Abstract
Purpose
Previous studies have indicated an important role for pleckstrin homology domain-interacting protein (PHIP) as a marker and mediator of melanoma metastasis. Here we aimed to confirm the role of PHIP copy number in successive stages of melanoma progression.
Experimental Design
PHIP copy number was examined using fluorescence in situ hybridization (FISH) in three independent cohorts by recording the percentage of cells harboring ≥ 3 copies of PHIP. The impact of PHIP copy number on survival was assessed using Cox regression analysis. The enrichment of PHIP was assessed in various molecular melanoma subtypes. PHIP expression was analyzed in The Cancer Genome Atlas (TCGA) melanoma cohort.
Results
Elevated PHIP copy number was significantly predictive of reduced distant metastasis-free survival (DMFS) and disease-specific survival (DSS), and increased prevalence of ulceration in primary melanoma (cohort #1). By multivariate analysis, PHIP FISH scores were independently predictive of DMFS and DSS. PHIP copy number was enriched in metastatic melanomas harboring mutant NRAS or expressing PTEN protein (cohort #2). PHIP copy number was significantly elevated in metastatic melanomas when compared with matched primary tumors from the same patient (cohort #3). Several of these associations were replicated using TCGA cohort analysis.
Conclusions
These results underscore the important role of PHIP copy number elevation in melanoma progression, and identify molecular subtypes of melanoma in which PHIP is enriched. Finally, as elevated PHIP copy number appears to be selected for during the progression of primary to metastatic melanoma, these results confirm PHIP as a promising therapeutic target for melanoma.
Introduction
Melanoma represents the fifth most common malignancy in the United States (1). As the clinical behavior of melanoma is unpredictable, the development of prognostic markers has represented an important area of research. Histological prognostic factors are the mainstay of prognostic assessment for primary cutaneous melanoma (2). While many factors may play a role in refining melanoma prognosis, tumor thickness and ulceration are the only factors currently incorporated within the melanoma staging classification for primary melanoma (3).
Molecular factors represent the next frontier in prognostic factor research. The development of molecular prognostic factors for melanoma has been hampered largely by the lack of sufficient validation of promising markers in independent tissue sets. In addition, the role of a putative marker in melanoma progression has rarely been explored in the context of the same patient. Finally, given the identification of dominant mutational drivers of melanoma, information regarding the role of a given marker in the context of the known mutational landscape of melanoma would be important (4). As a result, no molecular markers are routinely utilized in the prognostic assessment of melanoma patients, and none are incorporated into its staging classification.
Our previous studies identified both a functional and a biomarker role for PHIP in melanoma progression, and as a molecular prognostic factor for melanoma. PHIP was identified as the top gene overexpressed in melanoma metastases when compared with primary tumors using cDNA microarray analysis (5). Subsequent studies showed an independent prognostic role for PHIP in primary melanoma (6,7). Functional studies demonstrated that increasing PHIP expression enhanced the distant metastatic potential of melanoma (6,7). Here, we aimed to confirm the prognostic role of PHIP in primary melanoma, to identify the molecular subtypes of melanoma in which PHIP is enriched, and to determine whether PHIP is selected for during the progression from primary to metastatic melanoma.
Materials and Methods
Study population
We developed three non-overlapping patient cohorts to assess PHIP’s biomarker role: (i) cohort #1- unstained primary melanoma tissue sections obtained from 204 stage I/II patients in the German Cancer Consortium (see Table 1 for characteristics); (ii) cohort #2- tissue microarrays (TMA) from a previously described cohort of 130 stage III melanoma patients in the MD Anderson Cancer Center (8); and (iii) cohort #3- 37 matched primary and metastatic melanomas obtained from the same patient (N=15) who developed both regional lymph node and distant metastasis, identified from the CPMC Center for Melanoma Research and Treatment archives (see Supplemental Table 1 for characteristics). In cohort #3, 21 samples were from 7 patients with triple-matched primary melanoma, regional lymph node and distant metastasis, and 16 samples were from 8 patients where the primary melanoma was matched either with a regional lymph node or distant metastasis. The mean follow up of cohort #1 was 4.58 years. Information regarding mitotic rate was not available in cohort #1. These analyses were complemented by analysis of PHIP expression levels obtained from TCGA (4), available from 66 primary tumors and 264 metastases. The investigations described here were conducted in accordance with recognized ethical guidelines (i.e., Declaration of Helsinki), were approved by the Sutter Health Institutional Review Board, and informed consent was obtained from each subject.
Table 1.
Characteristics of cohort #1 (N=204)
| Male gender | 111 (54.4%) |
| Age over 50 (years) | 155 (76.0%) |
| Ulceration present | 53 (26.0%) |
| T category | |
| T1 | 56 (27.5%) |
| T2 | 51 (25.0%) |
| T3 | 59 (28.9%) |
| T4 | 30 (14.7%) |
| Unknown | 8 (3.9%) |
| Median thickness | 1.85 mm |
| Clark level | |
| II/III | 84 (41.2%) |
| IV/V | 100 (49.0%) |
| Unknown | 20 (9.8%) |
| Tumor site | |
| Trunk | 100 (49.0%) |
| Extremity | 74 (36.3%) |
| Head and neck | 24 (11.8%) |
| Unknown | 6 (2.9%) |
FISH analysis
FISH for PHIP copy number was performed as previously described (6,7) using bacterial artificial chromosome (BAC) clones RP11-767O1and CTD-2297E14 to detect the PHIP locus and clones RP11-26M18 and RP11-136K2 to detect 6q11.1 and 6p11.1, respectively (interpreted as chromosome 6 centromere). FISH for PTEN copy number was performed using BAC clones RP11-79A15 and RP11-813O3. For a chromosome 10-specific probe, BAC clone RP11-96F8 was used to detect 10p11.1 (all BAC clones were obtained from the Children’s Hospital Oakland Research Institute). The quality and mapping of all probes were verified by hybridization to normal metaphase spreads in combination with a commercially available centromeric probe for chromosomes 6 and 10 (Open Biosystems, Lafayette, CO) before tissue analysis. Z-stacked images were acquired using a Zeiss Axio Image Z2 microscope controlled by AxioVision software (Zeiss, Jena, Germany). At least 30 nuclei from each case were evaluated, and the signals were interpreted according to guidelines described previously (9). Signals from BAC clones detecting 6q11.1, 6p11.1 and 10p11.1 were interpreted as the centromeric signal for chromosome 6 and 10, respectively.
Statistical Analysis
The impact of elevated PHIP copy number on melanoma progression was analyzed in several ways. In cohort #1, consistent with our previous analysis (6), elevated PHIP copy number was measured by recording the percentage of cells harboring 3 or more copies of the PHIP locus. The cut-point utilized to define high PHIP copy number was that which maximized the average of sensitivity and specificity for predicting DSS (determined to be greater than or equal to 19%). The same cut-point was uniformly applied to analyses of DMFS and ulceration status. Subsequently, a previously identified cut-point for PHIP copy number (greater than or equal to 16%) (6) was applied to the analysis of DMFS, DSS, and ulceration in this cohort. The association between elevated PHIP copy number and ulceration status was determined using logistic regression analysis. The association between elevated PHIP copy number and survival outcomes (i.e., DMFS and DSS) was determined using Kaplan-Meier analysis, and univariate and multivariate Cox regression analyses. In cohort #2, the enrichment of PHIP in various molecular subtypes of melanoma was assessed using the T test or the one-way analysis of variance (ANOVA). In cohort #3, for the analysis of 7 triple-matched tissue specimens, the significance of monotonically increasing PHIP copy number in melanoma progression was assessed using the Friedman two-way analysis of variance by ranks test. The significance of increasing PHIP copy number between primary and metastatic melanoma in the 8 double-matched specimens was assessed using the binomial sign test. As a control, identical analyses were performed for PTEN copy number in the same 15-patient matched cohort. Except for the directional analyses just described, all P values reported are two-sided.
Results
In this study, we aimed to perform a comprehensive analysis of the role of PHIP levels in distinct, sequential stages of melanoma progression, and to confirm its role as a prognostic marker for primary melanoma. Initially, we aimed to replicate the prognostic role of elevated PHIP copy number (assessed as percentage of cells harboring 3 or more copies of PHIP) in primary melanoma in cohort #1. We assessed the potential association between PHIP copy number and survival using univariate Cox regression. Increasing PHIP FISH scores (as a continuous variable, using the entire scale) were significantly predictive of distant metastasis-free survival (DMFS, P<0.03) and disease-specific survival (DSS, P=0.025). By Kaplan-Meier analysis, high PHIP scores (assessed as a dichotomous variable) were also significantly predictive of DMFS (P=0.0017, Fig. 2A) and DSS (P=0.0002, Fig. 2B). In addition, we assessed the association between PHIP FISH scores and both tumor thickness and ulceration status (6). Increasing PHIP copy number (using the entire scale) was associated separately with increased tumor thickness (P=0.0001) and with a higher prevalence of ulceration (P<0.0001) by univariate logistic regression analysis. High PHIP FISH scores (assessed as a dichotomous variable) were significantly associated with increased prevalence of ulceration (P<0.00005).
Fig. 2.
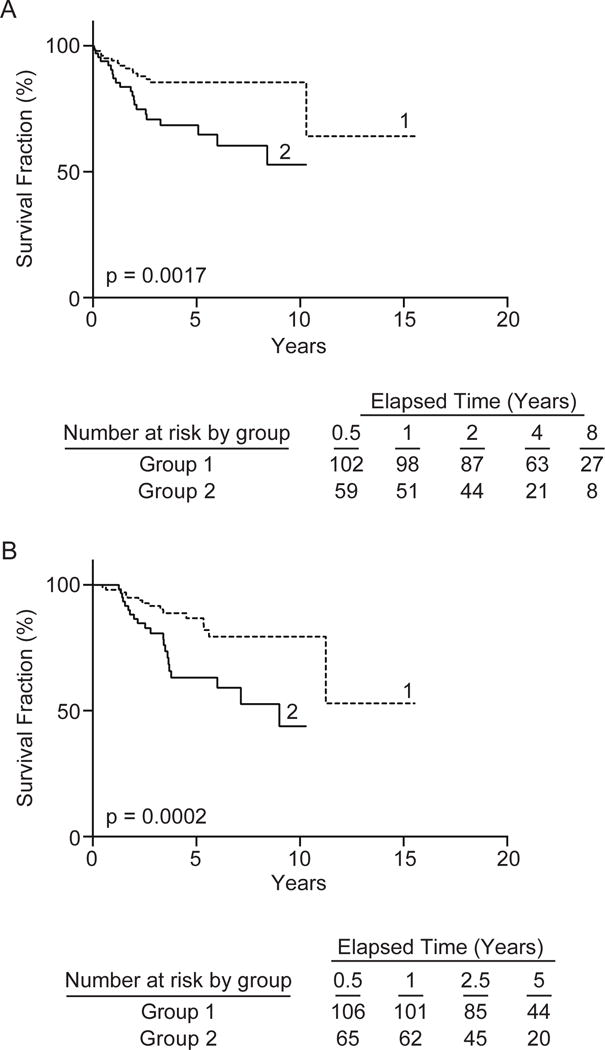
Kaplan-Meier analysis of the prognostic significance of PHIP copy number in cohort #1. Analysis of DMFS (panel A) and DSS (panel B) in patients with low PHIP copy number (curve 1) versus patients with high PHIP copy number (curve 2).
We then assessed the independent impact of elevated PHIP copy number on survival associated with melanoma using multivariate stepwise Cox regression analysis with backward elimination. We included in the model the following covariates included in the legacy prognostic analyses performed by the AJCC melanoma committee (10): tumor thickness (assessed by T category), ulceration status, Clark level, patient age, patient gender, and tumor site. In the analysis of DMFS, elevated PHIP copy number (as a dichotomous variable) emerged as significantly predictive of survival (P=0.025), along with tumor thickness and gender (Table 2). In the analysis of DSS, PHIP FISH scores were also significantly predictive of survival (P=0.01), along with tumor thickness (Table 2). Thus, elevated PHIP copy number was independently predictive of both DMFS and DSS, replicating our original analysis (6).
Table 2.
Results of stepwise multivariate Cox regression analysis of the impact of various factors on DMFS and DSS
| DMFS | ||
|---|---|---|
| Covariate | Risk ratio | P value |
| Tumor thickness | 2.88 | < 0.0001 |
| Gender | 3.19 | 0.0045 |
| PHIP copy number | 1.51 | 0.025 |
| DSS | ||
| Covariate | Risk ratio | P value |
| Tumor thickness | 2.61 | < 0.0001 |
| PHIP copy number | 1.68 | 0.01 |
In addition, given that the cut-point identified in this analysis (19%) was very close to that identified in our original analysis (16%) (6), we examined whether use of the previously-identified cut-point would be predictive of outcome in cohort #1. High PHIP scores using the original cut-point were also significantly predictive of DMFS (P=0.03), DSS (P=0.0069), and ulceration status (P<0.00005) by univariate analysis.
Next, we assessed the role of PHIP in melanoma nodal metastases in cohort #2. We initially assessed the potential prognostic role of elevated PHIP copy number in this cohort. There was no significant association between PHIP copy number (assessed as a continuous variable) in metastatic tumors and survival in stage III melanoma. The dataset from this cohort contained information regarding three important molecular markers in melanoma: BRAF and NRAS mutation status, and PTEN expression level (8). Our analysis identified a significant association between elevated PHIP copy number and NRAS mutation status in melanoma. The mean percentage of melanoma cells harboring ≥ 3 copies of PHIP was 45.7% in NRAS-mutant melanomas, compared with 19.3% in BRAF-mutant melanomas, and 15.4% in BRAF/NRAS-wild-type melanomas to (P<0.00005, ANOVA, Fig. 3A). Separately, our analysis indicated that PHIP copy number was significantly higher in melanomas with intact PTEN protein expression versus those with PTEN loss. Specifically, the mean percentage of melanoma cells harboring ≥ 3 copies of PHIP increased from 13.5% in melanomas with PTEN loss to 22.6% in PTEN-expressing melanomas (P=0.004, T test, Fig. 3B). In addition, in BRAF-mutant melanomas, there was a trend toward increased PHIP copy number in PTEN-expressing melanomas vs. those with PTEN loss (P=0.086, T test, Fig. 3C). Finally, cases with a mean PHIP copy number of ≥ 3 were present in each molecular subtype of melanoma investigated, including in wild-type BRAF/wild-type NRAS/PTEN-expressing melanomas. Taken together, these results indicate that PHIP levels are enriched in NRAS-mutant and PTEN-expressing melanoma, and that PHIP copy number elevations are observed in both BRAF-mutant and BRAF wild-type/NRAS wild-type/PTEN-expressing melanomas.
Fig. 3.
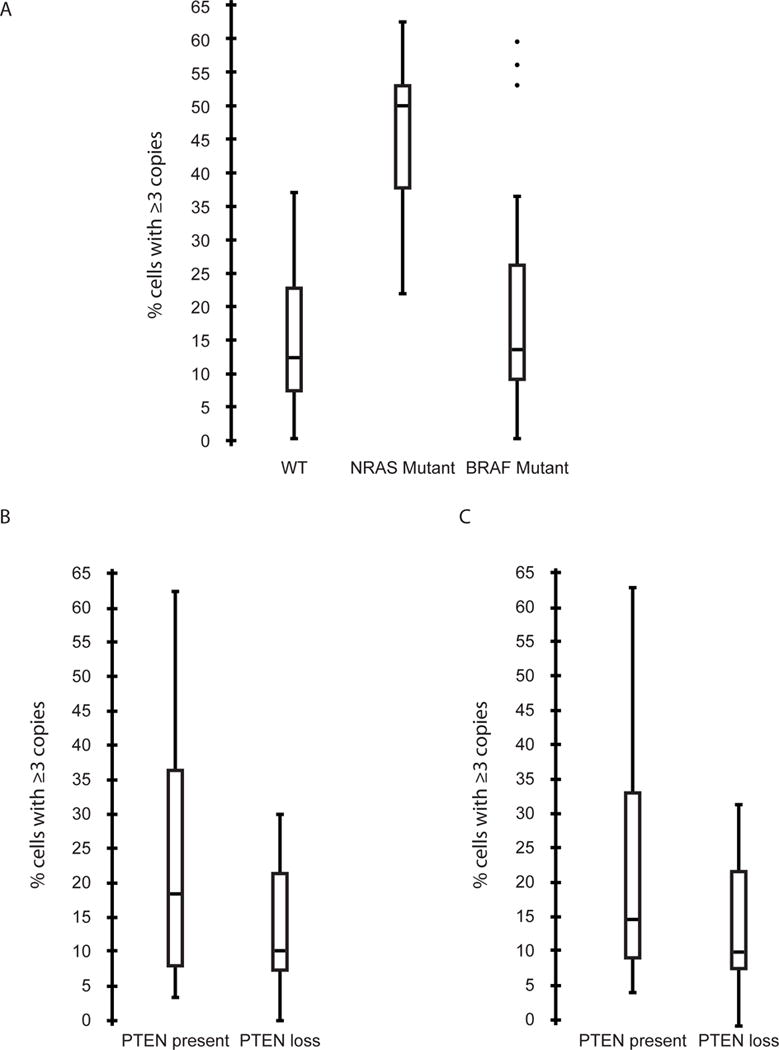
Comparison of PHIP copy number in different molecular subgroups of melanoma in cohort #2. (A) Box plots showing PHIP copy number in melanomas with wild-type BRAF/NRAS vs. mutant BRAF or mutant NRAS. (B) Box plots showing PHIP copy number in melanomas expressing PTEN protein versus melanomas with PTEN loss. (C) Box plots showing PHIP copy number in BRAF-mutant melanomas expressing PTEN protein versus melanomas with PTEN loss.
Subsequently, we assessed the role of PHIP as a progression marker in melanoma. We performed FISH analysis of PHIP copy number on a matched cohort of 15 patients from whom the primary tumor and at least one metastatic tumor were available (cohort #3, Supplemental Table 1). Among the 7 triple-matched patients (in whom the primary tumor, a lymph node metastasis, and a distant metastasis were available for analysis), PHIP copy number increased monotonically in the transition from primary to lymph node to distant metastasis in 4 patients (Fig. 4). In two patients, the lymph node metastasis had a lower copy number than the primary tumor, but the distant metastasis had a higher copy number than both the primary tumor and the lymph node metastasis. In one patient, the lymph node metastasis had a slightly higher copy number than the distant metastasis, but both the lymph node and distant metastasis had higher copy numbers than the primary tumor. There was a significant increase in PHIP copy number in the transition from primary tumor to lymph node to distant metastasis (P=0.008, Friedman test). The mean percentage of cells with elevated PHIP copy number increased monotonically from 13.5% in the primary tumor to 21.6% in the lymph node metastases, and to 55.5% in the distant metastases. In the matched-pair analysis, for each of the 8 cases, PHIP copy number was higher in the metastatic lesion when compared to the corresponding primary tumor (P=0.0039, binomial sign test, Fig. 4). To determine whether this systematic change was unique to PHIP, we performed an identical analysis of PTEN copy number on the same matched-specimen cohort. There was no significant change in PTEN copy number in the transition from primary tumors to either lymph node or distant metastases or both (Supplemental Fig. 1). By comparison, the mean percentage of cells with elevated PTEN copy number was 9.1% in the primary tumor, 12.7% in the lymph node metastases, and 11.8% in the distant metastases (P=0.19, Friedman test applied to the same 7 matched triplets).
Fig. 4.
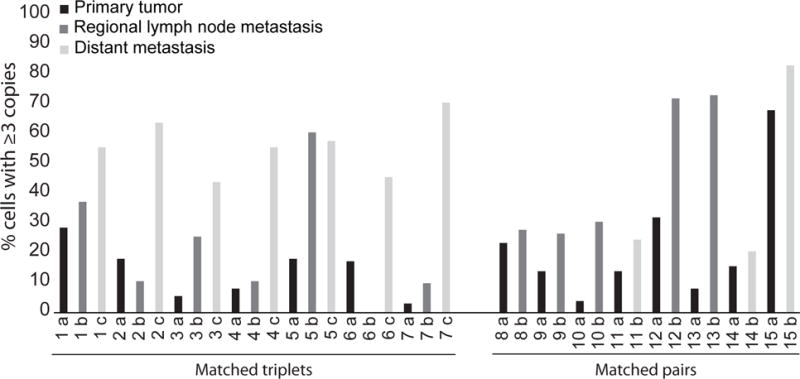
PHIP copy number in a matched cohort of 15 patients, including the primary and at least one metastatic tumor (cohort #3). Bar graphs present the percentage of cells with at least 3 copies of PHIP.
Finally, we examined several of these associations by analyzing PHIP transcript levels in the TCGA cutaneous melanoma cohort (4). Increasing PHIP expression (assessed as a continuous variable) in the primary tumor was associated with significantly reduced relapse-free (RFS, P=0.02) and overall survival (OS, P=0.018) by univariate Cox regression analysis. By multivariate analysis, PHIP expression levels remained significantly predictive of RFS (P<0.05) and OS (P=0.015), even with the inclusion of AJCC stage. Similar to the FISH analysis, there was no significant association between increasing PHIP expression levels in metastatic tumors and survival. In addition, mean PHIP expression levels were significantly (4.3-fold) higher in NRAS-mutant melanomas when compared with NRAS-wild-type cases (P=0.02, Mann-Whitney test; Supplemental Fig. 2A). Finally, mean PHIP transcript levels were significantly (3.7-fold) higher in melanoma metastases when compared to primary tumors (P<0.00005; Supplemental Fig. 2B).
Discussion
We have performed a rigorous analysis to elucidate the role of PHIP copy number elevation in sequential stages of melanoma progression, and as a prognostic marker for primary melanoma. We analyzed the role of PHIP copy number in three independent patient cohorts from different centers. In cohort #1, we show that elevated PHIP copy number is an independent prognostic marker for primary cutaneous melanoma in a cohort of patients derived from a distinct population in Germany, and confirm a significant association with ulceration status, an important prognostic factor incorporated into the AJCC melanoma staging system. In cohort #2, we identify distinct molecular subtypes of melanoma in which PHIP is enriched. In cohort #3, we determine that the degree of PHIP copy number elevation is uniformly increased in the progression from primary to metastatic melanoma. Several of the key findings from these three cohorts were replicated using TCGA cohort analysis.
The independent prognostic role of PHIP levels in primary cutaneous melanoma has now been shown using FISH analysis in two distinct cohorts, replicated at the protein level using immunohistochemical analysis (7), and replicated at the RNA level using TCGA cohort analysis. In addition, our prior studies demonstrated a significant correlation between PHIP copy number and expression in primary melanoma (6). There are few (if any) molecular markers whose prognostic impact has been demonstrated both in distinct cohorts of melanoma patients and using several distinct platforms. These studies advance PHIP as an important molecular marker of melanoma outcome, though additional studies in larger cohorts will be required in order for it be routinely used as a prognostic factor.
Beyond its independent prognostic role, our studies of cohort #1 confirmed a powerful association between PHIP copy number and ulceration. While the prognostic significance of ulceration is well appreciated, the biologic basis for ulceration development in melanoma has been poorly understood. Our previous studies showed that PHIP mediates some of its effects on melanoma metastasis by virtue of activation of the IGF1R-PI3K pathway. PHIP is a potent activator of AKT (7), which drives both glucose metabolism and angiogenesis (11,12). Our studies in melanoma cell lines showed that PHIP gene silencing results in decreased glycolysis (including reduced LDH expression) in vitro and decreased angiogenesis in vivo (6). These results are consistent with a model in which PHIP activation, in part through elevated copy number, promotes glycolysis and angiogenesis, resulting in increased risk of development of ulceration and distant metastasis. Intriguingly, this model suggests that a common signaling pathway may underlie the development of and explain the biological basis of ulceration, a key prognostic marker in stage I-III melanoma, and LDH levels, a prognostic marker for stage IV disease.
In addition, our analyses of cohort #2 identified the molecular subtypes of melanoma in which PHIP copy number elevations are present, and in which they are specifically enriched. Elevated PHIP copy number was present in each molecular subtype of melanoma examined, including melanomas with BRAF mutation, NRAS mutation, as well as in melanomas without any alterations in these markers. Moreover, we found that PHIP copy number was significantly higher in melanomas with NRAS mutation or with intact PTEN expression. The observation that PHIP participates in the IGF1R-PI3K signal transduction pathway in which PTEN also operates provides a rational basis for these results, suggesting that either molecular alteration (PHIP copy number gain or PTEN loss) may be sufficient to activate this pathway in melanoma. Given the importance of PTEN loss specifically in BRAF-mutant melanomas (4), our analysis also identified a trend toward higher PHIP copy number in PTEN-expressing BRAF-mutant tumors, introducing PHIP as a novel potential driver of this molecular subtype of melanoma. An intriguing and unexpected finding was the enrichment of PHIP copy number in NRAS-mutant melanoma. Given that NRAS mutations are thought to activate both the MAPK and PI3K pathways, our results suggest the intriguing possibility of novel biochemical functions of PHIP beyond its participation in the IGF1R-PI3K axis. However, our results are consistent with the observation that PTEN loss is essentially mutually exclusive with NRAS mutation. In addition, they suggest PHIP targeting as a potential strategy for the treatment of NRAS-mutant melanoma, which has represented an important therapeutic challenge given the modest activity of MEK inhibitors (13). Recent studies describing the development of a small molecular inhibitor of PHIP (14) demonstrate its druggability, thereby highlighting its role as a potential therapeutic target.
Our studies of cohort #3 analyzed the role of PHIP in the progression cascade of melanoma by comparing copy number changes in matched primary versus metastatic tumors from the same patient. Remarkably, the matched-specimen analysis revealed elevated PHIP copy number in at least one metastatic lesion in every case analyzed when compared with its corresponding primary tumor. In many instances, the copy number in the metastasis was extremely high, with three or more copies of PHIP observed in greater than 50% of cells in 40% of the metastases, including 56% of distant metastases examined. In addition, in the majority of cases where we had access to the primary tumor, and both lymph node and distant metastases, there was a monotonic increase in PHIP copy number elevation in the transition from primary melanoma to lymph node metastasis, and then to distant metastasis. These results strongly suggest that elevated PHIP copy number is selected for in melanoma progression, supporting our original cDNA microarray analysis in which PHIP was identified, and which was further supported by TCGA cohort analysis, showing significantly increased PHIP expression in metastatic versus (unmatched) primary melanomas. We are unaware of any similar studies analyzing the role of a given molecular marker in successive stages of melanoma progression using tissues from the same patient. Similarly, a recent analysis of colon cancer assessed the somatic variants in hypermutable DNA regions in a cohort of 19 patients with primary colon cancer and matched lymph node and distant metastasis, enabling a conclusion that two-thirds of distant metastases have a profile distinct from that of the lymph node metastasis (15). Our analysis underscores the potency of these matched-tissue comparisons to determine the role of a given gene in cancer progression within individual patients.
In conclusion, our studies to date have shown that increased PHIP levels are predictive of melanoma distant metastasis, promote distant metastasis in mouse models of melanoma, and are enriched in melanoma metastases. The presence of elevated PHIP copy number in distant melanoma metastases as shown in this study indicates that this molecular target is present and enriched in the setting of advanced melanoma. Taken together with functional studies demonstrating an important role for PHIP in promoting melanoma progression, these studies describe an unprecedented role for PHIP in melanoma progression, suggesting it as a promising target for therapy.
Supplementary Material
Fig. 1.
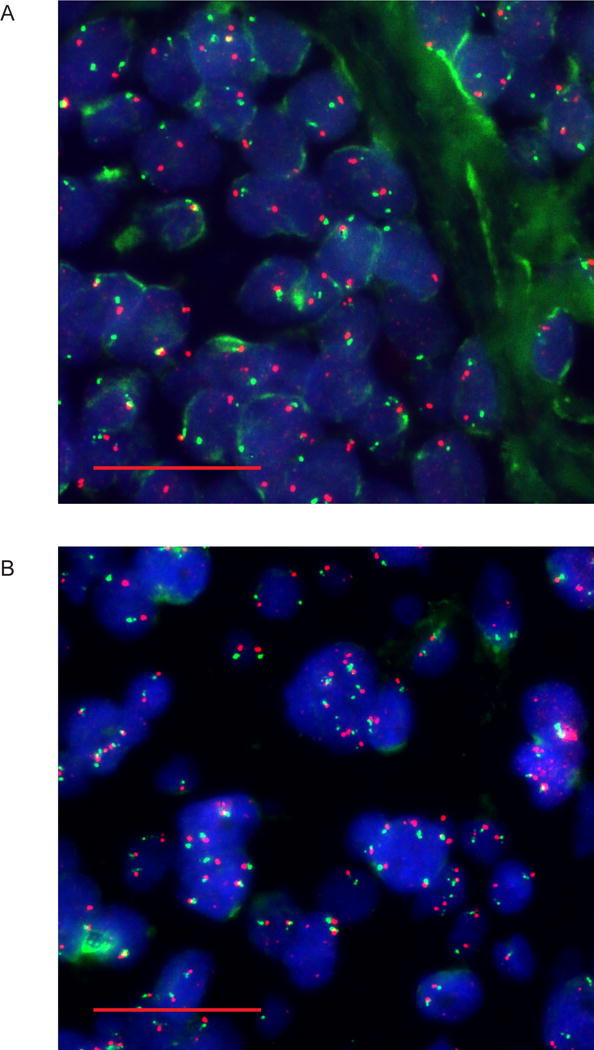
PHIP copy number analysis by FISH. Representative images of melanoma samples with normal (panel A) versus elevated (panel B) PHIP copy number as detected using probes for PHIP (red) and centromere of chromosome 6 (green). Scale bars, 20 μm.
Translational Relevance.
This manuscript presents a comprehensive analysis of the role of PHIP copy number in sequential stages of melanoma progression. We confirm that elevated PHIP copy number is an independent prognostic marker for primary cutaneous melanoma. In addition, we identify distinct molecular subtypes of melanoma in which PHIP is enriched. Finally, we show that the percentage of melanoma cells with elevated PHIP copy number is increased in the progression from primary to metastatic melanoma. Taken together, these studies describe a key role for PHIP in melanoma progression, confirming it as a promising target for therapy.
Acknowledgments
This work by supported by a grant from the National Institutes of Health (R01CA175768) to MKS. The collection of the tissue set at MD Anderson was supported by the following: The Melanoma Specialized Program of Research Excellence of the University of Texas MD Anderson Cancer Center (P50 CA093459); and philanthropic contributions from the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, the AIM at Melanoma Foundation, and the MD Anderson Melanoma Moon Shot Program.
J.R.M. has ownership interests (including patents) at MDMS LLC. J.E.G. has served on advisory boards for Merck, Syndax, and Castle Biosciences. M.A.D. has served on advisory boards for Bristol-Myers Squibb, Roche/Genentech, Novartis, Sanofi-Aventis, Syndax, and Vaccinex, as a consultant for Nanostring (without compensation), and has received grant support from Bristol-Myers Squibb, Roche/Genentech, GSK, Sanofi-Aventis, Oncothyreon, Merck, and Astra Zeneca. D.S. has served on advisory boards for and received honoraria from Bristol-Myers Squibb, Roche, Merck, Incyte, Amgen, Novartis, and EMD. M.K.S. has ownership interest in Melanoma Diagnostics, has received honoraria from Cepheid, and has received grant support from Merck.
Footnotes
Conflicts of interest: All other authors declare no conflicts.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Zettersten E, Shaikh L, Ramirez R, Kashani-Sabet M. Prognostic factors in primary cutaneous melanoma. Surg Clin North Am. 2003;83(1):61–75. doi: 10.1016/s0039-6109(02)00094-4. [DOI] [PubMed] [Google Scholar]
- 3.Gershenwald J, Scolyer R, Hess K, Thompson J, Long G, Ross M. Melanoma of the skin. AJCC Cancer Staging Manual. 2017 [Google Scholar]
- 4.Cancer Genome Atlas N. Genomic Classification of Cutaneous Melanoma. Cell. 2015;161(7):1681–96. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haqq C, Nosrati M, Sudilovsky D, Crothers J, Khodabakhsh D, Pulliam BL, et al. The gene expression signatures of melanoma progression. Proc Natl Acad Sci U S A. 2005;102(17):6092–7. doi: 10.1073/pnas.0501564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bezrookove V, De Semir D, Nosrati M, Tong S, Wu C, Thummala S, et al. Prognostic impact of PHIP copy number in melanoma: linkage to ulceration. J Invest Dermatol. 2014;134(3):783–90. doi: 10.1038/jid.2013.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Semir D, Nosrati M, Bezrookove V, Dar AA, Federman S, Bienvenu G, et al. Pleckstrin homology domain-interacting protein (PHIP) as a marker and mediator of melanoma metastasis. Proc Natl Acad Sci U S A. 2012;109(18):7067–72. doi: 10.1073/pnas.1119949109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bucheit AD, Chen G, Siroy A, Tetzlaff M, Broaddus R, Milton D, et al. Complete loss of PTEN protein expression correlates with shorter time to brain metastasis and survival in stage IIIB/C melanoma patients with BRAFV600 mutations. Clin Cancer Res. 2014;20(21):5527–36. doi: 10.1158/1078-0432.CCR-14-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bayani J, Squire J. Multi-color FISH techniques. Curr Protoc Cell Biol. 2004:5. doi: 10.1002/0471143030.cb2205s24. Chapter 22: Unit 22. [DOI] [PubMed] [Google Scholar]
- 10.Balch CM, Soong SJ, Gershenwald JE, Thompson JF, Reintgen DS, Cascinelli N, et al. Prognostic factors analysis of 17,600 melanoma patients: validation of the American Joint Committee on Cancer melanoma staging system. J Clin Oncol. 2001;19(16):3622–34. doi: 10.1200/JCO.2001.19.16.3622. [DOI] [PubMed] [Google Scholar]
- 11.Arsham AM, Plas DR, Thompson CB, Simon MC. Akt and hypoxia-inducible factor-1 independently enhance tumor growth and angiogenesis. Cancer Res. 2004;64(10):3500–7. doi: 10.1158/0008-5472.CAN-03-2239. [DOI] [PubMed] [Google Scholar]
- 12.Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR, et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64(11):3892–9. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 13.Dummer R, Schadendorf D, Ascierto PA, Arance A, Dutriaux C, Di Giacomo AM, et al. Binimetinib versus dacarbazine in patients with advanced NRAS-mutant melanoma (NEMO): a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol. 2017;18(4):435–45. doi: 10.1016/S1470-2045(17)30180-8. [DOI] [PubMed] [Google Scholar]
- 14.Cox OB, Krojer T, Collins P, Monteiro O, Talon R, Bradley A, et al. A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain. Chemical Science. 2016;7(3):2322–30. doi: 10.1039/C5SC03115J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naxerova K, Reiter JG, Brachtel E, Lennerz JK, van de Wetering M, Rowan A, et al. Origins of lymphatic and distant metastases in human colorectal cancer. Science. 2017;357(6346):55–60. doi: 10.1126/science.aai8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


