Summary
Skeletal muscles undergo atrophy in response to diseases and aging. Here we report mitofusin 2 (Mfn2) acts as a dominant suppressor of neuromuscular synaptic loss to preserve skeletal muscles. Mfn2 is reduced in spinal cords of transgenic SOD1G93A and aged mice. Through preserving neuromuscular synapses, increasing neuronal Mfn2 prevents skeletal muscle wasting in both SOD1G93A and aged mice, whereas deletion of neuronal Mfn2 produces neuromuscular synaptic dysfunction and skeletal muscle atrophy. Neuromuscular synaptic loss after sciatic nerve transection can also be alleviated by Mfn2. Mfn2 coexists with calpastatin largely in mitochondria-associated membranes (MAMs) to regulate its axonal transport. Genetic inactivation of calpastatin abolishes Mfn2-mediated protection of neuromuscular synapses. Our results suggest that as a potential key component of a novel and heretofore unrecognized mechanism of cytoplasmic protein transport, Mfn2 may play a general role in preserving neuromuscular synapses, and serve as a common therapeutic target for skeletal muscle atrophy.
Keywords: Mfn2, Calpastatin, mitochondria-associated membranes, axonal transport, mitochondria, neuromuscular synapse, amyotrophic lateral sclerosis, sarcopenia, nerve injury, skeletal muscle atrophy
Introduction
Amyotrophic lateral sclerosis (ALS) is the most common neuromuscular disease primarily characterized by progressive motor neuron degeneration and skeletal muscle wasting, while sarcopenia is an aging-associated condition showing gradual loss of skeletal muscle mass and strength. Currently, there is no cure or effective treatment for skeletal muscle loss in ALS or sarcopenia. Skeletal muscles are innervated and supplied by the axon branches of motor neurons in the spinal cord. As a prominent feature, the decline in neuromuscular junctions (NMJs), the communications between motor neuron axonal terminals (neuromuscular synapses) and muscle fibers, has long been implicated in the onset and progression of both ALS and sarcopenia (Deschenes, 2011; Dupuis and Loeffler, 2009). However, the cellular and molecular mechanisms underlying the alterations in NMJs in ALS and during aging are largely unknown.
Mitofusin2 (Mfn2) is a conserved dynamin-like GTPase protein localized in the mitochondrial outer membrane regulating mitochondrial fusion (Chen et al., 2003). Mfn2 has also been reported to be present in the endoplasmic reticulum (ER) or mitochondria-associated membranes (MAMs) and to regulate ER and mitochondria tethering (de Brito and Scorrano, 2008; Sebastian et al., 2012; Sugiura et al., 2013), autophagosome formation (Hailey et al., 2010) and autophagosome-lysosome fusion (Zhao et al., 2012). Mutations in Mfn2, rather than other mitochondrial fusion regulators such as Mfn1 or OPA1, are directly associated with neuromuscular diseases, i.e. Charcot Marie Tooth type 2A (CMT2A) diseases, that are also characterized by neuromuscular dysfunction and skeletal muscle loss (Chen et al., 2003). Although previous studies demonstrated the essential role of Mfn2 for survival of Purkinje cells (Chen et al., 2007) and dopaminergic neurons (Pham et al., 2012), the physiological role of Mfn2 in the regulation of neuromuscular function has not been investigated.
Results
Mfn2 deficiency in motor neurons induces neuromuscular synaptic loss and skeletal muscle atrophy
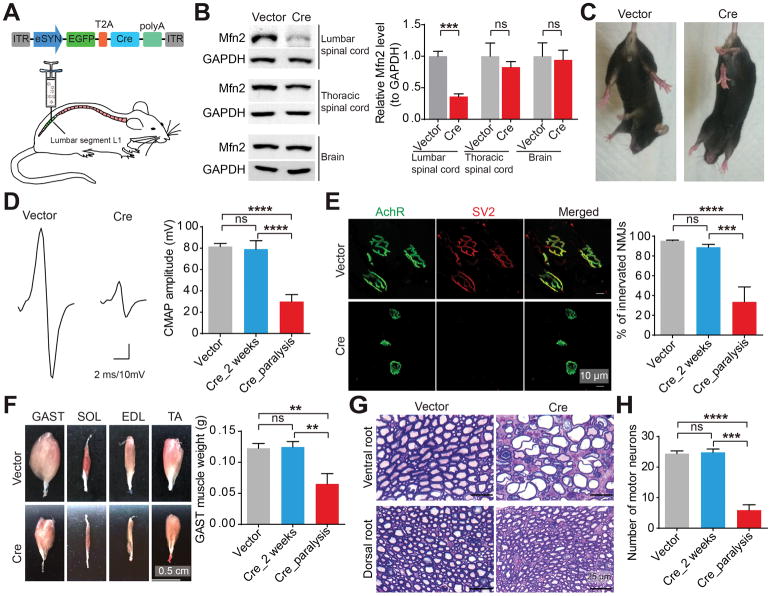
In light of the previous study implicating the dispensable role of Mfn2 for skeletal muscle integrity (Chen et al., 2010), we first examined the impact of Mfn2 loss in spinal cord motor neurons. To bypass embryonic lethality (Chen et al., 2003) and restrict the ablation of Mfn2 to motor regions as described before (Raoul et al., 2005), we bilaterally injected adeno-associated virus serotype 1 encoding Cre recombinase under the neuron specific promoter eSYN (AAV1-Cre) into the lumbar spinal cord of adult Mfn2 floxed mice (Mfn2fl/fl mice) (Chen et al., 2007) (Figures 1A, S1A and S1B). The intraspinal delivery of AAV1-Cre lead to significant loss of Mfn2 in the lumbar spinal cord but not the thoracic spinal cord or the brain (Figure 1B). 3 weeks after injection, compared to mice injected with AAV1 encoding green fluorescent protein only (AAV1-GFP), Cre-injected mice began to display hindlimb clasping, foot-dragging and paralysis (Figures 1C, S1C, S1D and Movies S1–4), characteristic signs of motor deficits. Electrophysiological recording in gastrocnemius (GAST) muscles with paralysis showed reduced compound muscle action potential (CMAP) evoked by supramaximal stimulation of sciatic nerves (Figure 1D), indicating the attenuated NMJ function in response to Mfn2 reduction. Consistently, paralyzed mice showed significant NMJ denervation and atrophy of hindlimb skeletal muscles innervated by motor neurons (Figures 1E and 1F).
Figure 1. Mfn2 deficiency in motor neurons results in marked neuromuscular synapse loss and skeletal muscle atrophy.
(A) Schematic diagram of lumbar spinal cord injection of AAV1-Cre in Mfn2fl/fl mice. ITR, inverted terminal repeats; eSYN, a hybrid promoter consisting of cytomegalovirus enhancer and human Synapsin I promoter; EGFP, enhanced green fluorescent protein; T2A; thoseaasigna virus 2A peptide sequence; Cre, recombinase. (B–H) Representative western blot detection of Mfn2 in lumbar spinal cord, thoracic spinal cord and brain (B, n = 3 or 4 mice per group ), tail suspension test (C), images and quantification of CMAPs evoked by supramaximal stimulation of sciatic nerves (D, n = 9, 6 and 6 mice per group), images and quantification of NMJ innervation (E, green, AchR stained by α-bungarotoxin for motor endplates; red, SV2 for neuromuscular synapse, n = 5, 6 and 3 mice per group), images and quantification of hindlimb skeletal muscles (F, n = 7, 6 and 3 mice per group), toluidine blue staining of Epon-embedded sections of L3–5 dorsal and ventral nerve roots (G, quantification shown in Figure S1E) and quantification of lumbar spinal cord motor neurons (H) of 4–6 month old Mfn2fl/fl mice at 2 weeks or the paralyzed stage after injection. Vector: AAV1-GFP (3–4 weeks after injection); Cre: AAV1-Cre. Soleus (SOL), extensor digitorum longus (EDL) and tibialis anterior (TA) muscles. Equal protein amounts of 20 μg were loaded for all blots. Data are means ± s.e.m., representative of triplicate experiments. Student’s t test or one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. **P < 0.01, ***P < 0.001, ****P < 0.0001. ns, not significant.
Further histological analysis revealed substantial loss of motor neuron axons and cell bodies but very mild change of sensory neuron axons in paralyzed Cre-injected mice (Figures 1G, 1H and S1E). At week 2 post-injection, i.e. at least 1 week prior to onset of paralysis, NMJ function and innervation, skeletal muscle weight and motor neuron number in Cre-injected mice were all comparable to vector-injected mice even though Mfn2 was reduced in motor neuron cell bodies (Figures 1D–F, 1H, S1F and S1G), implying the likely concurrent and late loss of motor neurons and neuromuscular synapses in the paralysis stage. As Mfn2 is a key regulator of mitochondrial morphology, we also investigated mitochondria in neuromuscular synapses by co-injecting AAV1-Cre and AAV1-mitoDsRed encoding a mitochondrial specifically localized red fluorescent protein into the lumbar spinal cord of Mfn2fl/fl mice. After paralysis, AAV1-Cre caused substantial mitochondrial depletion and fragmentation in neuromuscular synapses or neurites nearby them (Figure S1H). 2 weeks after injection, the intensity of mitoDsRed fluorescence was only strong enough to label mitochondria in spinal cord motor neurons but not neuromuscular synapses. As expected, both spinal cord motor neuron cell bodies and proximal motor axons exhibited significant mitochondrial fragmentation well preceding paralysis (Figure S1I).
Forced expression of Mfn2 in neurons abolishes neuromuscular synaptic loss and skeletal muscle atrophy in transgenic mice expressing SOD1 G93A
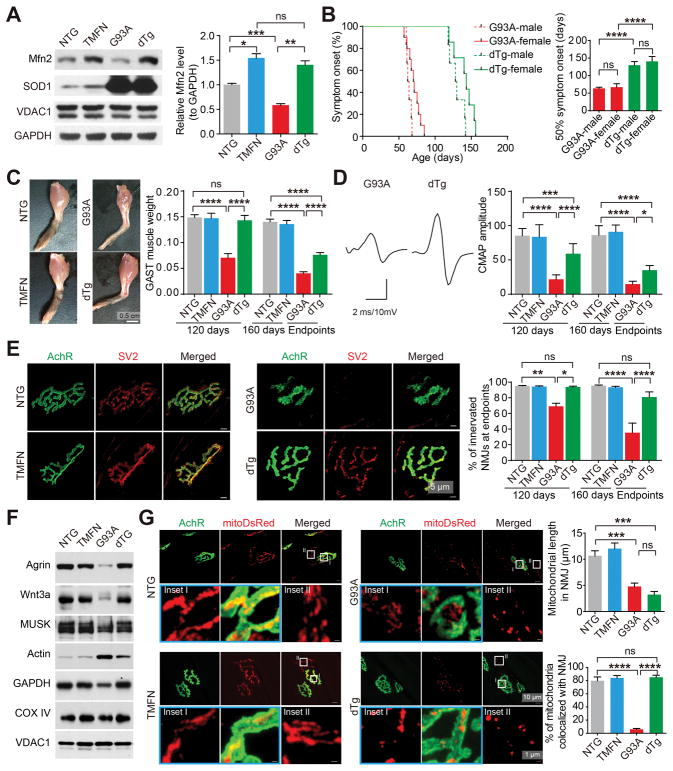
Transgenic SOD1 G93A mice (G93A mice), ubiquitously expressing ALS-associated enzyme superoxide dismutase 1, are the most widely used mouse model associated with neuromuscular synaptopathy. This mouse model has an onset of symptoms at ~60 days and the survival endpoint at ~150 days. Compared with non-transgenic wild type mice (NTG mice), Mfn2 was significantly decreased in spinal cords but not skeletal muscles or brains of G93A mice, and the reduction of Mfn2 in spinal cord motor neurons coincided with the onset of symptoms (Figures S2A and S2B). Consistent with this, ALS patients showed reduced expression of Mfn2 in the spinal cords (Figures S2C and S2D). We then tested whether neuronal Mfn2 upregulation could preserve the neuromuscular function in G93A mice by crossing G93A mice with transgenic mice with the forced expression of Mfn2 specifically in neurons (TMFN mice) (Wang et al., 2015) to obtain double transgenic (dTg) mice. Two independent lines of TMFN mice with similar transgenic expression were tested and there was no phenotypic difference between them. Mfn2 was confirmed to be upregulated in the spinal cords but not hindlimb skeletal muscles of dTg mice even at the disease endpoints (Figures 2A and S2E). There was no statistically significant difference between males and females for any parameter other than body weight (data not shown). Surprisingly, despite similar survival time, body weight and overall appearance between G93A and dTg mice at endpoints (Figures S2F–H), symptom onset determined by skeletal muscle weakness was substantially delayed in dTg mice, with a mean extension of more than 60 days (Figure 2B). Remarkably, endpoint dTg mice displayed well preserved hindlimb skeletal muscles (Figures 2C and S2I). At 120-days old, a very late stage immediately before the paralysis endstage, while G93A mice exhibited significant body weight loss, impaired grip strength, gait abnormalities and muscle atrophy (Figure 2C, S2H and S2J–L), dTg mice showed no change in these parameters compared with NTG or TMFN mice.
Figure 2. Mfn2 upregulation in motor neurons dramatically delays symptom onset and abolishes skeletal muscle atrophy and neuromuscular synapse loss in G93A mice.
(A) Representative western blot and quantification of Mfn2 levels in spinal cords of endpoint G93A and dTg mice and aged-matched 160-day old NTG and TMFN mice (n = 4 mice per group). (B) Age of disease onset determined by hindlimb weakness for G93A (female = 14, male = 6) and dTg mice (female = 6, male = 7). (C) Representative images of endpoint skeletal muscles and quantification of GAST muscle weight (n=6, 8, 5 and 6 mice per group for 120-day old; n=13, 11, 16 and 13 mice per group for 160-day old or endpoints). (D) Representative images (endpoint) and quantification of CMAPs evoked by supramaximal stimulation of sciatic nerves (n=10, 8, 12 and 8 mice per group for 120-day old; n=8, 5, 9 and 6 mice per group for 160-day old or endpoints). (E) Representative images (endpoint) and quantification of NMJ innervation and collapse (n=5, 5, 3 and 3 mice per group for 120-day old; n=6, 5, 3 and 5 mice per group for 160-day old or endpoints). (F) Representative immunoblot of Agrin or Wnt3a in GAST muscles of endpoint G93A and dTg mice and age-matched NTG and TMFN mice (quantification shown in Figure S2M). (G) Representative images and quantification of mitochondrial length and colocalization with neuromuscular synapses in GAST muscles of endpoint G93A and dTg mice and age-matched NTG and TMFN mice injected with AAV1-mitoDsRed at 60-day old (n = 11–14 neurites or NMJs from 3 mice per group). Equal protein amounts of 20 μg were loaded for all blots. Data are means ± s.e.m., representative of triplicate experiments. One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. ns, not significant.
CMAP reduction observed in 120-day old or endpoint G93A mice was also significantly mitigated in endpoint dTg mice (Figure 2D). Remarkably, NMJ denervation was completely abolished in dTg mice even at the disease endpoint (Figure 2E). Agrin, Wnt3 and muscle-specific kinase (MuSK) are key regulators of NMJs, in which Agrin and Wnt3 are secreted by motor neurons whereas MuSK is mainly expressed in skeletal muscles (Koles and Budnik, 2012). G93A mice exhibited greatly reduced expression of these proteins in skeletal muscles but not in spinal cords at age 120 days and disease endpoints (Figures 2F and S2M). By striking contrast, the expression of Agrin, Wnt3 and MuSK all remained unchanged in skeletal muscles of dTg mice (Figures 2F and S2M). Together, these data demonstrate that, despite the ubiquitous expression of mutant SOD1, the neuro-specific expression of Mfn2 is sufficient to greatly protect neuromuscular synapses in G93A mice.
Mitochondrial fragmentation has been observed in neurons expressing ALS-associated mutant SOD1 (Magrane et al., 2009; Song et al., 2013; Vande Velde et al., 2011). To determine whether overexpression of neuronal Mfn2 protected NMJ by preventing mitochondrial fragmentation, in addition to electron microscopic (EM) analysis of mitochondria in motor neuron cell bodies, we performed the intraspinal injection of AAV1-mitoDsRed to label mitochondria in neuromuscular synapses. TMFN mice showed elongated mitochondria whereas G93A displayed fragmented mitochondria in neuromuscular synapses and motor neuron cell bodies and neurites (Figures 2G and S3A–D). Surprisingly, in dTg mice, mitochondrial morphology was only rescued in motor neuron cell bodies and proximal neurites, but not in neuromuscular synapses and distal neurites, even though mitochondria in neuromuscular synapses were still highly colocalized with NMJ (Figures 2G and S3A–D). In an attempt to further investigate how Mfn2 maintained NMJ, we performed histological, biochemical and genome-wide gene expression analyses of spinal cords, nerves and skeletal muscles of G93A and dTg mice. Unexpectedly, motor neuron loss, activated apoptotic signals and systemic inflammation in spinal cords were not suppressed in dTg mice, even though motor axon demyelination and degradation were alleviated (Figures S3E–I), suggesting minimal effects of mitochondrial fragmentation inhibition on motor neuron loss or neuropathologies.
Forced expression of Mfn2 in neurons abolishes neuromuscular synaptic loss and skeletal muscle atrophy in aged mice
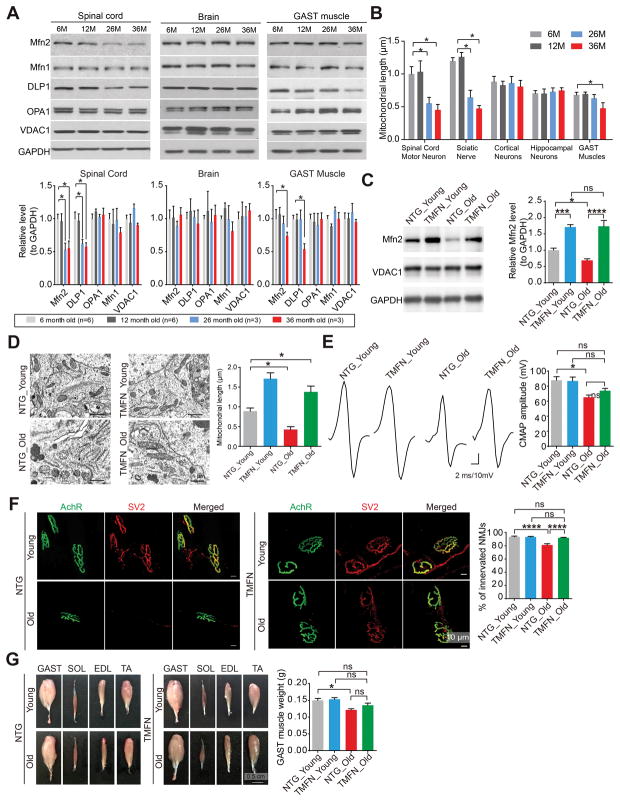
The decline in NMJs is generally believed to be the major contributor to age-related muscle wasting, i.e. sarcopenia (Deschenes, 2011). Like G93A mice, aged mice also showed neuronal mitochondrial fragmentation and decreased Mfn2 expression in spinal cords but not cortex or hippocampus when compared to young mice (Figures 3A and 3B). Given the remarkable Mfn2-mediated NMJ protection in G93A mice, we further tested whether age-related NMJ decline and skeletal muscle wasting was alleviated in TMFN mice. We confirmed that age-related neuronal mitochondrial fragmentation and Mfn2 reduction in spinal cords were indeed abolished in TMFN mice (Figures 3C and 3D). During aging, NTG and TMFN mice had similar body weight and survival. Neither NTG nor TMFN mice exhibited significant reduction in grip strength or rotarod performance, the loss of spinal cord motor neurons or motor axons or the decreased expression of Agrin, Wnt3 and MuSK in spinal cords or skeletal muscles even at 22 months of age (data not shown). At 22 months old, NTG mice demonstrated significantly decreased CMAP, NMJ innervation and skeletal muscle weight when compared to young adult NTG mice (Figures 3E–G). Remarkably, in 22-month old TMFN mice, the CMAP, NMJ innervation and skeletal muscle weight were all sustained at a level comparable to those of young NTG or TMFN mice (Figures 3E–G). These data underscore the critical role of NMJ in age-related skeletal muscle loss, and demonstrate that maintaining neuronal Mfn2 levels protects neuromuscular synapses against aging.
Figure 3. Mfn2 upregulation in motor neurons protects neuromuscular synapses in aged mice.
(A) Representative immunoblot and quantification of Mfn2, DLP1, OPA1, Mfn1 and VDAC1 levels in spinal cords, brains and GAST muscles of 6, 12, 26 and 36-month old wild type mice. Relative levels are all adjusted by GAPDH. n = 3 per group. (B) Quantification of mitochondrial length in spinal cord motor neurons, sciatic nerves, hippocampal neurons, cortical neurons and GAST muscle based on EM micrographs. n > 10 per group. (C) Representative immunoblot and quantification of Mfn2 levels in spinal cords of young (6-month old) and aged (22-month old) NTG and TMFN mice (n = 8, 4, 6 and 4 per group). Equal protein amounts of 20 μg were loaded. (D) Representative EM and quantification of mitochondrial length in spinal cords motor neurons of young (6-month old) and aged (22-month old) NTG and TMFN mice. n > 10 neurons per group. (E) Representative images and quantification of CMAPs evoked by supramaximal stimulation of sciatic nerves in young (6-month old) and aged (22-month old) NTG and TMFN mice (n = 8, 8, 7 and 8 per group). (F) Representative images and quantification of NMJ innervation in GAST muscles of young (6-month old) and aged (22-month old) NTG and TMFN mice (n = 3, 4, 5 and 8 per group). (G) Representative images of skeletal muscles and quantification of GAST muscle weight in young (6-month old) and aged (22-month old) NTG and TMFN mice (n = 12, 16, 9 and 16 per group). Data are means ± s.e.m., representative of triplicate experiments. One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. *P < 0.05, ***P < 0.001, ****P < 0.0001. ns, not significant.
Forced expression of Mfn2 in neurons delays neuromuscular synaptic loss upon denervation
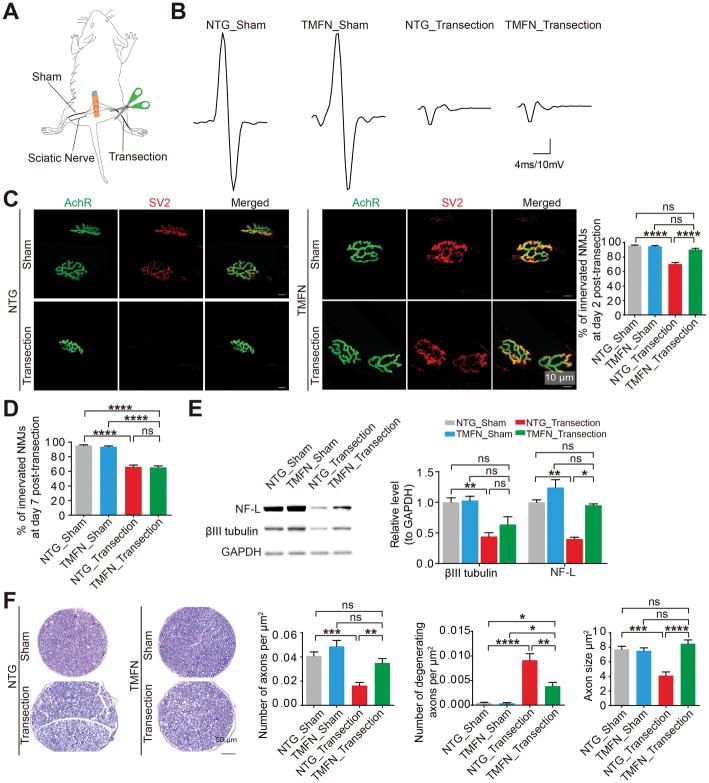
We have showed that neuronal Mfn2 maintains neuromuscular synapses not only in G93A mice but also in aged mice. To further test the likely generalized function of Mfn2 in preserving neuromuscular synapses, we analyzed NMJ innervation in NTG and TMFN mice after severing their sciatic nerves (Figure 4A). In NTG and TMFN mice, the sciatic nerve-transected hindlimb, but not sham-operated hindlimb, showed no evoked CMAP (Figure 4B). At day 2 post-transection, NTG mice displayed greatly reduced NMJ innervation, yet TMFN mice still maintained NMJ innervation comparable to sham controls (Figure 4C). By 7 days following transection, both groups of mice showed similar NMJ denervation (Figure 4D), suggesting that Mfn2 could delay but not abolish the loss of neuromuscular synapses upon nerve injury. Consistently, axon degradation induced by nerve cut was also delayed in TMFN mice (Figures 4E and 4F), indicating the similar protective effect of neuronal Mfn2 on axons. Agrin, Wnt3 and MuSK remained unchanged in skeletal muscles after sciatic nerve cut (data not shown). Neuronal Mfn2 upregulation had no effect on Agrin, Wnt3 and or MuSK expression in spinal cords and skeletal muscles (Figure S2M), excluding the possibility of Mfn2 promoting new neuromuscular synapse formation. As sciatic nerve transection blocks the transport of newly synthesized proteins into neuromuscular synapses, and the degradation of axonal proteins such as βIII tubulin and NF-L upon sciatic nerve transection is suppressed by neuronal Mfn2 (Figure 4E), these results imply that Mfn2 may protect neuromuscular synapses through skeletal muscle independent mechanisms involving the localized protein degradation inhibition.
Figure 4. Mfn2 upregulation in motor neurons protects neuromuscular synapses and axons against sciatic nerve transection.
(A) Schematic of mice with right sciatic nerve cut (transection) and left sciatic nerve sham-operated (sham). (B) Representative images of CMAPs evoked by supramaximal stimulation of sciatic nerves in transected or sham-operated hindlimbs of 4–6 month old NTG and TMFN mice 2 days after sciatic nerve transection. (C) Representative images and quantification of NMJ innervation in transected or sham-operated GAST muscles of 4–6 month old NTG and TMFN mice 2 days after right sciatic nerve cut. n = 4 mice per group. (D) Quantification of NMJ innervation in transected or sham-operated GAST muscles of 4–6 month old NTG and TMFN mice 7 days after right sciatic nerve cut. n = 3 mice per group. (E) Representative immunoblot and quantification of βIII tubulin and NF-L (neurofilament light subunit) in sciatic nerves in transected or sham-operated hindlimbs of 4–6 month old NTG and TMFN mice 2 days after surgery. n = 3 mice per group. Equal protein amounts of 20 μg were loaded for all blots. (F) Representative toluidine blue staining and quantification of axon number, degeneration and size in Epon-embedded sciatic nerves sections of hindlimb transected or sham-operated 4–6 month old NTG and TMFN mice at day 2 after surgery. n = 5, 6, 7 and 6 per group. Data are means ± s.e.m., representative of triplicate experiments. One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. ns, not significant.
Mfn2 regulates the active axonal transport of calpastatin into motor axons
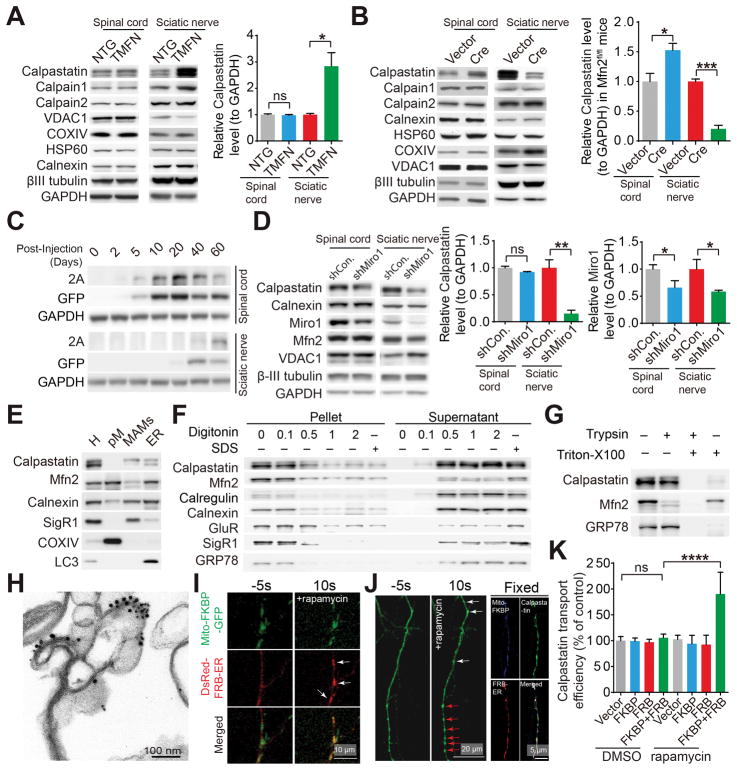
Calpastatin, an endogenous inhibitor of calcium-dependent cysteine protease calpain, is one of the few identified proteins that are involved in protein degradation, neuromuscular function regulation (Diepenbroek et al., 2014; Rao et al., 2014; Simoes et al., 2012; Spencer and Mellgren, 2002; Tidball and Spencer, 2002) and axon survival (Yang et al., 2013). Interestingly, compared with NTG mice, TMFN mice exhibited significantly increased calpastatin staining in neuromuscular synapses (Figure S4A). Consistently, the level of calpastatin was greatly increased in sciatic nerves but not spinal cords of TMFN mice (Figure 5A). To further test whether Mfn2 was required for calpastatin expression in motor axons, we injected AAV1-Cre into the lumbar spinal cord of Mfn2fl/fl mice. Remarkably, at week 2 after injection, mild Mfn2 deficiency caused substantial loss of calpastatin in sciatic nerves and accumulation of calpastatin in spinal cords (Figures 5B, S1F and S1G). Mfn2 overexpression or mild Mfn2 deficiency did not cause axon degeneration or neuromuscular synaptic dysfunction. Further, the levels of two large catalytic subunits of calpain, i.e. calpain 1 or calpain 2, mitochondrial marker VDAC1 and βIII tubulin in nerves of TMFN mice or Cre-injected Mfn2fl/fl mice all remained unchanged (Figures 5A, 5B, S4B and S4C), further indicating the specific change of calpastatin in sciatic nerves. Axonal proteins are largely synthesized in the neuronal cell body and moved into axons by either active transport, passive diffusion or both. Of note, calpastatin in the proximal neurites but not cell bodies of spinal cord motor neurons was also greatly depleted in Cre-injected Mfn2fl/fl mice (Figure S4D), thus suggesting the minimal contribution of passive diffusion to calpastatin axonal transport. Together, these results suggest that Mfn2 may selectively control the active axonal transport of calpastatin.
Figure 5. Mfn2 regulates the level of calpastatin in motor axons through the mitochondrial trafficking dependent active axonal transport.
(A and B) Representative immunoblot and quantification of calpastatin in endpoint G93A and dTg mice and age-matched NTG and TMFN mice (A) or 4–6 month old Mfn2fl/fl mice 12 days after injection with AAV1-GFP (Vector) or AAV1-Cre (Cre) (B). n = 3 or 4 mice per group. (C) Representative western blot detection of T2A tagged human calpastatin in lumbar spinal cords and sciatic nerves of 3–4 month old NTG mice at different days after AAV1-calpastatin injection. (D) Representative immunoblot and quantification of calpastatin in 3–4 month old NTG mice 12 days after injection of AAV1 encoding control shRNA (AAV1-shCon.) or AAV1-shMiro-1. n = 3 or 4 mice per group. (E–H) Representative western blot detection (E–G) and immunoelectron-microscopic analysis (H) of calpastatin (labeled by 15 nm gold particles) and Mfn2 (labeled by 10 nm gold particles) in MAMs isolated from pooled spinal cords of 4 NTG mice at 3–4 month old with or without indicated digitonin (w/v), triton X-100 (0.5% v/v) or trypsin treatment (F and G). H: total spinal cord homogenate; pM: highly purified mitochondria isolated from spinal cords. The purity of pM and MAMs was confirmed by EM (Figure S5A and S5B). (I and J) Representative confocal images of mitochondria and ER (I) or GFP tagged calpastatin in live cultured (I and J left panel) or fixed (J right panel) neurons 2 days after co-transfection with mito-FKBP-GFP and DsRed-FRB-ER (I, white arrows point ER recruited to mitochondria) or mito-FKBP-myc, Flag-FRB-ER and GFP tagged calpastatin (J, white arrows point newly formed calpastatin puncta while red arrows point enlarged pre-existing calpastatin puncta) before and after rapamycin treatment. (K) Quantification of calpastatin axonal transport (indexed by the relative ratio of the mean gray value of calpastatin in a segment of neuronal process 200 μm length beginning from the cell body of neurons to the mean gray value of calpastatin in the cell body of neurons) in neurons 2 days after transfection or co-transfection with mito-FKBP-myc, Flag-FRB-ER and GFP tagged with or without rapamycin treatment for 30 min. n = 15–24 neurons per group. Equal protein amounts of 20 μg were loaded for all blots. Data are means ± s.e.m., representative of triplicate experiments. Student’s t test or one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. *P < 0.05, **P < 0.01, ***P < 0.0001, ****P < 0.0001. ns, not significant.
Based on the speed of transport, anterograde active axonal transport from cell body to nerve terminals is divided into two major forms, i.e., fast transport for mostly transmembrane or secreted proteins traveling at a speed of 50–200 mm per day, and slow transport for cytoskeletal or most cytoplasmic proteins moving at a speed of 0.2–10 mm per day (Maday et al., 2014). To investigate the transport mode of calpastatin in vivo, we injected AAV1 encoding human calpastatin and GFP under the eSYN promoter (AAV1-calpastatin) into the mouse lumbar spinal cord (Figure S4E). Exogenously expressed human calpastatin (detected by 2A tag) and GFP were detectable in spinal cords at day 2–5 post-injection and reached their highest levels at day 20 post-injection (Figure 5C). However, in sciatic nerves, 2A tagged calpastatin first appeared 40 days after injection and further increased at day 60 post-injection, while GFP could be first detected 20 days after injection and then increased at day 40. Therefore, like GFP, calpastatin is largely conveyed by slow-going active transport in vivo. To further study the mobile behavior of calpastatin in live axons, we co-transfected in vitro cultured mouse primary cortical neurons with constructs encoding GFP tagged calpastatin and fast transport organelle markers mitoBFP (a mitochondrial specifically localized blue fluorescent protein) and ER-DsRed (an ER specifically localized red fluorescent protein). Interestingly, despite its largely diffusive pattern, calpastatin could form puncta highly co-localizing and moving together with mitochondria in axons (Figures S4F–H and Movie S5), but not closely associated with local axonal enlargements (data not shown). Of note, although Mfn2 overexpression had no significant effect on the velocity of calpastatin puncta or the transport of mitochondria and ER, Mfn2 overexpression consistently increased while Mfn2 knockdown decreased calpastatin transport in vitro (Figures S4H–K).
Mitochondria transports calpastatin via MAMs
Calpastatin puncta and mitochondria cotransport in axons. To further test the dependence of global calpastatin trafficking on mitochondria, we immobilized mitochondria by knocking down Miro-1, the key adapter linking mitochondria to microtubules (MacAskill and Kittler, 2010). In vitro cultured neurons expressing a short hairpin RNA targeting Miro-1 (shMiro1) showed significantly impaired calpastatin puncta movement, overall calpastatin transport and mitochondrial trafficking with unchanged ER transport (Figures S4H–K). Consistently, the intraspinal delivery of AAV1 encoding Miro-1 shRNAi (AAV1-shMiro1) significantly reduced Miro-1 in sciatic nerves but not spinal cords in vivo (Figure 5D). Of note, the knockdown of Miro-1 had no effect on Mfn2, VDAC1 and βIII tubulin (Figures 5D and S4L), further suggesting that the change of calpastatin is unlikely due to side effects of Miro-1 deficiency. Taken together, all these results imply that mitochondria trafficking is critical for the global axonal transport of calpastatin and that mitochondria may act as the conveyor for calpastatin.
Subcellular fractionation of mouse spinal cords revealed that calpastatin was not present in purified mitochondria, but rather was co-enriched with Mfn2 in the purified mitochondria-associated membrane fraction, i.e. MAMs (Figures 5E, S5A and S5B), therefore indicating the possibility that mitochondria may convey calpastatin “indirectly” via MAMs. As mitochondria-associated membranes are largely subdomains of ER, MAMs contained the ER markers such as Calnexin, Calregulin and GRP78 and its specific marker SigR1 (Figures 5E–G). Like Mfn2 and other ER proteins, permeabilization of MAMs by digitonin resulted in the extraction of some but not all calpastatin, suggesting the tight association of calpastatin with MAMs (Figure 5F). Exposure of MAMs to trypsin after Triton-X100 permeabilization completely digested calpastatin, which was expected (Figure 5G). Subsequent immuno-EM analysis of purified MAMs or spinal cords confirmed the localization of both calpastatin and Mfn2 in MAMs (Figures 5H, S5C and S5D). Interestingly, clusters of gold particle labeled calpastatin were readily observed (Figure 5H). Of note, MAMs isolated from either Cre-injected Mfn2fl/fl mice at week 2 after injection or endpoint G93A mice demonstrated greatly increased calpastatin, further supporting the impaired calpastatin delivery to nerve terminals in these mice (Figure S5E). Noteworthily, although calpastatin sometimes was observed to be in close proximity to Mfn2, immunoprecipitation of calpastatin failed to pull down Mfn2 (data not shown), suggesting no direct involvement of Mfn2 in recruiting calpastatin to MAMs. This is indeed in line with the facts that calpastatin is absent in mitochondria where Mfn2 predominantly resided, and that calpastatin remains unchanged in MAMs of TMFN mice when Mfn2 was upregulated (Figure S5E).
Consistent with previous findings showing Mfn2 as an ER-mitochondria tether (de Brito and Scorrano, 2008; Sebastian et al., 2012; Sugiura et al., 2013), the increased association between mitochondria and membranes or ER-derived membranes in axons was noted in TMFN mice or neurons overexpressing Mfn2 (Figures S5F and S5G). MAMs only represented a small portion of ER-derived membranes or vesicles in axons. Not surprisingly, Mfn2 or Miro1 did not show any significant effect on the global axonal transport of ER-derived membranes or the expression of ER marker Calnexin in sciatic nerves (Figures S4K and S5H). Mfn2ActA is a mitochondrial exclusively localized Mfn2 mutant that can fuse mitochondria but is unable to tether ER and mitochondria, whereas Mfn2IYFFT is an ER exclusively targeted Mfn2 mutant that can connect ER and mitochondria but lack the inability to promote mitochondrial fusion (de Brito and Scorrano, 2008). Unlike wild type Mfn2 or Mfn2IYFFT, Mfn2ΔRAS (lacking the p21-Ras-binding domain), Mfn2ActA and Mfn2IYFFTΔRAS mutants all failed to increase calpastatin transport (Figures S5I), implying that the Mfn2-mediated calpastatin transport depends on its ER-mitochondria tethering activity. To further investigate whether the enhanced ER-mitochondria tethering is sufficient to increase calpastatin transport, we transfected neurons with constructs encoding Flag tagged calpastatin and two synthetic rapamycin-inducible ER-mitochondria linkers as described (Csordas et al., 2010). Addition of low-dose rapamycin rapidly connected ER and mitochondria, promoted the quick formation of calpastatin puncta highly colocalized with ER and mitochondria and concurrently increased axonal calpastatin transport (Figures 5I–K, S5J and S5K). Of note, the tethering of ER and mitochondria for 30 min did not obviously impair mitochondria and ER dynamics or trafficking (Movies S6–8). Taken together, although it’s unlikely that mitochondria carry calpastatin directly, they may utilize Mfn2 as a cargo handler to specifically load calpastatin-enriched MAMs on them to further mediate calpastatin axonal transport.
The protective effect of Mfn2 on neuromuscular synapses depends on calpastatin
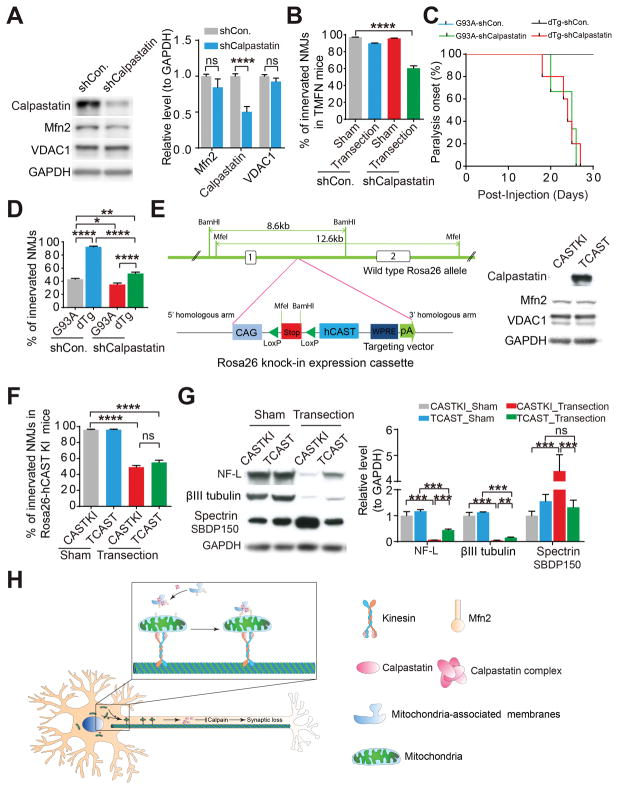
To investigate the necessity of calpastatin for Mfn2-mediated neuromuscular synapse protection, we injected AAV1 encoding a short hairpin RNA targeting calpastatin (AAV1-shCalpastatin) into the lumbar spinal cord of NTG or TMFN mice followed by sciatic nerve transection (Figure 6A). Consistent with previous studies (Yang et al., 2013), calpastatin reduction alone has no effect on CMAP (data not shown) or NMJ innervation (Figure 6B). Notably, upon calpastatin knocking down, TMFN mice showed NMJ denervation similar to that of NTG mice 2 days after sciatic nerve transfection (Figure 6B), indicating calpastatin as an indispensable factor for Mfn2-mediated motor synaptic protection. Consistent with this, dTg mice injected with AAV1-shCalpastatin at 60-days old demonstrated remarkable accelerated onset of paralysis and severe NMJ denervation comparable to age-matched G93A mice (Figures 6C and 6D). To test the sufficiency of calpastatin alone for neuromuscular synapse survival upon nerve injury, we generated a novel calpastatin transgenic mouse model, i.e. CASTKI mice, with targeted ROSA26 locus insertion of an expression cassette to drive tissue specific upregulation of calpastatin by Cre-mediated recombination (Figure 6E). We crossed CASTKI mice with transgenic mice expressing Cre under the same neuron-specific promoter thymus cell antigen 1 (Thy1-Cre mice) to obtain the double transgenic TCAST mice and also injected AAV1-Cre into the lumbar spinal cord of CASTKI mice (Figures 6E and S6A). Surprisingly, while neuronal calpastatin overexpression suppressed calpain activation and axon degradation, it did not protect neuromuscular synapses 2 days after sciatic nerve transection (Figures 6F, 6G and S6B–D). Consistently, the intraspinal delivery of AAV1-calpastatin failed to protect NMJs in G93A mice (Figures S6E and S6F). Mfn2 remained unchanged in TCAST mice or Cre-injected CASTKI mice, and compared with CASTKI or Thy1-Cre mice, TCAST mice or Cre-injected CASTKI mice did not show increased calpastatin staining in neuromuscular synapse (Figure S6G), thus implying that the inability of calpastatin to protect neuromuscular synapses in these mice is likely due to the insufficient calpastatin transport to neuromuscular synapses.
Figure 6. Calpastatin is necessary for Mfn2-mediated protection of neuromuscular synapses.
(A) Representative immunoblot and quantification of calpastatin in spinal cords of mice injected with AAV1-shCon. or shRNA targeting calpastatin (AAV1-shCalpastatin) into lumbar spinal cords of 4–6 month old TMFN mice for 2 weeks. n = 3 mice per group. (B) Quantification of NMJ innervation in transected or sham-operated GAST muscles of 4–6 month old TMFN mice 2 days after right sciatic nerve cut. 12 days before transection, mice were injected with indicated AAV1 into lumbar spinal cords. n = 3 mice per group. (C and D) Age of paralysis onset (C, n = 5–8 mice per group) and quantification of NMJ innervation (D, n = 5, 4, 3, 5 mice per group, 90-day old or paralyzed mice) in G93A and dTg mice injected with indicated AAV1 into lumbar spinal cords at 60-day old. (E) Schematic of generation of Rosa26-hCAST KI mice. Upon Cre recombination, STOP cassette is removed, resulting in human calpastatin (CAST) expression under CMV early enhancer/chicken beta actin (CAG) promoter. Representative western blot detection of human calpastatin and Mfn2 in spinal cords of TCAST mice shown in the right panel (note that all used commercially available calpastatin antibodies have much higher affinity to human calpastatin than mouse calpastatin. The immunoblot analysis does not reflect the actual level of human calpastatin overexpression). (F and G) Quantification of NMJ innervation (F) and representative immunoblot and quantification of βIII tubulin, NF-L and the calpain cleaved 150-kDa αII-spectrin breakdown product (SBDP150) in sciatic nerves (G) of 4–6 month old TCAST mice 2 days after right sciatic nerve cut. n = 3 mice per group. (H) Hypothetical model of Mfn2-mediated protective effects on neuromuscular synapses. First, calpastatin forms the complex on MAMs. Then, likely through the intermolecular interaction between MAM-Mfn2 and mitochondrial Mfn2, calpastatin-containing MAMs are connected to mitochondria and further transport along axons by moving mitochondria. The delivery of calpastatin to distal nerve endings leads to the localized inhibition of calpain, which finally prevents axon degeneration and neuromuscular synaptic loss. Equal protein amounts of 20 μg were loaded for all blots. Data are means ± s.e.m., representative of triplicate experiments. One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. *P < 0.05, **P < 0.01, ***P < 0.0001, ****P < 0.0001. ns, not significant.
Discussion
Here, we provide evidence supporting the necessity and sufficiency of Mfn2 for the maintenance of neuromuscular synapses in disease, during aging or upon nerve injury. As Mfn2 deficiency or mutations are commonly observed in patients with ALS, CMT2A, Alzheimer’s disease (AD) (Wang et al., 2009) and aged mice, in which synaptic loss has long been recognized as a prominent early feature, we speculate that Mfn2 may exert a generalized protective effect on synapses in a wide range of muscular disorders and neurodegenerative diseases associated with synaptic degeneration.
Neuronal Mfn2 upregulation is sufficient to greatly delay the symptom onset and remarkably prevents muscle atrophy in G93A mice. Due to the very mild overexpression of Mfn2 in TMFN mice and the high-copy transgene number and aggressive phenotypes in G93A mice, the unextended survival of dTg mice does not necessarily limit the translation of our findings into therapeutic strategies for ALS and especially sarcopenia. In fact, our preliminary studies have found that the intraspinal injection of AAV1 encoding Mfn2 in G93A mice after symptom onset protects NMJs and muscles (data not shown). On top of this, considering that mild Mfn2 upregulation significantly alleviates muscle atrophy in both G93A mice and aged mice, extended survival may be observed in G93A mice with low-copy transgene number or higher Mfn2 upregulation. dTg mice died in less than two weeks after the appearance of muscle weakness at ~130-days old with relatively well-preserved hindlimb skeletal muscles. One major cause of death in ALS patients is the respiratory insufficiency due to diaphragm muscle loss (de Carvalho et al., 1996), also found in G93A mice. Yet, significant change of diaphragm muscles was not noted in dTg mice. Nor was the loss of motor neurons and likely interneurons alleviated in dTg mice. As SOD1 G93A is expressed ubiquitously in all tissues and Mfn2 is only upregulated in neurons in dTg mice, future studies will be needed to explore the contribution of other organs especially autonomic organs to the rapid death.
Proteasome is impaired in ALS patients and G93A mice (Allen et al., 2003; Kabashi and Durham, 2006; Urushitani et al., 2002), excluding the possible involvement of the ubiquitin-proteasome system in Mfn2 reduction in G93A as reported in non-neuronal cells (Leboucher et al., 2012). Mfn2 can be cleaved by calpain in motor neurons during excitotoxicity (Wang et al., 2015). Interestingly, similar to Mfn2, calpain activation was reported in G93A mice at early disease stages (Stifanese et al., 2014). Of note, Mfn2 reduction in G93A mice is abolished by Mfn2 overexpression, thus suggesting a likely Mfn2-calpain feedback loop in mediating Mfn2 degradation. Along this line, we have found that Mfn2 selectively controls axonal transport of calpastatin into motor axons to exert its protective effect on neuromuscular synapses. As for the Mfn2 closely related protein Mfn1, the ablation of Mfn1 in spinal cord neurons by the intraspinal delivery of AAV1-Cre into the lumbar spinal cord of Mfn1 floxed mice (Mfn1fl/fl mice) (Chen et al., 2007) did not cause any behavior deficits, NMJ loss or axonal calpastatin reduction (data not shown). On the basis of additional facts that the inhibition of mitochondrial fission by DLP1 K38A or DLP1 RNAi or the promotion of mitochondrial fusion by OPA1 or Mfn1 has no effect on calpastatin transport (data not shown), and that the mitochondrial fusion deficient Mfn2IYFFT mutant is able to promote calpastatin transport, our data suggest that the Mfn2-mediated calpastatin axonal transport does not depend on its fusion activity. Moreover, Mfn2 upregulation or deficiency has no effect on autophagy marker LC3 expression in motor neurons (Wang et al., 2015) and Mfn2 fails to prevent mitochondrial fragmentation in neuromuscular synapses in G93A mice. Therefore, here we uncover an unexpected novel function of Mfn2 in the direct regulation of calpain–calpastatin proteolytic system.
The expression of mitochondria/ER markers or neuron-secreted Agrin and Wnt3 in nerves is not changed by Mfn2, suggesting that Mfn2 is unlikely to be involved in global fast transport of transmembrane/secreted proteins or organelles. To be conveyed by active axonal trafficking, the soluble protein is usually organized into protein complexes on vesicles and then moved by transient interactions with fast transport cargos (Maday et al., 2014). Calpastatin clusters were noted in MAMs, and calpastatin puncta and mitochondria can be co-transported. In addition, the ER-mitochondria tethering is necessary and sufficient for calpastatin transport. Therefore, calpastatin may use a similar route as other reported soluble proteins, i.e., a portion of calpastatin forms protein complexes on MAMs, thereby generating a pool of fast-moving calpastatin-enriched MAMs that are transiently connected to and conveyed by mitochondria through Mfn2-mediated MAMs-mitochondria tethering. Of note, Mfn2 knockout reduces but cannot abolish ER and mitochondria tethering. And, the association of MAM with mitochondria is regulated by many factors including but not limited to Mfn2. Therefore, the accumulation of calpastatin in the MAM fraction isolated from Mfn2-deficient spinal cords further supports the likely presence of a reservoir for calpastatin-containing MAMs, the transport but not formation of which is largely regulated by Mfn2. Apart from its MAMs localization, the majority of calpastatin resides in cytosol. Of note, calpastatin puncta are usually moved rapidly over short distances followed by a relatively long process of disassembly. Some calpastatin in MAMs are not clustered or tightly associated with the membrane surface. Therefore, depending on the distance that calpastatin puncta can be moved, the speed of their recruitment to or fall off from MAMs and the likely dynamic connection between MAMs and mitochondria, most calpastatin may globally appear to be diffusive and moved along axons in a slowly transported pool. Nevertheless, although it is still possible that calpastatin is transported by mixed populations of fast and slow cargos, our study provides strong evidence supporting the critical role of mitochondria-MAMs mediated fast trafficking for calpastatin axonal transport.
The relatively tight association of some calpastatin with MAMs enables the possible long-distance axonal transport of calpastatin into distal motor axons and neuromuscular synapses before degradation or disassembly. On the other side, calpastatin puncta usually does not move together with mitochondria over long distances. Considering the relatively loose association between MAMs and mitochondria, mitochondria may use a sliding mode to dynamically load, move and unload calpastatin-enriched MAMs to the target site without the full stop of mitochondria. It is worth noting that levels of calpain1/2 or ER markers in nerves remain unchanged by Mfn2. Also, the ER derived vesicle transport is not altered by Mfn2 or Miro-1 either, indicating the selectivity of Mfn2-mediated mobile of calpastatin-enriched MAMs. Even though some ER derived vesicles co-traffic with mitochondria or calpastatin, they do not form puncta. On top of this, there is no specific MAM marker available for live imaging and the MAM specific marker Sigma-1 receptor in axons could not be reliably detected by immunoblot or immunofluorescent staining (data not shown). Due to these technical challenges, future studies using different approaches are needed to investigate whether Mfn2 indeed regulates the transport of global MAMs or other MAM-associated proteins. While this and other aspects such as the mechanism underlying calpastatin recruitment to MAMs are still under investigation, our study has proposed a novel mechanism by which mitochondria act like miniature conveyors to transiently carry calpastatin-enriched MAMs to mediate their transport along axons.
Calpastatin is required for Mfn2-mediatated neuromuscular synaptic protection. However, unlike Mfn2, calpastatin overexpression alone is not sufficient to protect neuromuscular synapses following transection or in G93A mice. Although this could be due to the limited transport of exogenous calpastatin into neuromuscular synapses, we could not rule out the possibility that in addition to calpastatin, Mfn2 upregulation may also regulate the transport of other protective factors on MAMs to preserve neuromuscular synapses. Our transcriptional profiling using genome-wide RNA sequencing (RNA-seq) analysis to characterize the transcriptomes of TMFN and NTG spinal cords has revealed the metabolism of lipids and lipoproteins as one of the most significant pathways in TMFN vs NTG spinal cords (Figure S3I). Thus, it is also worth investigating whether Mfn2 indirectly regulates calpastatin trafficking or protects NMJs through other mechanisms such as lipid metabolism.
Following nerve transection, axons are degraded via anterograde degeneration (i.e., Wallerian degeneration). The entry of Ca2+ to activate calpain is generally believed to trigger axonal degeneration, whereas bioenergetic failure is usually considered as one of the final steps (Coleman, 2005). Synaptic mitochondria isolated from TMFN mice do not show altered function (data not shown), and Mfn2 preserved NMJs but not mitochondrial morphology or density in G93A mice. Therefore, it’s unlikely that Mfn2 protects neuromuscular synapse via promoting bioenergetics during Wallerian degeneration. Along this line, neither G93A mice crossed with slow Wallerian degeneration (WldS) mutant mice nor aged WldS mice demonstrate the same remarkable neuromuscular synapse protection as that of TMFN mice or aged TMFN mice (Fischer et al., 2005; Gillingwater et al., 2002). Considering Mfn2-meidated calpastatin transport and the likely multifaceted role of the calpain–calpastatin proteolytic system in axon degeneration and neuronal death, Mfn2, at least partially, protects neuromuscular synapse through mechanisms other than the inhibition of Wallerian degeneration in G93A mice or aged mice. Noteworthily, high concentrations of Ca2+ are required for calpain activation (Doshi and Lynch, 2009). It could be expected that calpastatin depletion alone does not cause calpain activation or neuromuscular synaptic loss. Interestingly, calpastatin deficiency greatly accelerated neuromuscular synaptic loss and disease progression in both G93A and dTg mice. As neurons expressing CMT2A-associated Mfn2 mutant also showed greatly impaired calpastatin transport (Figure S5I), our studies may imply an unrecognized important role of the calpain–calpastatin proteolytic system and Mfn2-mediated calpastatin transport in the disease progression of ALS or CMT2A. Mitochondrial dysfunction is a prominent feature repeatedly reported in various major neurodegenerative diseases and aged animals (Gonzalez-Freire et al., 2014; Ibebunjo et al., 2013; Li et al., 2013). Even though this study implies an unexpected yet important role of Mfn2-mediated calpastatin transport, it might still be worth investigating the likely interplay between mitochondrial dysfunction, calpastatin transport, the calpain–calpastatin proteolytic system and even mitochondrial dynamics.
It is still unclear how calpastatin is transported by Mfn2 without effect on calpain. Interestingly, one previous study has reported that membrane-associated calpastatin is more phosphorylated than cytosolic calpastatin (Adachi et al., 1991), indicating phosphorylation as a likely mechanism regulating calpastatin recruitment to MAMs and axonal transport. This notion is further supported by the fact that calpastatin phosphorylation is closely associated with intracellular Ca2+ (Averna et al., 2001; Averna et al., 1999), the ablation of which inhibits Mfn2-mediated calpastatin axonal transport (Figure S4I). Although these results support the role of Ca2+ in calpastatin axonal transport, its potential direct contribute to motor protein regulation or Mfn2-mediated ER and mitochondria tethering could not be ruled out, and future studies are needed to clarify these aspects. Unfortunately, due to the limited amount of MAM isolated from pooled spinal cords and the relative low level of calpastatin in MAMs, the previously used immunoprecipitation-based approach cannot not be used to assess calpastatin phosphorylation in MAMs (Adachi et al., 1991; Averna et al., 2001; Averna et al., 1999). On top of this, the antibody specific to phosphorylated calpastatin is not available and the calpastatin phosphorylation sites crucial for the calpain-calpastatin colocalization have not been identified so far, making further investigation of the role of calpastatin phosphorylation in its axonal transport or nerve terminal localization even difficult.
Taken together, this study provides the first attractive framework for further understanding the role of Mfn2 and mitochondria in the axonal transport of calpastatin, MAMs and possibly other cytosolic proteins to inhibit localized calpain activation, axon degradation and neuromuscular synaptic loss (Figure 6H). A single protein such as Mfn2 that can remarkably prevent neuromuscular synapse loss in disease, during aging and upon nerve injury suggest that, like programed cell death, i.e. apoptosis, the programed synaptic elimination (we term as synaptosis) may exist and be a potential common therapeutic target in the treatment of a wide range of diseases including, but not limited to aging and disease related skeletal muscle atrophy.
Limitations of Study
Our work demonstrates the novel role of Mfn2 to regulate axonal transport of calpastatin and likely other proteins to preserve synapses in ALS, during aging and upon nerve injury. However, while our study suggests that Mfn2 regulates the transport but not the formation of calpastatin-containing MAMs, the recruitment of calpastatin to MAMs and the formation of calpastatin-containing MAMs need to be further explored to clarify our proposed model. And, whether Mfn2 globally mediates the transport of MAMs or other MAM-associated proteins remains unclear. Although calpastatin overexpression does not protect neuromuscular synapses in G93A mice, we could not rule out the possibility that the calpain–calpastatin proteolytic system contributes to disease progression. Finally, as neuronal Mfn2 upregulation only halts muscle atrophy, but fails to extend survival of G93A mice, other ALS transgenic mouse models with less aggressive disease progression may be more appropriate to translate this study to disease treatment development.
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Xinglong Wang (Xinglong.Wang@case.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Transgenic Mice
Mouse surgery and procedures were performed according to the NIH guidelines and were approved by the Institutional Animal Care and Use Committee (IACUC) at Case Western Reserve University. Mfn2 floxed mice were obtained from Dr. David Chan (California Institute of Technology). Mfn2 transgenic mice (TMFN mice) were generated by pronuclear injection of the murine Thy-1.2 genomic expression cassette (gift of Dr. Philip C. Wong, Johns Hopkins University) expressing human Mfn2 into C57BL/6 fertilized eggs. At least 4 independent lines with similar total Mfn2 expression were obtained and confirmed with same phenotype. Both transgenic SOD1 G93A mice (G93A mice) on C57BL/6 (B6.Cg-Tg(SOD1*G93A)1Gur/J, Stock No: 004435) and transgenic Thy1-Cre mice (FVB/N-Tg(Thy1-cre)1Vln/J, Stock No: 006143) were obtained from the Jackson laboratory. Two independent lines of TMFN mice were crossed with G93A mice to generate double transgenic mice (dTg mice). 6, 12, 26 and 36 month old male mice on C57BL/6 shown in Figure S7A and S7B were all obtained from NIA Aged Rodent Colonies. The Rosa26-hCAST KI conditional transgenic mice were generated by microinjection of the human calpastatin expression cassette, Cas9 mRNA (prepared by in vitro transcription) and sgRNA into zygotes (gene targeting into the Rosa26 locus). Microinjected zygotes were implanted into recipient C57Bl/6J followed by backcross and genotyping. The human calpastatin was assembled into mROSA KI-RR-LR-0512 vector by AIOTM Cloning (Biocytogen, Worcester, MA). The insertion of human calpastatin expression cassette was confirmed by PCR and Southern blotting. The primers are as follows: Rosa26-L-GT-F (CCTCAGAGAGCCTCGGCTAG) and CAG-L-GT-R (AGTCCCTATTGGCGTTACTATGG) for a product size of 1331bp or WPRE-R-GT-F (ACGAGTCGGATCTCCCTTTGG) and Rosa26-R-GT-R (CATGAGTCAAGCCAGTCCAAGAG) for a product of 1408bp.
Embryonic Primary Rat Cortical Neurons
Primary cortical neurons were isolated from E18 Sprague Dawley rats (Charles River, Wilmington, MA). Briefly, rat brains were dissected out in HBSS (Life Technologies, Grand Island, NY) and stored in Hibernate E (BrainBits, Springfield, IL) supplemented with 2% B27 (Life Technologies, Grand Island, NY). Under a dissecting microscope, the meninges were removed completely with fine forceps and cortices were dissected out. Cortices were then digested in 0.25% trypsin for 15 min at room temperature, followed by brief incubation in Opti-MEM (Life Technologies, Grand Island, NY) supplemented with 10% FBS and 50 units per ml DNAse I (Worthington-Biochem, Lakewood Township, NJ). Digested cortices were further dissociated by gentle trituration with pipette until the cell suspension was homogenous and no large pieces of tissue remain visible. Cortical neurons were finally collected and seeded on poly-L-lysine and laminin-coated coverslips or chamber slides (BD, Franklin Lakes, NJ), 35-mm dishes or 24-well plates, and cultured as we described before(Wang et al., 2016). Neurons were transfected at DIV5 or 7 with lipofectamine 2000 (Life Technologies, Grand Island, NY) according to the manufacturer’s protocol.
Human Tissue Samples
Human spinal cord frozen tissues were obtained from Eunice Kennedy Shriver National Institute of Child Health and Human Development Brain and Tissue Bank for Developmental Disorders at the University of Maryland, Baltimore (UMB), Maryland, USA, contract HHSN275200900011C, ref. no. N01-HD-9-0011. Fixed human spinal cord tissues used for immunocytochemistry were obtained from University Hospitals of Cleveland and 6 μm-thick consecutive sections were prepared as we described before(Zhu et al., 2000). All ALS cases used are sporadic and no known genetic mutation in any gene is noted.
METHOD DETAILS
Lumbar spinal cord injection of AAV
Mouse surgery and procedures were performed according to the NIH guidelines and were approved by the Institutional Animal Care and Use Committee (IACUC) at Case Western Reserve University. Adeno-associated virus serotype 1 encoding GFP, Cre recombinase, Mfn2, calpastatin or syntaphilin under the neuron specific promoter eSYN (AAV1-GFP, AAV1-Cre, AAV1-Mfn2, AAV1-calpastatin or AAV1-syntaphilin), control short hairpin RNA (AAV1-control shRNAi), or a short hairpin targeting calpastatin (AAV1-calpastatin shRNAi), KIF5C (AAV1 KIF5C shRNAi) or Miro-1 (AAV1 Miro-1 shRNAi) with 1013 genome copies per mL (GC/ml) were obtained from Vector BioLabs (Malvern, PA). For AAV1-Cre injection, mice were anaesthetized with isoflurane followed by hair removal in the lumbar area. With the support of a heating pad, the spines of the mouse were then immobilized using the stereotactic frame. After the cleaning of the shaved area with betadine and alcohol, and the use of eye ointment to protect the eyes, bupivacaine/lidocaine (1:1 v/v) was injected and the skin was incised at the caudal end of the rib cage along the midline (0.5–1 cm) to separate the fascia covering the spine. Vertebra L1 was identified and exposed by moving the small spinal muscles and ligaments attached to its dorsal surface. Dedicated laminectomy forceps were then used to remove the dorsal portion of the vertebra (spine and lamina) to expose the spinal cord. 3 μl of 1013 GC/ml AAV were slowly injected into two different locations at the depth of 1 mm rostral to the transection using a glass micropipette attached to a Hamilton syringe (bilaterally injected just medial to the lateral edge of the spinal cord at a depth of 1 mm and 1 mm lateral to midline). Each injection lasted for 5 minutes, and remained for another 5 minutes for absorption. After injection, the fascia and the skin were sutured with nylon suture and with an injection of Carprofen, the mice were placed back in a clean cage for complete recovery on the warming pad.
Sciatic nerve transection
Mice were anaesthetized with isoflurane and oxygen inhalation followed by shaving the leg area. With the support of a heating pad, the anesthetized mouse was placed in dorsal recumbency and the paws were gently immobilized on the surgery platform with a piece of tape with the paw-palm facing up. After marcaine injection, the skin and then fascial plane between the gluteus maximus and the anterior head of the biceps femoris were opened to reveal the sciatic nerve. The sciatic nerve segment proximal to spinal cord was located. A 3mm segment of the right sciatic nerve was removed with scissors, while the left sciatic nerve was exposed but not severed as sham control. After skin suture, the mice recovered on a warming pad and were subjected to carprofen injection to relieve pain.
Grip strength and footprint test
Muscular strength of mice was measured using a grip strength test meter from Bioseb (Vitrolles, France). First, two hind paws of the mouse were placed on a bar connected to the meter. After trials, the single best recorded value was used for statistical analysis. Footprint analyses were performed using a customized runway (50 cm long, 5 cm wide and both sides bordered by walls which led to an enclosed chamber). The forepaws and hindpaws of mice were first dipped into red, purple or orange non-toxic paints. Mice were placed on the runway covered by white paper where they ran toward the enclosed dark box. Stride length is presented as the mean distance of forward movement between each stride.
Electromyography recordings of skeletal muscles
Electrophysiological recording of CMAP in hindlimb muscles evoked by supramaximal stimulation of sciatic nerves was performed using the PowerLab 4/35 data acquisition system, the FE155 Stimulator HC, the FE136 Animal Bio Amp and needle electrodes from ADInstruments (Colorado Springs, CO) and ring surface electrodes from Natus Neurology (Middleton, WI) as described(Arnold et al., 2014; Arnold et al., 2015; Major et al., 2007). Taken briefly, mice were anaesthetized with isoflurane and oxygen inhalation and immobilized to the table with a piece of tape with the paw-palm facing down. Two reference and ground needle electrodes were inserted into the sacrum location and tail end. After the removal of hind limb fur, two ring positive and negative electrodes were placed on the gastrocnemius muscle with contact gel (the positive placed on Achille’s tendon and the negative on top of the GAST muscle). The stimulus needle electrode was inserted into muscle near the sciatic nerve. 1 to 10 mV stimulus with 0.1 ms duration were applied at a frequency of 10 Hz.
Cellular, mitochondrial and MAMs fractionation
Mice were sacrificed using CO2 or isoflurane followed by cervical dislocation. Spinal cord or brain tissues were collected and homogenized in the solution with 225 mM mannitol, 75 mM sucrose, 0.1 mM EGTA, 1M Tris-HCl (pH =7.4). After centrifugation of total homogenate at 740 g at 4°C for 5 min twice to separate the cell fragments, the supernatant was collected, further centrifuged at 9,000g for 10 min and suspended in the buffer solution with 250 mM Mannitol, and 5 mM HEPES (pH 7.4) to obtain the crude mitochondrial fraction. To further obtain purified mitochondria, the crude mitochondrial fraction was overlaid on top of Percoll medium with 225 mM mannitol, 35 mM HEPES, and 1.4 mM EGTA and 30% Percoll (vol/vol) (pH = 7.4) followed by centrifugation at 95,000 g for 30 min at 4°C in a SW50.1 rotor and pellets were washed by the buffer solution again followed by centrifugation at 18,200 g for 15 min at 4°C to obtain purified mitochondria. MAMs-containing fractionation were visible as diffused white bands located in the middle of the ultracentrifuge tube. To obtain MAMs, white bands were collected and diluted 10 times with mitochondria resuspending buffer (MRB; 250 mM mannitol, 5 mM HEPES, 0.5 mM EGTA, pH = 7.4). After centrifugation at 63,000 g for 10 min at 4°C, the supernatant was collected and further centrifuged at 100,000 g for 60 min. The pellet containing MAMs was finally washed by MRB buffer followed by centrifugation at 18,200 g for 15 min at 4°C.
To test whether calpastatin was tightly associated with MAMs, purified MAMs fraction was resuspended in 30ul MRB buffer with different concentrations of digitonin (w/v). After a 30-min incubation on ice, samples were spun down for 30 min. The 30 ul supernatant was collected and 10 ul 4x Laemmli Sample Buffer (Bio-Rad, Hercules, CA) was added. The pellets were dissolved in the same volume of 40ul 1 x Laemmli Sample Buffer. For trypsin digestion, MAMs fraction were resuspended in HBSS buffer or 0.125 mg/ml trypsin solution with or without 0.2% Triton-X100, followed by incubation on ice for 30 min. The reaction was terminated by adding 4 x Laemmli Sample Buffer.
Expression vectors, chemicals, immunoblot and immunoprecipitation
MitoDsRed2-ER (Clontech, Mountain View, CA) and mito-TagBFP2 (Evrogen JSC, Moscow, Russia) were obtained. GFP tagged calpastatin constructs were generated by cloning human calpastatin and GFP into PCMV-3Tag-1a (Agilent, Santa Clara, CA). The EGFP or Flag tagged calpastatin expression construct was generated by Gibson assembly with PCR-amplified EGFP or 4 x Flag tag fragment, calpastatin fragment and linearized pcDNA3.1(+) backbone plasmid (Invitrogen, Carlsbad, CA). For targeting the FKBP12 protein to the cytoplasmic surface of the mitochondria (OMM), the N-terminal sequence of the mAKAP1 (34–63) was fused to the N-terminus of human FKBP12 protein through a linker (DPTRSANSGAGAGAGAILSR). The fusion proteins were tagged with TagGFP or Myc at the C terminus. For targeting the TagRFP or Flag-tagged FRB protein to the cytoplasmic surface of the ER, the targeting sequence of Sal protein was fused to the C-terminus of these fusion proteins. All these plasmids were cloned into pcDNA3.1(+) vector. Primary antibodies used in this study are listed. All chemicals were obtained from Sigma (Sigma St. Louis, MO). Recombinant human calpastatin was obtained from Abnova (Walnut, CA). Mouse or human tissue samples were all lysed with Cell Lysis Buffer (Cell Signaling, Danvers, MA) plus 1 mM PMSF (Sigma-Aldrich, St. Louis, MO) and Protease Inhibitor Cocktail (Roche, Indianapolis, IN). Equal amounts of total protein extract were resolved by SDS-PAGE and transferred to Immobilon-P (EMD Millipore, Billerica MA). Following blocking with 10% nonfat dry milk, primary and secondary antibodies were applied as previously described(Wang et al., 2016) and the blots developed with Immobilon Western Chemiluminescent HRP Substrate (EMD Millipore, Billerica MA or Santa Cruz, Dallas, TX). For immunoprecipitation, isolated mitochondria were first lysed by immunoprecipitation lysis buffer (50 mM Tris, pH 7.4, 250 mM NaCl, 5 mM EDTA, 50 mM NaF, 1 mM Na3VO4, 1% Nonidet P40 and 0.02% NaN3) containing protease inhibitor cocktail and phosphatase inhibitor cocktail (Roche, Indianapolis, IN). The homogenate was then centrifuged at 18,000 g at 4°C for 30 min. The supernatant was collected and incubated with magnetic beads bound by Mfn2 antibody overnight at 4°C. Magnetic beads were then washed with immunoprecipitation lysis followed by immunoblot.
Immunocytochemistry and immunofluorescence
Immunocytochemistry was performed by the peroxidase anti-peroxidase protocol as we described(Wang et al., 2016). For immunofluorescent staining of NMJs, GAST muscles were isolated and fixed in 10% formalin (Sigma-Aldrich, St. Louis, MO) for 30 minutes. Following washing 3 times with PBS (137mM NaCl, 2.7mM KCl, 10mM Na2HPO4, 17.6mM KH2PO4, pH = 7.4), muscles were immersed into 30% (w/v) sucrose in PBS at 4°C until muscles sunk to the bottom. After embedding in Tissue-Plus O.C.T Compound (Fisher, Pittsburgh, PA), the GAST muscle was longitudinally sectioned into 20 μm cryosections. Followed by drying at 37°C, the cryosections were washed 3 times with PBS shaking. After incubation overnight at 4°C in blocking buffer (2% BSA, 0.2% non-fat dry milk, 5% normal goat serum and 0.5% Triton X-100 in PBS), primary antibody in PBS with 2% BSA and 5% normal goat serum was applied for 48 hours at 4°C. The slides were then washed with PBS, followed by 2-hour incubation with Alexa Fluor 488 or 568 dye labeled second antibody and α-bungarotoxin conjugated to either Alex Fluor 488 or 555 (to label AchR, Invitrogen, Carlsbad, CA) in room-temperature and subsequent PBS wash. All slides were mounted with Fluoromount-G mounting medium (SouthernBiotech, Birmingham, AL).
Confocal microscopy and electron microscopy
All confocal images were captured with a Leica SP8 confocal microscopy equipped with confocal class objectives (10x, 20x, 40x, 60x, 100x), a motorized super Z galvo stage, two PMTs, 3 Hyd SP GaAsP detectors for gated imaging, and the AOBS system lasers including a 405 nm, Argon (458, 476, 488, 496, 514 nm), a tunable white light (470 to 670 nm), and a 592 nm STED depletion laser. For time-lapse imaging, neurons were seeded in glass bottomed 35mm dishes, and co-transfected with indicated plasmids. Neurons were imaged by the Leica SP8 confocal microscopy well-equipped with environmental chamber with controlled CO2 content, humidity and temperature (37°C). Images were captured with lowest intensity to avoid photobleaching or signal oversaturation. During time-lapse imaging, frames were captured every 15 s for at least 1 h without phototoxicity or photobleaching.
For electron microscopy (EM), mice were transcardially perfused with EM fixative (quarter strength Karnovsky-1.25% DMSO mixture) for 5 minutes and then spinal cords, sciatic nerves and skeletal muscles were removed quickly and placed in fresh EM fixative solution for an additional 15 mins at RT followed by additional fixation in fresh EM fixative for another 2 hours at RT. Spinal cords or spinal nerve roots were then dissected out. After washing, tissue blocks were postfixed in 1% osmium–1.25 % ferrocyanide mixture for two changes of solution of one hour each (total postfixation time is two hours) at RT. Then the specimens were rinsed, and soaked overnight in acidified 0.5% uranylacetate. After another wash, the tissue blocks were dehydrated in ascending concentrations of ethanol, passed through propylene oxide, and embedded in Poly/Bed 812 embedding resin (Polysciences, PA). Thin sections were sequentially stained with 2 % acidified uranyl acetate followed by Sato’s triple lead staining as modified by Hanaichi et al.(Hanaichi et al., 1986) and examined in an FEI Tecnai T12 electron microscope equipped with a Gatan single tilt holder and a Gatan US4000 4kx4k CCD camera. EM thin sections (1 μm) of spinal cords and nerve roots were also stained with toluidine blue and examined with a light microscope.
For immuno-EM, isolated MAMs were immunolabeled with antibodies (anti-Mfn2 or anit-CAST) at 1:10~1:30 dilution in PBS containing 1% w/v bovine serum albumin (BSA), 1% v/v normal goat serum and 0.01% v/v Tween 20 (PBGT) for 12 h at 4°C. The MAMs were then washed 3 times in PBS containing 5 % v/v normal goat serum (PBSG) and were incubated for 2 h in 10nm gold-conjugated goat anti-rabbit IgG and 15 nm gold-conjugated goat anti-mouse IgG (British BioCell International, Ted Pella, Inc., Redding, CA) diluted 1:50 in PBSG, and rinsed with PBSG. After rinsing 2 times in PBS, the pellet was fixed with glutaraldehyde to stabilize the gold particles. The MAMs were postfixed in 1% osmium–1.25 % ferrocyanide mixture, and rinsed in distilled water. The postfixed pellets were embedded in 1.5 % low gel temperature agarose. Another water rinse was followed by an overnight soak in acidified 0.5 % uranyl acetate. After rinsing in distilled water, the block was processed as for the regular EM (see above).
RNA-seq and bioinformatics
For mRNA-seq analyses, total RNA samples prepared from Transgenic SOD1 G93A mice (G93A mice), the double transgenic mice (dTg mice) and the non-transgenic mic (NTG) were submitted to the Mayo Clinic Medical Genome Core Facility (Rochester, MN) for library construction and sequencing. Total RNA was extracted using the RNAeasy Plus Mini Kit (QIAGEN, Germantown, MD, USA), as per manufacturer’s instructions. Paired-end sequencing libraries were prepared using the TruSeq Stranded Total Sample Preparation kit (Illumina) by the Mayo Clinic sequencing core facilities followed by quality control, cluster generation and sequencing on the Illumina HiSeq 2000 platform. Sequence alignment was performed using TopHat v2.0.12 against the UCSC mm10 Assembly. Expression values were calculated with featureCounts v1.4.6-p2, and differential expression analysis was determined by DESeq2 (PMID 25516281) and the downstream statistical analyses and plots were made in R (v3.1.1; http://www.r-project.org/). Pathway overrepresentation analysis was performed using R package ReactomePA (Subramanian et al., 2005).
Image analysis
Image analysis was performed with open-source image analysis programs WCIF ImageJ (developed by W. Rasband) or Imaris (Bitplane, CT). All confocal images of mitochondria and NMJs were reconstructed using “easy 3D” module in Imaris. For each mouse, at least 200 NMJs were counted for quantification. Mitochondria morphology was quantified as we described (Wang et al., 2008). Taken briefly, a series of z-stacks of raw images were background corrected, linearly contrast optimized, applied with a 7 × 7 ‘top hat’ filter, subjected to a 3 × 3 median filter and then thresholded to generate binary images. Most mitochondria were well separated in binary images and large clusters of mitochondria were excluded automatically. All binary images were further analyzed by Image J. Mitochondrial morphology in EM micrographs were directly analyzed by image J.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical Analysis
Statistical analysis was done with one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test or student-t-test using GraphPad Prism (GraphPad, CA). Data are means ± s.e.m. n represents number of neurons, cells or mice per experiment. p < 0.05 was considered to be statistically significant.
Key Resource Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-2A peptide | EMD Millipore | Cat# ABS31, RRID: AB_11214282 |
| Mouse monoclonal anti-Neurofilament (clone 2H3) | Developmental Studies Hybridoma Bank, University of Iowa | Cat# 2H3, RRID: AB_531793 |
| Mouse monoclonal anti-Myosin heavy chain (clone 6H1) | Developmental Studies Hybridoma Bank, University of Iowa | Cat# 6H1, RRID: AB_1157897 |
| Mouse monoclonal anti-Actin (clone C4) | EMD Millipore | Cat# MAB1501, RRID: AB_2223041 |
| Rabbit polyclonal anti-Agrin | Abcam | Cat# ab85174, RRID: AB_1860988 |
| Mouse monoclonal anti-ATP5A (clone 15H4C4) | Abcam | Cat# ab14748, RRID: AB_301447 |
| Rabbit polyclonal anti-Bax | Cell Signaling Technology | Cat# 2772S, RRID: AB_10695870 |
| Rabbit polyclonal anti-Bim | Cell Signaling Technology | Cat# 2819S, RRID: AB_10692515 |
| Rabbit polyclonal anti-Calnexin | Enzo Life Sciences | Cat# ADI-SPA-860-F, RRID: AB_11178981 |
| Rabbit polyclonal anti-Calpain1 Large Subunit (Mu-type) | Cell Signaling Technology | Cat# 2556S, RRID: AB_10692800 |
| Rabbit polyclonal anti-Calpain2 Large Subunit (M-type) | Cell Signaling Technology | Cat# 2539S, RRID: AB_10694687 |
| Mouse monoclonal anti-Calpastatin (clone PI-11) | Santa Cruz Biotechnology | Cat# sc-32324, RRID: AB_831042 |
| Rabbit polyclonal anti-Calpastatin | Cell Signaling Technology | Cat# 4146S, RRID: AB_2244162 |
| Mouse monoclonal anti-Calregulin (clone F-4) | Santa Cruz Biotechnology | Cat# sc-373863, RRID: AB_10915425 |
| Rabbit monoclonal anti-cleaved Caspase-3 (Asp175) (clone 5A1E) | Cell Signaling Technology | Cat# 9664, RRID: AB_2070042 |
| Mouse monoclonal anti-COX IV (clone 4D11-B3-E8) | Cell Signaling Technology | Cat# 11967 |
| Mouse monoclonal anti-DLP1 | BD Biosciences | Cat# 611112, RRID: AB_398423 |
| Rabbit polyclonal anti-p44/42 MAPK (Erk1/2) | Cell Signaling Technology | Cat# 9102, RRID: AB_330744 |
| Rabbit polyclonal anti-p44/42 MAP kinase (phosphorylated Erk1/2) | Cell Signaling Technology | Cat# 9101, RRID: AB_331646 |
| Rabbit monoclonal anti-FACL4 (clone EPR8640) | Abcam | Cat# ab155282, RRID: AB_2714020 |
| Rabbit monoclonal anti-GAPDH (clone 14C10) | Cell Signaling Technology | Cat# 2118, RRID: AB_561053 |
| Rabbit monoclonal anti-GFP (clone E385) | Abcam | Cat# ab32146, RRID: AB_732717 |
| Rabbit monoclonal anti-AMPA Receptor 1 (GluA1) (clone D4N9V) | Cell Signaling Technology | Cat# 13185 |
| Rat monoclonal anti-GRP78 (76-E6) | Santa Cruz Biotechnology | Cat# sc-13539, RRID: AB_627698 |
| Rabbit monoclonal anti-GSK3β (clone D5C5Z) | Cell Signaling Technology | Cat# 12456, RRID: AB_2636978 |
| Rabbit monoclonal anti-phospho-GSK-3 (Ser9) (clone D85E12) | Cell Signaling Technology | Cat# 5558, RRID: AB_10013750 |
| Rabbit polyclonal anti-Histone deacetylase 1 (HDAC1) | Cell Signaling Technology | Cat# 2062S, RRID: AB_10698751 |
| Rabbit polyclonal anti-Hsp60 | Abcam | Cat# ab46798, RRID: AB_881444 |
| Mouse monoclonal anti-JNK1 (clone 2C6) | Cell Signaling Technology | Cat# 3708, RRID: AB_1904132 |
| Rabbit polyclonal anti-phospho-SAPK/JNK (Thr183/Tyr185) | Cell Signaling Technology | Cat# 9251, RRID: AB_331659 |
| Rabbit polyclonal anti-MEK1/2, phospho (Ser217/Ser221) | Cell Signaling Technology | Cat# 9121S, RRID: AB_331648 |
| Rabbit monoclonal anti-LC3A/B (clone D3U4C) | Cell Signaling Technology | Cat# 12741, RRID: AB_2617131 |
| Rabbit polyclonal anti-MFF | Abcam | Cat# ab81127, RRID: AB_1860496 |
| Rabbit polyclonal anti-Mfn1 (clone H-65) | Santa Cruz Biotechnology Cat# sc-50330, RRID: AB_2250540 | |
| Mouse monoclonal anti-Mfn2 (clone XX-1) | Santa Cruz Biotechnology | Cat# sc-100560, RRID: AB_2235195 |
| Mouse monoclonal anti-Mitofusin 2 | Abcam | Cat# ab56889, RRID: AB_2142629 |
| Rabbit monoclonal anti-Mitofusin 2 (clone D1E9) | Cell Signaling Technology | Cat# 11925 |
| Mouse monoclonal anti-Miro1 (clone 4h4) | Novus Biologicals | Cat# H00055288-M01, RRID: AB_2179528 |
| Rabbit polyclonal anti-MuSK | Abcam | Cat# ab5510, RRID: AB_304946 |
| Mouse monoclonal anti-NDUFB8 (clone 20E9DH10C12) | Abcam | Cat# ab110242, RRID: AB_10859122 |
| Mouse monoclonal anti-Neurofilament-H (clone RMdO 20) | Cell Signaling Technology | Cat# 2836S, RRID: AB_10694081 |
| Rabbit monoclonal anti-Neurofilament-L (clone C28E10) | Cell Signaling Technology | Cat# 2837, RRID: AB_823575 |
| Mouse monoclonal anti-OPA1 (Clone 18) | BD Biosciences | Cat# 612606, RRID: AB_399888 |
| Mouse monoclonal anti-Myosin | Developmental Studies | Cat# SC-71, RRID: AB_2147165 |
| Heavy Chain Type IIA (clone SC-71) | Hybridoma Bank, University of Iowa | |
| Mouse monoclonal anti-SDHB (clone 21A11) | Abcam | Cat# ab14714, RRID: AB_301432 |
| Mouse monoclonal anti-Spectrin (clone 17C7) | Abcam | Cat# ab11751, RRID: AB_2194328 |
| Rabbit polyclonal anti-Sigma1 receptor (SigR1) C-terminal | Abcam | Cat# ab53852, RRID: AB_881796 |
| Rabbit polyclonal anti-SOD1 | Santa Cruz Biotechnology Cat# sc-11407, RRID: AB_2193779 | |
| Mouse monoclonal anti-Synaptic vesicle glycoprotein 2A (SV2) | Developmental Studies Hybridoma Bank, University of Iowa | Cat# SV2, RRID: AB_2315387 |
| Mouse monoclonal anti-α-Tubulin (clone B-5-1-2) | Thermo Fisher Scientific | Cat# 32-2500, RRID: AB_2533071 |
| Rabbit monoclonal anti-βIII-Tubulin (clone D65A4) | Cell Signaling Technology | Cat# 5666S, RRID: AB_10694388 |
| Mouse monoclonal anti-Tau (clone Tau46) | Cell Signaling Technology | Cat# 4019S, RRID: AB_10695394 |
| Mouse monoclonal anti-Tom20 (clone 29) | BD Biosciences | Cat# 612278, RRID: AB_399595 |
| Mouse monoclonal anti-UQCRC2 (clone 13G12) | Abcam | Cat# ab14745, RRID: AB_2213640 |
| Mouse monoclonal anti-VDAC1 (clone 20B12) | Abcam | Cat# ab14734, RRID: AB_443084 |
| Rabbit monoclonal anti-Wnt3a (clone C64F2) | Cell Signaling Technology | Cat# 2721S, RRID: AB_2215411 |
| Anti-rabbit IgG, HRP-linked | Cell Signaling Technology | Cat# 7074S, RRID: AB_2099233 |
| Anti-mouse IgG, HRP-linked | Cell Signaling Technology | Cat# 7076S, RRID: AB_330924 |
| Goat anti-Rabbit IgG (H+L) Secondary Antibody, Alexa Fluor 488 conjugate | Thermo Fisher Scientific | Cat# A-11034, RRID: AB_2576217 |
| Goat anti-Rabbit IgG (H+L) Cross- Adsorbed Secondary Antibody, Alexa Fluor 568 | Thermo Fisher Scientific | Cat# A-11011, RRID: AB_143157 |
| Goat anti-Mouse IgG (H+L) Cross- Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-11001, RRID: AB_2534069 |
| Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Thermo Fisher Scientific | Cat# A-11031, RRID: AB_144696 |
| Adeno-Associated Virus Serotype 1 | ||
| AAV1-calpastatin (AAV1-eSYN-hCAST-T2A-EGFP) | Vector BioLabs | Cat# 2016-1226 |
| AAV1-Cre (AAV1-eSYN-eGFP-T2A-iCre) | Vector BioLabs | Cat# 160411-171208A |
| AAV1-GFP (AAV1-eSYN-EGFP- WPRE) | Vector BioLabs | Cat# 150608-160726 |
| AAV1-shCon. (AAV1-GFP-U6- Scrmb-shRNA) | Vector BioLabs | Cat# 170522-171208 |
| AAV1-shMiro1 (AAV1-GFP-U6-m-Miro1-shRNA) | Vector BioLabs | Cat# 170501-17 |
| Biological Samples (Human Spinal Cord Samples) | ||
| Frozen ALS sample (Age: 41 year old; Sex: male; Postmortem interval: 21 hours) | University of Maryland, Baltimore (UMB) | UMB Case# 4627 |
| Frozen ALS sample (Age: 45 year old; Sex: male; Postmortem interval: 19 hours) | University of Maryland, Baltimore (UMB) | UMB Case# 4768 |
| Frozen ALS sample (Age: 49 year old; Sex: male; Postmortem interval: 9 hours) | University of Maryland, Baltimore (UMB) | UMB Case# 4528 |
| Frozen ALS sample (Age: 59 year old; Sex: male; Postmortem interval: 6 hours) | University of Maryland, Baltimore (UMB) | UMB Case# 4762 |
| Frozen ALS sample (Age: 61 year old; Sex: female; Postmortem interval: 22 hours) | University of Maryland, Baltimore (UMB) | UMB Case# 5004 |
| Frozen ALS sample (Age: 61 year old; Sex: male; PMI: 8 hours) | University of Maryland, Baltimore (UMB) | UMB Case# 5388 |
| Frozen ALS sample (Age: 70 year old; Sex: male; Postmortem interval: 14 hours) | University of Maryland, Baltimore (UMB) | UMB Case# 4871 |
| Frozen ALS sample (Age: 71 year old; Sex: female; Postmortem interval: 6 hours) | University of Maryland, Baltimore (UMB) | UMB Case# 4872 |
| Frozen control Sample (Age: 48 year old; Sex: male; Postmortem interval: 21 hours) | University of Maryland, Baltimore (UMB) | UMB Case# 145 |
| Frozen control Sample (Age: 50 year old; Sex: male; Postmortem interval: 17 hours) | University of Maryland, Baltimore (UMB) | UMB Case# 5399 |
| Frozen control Sample (Age: 55 year old; Sex: male; Postmortem interval: 24 hours) | University of Maryland, Baltimore (UMB) | UMB Case# 1471 |
| Frozen control Sample (Age: 58 year old; Sex: male; Postmortem interval: 18 hours) | University of Maryland, Baltimore (UMB) | UMB Case# 840 |
| Frozen control Sample (Age: 69 year old; Sex: male; Postmortem interval: 12 hours) | University of Maryland, Baltimore (UMB) | UMB Case# 946 |
| Frozen control Sample (Age: 70 v; Sex: female; Postmortem interval: 24 hours) | University of Maryland, Baltimore (UMB) | UMB Case# 5572 |
| Fixed ALS sample (Age: 80 year old; Sex: N/A; Postmortem interval: N/A) | University Hospitals of Cleveland | N/A |
| Fixed ALS sample (Age: 49 year old; Sex: N/A; Postmortem interval: N/A) | University Hospitals of Cleveland | N/A |
| Fixed ALS sample (Age: N/A; Sex: N/A; Postmortem interval: N/A) | University Hospitals of Cleveland | N/A |
| Fixed control sample (Age: 52 year old; Sex: N/A; Postmortem interval: N/A) | University Hospitals of Cleveland | N/A |
| Fixed control sample (Age: 63 year old; Sex: N/A; Postmortem interval: N/A) | University Hospitals of Cleveland | N/A |
| Fixed control sample (Age: 64 year old; Sex: N/A; Postmortem interval: N/A) | University Hospitals of Cleveland | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Artificial Tears Ointment 1/8 oz | Rugby Laboratories Inc | Cat# 05366550 |
| Bovine Serum Albumins (BSA) | Sigma-Aldrich | Cat# A7030-100G |
| α-Bungarotoxin, Alexa Fluor™ 488 conjugate | Thermo Fisher Scientific | Cat# B13422 |
| α-Bungarotoxin, Alexa Fluor™ 555 conjugate | Thermo Fisher Scientific | Cat# B35451 |
| Calpastatin recombinant protein (human) | Abnova | Cat# H00000831-P01 |
| Cell Lysis Buffer (10X) | Cell Signaling Technology | Cat# 9803 |
| Digitonin | Sigma-Aldrich | Cat# D141-100MG, EC Number: 234-255-6 |
| DNase I | Worthington Biochemical Corporation | Cat# LS006362 |
| Fluoromount-G™ Slide Mounting Medium | SouthernBiotech | Cat# 0100-01 |
| Immobilon Western Chemiluminescent HRP Substrate | EMD Millipore or Santa Cruz | Cat# WBKLS0500 or Cat#sc-2048 |
| Normal Goat Serum | Thermo Fisher Scientific | Cat# 50062Z |
| Lipofectamine 2000 Transfection Reagent | Thermo Fisher Scientific | Cat# 11668-019 |
| Phenylmethanesulfonyl fluoride (PMSF) | Sigma-Aldrich | Cat# 329-98-6 |
| PhosSTOP Phosphatase Inhibitor Cocktail | Roche | Cat# 04906845001 |
| Poly-L-Lysine Hydrobromide | Sigma-Aldrich | Cat# 9155-5MG |
| Premixed 4 x Laemmli protein sample buffer for SDS-PAGE | Bio-Rad Laboratories | Cat# 1610747 |
| Protease Inhibitor Cocktail Tablets | Roche | Cat# 05892791001 |
| Tissue-Plus O.C.T. Compound | Thermo Fisher Scientific | Cat# 23730571 |
| Triton™ X-100 | Sigma-Aldrich | Cat# X100-500ML |
| Tween™ 20 | Thermo Fisher Scientific | Cat# 85113 |
| Culture Medium | ||
| B-27™ Supplement (50X) | Thermo Fisher Scientific | Cat# 17504044 |
| Fetal Bovine Serum | Thermo Fisher Scientific | Cat# 10438026 |
| Hank’s Balanced Salt Solution | Thermo Fisher Scientific | Cat# 14170112 |
| Hibernate E | BrainBits, LLC | Cat# 1835267 |
| Opti-MEM™ I Reduced Serum Medium | Thermo Fisher Scientific | Cat# 31985-070 |
| Trypsin-EDTA (0.25%) with phenol red | Thermo Fisher Scientific | Cat# 25200-072 |
| Critical Commercial Assays | ||
| DAB Chromogen Kit | Biocare Medical | Cat# DB801L |
| QIAprep Spin Miniprep Kit | QIAGEN | Cat# 27106 |
| RNAeasy Mini Kit | QIAGEN | Cat# 74104 |
| TruSeq Stranded Total Sample Preparation Kit | Illumina | Cat# RS-122-2001 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Mfn2flox/flox mice | David Chan | |
| Mouse: TMFN mice (transgenic mice overexpressing Mfn2 in neurons) | This paper | N/A |
| Mouse: G93A mice (B6.Cg-Tg(SOD1*G93A)1Gur/J) | the Jackson Laboratory | Cat# 004435 |
| Mouse: dTg mice (double transgenic mice by crossing TMFN mice with G93A mice) | This paper | N/A |
| Mouse: CASTKI mice | This paper | N/A |
| Mouse: Thy1-Cre mice (FVB/N-Tg(Thy1-cre)1Vln/J) | the Jackson Laboratory | Cat# 006143 |
| Mouse: TCAST mice (double transgenic mice by crossing CASTKI mice with Thy1-Cre mice) | This paper | N/A |
| Mouse: Aged C57BL/6J mice | NIA Aged Rodent Colonies | N/A |
| Mouse: C57BL/6J mice | the Jackson Laboratory | Cat# 000664 |
| Rat: Sprague Dawley rats | Charles River | N/A |
| Oligonucleotides | ||
| Rosa26-L-GT-F CCTCAGAGAGCCTCGGCTAG |
This paper | N/A |
| Rosa26-R-GT-R CATGAGTCAAGCCAGTCCAAGAG |
This paper | N/A |
| CAG-L-GT-R AGTCCCTATTGGCGTTACTATGG |
This paper | N/A |
| WPRE-R-GT-F ACGAGTCGGATCTCCCTTTGG |
This paper | N/A |
| Recombinant DNA | ||
| pDsRed2-Mito | Clontech | Cat# 632421 |
| pDsRed2-ER | Clontech | Cat# 632409 |
| pcDNA-3.1-Flag-MFN2 | This paper | N/A |
| pcDNA-3.1-Flag-MFN2ActA | This paper | N/A |
| pcDNA-3.1-Flag-MFN2IYFFT | This paper | N/A |
| pcDNA-3.1-Flag-MFN2IYFFTΔRAS | This paper | N/A |
| pcDNA-3.1-Flag-MFN2ΔRAS | This paper | N/A |
| pcDNA-3.1-Flag-MFN2R94Q | This paper | N/A |
| pcDNA-3.1-EGFP-Calpastatin | This paper | N/A |
| pcDNA-3.1-DsRed-FRB-ER | This paper | N/A |
| pcDNA-3.1-Mito-GFP | This paper | N/A |
| pcDNA-3.1-Mito-FKBP-GFP | This paper | N/A |
| pcDNA-3.1-DsRed-FRB-ER | This paper | N/A |
| mito-TagRFP2 | Evrogen | Cat# FP148 |
| PCMV-3Tag-1a | Agilent Technologies | Cat# 240195 |
| pcDNA3.1(+) | Thermo Fisher Scientific | Cat# V790-20 |
| Software and Algorithms | ||
| Bitplane Imaris 7.2.3 | Bitplane | http://www.bitplane.com/ |
| DESeq2 | Bioconductor | http://www.bioconductor.org/ |
| FeatureCount v1.4.6-p2 | The Walter and Eliza Hall Institute of Medical Research | http://subread.sourceforge.net/ |
| GraphPad Prism | GraphPad | http://www.graphpad.com/ |
| Primer-3 v.0.4.0 | Bioinformatics 23(10):1289-91 | http://bioinfo.ut.ee/primer3-0.4.0/ |
| Primer-BLAST | National Institutes of Health | https://www.ncbi.nlm.nih.gov/tools/primer-blast/ |
| R v3.1.1 | The R Foundation | https://www.r-project.org/ |
| TopHat v2.0.12 | Johns Hopkins University | http://bowtie-bio.sourceforge.net/index.shtml |
| WCIF ImageJ | University Health Network, Toronto, Ontario | http://wwwfacilities.uhnresearch.ca/wcif/fdownload.html |
| Other | ||
| Bupivicanie 1:1 (2.5 mg/ml = 0.25%) | Institutional Animal Care and Use Committee (IACUC) at Case Western Reserve University | N/A |
| Carprofen 0.5 mg/ml | Institutional Animal Care and Use Committee (IACUC) at Case Western Reserve University | N/A |
| Isoflurane (100%) | Institutional Animal Care and Use Committee (IACUC) at Case Western Reserve University | N/A |
| Lidocaine (0.5%) | Institutional Animal Care and Use Committee (IACUC) at Case Western Reserve University | N/A |
Supplementary Material
Data S1. Representative movies of Mfn2fl/fl mice after AAV1 injection or calpastatin, ER and mitochondria trafficking in in vitro cultured primary cortical neurons (Related to Figures 1 and 5).
A 5-month old Mfn2fl/fl mouse injected with AAV1-GFP for 3 weeks demonstrates no hindlimb clasping (Movie S1) and normal movement (Movie S2), whereas a 5-month old Mfn2fl/fl mouse injected with AAV1-Cre for 3 weeks demonstrates severe hindlimb clasping (Movie S3) and hindlimb paralysis (Movie S4). Representative movie of calpastatin puncta (green) and mitochondria (red, mito-DsRed2) trafficking in axons of in vitro cultured primary cortical neurons is shown in Movie S4. Representative movies of mitochondria (green, Movie S6), ER (red, Movie S7), and merged mitochondria and ER (Movie S8) 1 min after ER and mitochondria tethering in neurons transfected with mito-FKBP-GFP and DsRed-FRB-ER are also shown.
Figure S1. Characterization of Mfn2fl/fl mice after Cre injection (related to Figure 1).
(A and B) Representative fluorescent images of the entire brain and spinal cord (A) and cross sections of spinal cord ventral horn (B) of Mfn2fl/fl mice at 2 weeks after injection.
(C) Kaplan-Meier log rank test for the onset of paralysis in Cre-injected Mfn2fl/fl mice. n = 7.
(D) Representative walking footprint patterns (arrow shows the walking direction) of 4 6 month old Mfn2fl/fl mice 3–4 weeks after injection with AAV1-GFP (Vector) or paralyzed Mfn2fl/fl mice after injection AAV1-Cre (Cre).
(E) Quantification of axons in toluidine blue staining of dorsal and ventral nerve roots of Cre-injected paralyzed Mfn2fl/fl mice or age-matched vector injected control Mfn2fl/fl mice.
(F) Representative immunoblot and quantification of Mfn2 levels in lumbar spinal cord of Mfn2fl/fl mice 2 weeks after vector or Cre injection (n = 3 mice per group).
(G) Representative immunohistochemistry and quantification of Mfn2 immunoreactivity in spinal cord motor neurons of Mfn2fl/fl mice 2 weeks after vector or Cre injection (n = 3 mice per group).
(H) Representative images and quantification of mitochondria in neuromuscular synaptic terminals in GAST muscles of paralyzed Mfn2fl/fl mice after co-injection with AAV1-mitoDsRed and AAV1-Cre (n = 13 and 11 NMJs for coverage; n = 14 and 9 neurites for length).
(I) Representative images and quantification of mitochondrial length in the cell bodies and proximal neurites of Mfn2fl/fl mice after co-injection with AAV1-mitoDsRed and AAV1-Cre or AAV1-GFP (n = 14, 16, 10 neurons for cell body and n = 22, 17 neurites for proximal motor axons).
Data are means ± s.e.m., representative of triplicate experiments. Student’s t test. **P < 0.01, ***P < 0.001, ****P < 0.0001. ns, not significant.
Figure S2. Reduced expression of Mfn2 in G93A mice and ALS patients and well-preserved skeletal muscles in dTg mice (related to Figure 2).
(A) Representative immunoblot and quantification of Mfn2 levels in endpoint G93A mice and age-matched control mice. Relative Mfn2 levels are all adjusted by GAPDH. n = 4 per group.
(B) Representative images and quantification of Mfn2 immunoreactivity in spinal cord motor neurons of young (60 day) and old (>180 day, not shown) non-transgenic (NTg) mice and transgenic mice overexpressing wild type SOD1 (“WT”) or G93A mice. ALS disease progression goes through four stages in SOD1 G93A mice based on muscle strength: pre-muscle weakness stage (PMW), a rapid declining stage (RD), a slow declining stage (SD) and a paralysis stage (PARA) (Kong and Xu, 1998). The immunoreactivity of Mfn2 in spinal cord motor neurons began to decrease significantly during the PMW stage, which preceded the onset of the rapid decline of muscle strength (RD stage). There was a further decrease of Mfn2 immunoreactivity to very low level during the RD and SD stage. However, no further reduction was observed thereafter. Not surprisingly, concurrent with Mfn2 reduction, mitochondrial length also began to decrease during the PMW stage and then declined to minimal during the RD, SD or PARA stage (data not shown). n > 20 neurons per group.
(C) Representative immunoblot and quantification of mitochondrial fission and fusion proteins in human thoracic spinal cord tissues of sporadic ALS patients (n=8) and age-matched control subjects (n=6). Equal protein amounts (20 μg) were loaded and confirmed by actin. Tom20 was used as a mitochondrial specific marker.
(D) Representative immunocytochemistry of DLP1, OPA1, Mfn1, Mfn2 and Mff in human spinal cord from sALS patient (lower panel) and age-matched control (upper panel).
(E) Quantification of Mfn2, SOD1 and VDAC1 (mitochondrial marker) levels in spinal cords, brains and GAST muscles of endpoint G93A and dTg mice and aged-matched NTG and TMFN mice. Relative Mfn2, SOD1 or VDAC1 levels are adjusted by GAPDH or VDAC1. n = 4 per group.
(F) Kaplan-Meier log rank test for survival in G93A (female = 5, male = 6) and dTg mice (female = 5, male = 8).
(G) Representative images of endpoint G93A and dTg mice and aged-matched 5–6 month old NTG and TMFN mice.
(H) Body weight of 5–6 month old NTG (female = 3, male = 5) and TMFN mice (female = 5, male = 5) and endpoint G93A (female = 3, male = 4) and dTg mice (female = 5, male = 6), or NTG (female = 9, male = 12), TMFN mice (female = 7, male = 9), G93A (female = 13, male = 14) and dTg mice (female = 9, male = 9) at 120-day old.
(I) Representative images of GAST muscles of endpoint G93A and dTg mice and age-matched NTG and TMFN mice stained by hematoxylin and eosin (H&E), ATPase (pH 4.6; dark stain for type1 fibers, light stain for type 2A fibers, and intermediate stain for type 2B/2D fibers) and antibodies against SC-71 (green, for type 2A fibers) and 6H1 (red, for type 2X fibers as described (Bloemberg and Quadrilatero, 2012)). In H&E staining, arrows point to dystrophic muscle fibers in G93A, which are absent in dTg mice. In ATPase pH 4.6 staining, there is a marked increase of slow twitch type1 fibers in G93A but not in dTg, NTG and TMFN mice. Consistent with this, G93A but not dTg mice exhibited significant loss of fast twitch type 2 fibers (light staining in ATPase pH 4.6 staining) especially type 2X fibers (6H1 immunofluorescence staining), therefore suggesting the recovery of short-duration high intensity bursts of power in dTg mice.
(J) Grip strength of hindlimbs of NTG (n = 11), TMFN mice (n = 7), G93A (n = 17) and dTg mice (n = 8) at 120-day old.
(K) walking footprint patterns and quantification of stride length of NTG (n = 4), TMFN mice (n = 4), G93A (n = 3) and dTg mice (n = 4) at 120-day old.
(L) Representative images of GAST muscle weight in NTG, TMFN mice, G93A and dTg mice at 120-day old.
(M) Representative immunoblot and/or quantification of Agrin or Wnt3a in spinal cords and/or GAST muscles of endpoint and 120-day old G93A and dTg mice and age-matched NTG and TMFN mice. n = 3 mice group. Equal protein amounts of 20 μg were loaded for all blots.
Data are means ± s.e.m., representative of triplicate experiments. One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. ns, not significant.
Figure S3. Neuronal Mfn2 protects mitochondria in motor cell bodies and alleviates motor axon degradation in G93A mice (related to Figure 2).
(A) Representative images of mitochondria in cell bodies (Inset I) and proximal neurites (Inset II) of spinal cord motor neurons of endpoint G93A and dTg mice and age-matched NTG and TMFN mice.
(B) Representative electron microscopic (EM) images and quantification of mitochondria in cell bodies of spinal cord motor neurons of endpoint G93A and dTg mice and age-matched NTG and TMFN mice. n = 7, 6, 5, 9 neurons per group.
(C) Quantification of the percentage of NMJ area covered by mitochondria in endpoint G93A and dTg mice and age-matched NTG and TMFN mice. n = 20 neurons or NMJs per group.
(D) Quantification of mitochondrial length in proximal motor neurites in spinal cords and distal motor neurites in GAST muscles. n = 11 14 motor neurons or NMJs from 3 mice per group.
(E) Representative H&E staining and quantification of lumbar spinal cord motor neurons of endpoint G93A (n = 6) and dTG (n = 6) mice and age-matched NTG (n = 5) and TMFN (n = 3) mice. Despite unalleviated spinal cord motor neuron loss, the vacuolar pathology was decreased in spinal cords in dTg mice.
(F and G) Representative EM images (F) and toluidine blue staining (G) of Epon-embedded sections of L3–5 ventral nerve roots of endpoint G93A and dTG mice and age-matched NTG and TMFN mice.
(H) Representative western blot of proteins involved in apoptosis, MAPK/GSK3β signaling, mitochondrial bioenergetics (OXPHOS: oxidative phosphorylation) and cytoskeleton formation in spinal cords of endpoint G93A and dTG mice and age-matched NTG and TMFN mice. Equal protein amounts of 20 μg were loaded for all blots.
(I) Transcriptome analysis of spinal cords of NTG, TMFN, G93A and dTg mice. Pathway overrepresentation analysis was performed using upregulated genes with fold changes no less than 2 from G93A mice versus NTG mice, dTg mice versus NTG mice and TMFN mice versus NTG mice respectively. The length of each bar indicates the number of upregulated genes found in each pathway, while the color indicates the p-value of each overrepresentation test.
Data are means ± s.e.m., representative of triplicate experiments. One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.001. ns, not significant.
Figure S4. Calpastatin expression and trafficking in mice and cultured primary neurons (related to Figure 5).
(A) Representative images and quantification of calpastatin (green), AchR (red) and SV2/2H3 (white) in hindlimb skeletal GAST muscles of 4 6 month old NTG and TMFN mice. n = 14 or 15 NMJs per group. The specificity of antibody to was confirmed by the absence of calpastatin immunoreactivity in TMFN mouse GAST muscles stained by the calpastatin antibody pre-adsorbed with calpastatin recombinant protein “TMFN (Pre-adsorbed)”.
(B and C) Quantification of the relative levels of Calpain1, Calpain2 or VDAC1 to GAPDH in 4 6 month old NTG and TMFN mice (B) or 4 6 month old Mfn2fl/fl mice 12 days after injection with AAV1-GFP (Vector) or AAV1-Cre (Cre) (C) (n = 3 or 4 mice per group).
(D) Representative immunocytochemistry of calpastatin in lumbar spinal cords of 4 6 month old Mfn2fl/fl mice 12 days after injection with AAV1-GFP (Vector) or AAV1-Cre (Cre). Note that the immunocytochemistry of Mfn2 is shown in Figure S1G.
(E) Schematic diagram of lumbar spinal cord injection of AAV1-calpastatin in NTG mice. T2A autocleavage generates T2A tagged calpastatin separately from GFP.
(F) Representative confocal images and line-scan analysis of calpastatin (green), mitochondria (blue) and ER (red) in rat primary cortical neurons (DIV7) co-transfected with GFP tagged calpastatin (GFP-calpastatin), mitoBFP and ER-DsRed at DIV5. Representative line-scan analysis of the co-localization between Calpastatin and mitochondria in white solid lines with “2” width shown in the merged low panel with light exposure. The line-scan analysis was performed by Image J RGB Profile Plot plugin as described before(Wang et al., 2016) using high resolution images shown in the enlargements. Further quantification of degree of co-localization between calpastatin and mitochondrial marker mitoBFP in axons shows a Manders’s coefficient of 0.76 ± 0.124 (n = 8 neurons). Manders’s coefficient is % of both red and green signal co-localize (range from 0–1) and independent of fluorescence intensity. A Mander’s coefficient greater than 0.5 is considered significant co-localization (“0” means non-overlapping while “1” indicates complete overlapping).
(G) Time lapse imaging of calpastatin (green) and mitochondria (red) in live cultured rat primary cortical neurons (DIV7) co-transfected with GFP-calpastatin and mitoDsRed at DIV5. calpastatin puncta (arrowhead) could move in a short distance in the fast transport mode and dissemble slowly. The time lapse images here or Movie S5 clearly show the continuous moving pattern of the puncta (it is more obvious when Movie S5 is played in slower speeds).
(H K) Quantification of the velocity of calpastatin moving puncta (H), calpastatin axonal transport (indexed by the relative ratio of the mean gray value of calpastatin in a segment of neuronal process 200 μm length beginning from the cell body of neurons to the mean gray value of calpastatin in the cell body of neurons, I), the proportion of mobile mitochondria (J) and ER axonal transport (indexed by the relative ratio of the mean gray value of ER-DsRed in a segment of neuronal process 200 μm length beginning from the cell body of neurons to the mean gray value of calpastatin in the cell body of neurons, K) in neurons (DIV7) overexpressing or knocking down Mfn2 or Miro1 (Mfn2 or Miro1 constructs also encoding GFP for the identification of positively transfected neurons). Neurons were co-transfected with RFP tagged Calpastatin, ER-DsRed2 or mitoDsRed at DIV5 for the live imaging of calpastatin, mitochondria and ER. For BAPTA/AM-related experiments, neurons were also transfected with/without Mfn2 overexpression constructs at DIV5, treated with 1μM BAPTA/AM at DIV6 for 24 hours, and fixed at DIV7 for quantification. n > 20 neurons per group.
(L) Quantification of the relative levels of Mfn2 or VDAC1 to GAPDH in 3–4 month old NTG mice 12 days after injection of AAV1-shMiro-1. n = 3 or 4 mice per group.
Data are means ± s.e.m., representative of triplicate experiments. One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (compared with control neuron), ####P < 0.0001 (compared with neurons overexpressing Mfn2). ns, not significant.
Figure S5. EM, immuno-EM, immunoblot and confocal microscopic analysis of calpastatin, mitochondria, MAMs and/or ER (related to Figure 5).
(A and B) Representative EM micrographs of purified mitochondria (A) or MAMs (B) isolated from spinal cords of NTG mice. The isolation and purification of highly purified mitochondria were based on percoll gradient as we described(Wang et al., 2016).
(C and D) Representative immuno-EM analysis of calpastatin (labeled by 15 nm gold particles) and Mfn2 (labeled by 10 nm gold particles) in crude mitochondria (containing both mitochondria and MAMs, C) isolated from spinal cords or motor neurons in spinal cords (D) of NTG mice at 3–4 month old.
(E) Representative western blot detection of calpastatin in MAMs isolated from pooled spinal cords of NTG and TMFN mice at 3–4 month old, 4 6 month old Mfn2fl/fl mice 12 days after injection with AAV1-GFP (Vector) or AAV1-Cre (Cre), or 160-day old NTG and TMFN mice. H: total spinal cord homogenate; pM: highly purified mitochondria isolated from spinal cords; S: post mitochondrial supernatant. Equal protein amounts of 20 μg were loaded for all blots. All data are representative of triplicate experiments.
(F) Representative EM micrographs and quantification of membranes associated with mitochondria in sciatic nerves of NTG and TMFN mice at 3 4 month old showing the increased association of membranous structures with mitochondria. Red arrow heads point MAMs associated with mitochondria. n = 12, 13 per group.
(G) Representative image and quantification of the localization of ER-derived vesicles on mitochondria in a segment of neuronal process 200 μm length beginning from the cell body of neurons in neurons (DIV7) overexpressing or knocking down Mfn2 or Miro1. Neurons were co-transfected with ER-DsRed2 at DIV5 and fixed for imaging at DIV7. n > 20 neurons per group.
(H) Quantification of the relative levels of Calnexin to GAPDH in 4 6 month old NTG and TMFN mice, 4 6 month old Mfn2fl/fl mice 12 days after injection with AAV1-GFP (Vector) or AAV1-Cre (Cre) or 3 4 month old NTG mice 12 days after injection of AAV1-shMiro-1. n = 3 or 4 mice per group.
(I) Quantification of the calpastatin axonal transport (indexed by the relative ratio of the mean gray value of calpastatin in a segment of neuronal process 200 μm length beginning from the cell body of neurons to the mean gray value of calpastatin in the cell body of neurons) in neurons (DIV7) co-expressing indicated Mfn2 mutants with GFP-tagged calpastatin. n > 20 neurons per group.
(J and K) Representative large field confocal images of mitochondria and ER in live cultured neurons 2 days after co-transfection with mito-FKBP-GFP and DsRed-FRB-ER before and after rapamycin treatment (related to Figure 5I).
Data are means ± s.e.m., representative of triplicate experiments. One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. **P < 0.01, ***P < 0.001, ****P < 0.0001 (G and I; compared with control neuron). ns, not significant.
Figure S6. Calpastatin protects axons but not NMJs in mice with sciatic nerve transection or G93A mice (related to Figure 6).
(A) Representative western blot detection of human calpastatin and Mfn2 in spinal cords of Rosa26-hCAST KI mice 30–40 days after injection with AAV1-GFP (Vector) or AAV1-Cre (Cre).
(B D) Quantification of NMJ innervation (B), representative immunoblot of βIII tubulin, NF-L and the calpain cleaved 150-kDa αII-spectrin breakdown product (SBDP150) in tibial nerves (C) and representative toluidine blue staining of Epon-embedded sections of tibial nerves (D) in 4 6 month old Rosa26-hCAST KI mice 2 days after right sciatic nerve cut. 30–40 days before transection, mice were injected with AAV1-GFP (Vector) or AAV1-Cre (Cre). n = 3 or 4 per group. Data are means ± s.e.m., representative of triplicate experiments.
(E) Representative western blot detection and quantification of calpastatin in spinal cords of 100-day old G93A mice 60 days after injection (n = 4 mice per group). 40-day old G93A mice were bilaterally injected with AAV1-calpastatin or AAV1-GFP into the lumbar spinal cord and sacrificed at 100-day old for NMJ innervation assessment.
(F) Quantification of NMJ innervation in GAST muscles of 100-day old G93A mice with indicated AAV1 injection at 40-day old (D, n = 6 and 4 mice per group).
(G) Representative images and quantification of calpastatin (green), AchR (red) and SV2/2H3 (white) in hindlimb skeletal GAST muscles of 3-month old CASTKI and TCAST mice.
Data are means ± s.e.m., representative of triplicate experiments. One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. *P < 0.05, ***P < 0.001. ns, not significant.
Acknowledgments
This study is supported by grants from US National Institutes of Health (1R01NS097679 and 1R01NS089604), US Alzheimer’s Association (AARG-17-499682) and US Association for Frontotemporal Degeneration and Alzheimer’s Drug Discovery Foundation (20161206). H.L. was supported by US National Institutes of Health (R01CA196631 and R01CA208517) and Glenn Foundation for Medical Research.
Footnotes
Author contributions
X.W. conceived and directed the project. X.W., L.W., and J. G. designed experiments, interpreted results and wrote the manuscript. X.W., L.W., J.G., J.L., S.L.S, S.T., M.L.H, Y.J., H.J., T.Y., M.H., P.T., S.Z. and Z.J. contributed to experiments, data analysis and manuscript preparation. H.F. contributed to electron microscopy study. J.L., C.Z. and H.L contributed to RNA-seq study. C.H. contributed to mitochondrial function study.
Declaration of interests
The authors declare no competing interests.
References
- Adachi Y, Ishida-Takahashi A, Takahashi C, Takano E, Murachi T, Hatanaka M. Phosphorylation and subcellular distribution of calpastatin in human hematopoietic system cells. The Journal of biological chemistry. 1991;266:3968–3972. [PubMed] [Google Scholar]
- Allen S, Heath PR, Kirby J, Wharton SB, Cookson MR, Menzies FM, Banks RE, Shaw PJ. Analysis of the cytosolic proteome in a cell culture model of familial amyotrophic lateral sclerosis reveals alterations to the proteasome, antioxidant defenses, and nitric oxide synthetic pathways. The Journal of biological chemistry. 2003;278:6371–6383. doi: 10.1074/jbc.M209915200. [DOI] [PubMed] [Google Scholar]
- Arnold WD, Porensky PN, McGovern VL, Iyer CC, Duque S, Li X, Meyer K, Schmelzer L, Kaspar BK, Kolb SJ, et al. Electrophysiological Biomarkers in Spinal Muscular Atrophy: Preclinical Proof of Concept. Annals of clinical and translational neurology. 2014;1:34–44. doi: 10.1002/acn3.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold WD, Sheth KA, Wier CG, Kissel JT, Burghes AH, Kolb SJ. Electrophysiological Motor Unit Number Estimation (MUNE) Measuring Compound Muscle Action Potential (CMAP) in Mouse Hindlimb Muscles. Journal of visualized experiments : JoVE. 2015 doi: 10.3791/52899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averna M, de Tullio R, Passalacqua M, Salamino F, Pontremoli S, Melloni E. Changes in intracellular calpastatin localization are mediated by reversible phosphorylation. Biochem J. 2001;354:25–30. doi: 10.1042/0264-6021:3540025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averna M, De Tullio R, Salamino F, Melloni E, Pontremoli S. Phosphorylation of rat brain calpastatins by protein kinase C. FEBS Lett. 1999;450:13–16. doi: 10.1016/s0014-5793(99)00461-5. [DOI] [PubMed] [Google Scholar]
- Bloemberg D, Quadrilatero J. Rapid Determination of Myosin Heavy Chain Expression in Rat, Mouse, and Human Skeletal Muscle Using Multicolor Immunofluorescence Analysis. PloS one. 2012:7. doi: 10.1371/journal.pone.0035273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. The Journal of cell biology. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, McCaffery JM, Chan DC. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130:548–562. doi: 10.1016/j.cell.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Chen HC, Vermulst M, Wang YE, Chomyn A, Prolla TA, McCaffery JM, Chan DC. Mitochondrial Fusion Is Required for mtDNA Stability in Skeletal Muscle and Tolerance of mtDNA Mutations. Cell. 2010;141:280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman M. Axon degeneration mechanisms: commonality amid diversity. Nat Rev Neurosci. 2005;6:889–898. doi: 10.1038/nrn1788. [DOI] [PubMed] [Google Scholar]
- Csordas G, Varnai P, Golenar T, Roy S, Purkins G, Schneider TG, Balla T, Hajnoczky G. Imaging interorganelle contacts and local calcium dynamics at the ER-mitochondrial interface. Mol Cell. 2010;39:121–132. doi: 10.1016/j.molcel.2010.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- de Carvalho M, Matias T, Coelho F, Evangelista T, Pinto A, Luis ML. Motor neuron disease presenting with respiratory failure. Journal of the neurological sciences. 1996;139(Suppl):117–122. doi: 10.1016/0022-510x(96)00089-5. [DOI] [PubMed] [Google Scholar]
- Deschenes MR. Motor unit and neuromuscular junction remodeling with aging. Current aging science. 2011;4:209–220. doi: 10.2174/1874609811104030209. [DOI] [PubMed] [Google Scholar]
- Diepenbroek M, Casadei N, Esmer H, Saido TC, Takano J, Kahle PJ, Nixon RA, Rao MV, Melki R, Pieri L, et al. Overexpression of the calpain-specific inhibitor calpastatin reduces human alpha-Synuclein processing, aggregation and synaptic impairment in [A30P]alphaSyn transgenic mice. Human molecular genetics. 2014;23:3975–3989. doi: 10.1093/hmg/ddu112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doshi S, Lynch DR. Calpain and the glutamatergic synapse. Frontiers in bioscience. 2009;1:466–476. doi: 10.2741/s38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis L, Loeffler JP. Neuromuscular junction destruction during amyotrophic lateral sclerosis: insights from transgenic models. Current opinion in pharmacology. 2009;9:341–346. doi: 10.1016/j.coph.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Fischer LR, Culver DG, Davis AA, Tennant P, Wang M, Coleman M, Asress S, Adalbert R, Alexander GM, Glass JD. The WldS gene modestly prolongs survival in the SOD1G93A fALS mouse. Neurobiol Dis. 2005;19:293–300. doi: 10.1016/j.nbd.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Gillingwater TH, Thomson D, Mack TG, Soffin EM, Mattison RJ, Coleman MP, Ribchester RR. Age-dependent synapse withdrawal at axotomised neuromuscular junctions in Wld(s) mutant and Ube4b/Nmnat transgenic mice. J Physiol. 2002;543:739–755. doi: 10.1113/jphysiol.2002.022343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Freire M, de Cabo R, Studenski SA, Ferrucci L. The neuromuscular junction: aging at the crossroad between nerves and muscle. Front Aging Neurosci. 2014:6. doi: 10.3389/fnagi.2014.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaichi T, Sato T, Iwamoto T, Malavasi-Yamashiro J, Hoshino M, Mizuno N. A stable lead by modification of Sato’s method. Journal of electron microscopy. 1986;35:304–306. [PubMed] [Google Scholar]
- Ibebunjo C, Chick JM, Kendall T, Eash JK, Li C, Zhang Y, Vickers C, Wu Z, Clarke BA, Shi J, et al. Genomic and proteomic profiling reveals reduced mitochondrial function and disruption of the neuromuscular junction driving rat sarcopenia. Molecular and cellular biology. 2013;33:194–212. doi: 10.1128/MCB.01036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E, Durham HD. Failure of protein quality control in amyotrophic lateral sclerosis. Biochimica et biophysica acta. 2006;1762:1038–1050. doi: 10.1016/j.bbadis.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Koles K, Budnik V. Wnt signaling in neuromuscular junction development. Cold Spring Harbor perspectives in biology. 2012:4. doi: 10.1101/cshperspect.a008045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong J, Xu Z. Massive mitochondrial degeneration in motor neurons triggers the onset of amyotrophic lateral sclerosis in mice expressing a mutant SOD1. J Neurosci. 1998;18:3241–3250. doi: 10.1523/JNEUROSCI.18-09-03241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leboucher GP, Tsai YC, Yang M, Shaw KC, Zhou M, Veenstra TD, Glickman MH, Weissman AM. Stress-induced phosphorylation and proteasomal degradation of mitofusin 2 facilitates mitochondrial fragmentation and apoptosis. Mol Cell. 2012;47:547–557. doi: 10.1016/j.molcel.2012.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Kumar Sharma L, Li Y, Hu P, Idowu A, Liu D, Lu J, Bai Y. Comparative bioenergetic study of neuronal and muscle mitochondria during aging. Free radical biology & medicine. 2013;63:30–40. doi: 10.1016/j.freeradbiomed.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAskill AF, Kittler JT. Control of mitochondrial transport and localization in neurons. Trends in cell biology. 2010;20:102–112. doi: 10.1016/j.tcb.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Maday S, Twelvetrees AE, Moughamian AJ, Holzbaur EL. Axonal transport: cargo-specific mechanisms of motility and regulation. Neuron. 2014;84:292–309. doi: 10.1016/j.neuron.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrane J, Hervias I, Henning MS, Damiano M, Kawamata H, Manfredi G. Mutant SOD1 in neuronal mitochondria causes toxicity and mitochondrial dynamics abnormalities. Human molecular genetics. 2009;18:4552–4564. doi: 10.1093/hmg/ddp421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major LA, Hegedus J, Weber DJ, Gordon T, Jones KE. Method for counting motor units in mice and validation using a mathematical model. Journal of neurophysiology. 2007;97:1846–1856. doi: 10.1152/jn.00904.2006. [DOI] [PubMed] [Google Scholar]
- Pham AH, Meng S, Chu QN, Chan DC. Loss of Mfn2 results in progressive, retrograde degeneration of dopaminergic neurons in the nigrostriatal circuit. Human molecular genetics. 2012;21:4817–4826. doi: 10.1093/hmg/dds311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MV, McBrayer MK, Campbell J, Kumar A, Hashim A, Sershen H, Stavrides PH, Ohno M, Hutton M, Nixon RA. Specific calpain inhibition by calpastatin prevents tauopathy and neurodegeneration and restores normal lifespan in tau P301L mice. J Neurosci. 2014;34:9222–9234. doi: 10.1523/JNEUROSCI.1132-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoul C, Abbas-Terki T, Bensadoun JC, Guillot S, Haase G, Szulc J, Henderson CE, Aebischer P. Lentiviral-mediated silencing of SOD1 through RNA interference retards disease onset and progression in a mouse model of ALS. Nature medicine. 2005;11:423–428. doi: 10.1038/nm1207. [DOI] [PubMed] [Google Scholar]
- Sebastian D, Hernandez-Alvarez MI, Segales J, Sorianello E, Munoz JP, Sala D, Waget A, Liesa M, Paz JC, Gopalacharyulu P, et al. Mitofusin 2 (Mfn2) links mitochondrial and endoplasmic reticulum function with insulin signaling and is essential for normal glucose homeostasis. P Natl Acad Sci USA. 2012;109:5523–5528. doi: 10.1073/pnas.1108220109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes AT, Goncalves N, Koeppen A, Deglon N, Kugler S, Duarte CB, Pereira de Almeida L. Calpastatin-mediated inhibition of calpains in the mouse brain prevents mutant ataxin 3 proteolysis, nuclear localization and aggregation, relieving Machado-Joseph disease. Brain : a journal of neurology. 2012;135:2428–2439. doi: 10.1093/brain/aws177. [DOI] [PubMed] [Google Scholar]
- Song W, Song Y, Kincaid B, Bossy B, Bossy-Wetzel E. Mutant SOD1G93A triggers mitochondrial fragmentation in spinal cord motor neurons: neuroprotection by SIRT3 and PGC-1alpha. Neurobiol Dis. 2013;51:72–81. doi: 10.1016/j.nbd.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer MJ, Mellgren RL. Overexpression of a calpastatin transgene in mdx muscle reduces dystrophic pathology. Human molecular genetics. 2002;11:2645–2655. doi: 10.1093/hmg/11.21.2645. [DOI] [PubMed] [Google Scholar]
- Stifanese R, Averna M, De Tullio R, Pedrazzi M, Milanese M, Bonifacino T, Bonanno G, Salamino F, Pontremoli S, Melloni E. Role of calpain-1 in the early phase of experimental ALS. Archives of biochemistry and biophysics. 2014;562:1–8. doi: 10.1016/j.abb.2014.08.006. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura A, Nagashima S, Tokuyama T, Amo T, Matsuki Y, Ishido S, Kudo Y, McBride HM, Fukuda T, Matsushita N, et al. MITOL regulates endoplasmic reticulum-mitochondria contacts via Mitofusin2. Mol Cell. 2013;51:20–34. doi: 10.1016/j.molcel.2013.04.023. [DOI] [PubMed] [Google Scholar]
- Tidball JG, Spencer MJ. Expression of a calpastatin transgene slows muscle wasting and obviates changes in myosin isoform expression during murine muscle disuse. The Journal of physiology. 2002;545:819–828. doi: 10.1113/jphysiol.2002.024935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urushitani M, Kurisu J, Tsukita K, Takahashi R. Proteasomal inhibition by misfolded mutant superoxide dismutase 1 induces selective motor neuron death in familial amyotrophic lateral sclerosis. Journal of neurochemistry. 2002;83:1030–1042. doi: 10.1046/j.1471-4159.2002.01211.x. [DOI] [PubMed] [Google Scholar]
- Vande Velde C, McDonald KK, Boukhedimi Y, McAlonis-Downes M, Lobsiger CS, Bel Hadj S, Zandona A, Julien JP, Shah SB, Cleveland DW. Misfolded SOD1 associated with motor neuron mitochondria alters mitochondrial shape and distribution prior to clinical onset. PLoS One. 2011;6:e22031. doi: 10.1371/journal.pone.0022031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Wang L, Lu J, Siedlak SL, Fujioka H, Liang J, Jiang S, Ma X, Jiang Z, da Rocha EL, et al. The inhibition of TDP-43 mitochondrial localization blocks its neuronal toxicity. Nat Med. 2016;22:869–878. doi: 10.1038/nm.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zhang F, Li L, Tang F, Siedlak SL, Fujioka H, Liu Y, Su B, Pi Y, Wang X. MFN2 couples glutamate excitotoxicity and mitochondrial dysfunction in motor neurons. The Journal of biological chemistry. 2015;290:168–182. doi: 10.1074/jbc.M114.617167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Su B, Lee HG, Li X, Perry G, Smith MA, Zhu X. Impaired balance of mitochondrial fission and fusion in Alzheimer’s disease. J Neurosci. 2009;29:9090–9103. doi: 10.1523/JNEUROSCI.1357-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Su B, Siedlak SL, Moreira PI, Fujioka H, Wang Y, Casadesus G, Zhu X. Amyloid-beta overproduction causes abnormal mitochondrial dynamics via differential modulation of mitochondrial fission/fusion proteins. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19318–19323. doi: 10.1073/pnas.0804871105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Weimer RM, Kallop D, Olsen O, Wu Z, Renier N, Uryu K, Tessier-Lavigne M. Regulation of axon degeneration after injury and in development by the endogenous calpain inhibitor calpastatin. Neuron. 2013;80:1175–1189. doi: 10.1016/j.neuron.2013.08.034. [DOI] [PubMed] [Google Scholar]
- Zhao T, Huang X, Han L, Wang X, Cheng H, Zhao Y, Chen Q, Chen J, Cheng H, Xiao R, et al. Central role of mitofusin 2 in autophagosome-lysosome fusion in cardiomyocytes. J Biol Chem. 2012;287:23615–23625. doi: 10.1074/jbc.M112.379164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XW, Rottkamp CA, Boux H, Takeda A, Perry G, Smith MA. Activation of p38 kinase links tau phosphorylation, oxidative stress, and cell cycle-related events in Alzheimer disease. Journal of neuropathology and experimental neurology. 2000;59:880–888. doi: 10.1093/jnen/59.10.880. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Representative movies of Mfn2fl/fl mice after AAV1 injection or calpastatin, ER and mitochondria trafficking in in vitro cultured primary cortical neurons (Related to Figures 1 and 5).
A 5-month old Mfn2fl/fl mouse injected with AAV1-GFP for 3 weeks demonstrates no hindlimb clasping (Movie S1) and normal movement (Movie S2), whereas a 5-month old Mfn2fl/fl mouse injected with AAV1-Cre for 3 weeks demonstrates severe hindlimb clasping (Movie S3) and hindlimb paralysis (Movie S4). Representative movie of calpastatin puncta (green) and mitochondria (red, mito-DsRed2) trafficking in axons of in vitro cultured primary cortical neurons is shown in Movie S4. Representative movies of mitochondria (green, Movie S6), ER (red, Movie S7), and merged mitochondria and ER (Movie S8) 1 min after ER and mitochondria tethering in neurons transfected with mito-FKBP-GFP and DsRed-FRB-ER are also shown.
Figure S1. Characterization of Mfn2fl/fl mice after Cre injection (related to Figure 1).
(A and B) Representative fluorescent images of the entire brain and spinal cord (A) and cross sections of spinal cord ventral horn (B) of Mfn2fl/fl mice at 2 weeks after injection.
(C) Kaplan-Meier log rank test for the onset of paralysis in Cre-injected Mfn2fl/fl mice. n = 7.
(D) Representative walking footprint patterns (arrow shows the walking direction) of 4 6 month old Mfn2fl/fl mice 3–4 weeks after injection with AAV1-GFP (Vector) or paralyzed Mfn2fl/fl mice after injection AAV1-Cre (Cre).
(E) Quantification of axons in toluidine blue staining of dorsal and ventral nerve roots of Cre-injected paralyzed Mfn2fl/fl mice or age-matched vector injected control Mfn2fl/fl mice.
(F) Representative immunoblot and quantification of Mfn2 levels in lumbar spinal cord of Mfn2fl/fl mice 2 weeks after vector or Cre injection (n = 3 mice per group).
(G) Representative immunohistochemistry and quantification of Mfn2 immunoreactivity in spinal cord motor neurons of Mfn2fl/fl mice 2 weeks after vector or Cre injection (n = 3 mice per group).
(H) Representative images and quantification of mitochondria in neuromuscular synaptic terminals in GAST muscles of paralyzed Mfn2fl/fl mice after co-injection with AAV1-mitoDsRed and AAV1-Cre (n = 13 and 11 NMJs for coverage; n = 14 and 9 neurites for length).
(I) Representative images and quantification of mitochondrial length in the cell bodies and proximal neurites of Mfn2fl/fl mice after co-injection with AAV1-mitoDsRed and AAV1-Cre or AAV1-GFP (n = 14, 16, 10 neurons for cell body and n = 22, 17 neurites for proximal motor axons).
Data are means ± s.e.m., representative of triplicate experiments. Student’s t test. **P < 0.01, ***P < 0.001, ****P < 0.0001. ns, not significant.
Figure S2. Reduced expression of Mfn2 in G93A mice and ALS patients and well-preserved skeletal muscles in dTg mice (related to Figure 2).
(A) Representative immunoblot and quantification of Mfn2 levels in endpoint G93A mice and age-matched control mice. Relative Mfn2 levels are all adjusted by GAPDH. n = 4 per group.
(B) Representative images and quantification of Mfn2 immunoreactivity in spinal cord motor neurons of young (60 day) and old (>180 day, not shown) non-transgenic (NTg) mice and transgenic mice overexpressing wild type SOD1 (“WT”) or G93A mice. ALS disease progression goes through four stages in SOD1 G93A mice based on muscle strength: pre-muscle weakness stage (PMW), a rapid declining stage (RD), a slow declining stage (SD) and a paralysis stage (PARA) (Kong and Xu, 1998). The immunoreactivity of Mfn2 in spinal cord motor neurons began to decrease significantly during the PMW stage, which preceded the onset of the rapid decline of muscle strength (RD stage). There was a further decrease of Mfn2 immunoreactivity to very low level during the RD and SD stage. However, no further reduction was observed thereafter. Not surprisingly, concurrent with Mfn2 reduction, mitochondrial length also began to decrease during the PMW stage and then declined to minimal during the RD, SD or PARA stage (data not shown). n > 20 neurons per group.
(C) Representative immunoblot and quantification of mitochondrial fission and fusion proteins in human thoracic spinal cord tissues of sporadic ALS patients (n=8) and age-matched control subjects (n=6). Equal protein amounts (20 μg) were loaded and confirmed by actin. Tom20 was used as a mitochondrial specific marker.
(D) Representative immunocytochemistry of DLP1, OPA1, Mfn1, Mfn2 and Mff in human spinal cord from sALS patient (lower panel) and age-matched control (upper panel).
(E) Quantification of Mfn2, SOD1 and VDAC1 (mitochondrial marker) levels in spinal cords, brains and GAST muscles of endpoint G93A and dTg mice and aged-matched NTG and TMFN mice. Relative Mfn2, SOD1 or VDAC1 levels are adjusted by GAPDH or VDAC1. n = 4 per group.
(F) Kaplan-Meier log rank test for survival in G93A (female = 5, male = 6) and dTg mice (female = 5, male = 8).
(G) Representative images of endpoint G93A and dTg mice and aged-matched 5–6 month old NTG and TMFN mice.
(H) Body weight of 5–6 month old NTG (female = 3, male = 5) and TMFN mice (female = 5, male = 5) and endpoint G93A (female = 3, male = 4) and dTg mice (female = 5, male = 6), or NTG (female = 9, male = 12), TMFN mice (female = 7, male = 9), G93A (female = 13, male = 14) and dTg mice (female = 9, male = 9) at 120-day old.
(I) Representative images of GAST muscles of endpoint G93A and dTg mice and age-matched NTG and TMFN mice stained by hematoxylin and eosin (H&E), ATPase (pH 4.6; dark stain for type1 fibers, light stain for type 2A fibers, and intermediate stain for type 2B/2D fibers) and antibodies against SC-71 (green, for type 2A fibers) and 6H1 (red, for type 2X fibers as described (Bloemberg and Quadrilatero, 2012)). In H&E staining, arrows point to dystrophic muscle fibers in G93A, which are absent in dTg mice. In ATPase pH 4.6 staining, there is a marked increase of slow twitch type1 fibers in G93A but not in dTg, NTG and TMFN mice. Consistent with this, G93A but not dTg mice exhibited significant loss of fast twitch type 2 fibers (light staining in ATPase pH 4.6 staining) especially type 2X fibers (6H1 immunofluorescence staining), therefore suggesting the recovery of short-duration high intensity bursts of power in dTg mice.
(J) Grip strength of hindlimbs of NTG (n = 11), TMFN mice (n = 7), G93A (n = 17) and dTg mice (n = 8) at 120-day old.
(K) walking footprint patterns and quantification of stride length of NTG (n = 4), TMFN mice (n = 4), G93A (n = 3) and dTg mice (n = 4) at 120-day old.
(L) Representative images of GAST muscle weight in NTG, TMFN mice, G93A and dTg mice at 120-day old.
(M) Representative immunoblot and/or quantification of Agrin or Wnt3a in spinal cords and/or GAST muscles of endpoint and 120-day old G93A and dTg mice and age-matched NTG and TMFN mice. n = 3 mice group. Equal protein amounts of 20 μg were loaded for all blots.
Data are means ± s.e.m., representative of triplicate experiments. One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. ns, not significant.
Figure S3. Neuronal Mfn2 protects mitochondria in motor cell bodies and alleviates motor axon degradation in G93A mice (related to Figure 2).
(A) Representative images of mitochondria in cell bodies (Inset I) and proximal neurites (Inset II) of spinal cord motor neurons of endpoint G93A and dTg mice and age-matched NTG and TMFN mice.
(B) Representative electron microscopic (EM) images and quantification of mitochondria in cell bodies of spinal cord motor neurons of endpoint G93A and dTg mice and age-matched NTG and TMFN mice. n = 7, 6, 5, 9 neurons per group.
(C) Quantification of the percentage of NMJ area covered by mitochondria in endpoint G93A and dTg mice and age-matched NTG and TMFN mice. n = 20 neurons or NMJs per group.
(D) Quantification of mitochondrial length in proximal motor neurites in spinal cords and distal motor neurites in GAST muscles. n = 11 14 motor neurons or NMJs from 3 mice per group.
(E) Representative H&E staining and quantification of lumbar spinal cord motor neurons of endpoint G93A (n = 6) and dTG (n = 6) mice and age-matched NTG (n = 5) and TMFN (n = 3) mice. Despite unalleviated spinal cord motor neuron loss, the vacuolar pathology was decreased in spinal cords in dTg mice.
(F and G) Representative EM images (F) and toluidine blue staining (G) of Epon-embedded sections of L3–5 ventral nerve roots of endpoint G93A and dTG mice and age-matched NTG and TMFN mice.
(H) Representative western blot of proteins involved in apoptosis, MAPK/GSK3β signaling, mitochondrial bioenergetics (OXPHOS: oxidative phosphorylation) and cytoskeleton formation in spinal cords of endpoint G93A and dTG mice and age-matched NTG and TMFN mice. Equal protein amounts of 20 μg were loaded for all blots.
(I) Transcriptome analysis of spinal cords of NTG, TMFN, G93A and dTg mice. Pathway overrepresentation analysis was performed using upregulated genes with fold changes no less than 2 from G93A mice versus NTG mice, dTg mice versus NTG mice and TMFN mice versus NTG mice respectively. The length of each bar indicates the number of upregulated genes found in each pathway, while the color indicates the p-value of each overrepresentation test.
Data are means ± s.e.m., representative of triplicate experiments. One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.001. ns, not significant.
Figure S4. Calpastatin expression and trafficking in mice and cultured primary neurons (related to Figure 5).
(A) Representative images and quantification of calpastatin (green), AchR (red) and SV2/2H3 (white) in hindlimb skeletal GAST muscles of 4 6 month old NTG and TMFN mice. n = 14 or 15 NMJs per group. The specificity of antibody to was confirmed by the absence of calpastatin immunoreactivity in TMFN mouse GAST muscles stained by the calpastatin antibody pre-adsorbed with calpastatin recombinant protein “TMFN (Pre-adsorbed)”.
(B and C) Quantification of the relative levels of Calpain1, Calpain2 or VDAC1 to GAPDH in 4 6 month old NTG and TMFN mice (B) or 4 6 month old Mfn2fl/fl mice 12 days after injection with AAV1-GFP (Vector) or AAV1-Cre (Cre) (C) (n = 3 or 4 mice per group).
(D) Representative immunocytochemistry of calpastatin in lumbar spinal cords of 4 6 month old Mfn2fl/fl mice 12 days after injection with AAV1-GFP (Vector) or AAV1-Cre (Cre). Note that the immunocytochemistry of Mfn2 is shown in Figure S1G.
(E) Schematic diagram of lumbar spinal cord injection of AAV1-calpastatin in NTG mice. T2A autocleavage generates T2A tagged calpastatin separately from GFP.
(F) Representative confocal images and line-scan analysis of calpastatin (green), mitochondria (blue) and ER (red) in rat primary cortical neurons (DIV7) co-transfected with GFP tagged calpastatin (GFP-calpastatin), mitoBFP and ER-DsRed at DIV5. Representative line-scan analysis of the co-localization between Calpastatin and mitochondria in white solid lines with “2” width shown in the merged low panel with light exposure. The line-scan analysis was performed by Image J RGB Profile Plot plugin as described before(Wang et al., 2016) using high resolution images shown in the enlargements. Further quantification of degree of co-localization between calpastatin and mitochondrial marker mitoBFP in axons shows a Manders’s coefficient of 0.76 ± 0.124 (n = 8 neurons). Manders’s coefficient is % of both red and green signal co-localize (range from 0–1) and independent of fluorescence intensity. A Mander’s coefficient greater than 0.5 is considered significant co-localization (“0” means non-overlapping while “1” indicates complete overlapping).
(G) Time lapse imaging of calpastatin (green) and mitochondria (red) in live cultured rat primary cortical neurons (DIV7) co-transfected with GFP-calpastatin and mitoDsRed at DIV5. calpastatin puncta (arrowhead) could move in a short distance in the fast transport mode and dissemble slowly. The time lapse images here or Movie S5 clearly show the continuous moving pattern of the puncta (it is more obvious when Movie S5 is played in slower speeds).
(H K) Quantification of the velocity of calpastatin moving puncta (H), calpastatin axonal transport (indexed by the relative ratio of the mean gray value of calpastatin in a segment of neuronal process 200 μm length beginning from the cell body of neurons to the mean gray value of calpastatin in the cell body of neurons, I), the proportion of mobile mitochondria (J) and ER axonal transport (indexed by the relative ratio of the mean gray value of ER-DsRed in a segment of neuronal process 200 μm length beginning from the cell body of neurons to the mean gray value of calpastatin in the cell body of neurons, K) in neurons (DIV7) overexpressing or knocking down Mfn2 or Miro1 (Mfn2 or Miro1 constructs also encoding GFP for the identification of positively transfected neurons). Neurons were co-transfected with RFP tagged Calpastatin, ER-DsRed2 or mitoDsRed at DIV5 for the live imaging of calpastatin, mitochondria and ER. For BAPTA/AM-related experiments, neurons were also transfected with/without Mfn2 overexpression constructs at DIV5, treated with 1μM BAPTA/AM at DIV6 for 24 hours, and fixed at DIV7 for quantification. n > 20 neurons per group.
(L) Quantification of the relative levels of Mfn2 or VDAC1 to GAPDH in 3–4 month old NTG mice 12 days after injection of AAV1-shMiro-1. n = 3 or 4 mice per group.
Data are means ± s.e.m., representative of triplicate experiments. One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (compared with control neuron), ####P < 0.0001 (compared with neurons overexpressing Mfn2). ns, not significant.
Figure S5. EM, immuno-EM, immunoblot and confocal microscopic analysis of calpastatin, mitochondria, MAMs and/or ER (related to Figure 5).
(A and B) Representative EM micrographs of purified mitochondria (A) or MAMs (B) isolated from spinal cords of NTG mice. The isolation and purification of highly purified mitochondria were based on percoll gradient as we described(Wang et al., 2016).
(C and D) Representative immuno-EM analysis of calpastatin (labeled by 15 nm gold particles) and Mfn2 (labeled by 10 nm gold particles) in crude mitochondria (containing both mitochondria and MAMs, C) isolated from spinal cords or motor neurons in spinal cords (D) of NTG mice at 3–4 month old.
(E) Representative western blot detection of calpastatin in MAMs isolated from pooled spinal cords of NTG and TMFN mice at 3–4 month old, 4 6 month old Mfn2fl/fl mice 12 days after injection with AAV1-GFP (Vector) or AAV1-Cre (Cre), or 160-day old NTG and TMFN mice. H: total spinal cord homogenate; pM: highly purified mitochondria isolated from spinal cords; S: post mitochondrial supernatant. Equal protein amounts of 20 μg were loaded for all blots. All data are representative of triplicate experiments.
(F) Representative EM micrographs and quantification of membranes associated with mitochondria in sciatic nerves of NTG and TMFN mice at 3 4 month old showing the increased association of membranous structures with mitochondria. Red arrow heads point MAMs associated with mitochondria. n = 12, 13 per group.
(G) Representative image and quantification of the localization of ER-derived vesicles on mitochondria in a segment of neuronal process 200 μm length beginning from the cell body of neurons in neurons (DIV7) overexpressing or knocking down Mfn2 or Miro1. Neurons were co-transfected with ER-DsRed2 at DIV5 and fixed for imaging at DIV7. n > 20 neurons per group.
(H) Quantification of the relative levels of Calnexin to GAPDH in 4 6 month old NTG and TMFN mice, 4 6 month old Mfn2fl/fl mice 12 days after injection with AAV1-GFP (Vector) or AAV1-Cre (Cre) or 3 4 month old NTG mice 12 days after injection of AAV1-shMiro-1. n = 3 or 4 mice per group.
(I) Quantification of the calpastatin axonal transport (indexed by the relative ratio of the mean gray value of calpastatin in a segment of neuronal process 200 μm length beginning from the cell body of neurons to the mean gray value of calpastatin in the cell body of neurons) in neurons (DIV7) co-expressing indicated Mfn2 mutants with GFP-tagged calpastatin. n > 20 neurons per group.
(J and K) Representative large field confocal images of mitochondria and ER in live cultured neurons 2 days after co-transfection with mito-FKBP-GFP and DsRed-FRB-ER before and after rapamycin treatment (related to Figure 5I).
Data are means ± s.e.m., representative of triplicate experiments. One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. **P < 0.01, ***P < 0.001, ****P < 0.0001 (G and I; compared with control neuron). ns, not significant.
Figure S6. Calpastatin protects axons but not NMJs in mice with sciatic nerve transection or G93A mice (related to Figure 6).
(A) Representative western blot detection of human calpastatin and Mfn2 in spinal cords of Rosa26-hCAST KI mice 30–40 days after injection with AAV1-GFP (Vector) or AAV1-Cre (Cre).
(B D) Quantification of NMJ innervation (B), representative immunoblot of βIII tubulin, NF-L and the calpain cleaved 150-kDa αII-spectrin breakdown product (SBDP150) in tibial nerves (C) and representative toluidine blue staining of Epon-embedded sections of tibial nerves (D) in 4 6 month old Rosa26-hCAST KI mice 2 days after right sciatic nerve cut. 30–40 days before transection, mice were injected with AAV1-GFP (Vector) or AAV1-Cre (Cre). n = 3 or 4 per group. Data are means ± s.e.m., representative of triplicate experiments.
(E) Representative western blot detection and quantification of calpastatin in spinal cords of 100-day old G93A mice 60 days after injection (n = 4 mice per group). 40-day old G93A mice were bilaterally injected with AAV1-calpastatin or AAV1-GFP into the lumbar spinal cord and sacrificed at 100-day old for NMJ innervation assessment.
(F) Quantification of NMJ innervation in GAST muscles of 100-day old G93A mice with indicated AAV1 injection at 40-day old (D, n = 6 and 4 mice per group).
(G) Representative images and quantification of calpastatin (green), AchR (red) and SV2/2H3 (white) in hindlimb skeletal GAST muscles of 3-month old CASTKI and TCAST mice.
Data are means ± s.e.m., representative of triplicate experiments. One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. *P < 0.05, ***P < 0.001. ns, not significant.