Summary
Long chain fatty acid (LCFA) oxidation has been shown to play an important role in IL-4-mediated macrophage polarization [M(IL-4)]. However, many of these conclusions are based on the inhibition of carnitine palmitoyltransferase-1 with high concentrations of etomoxir that far exceed what is required to inhibit enzyme activity (EC90 < 3 μM). We employ genetic and pharmacologic models to demonstrate that LCFA oxidation is largely dispensable for IL-4-driven polarization. Unexpectedly, high concentrations of etomoxir retained the ability to disrupt M(IL-4) polarization in the absence of Cpt1a or Cpt2 expression. Although excess etomoxir inhibits the adenine nucleotide translocase, oxidative phosphorylation is surprisingly dispensable for M(IL-4). Instead, the block in polarization was traced to depletion of intracellular free CoA, likely resulting from conversion of the pro-drug etomoxir into active etomoxiryl-CoA. These studies help explain the effect(s) of excess etomoxir on immune cells, and reveal an unappreciated role for CoA metabolism in macrophage polarization.
TOC image
The CPT-1 inhibitor etomoxir has been used to suggest long chain fatty acid (LCFA) oxidation is necessary for alternative macrophage activation. Divakaruni and colleagues now show that LCFA oxidation is dispensable. They demonstrate multiple off-target effects of etomoxir, and show that depletion of coenzyme A by etomoxir blocks M(IL-4) differentiation.
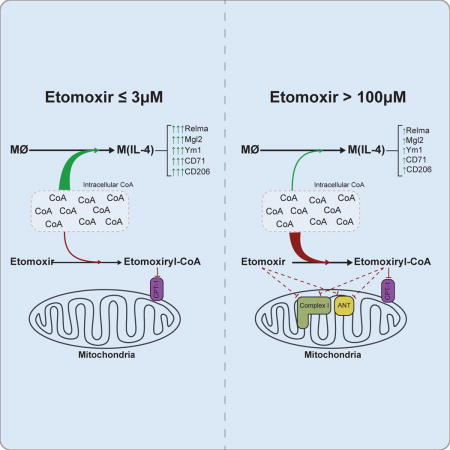
Introduction
Macrophages are innate immune cells responsible for a broad range of processes such as detecting and killing pathogens, clearing apoptotic cells and subcellular debris, initiating wound-healing programs, and presenting antigens to T cells. To achieve these different functions, macrophages possess the capacity to acquire pro- and anti-inflammatory programs in response to extrinsic signals. Accumulating evidence indicates that alterations in cellular metabolism are critical for supporting these varied roles (O’Neill et al., 2016). For example, activation of Toll-like receptor 4 signaling with the pro-inflammatory stimulus lipopolysaccharide (LPS) results in profound changes to both glycolytic and mitochondrial metabolism (Tannahill et al., 2013). Flux through glycolysis and the pentose phosphate pathway are substantially increased to support energy metabolism, anabolism, and cytoplasmic NADPH production (Van den Bossche et al., 2017), while oxidative phosphorylation is inhibited, presumably to reprogram mitochondria for the production of pro-inflammatory metabolites and redox signals (Mills et al., 2016). Conversely, macrophages differentiated with the anti-inflammatory cytokine interleukin 4 (IL-4) (± IL-13) induce transcriptional programs associated with mitochondrial biogenesis and oxidative mitochondrial metabolism (Vats et al., 2006).
Importantly, genetic inhibition of these metabolic pathways attenuates the acquisition of effector responses. This demonstrates a fundamental role for metabolism in macrophage function and informs the hypothesis that pharmacologically targeting these pathways can influence macrophage biology. In particular, the acquisition of anti-inflammatory phenotypes and/or wound healing functions can be inhibited by perturbing the reprogramming of oxidative metabolism (Odergaard et al., 2007). However, it remains unclear precisely which metabolic pathways or signals governed by these larger transcriptional programs are indispensable for (rather than merely associated with) IL-4-induced alternative macrophage activation.
It is well accepted that differentiation of macrophages with IL-4 [referred to herein as M(IL-4) polarization or alternative activation] is associated with enhanced fatty acid oxidation (O’Neill et al., 2016; Van den Bossche et al., 2017). Long-chain fatty acid (LCFA) oxidation is demonstrably increased in macrophages upon IL-4 stimulation (Gonzalez-Hurtado et al., 2017; Namgaladze and Brüne, 2014), and PPARγ, a nuclear receptor controlling gene expression for lipid metabolism-related genes, is associated with and essential for alternative macrophage activation (Odegaard et al., 2007; Szanto et al., 2010; Vats et al., 2006). Whether LCFA oxidation is obligatory for M(IL-4) polarization, though, remains controversial. Supportive evidence comes largely from studies using etomoxir (Covarrubias et al., 2016; Huang et al., 2014; Huang et al., 2016), an inhibitor of carnitine palmitoyl transferase-1 (CPT-1). CPT-1 is a mitochondrial outer membrane enzyme that conjugates long chain fatty acyl CoAs to carnitine and facilitates uptake into the mitochondrial matrix for oxidation.
Etomoxir is a small molecule developed for metabolic and cardiovascular disease that exhibits nanomolar potency towards CPT-1a and CPT-1b upon enzymatic conversion to the active inhibitor etomoxiryl CoA (IC50 = 0.01 – 0.70 μM; Bentebibel et al., 2006; Ceccarelli et al., 2011; Declercq et al., 1987). Therefore, the specificity of etomoxir at 200 μM, a concentration frequently used to elicit inhibition of alternative macrophage activation, has been questioned. For example, multiple studies have shown that lower concentrations of etomoxir fail to affect IL-4-associated markers [Namgaladze and Brüne, 2014 (10 μM); Tan et al., 2015 (100 μM); Van den Bossche et al., 2016 (100 μM)]. Similar to pharmacologic inhibition of CPT-1, genetic studies have also yielded inconclusive results. Although viral knockdown of Cpt1a reduced expression of IL-4-associated cell surface markers (Huang et al., 2016), an integrative systems approach merging metabolomics and transcriptomics indicated Cpt1 expression is more closely associated with pro-inflammatory macrophage polarization rather than an anti-inflammatory program (Jha et al., 2015). Moreover, myeloid-specific knockout of Cpt2, the enzyme required to convert fatty acyl carnitines back to fatty acyl CoAs in the mitochondrial matrix, does not impair macrophage differentiation in vitro or in vivo despite significant reductions in LCFA oxidation (Gonzalez-Hurtado et al., 2017). When used at 200 μM, etomoxir was equally effective in blocking M(IL-4) polarization in Cpt2−/− bone marrow-derived macrophages (BMDMs) as in wild-type (WT) cells (Nomura et al., 2016). This aligns with literature suggesting etomoxir may have CPT-1-independent effects on metabolism at high concentrations (Agius et al., 1991; Ceccarelli et al., 2011; O’Connor et al., 2018), including inhibition of mitochondrial complex I (Yao et al., 2018).
To address these discrepancies, we re-examined the role of LCFA oxidation in macrophage polarization and the inhibitory effects of etomoxir on cellular metabolism. We find that loss of Cpt1a or Cpt2 in myeloid cells has little impact on IL-4-mediated polarization despite significant decreases in LCFA oxidation. Likewise, inhibition of CPT-1 enzymatic activity with a low yet effective concentration of etomoxir (3 μM) did not influence M(IL-4) polarization, suggesting that LCFA oxidation is dispensable for this process. Nevertheless, we observed that treatment of macrophages with a higher concentration of etomoxir (200 μM) inhibited polarization even in the absence of Cpt1a. This indicated that the inhibitory effect of excess etomoxir on alternative macrophage activation is likely an off-target effect. Mechanistic studies revealed that high concentrations of etomoxir inhibit the adenine nucleotide translocase (ANT) and respiratory complex I, suggesting that perturbations in mitochondrial function could be responsible for this effect on macrophages. However, we observed that pharmacologic inhibition of oxidative phosphorylation with a variety of inhibitors had no significant effect on M(IL-4) polarization, although inhibition of the ANT is likely relevant to the mechanism by which etomoxir inhibits T cell activation and differentiation (Raud et al., 2018). Metabolomics studies on IL-4-differentiated macrophages treated with 3 μM or 200 μM etomoxir suggested that the inhibitory effects on macrophage polarization were a function of perturbed intracellular coenzyme A homeostasis. Consistent with this, CoA supplementation was sufficient to overcome the etomoxir-mediated blockade of alternative activation. Moreover, provision of exogenous CoA was able to augment M(IL-4) polarization even in the absence of etomoxir. Taken together, these findings suggest that LCFA oxidation is largely dispensable for IL-4-driven macrophage polarization, and identify CoA metabolism as an important regulator for macrophage fate.
Results
Low concentrations of etomoxir effectively inhibit CPT-1 but do not affect M(IL-4) polarization
Prior to localizing potential off-target effects of etomoxir, we first confirmed an appropriate on-target concentration at CPT-1 in a variety of cell types. We measured oxygen consumption rates in cultured cells offered albumin-buffered palmitate as a respiratory substrate (Rogers et al., 2014). HepG2 cells could sustain robust rates of FCCP-stimulated respiration that were largely sensitive to 1 μM etomoxir (Fig. 1A). Under these conditions, concentration-response curves showed etomoxir had a sub-micromolar half-maximal inhibitory concentration (EC50) for uncoupler-stimulated respiration in both HepG2 and A549 cells (Fig. 1B). To corroborate this finding with an orthogonal measurement of LCFA oxidation in BMDMs, we observed that 3 μM etomoxir can effectively suppress 3H2O release from tritiated palmitate (Fig. 1C). The results confirm etomoxir is a potent, saturable inhibitor of LCFA oxidation in whole cells. Importantly, these data align with enzymology, cell-based assays, and tissue studies showing the inhibitor works with nanomolar or low micromolar efficacy depending upon the biological model (Ceccarelli et al., 2011; Declercq et al., 1987; Lopaschuk et al., 1988; Namgaladze and Brüne, 2014).
Figure 1. Etomoxir concentrations that specifically inhibit CPT-1 do not inhibit M(IL-4) polarization.
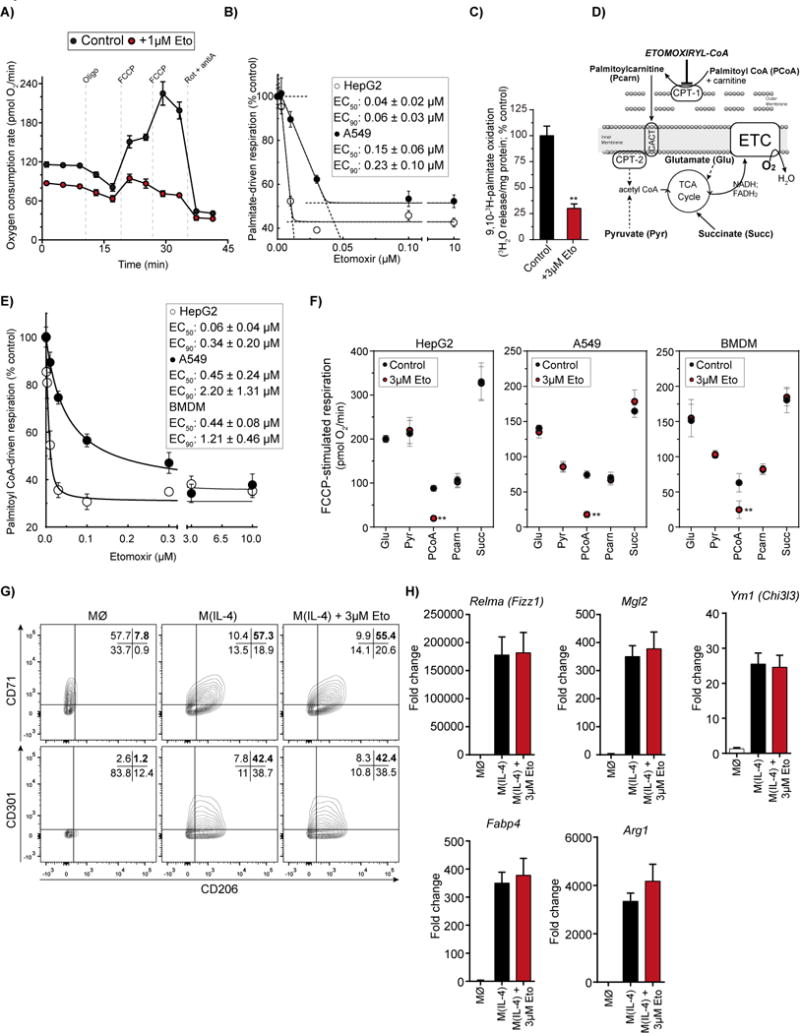
(A) Respirometry trace with intact HepG2 cells ± 1 μM etomoxir (Eto) offered albumin-buffered palmitate as substrate. (n = 6 technical replicates)
(B) Concentration-response curve of FCCP-stimulated respiration in intact HepG2 and A549 cells [as in (A)] in response to increasing concentrations of etomoxir. (n = 6 technical replicates). Inset: Aggregate values for EC50 (concentration required for 50% inhibition) and EC90 (90% inhibition) from n ≥ 4 independent biological replicates.
(C) Fatty acid oxidation as measured by 3H2O release from 9,10-3H-palmitate ± 3 μM etomoxir (Eto). (n = 4 independent biological replicates).
(D) Schematic depicting respiratory substrates used for permeabilized cell respirometry. CPT, carnitine palmitoyl transferase; CACT, carnitine acyl carnitine transferase; ETC, electron transport chain.
(E) Sample concentration-response curve of FCCP-stimulated oxygen consumption in permeabilized HepG2 and A549 cells (BMDMs omitted for clarity) in response to increasing concentrations of etomoxir (n=6 technical replicates). Cells were offered palmitoyl CoA with carnitine and malate as substrates. Inset: Aggregate values for EC50 and EC90 in all cell types tested. (n ≥ 4 independent biological replicates).
(F) Effect of etomoxir on various respiratory substrates in permeabilized cells. Glu, glutamate/malate; Pyr, pyruvate/malate; PCoA, palmitoyl CoA/carnitine/malate; Pcarn, palmitoylcarnitine/malate; Succ, succinate/rotenone. (n = 3 independent biological replicates)
(G) Flow cytometric analysis of the IL-4-associated cell surface markers CD206, CD301, and CD71 in BMDMs 24 hr after the indicated treatment. Cells were co-treated with IL-4 (20 ng/mL) and etomoxir (3 μM). Numbers in the top-right quandrant indicate cells positive for both markers measured. The data shown are from one experiment and representative of a total of six independent biological replicates.
(H) qPCR analysis of the IL-4-associated genes Relma, Mgl2, Ym1, Fabp4, and Arg1 after 24 hr of IL-4 ± 3 μM etomoxir co-treatment. (n = 4 independent biological replicates)
All data are presented as mean ± S.E.M. *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001
Although experiments in intact cells address the potency of etomoxir, they do not demonstrate specificity. We therefore used substrate-specific respirometry in permeabilized cells (Divakaruni et al., 2013; Divakaruni et al., 2014b) to determine a concentration of etomoxir that produced a saturable, specific inhibitory effect at CPT-1. Permeabilized cells were offered palmitoyl-CoA with carnitine to directly interrogate respiration mediated by CPT-1 (Fig. 1D). The CoA thioester of etomoxir is the active, irreversible inhibitor of CPT-1 (Bartlett et al., 1981; Ceccarelli et al., 2011; Kiorpes et al. 1984), so cells were pretreated with etomoxir to allow formation of etomoxiryl-CoA prior to plasma membrane permeabilization. In permeabilized HepG2s, A549s, and BMDMs, etomoxir inhibited FCCP-stimulated, palmitoyl CoA-driven respiration with nanomolar EC50 values, and had a saturable effect at less than 3 μM (Fig. 1E).
To verify this was a specific effect at CPT-1 rather than another downstream enzyme, permeabilized cells were offered other respiratory substrates in the presence of 3 μM etomoxir. Etomoxir had a significant effect on palmitoyl CoA oxidation, but no effect on respiration driven by substrates which do not require CPT-1: glutamate/malate, pyruvate/malate, palmitoylcarnitine/malate, and succinate/rotenone (Fig 1F). Residual respiratory rates in the presence of saturating etomoxir may be driven by endogenous substrates and/or exogenously added malate. Nonetheless, these substrate specificity experiments demonstrate that 3 μM etomoxir is a specific, saturable inhibitor of CPT-1 in BMDMs, and reinforce data showing this concentration specifically inhibits CPT-1 in regulatory T (Treg) cells (Raud et al., 2018).
Having established the efficacy of low concentrations of etomoxir in blocking CPT-1, we then examined whether 3 μM etomoxir impacts IL-4-mediated polarization of macrophages. FACS analysis revealed that 3 μM etomoxir had no effect on the expression levels of IL-4-induced CD206, CD71, or CD301 (Fig. 1G), markers of alternative macrophage activation (Misharin A.V. et al., 2013; Van den Bossche et al., 2016). Moreover, no change was observed in the gene expression of Relma, Mgl2, Ym1, Fabp4, and Arg1, key markers of M(IL-4) polarization (Fig. 1H) (Szanto et al., 2010; Vats et al., 2006). Taken together, the data indicate pharmacologic inhibition of CPT-1 by etomoxir inhibits LCFA oxidation but does not block IL-4-induced macrophage polarization. They also support similar observations showing the IL-4 response is unaffected by etomoxir concentrations ranging from 10 μM (Namgaladze and Brüne, 2014) to 100 μM (Tan et al., 2015; Van den Bossche et al., 2016).
High concentrations of etomoxir inhibit M(IL-4) polarization independently of CPT-1 activity
Having established that concentrations of etomoxir that maximally inhibit CPT-1 do not block IL-4-mediated responses in BMDMs, we then hypothesized that promiscuous effects of the drug were responsible for its well-established inhibition of alternative macrophage activation (Covarrubias et al., 2016; Huang et al., 2014; Huang et al., 2016; Nomura et al., 2016). As previously reported, 200 μM etomoxir strongly reduced the population of CD206+/CD71+ cells in response to IL-4-driven polarization (Fig. 2A). However, significant changes in macrophage polarization were only observed at concentrations >100 μM (Fig. S1A), orders of magnitude greater than the EC50 for CPT-1 inhibition and suggestive of a mechanism discrete from LCFA oxidation. 200 μM etomoxir also had little effect on the population of CD301+ cells (Fig. S1B), suggesting that etomoxir affects only a subset of the IL-4 responses. Additional evidence supporting a mosaic-like effect of etomoxir on M(IL-4) polarization is given by gene expression patterns: 200 μM etomoxir blocked the induction of Relma, Mgl2, and Ym1 expression, but no change was observed for Fabp4 and Arg1 (Fig. 2B). Arginase activity was also unaffected by high etomoxir (Fig. S1C), reinforcing previous findings (Van den Bossche et al., 2016).
Figure 2. High concentrations of etomoxir block M(IL-4) polarization independently of CPT-1 activity.
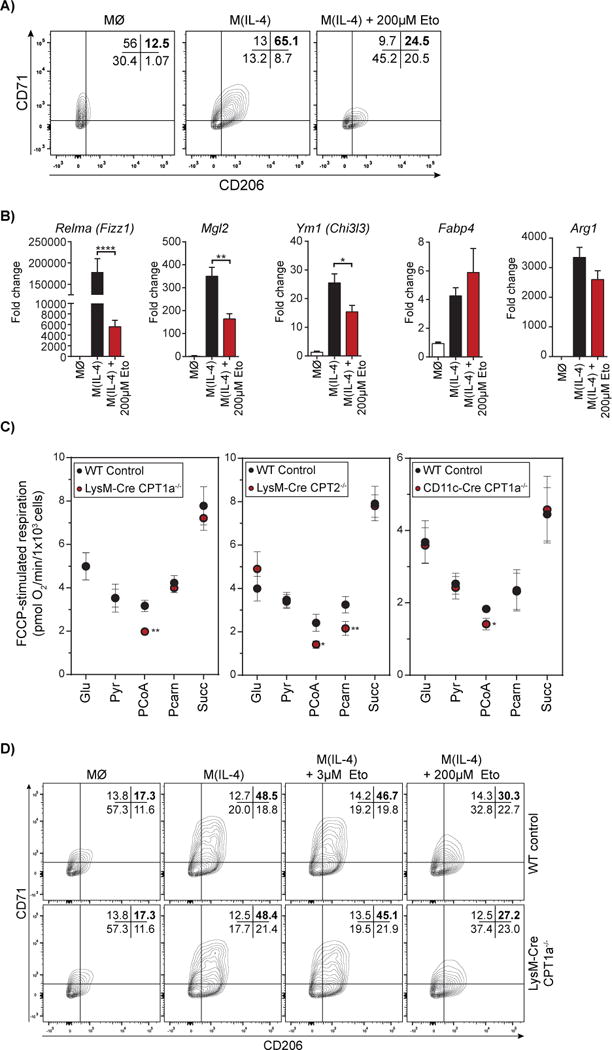
(A) Flow cytometric analysis of CD206 and CD71 on macrophages treated with IL-4 ± 200 μM etomoxir co-treatment after 24 hr. The data shown are from one experiment representative of a total of six independent biological replicates.
(B) qPCR analysis of Relma, Mgl2, Ym1, Fabp4, and Arg1 in BMDM with IL-4 ± 200 μM etomoxir co-treatment after 24 hr. (n = 6 independent biological replicates)
(C) Effect of various respiratory substrates in permeabilized WT, Cpt1−/−, and Cpt2−/− BMDMs. Glu, glutamate/malate; Pyr, pyruvate/malate; PCoA, palmitoyl CoA/carnitine/malate; Pcarn, palmitoylcarnitine/malate; Succ, succinate/rotenone. (n = 4 independent biological replicates)
(D) Flow cytometric analysis for the CD206+/CD71+ population in WT and Cpt1a−/− BMDMs differentiated with IL-4 ± 3 μM or 200 μM etomoxir. The data shown are from one experiment representative of a total of four independent biological replicates.
See also Supplementary Fig. 1
To further confirm that the effect of excess etomoxir on M(IL-4) polarization was independent of LCFA oxidation, we characterized macrophage polarization in BMDMs with genetic disruption of CPT-1a or CPT-2. BMDMs with a myeloid-specific deletion of Cpt1a were generated by breeding LoxP-Cpt1a mice with either LysM-Cre or CD11c-Cre mice, and a similar strategy was used for generation of Cpt2−/− mice (Nomura et al., 2016). In addition to reduced expression at the transcript and protein level (Fig. S1D), functional deficiency of CPT1a was characterized by substrate-specific respirometry in permeabilized BMDMs (Fig 2C). Analogous to the specificity of 3 μM etomoxir (Fig. 1F), decreased rates of palmitoyl CoA-driven respiration were observed in the two conditional Cpt1a−/− strains while respiration on other mitochondrial substrates was unchanged relative to controls (Fig. 2C). Moreover, since CPT-2 is required for the oxidation of both palmitoyl CoA and palmitoylcarnitine (Fig 1C; Ceccarelli et al., 2011), BMDMs from Cpt2−/− mice demonstrated compromised rates when oxygen consumption was driven by these substrates but not others (Fig. 2C), aligning with radiometric measurements of LCFA oxidation in these cells (Gonzalez-Hurtado et al., 2017). Sensitivity of respiration to 3 μM etomoxir, a concentration shown to be specific to CPT-1 (Fig. 1), was largely absent in all three conditional CPT KO cells, further confirming a specific reduction in LCFA oxidation (Fig. S1E,F).
Similar to pharmacologic inhibition of CPT-1 using 3 μM etomoxir, genetic disruption of LCFA oxidation did not affect the response to IL-4. The population of CD206+/CD71+ in LysM-Cre Cpt1a−/− macrophages was unchanged upon M(IL-4) polarization (Figure 2D). Moreover, the lack of an effect from 3 μM etomoxir but a robust block of CD206+/CD71+ expression from 200 μM was observed in both WT and Cpt1a−/− macrophages. A similar result was observed with the Cpt2−/− (Fig. S1G) and the CD11c-Cre Cpt1a−/− BMDMs (Fig. S1H). IL-4-stimulated lactate efflux was also unchanged despite genetic disruption of LCFA oxidation in all three cell types (Fig. S1I). In summary, these data reinforce the concept that LCFA oxidation is dispensable for alternative macrophage activation (Gonzales-Hurtado et al, 2017; Namgaladze and Brüne, 2014; Nomura et al., 2016), and show the effects of excess etomoxir on M(IL-4) polarization are independent of CPT activity.
High concentrations of etomoxir inhibit the adenine nucleotide translocase
Having shown LCFA oxidation does not control alternative macrophage activation, we then sought to determine the off-target effect by which etomoxir was blocking M(IL-4) polarization. A growing body of literature has linked M(IL-4) polarization with genetic programs supporting mitochondrial biogenesis (Vats et al., 2006; Odegaard et al., 2007) and aerobic glucose oxidation (Covarrubias et al., 2016; Huang et al., 2016; Tan et al., 2015; Van den Bossche et al., 2016). Thus, we hypothesized that the off-target effect(s) of etomoxir on immune cell phenotypes could be via perturbed mitochondrial metabolism. Indeed, the extent of respiratory inhibition by 200 μM etomoxir was similar in WT and Cpt1a−/− or Cpt2−/− BMDMs (Fig. 3A). The observation is similar to findings in Treg cells (Raud et al., 2018), and suggests the reductions in respiration are attributable to effects on energy metabolism independent of LCFA oxidation.
Figure 3. Excess etomoxir has off-target effects on mitochondrial bioenergetics, but these cannot explain its inhibition of M(IL-4).
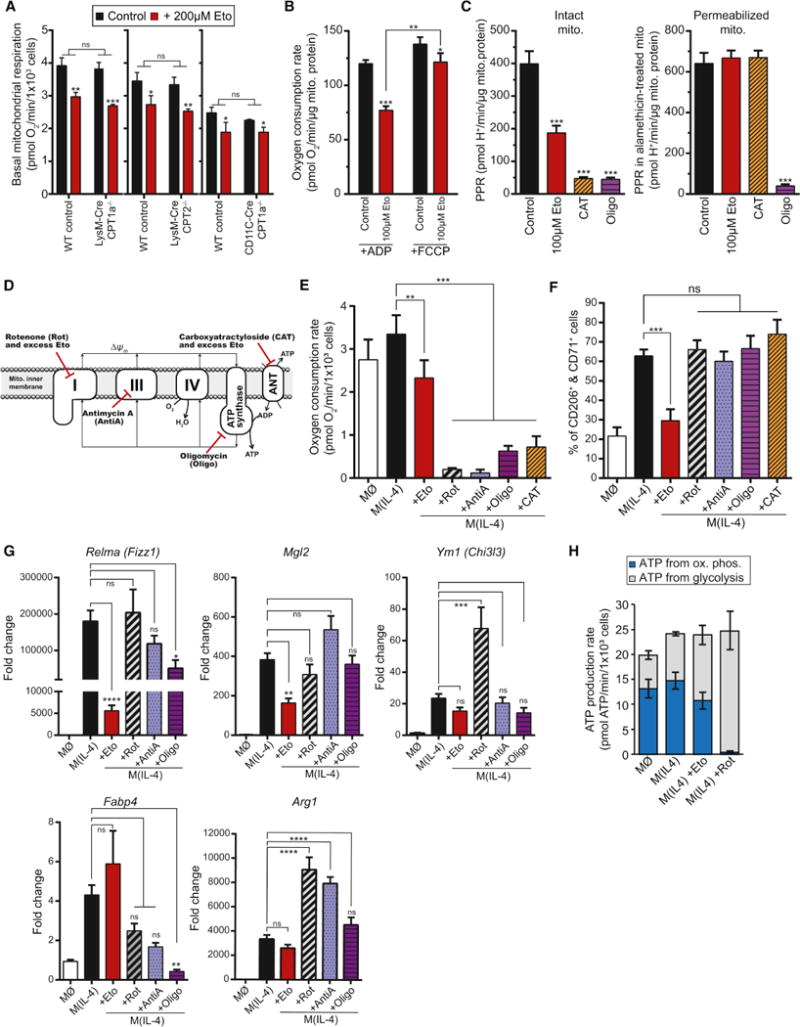
(A) Effects of 200 μM etomoxir on basal mitochondrial respiration in intact WT, Cpt1a−/−, and Cpt2−/− BMDMs. Cells were offered 8 mM glucose, 2 mM glutamine, and 2 mM pyruvate (no fatty acids or carnitine) in the experimental medium. (n = 4 independent biological replicates)
(B) ADP-stimulated (State 3) or maximal FCCP-stimulated respiration was measured ± 100 μM etomoxir in isolated rat liver mitochondria. Mitochondria were offered pyruvate/malate as respiratory substrates. (n = 4 independent biological replicates)
(C) Inhibition of ANT activity was assessed by measuring the rate of ATP hydrolysis in both intact (left) and permeabilized (right) rat liver mitochondria. ATP hydrolysis acidifies the experimental medium (see Supplemental Figure S3). Mitochondria were treated with alamethicin to permeabilize the inner membrane to small solutes such as ATP to discriminate between effects of ATP transport versus hydrolysis, Eto., etomoxir (100 μM); CAT, carboxyatractyloside (7.5 ng/mg mito. protein); Oligo., oligomycin (3 ng/mg mito. protein). (n = 4 independent biological replicates)
(D) Scheme depicting sites of action for mitochondrial inhibitors used in (E-G).
(E) Basal mitochondrial respiration in intact BMDMs after 24 hr treatment with 200 μM etomoxir (Eto), 200 nM rotenone (Rot), 200 nM antimycin A (antiA), 1.2 μM oligomycin (Oligo), or 5 μM carboxyatractyloside (CAT). (n = 4 independent biological replicates)
(F) Flow cytometric analysis of the CD206+/CD71+ population after 48 hr treatment with 200 μM etomoxir or mitochondrial effectors as in (E). (n = 6 independent biological replicates)
(G) qPCR analysis of Relma, Mgl2, Ym1, Fabp4, and Arg1 after 24 hr treatment of compounds used in E & F. (n = 4 independent biological replicates)
(H) Cellular ATP production rates were estimated as the sum of ATP generated from oxidative phosphorylation and glycolysis. BMDMs were treated with IL-4 for 24 hr ± 200 μM etomoxir or 200 nM rotenone. (n = 4 independent biological replicates)
See also Supplementary Figs. 2–4
To localize the specific protein(s) inhibited, we first identified assay conditions and cell types in which CPT-1 inhibition with 3 μM etomoxir had no effect on the maximal respiratory rate (Fig. S2A). In six different cell types, this low concentration of etomoxir did not inhibit respiration when intact cells were offered glucose, glutamine, and pyruvate (with no fatty acids or carnitine). This panel included A549s and BMDMs, in which 3 μM etomoxir blocks CPT-1 activity (Fig. 1F). It therefore stands to reason that any effects on respiration from concentrations above 3 μM etomoxir in these cells are attributable to off-target interactions. Indeed, other laboratories have observed a lack of significant changes in respiration under similar conditions in BMDMs at concentrations as high as 100 μM etomoxir (Tan et al., 2015; Van den Bossche et al., 2016), far exceeding the concentration required to maximally inhibit CPT-1. As an important positive control, 3 μM etomoxir was sufficient to inhibit respiration in intact HepG2 hepatoma cells and 3T3-L1 adipocytes (Fig. S2B), cells that can oxidize endogenous LCFAs under certain experimental conditions.
Indeed, in all six cell types tested in Fig. S2A, high concentrations of etomoxir inhibited respiration with a uniform and instructive profile (Fig. S2C-E). Inhibitors of the electron transport chain often have greater effects on the maximal FCCP-stimulated rate of respiration (when oxygen consumption is uncoupled from energy metabolism and can approach a maximal rate) compared to respiration coupled to ATP synthesis (when rates are limited by the energy demands of the cell) (Divakaruni et al., 2014a). High concentrations of etomoxir, however, showed the opposite effect: increasing concentrations of etomoxir disproportionately inhibited ATP-linked respiration relative to uncoupler-stimulated respiration (Fig. S2C-E). We therefore hypothesized that the primary off-target effect of etomoxir was inhibition of a protein directly involved in ATP synthesis: the ATP synthase, ANT, or the phosphate (Pi) transporter.
To more directly examine the effect of etomoxir on the enzymes and transporters involved in oxidative phosphorylation, we measured respiration in isolated mitochondria and plasma membrane-permeabilized cells. As expected, high concentrations of etomoxir had a greater inhibitory effect on respiration driven by ADP versus FCCP in both isolated rat liver mitochondria (Fig. 3B) as well as permeabilized cells (Fig. S3A,B), supporting a direct effect of etomoxir on mitochondrial proteins rather than an effect on cytoplasmic ATP-consuming reactions (Divakaruni et al., 2014a). After showing high etomoxir does not inhibit the Pi transporter (Fig. S3C), we used ATP hydrolysis measurements in isolated mitochondria to discriminate between effects on the ATP synthase versus the ANT (Fig. 3C; Fig. S3D,E) (Divakaruni et al., 2017a; McQuaker et al., 2013). In intact isolated mitochondria, etomoxir inhibited ATP hydrolysis, as did both oligomycin (ATP synthase inhibitor) and carboxyatractyloside (CAT; ANT inhibitor) (Fig. 3C, left; Fig. S3D). However, when alamethicin was used to permeabilize mitochondria to small solutes and eliminate the need for nucleotide transport across the inner membrane, both etomoxir and CAT showed no effect while oligomycin remained inhibitory (Fig. 3C, right; Fig. S3D). Thus, high concentrations of etomoxir inhibit the ANT.
Additionally, oxygen consumption measurements with double-permeabilized cells (Divakaruni et al., 2017a) show high concentrations of etomoxir inhibit oxidation of NADH but not succinate (Fig. S3F). This demonstrates a secondary off-target effect at respiratory complex I, which has been noted by others (Yao et al., 2018) and provides an explanation for why excess etomoxir also inhibits uncoupler-stimulated respiration (Fig. 3B, S3A, S3B). A similar profile of a pronounced off-target effect on phosphorylating respiration with a secondary effect at uncoupled respiration is apparent in both BMDMs and Treg cells, and is independent of CPT-1 activity in both cell types (Fig. S3G; Raud et al., 2018). As the inhibition of the ANT is likely essential to the mechanism by which etomoxir inhibits T cell function (Raud et al., 2018), we then examined whether off-target effects on oxidative phosphorylation can also explain why etomoxir blocks M(IL-4) polarization.
Oxidative phosphorylation does not regulate many markers of M(IL-4) polarization
To test whether the inhibitory effects at the ANT and complex I could explain inhibition of IL-4-driven macrophage polarization by 200 μM etomoxir, we measured whether other inhibitors of oxidative phosphorylation could phenocopy this effect. Specifically, we used compounds that block both ATP synthesis (CAT or oligomycin) and respiratory chain activity (rotenone, piericidin A, or antimycin A; Fig 3D). Indeed, each of the five compounds strongly reduced respiration at 24 hr., well beyond the extent of inhibition by etomoxir (Fig. 3E, S4A). Additionally, and as is expected with potent respiratory chain blockers, rates of 3H2O release from 9,10-3H-palmitate showed rotenone or antimycin A suppressed LCFA oxidation to levels in line with 200 μM etomoxir (Fig. S4B). Remarkably, however, none of the inhibitors of ATP synthesis or the electron transport chain significantly lowered the population of CD206+/CD71+ cells (Fig. 3F, Fig. S3C,D), suggesting that inhibition of oxidative phosphorylation cannot explain the effects of excess etomoxir on M(IL-4) polarization.
Gene expression results further confirmed that the mechanism by which etomoxir blocks alternative macrophage activation is distinct from electron transport chain activity and oxidative mitochondrial metabolism (Fig. 3G, S4E). In the same way that only a subset of IL-4-associated genes was affected by high concentrations of etomoxir (Fig. 2B), inhibitors of oxidative phosphorylation had a mosaic-like effect on IL-4-responsive genes, and varied based on the site of inhibition. The most striking observation was that rotenone, piericidin A, and antimycin A, potent inhibitors of the electron transport chain, failed to reduce IL-4-induced expression of Relma, Ym1, and Mgl2, all of which were significantly decreased (relative to IL-4) by 200 μM etomoxir (Fig. 2B, 3G). Moreover, the complex I inhibitors rotenone and piericidin A actually increased expression of Ym1 and Arg 1, but similar changes were not observed with the ATP synthase inhibitor oligomycin (Fig. 3G, S4E). Further separating the effects of etomoxir from respiratory chain activity, Fabp4 expression was unresponsive to etomoxir despite being sensitive to other mitochondrial inhibitors (Fig. 3G, S4E).
Finally, to demonstrate that rotenone-treated BMDMs were not bioenergetically compromised, we estimated rates of ATP synthesis using respirometry and matched enzymatic lactate assays (Divakaruni et al., 2017b). Although complex I inhibition with rotenone predictably limited ATP production from oxidative phosphorylation, IL-4-treated BMDMs sufficiently increased glycolytic ATP production to maintain overall rates of energy metabolism (Fig. 3H). Thus, high concentrations of etomoxir inhibit alternative macrophage activation by a mechanism independent from its off-target effects on mitochondrial ATP synthesis. Moreover, the results show excess etomoxir inhibits M(IL-4) polarization by a mechanism distinct from the off-target effect relevant for blocking T cell activation/differentiation (Raud et al., 2018).
High concentrations of etomoxir disrupt CoA homeostasis
Having eliminated reduced oxidative phosphorylation as the mechanism by which etomoxir inhibits M(IL-4) polarization, we then searched for other off-target effects that could explain its blockade of alternative macrophage activation. Given the well-documented polypharmacology of etomoxir (Ceccarelli et al., 2011), several promiscuous effects of the drug on metabolism have been identified in the literature, including inhibition of fatty acid and cholesterol synthesis (Agius et al., 1991), off-target transcriptional agonism (Rupp et al., 2003), and depletion of cytoplasmic CoA (Agius et al., 1991). We therefore conducted a metabolomics screen for an unbiased analysis of what pathways might be most differentially affected between a low, specific concentration of etomoxir and a high, non-specific concentration during M(IL-4) polarization. Indeed, a principal component analysis (PCA) showed that treatment of IL-4 stimulated macrophages with 200 μM etomoxir caused a profound change in the metabolite composition of BMDMs relative to untreated or IL-4-stimulated macrophages (Fig. 4A). As important controls, the PCA showed not only that IL-4 stimulation created a shift in the metabolite profile of BMDMs relative to untreated cells, but also that there was no appreciable change in IL-4-treated cells upon CPT-1 inhibition with 3 μM etomoxir.
Figure 4. Etomoxir disrupts intracellular CoA homeostasis.
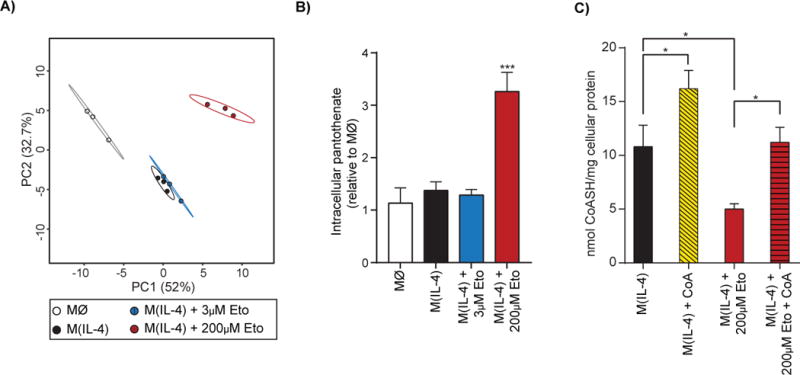
(A) Principal component analysis of untargeted metabolomics data from WT BMDM after 24 hr. IL-4-treatment ± 3 μM or 200 μM etomoxir (co-treated with IL-4; n = 3 technical replicates). The percent variance explained is indicated. Prediction ellipses are drawn such that a new observation from the same group will fall inside the ellipse with 95% certainty.
(B) Intracellular pantothenate levels as measured by LC/MS in conditions as in (A).
(C) Intracellular free coenzyme A (CoASH) measurement in BMDMs after 20 hr treatment with IL-4 ± 200 μM etomoxir and ± 500 μM CoA (all co-treatments with IL-4). The data shown are three independent biological replicates.
See also Supplementary Fig. 5
We then identified which metabolites were most altered by treatment with 200 μM etomoxir. The largest change observed was in pantothenate: excess etomoxir raised intracellular levels by three-fold whereas there was no change among untreated controls, IL-4-stimulated macrophages, and BMDMs treated with IL-4 and 3 μM etomoxir (Fig. 4B; S5A). Pantothenate (also known as vitamin B5) is the precursor for the biosynthesis of coenzyme A, which is an obligatory co-factor central to a broad range of metabolic pathways (Leonardi et al., 2005; Pietrocola et al, 2015). This data, coupled with the fact that the active inhibitor against CPT-1 is the etomoxiryl-CoA thioester (a palmitoyl CoA mimetic generated by long chain acyl CoA synthetase; Ceccarelli et al., 2011), led us to explore whether excess etomoxir caused dysregulated CoA homeostasis.
Specifically, we hypothesized that high concentrations of etomoxir could sequester appreciable amounts of CoA as etomoxiryl-CoA (Agius et al., 1991). Free cytoplasmic CoA levels in mammalian tissues are estimated to range between 20-230 μM, though intramitochondrial concentrations are generally an order of magnitude greater (Idell-Wenger et al., 1978; Williamson and Corkey, 1979). As such, if cytoplasmic levels of free CoA broadly match the concentrations of etomoxir required to block macrophage polarization, then formation of etomoxiryl-CoA from excess etomoxir could deplete free CoA below threshold levels necessary for IL-4-driven responses. As predicted, enzymatic measurements of free CoA showed that 200 μM significantly dropped intracellular CoA levels of IL-4-treated BMDMs (Fig. 4C). This loss of intracellular free CoA, however, could be restored by supplementation of CoA to the medium in these phagocytic cells. Additionally, provision of CoA in the absence of etomoxir significantly increased intracellular levels beyond IL-4-treated controls.
Exogenous CoA restores expression of IL-4-associated markers
To test whether depletion of intracellular CoA is the mechanism by which etomoxir blocks M(IL-4) polarization, we measured IL-4-associated cell surface markers in response to 200 μM etomoxir with or without exogenously added CoA. As predicted, the reduction of CD206+/CD71+ cells upon etomoxir treatment can be rescued by provision of free CoA to the experimental medium (Fig. 5A,B). Importantly, free CoA significantly increased IL-4-induced expression of CD206 and CD71 in the absence of etomoxir, and had no effect on polarization in the absence of IL-4 (Fig. 5B). Indeed, the CD206+/CD71+ population can be adjusted as a function of the [etomoxir]:[CoA] ratio – the rescue of cell surface marker expression in the presence of 200 μM etomoxir is a concentration-dependent function of exogenously added CoA (Fig. 5A,C). Similarly, in the presence of 500 μM CoA, the population of CD206+/CD71+ cells can be proportionally decreased with increasing concentrations of etomoxir (Fig. 5A,D). CoA supplementation also did not alter basal respiration in BMDMs treated with excess etomoxir, further demonstrating that the effects of CoA are independent of the off-target effects of etomoxir on the ANT and complex I (Fig. S5B).
Figure 5. Coenzyme A (CoA) rescues inhibition of M(IL-4) polarization by excess etomoxir.
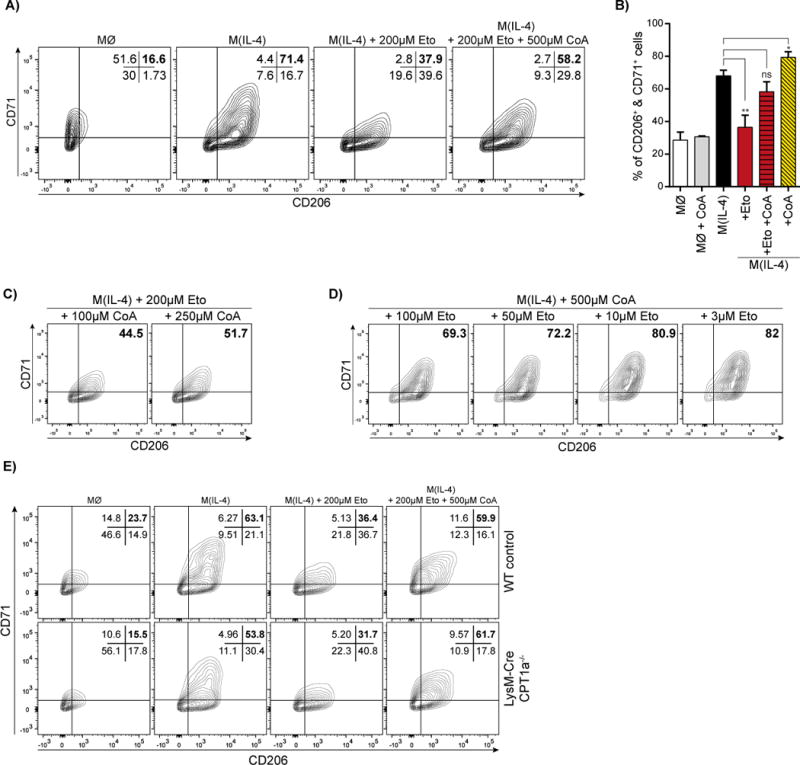
(A) Flow cytometric analysis of CD206+/CD71+ BMDMs after 24 hr treatment with 200 μM etomoxir ± 500 μM CoA. The data shown are from one experiment representative of a total of six independent biological replicates.
(B) Aggregate data of the CD206+/CD71+ population in response to 200 μM etomoxir with or without 500 μM CoA. (n = 6 independent biological replicates)
(C) Flow cytometric analysis of BMDMs showing CD206+/CD71+ in the presence of 200 μM etomoxir and varying the concentration of added CoA. The data shown are from one experiment representative of a total of three independent biological replicates.
(D) Flow cytometric analysis of CD206+/CD71+ BMDMs in the presence of 500 μM CoA and varying the etomoxir concentration between 3-100 μM etomoxir. The data shown are from one experiment representative of a total of three independent biological replicates.
(E) Flow cytometric analysis of the CD206+/CD71+ population in WT and Cpt1a−/− BMDMs in response to IL-4 ± 200 μM etomoxir ± 500 μM CoA. The data shown are from one experiment representative of a total of four independent biological replicates.
See also Supplementary Fig. 5
Finally, to further demonstrate that etomoxir influences macrophage polarization independently of LCFA oxidation, we measured whether CoA supplementation can overcome the effects of excess etomoxir on M(IL-4) polarization in Cpt1a−/− and Cpt2−/− cells. Indeed, the etomoxir-induced reduction in CD206+/CD71+ cells could be rescued by CoA supplementation in all three models with disrupted LCFA oxidation used in this study: LysM-Cre Cpt1a−/− (Fig. 5E), LysM-Cre Cpt2−/−(Fig. S5C), and CD11c-Cre Cpt1a−/− BMDMs (Fig. S5D). The result is consistent with our findings throughout the study demonstrating that high concentrations of etomoxir block alternative macrophage activation independently of CPT-1 and LCFA oxidation.
Discussion
LCFA Oxidation is dispensable for M(IL-4) polarization
Our results demonstrate that LCFA oxidation is not required for IL-4-mediated macrophage M(IL-4) polarization, and help to resolve the controversy surrounding whether CPT-1 activity is obligatory for alternative macrophage activation (Namgaladze and Brüne, 2016; Van den Bossche et al., 2017). Previous work has shown that pharmacologic inhibition of CPT-1 enzymatic activity with etomoxir (Covarrubias et al., 2016; Huang et al., 2014) or viral knockdown of Cpt1a (Huang et al., 2016) attenuated M(IL-4) polarization, suggesting an essential role for the LCFA oxidation and CPT enzymes in this process. Conversely, IL-4-associated gene expression in Cpt2−/− macrophages was found to be indistinguishable from controls both in vitro (Nomura et al., 2016) and in vivo (Gonzalez-Hurtado et al., 2017). These observations raise the possibility that CPT-1 could regulate alternative macrophage activation by a mechanism unrelated to its function of conjugating long-chain fatty acyl CoAs with carnitine (Nomura et al., 2016). Recent evidence even suggests that CPT-1 may have additional physiological functions (Kurmi et al., 2018; Yao et al., 2018). However, genetic deletion of Cpt1a had little impact on macrophage polarization, and loss of CPT-1a did not influence either Treg differentiation or T cell memory formation (Raud et al. 2018). Thus, we conclude that CPT-1a-dependent LCFA oxidation is largely dispensable for immune cell differentiation and polarization, and that the effects of etomoxir are due to the multiple off-target effects described in this manuscript.
Oxidative Phosphorylation is dispensable for M(IL-4) polarization
Surprisingly, in attempting to discover which of the multiple off-target effects of etomoxir was responsible for blocking M(IL-4) polarization, we also determined that oxidative phosphorylation was not required for IL-4-associated expression of genes or cell surface markers. It is unquestioned that oxidative mitochondrial metabolism is closely associated with anti-inflammatory macrophage polarization (Huang et al., 2014; Van den Bossche et al., 2017). In fact, a large body of work links IL-4 stimulation to mitochondrial ATP production, often with a specific focus on aerobic glucose oxidation (Huang et al., 2014; Huang et al., 2016; Tan et al., 2015; Vats et al., 2006; van den Bossche et al., 2016). The ATP synthase inhibitor oligomycin has been frequently used to demonstrate an obligatory role for mitochondrial ATP production in alternative activation (Tan et al., 2015; Vats et al., 2006; van den Bossche et al., 2016). However, the inability of rotenone, piericidin A, or antimycin A (all potent inhibitors of oxidative phosphorylation) to block IL-4-associated genes or cell surface markers suggests our mechanistic understanding of how mitochondrial metabolism regulates M(IL-4) polarization is incomplete. Indeed, we also observe an oligomycin-induced reduction in expression for some IL-4-associated genes including Relma, Fabp4, and, to an extent, Ym1. This suggests that ATP synthase inhibition affects a subset of IL-4-stimulated genes differently than electron transport chain inhibition.
The effect of the complex I inhibitors rotenone and piericidin A on M(IL-4) polarization is therefore noteworthy, particularly given recent studies into the metabolic requirements of pro-inflammatory macrophage polarization. Expression of pro-IL-1β in LPS-stimulated macrophages can be inhibited by rotenone as well as by dissipating the mitochondrial membrane potential (Mills et al., 2016). The results suggest a complex-I-mediated redox signal, perhaps reactive oxygen species from reverse electron transport (Chouchani et al., 2016), is a pro-inflammatory trigger during M(LPS) polarization. As such, it is striking that complex I inhibition can enhance expression of some IL-4-associated genes and cell surface markers. This may suggest that signals similar to those that suppress pro-inflammatory programs might also be used to promote anti-inflammatory programs. Such a model could also explain why oligomycin, but not rotenone, can affect some M(IL-4) markers: both compounds will inhibit mitochondrial ATP production but have opposing effects on the mitochondrial membrane potential (Divakaruni and Brand, 2011).
Etomoxir Inhibits the Adenine Nucleotide Translocase (ANT)
In support of the novel demonstration that etomoxir inhibits the ANT, palmitoyl CoA has also been reported to have similar activity (Pande and Blanchaer, 1971; Shug et al, 1971). A common structural mechanism may therefore underlie ANT inhibition by both compounds since etomoxiryl-CoA inhibits CPT-1 by acting as a mimetic of palmitoyl CoA and the endogenous CPT-1 inhibitor malonyl CoA (Bartlett et al., 1981; Ceccarelli et al., 2011; Declercq et al., 1987; Kiorpes et al. 1984). This mechanism appears to at least partially mediate the block of TH17 and Treg viability and expansion, though other off-target effects may contribute to this phenotype as well (Raud et al, 2018). We also identify a weak effect at complex I, although measurements of membrane potential in intact Treg cells and respiration in permeabilized Treg cells suggest ANT inhibition is the relevant target of excess etomoxir in this system.
Recent work has highlighted off-target effects of excess etomoxir. In tumor cells, etomoxir inhibited cell growth at 200 μM but not 10 μM, and the effect was attributed to complex I inhibition (Yao et al., 2018). However, etomoxir more potently inhibited phosphorylating respiration relative to uncoupler-stimulated respiration in this study as well, suggesting a primary effect at ATP synthesis rather than the respiratory chain. Others using human T cells cultured ex vivo have shown that etomoxir loses specificity to CPT-1 at concentrations >5 μM, depletes reduced glutathione, and increases oxidative stress (O’Connor et al., 2018). The oxidative stress induced by excess etomoxir could result from both ANT inhibition, which increases the mitochondrial membrane potential (Chouchani et al., 2016), as well as by CoA depletion, which could repartition cysteine as a biosynthetic substrate away from glutathione and towards CoA synthesis.
Perturbation of intracellular CoA by etomoxir and the effect of CoA on M(IL-4) polarization
Although LCFA oxidation and oxidative phosphorylation are dispensable for M(IL-4) polarization, we demonstrate an essential role for homeostatic CoA metabolism. We find that use of etomoxir at high concentrations (200 μM) depletes intracellular CoA and results in the accumulation of the CoA precursor pantothenate. The concentrations of etomoxir required to inhibit M(IL-4) polarization match or exceed cytoplasmic concentrations (Idell-Wenger et al., 1978; Williamson and Corkey, 1979), and the drug is readily converted to its active CoA thioester by long chain acyl CoA synthetases (Ceccarelli et al., 2011). Importantly, not only does medium supplementation with exogenous CoA restore polarization in etomoxir-treated cells, but elevating intracellular CoA levels in control BMDMs is also sufficient to significantly increase the population of CD206+/CD71+ cells. This suggests CoA availability may be a limiting and targetable factor in regulating M(IL-4) polarization.
A natural question arising from our findings is how CoA levels facilitate alternative macrophage activation. Cytoplasmic CoA is required for processes as varied as lipid synthesis, vesicular trafficking, fatty acid oxidation and desaturation, and post-translational modification of macromolecules (Leonardi et al., 2005; Pietrocola et al, 2015). Additionally, long chain fatty acyl CoAs are highly compartmentalized, with specific regulatory roles in carbohydrate metabolism, lipid synthesis and degradation, and even ion homeostasis via regulation of potassium channels (Neess et al, 2015). A critical role for long chain acyl CoA metabolism in anti-inflammatory macrophage polarization is supported by studies of fatty acid transport 1 (FATP1), an enzyme with long chain acyl CoA synthase activity involved in lipid trafficking. Knockout of FATP1 caused increased adipose tissue inflammation in vivo, and in vitro overexpression of FATP1 blocks pro-inflammatory macrophage programs (Johnson et al., 2016).
Others have noted that CoA depletion could compromise de novo lipid synthesis and potentially explain the CPT-1-independent mechanism by which etomoxir inhibits fatty acid and cholesterol synthesis (Agius et al., 1991). While these effects could be relevant to macrophages, our data show that M(IL-4) polarization can proceed even in the presence of respiratory chain inhibitors that compromise flux of mitochondrial precursors for de novo fatty acid synthesis. However, a requirement for complex lipids or sterols synthesized in the presence of CoA from cytoplasmic/extracellular precursors is a possible explanation for the importance of CoA levels.
Depletion of CoA could also limit M(IL-4) polarization by depleting CoA esters that are involved in post-translational or epigenetic modification. Sequestration of CoA in excess etomoxiryl-CoA could limit availability of acetyl CoA, which reportedly regulates a subset of IL-4-responsive genes through histone acetylation (Covarrubias et al., 2016). This mechanism would be consistent with the tight regulation of cytoplasmic concentrations of CoA and CoA esters, and aligns well with the finding that nutrient-dependent signaling pathways that adjust acetyl CoA levels are associated with altered epigenetic signatures in BMDMs (Covarrubias et al., 2016). It is worth noting, though, that metabolic regulation of IL-4-dependent transcription is likely a composite effect of multiple factors. For example, Relma expression can be inhibited by both restrictions in histone acetylation (Covarrubias et al., 2016) as well as by inhibition of mitochondrial ATP synthesis with oligomycin (Fig. 3G; Van den Bossche, 2016). To that end, the strong inhibition of Relma by excess etomoxir may be a compounded effect from both CoA depletion as well as ANT inhibition.
In conclusion, our results along with those of the companion manuscript (Raud et al., 2018) demonstrate that LCFA oxidation and CPT-1 are dispensable for many types of immune cell function. Although high concentrations of etomoxir can inhibit M(IL-4) polarization as well as Treg and Tmem formation, different off-target effects from excess etomoxir drive the observed phenotypes in macrophages (CoA depletion) versus T cells (ANT inhibition). Together, these studies highlight the well-established polypharmacology of etomoxir (Ceccarelli et al., 2011), and reinforce the need for caution when interpreting effects of metabolic inhibitors in the absence of established specificity.
Limitations of Study
The present study examined the role of LCFA oxidation in alternative macrophage activation and determined that etomoxir inhibited M(IL-4) by depletion of intracellular CoA. Some limitations are as follows:
One limitation stems from the lack of a well-defined in vitro functional assay to characterize M(IL-4) in BMDMs. We characterized macrophage polarization with multiple IL-4-induced genes and cell surface markers, as well as an arginase activity assay. However, this stands in contrast to the variety of functional assays for LPS-stimulated macrophages, such as NO generation or release of various cytokines. Assays for the nuanced behavior of alternatively activated macrophages that parallel in vivo responses would benefit the study.
We show that high etomoxir depletes cells of free CoA but we did not measure levels of etomoxiryl CoA. Studies explicitly measuring the effects of etomoxiryl CoA would also help determine whether etomoxir or its CoA derivative is the active inhibitory molecule of the ANT and complex I.
Our experiments enhancing the intracellular CoA pool by supplementing medium with free CoA are likely not applicable to a broad range of cell types, and might only be effective in phagocytic cells.
Our findings do not address a potential role in alternative macrophage activation for peroxisomal β-oxidation, which requires neither CPT-1 activity (Visser et al., 2007) nor mitochondrial oxidation.”
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ajit Divakaruni (adivakaruni@mednet.ucla.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All animal protocols were approved by either the UCSD Institutional Animal Care and Use Committee or the UCLA Animal Research Committee.
Mouse bone marrow-derived (BMDM) and peritoneal macrophages
WT
Bone marrow cells from WT control, Cpt1−/−, and Cpt2−/− were isolated from femurs of male mice. Cells were treated with RBC lysis buffer to remove red blood cells for five minutes, centrifuged at 386g for 5 minutes, and resuspended in BMDM culture medium. Culture medium consisted of high-glucose DMEM (Gibco 11965) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM L-glutamine, 100 units/mL, 100 μg/mL penicillin/streptomycin, 500 μM sodium pyruvate, and 5% v/v conditioned medium containing macrophage colony stimulating factor (M-CSF) produced by CMG cells to induce differentiation to BMDMs (Takeshita et al., 2000). BMDMs were maintained at 37°C in a humidified 5% CO2 incubator. BMDMs were differentiated for 6 days prior to harvesting and re-plating for experiments, and medium was changed at Day 4 of differentiation.
Cpt−/−
CD11c-Cre Cpt1a−/− BMDMs were generated from crossing Cpt1a flox/flox mice [generously provided by Dr. Peter Carmeliet (Schoors et al., 2015)] with Itgax-cre (CD11c-cre) mice (Caton et al., 2007) and maintained on a C57BL/6 genetic background. LysM-Cre Cpt1a−/− macrophages were generated from Cpt1a flox/flox mice with LysM-Cre mice and maintained on a C57BL/6 genetic background (Clausen et al., 1999). Mice were bred under specific pathogen-free conditions at the animal facility of the Helmholtz Centre for Infection Research (HZI, Braunschweig, Germany) or at TWINCORE (Hannover, Germany). WT control and CPT1a−/− BMDMs were isolated and differentiated as described above. Cpt2−/− BMDMs were generated as previously described (Gonzalez-Hurtado et al., 2017; Nomura et al., 2016).
Peritoneal macrophages
Male mice were euthanized with CO2 and abdominal skin was retracted to expose the intact peritoneal wall. 5 mL of ice-cold PBS with 2 mM EDTA and 2% FCS (Biochrom) were injected into the peritoneal cavity using a syringe with a 20-G needle. The fluid was then aspirated from the peritoneal cavity using the same syringe and collected in a 15 mL tube, and the procedure was repeated twice to obtain a final volume of 10 mL. The cell suspension was centrifuged for 7 min at 400 g at 4°C, and pellets were incubated with anti-CD16/CD32 (homegrown) for 5 min on ice to prevent non-specific antibody binding. A staining cocktail containing the following antibodies was then added: anti-F4/80 eF660, anti CD11b-FITC, anti-CD19 eF450, anti NK1.1 eF450, anti CD3 eF450. After 20 min of incubation on ice, cells were washed and stained with DAPI for dead cell exclusion. Cells were sorted at the MHH cell sorting facility using a MoFlo XDP (Beckman-Coulter) with a 100μm nozzle. Peritoneal macrophages were gated as DAPI− Lin (CD3, CD19, NK1.1)−, F4/80+, CD11b+.
Rat primary cell cultures
Rat cortical neurons (Kushnareva et al., 2005) and cortical astrocytes (Kim and Magrane, 2011) were prepared according to established methods. Primary cultures of cortical neurons were prepared from embryonic day 18 (E18) Sprague Dawley rats without determination of sex. The cerebral cortices were collected and triturated gently (3-4 times) in ice-cold Hibernate E (Gibco) medium with 1X B27 supplement (Invitrogen), 100 U/ml penicillin, and 100 μg/ml streptomycin. After the tissue settled, the Hibernate E medium was aspirated and the tissue was triturated for approximately 2 min in trypsin [0.1% (w/v); Sigma-Aldrich] in Ca2+/Mg2+-free phosphate-buffered saline solution (PBS) supplemented with glucose (5 mM). After these 2 min, trypsin was inactivated by addition of soybean trypsin inhibitor (0.1 mg/mL; Sigma-Aldrich). The mixture was transferred into Hibernate E medium containing 20 U/mL DNAse (Promega) in 0.2X reaction buffer (Promega) and the cells were centrifuged at 200 g for 1.5 min. The supernatant was quickly aspirated and the cells were resuspended in 10 mL Neurobasal (E) medium (Invitrogen) plus glutamate (0.4 μg/mL), 0.5 mM L-glutaMAX, penicillin (100 U/mL), streptomycin (100 μg/mL), 1X B27 supplement, and 5 mM sodium pyruvate. Once in suspension, the cells were diluted into 30-45 mL of the same medium without pyruvate (plating medium) and the number of viable cells was determined by trypan blue exclusion using a Countess Automated Cell Counter (Thermo Fisher). Cells were plated on poly-D-lysine coated Agilent Seahorse XF96 plates at a concentration of 3 × 104 cells in a volume of 100 μL per per well. Cells were maintained at 37°C in a 5% CO2 incubator. After 4 days in vitro, the initial plating medium was diluted with an equal volume of maintenance medium of the same composition lacking glutamate and supplemented with 2mM GlutaMAX (Thermo Fisher). Half the medium was replaced every 3–4 days. All experiments were performed with cultures that were 13-15 days in vitro.
For primary cortical astrocytes, cortices were collected from E18 Sprague Dawley rat embryos without determination of sex. Cortical pieces were then pushed through a Corning 100 m cell strainer into a sterile 50 mL centrifuge tube using a sterile glass rod, and the strainer was washed with 8 mL of glial culture medium [DMEM supplemented with 10% FBS, 2mM GlutaMAX, penicillin (100 U/mL), and streptomycin (100 μg/mL)]. This suspension was then filtered through a Corning 70 m cell strainer. After centrifugation at 800g, the supernatant was aspirated, and the cells were resuspended with glia culture media, counted, and seeded in T-75 culture flasks at 3 × 105 cells/cm2. Medium was replaced the following day, and after 7-9 days the cultures were washed, the solid caps were tightened, and the flasks were shaken in an orbital shaker at 250 rpm at 37°C overnight. Oligodendrocytes were then removed by hand-shaking the flasks followed by five washes of the monolayers. Cells were then trypsinized, counted, and plated in glial culture medium in Agilent Seahorse XF96 plates and kept at 37°C in a 5% CO2 incubator for 24-48 hr prior to experiments.
Neonatal rat ventricular myocytes (NRVMs) were isolated according to established methods (Rubio et al., 2009). Briefly, NRVMs were isolated from Sprague-Dawley rat hearts (2-3 days old; mixed gender) by several rounds of digestion with collagenase type I (Worthington) and pancreatine (Sigma). Cell suspensions were pre-plated for 2 hours on 10 cm2 dishes in DMEM supplemented with 15% (v/v) FBS, 2 mM GlutaMAX, 100 U/ml penicillin, and 100 μg/ml streptomycin to reduce fibroblast contamination. Non-adherent cells were harvested, centrifuged (400 g for 3 min), and plated in gelatin-coated Seahorse XF96 plates in DMEM with 15% (v/v) FBS, 2 mM GlutaMAX, 100 U/ml penicillin, and 100 μg/ml streptomycin. After 24 hr., serum-containing medium was replaced with DMEM lacking FBS but supplemented with GlutaMAX, penicillin, and streptomycin and kept 24-48 hrs. before experiments were conducted. Cells were maintained at 37°C in a 5% CO2 incubator.
Cultured cell lines
All cultured cells were obtained from American Type Culture Collection (ATCC) and cultured as suggested by the supplier. Cell lines were maintained in the medium listed parenthetically: HepG2 [MEM (Gibco 11095)], A549 [DMEM/F12 (Gibco 11330)], and HCT116 [DMEM (Gibco 11965)]. Each formulation of medium was supplemented with 10% (v/v) fetal bovine serum (FBS), 2 mM GlutaMAX, 100 U/mL penicillin, and 100 μg/mL streptomycin. Cells were maintained at 37°C in a humidified 5% CO2 incubator.
METHOD DETAILS
Mitochondrial Isolation
Mouse liver mitochondria were isolated according to standard techniques (Rogers et al., 2011). The liver from a male mouse was minced and rinsed several times in MSHE [70 mM sucrose, 210 mM mannitol, 5 mM HEPES (pH 7.2), and 1 mM EGTA with 0.5% (w/v) fatty acid-free BSA] to remove blood. Tissue was disrupted using a drill-driven Teflon-on-glass Dounce homogenizer (between 2-4 strokes). The homogenate was centrifuged at 800g for 10 min and the supernatant was filtered through cheesecloth and centrifuged at 8000g for 10 min. The light layer from the pellet was removed and the remaining pellet was resuspended, centrifuged again, and resuspended in a minimal volume of MSHE.
Respirometry
All respirometry studies were conducted in either a Seahorse XF96 or XFe96 Analyzer. All experiments were conducted at 37°C, and at pH 7.4 (intact cells) or 7.2 (permeabilized cells and isolated mitochondria). Calculation of respiratory parameters was made according to standard protocols (Divakaruni et al., 2014a), and all rates were corrected for non-mitochondrial respiration/background signal by subtracting the oxygen consumption rate insensitive to rotenone plus antimycin A.
Intact Cells
Palmitate oxidation in intact cells (Fig. 1A,B) was assessed by measuring palmitate-driven respiration that was sensitive to low concentrations of etomoxir (Rogers et al., 2014). Cells were plated at either 3×104 cells/well (HepG2) or 1.5×104 cells/well (A549) for 48 hr. prior to the assay. 24 hr after plating, standard maintenance medium was replaced with a substrate-limited medium to deplete endogenous nutrient stores and prime cells for fatty acid oxidation. This substrate-limited medium was DMEM (Gibco A14430) supplemented with 0.5 mM glucose, 1% (v/v) FBS, 0.5 mM carnitine, 1 mM GlutaMAX, 100 U/mL penicillin, and 100 μg/mL streptomycin. On the day of the assay, the substrate-limited medium was replaced with assay medium composed of unbuffered DMEM (Sigma #D5030) supplemented with 0.2% (v/v) BSA-conjugated palmitate ([FFA ≥ 50 nM]; Seahorse XF Palmitate-BSA FAO Substrate, Agilent Technologies), 2.5 mM glucose, 0.5 mM carnitine, and 2 mM HEPES. Respiration was measured under basal conditions as well as after injection of 2 μM oligomycin, two sequential additions of 1 μM FCCP, followed by 0.2 μM rotenone with 1 μM antimycin A. Etomoxir was added to the assay medium 30 min. prior to measurements of basal respiratory rates.
Respiration in intact BMDMs was measured in medium containing 8 mM glucose, 2 mM glutamine, 2 mM pyruvate, and 5 mM HEPES. Cells were plated at 5.0×104 cells/well and assayed after 48 hr. When measuring the response to IL-4, cells were treated with 20 ng/mL of IL-4 24 hr. after initial plating, and respiration was measured the following day (~48 hr after initial seeding). Respiration was measured in response to oligomycin (1 μM), FCCP (1.2 μM), and rotenone (0.2 μM) with antimycin A (1 μM).
When diagnosing an off-target effect of etomoxir (Fig. S2A-C), all cells apart from primary neurons were offered 8 mM glucose, 2 mM glutamine, and 2 mM pyruvate in unbuffered DMEM supplemented with 5 mM HEPES. Cells were plated at the following densities: A549 (1.2×104 cells/well for 48 hr), HCT116 (1.5×104 cells/well for 48 hr.), cortical neurons (3.0×104 cells/well and assayed at day in vitro 13-15), cortical astrocytes (plated at 2.0×104 cells/well for 24 or 48 hr.), BMDM (plated at 5.0×104 cells/well for 48 hr), and NRVM (plated at 5.0×104 cells/well for 48 hr.). Primary neurons were assayed in artificial cerebrospinal fluid [aCSF; 120 mM NaCl, 3.5 mM KCl, 1.3 mM CaCl2, 0.4 mM KH2PO4, 1 mM MgCl2, 5 mM HEPES (pH 7.4)] and offered 10 mM glucose and 1 mM pyruvate as respiratory substrates. Respiration in all intact cells was measured in the basal state as well as in response to 2 μM oligomycin, FCCP (either 400 nM, 800 nM or 1.2 μM as appropriate and previously optimized), and 0.2 μM rotenone with 1 μM antimycin A. Etomoxir (at concentrations indicated in the text), UK5099 (3 μM), or BPTES (3 μM) were added to the assay medium 30 min prior to measurements.
Permeabilized Cells
The plasma membrane of cells was selectively permeabilized with recombinant, mutant perfringolysin O [rPFO; commercially XF Plasma Membrane Permeabilizer (XF PMP, Agilent Technologies, Divakaruni et al, 2013]. Experiments were conducted essentially as previously described except medium lacked BSA to align with intact cell experiments and avoid binding of etomoxir by BSA (Divakaruni et al, 2014b). Immediately prior to assay, cell media was replaced with MAS buffer (70 mM sucrose, 220 mM mannitol, 10 mM KH2PO4, 5 mM MgCl2, 2 mM Hepes, and 1 mM EGTA; pH 7.2) containing 3 nM rPFO, respiratory substrates, and 4 mM ADP. The ADP-stimulated respiration rate (referred to interchangeably as ‘phosphorylating’ or ‘State 3’ respiration) was measured, and rates were subsequently measured in response to 2 μM oligomycin, sequential additions of 400 nM FCCP, and 0.2 μM rotenone with 1 μM antimycin A. Substrate concentrations were as follows: Glu, 5 mM glutamate with 5 mM malate; Pyr, 10 mM pyruvate with 1 mM malate; PCoA, 40 μM palmitoyl CoA with 1 mM malate and 0.5 mM carnitine; Pcarn, 40 μM palmitoylcarnitine with 1 mM malate; Gln 5 mM glutamine (no malate was necessary for glutamine oxidation in permeabilized A549 cells); Succ, 10 mM succinate with 2 μM rotenone.
When assessing the effects of etomoxir on specific oxidative pathways in permeabilized cells (Fig. 1F), cells were pre-treated for 30 min in intact cell assay medium prior to permeabilization to allow formation of etomoxiryl-CoA, the active inhibitor of CPT-1. Etomoxiryl-CoA is an irreversible inhibitor of CPT-1 (Kiorpes et al, 1984), so the inhibition of the enzyme persists through medium changes and cell permeabilization. To measure the off-target effect of etomoxir in isolated mitochondria and permeabilized cells (Fig. 3B; Fig. S3A,B), etomoxir was added directly to the MAS medium 15 min prior to initial measurements.
When permeabilized cells were treated with alamethicin to form pores of 3-6 kDa in the mitochondrial inner membrane (“double-permeabilized” cells) and complex I-mediated respiration was directly assessed (Fig. S3F), 10 μg/mL alamethicin was added at 37°C 15 minutes prior to measurements (Divakaruni et al., 2017a). Double-permeabilized cells were offered 10 μM cytochrome c in the experimental medium and either 10 mM NADH or 10 mM succinate with 2 μM rotenone to drive respiration.
Isolated Mitochondria
Phosphorylating respiration was measured in isolated liver mitochondria (1.5 μg/well) as previously described (Rogers et al., 2011), and offered 10 mM pyruvate, 1 mM malate, and 4 mM ADP. Uncoupler stimulated respiration was measured after addition of oligomycin (3 ng/mg mitochondrial protein) and FCCP (4 μM). All rates were corrected for background by subtracting the oxygen consumption rate insensensitive to 0.5 μM rotenone and 1 μM antimycin A. Etomoxir was acutely added to the experimental medium 15 minutes prior to taking initial measurements.
ATP hydrolysis
ATP hydrolysis in isolated mitochondria (McQuaker et al., 2013; Divakaruni et al., 2017a) was performed in MAS buffer, and stimulated by respiratory chain inhibition (1 μM rotenone and 2 μM antimycin A) followed by addition of excess ATP (25 mM) and uncoupler (1 μM FCCP). Hydrolysis was inhibited by injection of either oligomycin (3 ng/mg mitochondrial protein) or carboxyatractyloside (7.5 ng/mg mitochondrial protein). Rates of extracellular acidification (ECAR, in mpH/min) were converted to proton production rates (PPR, in pmol H+/min) by determining the buffer capacity with sequential injections of HCl and H2SO4 (Mookerjee et al., 2015). 10 μg/mL alamethicin was used to permeabilize the mitochondrial inner membrane (1 μg/mg mitochondrial protein).
Western analysis
Cells were lysed in Pierce RIPA buffer (Thermo Scientific) supplemented with complete EASYpack Mini Protease Inhibitor Cocktail and PhosSTOP Phosphatase Inhibitor (Roche). Cell lysates were resolved by SDS-PAGE and transferred to PVDF membranes (Merck Millipore). Membranes were incubated with anti-CPT1A (clone 8F6AE9, Abcam) and anti-β-Actin (clone AC-15, Sigma-Aldrich).
9,10-3H-palmitate oxidation
BMDMs at day 6 of differentiation were seeded at 3×105 cells/well in 12 well plates in BMDM media. The next day, cells were treated in DMEM containing 25mM glucose supplemented with 5% FBS, 2 mM L-Glutamax, 100 units/mL, 100 μg/mL penicillin/streptomycin, 1 mM of sodium pyruvate, 1 mM carnitine, and 5% v/v conditioned media containing macrophage colony stimulating factor (M-CSF) under one of 5 conditions: control, 20 ng/mL of IL-4 ± 3 μM or 200 μM of etomoxir, or 200 nM rotenone, or 200nM antimycin A for 24 hr. Fatty acid oxidation was measured 24 hours later as previously described (Rognstad et al, 1991; Cha et al, 2005) following a fluid change to DMEM with components as described above except 10mM glucose and 0.2 μCi (final concentration of 5 μM) BSA-conjugated [9,10-3H]palmitic acid. No additional IL-4 was added at the time of this fluid change, but etomoxir, rotenone and antimycin A were added, and the plates were placed in a 95% O2-5% CO2 incubator at 37°C for 3 hr. After incubation, a 100 μL aliquot of the culture medium was passed over an ion-exchange resin, and the column was washed twice with 0.75 ml of water. Free fatty acids were retained by the resin, whereas the tritium hydrolyzed from radiolabeled palmitate catabolism was released as labeled water, which passes freely through the resin, and is subsequently counted for radioactivity. All data are corrected for protein content in each well. There were 3 technical replicates for each condition, and 4 biological replicates arising from 4 separate animals.
Intracellular free Coenzyme A levels
On Day 6 of differentiation, BMDMs were seeded at 1.2×106 cells/well in 6-well plates. 24 hr later, BMDMs were mock-treated or co-treated with 20 ng/mL IL-4 ± 200 μM etomoxir ± 500 μM CoA in DMEM supplemented with 5% FBS, 2 mM L-glutamine, 100 units/mL, 100 μg/mL penicillin/streptomycin, 500 μM of sodium pyruvate and 5% v/v conditioned media containing macrophage colony stimulating factor (M-CSF) for 20 hr. Prior to sample collections, cells were washed with 600 μL of 0.9% (w/v) NaCl twice. Three wells per replicate per treatment were combined in 2 mL centrifuge tubes and centrifuged at 500g for 5 mins at 4°C. The supernatant was removed, and the cell pellet was re-suspended in 75 μL of ice-cold 0.7% (v/v) perchloric acid for 5 min. Samples were then centrifuged at 10,000g for 7 mins at 4°C and the resulting supernatant wa s adjusted to pH 7 with 2M NaOH and 500 mM K2HPO4. The coenzyme A concentration was determined with Coenzyme A Assay Kit following manufacturer’s instructions (Sigma MAK034) using 40 μL of sample supernatant. Coenzyme A levels were normalized to total cellular protein (BCA method).
Arginase activity
On Day 6 of differentiation, BMDMs were seeded at 1×106 cells/well in 6-well plates. 24 hr later, BMDMs were mock-treated or co-treated with 20 ng/mL IL-4 ± 200 μM etomoxir in DMEM supplemented with 5% FBS, 2 mM L-glutamine, 100 units/mL, 100 μg/mL penicillin/streptomycin, 500 μM of sodium pyruvate and 5% v/v conditioned media containing macrophage colony stimulating factor (M-CSF) for 24 hr. Prior to sample collections, cells were washed with 600 μL of ice-cold PBS twice and scraped into 1.5 mL centrifuge tubes. Cell lysate was prepared according to Arginase Activity Assay Kit (Sigma MAK112) instructions and the 20 μL of lysate was used to measure arginase activity. Arginase activity was adjusted to total cell number.
Lactate assay
Under conditions matching intact cell respirometry for BMDMs, the extracellular medium was harvested and lactate was analyzed as previously described (Mookerjee et al., 2015; Divakaruni et al., 2017b). Briefly, the harvested medium was mixed 1:1 with a solution of 40 U/ml LDH (Sigma-Aldrich L3916), 1 μM Tris, pH 9.8, 20 mM EDTA, 400 mM hydrazine (Sigma-Aldrich 309400), and 4 mM NAD+. The reaction velocity was measured after 2 min (340/460 nm excitation/emission). Values were calibrated against known lactate concentrations (L7022; Sigma-Aldrich).
Cell counts and normalization
For BMDM respirometry experiments normalized to cell number, cells were fixed immediately upon completion of the assay with 4% formaldehyde for 20 min at room temperature and kept refrigerated between 1 and 21 days until assessment. The day prior to cell counting, cells were stained with Hoescht 33342 (Thermo Fisher) at 10 ng/mL overnight at 4°C. Cell counts were obtained using the Operetta High Content Imaging System (Perkin Elmer).
Phosphate transporter assay
Phosphate transport was measured by monitoring swelling of de-energized mitochondria in isotonic potassium phosphate medium based upon principles described by Coty and Pedersen (1975). As the phosphate carrier acts as electroneutral symporter with protons, the process is self-restricting due to generation of an opposing pH gradient. The restriction is relieved, and the full rate of phosphate transport revealed with addition of the chemical potassium/proton antiporter, nigericin. In the presence of nigericin, the proton fluxes cycle resulting in electro- and pH-neutral transport of potassium phosphate down its concentration gradient into the mitochondrial matrix, resulting in osmotic accumulation of water. Under these conditions, transport is limited by the activity of phosphate carrier, and can be recorded as mitochondrial swelling (a decrease in light scattering).
The measurements were performed in an LS-50B spectrofluorometer (Perkin-Elmer) at 540 nm. Assay medium contained 125 mM potassium phosphate, pH 7.2, 0.5 mM EGTA, and 2 μM rotenone. Mouse liver mitochondria (0.5 mg/ml) were treated with 250 μM etomoxir, 100 μM N-ethylmaleimide (NEM) (Klingenberg et al 1974) or 10 μM mersalyl (Paradies and Ruggiero, 1991) and the reaction was started with 5 μM nigericin. Typical traces from 3 experiments are shown. To establish the maximally swollen state of mitochondria, 80 μg/ml alamethicin was used.
Mitochondrial effector treatments for qPCR and flow cytometry
For all gene expression and flow cytometry experiments described below, day 6 BMDM were plated at 3×105 cell/well in 12-well plates. The next day, cells were treated with appropriate compounds (and duration as indicated in figures and below) in DMEM supplemented with 5% FBS, 2 mM L-glutamine, 100 units/mL, 100 μg/mL penicillin/streptomycin, 500 μM of sodium pyruvate and 5% v/v conditioned media containing macrophage colony stimulating factor (M-CSF).
For flow cytometry experiments involving etomoxir (Figs. 1G, 2A, 2D, S1A, S1B, S1G, S1H): BMDMs were mock-treated (media replacement only) or treated with 20 ng/mL of IL-4 ± 3 μM or 200 μM of etomoxir for 48 hr. When measuring expression of M(IL-4)-associated genes in response to etomoxir (Figs. 1H, 2B), BMDMs were mock-treated or treated with 20 ng/mL of IL4 ± etomoxir as indicated for 24 hr.
For experiments involving respiratory chain inhibitors: BMDMs were mock-treated (media replacement only) or treated with 20 ng/mL of IL4 ± 1.2 μM oligomycin (Oligo), 200 nM rotenone (Rot), 200 nM antimycin A (AntiA), 100 nM piericidin A (PierA), or 5 μM carboxyatractylate (CAT) for either 24 (Figs. 3G, 3H, S3G) or 48 hr (Figs. 3F, S4D). All inhibitors were given as co-treatments simultaneously with IL-4.
For experiments involving CoA supplementation (Figs. 5A-E, S5C-D): BMDMs were mock-treated (media replacement only) or treated with 20 ng/mL of IL4 ± etomoxir (concentration indicated in figures) ± CoA (concentration indicated in figures) for 48 hr. Etomoxir and CoA were given as co-treatments simultaneously with IL-4.
RNA Isolation and qPCR analysis for BMDM
RNA was isolated with TRIzol, then cDNA was synthesized with high-capacity cDNA reverse transcription kit. KAPA SYBR FAST qPCR Master Mix kit and a LightCycler 480 were used for quantitative RT-PCR. Fold change related to the control group was calculated using 2ΔΔCP method with 36b4 as the reference gene.
FACS analysis of BMDM polarization
Prior to FACS analysis, BMDMs were detached using 350 μL of Accutase, followed by two washes with FACS buffer (PBS + 1%(v/v) BSA + 0.05%(w/v) sodium azide). Anti-mouse CD16/32 antibody was used at a dilution of 1:500, while all other antibodies were used at a dilution of 1:250. All flow cytometry data were captured using BD FACSVerse flow cytometer and analyzed using the FlowJo X 10.0.7r2 software. In all, 2×104 live events were acquired for each sample (gated on DAPIneg). For analysis, CD206+/CD71+ and CD206+/CD301+ populations were gated based on Fluorescence Minus One (FMO) controls.
Mass spectrometry-based metabolomics analysis
The experiments were performed as described in (Xiao et al., 2018). Briefly, cells were seeded in 6-well dishes at 1×106 cells/well. Cells were treated with IL-4 with and without etomoxir co-treatments (3 μM or 200 μM) similarly to other experiments. To extract intracellular metabolites, cells were briefly rinsed with cold 150 mM ammonium acetate (pH 7.3), followed by addition of 1 ml cold 80% MeOH on dry ice. Cells were detached with cell scrapers and suspensions transferred into centrifuge tubes. After rigorous mixing, the suspension was pelleted by centrifugation (1.3×104 rpm, 4°C). The supernatant was transferred into a glass vial, metabolites were dried down under vacuum, and resuspended in 50% (v/v) acetonitrile including heavy isotope-labeled amino acids as internal standards (Chen et al., 2018).
For the mass spectrometry-based analysis of the sample, 5 μL were injected onto a Luna NH2 (150 mm × 2 mm, Phenomenex) column. The samples were analyzed with an UltiMate 3000RSLC (Thermo Scientific) coupled to a Q Exactive mass spectrometer (Thermo Scientific). The Q Exactive was run with polarity switching (+3.50 kV/−3.50 kV) in full scan mode with an m/z range of 65-975. Separation was achieved using A) 5 mM NH4AcO (pH 9.9) and B) ACN. The gradient started with 15% A) going to 90% A) over 18 min, followed by an isocratic step for 9 min and reversal to the initial 15% A) for 7 min. Metabolites were quantified with El Maven (v0.2.4) using accurate mass measurements (≤ 3 ppm) and retention times of pure standards. Data analysis was performed using the statistical language R. For the PCA and clustering analyses, only metabolites with an ANOVA p-value ≤ 0.05 were incorporated. For visualization using heatmaps, metabolites were scaled across samples but not across metabolites. To assess what metabolites contributed the most to a given metabolic phenotype, we calculated the correlation between the loading vector for a given metabolite and the score vector of a given sample for PCs 1 and 2.
Quantification and statistical analysis
All statistical parameters, including the number of replicates (n), can be found in the figure legends. Statistical analyses were performed using Graph Pad Prism 6 software. Data are presented as the mean ± SEM. Individual pairwise comparisons were performed using two-tailed Student’s t-test. For experiments involving two or more factors, data were analyzed by one-way, repeated measures ANOVA followed by Dunnett post-hoc multiple comparisons tests. Data were assumed to follow a normal distribution (no tests were performed). Values denoted as follows were considered significant: *,p < 0.05; **, p <0.01; ***, p<0.001; ****, p<0.0001.
Supplementary Material
Highlights.
Alternative macrophage activation by IL-4 does not require fatty acid oxidation
Etomoxir concentrations ≥10 μM inhibit the adenine nucleotide translocase
OXPHOS is dispensable for many markers of IL-4 activated macrophages [M(IL-4)]
200 μM etomoxir blocks M(IL-4) by perturbing intracellular coenzyme A homeostasis
Acknowledgments
This work was supported by U.S. National Institutes of Health (R01NS087611 to A.N.M., R01AI122282 to S.J.B., R01NS072241 to M.J.W.), American Heart Association Grant-in-Aid to S.J.B, and Agilent Technologies (to A.S.D. and A.N.M). L.B. was funded by HiLF and the Ellen-Schmidt Program from the Medical School Hannover and L.M. by a PhD fellowship from the Boehringer Ingelheim Fonds, Foundation for Basic Research in Medicine. This material is the result of work supported with resources and the use of facilities at the VA San Diego Medical Center. The contents do not represent the views of the U.S. Department of Veterans Affairs or the United States Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
Conceptualization, A.S.D., W.Y.H., S.J.B., and A.N.M.; Investigation, A.S.D., W.Y.H., L.M., T.N.D., K.K.O.K., B.R.D., G.W.R., A.Y.A., K.C., M.L., L.S., D.B., T.P.C., and A.N.M.; Resources, C.E.B., B.P.D., D.A.F., M.J.W., T.S., and L.B.; Writing and Visualization, A.S.D., W.Y.H., S.J.B., and A.N.M.; Supervision and Project Administration, A.S.D., S.J.B., and A.N.M.; Funding Acquisition, A.S.D., S.J.B., and A.N.M.
Declaration of Interests
K.C., B.P.D., D.A.F., and G.W.R. are employees and shareholders of Agilent Technologies.
References
- Agius L, Meredith EJ, Sherratt H, Stanley A. Stereospecificity of the inhibition by etomoxir of fatty acid and cholesterol synthesis in isolated rat hepatocytes. Biochem Pharmacol. 1991;42:1717–1720. doi: 10.1016/0006-2952(91)90507-2. [DOI] [PubMed] [Google Scholar]
- Bartlett K, Bone AJ, Koundakjian P, Meredith E, Turnbull DM, Sherratt HS. Inhibition of mitochondrial β-oxidation at the stage of carnitine palmitoyltransferase I by the coenzyme A esters of some substituted hypoglycaemic oxirane-carboxylic acids. Biochem Soc Trans. 1981;9:574–575. [Google Scholar]
- Bentebibel A, Sebastian D, Herrero L, Lopez-Vinas E, Serra D, Asins G, Gomez-Puertas P, Hegardt FG. Novel effect of C75 on carnitine palmitoyltransferase I activity and palmitate oxidation. Biochemistry. 2006;45:4339–4350. doi: 10.1021/bi052186q. [DOI] [PubMed] [Google Scholar]
- Caton ML, Smith-Raska MR, Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8-dendritic cells in the spleen. J Exp Med. 2007;204:1653–64. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli SM, Chomienne O, Gubler M, Arduini A. Carnitine Palmitoyltransferase (CPT) Modulators: A Medicinal Chemistry Perspective on 35 Years of Research. J Med Chem. 2011;54:3109–3152. doi: 10.1021/jm100809g. [DOI] [PubMed] [Google Scholar]
- Cha BS, Ciaraldi TP, Park KS, Carter L, Mudaliar SR, Henry RR. Impaired fatty acid metabolism in type 2 diabetic skeletal muscle cells is reversed by PPARgamma agonists. Am J Physiol Endocrinol Metab. 2005;289:E151–9. doi: 10.1152/ajpendo.00141.2004. [DOI] [PubMed] [Google Scholar]
- Chen WW, Freinkman E, Sabatini DM. Rapid immunopurification of mitochondria for metabolite profiling and absolute quantification of matrix metabolites. Nat Protoc. 2017;12:2215–2231. doi: 10.1038/nprot.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouchani ET, Pell VR, James AM, Work LM, Saeb-Parsy K, Frezza C, Krieg T, Murphy MP. A Unifying Mechanism for Mitochondrial Superoxide Production during Ischemia-Reperfusion Injury. Cell Metab. 2016;23:254–63. doi: 10.1016/j.cmet.2015.12.009. [DOI] [PubMed] [Google Scholar]
- Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–77. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- Coty WA, Pedersen PL. Phosphate transport in rat liver mitochondria. Kinetics, inhibitor sensitivity, energy requirements, and labeled components. Mol Cell Biochem. 1975;9:109–24. doi: 10.1007/BF01732202. [DOI] [PubMed] [Google Scholar]
- Covarrubias AJ, Aksoylar HI, Yu J, Snyder NW, Worth AJ, Iyer SS, Wang J, Ben-Sahra I, Byles V, Polynne-Stapornkul T, et al. Akt-mTORC1 signaling regulates Acly to integrate metabolic input to control of macrophage activation. Elife. 2016;19:e11612. doi: 10.7554/eLife.11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Declercq PE, Falck JR, Kuwajima M, Tyminski H, Foster DW, McGarry JD. Characterization of the mitochondrial carnitine palmitoyltransferase enzyme system: use of inhibitors. J Biol Chem. 1987;262:9812–9821. [PubMed] [Google Scholar]
- Divakaruni AS, Brand MD. The regulation and physiology of mitochondrial proton leak. Physiology. 2011;26:192–205. doi: 10.1152/physiol.00046.2010. [DOI] [PubMed] [Google Scholar]
- Divakaruni AS, Wiley SE, Rogers GW, Andreyev AY, Petrosyan S, Loviscach M, Wall EA, Yadava N, Heuck AP, Ferrick DA, et al. Thiazolidinediones are acute, specific inhibitors of the mitochondrial pyruvate carrier. Proc Natl Acad Sci USA. 2013;110:5422–7. doi: 10.1073/pnas.1303360110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divakaruni AS, Paradyse A, Ferrick DA, Murphy AN, Jastroch M. Analysis and interpretation of microplate-based oxygen consumption and pH data. Meth Enzym. 2014a;547:509–54. doi: 10.1016/B978-0-12-801415-8.00016-3. [DOI] [PubMed] [Google Scholar]
- Divakaruni AS, Rogers GW, Murphy AN. Measuring mitochondrial function in permeabilized cells using the Seahorse XF Analyzer of a Clark-type oxygen electrode. Curr Prot Tox. 2014b;60(25.2):1–16. doi: 10.1002/0471140856.tx2502s60. [DOI] [PubMed] [Google Scholar]
- Divakaruni AS, Andreyev AY, Rogers GW, Murphy AN. In situ measurements of mitochondrial matrix enzyme activities using plasma and mitochondrial membrane permeabilization agents. Anal Biochem. 2017a;17:30385–8. doi: 10.1016/j.ab.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divakaruni AS, Wallace M, Buren C, Martyniuk K, Andreyev AY, Li E, Fields JA, Cordes T, Reynolds IJ, Bloodgood BL, et al. Inhibition of the mitochondrial pyruvate carrier protects from excitotoxic neuronal death. J Cell Biol. 2017b;216:1091–1105. doi: 10.1083/jcb.201612067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc Natl Acad Sci U S A. 1997;94:4312–7. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Hurtado E, Lee J, Choi J, Selen Alpergin ES, Collins SL, Horton MR, Wolfgang MJ. Loss of macrophage fatty acid oxidation does not potentiate systemic metabolic dysfunction. Am J Physiol Endocrinol Metab. 2017;312:E381–E393. doi: 10.1152/ajpendo.00408.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SC, Everts B, Ivanova Y, O’Sullivan D, Nascimento M, Smith AM, Beatty W, Love-Gregory L, Lam WY, O’Neill CM, et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat Immunol. 2014;15:846–55. doi: 10.1038/ni.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SC, Smith AM, Everts B, Colonna M, Pearce EL, Schilling JD, Pearce EJ. Metabolic Reprogramming Mediated by the mTORC2-IRF4 Signaling Axis Is Essential for Macrophage Alternative Activation. Immunity. 2016;45:817–830. doi: 10.1016/j.immuni.2016.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idell-Wenger JA, Grotyohann LW, Neely JR. Coenzyme A and carnitine distribution in normal and ischemic hearts. J Biol Chem. 1978;253:4310–8. [PubMed] [Google Scholar]
- Jha AK, Huang SC, Sergushichev A, Lampropoulou V, Ivanova Y, Loginicheva E, Chmielewski K, Stewart KM, Ashall J, Everts B, et al. Network Integration of Parallel Metabolic and Transcriptional Data Reveals Metabolic Modules that Regulate Macrophage Polarization. Immunity. 2015;42:419–430. doi: 10.1016/j.immuni.2015.02.005. [DOI] [PubMed] [Google Scholar]
- Johnson AR, Qin Y, Cozzo AJ, Freemerman AJ, Huang MJ, Zhao L, Sampey BP, Milner JJ, Beck MA, Damania B, et al. Metabolic reprogramming through fatty acid transport protein 1 (FATP1) regulates macrophage inflammatory potential and adipose inflammation. Mol Metab. 2016;5:506–26. doi: 10.1016/j.molmet.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Magrané J. Isolation and culture of neurons and astrocytes from the mouse brain cortex. Methods Mol Biol. 2011;793:63–75. doi: 10.1007/978-1-61779-328-8_4. [DOI] [PubMed] [Google Scholar]
- Kiorpes TC, Hoerr D, Ho W, Weaner LE, Inman MG, Tutwiler GF. Identification of 2-tetradecylglycidyl coenzyme A as the active form of methyl 2-tetradecylglycidate (methyl palmoxirate) and its characterization as an irreversible, active site-directed inhibitor of carnitine palmitoyltransferase A in isolated rat liver mitochondria. J Biol Chem. 1984;259:9750–5. [PubMed] [Google Scholar]
- Klingenberg M, Durand R, Guerin B. Analysis of the reactivity of SH-reagents with the mitochondrial phosphate carrier. Eur J Biochem. 1974;42:135–150. doi: 10.1111/j.1432-1033.1974.tb03323.x. [DOI] [PubMed] [Google Scholar]
- Kurmi K, Hitosugi S, Wiese EK, Boakye-Agyeman F, Gonsalves WI, Lou Z, Karnitz LM, Goetz MP, Hitosugi T. Carnitine Palmitoyltransferase 1A Has a Lysine Succinyltransferase Activity. Cell Rep. 2018;22:1365–73. doi: 10.1016/j.celrep.2018.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnareva YE, Wiley SE, Ward MW, Andreyev AY, Murphy AN. Excitotoxic injury to mitochondria isolated from cultured neurons. J Biol Chem. 2005;280:28894–902. doi: 10.1074/jbc.M503090200. [DOI] [PubMed] [Google Scholar]
- Leonardi R, Zhang YM, Rock CO, Jackowski S. Coenzyme A: Back in action. Prog Lip Res. 2005;44:125–53. doi: 10.1016/j.plipres.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Lopaschuk GD, Wall SR, Olley PM, Davies NJ. Etomoxir, a carnitine palmitoyltransferase I inhibitor, protects hearts from fatty acid-induced ischemic injury independent of changes in long chain acylcarnitine. Circ Res. 1988;63:1036–43. doi: 10.1161/01.res.63.6.1036. [DOI] [PubMed] [Google Scholar]
- McQuaker SJ, Quinlan CL, Caldwell ST, Brand MD, Hartley RC. A prototypical small molecule modulator uncouples mitochondria in response to endogenous hydrogen peroxide production. Chembiochem. 2013;14:993–1000. doi: 10.1002/cbic.201300115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EL, Kelly B, Logan A, Costa ASH, Varma M, Bryant CE, Tourlomousis P, Däbritz JHM, Gottlieb E, Latorre I, et al. Succinate Dehydrogenase Supports Metabolic Repurposing of Mitochondria to Drive Inflammatory Macrophages. Cell. 2016;167:457–470. doi: 10.1016/j.cell.2016.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misharin AV, Morales-Nebreda L, Mutlu GM, Budinger GRS, Perlman H. Flow Cytometric Analysis of Macrophages and Dendritic Cell Subsets in the Mouse Lung. Am J Respir Cell Mol Biol. 2013;49:503–510. doi: 10.1165/rcmb.2013-0086MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mookerjee SA, Goncalves RL, Gerencser AA, Nicholls DG, Brand MD. The contributions of respiration and glycolysis to extracellular acid production. Biochim et Biophys Acta. 2015;1847:171–81. doi: 10.1016/j.bbabio.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Namgaladze D, Brüne B. Fatty acid oxidation is dispensable for human macrophage IL-4-induced polarization. Biochim Biophys Acta. 2014;1841:1329–35. doi: 10.1016/j.bbalip.2014.06.007. [DOI] [PubMed] [Google Scholar]
- Namgaladze D, Brüne B. Macrophage fatty acid oxidation and its roles in macrophage polarization and fatty acid-induced inflammation. Biochim Biophys Acta. 2016:1796–807. doi: 10.1016/j.bbalip.2016.09.002. [DOI] [PubMed] [Google Scholar]
- Neess D, Bek S, Engelsby H, Gallego SF, Færgeman NJ. Long-chain acyl-CoA esters in metabolism and signaling: Role of acyl-CoA binding proteins. Prog Lipid Res. 2015;59:1–25. doi: 10.1016/j.plipres.2015.04.001. [DOI] [PubMed] [Google Scholar]
- Nomura M, Liu J, Rovira II, Gonzalez-Hurtado E, Lee J, Wolfgang MJ, Finkel T. Fatty acid oxidation in macrophage polarization. Nat Immunol. 2016;17:216–7. doi: 10.1038/ni.3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor RS, Guo L, Ghassemi S, Snyder NW, Worth AJ, Weng L, Kam Y, Philipson B, Trefely S, Nunez-Cruz S, et al. The CPT1a inhibitor, etomoxir induces severe oxidative stress at commonly used concentrations. Sci Rep. 2018;8:6289. doi: 10.1038/s41598-018-24676-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, Morel CR, Subramanian V, Mukundan L, Red Eagle A, Vats D, Brombacher F, Ferrante AW, et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447:1116–20. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill LA, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol. 2016;66:553–65. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pande SV, Blanchaer MC. Reversible inhibition of mitochondrial adenosine diphosphate phosphorylation by long chain acyl coenzyme A esters. J Biol Chem. 1971;246:402–411. [PubMed] [Google Scholar]
- Paradies G, Ruggiero FM. Effect of aging on the activity of the phosphate carrier and on the lipid composition in rat liver mitochondria. Arch Biochem Biophys. 1991;284:332–337. doi: 10.1016/0003-9861(91)90304-2. [DOI] [PubMed] [Google Scholar]
- Pietrocola F, Galluzzi L, Bravo-San Pedro JM, Madeo F, Kroemer G. Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab. 2015;21:805–21. doi: 10.1016/j.cmet.2015.05.014. [DOI] [PubMed] [Google Scholar]
- Raud B, Roy DG, Divakaruni AS, Tarasenko TN, Franke R, Ma EH, Samborska B, Hsieh W, Wong AH, Stüve P, et al. Etomoxir actions on regulatory and memory T cells are independent of Cpt1a-mediated fatty acid oxidation. 2018 doi: 10.1016/j.cmet.2018.06.002. (Co-submission) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GW, Brand MD, Petrosyan S, Ashok D, Elorza AA, Ferrick DA, Murphy AN. High throughput microplate respiratory measurements using minimal quantities of isolated mitochondria. PLoS One. 2011;6:e21746. doi: 10.1371/journal.pone.0021746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GW, Nadanaciva S, Swiss R, Divakaruni AS, Will Y. Assessment of fatty acid beta oxidation in cells and isolated mitochondria. Curr Protoc Toxicol. 2014;60(25.3):1–19. doi: 10.1002/0471140856.tx2503s60. [DOI] [PubMed] [Google Scholar]
- Rognstad R. Estimation of peroxisomal and mitochondrial fatty acid oxidation in rat hepatocytes using tritiated substrates. Biochem J. 1991;279:147–150. doi: 10.1042/bj2790147. 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio M, Avitabile D, Fischer K, Emmanuel G, Gude N, Miyamoto S, Mishra S, Schaefer EM, Brown JH, Sussman MA. Cardioprotective stimuli mediate phosphoinositide 3-kinase and phosphoinositide dependent kinase 1 nuclear accumulation in cardiomyocytes. J Mol Cell Cardiol. 2009;47:96–103. doi: 10.1016/j.yjmcc.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoors S, Bruning U, Missiaen R, Queiroz KC, Borgers G, Elia I, Zecchin A, Cantelmo AR, Christen S, Goveia J, et al. Fatty acid carbon is essential for dNTP synthesis in endothelial cells. Nature. 2015;520:192–197. doi: 10.1038/nature14362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shug A, Lerner E, Elson C, Shrago E. The inhibition of adenine nucleotide translocase activity by oleoyl CoA and its reversal in rat liver mitochondria. Biochem Biophys Res Commun. 1971;43:557–563. doi: 10.1016/0006-291x(71)90650-4. [DOI] [PubMed] [Google Scholar]
- Szanto A, Balint BL, Nagy ZS, Barta E, Dezso B, Pap A, Szeles L, Poliska S, Oros M, Evans RM, et al. STAT6 Transcription Factor Is a Facilitator of the Nuclear Receptor PPARγ-Regulated Gene Expression in Macrophages and Dendritic Cells. Immunity. 2010;33:699–712. doi: 10.1016/j.immuni.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita S, Kaji K, Kudo A. Identification and characterization of the new osteoclast progenitor with macrophage phenotypes being able to differentiate into mature osteoclasts. J Bone Miner Res. 2000;15:1477–88. doi: 10.1359/jbmr.2000.15.8.1477. [DOI] [PubMed] [Google Scholar]
- Tan Z, Xie N, Cui H, Moellering DR, Abraham E, Thannickal VJ, Liu G. Pyruvate dehydrogenase kinase 1 participates in macrophage polarization via regulating glucose metabolism. J Immunol. 2015;194:6082–9. doi: 10.4049/jimmunol.1402469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013;496:238–42. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Bossche J, Baardman J, Otto NA, van der Velden S, Neele AE, van den Berg SM, Luque-Martin R, Chen HJ, Boshuizen MC, Ahmed M, et al. Mitochondrial Dysfunction Prevents Repolarization of Inflammatory Macrophages. Cell Rep. 2016;17:684–696. doi: 10.1016/j.celrep.2016.09.008. [DOI] [PubMed] [Google Scholar]
- Van den Bossche J, O’Neill LA, Menon D. Macrophage Immunometabolism: Where Are We (Going)? Trends Immunol. 2017;38:395–406. doi: 10.1016/j.it.2017.03.001. [DOI] [PubMed] [Google Scholar]
- Vats D, Mukundan L, Odegaard JI, Zhang L, Smith KL, Morel CR, Wagner RA, Greaves DR, Murray PJ, Chawla A. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell Metab. 2006;4:13–24. doi: 10.1016/j.cmet.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser WF, Van Roermund CWT, Ijlst L, Waterham HR, Wanders RJA. Metabolite transport across the peroxisomal membrane. Biochem J. 2007;401:365–75. doi: 10.1042/BJ20061352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KJ, Argus JP, Zhu Y, Wilks MQ, Marbois BN, York AG, Kidani Y, Pourzia AL, Akhavan D, Lisiero DN, et al. An essential requirement for the SCAP/SREBP signaling axis to protect cancer cells from lipotoxicity. Cancer Res. 2013;73:2850–62. doi: 10.1158/0008-5472.CAN-13-0382-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson JR, Corkey BE. Assay of citric acid cycle intermediates and related compounds–update with tissue metabolite levels and intracellular distribution. Methods Enzymol. 1979;55:200–22. doi: 10.1016/0076-6879(79)55025-3. [DOI] [PubMed] [Google Scholar]
- Xiao G, Chan LN, Klemm L, Braas D, Chen Z, Geng H, Zhang QC, Aghajanirefah A, Cosgun KN, Sadras T, et al. Cell-specific diversion of glucose carbon utilization reveals a unique vulnerability in B cell malignancies. Cell. 2018;173:470–484. doi: 10.1016/j.cell.2018.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao CH, Liu GY, Wang R, Moon SH, Gross RW, Patti GJ. Identifying off-target effects of etomoxir reveals that carnitine palmitoyltransferase I is essential for cancer cell proliferation independent of β-oxidation. PLoS Biol. 2018;16:e2003782. doi: 10.1371/journal.pbio.2003782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


