Abstract
AK3 compounds are mitotic-arrest agents that induce high levels of γH2AX during mitosis and apoptosis following release from arrest. We synthesized a potent AK3 derivative, AK306, that induced arrest and apoptosis of the HCT116 colon cancer cell line with an EC50 of ~50 nM. AK306 was active on a broad spectrum of cancer cell lines with total growth inhibition values ranging from ~25 nM to 25 μM. Using biotin and BODIPY-linked derivatives of AK306, binding to clathrin heavy chain (CLTC/CHC) was observed, a protein with roles in endocytosis and mitosis. AK306 inhibited mitosis and endocytosis, while disrupting CHC cellular localization. Cells arrested in mitosis by AK306 showed the formation of multiple microtubule organizing centers consisting of pericentrin (PCNT), γ-tubulin and Aurora A foci, without apparent centrosome amplification. Cells released from AK306 arrest were unable to form bipolar spindles, unlike nocodazole-released cells that reformed spindles and completed division. Like AK306, CHC siRNA knockdown disrupted spindle formation and activated p53. A short term (3 day) treatment of tumor-bearing APC-mutant mice with AK306 increased apoptosis in tumors, but not normal mucosa. These findings indicate that targeting the mitotic CHC complex can selectively induce apoptosis and may have therapeutic value.
STATEMENT OF IMPLICATION
Disruption of clathrin with a small-molecule inhibitor, AK306, selectively induces apoptosis in cancer cells by disrupting bi-polar spindle formation.
Keywords: Colorectal cancer, Spindle assembly, Clathrin Heavy Chain (CHC), Apoptosis, Cell cycle arrest
INTRODUCTION
Mitotic disruption can trigger a number of cell responses, ranging from cell cycle arrest and repair, to senescence, mitotic catastrophe and apoptosis (1,2). How the cell makes this decision is not entirely clear, but it appears to involve the nature of the damage and a range of cell-specific variables. One pathway that can link aberrant mitosis with cell death is the p53 pathway (3–7). Cells arrested in mitosis can activate the ATM-p53 pathway, which can promote apoptosis directly, or after an aberrant cell division. Cells stalled in mitosis can also become more sensitive to death ligands, which may provide an additional means of eliminating cells at risk of undergoing an aberrant division (8,9). A better understanding of how mitotic disruption can lead to apoptosis could provide insight into the mechanisms protecting cells from aneuploidy.
Cancer cells often carry mutations and other defects that debilitate the mitotic checkpoint. Mutations in genes for BUB1, BUBR1, MAD1 and MAD2 can directly affect the ability of cells to sense the spindle assembly checkpoint, whereas mutations in ATM, ATR and p53 can interfere with a cell’s ability to translate this checkpoint into apoptosis (10–17). In contrast to the loss of checkpoint proteins, cancer cells frequently overexpress mitotic kinases such as the Aurora and Polo-like kinases (18–21). The overexpression of these kinases may serve to drive mitosis, even in the presence of cellular damage (22). Aberrant mitoses can result directly in the generation of aneuploid cells. In addition, failed cytokinesis can result in the generation of tetraploid cells, which are at an elevated risk of erroneous chromosome segregation (23–25). Although the mitotic defects in cancer cells can lead to chromosomal instability, these defects may also increase cancer cell sensitivity to agents that interfere with mitosis (26–28).
We have been studying a class of compounds selected for their ability to arrest cells in a mitotic state that transitions to apoptosis with high efficiency. These “AK3” compounds induce a mitotic arrest state with elevated levels of γH2Ax that resolves into a p53-facilitated cell death following release from arrest (8,9,29). These compounds can also enhance ligand-induced cell death in p53-mutant cells through a mechanism that involves a tighter coupling between the death receptors and caspase-8. Although the AK3 compounds have an affinity for tubulin, their apoptotic potency cannot be easily explained by targeting tubulin alone (8); traditional spindle poisons do not induce βH2Ax signaling as robustly as the AK3 compounds and are less apoptotic (29). Since βH2AX levels are increased specifically in mitotically arrested cells, we have proposed that the AK3 compounds cause DNA shearing through aberrant spindle pulling (as previously described)(30), although we cannot rule out other mechanisms such as DNA/chromatin binding or DNA repair enzyme inhibition.
To better understand how this class of molecules works, we generated a series of AK3 derivatives for structure-activity relationship (SAR) studies. Based on these finding, we constructed affinity and fluorescent probe molecules, and found selective binding to clathrin heavy chain (CHC), a protein involved in endocytosis and mitosis. Our findings support a central role of CHC in centrosome complex integrity and reveal a close linkage between CHC-directed centrosome complex maintenance and the regulation of cell death signaling. Given the range of centrosome defects found in cancer cells, our data indicate that CHC-directed centrosome assembly pathways are a potentially fruitful target for novel therapeutic agent development (26,31,32).
MATERIALS AND METHODS
Cell Culture
HCT116 colon cancer cell line was obtained from the American Type Culture Collection (Manassas, VA). The p53-mutant HCT116 cell line was the kind gift of Dr. Bert Vogelstein. The p53-null status of these cells was confirmed by Western blotting. Cells were cultured in McCoy’s 5A medium, with 10% (v/v) fetal bovine serum, non-essential amino acids, and antibiotic/antimycotic (Gibco ThermoFisher, Waltham, MA). The immortalized primary colon cell line, Young Adult Mouse Colonocytes (YAMCs), were a gift from Dr. R Whitehead (Vanderbilt University, Nashville, TN). YAMCs were cultured in RPMI medium containing 5% fetal bovine serum, non-essential amino acids, antibiotic/antimycotic, insulin-transferrin-selenium, and 5 units of murine gamma interferons (Gibco), and were grown at 33°C. Cell lines were expanded and frozen upon receipt with fresh cultures started every 3–4 months. Cells are examined for mycoplasma contamination at least monthly by DAPI staining.
Chemicals and Reagents
Compounds were synthesized as described below. Nocodazole was purchased from Sigma (St. Louis, MO). Transferrin-Alexa Fluor 488 was purchased from Molecular Probes ThermoFisher. All treatments with chemicals were performed approximately 24 h after passage unless otherwise indicated. AK306 was used at a concentration of 100–200 nM, unless otherwise indicated.
Chemical synthesis
All reactions, unless specified, were conducted under an atmosphere of Argon in glassware that had been flame dried. Methylene chloride (CH2Cl2) was bought from Baker Cycle-Tainers. Anhydrous toluene, triethylamine and dimethylformamide (DMF) were purchased from Sigma-Aldrich. N-Boc-piperazine, 2-ethoxy benzoic acid, 2-propoxy benzoic acid, and 1,4-benzodioxan-5-carboxylic acid were purchased from AK Scientific. 2-ethoxybenzoyl chloride and 1-bromo-3(trifluoromethoxy)benzene were bought from Alfa Aesar. 1-bromo-3,5-dichlorobenze was bought from Acros Organics. 2-ethoxy-4-nitro benzoic acid was purchased from Sigma-Aldrich. Boc-5-aminovaleric acid was purchased from Chem-Impex Int’L INC. NHS-Biotin was purchased from APExBIO. BDP FL NHS ester was bought from Lumiprobe Life Science Solutions. Where appropriate, control of temperature was achieved with a Neslab Cryocool CC-100 II immersion cooler, ice-bath or a heated oil bath. Flash chromatography was performed on Silica Gel, 40 microns, 32-63 flash silica and/or -NH2 capped spherical silica gel. High Performance Liquid Chromatography was performed on Phenomenex C18 silica gel column, 5 microns 250 × 4.60 mm, and monitored using the Shimadzu SIL-20AC equipped with a UV detector. Thin layer chromatography was performed on silica gel (Silica Gel 60 F254) glass plates and the compounds were visualized by UV and/or potassium permanganate stain. Details on specific compounds is provided as supplemental information file 2.
Flow cytometry
Cells were analyzed for DNA content by ethanol fixation and staining with propidium iodide as previously described (33). The stained cells were filtered prior to analysis on FACSCalibur flow cytometer (BD Biosciences) using Cell Quest software (BD Biosciences). The data were analyzed using FlowJo (TreeStar Inc.).
Affinity Chromatography and BODIPY Probing
Pierce™ Avidin Agarose (Thermo Scientific) slurry was added to two tubes, washed with 10 column volumes of PBS to remove preservative sodium azide, and suspended in one column volume of Wash Buffer (PBS, 2 mM MgCl2, 0.1% protease inhibitor cocktail, 0.1% Triton X-100, 1 mM DTT). Molar excess of biotinylated AK328 (relative to the avidin bead capacity) was added to the first tube. As a control, an equimolar amount of D-(+)-biotin (Sigma Aldrich) was added to the second tube. The mixtures were then incubated for 1 hour at room temperature to allow binding of biotin to avidin and washed twice with 10 column volumes of PBS to remove unbound material. Whole cell extract of HCT116 cells prepared with M-PER buffer was added to the tubes and the solutions were incubated at 4°C overnight under mild agitation to allow the compound to bind to cellular proteins (~15 mgs of protein per column). The slurries were then packed into two Gravity Flow Poly-Prep Chromatography columns (BIO-RAD) and the flow through was collected. Columns were washed with 10 column volumes of Wash Buffer with the last wash fraction collected to check for efficiency of the washing step. The proteins were eluted with 5 sequential column volumes of the Elution Buffer (PBS, 2 mM MgCl2, 1× protease inhibitor cocktail, 0.1% Triton X-100, 1 mM DTT). For SDS elution, the slurry was heated to 90°C for 5 minutes in the presence of 1% SDS prior to elution. The proteins in each elution fraction were concentrated using PierceTM 3K MWCO PES Protein Concentrators (Thermo Scientific). Before loading in the concentrator chamber, fractions were adjusted to 1% SDS to prevent protein binding and clogging of the membrane. Concentrated eluates, cell extract, flow through, and wash fractions from each column were then analyzed using SDS-PAGE. Proteins were separated on SDS polyacrylamide gels (Bio-Rad) and stained with SYPRO Ruby Red Gel stain (Bio-Rad). Bands were excised from the gel and sent to the Yale/Keck MS & Proteomics Resource facility for mass spectrometry. To confirm specific CHC binding to the column, the column was eluted with 1 μM AK306 (at room temperature) with the resulting fractions analyzed by western blotting using a CHC primary antibody (P1663; Cell Signaling Technology) and a IRDye® 680RD Goat anti-Rabbit secondary antibody for detection (LI-COR Biotechnology, Lincoln, Nebraska). Images were acquired and analyzed using the LiCor Odyssey® CLx Imaging System.
To determine the affinity of purified clathrin for the affinity column, clathrin was purified from mouse liver by differential centrifugation methods as previously described (34). Purified protein (~70 μg) was loaded on to the affinity and control columns as described above, and eluted sequentially with AK306 and 1% SDS. The resulting fractions were analyzed by SDS PAGE and SYPRO gel staining.
In-gel staining using AK327-BODIPY was performed by running 2 μg of purified clathrin, BSA (Sigma) and tubulin (Cytoskeleton, Inc., Denver, CO) on duplicate SDS PAGE gels. One gel was stained with SYPRO. The other was washed in 25 mM Tris, 192 mM glycine buffer to remove SDS, followed by incubation in HKM buffer (25 mM Hepes, pH 7.4, 125 mM potassium acetate, and 5 mM magnesium acetate with 1 mM DTT) with 500 nM AK327-BODIPY overnight at 4°C. BODIPY was then detected by illumination on a UV light box.
Immunofluorescence microscopy
Cells were seeded on glass coverslips coated with 1% gelatin in 24-well plates at 1 × 105 cells per well. After 24 h cells were fixed with 100% ice cold methanol for 5 min at −20°C or 4% paraformaldehyde for 15 min at room temperature followed by permeabilization with 0.5% Triton X-100 in PBS. Next, cells were blocked in 5% serum in PBS and then incubated with primary antibody for 1 h at room temperature. The following antibodies were used for these studies: β-tubulin (E7 monoclonal antibody, Developmental Studies Hybridoma Bank, IA), γ-tubulin (ab11316, Abcam), Aurora A (630938, BD Transduction Laboratories), centrin-3 (PA5-66623, Invitrogen), Clathrin Heavy Chain, s-20 (sc-6579, Santa Cruz Biotechnology), Pericentrin (ab4448, Abcam), Phospho p53, s-15 (9284, Cell Signaling Technology). Appropriate secondary antibodies (Life Technologies or Jackson ImmunoResearch) were used for 45 min incubation. Nuclei were visualized using DAPI (5 μg/ml in PBS; DI306, Life Technologies). Coverslips were mounted on slides using ProLong Gold Antifade Reagent (Life Technologies). For experiments in which cells were stained with AK327-BODIPY, antibody staining was performed first, followed by incubation with 500 nM AK327-BODIPY in PBS and direct imaging (without mounting).
Images were acquired using Nikon A1R confocal laser microscope and NIS-Elements Advanced Research Software (version 4.4, Nikon Instruments Inc.). Image brightness and contrast were modified when appropriate. Background subtraction was accomplished by setting the minimum displayed brightness/contrast value to the value of a region of known background. Quantification of immunostaining was performed using ImageJ/FIJI image processing software (http://fiji.sc/). The pixel value thresholds were set manually at identical ranges. Menu command Analyse/Analyse particles was used to generate the Integrated Density value (integrated fluorescence intensity) of the area of interest, which was then divided by the number of nuclei in the area to obtain Integrated Density value per cell. Calculations were conducted in MS Excel.
Endocytosis Assay
HCT116 cells plated onto glass cover slips were serum starved overnight in McCoy’s media plus 0.5% BSA. Cells were then treated with DMSO or varying concentrations of AK306 dissolved in DMSO for 30 minutes at 37°C. AK306 treatment was limited to 30 minutes to limit the mitotic arrest action of the compound. Media was then removed and fresh media containing 0.5% Transferrin (Tfn) Alexa Fluor® 488 Conjugate (Thermo Fischer) was added. Cells were then placed at 37°C for 30 minutes. After exposure to transferrin, media was removed, cells were washed once with PBS, and then fixed with 4% PFA for 10 minutes. Cells were then imaged as described above. Following background subtraction and image stacking, both DAPI and fluorescent Tfn channels were merged. Images were processed using FIJI. Cells were analyzed using the particle analysis plugin, standard with ImageJ image analysis software (http://rsb.info.nih.gov/ij). Image brightness and contrast was modified with Adobe Photoshop software CS6 (Adobe Systems).
Live cell imaging
Cells were plated on 8-well Nunc Lab-Tek II Chambered Coverglass slides at 2 × 104 cells per well. To visualize microtubules, cells were incubated with CellLight Tubulin-GFP construct (Invitrogen) for 24 h. Time-lapse imaging was performed in a humidified CO2 chamber at 37°C using the Nikon A1R confocal microscope and NIS-Elements software described above. Images were acquired every 45 sec as z-stacks with the spacing between slices 0.3μm. Videos were projected and adjusted for background, brightness/contrast using ImageJ/FIJI software as described above.
siRNA transfection
ON-TARGET plus SMART pool siRNA duplexes against CHC and nontargeting siRNA were obtained from GE Dharmacon (Lafayette, CO). Double reverse siRNA transfection was performed in a 24-well plate using Lipofectamine RNAiMAX (Invitrogen). Briefly: cells were lifted using trypsin-EDTA, diluted in OPTI-MEM medium (Life Technologies), pelleted, and resuspended in OPTI-MEM medium. siRNA duplexes diluted in the same medium to achieve final concentration of 100 nM and 4μl of Lipofectamine RNAiMAX were added to each well. 500 μl of resuspended cells were added on top of the siRNA- Lipofectamine mixture. Six hours later the OPTI-MEM medium was replaced with the regular McCoy’s 5A. After 48 h the same transfection procedure was repeated. The cells were then incubated for 36 h before proceeding with immunofluorescence.
Mouse treatment and tissue analysis
ApcΔ14/+ mice were kindly provided by Dr. Christine Perret at the Universite’ Paris and maintained in the animal facility at UConn Health (35). All mice were maintained in a light-cycled, temperature-controlled room and allowed free access to drinking water and diet ad libitum. All animal experiments were conducted with approval from the Center for Comparative Medicine (CCM) at UConn Health. Genotyping for Apc was performed using tail biopsies (35). Sixteen-week-old male ApcΔ14/+ mice were administered five i.p. injections AK306 or vehicle over the course of 3 days: injections were performed at 10 am and 5 pm on days one and two, and at 10 am on day three. Animals were sacrificed at 3 pm on day 3. Each dose of AK306 was 30 mg/kg in 200 μl. Control animals received 200 μl of vehicle (saline with 20% DMSO cosolvent). Three animals were in the vehicle group and four were in the treated group.
After dosing, intestinal tissue was laid flat on filter paper, fixed in formalin overnight, and switched to 70% ethanol in the morning. Small intestines were rolled, embedded in paraffin, and sectioned onto glass slides (20 μ). For immunostaining, tissue samples were deparaffinized and rehydrated with antigen retrieval performed in a pressure cooker using sodium citrate pH 6.0. Tissues were blocked with 1% donkey serum in PBS, and primary antibodies were applied in incubation buffer (1% BSA, 1% donkey serum, 0.3% Triton X-100, 0.01% sodium azide in PBS). Cy-3 conjugated secondary antibodies were used for detection, followed by DAPI staining, and mounting. Samples were imaged using a Leica SP8 confocal microscope. Z-stack images were captured and processed using ImageJ/FIJI software. Stacks were first separated by color and then averaged before being measured. Each color channel was thresholded using the Yen algorithm. Menu command Analyze particles was used to count the number of stained nuclei.
For TUNEL staining, sections were deparaffinized, rehydrated and stained using the Trevigen TACS TdT kit (Gaithersburg, MD). Tissue sections were counterstained with DAPI and mounted. Samples were imaged using Nikon A1R confocal microscope, stacked and processed with ImageJ/FIJI using the same parameter settings. Selective borders were manually drawn around tumor area, color channels were separated, background was subtracted, images were filtered and automatically thresholded (the Otsu algorithm was used for green channel and the Percentile - for DAPI channel), merged cells were separated using Watershed function, Cells were counted using particle analyzing tool.
Statistical analyses
A Student’s t-test was used for comparing two treatment groups. An analysis of variance test (ANOVA) when comparing more than two groups. A Tukey’s post-hoc test was employed to determine the significance of differences between multiple groups. Significance was calculated at an alpha of 0.05.
RESULTS
SAR Study
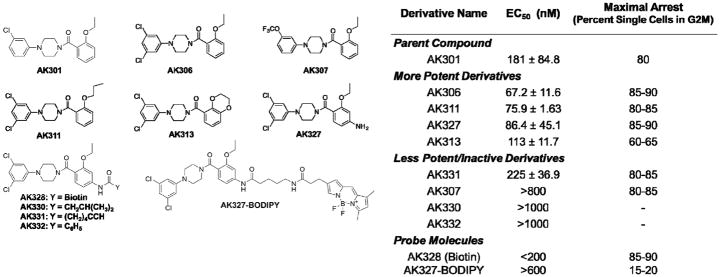
We performed SAR studies on the AK301 compound with the intention of synthesizing molecular probes to identify possible cellular targets. Our goal was to create more potent compounds to: 1) offset the loss in efficacy that typically accompanies the attachment of biotin or fluorescent molecules, and 2) identify sites for attachment of these probe molecules. From previous studies we knew that the position of the chlorine on the phenyl ring and the position and length of the ether group on the benzoyl ring were important for maximal activity (8,29). We therefore focused on these two areas of the molecule. Figure 1 shows the new derivatives generated from this study. A cell cycle assay was used to measure the percentage of cells arrested in G2/M. EC50 values were obtained from titration curves plotting the percent of arrested HCT116 colon cancer cells (Figure 1).
Figure 1.
Compounds synthesized and tested in this study. The table indicates the EC50 concentration required to induce a G2/M arrest in 50% of HCT116 cells, following an overnight/16-hour exposure. The maximum percent of cells arrested in G2/M was determined at approximately 4-times the EC50.
We first manipulated the phenyl ring of AK301. Since having one chloro-group in the meta-position decreased EC50, we decided to incorporate another chloro at the other meta-position. Because the chlorophenyl ring can rotate about its bond to the piperazine, we reasoned that placement of another chlorine could increase the number of configurations that interact with the cellular target. Alternatively, the additional chlorine could result in additional interactions within a hydrophobic space. This hypothesis lead to the development of a dichlorobenzene derivative named AK306. The EC50 was decreased by this modification from ~150 nM to ~75 nM. Our next compound aimed to examine the size of the hydrophobic pocket at this position. We developed a molecule with a trifluoromethyl ether (−OCF3) replacing the chlorine on AK301. OCF3 encompasses more space than a chloro group, but is similarly hydrophobic. Interestingly, the OCF3 derivative, called AK307, was significantly less potent than AK301 with an EC50 near 800 nM. This finding implies that the hydrophobic pocket in which the chlorine is interacting is relatively small.
With the success of AK306 we decided to fix the dichloro phenyl group and continue SAR with the benzoyl ring. From previous work, we knew that the length of the ether contributed to efficacy, with an ethoxy group being more effective than a methoxy group (8,29). To test whether increasing the length of this group increased the activity further, a derivative was synthesized with a propoxy group. This derivative, AK311, did not have significantly higher potency than AK306. We therefore did not pursue a 4 or 5 carbon ether functionality. A second modified benzoyl derivative was synthesized to determine whether decreasing the flexibility of the ethoxy group improved activity. We replaced the ether moiety with 1,4 dioxane, a cyclic acetal. This derivative, named AK313, had a higher EC50 than AK306, indicating that flexibility of the ether was beneficial for activity. We did not pursue further modifications on this site.
To find an attachment site for biotin and other probe molecules we first attached an amino group, which can readily form a peptide bond with NHS esters. We decided to introduce the amino group to the benzoyl ring because it appeared that the dichloro phenyl ring was important for activity. This aniline derivative, called AK327, was tested in HCT116 colon cancer and found to have similar mitotic arresting activity to AK306. Once AK327 was synthesized we began attaching groups that could aid in the biochemical identification of target proteins. AK328, which has a biotin attached to AK327, showed activity in HCT116 colon cancer cells and was therefore deemed useful for target identification purposes.
Finally, we tested the solvent accessibility of the benzoyl group of AK327 at the amino group. The biotin group attached to this site in AK328 includes a flexible carbon linker that may be solvent accessible. To determine how much space was available in this part of the molecule, an isovaleric acid and benzoic acid derivative were synthesized (AK330 and AK332, respectively). Isovaleric acid incorporates steric hindrance due to a branch point two carbons from the acid carbonyl. Interestingly, this derivative was inactive. Next, we tested AK332. Like with AK330, AK332 was also inactive at concentrations up to 4 μM. However, if four methylenes were included before the branch point, activity of the compound was improved (AK331). These data indicated that a carbon linker of at least four methylene groups was necessary for solvent accessibility at this location.
Effect of compounds on G2/M arrest and apoptosis
We moved forward with an analysis of AK306, since it had high potency, good solubility, and reproducible activity. Previous work with this family of compounds indicated that they induced mitotic arrest that resolved into apoptosis with relatively high efficiency following compound withdrawal (29). Cell cycle distribution analyzed by flow cytometry showed that both p53-null and wild type HCT116 cells arrested in G2/M at similar AK306 concentrations (Figure 2A). However, release of cells from arrest by compound withdrawal lead to significantly more apoptosis of p53-normal cells over a 24-hour period following release (Figure 2B). In addition to inducing apoptosis following release, titration experiments with AK306 showed that apoptosis could also be observed at concentrations just below that required to induce a mitotic arrest (Figure 2C). This finding suggests that mitotic arrest and the apoptotic response are not strictly linked and that arrest in mitosis may preclude cells from entering apoptosis.
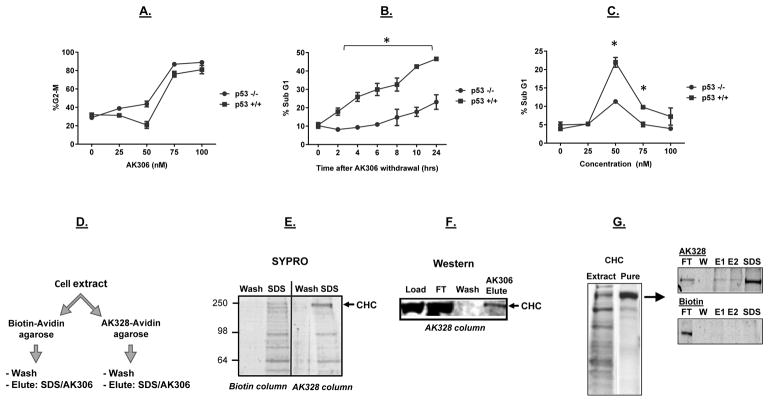
Figure 2.
The impact of p53 on AK306-induced G2/M arrest and apoptosis. A) A similar dose-dependent G2/M arrest in p53 wild type and mutant HCT116 cells. G2/M arrest was determined by flow cytometry. B) HCT116 cells were arrested with AK306, and then released from arrest by compound withdrawal (media change). The p53 wild type cells showed higher levels of apoptosis between 2 and 24 hours, as quantified by cell fragmentation. C) A spike in p53-promoted apoptosis was observed a 50 nM AK306. At higher concentrations, cells arrested in G2/M. Asterisks indicate a significant increase in apoptosis in p53-wild type treated cells (p < 0.01). D) Schematic of affinity chromatography experiments to identify cellular targets. HCT116 cell extracts were passed over an avidin column bound with AK328-biotin or biotin (a negative control). Bound proteins were then eluted with SDS or AK306. E) Columns were run as described in 2D. Shown are the final wash and the SDS elution from a biotin and an AK328-biotin column analyzed by SDS PAGE and SYPRO staining. The 200 kDa band enriched in the AK328-biotin eluate was excised and identified as CHC by LC-MS. F) HCT116 cell extract was loaded on the AK328-biotin column, and then eluted with 1 μM AK306. CHC was detected by western blotting in the load, flow through (FT), final wash and AK306 eluate. G) CHC purified from mouse liver extract was analyzed by SYPRO staining (left panel). Purified CHC was loaded on an AK328-biotin or biotin control columns, eluted with to sequential AK306 washes (1 μM, E1 and E2), followed by an SDS elution. The resulting fractions were analyzed by SDS PAGE followed by SYPRO staining (right panels).
To assess the sensitivity of other cancer cell lines to AK306, the NCI-60 cancer cell line panel was analyzed (supplementary Figure 1). A wide range of sensitivities was observed. Of note, direct lethality was observed in some p53-mutant cell lines (at sub-micromolar concentrations), including HL-60 (leukemia), NCI-H522 (lung cancer), COLO205 and HT-29 (colon cancer), and MDA-MB-435 and M14 (melanoma). These data indicate that cell sensitivity to AK306 is impacted by a range of factors, and does not always require p53 for cell killing.
Identification of clathrin heavy chain as a target of AK3 compounds
We took advantage of our biotin-linked compound, AK328, to identify potential cellular targets. Figure 2D shows the general strategy of our approach. Whole-cell extracts from HCT116 cells were loaded on streptavidin columns coupled to biotin or the biotinylated AK328 compound. Following extensive washing, elution was performed with either SDS or AK306. Figure 2E shows the results when elution was done with SDS and the resulting fractions analyzed on an SDS gel. A number of proteins were found to elute from the columns, but a protein at ~200 kDa appeared to be selectively retained on the AK328 column. This ~200 kDa protein was excised from the gel and analyzed by LC-MS, which identified the192 kDa clathrin heavy chain (CHC). To confirm the presence of CHC, and to obtain additional evidence for specificity, cell extract was again loaded on the AK328 column and elution was performed with AK306 (1 μM). CHC was present in this elution, as determined by immunoblotting (Figure 2F). Finally, to ensure the compounds were binding directly to CHC, CHC was purified from mouse liver and tested for AK328 column binding. As shown in Figure 2G, purified CHC bound to the AK328 column and could be eluted with AK306 (with residual binding eluted with SDS), whereas CHC binding to the biotin control column was not detected.
We also took advantage of a BODIPY linked compound, AK327-BODIPY. Although this compound was insufficiently potent to induce G2/M arrest in the HCT116 cell line, it was able to arrest the YAMC mouse colonocyte cell line at concentrations above ~400 nM (Supplementary Figure 2). We therefore determined the ability of AK327-BODIPY to bind mouse CHC in an in-gel staining experiment. Purified CHC (2 μg) was run on an SDS PAGE gel, along with comparable amounts of BSA and tubulin, as determine by SYPRO Ruby Gel Stain (Figure 3A, upper panel). A duplicate gel was then soaked in Tris-glycine buffer to remove SDS, and then in HKM buffer to stabilize CHC. AK327-BODIPY was then added to the gel to determine protein binding (500 nM, overnight). As shown in Figure 3A, AK327-BODIPY bound to CHC. There was also detectable binding to tubulin, but less to BSA. This finding supports the ability of the AK327-BODIPY to bind CHC. We also determined the cell-staining capabilities of AK327-BODIPY. YAMC cells were first fixed and immunostained for CHC, followed by treatment with 200 nM AK327-BODIPY. As shown in Figure 3B, considerable overlap between CHC and AK327-BODIPY is observed, consistent with AK327-BODIPY binding CHC.
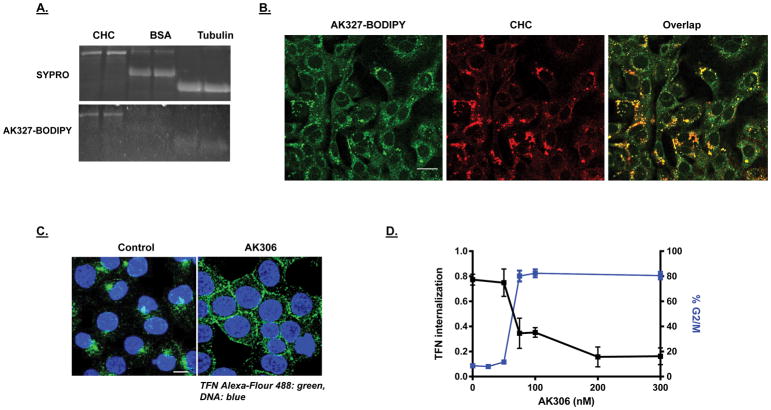
Figure 3.
A) AK3-BODIPY binding to CHC. Purified CHC, BSA and tubulin were run on duplicate SDS gels. The top gel was stained with SYPRO and the bottom gel was incubated with AK327-BODIPY (500 nM overnight in protein renaturation buffer). Both gels were viewed under UV/blue light illumination. B) Colocalization of CHC and AK3-BODIPY staining. YAMC cells were fixed and stained for CHC using Cy3 (red fluorescence). AK3-BODIPY was applied approximately 30 minutes prior to cell imaging. The BODIPY signal was found to concentrate in areas where CHC was highest. The bar shown is 20 μm. C) Endocytosis assay. HCT116 cells were treated with AK306 for thirty minutes, followed by addition of transferrin (TFN) conjugated to Alexa-488 (for thirty minutes). Cells were then fixed and analyzed by confocal microscopy. Bar indicates 20 μm. D) Dose response comparing the inhibition of TFN-Alexa 488 internalization and G2/M arrest.
AK306 effects on cellular CHC
CHC is important for many cell functions, including endocytosis and mitosis (36,37). To determine if AK306 could also affect endocytosis, we analyzed the impact of AK306 on fluorogenic transferrin uptake. As shown in Figure 3C, HCT116 cells take up GFP-labeled transferrin and rapidly transport it to the perinuclear region. AK306 prevented the trafficking of transferrin, leaving much of the signal at the plasma membrane (Figure 3C). An AK306 titration shows that endocytosis disruption and mitotic arrest occurs at similar concentrations (Figure 3D). These data support the possibility that both the mitotic and endocytic effects of AK306 occur via the same target, potentially CHC, although indirect affects through the interaction with other targets cannot yet be ruled out.
We also determined the effect of AK306 on the cellular localization of CHC. Figure 4A shows the results of an immunostaining experiment performed on cells treated with AK306 for 1 and 2 hours. These images reveal a rapid dispersal of the CHC away from the perinuclear region by 1 hour, and a significant reduction in cell staining at 2 hours. These findings indicate a rapid effect of AK306 on CHC cellular localization consistent with a direct binding of CHC. The decrease in immunofluorescent staining was not mirrored by a decrease in CHC levels, as determined by Western blotting (Figure 4B). The effect of AK306 on CHC immunofluorescence is therefore more likely the result of epitope masking. Figure 4C shows the quantification of CHC immunofluorescence (IF) and Western blotting.
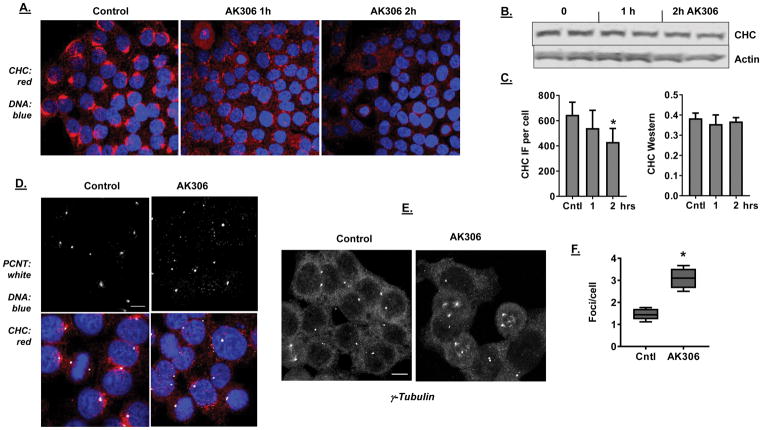
Figure 4.
A) Dispersion and reduction in CHC staining in response to AK306 treatment. HCT116 cells were treated with AK306 for 1 or 2 hours. Treated cells were fixed and stained with CHC antibody. Confocal images show disrupted CHC distribution and reduced CHC staining in AK306-treated cells. B) Western blot of CHC expression after 1 or 2 hours AK306 treatment. C) Quantification of CHC fluorescent staining and expression. The left panel quantifies the data from 4A and shows a reduction in CHC staining after 2 hours of treatment (p < 0.01). The right panel quantifies the Western blot data from 4B. D) Effect of AK306 on pericentrin. HCT 116 cells treated with AK306 (for two hours) were immunostained for pericentrin (PCNT) and analyzed confocal microscopy. Representative images show pericentrin localized at centrosomal foci in control cells, while AK306 caused the formation of multiple additional foci dispersed throughout the cells. Bar indicates 10 μm. E) Representative confocal images of cells stained for γ-tubulin before and after treatment with AK306. Cells were treated as in 4D, with the exception that 50 nM AK306 was used. F) The number of pericentrin foci per cell was quantified using FIJI particle counter. Quantification of large pericentrin foci (at least half the integrated fluorescence intensity of foci in control cells). The quantification indicated a significant increase in the number of foci after AK306 treatment (p < 0.01).
Mitotic effects of AK306
CHC knockdown has been reported to disrupt mitosis through a number of mechanisms, including the assembly and stability of the centrosome complex (38–41). We therefore examined the impact of AK306 on centrosome complex assembly by immunofluorescent staining for pericentrin and γ-tubulin. Treatment of HCT116 cells with AK306 resulted in the fragmentation of pericentrin and γ-tubulin staining, consistent with centrosome complex disruption (Figures 4D and 4E). Whereas control cells displayed normal bipolar spindles with two pericentrin foci or two γ-tubulin foci, AK306 cells typically displayed multiple puncta. Quantification of pericentrin staining indicated a significant increase in the number of pericentrin foci after AK306 treatment (Figure 4F).
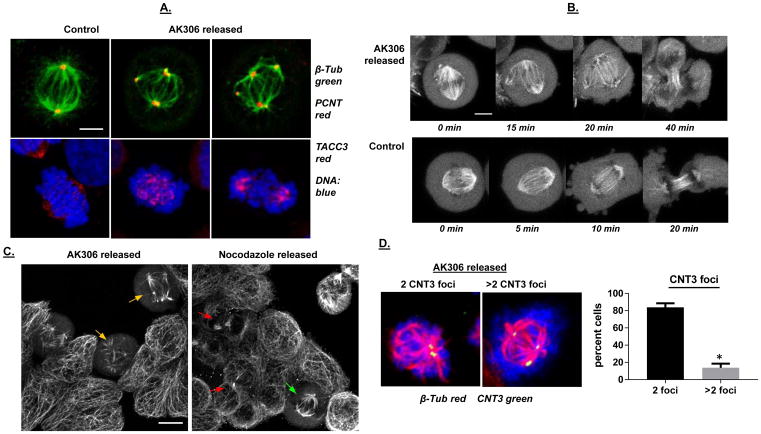
The disrupted centrosome complex assembly was not repaired after AK306 withdrawal. Figure 5A shows the formation of multipolar centrosome assemblies following removal of AK306, based on pericentrin staining. In addition, TACC3, a protein recruited to the centrosome and mitotic spindle in a complex with CHC, was also mislocalized in released cells (Figure 5A). Analysis of cells expressing GFP-tagged β-tubulin likewise showed multipolar spindle formation following AK306 withdrawal (Figure 5B). This response can be compared to cells released from a reversible arrest induced by the microtubule disruptor nocodazole (Figure 5C). Whereas AK306-released cells displayed multipolar spindles (yellow arrows), cells released from nocodazole treatment displayed normal bipolar spindles (green arrows) and midbodies indicative of a successful division (red arrows). Thus, AK306 treated cells appear to be unable to reform bipolar spindle assemblies even after the compound is withdrawn. This defect appears to be the result of acentrosomal MTOC formation rather than centriole over-replication, as centrin-3 staining showed that the majority of cells exiting AK306-induced arrest with aberrant spindles showed only two pairs of centrin-3 foci (Figure 5D).
Figure 5.
A) Analysis of mitotic structures following release from AK306 arrest. Release from AK306 arrest causes ectopic localization of centrosome-related proteins and abnormal mitosis. Confocal images of representative cells were collected after immunostaining for pericentrin and TACC3. Mitotic abnormalities include multipolar spindles, an inability to line up chromosomes at the metaphase plate, and multipolar cell division. The bar shown is 5 μm. B) Time-lapse images showing tripolar mitosis after removal of AK306, compared to normal bipolar mitosis. Cells were transduced with CellLightTubulin-GFP construct to label microtubules. Cells were then treated with AK306 and released. The top row shows the formation of aberrant tripolar spindle and abnormal cytokinesis in an AK306 released cell. The bottom row shows normal mitotic progression in a non-treated cell. The bar indicates 5 μm. C) Impeded cell division after AK306 withdrawal in contrast to nocodazole withdrawal. Cells were treated with either AK306 or nocodazole and then released for two hours. Cells were fixed and stained with antibody to tubulin. Cells released from AK306 formed aberrant multipolar spindles (yellow arrows), whereas cells released from nocodazole were able to achieve metaphase (green arrow) and complete cytokinesis (as indicated by midbody formation; red arrows). The bar shown indicates 10 μm. D) Cell were released from AK306 arrest as in 5C, and then stained for tubulin (red) and centrin-3 (green). Cell with aberrant spindles were then scored for the number of centrin-3 foci. Representative images are shown in the left panels and the quantified data in the graph. Most of the cells showed aberrant spindles in the absence of centrin over-replication.
Effects of CHC knockdown
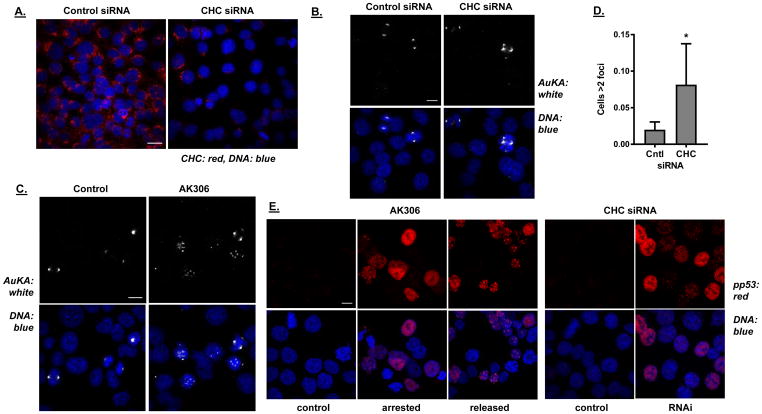
To further examine the relationship between clathrin and the cellular effects of AK306, we utilized siRNA to knockdown CHC expression in HCT116 cells. Figure 6A shows that CHC siRNA reduced CHC expression in HCT116 cells, relative to control, non-targeting siRNA. Cells were then analyzed by Aurora A staining, a protein that normally associates with centrosomes and the mitotic spindle (42). As shown in Figure 6B and 6C, both CHC siRNA and AK306 disrupted Aurora A localization, leading to the formation of multiple foci. The number of Aurora A foci was significantly increased following CHC siRNA knockdown, compared to control siRNA (Figure 6D).
Figure 6.
A) HCT116 cells were transfected with CHC siRNA or control (nontargeting) siRNA. After two rounds of transfection, immunostaining was performed with antibodies to CHC. Efficiency of CHC inhibition was assessed by immunofluorescence visualization using confocal microscope. Left image shows expression of CHC in the control cells and right image shows expression after knockdown with CHC siRNA. The bar shown indicates 20 μm. B) The effect of CHC knockdown on centrosome complex assembly was investigated by immunostaining for centrosome-associated Aurora kinase A (AuKA). Compared to control cells (left panel), an increase in the number of mitotic cells with multiple AuKA foci appear after CHC knockdown (two right panels). The bar shown indicates 20 μm. C) Representative images of AK306-treated cells (50nM) showing multiple Aurora A foci. The bar shown indicates 20 μm. D) Quantification of the increase in AuKA foci in CHC siRNA transfected cells. Cells with more than two AuKA foci were counted using the A1R confocal microscope 60× objective. A significantly greater number of cells with multiple AuKA foci appears after CHC siRNA transfection (Student’s t-test; p < 0.01). E) CHC depletion leads to p53 phosphorylation at serine-15. Left panel shows phosphorylation of p53 in response to AK306 treatments. HCT116 cells were treated with AK306 overnight (16 hours) and then released for 3 hours, as indicated. Cells were then stained for phospho-p53 (ser-15). Nuclear staining is seen in treated and released cells, but not in control cells. Right panels similarly show phospho-p53 nuclear staining in CHC siRNA knockdown cells, which is absent in the non-targeting siRNA controls. The bar shown indicates 10 μm.
Since AK306 induced a p53-dependent cell death, and centrosome disruption has been shown to activate a DNA damage response pathway, we analyzed the level of p53 phosphorylation at serine 15 (the ATM/ATR site) following AK306 treatment (Figure 6E, left panels) and after CHC knockdown (Figure 6E, right panels)(4,5,7,29,43,44). Control cells had low levels of phospho-p53 whereas AK306 treatment resulted in dispersed nuclear staining of phospho-p53. This staining concentrating into nuclear foci following release from AK306. CHC knockdown likewise increased phospho-p53 staining, with dispersed nuclear staining observed. These data are consistent with AK306 and CHC siRNA activating p53 though centrosome disruption.
Cancer-targeting by AK306
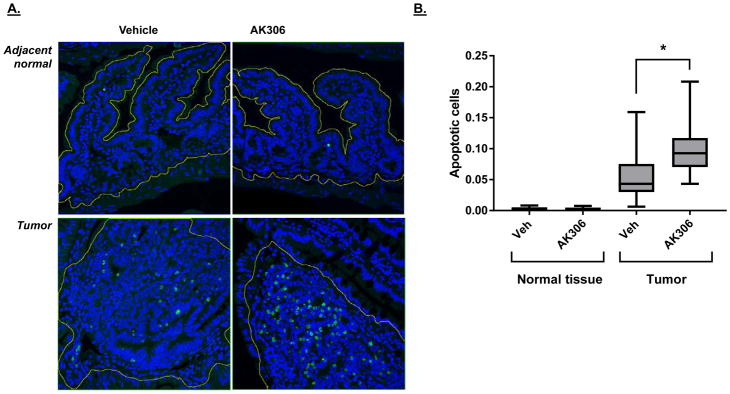
To assess the potential anticancer effect of AK306, sixteen-week-old ApcΔ14/+ mice received five doses of AK306 over the course of three days (30 mg/kg per dose). On day three, mice were euthanized, and intestinal tissue was analyzed for apoptosis by TUNEL staining. As shown in Figure 7, the apoptosis rate in tumors was approximately doubled by AK306 administration, whereas no significant induction of apoptosis was observed in normal tissue. No effect of AK306 on proliferation or mitosis was observed (supplementary Figure 3). Although the apoptosis induction in the tumors was modest, these results support the idea that disrupting the CHC complexes may be a fruitful approach for targeting cancer cells.
Figure 7.
Tumor-bearing ApcΔ14/+ mice were treated with five injections of AK306 (30 mg/kg) over the course of three days. Animals were sacrificed and tissue analyzed for apoptosis using TUNEL. A) Representive images of cancerous and normal tissue from animals treated with AK306 or vehicle (as indicated). Normal mucosa and cancerous tissue was demarcated for the analysis. B) The fraction of TUNEL positive cells in the tissue was quantified using ratio imaging with DAPI staining used to determine total cell number. Significant differences were observed between normal tissue and tumors, and in vehicle and AK306 treated tumors (ANOVA, p < 0.001).
DISCUSSION
In previous work, we described the effects of the AK301 compound that arrests cells in a mitotic state characterized by the formation of multipolar spindles and elevated γH2AX expression (8,29). Upon AK301 withdrawal, cells exit mitotic arrest with a significant portion undergoing apoptosis (up to 40%). Classic spindle poisons vincristine and colchicine were found to be less apoptotic in this treatment procedure (8,29). Although AK301 was found to moderately suppress microtubule polymerization, we reasoned that additional cellular targets were likely being affected to account for the strong apoptotic effect. We therefore performed a focused SAR study to increase compound potency and to design probe compounds for target identification. The most potent derivative synthesized was AK306, which has an EC50 of ~75 nM in HCT116 colon cancer cells. AK306 was then used as a scaffold for biotinylated and BODIPY-labeled probe molecules, with attachments made to a region of the molecule that did not interfere with its mitotic arrest activity. Affinity chromatography revealed that clathrin heavy chain (CHC) was a potential target for the AK3 compounds. In addition, a BODIPY-labeled AK3 compound bound to CHC in an in-gel binding assay. These findings, together with the similar cellular effects caused by AK306 and CHC siRNA knockdown, support CHC as an important cellular target for the AK3 family of compounds. Although other CHC-binding compounds have been described, the AK3 compounds are structurally distinct with nanomolar-level activity (45).
Clathrin plays a number of roles in mitosis. In relation to its membrane transport function, clathrin promotes cytokinesis and cell abscission (46,47). Clathrin also forms a complex with TACC3 and chTOG, two non-motor proteins involved in regulating mitotic spindle dynamics (39,48–51). CHC and TACC3 form a composite microtubule binding domain that serves to stabilize K-fibers attaching the centrosome to the kinetochore (52). In this complex, the trimeric clathrin structure is envisioned to bundle and stabilize microtubules. Interestingly, CHC in conjunction with chTOG also plays a critical role in centrosome complex stability, with knockdown of either protein leading to the formation of extra MTOCs in the cell (39). Our findings indicate that AK306 binding to CHC may lead to a rapid and irreversible fragmentation of the centrosome complex, resulting in the formation of acentrosomal MTOCs (which are observed within an hour of AK306 treatment). Even after AK306 is withdrawn, centrosome components (such as γ-tubulin and PCNT) remain scattered throughout the cell. An important cellular outcome of centrosome complex disruption in this manner is p53 activation, and apoptosis after release from arrest.
The cellular events linking AK306 treatment and CHC siRNA knockdown to p53 phosphorylation/activation and apoptosis are presently unclear. One possibility is that aberrant spindles generated by centrosome complex fragmentation shear the DNA and activate ATM and p53. However, ATM has also been shown to interact directly with the centrosome and can be activated during mitosis (4,53). ATM activated during mitosis phosphorylates p53, which then also localizes to the centrosome and can be released when the centrosome is heavily damaged (4,5,7). Our data suggest that AK306 (and CHC siRNA) activates p53 through a centrosome-based pathway and/or spindle-promoted DNA shearing, since DNA damage signaling is observed primarily during mitosis. However, we cannot rule out AK306 interactions with chromatin DNA or the inhibition of cellular DNA repair enzymes.
Experiments with an APC-mutant model of intestinal cancer showed that cancers are more sensitive to the apoptotic effects of AK306 than normal tissue. The mechanisms accounting for this increased sensitivity are not entirely clear. Like many cancers, APC-mutant cancers have centrosomal defects; the normal APC protein stabilizes the centrosome and facilitates microtubule growth, whereas the truncated APC proteins disrupts these activities (26,38,39,54,55). Since CHC has functions at the centrosome similar to APC, AK306 may provide a second centrosome hit in APC-mutant cancers. Other centrosomal defects in cancer cells may likewise affect their sensitivity to AK306. The HCT116 cells used in this study are APC-normal, but have an activated β-catenin oncogene that promotes the formation of aberrant MT structures consisting of a subset of centrosome proteins, including γ-tubulin (56). The propensity of HCT116 cells to form aberrant MTOC nucleating centers may underlie their sensitivity to AK306. Understanding the cellular defects that sensitize cancer cells to AK306 could provide insight into how centrosome defects might be utilized for therapeutic targeting.
Although our SAR study was limited, we were able to obtain important information regarding the potential binding pocket on CHC. The pocket surrounding the phenyl group is large enough to accommodate chlorines on the ring’s meta positions, but not the larger -OCF3 substituent. The two chlorines of AK306 may bind to separate pockets, or may provide two configurations with which the compound can bind CHC. AK306’s second ring, the benzoyl group, appears to lie in a portion of the protein that is partially solvent accessible. Specifically, the aniline derivative with an NH2 para to the benzoyl carbonyl allowed the attachment of straight carbon chains, but not aryl or isovaleryl substituents, while maintaining nanomolar activity. This portion of the molecule most likely sits at the edge of the protein. AK306 therefore appears to be a good scaffold for the construction of novel molecular probes to study the role of CHC in mitosis and endocytosis. Future studies of the AK306 binding site on CHC may reveal adjacent pockets that could be exploited to increase the compound’s effectiveness or allow for the design of molecules that selectively inhibit clathrin’s mitotic function.
Small molecules that bind CHC should prove useful for studying clathrin’s cellular roles and to provide information on the value of CHC as a therapeutic target. In addition to their potential anticancer activity, CHC inhibitors may also be useful as therapeutics by affecting plasma membrane receptor expression and trafficking. For example, AK306 can increase apoptotic signaling from death receptors by altering their trafficking, providing a potential means for enhancing apoptosis in TNF-rich microenvironments (8,9). CHC-targeting drugs may also be useful for suppressing infection by viruses that utilize clathrin mediated endocytosis for cell entry, or for limiting the up-take of some bacterial toxins. The work described here focuses on the potential of cancer therapeutics, but other applications are also possible.
Supplementary Material
Acknowledgments
Financial Support – R21CA208638 to CG and a SPARK Technology Commercialization Award to CG, DLW and DWR
This work was funded by an NCI grant R21CA208638 to CG and a SPARK Technology Commercialization award to CG, DLW and DWR.
D.L Wright has ownership interest (including patents) in QMD LLC. No potential conflicts of interest were disclosed by the other authors.
Footnotes
Conflict of interest statement – The authors report no potential conflict of interest.
References
- 1.Vitale I, Galluzzi L, Castedo M, Kroemer G. Mitotic catastrophe: a mechanism for avoiding genomic instability. Nature reviews Molecular cell biology. 2011;12(6):385–92. doi: 10.1038/nrm3115. [DOI] [PubMed] [Google Scholar]
- 2.Tovar C, Higgins B, Deo D, Kolinsky K, Liu JJ, Heimbrook DC, et al. Small-molecule inducer of cancer cell polyploidy promotes apoptosis or senescence: Implications for therapy. Cell cycle. 2010;9(16):3364–75. doi: 10.4161/cc.9.16.12732. [DOI] [PubMed] [Google Scholar]
- 3.Kuffer C, Kuznetsova AY, Storchova Z. Abnormal mitosis triggers p53-dependent cell cycle arrest in human tetraploid cells. Chromosoma. 2013;122(4):305–18. doi: 10.1007/s00412-013-0414-0. [DOI] [PubMed] [Google Scholar]
- 4.Shen K, Wang Y, Brooks SC, Raz A, Wang YA. ATM is activated by mitotic stress and suppresses centrosome amplification in primary but not in tumor cells. Journal of cellular biochemistry. 2006;99(5):1267–74. doi: 10.1002/jcb.20848. [DOI] [PubMed] [Google Scholar]
- 5.Oricchio E, Saladino C, Iacovelli S, Soddu S, Cundari E. ATM is activated by default in mitosis, localizes at centrosomes and monitors mitotic spindle integrity. Cell cycle. 2006;5(1):88–92. doi: 10.4161/cc.5.1.2269. [DOI] [PubMed] [Google Scholar]
- 6.Vogel C, Kienitz A, Hofmann I, Muller R, Bastians H. Crosstalk of the mitotic spindle assembly checkpoint with p53 to prevent polyploidy. Oncogene. 2004;23(41):6845–53. doi: 10.1038/sj.onc.1207860. [DOI] [PubMed] [Google Scholar]
- 7.Tritarelli A, Oricchio E, Ciciarello M, Mangiacasale R, Palena A, Lavia P, et al. p53 localization at centrosomes during mitosis and postmitotic checkpoint are ATM-dependent and require serine 15 phosphorylation. Molecular biology of the cell. 2004;15(8):3751–7. doi: 10.1091/mbc.E03-12-0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chopra A, Anderson A, Giardina C. Novel piperazine-based compounds inhibit microtubule dynamics and sensitize colon cancer cells to tumor necrosis factor-induced apoptosis. The Journal of biological chemistry. 2014;289(5):2978–91. doi: 10.1074/jbc.M113.499319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chopra AS, Kuratnik A, Scocchera EW, Wright DL, Giardina C. Identification of novel compounds that enhance colon cancer cell sensitivity to inflammatory apoptotic ligands. Cancer biology & therapy. 2013;14(5):436–49. doi: 10.4161/cbt.23787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mimori K, Inoue H, Alder H, Ueo H, Tanaka Y, Mori M. Mutation analysis of hBUB1, human mitotic checkpoint gene in multiple carcinomas. Oncology reports. 2001;8(1):39–42. [PubMed] [Google Scholar]
- 11.Gemma A, Seike M, Seike Y, Uematsu K, Hibino S, Kurimoto F, et al. Somatic mutation of the hBUB1 mitotic checkpoint gene in primary lung cancer. Genes, chromosomes & cancer. 2000;29(3):213–8. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1027>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 12.Ohshima K, Haraoka S, Yoshioka S, Hamasaki M, Fujiki T, Suzumiya J, et al. Mutation analysis of mitotic checkpoint genes (hBUB1 and hBUBR1) and microsatellite instability in adult T-cell leukemia/lymphoma. Cancer letters. 2000;158(2):141–50. doi: 10.1016/s0304-3835(00)00512-7. [DOI] [PubMed] [Google Scholar]
- 13.Yamaguchi K, Okami K, Hibi K, Wehage SL, Jen J, Sidransky D. Mutation analysis of hBUB1 in aneuploid HNSCC and lung cancer cell lines. Cancer letters. 1999;139(2):183–7. doi: 10.1016/s0304-3835(99)00035-x. [DOI] [PubMed] [Google Scholar]
- 14.Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, et al. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392(6673):300–3. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 15.Schuyler SC, Wu YF, Kuan VJ. The Mad1-Mad2 balancing act--a damaged spindle checkpoint in chromosome instability and cancer. Journal of cell science. 2012;125(Pt 18):4197–206. doi: 10.1242/jcs.107037. [DOI] [PubMed] [Google Scholar]
- 16.Eyfjord JE, Bodvarsdottir SK. Genomic instability and cancer: networks involved in response to DNA damage. Mutation research. 2005;592(1–2):18–28. doi: 10.1016/j.mrfmmm.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 17.Motoyama N, Naka K. DNA damage tumor suppressor genes and genomic instability. Current opinion in genetics & development. 2004;14(1):11–6. doi: 10.1016/j.gde.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi T, Sano B, Nagata T, Kato H, Sugiyama Y, Kunieda K, et al. Polo-like kinase 1 (PLK1) is overexpressed in primary colorectal cancers. Cancer science. 2003;94(2):148–52. doi: 10.1111/j.1349-7006.2003.tb01411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, et al. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nature genetics. 1998;20(2):189–93. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 20.Bischoff JR, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. The EMBO journal. 1998;17(11):3052–65. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weichert W, Kristiansen G, Schmidt M, Gekeler V, Noske A, Niesporek S, et al. Polo-like kinase 1 expression is a prognostic factor in human colon cancer. World journal of gastroenterology : WJG. 2005;11(36):5644–50. doi: 10.3748/wjg.v11.i36.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marumoto T, Hirota T, Morisaki T, Kunitoku N, Zhang D, Ichikawa Y, et al. Roles of aurora-A kinase in mitotic entry and G2 checkpoint in mammalian cells. Genes to cells : devoted to molecular & cellular mechanisms. 2002;7(11):1173–82. doi: 10.1046/j.1365-2443.2002.00592.x. [DOI] [PubMed] [Google Scholar]
- 23.Selmecki AM, Maruvka YE, Richmond PA, Guillet M, Shoresh N, Sorenson AL, et al. Polyploidy can drive rapid adaptation in yeast. Nature. 2015;519(7543):349–52. doi: 10.1038/nature14187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Current opinion in genetics & development. 2007;17(2):157–62. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 25.Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437(7061):1043–7. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 26.Harrison LE, Bleiler M, Giardina C. A look into centrosome abnormalities in colon cancer cells, how they arise and how they might be targeted therapeutically. Biochemical pharmacology. 2018;147:1–8. doi: 10.1016/j.bcp.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olziersky AM, Labidi-Galy SI. Clinical Development of Anti-mitotic Drugs in Cancer. Advances in experimental medicine and biology. 2017;1002:125–52. doi: 10.1007/978-3-319-57127-0_6. [DOI] [PubMed] [Google Scholar]
- 28.Penna LS, Henriques JAP, Bonatto D. Anti-mitotic agents: Are they emerging molecules for cancer treatment? Pharmacology & therapeutics. 2017;173:67–82. doi: 10.1016/j.pharmthera.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 29.Chopra A, Bond MJ, Bleiler M, Yeagley M, Wright D, Giardina C. Efficient Activation of Apoptotic Signaling during Mitotic Arrest with AK301. PloS one. 2016;11(4):e0153818. doi: 10.1371/journal.pone.0153818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guerrero AA, Gamero MC, Trachana V, Futterer A, Pacios-Bras C, Diaz-Concha NP, et al. Centromere-localized breaks indicate the generation of DNA damage by the mitotic spindle. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(9):4159–64. doi: 10.1073/pnas.0912143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cosenza MR, Kramer A. Centrosome amplification, chromosomal instability and cancer: mechanistic, clinical and therapeutic issues. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2016;24(1):105–26. doi: 10.1007/s10577-015-9505-5. [DOI] [PubMed] [Google Scholar]
- 32.Gonczy P. Centrosomes and cancer: revisiting a long-standing relationship. Nature reviews Cancer. 2015;15(11):639–52. doi: 10.1038/nrc3995. [DOI] [PubMed] [Google Scholar]
- 33.Verma R, Rigatti MJ, Belinsky GS, Godman CA, Giardina C. DNA damage response to the Mdm2 inhibitor nutlin-3. Biochemical pharmacology. 2010;79(4):565–74. doi: 10.1016/j.bcp.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hood FE, Williams SJ, Burgess SG, Richards MW, Roth D, Straube A, et al. Coordination of adjacent domains mediates TACC3-ch-TOG-clathrin assembly and mitotic spindle binding. The Journal of cell biology. 2013;202(3):463–78. doi: 10.1083/jcb.201211127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Colnot S, Niwa-Kawakita M, Hamard G, Godard C, Le Plenier S, Houbron C, et al. Colorectal cancers in a new mouse model of familial adenomatous polyposis: influence of genetic and environmental modifiers. Laboratory investigation; a journal of technical methods and pathology. 2004;84(12):1619–30. doi: 10.1038/labinvest.3700180. [DOI] [PubMed] [Google Scholar]
- 36.Fu W, Jiang Q, Zhang C. Novel functions of endocytic player clathrin in mitosis. Cell research. 2011;21(12):1655–61. doi: 10.1038/cr.2011.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Royle SJ. The cellular functions of clathrin. Cellular and molecular life sciences : CMLS. 2006;63(16):1823–32. doi: 10.1007/s00018-005-5587-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thakur HC, Singh M, Nagel-Steger L, Prumbaum D, Fansa EK, Gremer L, et al. Role of centrosomal adaptor proteins of the TACC family in the regulation of microtubule dynamics during mitotic cell division. Biological chemistry. 2013;394(11):1411–23. doi: 10.1515/hsz-2013-0184. [DOI] [PubMed] [Google Scholar]
- 39.Foraker AB, Camus SM, Evans TM, Majeed SR, Chen CY, Taner SB, et al. Clathrin promotes centrosome integrity in early mitosis through stabilization of centrosomal ch-TOG. The Journal of cell biology. 2012;198(4):591–605. doi: 10.1083/jcb.201205116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spiro Z, Thyagarajan K, De Simone A, Trager S, Afshar K, Gonczy P. Clathrin regulates centrosome positioning by promoting acto-myosin cortical tension in C. elegans embryos. Development. 2014;141(13):2712–23. doi: 10.1242/dev.107508. [DOI] [PubMed] [Google Scholar]
- 41.Olszewski MB, Chandris P, Park BC, Eisenberg E, Greene LE. Disruption of clathrin-mediated trafficking causes centrosome overduplication and senescence. Traffic. 2014;15(1):60–77. doi: 10.1111/tra.12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dutertre S, Descamps S, Prigent C. On the role of aurora-A in centrosome function. Oncogene. 2002;21(40):6175–83. doi: 10.1038/sj.onc.1205775. [DOI] [PubMed] [Google Scholar]
- 43.Kramer A, Lukas J, Bartek J. Checking out the centrosome. Cell cycle. 2004;3(11):1390–3. doi: 10.4161/cc.3.11.1252. [DOI] [PubMed] [Google Scholar]
- 44.Morgan SE, Kastan MB. p53 and ATM: cell cycle, cell death, and cancer. Advances in cancer research. 1997;71:1–25. doi: 10.1016/s0065-230x(08)60095-0. [DOI] [PubMed] [Google Scholar]
- 45.von Kleist L, Stahlschmidt W, Bulut H, Gromova K, Puchkov D, Robertson MJ, et al. Role of the clathrin terminal domain in regulating coated pit dynamics revealed by small molecule inhibition. Cell. 2011;146(3):471–84. doi: 10.1016/j.cell.2011.06.025. [DOI] [PubMed] [Google Scholar]
- 46.Smith CM, Chircop M. Clathrin-mediated endocytic proteins are involved in regulating mitotic progression and completion. Traffic. 2012;13(12):1628–41. doi: 10.1111/tra.12001. [DOI] [PubMed] [Google Scholar]
- 47.Boucrot E, Kirchhausen T. Endosomal recycling controls plasma membrane area during mitosis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(19):7939–44. doi: 10.1073/pnas.0702511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Royle SJ. The role of clathrin in mitotic spindle organisation. Journal of cell science. 2012;125(Pt 1):19–28. doi: 10.1242/jcs.094607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Booth DG, Hood FE, Prior IA, Royle SJ. A TACC3/ch-TOG/clathrin complex stabilises kinetochore fibres by inter-microtubule bridging. The EMBO journal. 2011;30(5):906–19. doi: 10.1038/emboj.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fu W, Tao W, Zheng P, Fu J, Bian M, Jiang Q, et al. Clathrin recruits phosphorylated TACC3 to spindle poles for bipolar spindle assembly and chromosome alignment. Journal of cell science. 2010;123(Pt 21):3645–51. doi: 10.1242/jcs.075911. [DOI] [PubMed] [Google Scholar]
- 51.Lin CH, Hu CK, Shih HM. Clathrin heavy chain mediates TACC3 targeting to mitotic spindles to ensure spindle stability. The Journal of cell biology. 2010;189(7):1097–105. doi: 10.1083/jcb.200911120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheeseman LP, Harry EF, McAinsh AD, Prior IA, Royle SJ. Specific removal of TACC3-ch-TOG-clathrin at metaphase deregulates kinetochore fiber tension. Journal of cell science. 2013;126(Pt 9):2102–13. doi: 10.1242/jcs.124834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang S, Hemmerich P, Grosse F. Centrosomal localization of DNA damage checkpoint proteins. Journal of cellular biochemistry. 2007;101(2):451–65. doi: 10.1002/jcb.21195. [DOI] [PubMed] [Google Scholar]
- 54.Lui C, Mok MT, Henderson BR. Characterization of Adenomatous Polyposis Coli Protein Dynamics and Localization at the Centrosome. Cancers (Basel) 2016;8(5) doi: 10.3390/cancers8050047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lui C, Ashton C, Sharma M, Brocardo MG, Henderson BR. APC functions at the centrosome to stimulate microtubule growth. The international journal of biochemistry & cell biology. 2016;70:39–47. doi: 10.1016/j.biocel.2015.10.028. [DOI] [PubMed] [Google Scholar]
- 56.Bahmanyar S, Guiney EL, Hatch EM, Nelson WJ, Barth AI. Formation of extra centrosomal structures is dependent on beta-catenin. Journal of cell science. 2010;123(Pt 18):3125–35. doi: 10.1242/jcs.064782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.