, chromosomal microarray, and/or Sanger sequencing of select genes. The recent increased availability of next generation sequencing has changed the molecular diagnostic landscape for HPE, associating new genes with this disorder such as FGFR1. We conducted a systematic review of the medical literature for the molecular testing of HPE for studies published in the last 20 years. We also queried known commercial diagnostic laboratories and used information on their websites to construct a list of available commercial testing. Our group released its first recommendations in 2010 and this update incorporates the technology shifts and gene discoveries over the last decade. These recommendations provide a guide for genetic diagnosis of HPE, which is paramount for patients and their families for prognosis, treatment, and genetic counseling.
INTRODUCTION
Holoprosencephaly (HPE) is a structural brain anomaly characterized by failure of the forebrain to separate during early embryogenesis. HPE spans a spectrum from alobar (no separation of the forebrain) to middle hemispheric variant (MIHV) type (partial separation). There is also a type of HPE termed microform where there are no brain malformations seen on neuroimaging, but facial characteristics typical of HPE are seen such as hypotelorism, single central maxillary incisor, retinal coloboma, or clefting (Kruszka, Hart, Hadley, Muenke, & Habal, 2015). As HPE is a rare condition without available evidence-based testing guidelines, we have summarized lessons from our three decades of molecular testing experience for HPE. Our group released its first recommendations in 2010 (Pineda-Alvarez, Dubourg, David, Roessler, & Muenke, 2010) and this update incorporates the technology shifts and gene discoveries over the last decade. The availability of next-generation sequencing (NGS) has changed the molecular diagnostic landscape for HPE, allowing for interrogation of genes associated with HPE in large cohorts of patients (Dubourg et al., 2016) and associating new genes with this disorder, such as FGFR1 (Hong et al., 2016; Roessler, Hu, & Muenke, 2018; Simonis et al., 2013).
METHODS
We conducted a systematic review of the medical literature for the molecular testing of HPE for studies published in the last 20 years. We searched PubMed, Embase, and Google Scholar using the search terms “holoprosencephaly”, “genetic testing”, “next-generation sequencing”, “whole exome sequencing”, “whole genome sequencing”, and “chromosomal microarray”. Journal articles included were case reports, cohort studies, expert consensus, and review studies. Additional studies were ascertained from reference lists in these studies. We also queried known commercial diagnostic laboratories and used information on their websites to construct a list of available commercial testing (Table I).
Table I.
Genetic testing panels for holoprosencephaly*.
| Diagnostic center | Method | Genes tested |
|---|---|---|
| ARUP Laboratories | Targeted capture NGS panel; CGH | DISP1, FGF8, FOXH1, GLI2, NODAL, PTCH1, SHH, SIX3, TDGF1, TGIF1, ZIC2, FGF8 |
| Blueprint Genetics | WES panel | CDON, FGF8, FGFR1, FOXH1, GLI2, GLI3, NODAL, PTCH1, SHH, SIX3, TGIF1, ZIC2 |
| GeneDx | Sanger sequencing/MLPA | SHH, SIX3, TGIF1, ZIC2 |
| Invitae | Targeted capture NGS panel | primary panel: GLI2, SHH, SIX3, TGIF1, ZIC2; add-on preliminary-evidence genes: CDON, FOXH1, NODAL, PTCH1 |
| Muenke Laboratory at NIH | Sanger sequencing | SHH, SIX3, TGIF1, ZIC2 |
| Prevention Genetics | Targeted capture NGS panel; CGH | CDON, DLL1, DISP1, FGF8, FOXH1, GAS1, GLI2, NODAL, PTCH1, SHH, SIX3, TDGF1, TGIF1, ZIC2 (TDGF1 and DLL1 not tested with CGH) |
| The University of Chicago Genetic Services Laboratories | Targeted capture NGS panel | CDON, FGFR1, PTCH1, SIX3, TGIF1, FGF8, GLI2, SHH, STIL, ZIC2 |
| OSHU Knight Diagnostic Laboratories | Targeted capture NGS panel | CDON, DISP1, DLL1, FGF8, FOXH1, GAS1, GLI2, NODAL, PTCH1, SHH, SIX3, TGIF1, ZIC2 |
NGS: next generation sequencing
CGH: comparative genomic hybridization
WES: whole exome sequencing
list of laboratories may not be complete
RESULTS AND TESTING RECOMMENDATIONS
Clinical evaluation
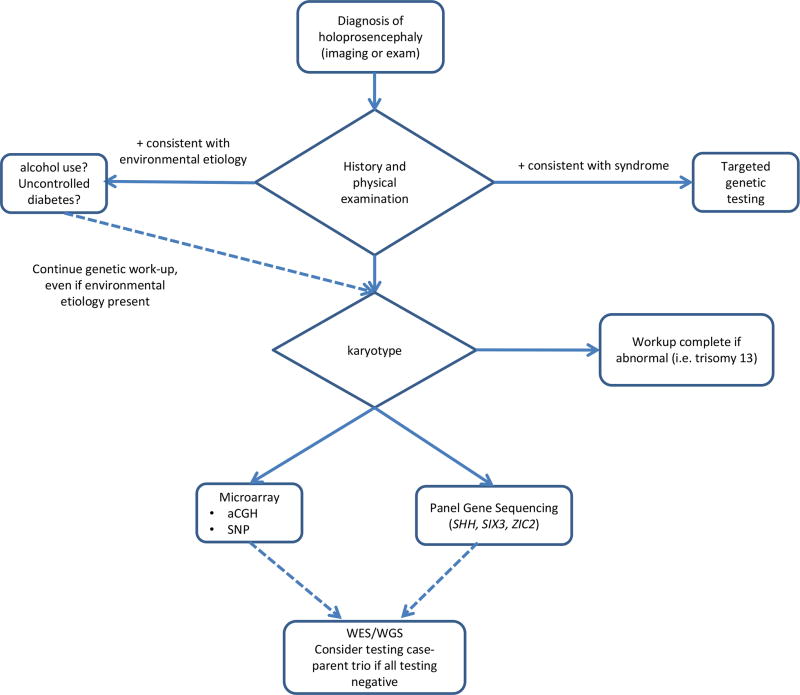
Molecular testing begins with the clinician collecting detailed phenotype information and pursuing genetic/genomic testing based on these findings. Simply ordering an HPE panel (Table I) may not evaluate for syndromic HPE types, which make up over one half of all HPE cases (Kruszka & Muenke, 2018). And as noted above, a pathogenic variant for HPE may not present with a brain anomaly, but as microform HPE (single central maxillary incisor, retinal coloboma, cleft palate/lip, hypotelorism, or microcephaly) (Kruszka et al., 2015). Additionally, it is not uncommon for members of the same family carrying the same variant to present very differently (Kruszka et al., 2015; Stokes et al., 2017). Thus, variable expression and incomplete penetrance in HPE can make diagnosis difficult. Once a clinical diagnosis of HPE spectrum is made, the molecular testing pathway (Figure I) is relatively simple and straightforward.
Figure 1.
Recommended genetic testing for individual with holoprosencephaly. Targeted testing should be done on cases with clincal exam findings consistent with a syndromic etiology. The last step of trio-case whole exome sequencing or whole genome sequencing has not been evaluated critically in the medical literature but is a consideration based on experience of clinician.
Karyotype
In the absence of known family mutations, the first test ordered after either a prenatal or postnatal diagnosis of HPE is a karyotype. Given that over half of all cases of HPE are related to aneuploidy (Kagan, Staboulidou, Syngelaki, Cruz, & Nicolaides, 2010), especially trisomy 13, a karyotype should be the first test. Aneuploidy can also be evaluated with chromosomal microarray and recently NGS (de Ligt et al., 2013; Li et al., 2014; Noll et al., 2016); however, these technologies in their current state may not be able to find structural anomalies such as balanced translocations. A review of chromosomal disorders is reviewed by Kruszka and Muenke in this issue (Kruszka & Muenke, 2018).
Microdeletion testing
In patients with a normal karyotype, genes known to cause HPE (see below) should be evaluated for deletions or duplications by one of the many available assays, such as microarray or multiplex ligation-dependent probe amplification (MLPA). Deletion/duplication testing is available on most commercial HPE testing panels (Table I). Single gene or multi-gene deletions account for a significant fraction of HPE etiologies (Bendavid, Dubourg, et al., 2006; Bendavid, Haddad, et al., 2006; Stokes et al., 2017). Bendavid et al. found that in cases with a normal karyotype and a normal screen for variants in SHH, TGIF1, SIX3, and ZIC2, 16 of 339 patients (4.7%) had a deletion in one of these genes (Bendavid, Haddad, et al., 2006). In another study evaluating fetuses with HPE and normal karyotypes, 13 of 94 (8.5%) fetuses had microdeletions involving SHH, TGIF1, SIX3, or ZIC2 (Bendavid, Dubourg, et al., 2006). If a pathogenic deletion is found in a proband, the parents should also be tested as there are multiple reports of microdeletion inheritance (Stokes et al., 2017).
Single gene testing
HPE is a genetically heterogeneous condition with many implicated genes. Traditionally, testing for HPE single gene variants has focused on Sanger sequencing technology. Recently, NGS has become part of clinical diagnostic practice, allowing for the simultaneous sequencing of multiple genes or entire genomes (Table I). Current NGS capture technologies have problems sequencing complete coding regions and are sensitive to GC content (Meienberg, Bruggmann, Oexle, & Matyas, 2016). Certainly, as technology advances, especially to whole genome sequencing, capture issues will be resolved. Although HPE research and next-generation sequencing is beyond the scope of this guideline, Roessler et al. reviews genomic research in HPE in this issue (Roessler et al., 2018). We have attached our Sanger sequencing procedure and primers for the genes SHH, SIX3, ZIC2, and TGIF1 (Supplementary tables I and II) that have been optimized for GC areas of genes. As noted above, NGS can also be used to evaluate for copy number variations (CNVs) (de Ligt et al., 2013; Li et al., 2014; Noll et al., 2016). Gene panels sequenced at high read depths are particularly valuable at detecting small exon deletions that will be missed with chromosomal microarray (de Ligt et al., 2013).
The most common and consistent genes associated with HPE are SHH, ZIC2, and SIX3. This has been supported by Dubourg et al. in an analysis of 257 patients with HPE that used an NGS panel and found the top genes associated with HPE to be SHH (5.8%), ZIC2 (4.7%), GLI2 (3.1%), SIX3 (2.7%), FGFR1 (2.3%), FGF8 (2.3%), DISP1 (1.2%), DLL1 (1.2%), and SUFU (0.4%) (Dubourg et al., 2016). Many genes that are typically evaluated in HPE panels have not been associated with the classic forebrain malformations of HPE. Some of these genes are associated instead with “HPE-like” phenotypes where there are facial anomalies similar to those found in HPE or midline brain malformations that do not involve the cerebral hemispheres. Below, we review 18 genes that have been associated with HPE or HPE-like disorders and are available on commercial panels (Table I). Based on the clinical presentation of the patient, any one of these genes may be appropriate for testing. Many of the genes below are members of three pathways implicated in HPE including the Shh signaling pathway, Nodal signaling pathway, and the Bmp signaling pathway (Geng & Oliver, 2009). Based on the clinical presentation of the patient, any one of these genes may be appropriate for testing.
SHH, SIX3, ZIC2, and TGIF1
Based on our experience and review of the medical literature, SHH, SIX3, and ZIC2 variants are most commonly associated with HPE. In 200 HPE cases (fetuses and children) with normal karyotypes, 34 (17%) had variants in SHH, SIX3, ZIC2, and TGIF1, with SHH variants being most common, occurring in 13 of the 17 (76%) (Dubourg C, 2004). In another study of fetuses, children and adults with children, 21 variants in 86 cases (24%) were found to have variants in SHH, SIX3, ZIC2 (Paulussen, 2010). Although TGIF1 has been associated with HPE and TGIF1 is on most HPE testing panels (Table I), variants in this gene are rare (Gripp et al., 2000; Keaton et al., 2010). TGIF1 variants were not in the top 10 genes associated with HPE in the Dubourg et al. study of 257 patients. About 10% of all patients with partial monosomy 18p, including deletion of the entire TGIF1 locus, manifest HPE, which indicates that monoallelic mutations in this gene may contribute to pathogenicity, but are not sufficient to cause HPE (Roessler & Muenke, 1998; Turleau, 2008). Interesting, variants in SHH, ZIC2, and SIX3 are distributed throughout all domains of these genes, see Figure 1 in this issue of Roessler et al. (Roessler et al., 2018).
GLI2
More recently, it has become clear that mutations in GLI2 do not tend to result in frank HPE, but instead cause a distinct phenotype that includes pituitary insufficiency and/or polydactyly, as well as subtle facial features. Although found in patients that may have similar facial features as HPE (midface hypoplasia, hypotelorism, and cleft lip/palate), a recent study has now shown that variants in GLI2 do not cause classic HPE, defined as partial or complete failure of forebrain division (Bear et al., 2014). This phenotype associated with GLI2 variants is known as Culler-Jones syndrome (Bear & Solomon, 2015).
CDON
There are a few reports in medical literature connecting CDON variants and HPE. Bae et al. found CDON variants in four unrelated individuals with HPE (Bae et al., 2011). One report has connected Steinfeld syndrome (characterized by HPE and limb anomalies (Kruszka & Muenke, 2018)) to a variant in CDON (Jones et al., 2016); however, this was not a case of classic, but microform HPE.
DISP1
DISP1 is of great interest as it is part of the sonic hedgehog signaling pathway; however, multiple microdeletions and truncating variants involving DISP1 have failed to show brain malformations consistent with HPE (Roessler, Ma, et al., 2009; Roessler & Muenke, 1998; Shaffer et al., 2007). Roessler et al. reported two families with truncating variants in DISP1 and microform HPE (Roessler, Ma, et al., 2009). Dubourg et al. found DISP1 variants in 1.2% of 257 individuals with HPE (Dubourg et al., 2016). Interestingly, microdeletions located on 1q41, where DISP1 is located, have been associated with more serious forms of HPE. Our group recently reported a variant of unknown significance in DISP1 (c.743C>T:p.Ala248Val) in an adult with lobar HPE who inherited this variant from an unaffected mother (Weiss et al., 2018).
NODAL
Roessler et al. evaluated approximately 400 patients with HPE for variants in NODAL and found common variants in two individuals, p.H165R (rs1904589) and p.R302C (rs150819707), with ExAC allele frequencies of 62% and 0.06%, respectively (Lek et al., 2016; Roessler, Pei, et al., 2009). Remarkably, Roessler et al. found that both of these common polymorphisms had greater than 50% reduced bioactivity when using a zebrafish/luciferase assay (Roessler, Pei, et al., 2009). Given the allele frequency of these variants, they are not driver mutations but may be modifiers. Dubourg et al. found no pathogenic variants in NODAL in their large interrogation of 257 patients (Dubourg et al., 2016).
FOXH1
FOXH1 participates in the NODAL signaling pathway and therefore is of interest to HPE research. Variants in FOXH1 are not a commonly found in HPE (Dubourg et al., 2016). Roessler et al. found multiple variants in FOXH1 associated with HPE and demonstrated decreased activity of the protein in zebrafish assays (Roessler et al., 2008); however, there are no further reports in the literature linking FOXH1 and holoprosencephaly.
TDGF1
TDGF1, a member of the nodal signaling pathway, has been described in one patient with HPE (de la Cruz et al., 2002). The TDGF1 variant reported in this case, p.P125L (rs121909501), was found in 18 of 121398 alleles (no homozygotes) in the ExAC data base (Lek et al., 2016).
PTCH1
PTCH1 is a receptor for SHH and acts to repress SHH signaling. Although an attractive candidate for HPE, PTCH1 is not a commonly found variant in HPE (Dubourg et al., 2016). Multiple individuals in two reports (Ming et al., 2002; Ribeiro, Murray, & Richieri-Costa, 2006) have associated PTCH1 variants with HPE.
DLL1
Rarely associated with HPE, Delta-like 1 (DLL1) is a notch ligand and has been shown to be co-expressed with FGF8 in the developing chick forebrain and part of the FGF signaling pathway (Dupe et al., 2011). Dupe et al. additionally reported 4 patients with HPE with microdeletions containing DLL1, and one individual with HPE with a 3bp deletion inherited from an unaffected parent (Dubourg et al., 2016; Dupe et al., 2011).
GAS1
Pineda-Alvarez et all tested 394 individuals with HPE for variants in the coding and flanking regions of GAS1 and found five individuals with missense variants that also these variants impair the physical interaction with SHH (Pineda-Alvarez et al., 2012). In a targeted NGS panel of 257 patients with HPE, Dubourg et al. found no pathogenic variants in GAS1 (Dubourg et al., 2016; Ribeiro, Quiezi, Nascimento, Bertolacini, & Richieri-Costa, 2010).
GLI3
GLI3 variants have been associated Pallister-Hall syndrome, Greig cephalopolysyndactyly syndrome, and polydactyly (Biesecker, 2011; Kang, Graham, Olney, & Biesecker, 1997; Wild et al., 1997). Although GLI3 is part of the SHH signaling pathway, we do not know of variants in GLI3 being associated with classic HPE.
FGFR1
FGFR1 is associated with Hartsfield syndrome and HPE (Hong et al., 2016; Kruszka & Muenke, 2018). Dubourg et al. found 2.3% of the 257 patients with HPE had FGFR1 variants (Dubourg et al., 2016). In Roessler et al. of this issue (Roessler et al., 2018), Figure 1 shows that the distribution of FGFR1 variants span all domains.
SUFU
Like the other genes that encode components of the SHH signaling pathway, SUFU is of interest to HPE research. As noted above, Dubourg found 0.4% of their cohort to have variants in SUFU (Dubourg et al., 2016). To our knowledge, testing for SUFU is not commercially available (Table I).
Possible autosomal recessive genes
A homozygous missense variant FGF8 was identified in a proband from a consanguineous family who presented with semilobar HPE (McCabe et al., 2011). STIL is associated with autosomal recessive HPE in two unrelated consanguineous families (Kakar et al., 2015; Mouden et al., 2015).
The final step in testing: testing family members and genetic counseling
The final part of the molecular work-up is testing family members when a pathogenic variant is discovered in HPE. Although, most HPE variants can have devastating effects, these variants are incompletely penetrant and there are numerous examples of unaffected family members who are mildly affect or not affected (Kruszka et al., 2015; Stokes et al., 2017). Whether a positive or negative result, the results of the testing is complex and requires in depth counseling. In this issue, Hadlely et al. discuss genetic counseling in HPE (Hadley, Kruszka, & Muenke, 2018).
SUMMARY
In this review, we provide a guide (Figure 1) for molecular testing for clinicians taking care of patients affected by HPE. There are a number of factors that affect a clinician’s decision making in the molecular workup of HPE including ease and availability of commercial testing, evidence of pathogenicity of genetic variation, and the phenotype spectrum considered. In this review, we have addressed each of these aspects of testing. Starting with availability of testing (Table I), there are a number of commercial HPE testing panels available and this presents a starting point for molecular work-up. Here we have provided a table of most available testing laboratories at the time of this publication. We have also emphasized that molecular testing begins with a detailed clinical examination and that these panels will not find an etiology in the majority of HPE cases, in contrast to a karyotype, which will provide a molecular diagnosis for over half of all HPE cases. Secondly, this review examines the evidence of pathogenicity for genes associated with HPE, and for many of the genes tested (Table I) the evidence for a causal association with HPE is weak or non-existent. Certainly, there is substantial evidence to support the testing for the common genes (SHH, SIX3, and ZIC2), but evidence is lagging in other genes. And lastly, the phenotype must be considered carefully. There are a number of genes that are considered “HPE spectrum” but are not consistently associated with classic HPE. As an example, GLI2 is widely tested in HPE (Table I), but it is part of the spectrum and not classic HPE. Thus, many of the genes reviewed in this study may be important for phenotypes other than classic HPE.
The challenge with testing recommendations put forth in this article is that technology and genetic research change quickly. Testing for HPE is a moving target and more genes will be added to testing panels as gene discovery continues over the next few years. However, current NGS technology seems to consistently fail to capture certain chromosomal regions in genes such as SHH, SIX3 and ZIC2, making Sanger sequencing still the gold standard for complete coverage of coding regions in the routine HPE four-gene screening. This limitation may apply to other HPE genes as well. Rare variants in approximately 1,500 genes have be associated with diseases that affect brain development or embryonic development (Wright et al., 2015), and we predict that many of these genes will be linked to HPE in the future. As more of the etiology of HPE is understood going forward, our patients and their families will benefit in receiving a genetic diagnosis in the form of prognosis, treatment and genetic counseling.
Supplementary Material
Acknowledgments
P.K., A.F.M., and M.M are supported by the Division of Intramural Research at the National Human Genome Research Institute, NIH.
Biographies
Paul S. Kruszka, MD, MPH, is a board-certified family physician and clinical geneticist at the Medical Genetics Branch, National Human Genome Research Institute, National Institutes of Health (Bethesda, MD). His research interest includes early embryonic errors in brain, craniofacial, and heart development.
Ariel F. Martinez, PhD, is a research biologist at the Medical Genetics Branch, National Human Genome Research Institute, National Institutes of Health (Bethesda, MD) with eight years of experience working on human developmental diseases, including holoprosencephaly (HPE). He is an Active Candidate of the American Board of Medical Genetics and Genomics in the specialty of Clinical Molecular Genetics and Genomics. His current work in Dr. Muenke’s laboratory involves genetics testing of patients with HPE, where he serves as CLIA Laboratory Supervisor. His research is focused on understanding the molecular mechanisms underlying HPE pathogenicity.
Maximilian Muenke, MD, is a board-certified pediatric geneticist and the Branch Chief of the Medical Genetics Branch, National Human Genome Research Institute, National Institutes of Health, Bethesda, MD. He has worked on the genetics of HPE for the past 25 years. His group has discovered numerous genes contributing to HPE, including the first and most well-known gene, Sonic Hedgehog. His other research interests include cardiac disease, craniofacial malformation syndromes, attention-deficit-hyperactivity disorder (ADHD), and the genetics of diverse populations.
Footnotes
The authors have no conflicts of interest to declare.
References
- Bae GU, Domene S, Roessler E, Schachter K, Kang JS, Muenke M, Krauss RS. Mutations in CDON, encoding a hedgehog receptor, result in holoprosencephaly and defective interactions with other hedgehog receptors. Am J Hum Genet. 2011;89(2):231–240. doi: 10.1016/j.ajhg.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear KA, Solomon BD. GLI2 mutations typically result in pituitary anomalies with or without postaxial polydactyly. Am J Med Genet A. 2015;167A(10):2491–2492. doi: 10.1002/ajmg.a.37160. [DOI] [PubMed] [Google Scholar]
- Bear KA, Solomon BD, Antonini S, Arnhold IJ, Franca MM, Gerkes EH, Muenke M. Pathogenic mutations in GLI2 cause a specific phenotype that is distinct from holoprosencephaly. J Med Genet. 2014;51(6):413–418. doi: 10.1136/jmedgenet-2013-102249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendavid C, Dubourg C, Gicquel I, Pasquier L, Saugier-Veber P, Durou MR, David V. Molecular evaluation of foetuses with holoprosencephaly shows high incidence of microdeletions in the HPE genes. Hum Genet. 2006;119(1–2):1–8. doi: 10.1007/s00439-005-0097-6. [DOI] [PubMed] [Google Scholar]
- Bendavid C, Haddad BR, Griffin A, Huizing M, Dubourg C, Gicquel I, Muenke M. Multicolour FISH and quantitative PCR can detect submicroscopic deletions in holoprosencephaly patients with a normal karyotype. J Med Genet. 2006;43(6):496–500. doi: 10.1136/jmg.2005.037176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesecker LG. Polydactyly: how many disorders and how many genes? 2010 update. Dev Dyn. 2011;240(5):931–942. doi: 10.1002/dvdy.22609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz JM, Bamford RN, Burdine RD, Roessler E, Barkovich AJ, Donnai D, Muenke M. A loss-of-function mutation in the CFC domain of TDGF1 is associated with human forebrain defects. Hum Genet. 2002;110(5):422–428. doi: 10.1007/s00439-002-0709-3. [DOI] [PubMed] [Google Scholar]
- de Ligt J, Boone PM, Pfundt R, Vissers LE, Richmond T, Geoghegan J, Hehir-Kwa JY. Detection of clinically relevant copy number variants with whole-exome sequencing. Hum Mutat. 2013;34(10):1439–1448. doi: 10.1002/humu.22387. [DOI] [PubMed] [Google Scholar]
- Dubourg C, Lazaro L, Pasquier L, Bendavid C, Blayau M, Le Duff F, Durou MR, Odent S, David V. Molecular screening of SHH, ZIC2, SIX3, and TGIF genes in patients with features of holoprosencephaly spectrum: Mutation review and genotype-phenotype correlations. Hum Mutat. 2004;24:43–51. doi: 10.1002/humu.20056. [DOI] [PubMed] [Google Scholar]
- Dubourg C, Carre W, Hamdi-Roze H, Mouden C, Roume J, Abdelmajid B, David V. Mutational Spectrum in Holoprosencephaly Shows That FGF is a New Major Signaling Pathway. Hum Mutat. 2016;37(12):1329–1339. doi: 10.1002/humu.23038. [DOI] [PubMed] [Google Scholar]
- Dupe V, Rochard L, Mercier S, Le Petillon Y, Gicquel I, Bendavid C, David V. NOTCH, a new signaling pathway implicated in holoprosencephaly. Hum Mol Genet. 2011;20(6):1122–1131. doi: 10.1093/hmg/ddq556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X, Oliver G. Pathogenesis of holoprosencephaly. J Clin Invest. 2009;119(6):1403–1413. doi: 10.1172/JCI38937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gripp KW, Wotton D, Edwards MC, Roessler E, Ades L, Meinecke P, Elledge SJ. Mutations in TGIF cause holoprosencephaly and link NODAL signalling to human neural axis determination. Nat Genet. 2000;25(2):205–208. doi: 10.1038/76074. [DOI] [PubMed] [Google Scholar]
- Hadley D, Kruszka P, Muenke M. Genetic Counseling and Holoprosencephaly. American Journal of Medical Genetics Part C (Seminars in Medical Genetics) 2018 doi: 10.1002/ajmg.c.31627. [DOI] [PubMed] [Google Scholar]
- Hong S, Hu P, Marino J, Hufnagel SB, Hopkin RJ, Toromanovic A, Muenke M. Dominant-negative kinase domain mutations in FGFR1 can explain the clinical severity of Hartsfield syndrome. Hum Mol Genet. 2016;25(10):1912–1922. doi: 10.1093/hmg/ddw064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones GE, Robertson L, Maniyar A, Shammas C, Phelan MM, Vasudevan PC, Tanteles GA. Microform holoprosencephaly with bilateral congenital elbow dislocation; increasing the phenotypic spectrum of Steinfeld syndrome. Am J Med Genet A. 2016;170(3):754–759. doi: 10.1002/ajmg.a.37511. [DOI] [PubMed] [Google Scholar]
- Kagan KO, Staboulidou I, Syngelaki A, Cruz J, Nicolaides KH. The 11–13-week scan: diagnosis and outcome of holoprosencephaly, exomphalos and megacystis. Ultrasound Obstet Gynecol. 2010;36(1):10–14. doi: 10.1002/uog.7646. [DOI] [PubMed] [Google Scholar]
- Kakar N, Ahmad J, Morris-Rosendahl DJ, Altmuller J, Friedrich K, Barbi G, Borck G. STIL mutation causes autosomal recessive microcephalic lobar holoprosencephaly. Hum Genet. 2015;134(1):45–51. doi: 10.1007/s00439-014-1487-4. [DOI] [PubMed] [Google Scholar]
- Kang S, Graham JM, Jr, Olney AH, Biesecker LG. GLI3 frameshift mutations cause autosomal dominant Pallister-Hall syndrome. Nat Genet. 1997;15(3):266–268. doi: 10.1038/ng0397-266. [DOI] [PubMed] [Google Scholar]
- Keaton AA, Solomon BD, Kauvar EF, El-Jaick KB, Gropman AL, Zafer Y, Muenke M. TGIF Mutations in Human Holoprosencephaly: Correlation between Genotype and Phenotype. Mol Syndromol. 2010;1(5):211–222. doi: 10.1159/000328203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruszka P, Hart RA, Hadley DW, Muenke M, Habal MB. Expanding the phenotypic expression of Sonic Hedgehog mutations beyond holoprosencephaly. J Craniofac Surg. 2015;26(1):3–5. doi: 10.1097/SCS.0000000000001377. [DOI] [PubMed] [Google Scholar]
- Kruszka P, Muenke M. Syndromes Associated with Holoprosencephaly. American Journal of Medical Genetics Part C. Seminars in Medical Genetics. 2018 doi: 10.1002/ajmg.c.31620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, Exome Aggregation C. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Xia Y, Wang C, Tang YT, Guo W, Li J, Li J. Identifying Human Genome-Wide CNV, LOH and UPD by Targeted Sequencing of Selected Regions. PLoS One. 2014;10(4):e0123081. doi: 10.1371/journal.pone.0123081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MJ, Gaston-Massuet C, Tziaferi V, Gregory LC, Alatzoglou KS, Signore M, Dattani MT. Novel FGF8 mutations associated with recessive holoprosencephaly, craniofacial defects, and hypothalamo-pituitary dysfunction. J Clin Endocrinol Metab. 2011;96(10):E1709–1718. doi: 10.1210/jc.2011-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meienberg J, Bruggmann R, Oexle K, Matyas G. Clinical sequencing: is WGS the better WES? Hum Genet. 2016;135(3):359–362. doi: 10.1007/s00439-015-1631-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming JE, Kaupas ME, Roessler E, Brunner HG, Golabi M, Tekin M, Muenke M. Mutations in PATCHED-1, the receptor for SONIC HEDGEHOG, are associated with holoprosencephaly. Hum Genet. 2002;110(4):297–301. doi: 10.1007/s00439-002-0695-5. [DOI] [PubMed] [Google Scholar]
- Mouden C, de Tayrac M, Dubourg C, Rose S, Carre W, Hamdi-Roze H, David V. Homozygous STIL mutation causes holoprosencephaly and microcephaly in two siblings. PLoS One. 2015;10(2):e0117418. doi: 10.1371/journal.pone.0117418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll AC, Miller NA, Smith LD, Yoo B, Fiedler S, Cooley LD, Kingsmore SF. Clinical detection of deletion structural variants in whole-genome sequences. NPJ Genom Med. 2016;1:16026. doi: 10.1038/npjgenmed.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulussen AD, Schrander-Stumpel CT, Tserpelis DC, Spee MK, Stegmann AP, Mancini GM, Brooks AS, Collée M, Maat-Kievit A, Simon ME, van Bever Y, Stolte-Dijkstra I, Kerstjens-Frederikse WS, Herkert JC, van Essen AJ, Lichtenbelt KD, van Haeringen A, Kwee ML, Lachmeijer AM, Tan-Sindhunata GM, van Maarle MC, Arens YH, Smeets EE, de Die-Smulders CE, Engelen JJ, Smeets HJ, Herbergs J. The unfolding clinical spectrum of holoprosencephaly due to mutations in SHH, ZIC2, SIX3 and TGIF genes. Eur J Hum Genet. 2010;18(9):999–1005. doi: 10.1038/ejhg.2010.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda-Alvarez DE, Dubourg C, David V, Roessler E, Muenke M. Current recommendations for the molecular evaluation of newly diagnosed holoprosencephaly patients. Am J Med Genet C Semin Med Genet. 2010;154C(1):93–101. doi: 10.1002/ajmg.c.30253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda-Alvarez DE, Roessler E, Hu P, Srivastava K, Solomon BD, Siple CE, Muenke M. Missense substitutions in the GAS1 protein present in holoprosencephaly patients reduce the affinity for its ligand, SHH. Hum Genet. 2012;131(2):301–310. doi: 10.1007/s00439-011-1078-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro LA, Murray JC, Richieri-Costa A. PTCH mutations in four Brazilian patients with holoprosencephaly and in one with holoprosencephaly-like features and normal MRI. Am J Med Genet A. 2006;140(23):2584–2586. doi: 10.1002/ajmg.a.31369. [DOI] [PubMed] [Google Scholar]
- Ribeiro LA, Quiezi RG, Nascimento A, Bertolacini CP, Richieri-Costa A. Holoprosencephaly and holoprosencephaly-like phenotype and GAS1 DNA sequence changes: Report of four Brazilian patients. Am J Med Genet A. 2010;152A(7):1688–1694. doi: 10.1002/ajmg.a.33466. [DOI] [PubMed] [Google Scholar]
- Roessler E, Hu P, Muenke M. Holoprosencephaly in the genomics era. American Journal of Medical Genetics Part C (Seminars in Medical Genetics) 2018 doi: 10.1002/ajmg.c.31615. [DOI] [PubMed] [Google Scholar]
- Roessler E, Ma Y, Ouspenskaia MV, Lacbawan F, Bendavid C, Dubourg C, Muenke M. Truncating loss-of-function mutations of DISP1 contribute to holoprosencephaly-like microform features in humans. Hum Genet. 2009;125(4):393–400. doi: 10.1007/s00439-009-0628-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Muenke M. Holoprosencephaly: a paradigm for the complex genetics of brain development. J Inherit Metab Dis. 1998;21(5):481–497. doi: 10.1023/a:1005406719292. [DOI] [PubMed] [Google Scholar]
- Roessler E, Ouspenskaia MV, Karkera JD, Velez JI, Kantipong A, Lacbawan F, Muenke M. Reduced NODAL signaling strength via mutation of several pathway members including FOXH1 is linked to human heart defects and holoprosencephaly. Am J Hum Genet. 2008;83(1):18–29. doi: 10.1016/j.ajhg.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Pei W, Ouspenskaia MV, Karkera JD, Velez JI, Banerjee-Basu S, Muenke M. Cumulative ligand activity of NODAL mutations and modifiers are linked to human heart defects and holoprosencephaly. Mol Genet Metab. 2009;98(1–2):225–234. doi: 10.1016/j.ymgme.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer LG, Theisen A, Bejjani BA, Ballif BC, Aylsworth AS, Lim C, Shaikh T. The discovery of microdeletion syndromes in the post-genomic era: review of the methodology and characterization of a new 1q41q42 microdeletion syndrome. Genet Med. 2007;9(9):607–616. doi: 10.1097/gim.0b013e3181484b49. doi:10.1097GIM.0b013e3181484b49. [DOI] [PubMed] [Google Scholar]
- Simonis N, Migeotte I, Lambert N, Perazzolo C, de Silva DC, Dimitrov B, Vilain C. FGFR1 mutations cause Hartsfield syndrome, the unique association of holoprosencephaly and ectrodactyly. J Med Genet. 2013;50(9):585–592. doi: 10.1136/jmedgenet-2013-101603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes B, Berger SI, Hall BA, Weiss K, Martinez AF, Hadley DW, Muenke M. SIX3 deletions and incomplete penetrance in families affected by holoprosencephaly. Congenit Anom (Kyoto) 2017 doi: 10.1111/cga.12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turleau C. Monosomy 18p. Orphanet J Rare Dis. 2008;3:4. doi: 10.1186/1750-1172-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss K, Kruszka P, Guillen Sacoto MJ, Addissie YA, Hadley DW, Hadsall CK, Muenke M. In-depth investigations of adolescents and adults with holoprosencephaly identify unique characteristics. Genet Med. 2018;20(1):14–23. doi: 10.1038/gim.2017.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild A, Kalff-Suske M, Vortkamp A, Bornholdt D, Konig R, Grzeschik KH. Point mutations in human GLI3 cause Greig syndrome. Hum Mol Genet. 1997;6(11):1979–1984. doi: 10.1093/hmg/6.11.1979. [DOI] [PubMed] [Google Scholar]
- Wright CF, Fitzgerald TW, Jones WD, Clayton S, McRae JF, van Kogelenberg M, study, D. D. D Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 2015;385(9975):1305–1314. doi: 10.1016/S0140-6736(14)61705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.