Abstract
Purpose
Prostate cancer translational research has been hampered by the lack of comprehensive and tractable models that represent the genomic landscape of clinical disease. Metastatic castrate-resistant prostate cancer (mCRPC) patient-derived xenografts (PDXs) recapitulate the genetic and phenotypic diversity of the disease. We sought to establish a representative, preclinical platform of PDX-derived organoids that is experimentally facile for high throughput and mechanistic analysis.
Experimental Design
Using 20 models from the LuCaP mCRPC PDX cohort, including adenocarcinoma and neuroendocrine lineages, we systematically tested > 20 modifications to prostate organoid conditions. Organoids were evaluated for genomic and phenotypic stability and continued reliance on the AR signaling pathway. The utility of the platform as a genotype-dependent model of drug sensitivity was tested with olaparib and carboplatin.
Results
All PDX models proliferated as organoids in culture. Greater than fifty percent could be continuously cultured long-term in modified conditions; however, none of the PDXs could be established long-term as organoids under previously-reported conditions. Additionally, the modified conditions improved the establishment of patient biopsies over current methods. The genomic heterogeneity of the PDXs, was conserved in organoids. Lineage markers and transcriptomes were maintained between PDXs and organoids. Dependence on AR signaling, was preserved in adenocarcinoma organoids, replicating a dominant characteristic of CRPC. Finally, we observed maximum cytotoxicity to the PARP inhibitor olaparib in BRCA2−/− organoids, similar to responses observed in patients.
Conclusions
The LuCaP PDX/organoid models provide an expansive, genetically-characterized platform to investigate mechanisms of pathogenesis as well as therapeutic responses and their molecular correlates in mCRPC.
Keywords: organoids, patient-derived-xenografts, LuCaP, prostate cancer, AR
Introduction
Progressive, recurrent and metastatic prostate cancers are treated with androgen deprivation therapy (ADT), leading to regression of almost all of these hormone-dependent cancers. Eventually, most re-emerge in the form of castrate resistant prostate cancer (CRPC) as a result of selection and acquired resistance, most often in the AR pathway (1). Profiling genomes, transcriptomes, and methylomes of clinical samples has revolutionized our knowledge of the molecular features underlying prostate cancers (2–5). Substantial genomic heterogeneity occurs between and within patients, and there are continuing efforts to classify molecular subtypes with respect to both drivers of the aggressive, metastatic phenotype, and as mechanisms contributing to acquired resistance (4,6). Major altered pathways enriched in CRPC relative to primary prostate cancer include AR, P53, PI3K, and RB as well as DNA damage repair and epigenetic modification (5). Improved treatments for mCRPC will require an approach informed by the genotype of the patient’s cancer and directed at the appropriate biological mechanism(s) (1). Investigations that translate molecular features into therapeutic vulnerabilities and mechanistic outcomes require a comprehensive set of preclinical models that faithfully reflect the broad genetic and phenotypic spectrums of the disease. Tractable, comprehensive platforms of CRPC have not been previously available, contributing to a major limitation in fully utilizing and generalizing patient molecular data.
Commonly-used CRPC models such as LnCaP have been effective for investigating AR signaling, but overall, they are lacking as representative models. Relatively few prostate cancer cell lines are available, and they do not reflect the diversity of tumors found in patients, nor do they accurately predict patient response to treatment (7,8). Although several genetically engineered mouse models exist (9), they fail to generally model clinical CRPC, as castration resistant tumors in mice do not depend upon AR signaling mechanisms (10).
Two preclinical models have emerged to meet the needs of translational prostate cancer research: patient-derived xenografts (PDX) and 3D organoids (11, 12). For many cancers, PDX lines are currently the best available representation of patient disease, with respect to genomic and phenotypic heterogeneity (11). They exhibit preservation of histologic features, and they have been shown to be highly predictive of genomically-matched patient responses to drug treatment, although prostate cancer has not been extensively tested (11, 13,14). The LuCaP PDXs are a large, well characterized, and clinically annotated cohort of advanced prostate cancer PDXs (15). Currently the set includes > 40 samples, representing much of the genomic heterogeneity, variety of subtypes, sites of metastasis, and treatment histories of prostate cancer (PC) (15,16). Unfortunately, the methodology to reproducibly grow prostate PDX-derived in vitro cultures in significant quantities has been elusive (17,18). Thus, although there is significant precedent showing that PDX cohorts effectively inform upon clinical disease, there are limitations to their use with respect to high throughput screens, genetic manipulation, and mechanistic analysis.
Organoid culture methods were developed to grow normal epithelial stem cells, which demonstrate self-renewal and differentiation activity as organoids, recapitulating tissue architecture (19). The rich growth media and 3D matrix of organoid culture conditions have also proven useful for increasing the efficiency and histological representation of cultures established from patient epithelial tumors including colorectal (20), liver (21), breast (22), pancreatic (23)and CRPC (17), among others. Organoids retain the genomic and phenotypic features of the patient tumor, and are well-suited for in vitro manipulations (12). Assembling a biobank of patient-derived organoids from CRPC has been particularly challenging due to the low success rate for establishing long-term, CRPC organoid cultures, compounded by the limited availability of patient material (17). Currently, the number of CRPC organoid lines available is small and under-represents the variance observed within the disease (17). To overcome these limitations, we have developed conditions for growing LuCaP PDX-derived cells as organoids that readily proliferate, yielding numbers sufficient for in vitro assays. Thus, the experimental flexibility enabled by organoid culturing allows genetic manipulations and in vitro mechanistic assays of PDX models, and in addition, broadens the naturally-occurring genomic mutations available for study in continuously passaged cultures. Our conditions have also allowed us to establish patient biopsy derived organoid lines that otherwise failed to grow in previously reported media conditions (17,18,24). Finally, the conservation of transcriptomic and genomic features between organoids and the originating PDX as well as our observation of genomically-predicted drug sensitivities, determined for the PARP inhibitor olaparib and carboplatin, supports the clinical relevance of the PDX-derived organoid platform described here.
Materials and Methods
Processing and plating of PDX and patient-derived tumor samples
LuCaP PDX tumors, collected at the University of Washington, were shipped overnight on ice, in DMEM/10% FBS and processed within 24 hours of the time the mouse was sacrificed. Alternatively, LuCaP PDX tumors were maintained at the NCI under an NCI Animal Care and Use Committee approved protocol. LuCaP PDX tumors were validated using STR analysis by Laragen, Inc. For biopsies, patients provided informed consent, and samples were procured from the NIH Clinical Center under NIH Institutional Review Board approval in accordance with U.S. Common Rule. Patient biopsy samples were processed immediately.
PDX and patient-derived samples were processed the same except where indicated. Tissue was cut into small pieces, 2–4 mm, with a scalpel blade. The tissue was then collected in Advanced DMEM/F12 with 10 mM HEPES and 2mM Glutamax, transferred to gentle MACS C tubes (Miltenyi Biotec cat. # 130-096-334) and digested using the Tumor Dissociation Kit for human tissue (Miltenyi Biotec cat. # 130-095-929) on a gentleMACS Octo Dissociator with heaters (Miltenyi Biotec cat. # 130-096-427), program 37C_h_TDK_2. Alternatively if a MACS Dissociator and reagents are not available, tissues can be digested in media plus collagenase type I and DNAse, at 37°C while rocking, for 2 hours, pipetting every 30’ to facilitate dissociation. Processed samples were centrifuged, resuspended and passed through a 100 µM cell strainer as needed to eliminate macroscopic tissue pieces. For PDXs, cell pellets were resuspended in 2–3 volumes of ACK lysis buffer (Lonza cat. # 10-548E) according to the manufacturers specifications, and washed cells were resuspended in media.
PDX tumors maintained at the NCI were passaged in athymic nude-Foxn1nu mice. After processing as described above, cell suspensions (~2 million cells) were mixed with growth factor reduced, phenol red-free Matrigel (BD Biosciences cat. # 356231), and injected subcutaneously.
To culture PDX-derived cells as organoids, cell suspensions, processed as described above, were plated in 3D on 12, 24, or 48-well plates. Growth media formulation for PrENR is described in Drost et al. (24). To plate in 12-well format, 1 × 105 cells were resuspended on ice in 20 µl of cold culture media and mixed with 80 µl of growth factor reduced, phenol red-free Matrigel. Cells were then plated around the circumference of the well and incubated for 45 minutes before adding media to the well. The procedure for plating on 24-well plates was scaled to 50% cell numbers and volumes as described for 12 well plates. To plate in 48-well format, 12,500 cells were resuspended on ice in 20 µl of Matrigel. Cells were plated as a drop in the center of the well.
Passing organoids
For passaging, organoids were incubated with dispase dissolved in Advanced DMEM/F12 to a final concentration of 1 mg/ml and incubated for 2 hours at 37° C. Pelleted organoids were resuspended in pre-warmed TrypLE Express (Thermo Fisher cat. # 12605028) for ~5 minutes at 37° C with periodic pipetting. The resulting cell suspensions were diluted in Advanced DMEM/F12, centrifuged, resuspended and plated.
Freezing organoids
Organoids were resuspended in freezing media (90% FBS/10% DMSO) in cryovials (Corning #375418), and immediately transferred to a CoolCell LX freezing container (Biocision #BCS-405G) and placed at −80° C for ≥ 2 hr.
Establishment of organoid-derived xenografts
Organoids cultured from as few as one to greater than ten generations were reinjected into mice to establish ODXs. One – two million partially dissociated organoids were mixed with Matrigel at a ratio of 1:1 and reinjected subcutaneously into athymic nude mice or into NOD scid gamma mice. For orthotopic injections, ~1×105 partially-dissociated organoids were mixed with Matrigel at a ratio of 1:1 in a final volume of 50 µl and injected into the prostate.
Systematic testing of culture conditions
Hypoxia chambers filled with 1% O2, 5% CO2, 94% N2 were used when culturing in a hypoxic atmosphere. The defined media formulation PrENR was systematically modified by the removal of some standard components and/or the addition of new factors to the media. Addition or subtraction of factors was first performed singly, and those modifications determined to have a positive impact on growth/survival were then combined with each other or with additional modifications.
Quantification assays
Cells were plated in 3D on 12 or 48-well plates and cultured for the indicated number of days. Quantification was performed using CellTiter-Glo 3D (Promega cat. # G9681) according to the manufacturer’s protocol.
Immunofluorescent staining of organoids
Following dispase treatment, washed organoids were fixed in 4% formaldehyde in PBS for 2 hours at room temp, washed, and resuspended in 70% ethanol. Spheres were paraffin-embedded and sectioned by Histoserve Inc., Germantown, MD. Images were taken with a Zeiss Axioscan.Z1 slide scanner.
Immunoblot assays
Organoids were lysed in RIPA buffer containing protease and phosphatase inhibitors. 15 – 30 µg of protein lysate was loaded onto 4–20% mini-gradient gels and transferred onto PVDF membranes. Membranes were probed overnight at 4° C.
Validation of human origin
Organoid cell suspensions were plated in 2D on Poly-D-Lysine coated 96 well plates and allowed to attach overnight and stained with anti-human nuclei antibody HuNu and DAPI. Additionally, mouse cells can be distinguished from human cells by distinct nuclear characteristics revealed by DAPI staining.
PSA quantification
0.5 – 1 ml of conditioned media was collected after 3 days of culture. Total PSA was quantified using a Roche Cobas 6000 Analyzer.
EdU incorporation assay
12,500 cells were plated in triplicate wells in 20 µl drops of Matrigel, on 48 well plates. After 48 hours, 10 µM EdU (Invitrogen cat. # C10338) was added to the wells and incubated for 24 hours. The organoids were then dissociated in TrypLE and replated in 2D on 96-well plates as described above and stained for EdU and DAPI according to the manufacturer’s protocol. DAPI-positive, and EdU-positive nuclei were then quantified using ImageJ.
RNA extraction
Organoids were grown in 12-well plates. Media was aspirated and 500 µl of Trizol was added directly to the organoids, mixed by pipette and incubated for 10 minutes at room temperature. RNA was extracted with 200 µl of chloroform/isoamyl alcohol, and then purified with a Qiagen RNeasy Mini Kit. PDX tissue was harvested immediately from euthanized mice. Tissue was cut into ~20 mg pieces, snap froze in liquid N2, and stored at −80° C. RNA was extracted with a Qiagen AllPrep DNA/RNA Mini Kit.
cDNA synthesis
100 ng of total RNA was converted to cDNA using SuperScript VILO MasterMix (Invitrogen cat. #11755) according to the manufacturer’s protocol.
Real-time qPCR
PCR was performed using FastStart Universal SYBR Green Master Mix (Roche # 04913850001) on an Applied Biosystems StepOnePlus Real-Time PCR machine. Relative quantification was performed using the 2−ΔΔCT method (25). Absolute quantification was calculated from a standard curve. To make the standard curve, known amounts of serially diluted λpolyA+ RNA-A (Takara cat. # 3789) was added to 100 ng of total RNA pooled from multiple LuCaP RNA extracts, then converted to cDNA as described for total RNA.
Lentiviral transduction
1 × 106 cells were transduced with lenti-pLVX-GFP by spin infection at 700 rpm for 1 hour at room temperature. Cells were incubated overnight in the presence of the virus. Cells were then collected and replated in 3D in Matrigel.
Dose response assays
Generation one organoids were plated in replicates of five, on 384-well plates: 4,000 cells in 40 µl of growth factor reduced Matrigel per well, overlaid with 40 µl of media (PrENR −p38i −NAC). Twenty-four hours after plating the media was replaced with fresh media containing olaparib or carboplatin. Media + drug was changed twice per week over the course of two weeks. Cell viability was quantified with CellTiter Glo 3D. Graph Pad Prism 7 was used to plot dose response curves and to calculate IC50.
Copy number analysis
Genomic DNA was extracted from fresh frozen PDX tissue, from fresh organoids, and from FFPE matched normal tissue, using a Qiagen AllPrep DNA/RNA Mini Kit. DNA from each sample was quality checked using an Agilent TapeStation 2200 and PicoGreen Assay, and an aliquot of 200 ng of DNA was fragmented and prepared into libraries for hybridization to Illumina OmniExpress BeadChip Arrays at the National Cancer Institute, Cancer Genomics Research Laboratory. Data was clustered against a panel of normal provided by Illumina using GenomeStudio (Illumina) and exported for analyses in Nexus Copy Number version 8 (BioDiscovery). SNPs were segmented along the genome using the SNP-FASST2 Hidden Markov Model algorithm wherein contiguity and B-allele frequency was considered significant at P < 5×10−7, allowing 15 minimum probes per segment and a maximum of 1Mb between discontinuous regions. Copy number gain and loss threshold values were determined empirically for each group of samples by visual inspection, with a range of log2 −0.4 to −0.45 for single copy losses, −1.1 for homozygous deletions, 0.27 to 0.4 for gains, and 0.41 to 7 for high-level amplifications. Regions of copy number gain or loss were processed with Bedtools version 2.25 to determine regions of shared or distinct copy number alterations between PDX and organoid lines from the same case. Total copy number aberrations and percentage of genome changed were determined with Nexus software.
Whole exome sequencing
At least 100 ng of genomic DNA, was fragmented, adaptor-ligated, hybridized in solution to biotinylated baits covering the exome and untranslated regions (Agilent v5 + UTR), and assembled into multiplexed barcoded libraries for sequencing on the Illumina platform. Library preparation and HiSeq 2500 sequencing was performed at the Frederick National Laboratory for Cancer Research Sequencing Facility. FastQ files were aligned using BWA-MEM version 0.7.10, deduplicated using PICARD version 2.6, and realigned around indels and quality recalibrated using GATK version 3.6. PICARD was used for calculating hybrid capture metrics.
Mutation calling
Somatic and germline (single-sample) mutation calling analyses were performed using MuTect2. For determining clonality, cancer cell fraction (CCF) (26) values were calculated using the MuTect2-provided variant allele fraction (VAF), the ploidy value for the corresponding interval of the genome as determined by Nexus Copy Number from Illumina OmniExpress array data, and an assumed purity value of 95% as follows: CCF = [VAF × ((0.95 × ploidy) + 0.1) ÷ 0.95]. Shared and distinct mutations were sorted by CCF for each case using in-house scripts and ranked based on overlap between PDX and organoid lines. Shared and distinct mutations were considered with copy number mutations for the total range of genomic sites that overlapped between PDX and organoid lines. Heatmaps were created using Gitools v. 2.2.2.
RNA sequencing
RNA-seq was performed on the Illumina HiSeq platform using rRNA-depleted RNA and paired end reads. Data were pre-processed for deconvolution of human- and mouse-genome aligning reads using the Disambiguate algorithm, which uses the highest quality alignment for each read pair to assign the correct species. After adaptor trimming, reads were simultaneously aligned to the hg19 genome (human) or mm10 genome (mouse) using Bowtie2 and Tophat version 2.1.1 without novel junction discovery, based on species-specific gene predictions downloaded from the UCSC genome table browser in January 2018. Each accepted-hits BAM file was sorted by read name and then processed by Disambiguate, which deposited species-specific read pairs into corresponding BAM files. Reads aligning equally well to both species' transcriptomes were discarded. The BAM file of reads preferentially aligning to the human transcriptome were processed by the PICARD SamToFastq tool to recover the FASTQ files, discarding all secondary alignments and singletons.
The pre-processed data were then filtered to remove low quality reads, followed by the trimming of low quality bases. RNA Sequence reads were aligned with STAR to the human genome, hg19. The RSEM algorithm was applied to compute the raw reads count. Data were normalized to convert the RSEM raw read count to CPM (counts per million) using EdgeR. The technical and biological replicates were merged.
Principal Component Analysis(PCA)
We applied PCA, unsupervised machine learning to visualize the sample-to sample distance. The principle components of the samples were determined using the normalized whole genome RNA-Seq data using the Singular Value Decomposition(SVD) algorithm. The PCA plot was generated with in-house customized R scripts. The heatmap was built with the HeatMapImage package from the GenePattern web portal by Broad Institute. We uploaded the gene expression(CPM) in gct format, utilized the default color scheme, and applied row normalization.
Accession Numbers
Segmented somatic copy number data and gene expression data has been deposited into the Gene Expression Omnibus (GEO), accession ID GSE113743.
Exome sequencing, whole transcriptome sequencing, and SNP array raw data has been deposited into the NCBI Database of Genotypes and Phenotypes (dbGaP), accession ID phs001587.v1.p1
Statistics
Graphed data is presented as standard error of the mean except for patient biopsy-derived PSA data where only single unique samples existed and replicates were not possible. P-values were calculated using two-tailed student’s t-test. P < 0.05 was considered significant.
Tables specifying antibodies and primers used, as well as the reagents used to test modified conditions are located in supplementary materials and methods.
Results
LuCaP PDX-derived cells grow as organoids in modified media
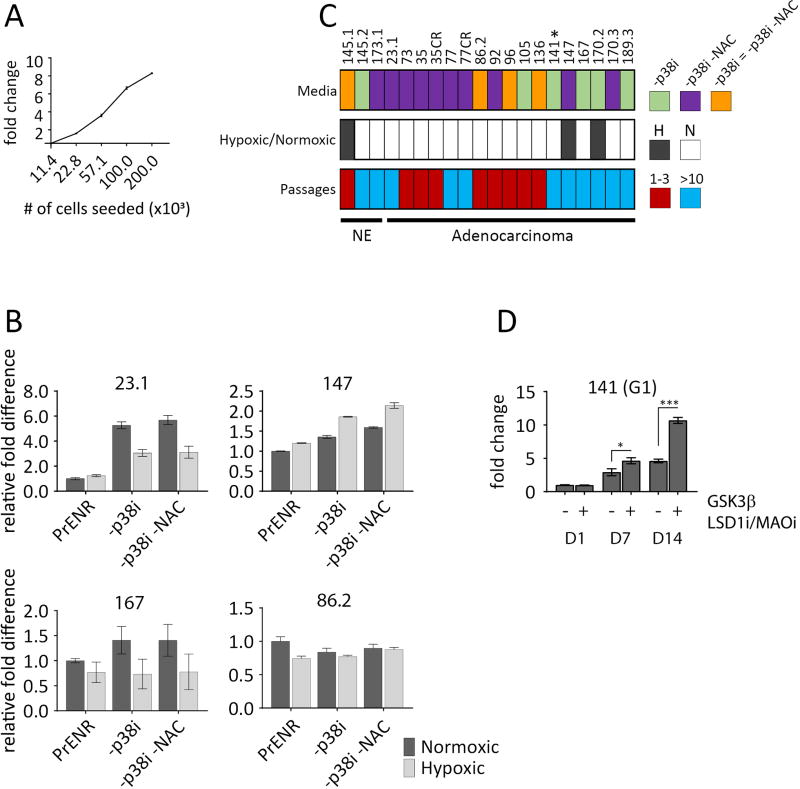
We found that organoid cultures derived from several different LuCaP PDX models, could be cultured in organoid conditions previously described (24) for metastatic prostate cancer biopsies, but the cultures quickly lost viability with passage. Given that organoids from metastatic PC biopsies establish at a modest frequency (17), we reasoned that the culture conditions may be selective and require modification. Using the previously described growth media (17,18), referred to here as prostate-specific ENR (PrENR: prostate EGF, Noggin, R-spondin), (see materials and methods), we tested > 20 modifications, including the addition of metabolic modulators, or growth and differentiation factors known to influence PC growth, as well as the systematic removal of selected components from the established media (Supplementary Table S1). To concentrate on those conditions that promoted growth, modifications that led to an inhibitory effect in at least two models were eliminated. Conditions with no obvious effect were assayed on average in five models before elimination, and conditions leading to enhanced growth were assayed in all models. Cells were seeded at densities within the linear range of the CellTiter Glo 3D assay (Fig. 1A), then growth and survival were measured following a two-week time course. Surprisingly, soluble factors such as IL-11, previously described to positively modulate the growth of highly-adapted, PC cell lines (27) had relatively little effect on a diverse group of organoids. The condition that influenced growth in all of the LuCaPs tested was removal of SB202190, a p38 MAPK inhibitor (p38i) from PrENR media. The removal of the anti-oxidant NAC also had a small positive effect, which was also observed when combined with the −p38i condition. A few models grew equally well in all conditions (e.g. LuCaP 86.2, Fig. 1b) in the two-week assays. In all, 20 models were compared in PrEN, PrEN −p38i, and PrEN −p38i/−NAC in both hypoxic and normoxic atmospheres. Remarkably, although the growth differences between conditions measured after two weeks in culture were often subtle, when applied over consecutive passages, a clear advantage in growth and survival was revealed for the −p38i and −p38i/−NAC conditions (Supplementary Fig. S1). No LuCaP model could be maintained long-term in PrENR, but 11 LuCaP models were grown for >10 generations (~ 6 months) in the −p38i or −p38/−NAC conditions (Fig. 1c and Supplementary Table 2). For logistical reasons, we did not continuously passage all models well-beyond 10 generations, but every indication was that most models that grow at least 10 generations, will grow continuously. There was no evidence of slowed growth after 10 generations, and a few models (e.g. LuCaP 23.1, LuCaP 145.2 and LuCaP 167) were passaged for > 20 generations (approximately 1 year). The remaining models proliferated between one and three generations. Although three models grew better in hypoxia, growth in hypoxia or normoxia had a relatively minor effect on the rate of continuous growth. Growing organoid cultures were validated for human origin by staining for a human-specific nuclear antigen (Supplementary Fig. S2).
Figure 1.
Comparison of culture conditions by relative cell viability. A, CellTiter Glo 3D assay is linear over a broad range of cell densities. Normal liver cells seeded in 3D in PrENR at 2-fold increasing numbers, assayed after 96hr by CellTiter Glo 3D. X axis is the number of cells seeded, Y axis is the fold difference relative to the lowest number of cells seeded. Error bars represent the standard error of the mean. B, An equal number of generation 1 (G1) cells were plated in 3D and then cultured for two weeks in either a normoxic or hypoxic atmosphere in standard PrENR, PrENR minus p38i, or PrENR minus p38i and NAC. Graphs represent a comparison of the viable organoids among each condition assayed by CellTiter Glo 3D. The Y axis indicates the fold difference set relative to the PrENR/normoxic condition. Error bars represent the standard error of the mean. Two independent experiments were performed. C, Summary of positive culture conditions for all LuCaPs tested. Top row, colored boxes indicate the media condition(s) that best supported growth for a given LuCaP. *LuCaP 141 requires additional components (see D). Middle row, filled boxes indicate that a given LuCaP grew best in a hypoxic atmosphere. Empty boxes indicate that a LuCaP grew best in a normoxic atmosphere. Bottom row, colored boxes indicate the number of generations that a given LuCaP could be grown in organoid culture. Red = 1–3 generations, Blue = at least 10 generations. D, Specific additional modifications allow for growth of LuCaP 141 organoids that do not otherwise survive in culture. G1 LuCaP 141 cells were plated, in −p38i or −p38i +GSK3β inhibitor (3µM) +LSD1/MAO inhibitor (2 µM) media, and assayed for proliferation by CellTiter Glo 3D at days 1, 7, and 14. Graphs show the change in cell number over time relative to day 1 in the −p38i condition. Error bars represent the standard error of the mean of three replicates. Student’s t-tests were performed; *P < 0.05, ***P < 0.0001. Representative figure of three independent experiments is shown
Further tailoring of growth media based on genotypic and phenotypic characteristics will likely influence the growth of individual LuCaPs. For example, LuCaP 141 did not survive beyond G1 in PrENR, PrENR −p38i, or PrENR −p38i −NAC. However, LuCaP 141 expresses high levels of the Wnt pathway antagonist GSK3β and weakly expresses various neuroendocrine-associated transcripts. Therefore, we used two additives that promote the growth of neural progenitors (10), a GSK3β inhibitor and an LSD1/MAO inhibitor, which improved the growth rate and allowed continuous culture of LuCaP 141 for more than 10 generations (Fig. 1D and Supplementary Table S2). Although GSK3β inhibitor improves the growth of several individual LuCaPs, it is not required for the growth of organoids, other than LuCaP 141. However, other modifications specific to individual LuCaP models may support continuous growth.
PDX-derived organoids proliferated in culture as determined by relative cell viability assays measured by CellTiter Glo, and EdU incorporation assays (Supplementary Fig. S3A and S3B). Doubling times determined from CellTiter Glo data, ranged from less than two days to just over twelve days with an average of five days. Importantly, PDX-derived organoids such as LuCaP 136 and LuCaP 96 that did not maintain proliferation over multiple generations, proliferated well in the first generation.
To summarize, there is significant heterogeneity in the in vitro growth properties of the LuCaP PDX cohort. There were no obvious genomic or phenotypic correlates of models that grew long term and those that did not. Nonetheless, an active p38 pathway was required for continuous growth of organoids and additional removal of the potent antioxidant NAC often enhanced this effect. These conditions were sufficient for long-term growth of 11 models, which significantly increases the genomic representation of continuously growing prostate cancer models. All PDX-derived organoids are initially proliferative, and therefore, are accessible to a variety of in vitro manipulations including drug response assays, biochemical assays, and genetic manipulation. For example, LuCaP organoids can be readily transduced with lentiviral vectors, enabling genetic modification and selection (Supplementary Fig. S4). Additionally these organoids can be viably-frozen and recovered with 100% efficiency so that continuous passage is not necessary.
Modified culture conditions improve long-term growth and survival of needle biopsy-patient-derived organoids
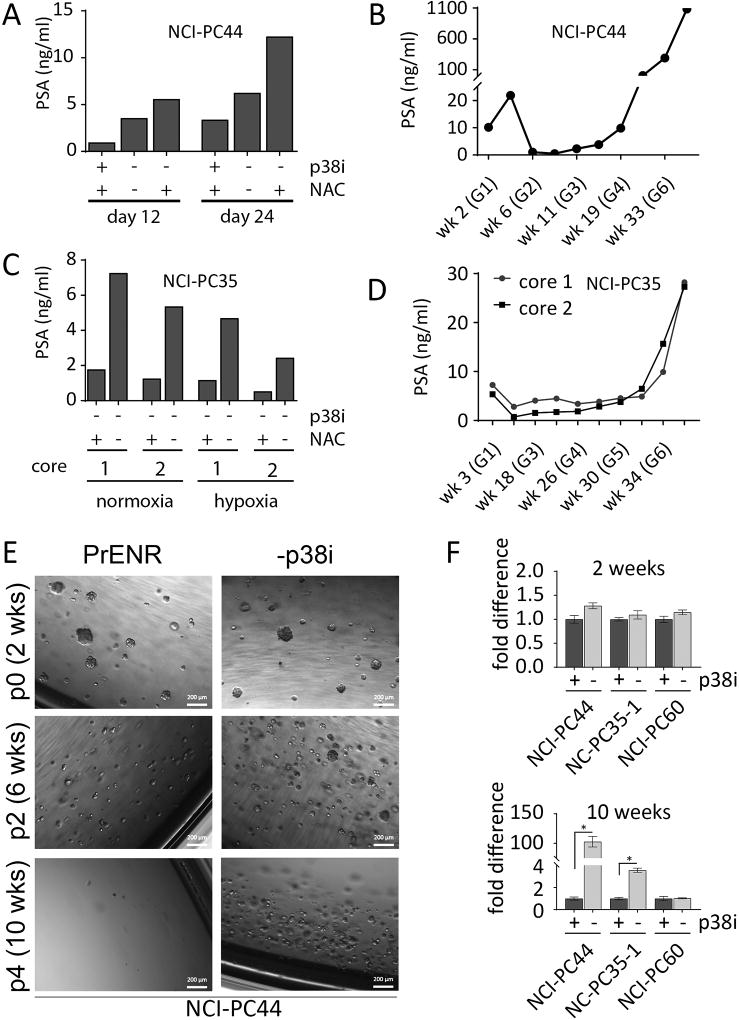
We also evaluated the effect of removing the p38i and NAC for growing patient-tissue-derived organoids. We have established four new patient-derived organoid lines of castration-resistant PC from three different patients (Supplementary Table S3). These include an AR+ adenocarcinoma (NCI-PC44), two independent samples of an AR+ carcinoma with NE features (NCI-PC35-1 & NCI-PC35-2), and an AR−/NE− (double negative) carcinoma (NCI-PC60). As the sources of these cultures were needle core biopsies, we obtained variable cell numbers, which were sufficient to test only limited conditions during the initial establishment phase of these lines. The order of priority for testing was: (1) −p38i −NAC, (2) −p38i, and (3) standard PrENR. Growth of NCI-PC44 and NCI-PC35, was assessed by PSA in culture supernatants as cell based proliferation assays require the destruction of cells. For NCI-PC44, there was a 4.7-fold difference between PSA production in the (−p38i/+NAC) and (PrENR) conditions, measured at 12 and 24 days (Fig. 2A). Continuous culture in −p38i/+NAC conditions led to expansion and establishment (Fig. 2B). In addition to the media conditions, growth in both normoxic and hypoxic atmospheres was compared for NCI-PC35. These organoids grew best in the −p38i −NAC condition in normoxia (Fig. 2C), and long-term cultures were established using those conditions (Fig. 2D). The NCI-PC60 organoids are negative for AR. Growth could not be tracked by PSA, however; these organoids began to proliferate immediately and robustly under both −p38i and −p38i −NAC conditions, as determined by observation.
Figure 2.
Modified culture conditions improve growth of metastatic prostate cancer cells cultured directly from patient samples. A, NCI-PC44 spinal metastasis G1 cultured in PrENR (+p38i), −p38i, and −p38i −NAC conditions. PSA was measured at days 12 and 24 after initial collection and plating of the biopsy sample. B, Growth of patient derived organoids tracked by secreted PSA at several time-points, starting at G1 and continuing until G7. C, NCI-PC35 neck metastasis G1 grown in −p38i and −p38i −NAC in both normoxic (20% O2) and hypoxic (1% O2) atmospheres. The graph for NCI-PC35 shows data for two spatially separate biopsy cores (1 and 2), taken from the same tumor at the same time and maintained separately in culture. D, Growth of NCI-PC35 organoids tracked by PSA as described for (B). E, Patient-derived organoids NCI-PC44 cultured in PrENR (+p38i), or −p38i. Organoids were passaged every two weeks for ten weeks. The passage numbers in this figure indicate the number of passages from the initiation of this experiment and are not an indication of the number of passages for the organoid line overall. Representative bright field images were taken every two weeks prior to each passage. F, Patient-derived organoids NCI-PC44, NCI-PC35-1, and NCI-PC60 were cultured in the optimal media condition determined for each, or the optimal media +p38i. Organoids were passaged every two weeks for ten weeks, and quantified by CellTiter Glo 3D at two and ten weeks. Results are shown as fold difference between the +p38i and −p38i conditions. Error bars represent the standard error of the mean of three replicates. Student’s t-tests were performed; *P < 0.004
To further evaluate the effect of the p38i on biopsy-derived organoid cultures, we analyzed the established NCI-PC44, NCI-PC35-1, and NCI-PC60 organoids for growth over five consecutive passages in optimal growth media with and without p38i (Fig. 2 E and F). Although growth was initially unaffected by the p38i, following further passages, growth of NCI-PC44 was strongly inhibited, growth of NCI-PC35-1 was partially inhibited, and NCI-PC60 was completely resistant to the presence of p38i. These results suggest that a high percentage of mCRPC-derived tumors require p38 activity for optimal establishment and long-term propagation in vitro.
The low cellular yield from biopsy samples limits the number of conditions that we can test simultaneously, when trying to establish new organoid lines. Therefore, we constructed a decision tree to guide the process (Supplementary Fig. S5). The priorities are based on our experience testing culture conditions for biopsy-derived and PDX-derived organoids as described (Figs. 1 and 2). The condition that supports growth most widely (-SB -NAC) is ranked at the top.
Inhibition of p38 causes a downregulation of AKT pathway activity in both adenocarcinoma and neuroendocrine mCRPC organoids
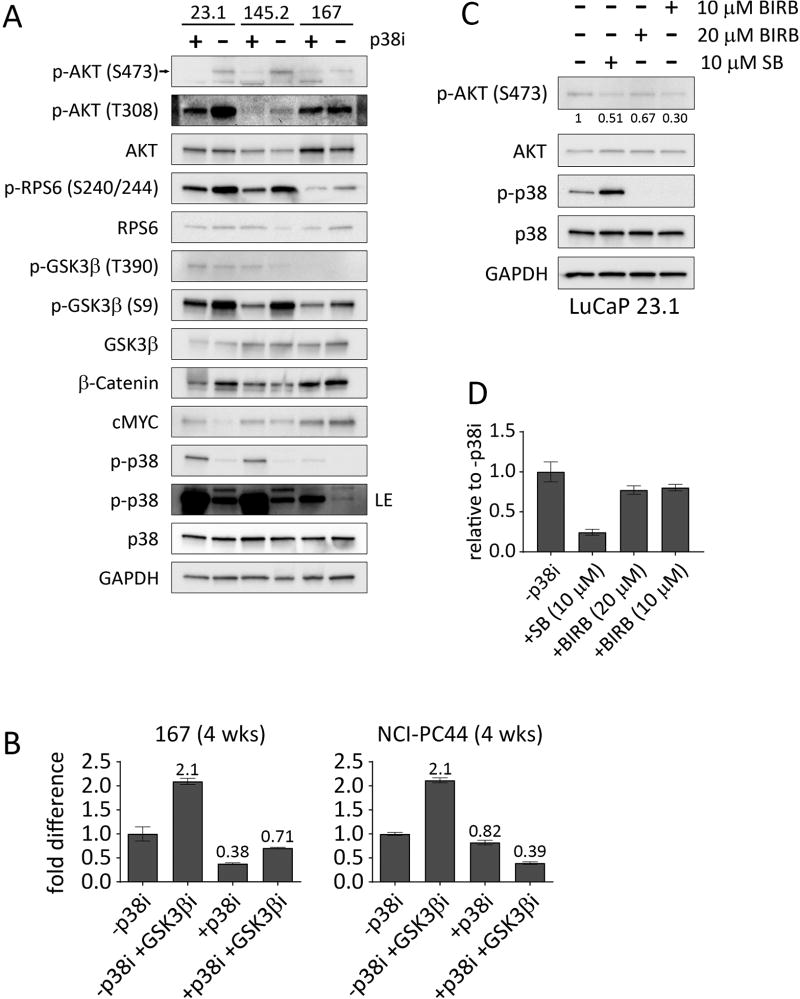
To validate the on-target effects of p38i, we investigated biochemical correlates of p38 activity using three LuCaP models, two adenocarcinomas and one NEPC, that establish in long-term culture. The Wnt pathway is important in PC and features as a prominent driver for organoid culture (17–19,24). p38 can positively regulate the Wnt pathway (28,29), in part as a result of an inactivating T390 phosphorylation on GSK3β (29). p38 is also required for phosphorylation of GSK3β at S9, although it has not been determined whether p38 phosphorylates this site directly (28). We determined the effect of the p38i, SB202190, on the phosphorylation state of GSK3β, by treating organoid cultures with or without the inhibitor for 24 hours (Fig. 3A). Interestingly we found that GSK3β T390 was not affected, but phosphorylation of GSK3β S9 was strongly decreased in the +p38i condition, without any obvious effect on total GSK3β levels (Fig. 3A). Consistent with an increase in GSK3β activity, total β-catenin was decreased in two of the three LuCaP organoids (LuCaP 23.1 & LuCaP 167) following treatment with SB202190, (Fig. 3A). Phosphorylation of MYC by GSK3β can lead to degradation (30); however, MYC expression levels did not correlate with changes to GSK3β phosphorylation status associated with p38i treatment. Also, we observed that the levels of phospho-p38 itself were increased in the p38i-treated organoids. This most likely reflects the fact that SB202190 is able to bind phosphorylated p38 and possibly interfere with phosphatase accessibility (31).
Figure 3.
Addition of p38i to the culture media downregulates the AKT pathway. A, Western blot analysis for the indicated proteins for LuCaPs 23.1, 145.2, and 167 treated with or without 10 µM p38i (SB202190) for 24 hours. B, CellTiter Glo assays for LuCaP 167 and patient-derived organoid line NCI-PC44, showing the relative fold difference between organoids grown for four weeks (passaged once after two weeks) with or without SB202190 and/or a GSK3βi (CHIR 99021). Error bars represent the standard error of the mean of two independent experiments. C, Western blot analysis of the indicated proteins for LuCaP 23.1, either untreated, treated for 24 hours with 10 µM SB202190, or treated with 10 or 20 µM BIRB 796. Quantification of phospho-AKT bands with ImageJ and normalized with total AKT are indicated below the bands. D, Relative fold difference of viability for untreated LuCaPs 23.1 compared to p38i-treated conditions. LuCaPs were grown for two weeks in the −p38i condition, with 10 µM SB202190, or with 10 or 20 µM BIRB 796, then quantified by CellTiter Glo 3D. Error bars represent the standard error of the mean. Two independent experiments were performed.
Since GSK3β S9 is a known site of phosphorylation by AKT (32), we evaluated the activity of AKT in the organoids treated with or without the p38i (Fig. 3A). The active form of AKT, phosphorylated at S473 and T308, was decreased in organoids treated with the p38i, but total AKT levels were unaffected. Phosphorylation of RPS6 at S240/244, a downstream target of AKT via mTOR, was lower in the p38i-treated organoids (Fig. 3A), consistent with p38 inhibition leading to decreased AKT activity.
Based on these results we evaluated if upregulation of GSK3β activity by inhibition of p38 is the mechanism that suppresses growth and survival of the organoids. Using direct inactivation of GSK3β with a chemical inhibitor (CHIR 99021), we found that inhibition of GSK3β had a positive effect on growth. However, it was not sufficient to rescue the inhibition of growth by SB202190, indicating an additional target beyond GSK3β by which p38i decreases growth (Fig. 3B).
To independently validate that the effects of SB202190 were due to inhibition of p38, we compared SB202190 to another specific p38 inhibitor, BIRB 796, that is structurally and mechanistically distinct from SB202190 (31,33). There is little overlap in the non-p38 targets of the two inhibitors (34,35). Treatment of LuCaP 23.1 with BIRB 796 decreased the level of phospho-AKT and decreased growth after two weeks compared to the −p38i condition (Fig. 3C and D). These data support inhibition of p38 as a necessary target for the decreased growth and survival of mCRPC organoids in the presence of SB202190.
Because inhibition of p38 has been reported to negatively affect AR signaling (36), we looked at the effect of SB202190 on AR responsive gene expression in three different adenocarcinoma LuCaP organoid cultures (LuCaP 167, LuCaP 189.3, and LuCaP77) (Supplementary Fig. S6). We did not find any consistent effect of p38 inhibition on AR signaling activity in these three models that are growth inhibited by SB202190. Thus, inhibition of p38 does not appear to act through AR to suppress growth and survival in the LuCaP organoids, nor through AR to indirectly effect the AKT pathway. In summary, consistent with the growth inhibitory effect of p38i, p38 activity is correlated with pathways that positively regulate growth, i.e. AKT and WNT/GSK3β, known targets of the p38 pathway. The specific mechanism by which p38 affects the AKT may be direct, or indirect
Genotypic and phenotypic characteristics are conserved between PDXs and organoids
An important consideration for PDX-derived organoids is how well they maintain the genomic features and subclonal diversity of the original PDX. We chose three LuCaPs (167, 170.2, and 189.3) that have not been genomically characterized previously, and compared PDX tissue to organoids. LuCaP 167 and LuCaP 170.2 were analyzed for copy number variations (CNV) in PDX, organoids, and matched normal tissue. Additionally, all three models (LuCaP 167, LuCaP 170.2, LuCaP 189.3) were assessed for somatic mutations by whole exome sequencing.
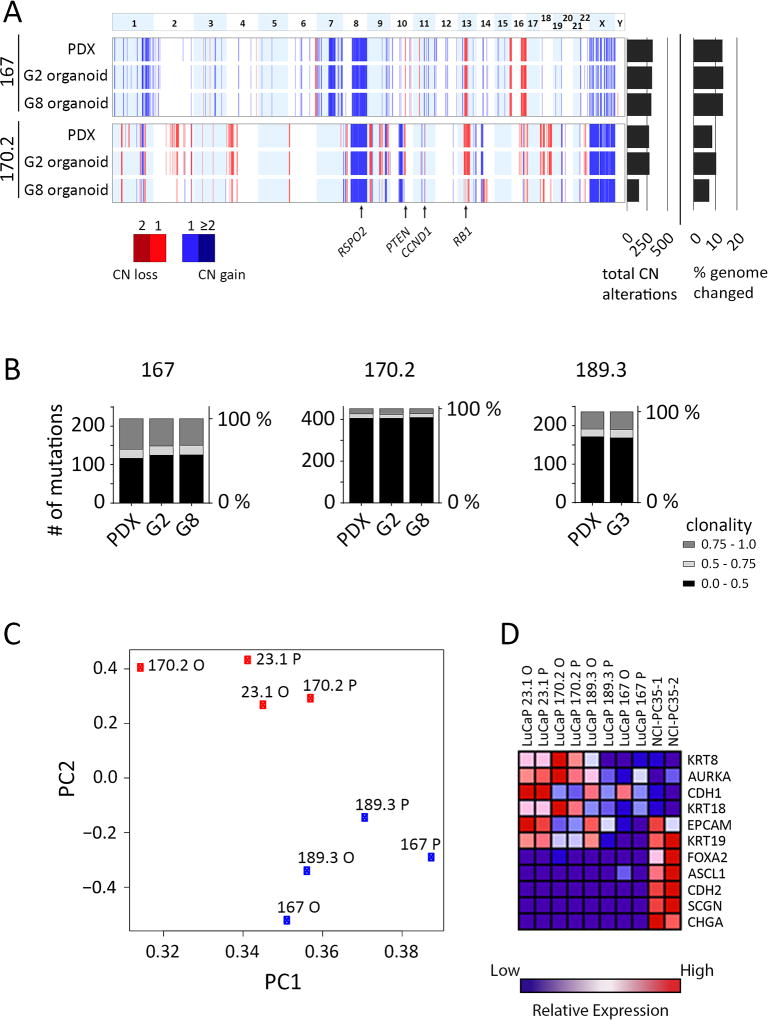
In summary, LuCaP 167 and LuCaP 170.2 showed CNVs characteristic of PC, including gains at chromosome 8q and losses at 8p and 13q (Fig. 4A) (5). LuCaP 167 harbors a hemizygous loss of PTEN, a commonly observed event in PC (3,5). The Wnt pathway is affected in all three models (see Supplementary Table S4): LuCaP 167 has a gain of RSPO2 and a somatic coding mutation in LRP5; LuCaP 170.2 has a gain of RSPO2; LuCaP 189.3 has somatic coding mutations in CTNNB1 and WNT1. LuCaP 167 and LuCaP 170.2 have alterations to the RB pathway. Each has a single copy loss of RB1 and a single copy gain of CCND1. Importantly all the alterations listed above are maintained from the PDX through the late generation organoids (Fig. 4A and Supplementary Table S4).
Figure 4.
Genomic and phenotypic comparisons of parental PDX tumor to organoids. A, Landscape view representing copy number alterations identified in the PDX and organoids for LuCaPs 167 and 170.2. Top panel represents chromosome number. Data for PDX, G2 and G8 organoids are shown. Total copy number aberrations and percentage of genome changed are shown at the right. The locations of specific genes are indicated by arrows below the figure. B, Comparison of exome somatic mutations between PDX and early and later generation organoids. Mutations were filtered so that at least one sample of a given LuCaP contained a minimum fraction of 0.2. Stacked column graphs show the total number of mutations binned by fraction (0.0–0.5, 0.5–0.75 and 0.75–1.0), for each sample. C, LuCaP PDXs and organoids cluster together by transcriptomic profile. Principle component analysis of PDX and organoid gene expression determined by RNA-seq. Proportion of variance PC1 = 0.64, PC2 = 0.07. D, Heatmap of normalized, relative gene expression comparing the indicated PDXs and organoids.
A comparison of PDX to early and late generation organoids by copy number revealed varying but relatively high levels of subclonal stability (Fig. 4A). LuCaP 167 maintained most of the copy number changes from the PDX through late generation organoids (Fig. 4A). LuCaP 170.2 was also highly stable between PDX and G2 organoids, with some apparent loss of clonality by G8, in part affecting losses in chromosomes 2q and 14q. These results were recapitulated by clonal and subclonal somatic mutations in organoids compared to the PDX for LuCaP 167, LuCaP 170.2, and LuCaP 189.3 (Fig. 4B and Supplementary Fig. S7). The existence of fractional variants underscores the presence of subclonal populations within the PDX that are maintained at a high frequency in the organoids. Although some degree of clonal dynamics or genomic instability can be discerned, de novo aberrations were infrequent. Thus, organoid populations preserve the driver mutations and clonal heterogeneity of PDX samples, especially in early generations.
We also compared phenotypic characteristics between PDXs and their matching organoids. Organoid cultures were routinely analyzed for lineage markers by immunohistological analysis, where we observed conservation with the matching PDX (Supplementary Table S5). We have not found organoid morphology to be a marker of PDX histological subtypes. Unlike primary prostate organoids (18), most PDX models grow as solid, filled 3D structures with lumen rarely observed. Additionally we compared organoid-derived xenograft (ODX) tumors to continually passaged PDX tumors and found that the tumor histology, and expression of marker proteins (AR, KRT8, E-cad, CHGA, p63) were the same in ODXs and PDXs (Supplementary Fig. S8). PDX-derived organoids can be reinjected into mice subcutaneously or orthotopically to establish ODXs with 100% efficiency.
To analyze the relatedness in phenotypes between PDXs and organoids, we compared gene expression profiles from four adenocarcinoma models (LuCaP 23.1, LuCaP 167, LuCaP 170.2, and LuCaP 189.3). Principal component analysis separated the models into two subgroups (Fig. 4C), and importantly, matching PDX and organoid models were grouped together. This demonstrates that global patters of gene expression are well-conserved between PDX tumors and organoids. When evaluating specific differentiation markers, we found that LuCaP 170.2 and LuCaP 23.1 demonstrated high relative expression of epithelial markers such as KRT8 and KRT18 compared to LuCaP 167 and LuCaP 189.3 (Fig. 4D), perhaps indicating some degree of epithelial dedifferentiation for LuCaP 167 and LuCaP 189.3. However, LuCaP 167 and LuCaP 189.3 have not progressed to the expression of neuroendocrine markers as shown by comparison to the NCI-PC35.1/.2 NEPC controls.
LuCaP-derived organoids represent an effective model to study mechanisms of resistance to ADT in PC
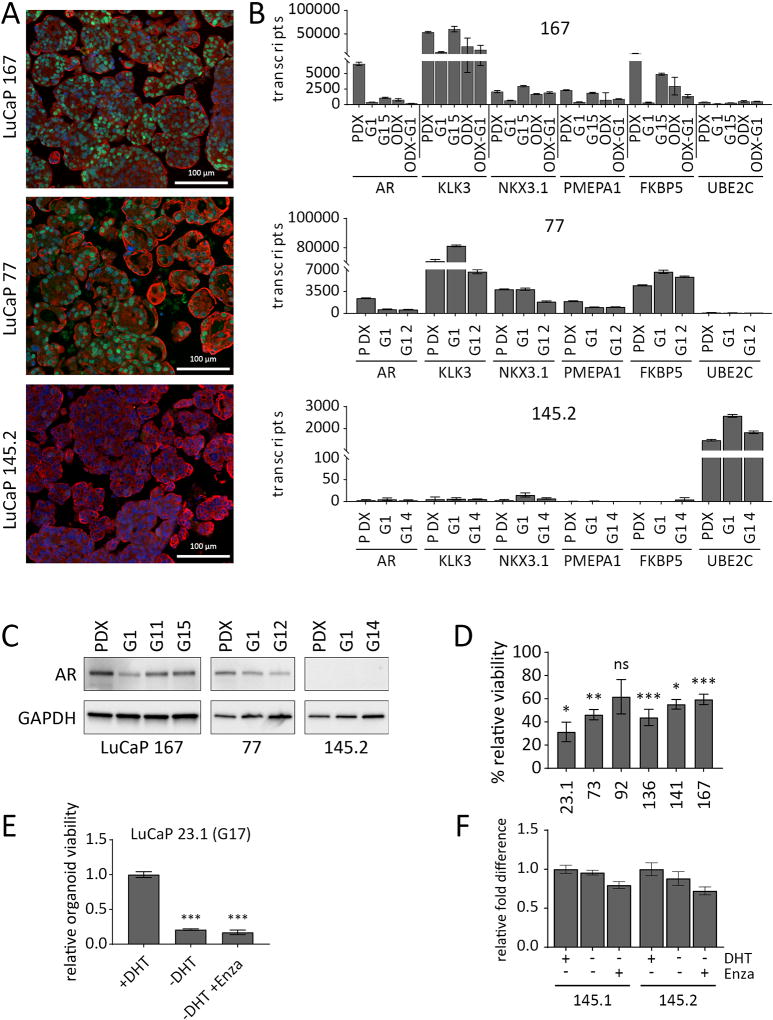
To determine the utility of the PDX-derived organoids as a model of AR-dependence, a cardinal feature of adenocarcinoma CRPC, we analyzed the dynamics of AR and AR-responsive gene expression in organoids derived from highly (LuCaP 167) and moderately (LuCaP 77) castration sensitive PDX models and from an AR-negative neuroendocrine control (LuCaP 145.2) (Fig. 5) (1,4,15). All three LuCaP models expressed cytokeratin 8. AR protein was expressed in the majority of cells within LuCaP 77 and LuCaP 167 organoids, but the level of expression was variable (Fig. 5A), similar to the heterogeneity described for CRPC metastatic tissue samples (37). AR and AR-responsive gene transcript levels were compared in the original LuCaP 77 and LuCaP 167 PDX tumors and early and late generation organoids. For LuCaP 167, organoid-derived tumors, and ODX tumors re-cultured as G1 organoids (ODX-G1) were also compared. AR target genes were expressed across the various LuCaP 77 and LuCaP 167 samples, indicating preservation of AR activity from PDX to organoid to ODX to ODX-G1 (Fig. 5B). Consistent with this, AR protein was expressed in both adenocarcinoma models in the PDXs and organoids, despite decreased AR transcript levels in the organoids (Fig. 5B and C). In accordance with a dependence on AR signaling, 5/6 G1 organoid cultures derived from PDXs that are highly responsive to castration (15), demonstrated significant inhibition by the AR antagonist, enzalutamide (Fig. 5D), which was maintained even after months of continuous passage for LuCaP 23.1 (Fig. 5E). As expected, AR-independent neuroendocrine LuCaPs 145.1 and 145.2 were resistant to enzalutamide inhibition (Fig. 5F). Thus, LuCaP adenocarcinoma organoids maintain the defining feature of AR-dependent gene expression and growth dependence.
Figure 5.
LuCaP organoids maintain AR-dependence over several generations in culture. A, Adenocarcinoma and neuroendocrine organoids stained for AR (green), KRT8 (red) and DAPI (blue). B, qPCR results for expression of AR and AR-response genes. The Y axis indicates absolute copy number. The X axis represents genes tested in PDX tissue, early and late generation organoids, organoid-derived xenograft (ODX) tissue, and ODX-derived G1 organoids (ODX-G1). Error bars represent the standard error of the mean. At least two independent experiments were performed. C, Western blot for AR expression comparing PDX to multiple organoid generations. D, CellTiter Glo assay for G1, LuCaP organoids treated with 10 µM enzalutamide for two weeks. Three independent experiments were performed. E, CellTiter Glo assay for G17, LuCaP 23.1 organoids, treated by DHT withdrawal, with or without 10 µM enzalutamide for two weeks. F, CellTiter Glo assay for LuCaP 145.1 and LuCaP 145.2 organoids grown for two weeks in +DHT, −DHT, or −DHT +enzalutamide (10 µM) conditions. Error bars represent the standard error of the mean of four replicates. *P = 0.01, **P < 0.01, ***P < 0.001 determined by Student’s t-test
LuCaP-derived organoid genotypes correlate with response to the PARP inhibitor olaparib
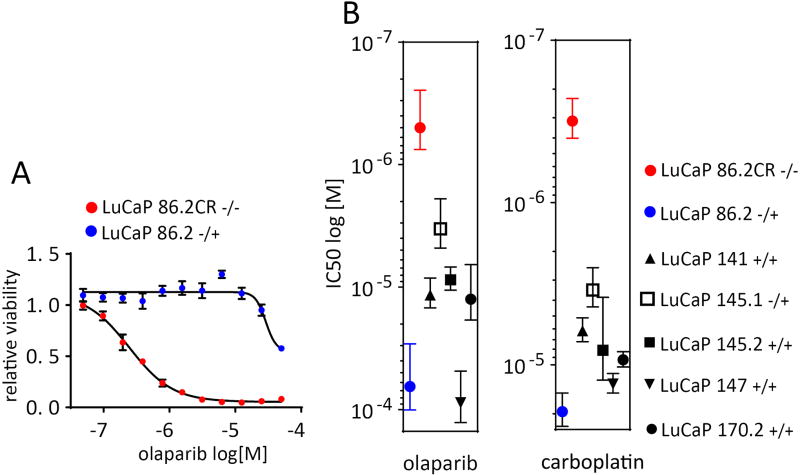
The genetic diversity of the LuCaP cohort provides an expansive preclinical tool. PDX-derived organoids offer a unified platform to study the genetic heterogeneity of CRPC as it relates to comparable drug responses. Recent clinical trials have demonstrated that CRPC patients whose tumors harbor homozygous deletions or loss of function mutations in BRCA2 respond to olaparib as determined by increased time to progression (38). To compare organoids to genotype-determined patient responses, we performed viability dose response assays to olaparib using several LuCaP-derived organoid lines representing three different BRCA2 genotypes (15). LuCaP 86.2 harbors a single copy loss of BRCA2, while LuCaP 86.2CR which is derived from LuCaP 86.2, harbors a two-copy loss. LuCaP 86.2CR was >100 fold more sensitive to olaparib than LuCaP 86.2, as determined by average IC50 (0.48 µM vs 65.0 µM) (Fig. 6A). Additionally, we found that all the LuCaPs that are either wild type or hemizygous for BRCA2, were markedly less sensitive to olaparib than LuCaP 86.2CR (Fig. 6B) paralleling the responses observed in patients. Given that BRCA2 deficient breast and ovarian cancers are sensitive to platinum-based chemotherapy, there is interest in determining the optimal application of platinum therapeutic regimens for CRPC (39). Although promising, the clinical data for genotype-determined platinum responses is limited to small patient numbers, and clinical data does not allow a direct comparison between PARP inhibitors and platinum responses. We performed a parallel dose response experiment using carboplatin in place of olaparib. Carboplatin, like olaparib, induces DNA damage that requires homologous recombination to repair (40). As shown in Fig. 6B, the relative responses of the LuCaP organoids to carboplatin were remarkably similar to olaparib, with 86.2CR clearly separated from the others. Overall these data point to the potential of the LuCaP-derived organoids as a preclinical model for drug screening.
Figure 6.
BRCA2 null LuCaP organoids are hypersensitive to the PARP inhibitor olaparib, and carboplatin. A, Dose response curves for LuCaP 86.2 and LuCaP 86.2CR treated for two weeks with olaparib as quantified by CellTiter Glo 3D. Two independent experiments were performed. A representative figure is shown with error bars showing the standard error of the mean of five replicates. B, IC50 values calculated from dose response curves for the indicated LuCaP organoids treated with either olaparib or carboplatin. Error bars represent the standard error of the mean of two independent experiments.
Discussion
The molecular profiling of CRPC has identified several pathways that are recurrently mutated and enriched relative to primary PC. However, the paucity of genomically-representative PC model systems has limited the ability to deduce general principles concerning the functionality of pathway alterations and their influence on treatment response. The methods to establish LuCaP-derived organoids described here immediately expand the number and diversity of experimentally tractable models. All the LuCaP tumors proliferated during the first generation of organoid culture, a property that makes the individual models amenable to nonbiased discovery screening (e.g. with RNAi or chemical libraries) as well as to genetic alterations and selection. In addition, more than half of the models analyzed can be passaged for at least 10 generations, allowing a variety of experimental manipulations and approaches. Metastatic clinical samples and PDX tumors are characterized by genomic heterogeneity, and some genetic drift is expected during the establishment and passage of PDX tumors (41). However, the similarities of genomic landscapes at the population level comparing CRPC metastatic biopsies and the LuCaP cohort suggest the preservation of driver events (15), which are also maintained in organoids (Fig. 4). Coupled with periodic genetic characterization, the experimental tractability of the PDX/organoid platform allows genetic manipulations to confirm mutation-dependent phenotypes/mechanisms, providing robust models.
A utility of the PDX/organoid platform described here is preclinical therapeutic investigation to address the genetic and phenotypic heterogeneity of CRPC. For a number of solid cancers, recent preclinical studies using PDX clinical trials clearly demonstrated that PDX responses in vivo were predictive of genomically-defined patient responses in co-clinical trials (13). Organoid cultures allow for rapid testing and comparison of multiple individual drugs among several models prior to in vivo analysis. For example, the presence of a homologous recombination deficiency genotype in CRPC and predicted responsiveness to olaparib is the first genomic biomarker driven therapy on track for FDA approval (38). Here we show that BRCA2 deficiency in organoid cultures correlates with sensitivity to olaparib, a result concordant with what is being observed in a clinical setting. Given the responsiveness of BRCA2-deficient breast and ovarian cancers to platinum chemotherapy (40), it is of interest to compare CRPC responses to PARP inhibitors and carboplatin, the latter of which is clinically used for CRPC with aggressive features, but in the absence of genomic biomarkers (39). We observed here remarkably similar responses to olaparib and carboplatin, with no obvious enhanced sensitivity to carboplatin in the LuCaP 145.1 and 145.2 neuroendocrine models. These data support the translational potential of the organoid platform for therapeutic testing.
Interestingly, when investigating the best growth conditions for PC organoids, we found that p38 activity was required for the growth of more than half of the LuCaP organoids as well as the majority of a small sample of CRPC biopsy-derived organoids. We also observed improved growth for some organoids in the absence of the anti-oxidant, NAC. Although the mechanism of this latter effect is unclear, increased reactive oxygen species, anticipated in the absence of NAC, are known to promote various signal transduction pathways, mediated by REDOX-dependent kinases (including p38), phosphatases, proteases, and transcription factors (42). Clearly, the presence of SB202190 and NAC do not entirely prevent prostate organoid establishment since a number of CRPC biopsy-derived organoid lines have been established in NAC/SB202190 containing media (17). However, our results show that the presence of p38i with or without NAC can contribute to selective growth, and we suggest an approach to potentially improve the efficiency of CRPC organoid establishment (Supplemental Fig. S5).
p38 activity correlated with AKT/mTOR pathway activity, a well-established driver of prostate cancer growth (3,5). Although this is a novel observation for prostate cancer signaling, p38-dependent AKT activation (43) or mTOR activation (44) have been described in other contexts. Further studies are required to determine the exact mechanisms of p38-dependent AKT activation in CRPC organoids. In PC cell line models, the influence of p38 activity on the AR pathway has been described both positively, as a result of HSP27 stabilization (36), and negatively, as a result of direct AR phosphorylation and nuclear exclusion (45). However, we found no apparent acute effect of SB202190 on AR target gene expression in three adenocarcinoma organoids.
In summary, the work described here immediately and significantly expands the number of clinically-relevant prostate cancer models available to the community by combining advantages of PDX and organoid formats. Increasing the number and genomic variability of available models will contribute to determining the generality of therapeutic responses and predictive molecular correlates, as well as mechanisms of pathogenesis. The preservation of phenotypic and genotypic characteristics between individual PDX tumors and the derived organoids as well as the experimental tractability of the system supports a robust approach to obtaining translatable pre-clinical data.
Supplementary Material
Translational Relevance.
Significant progress has been made in defining the genomic and molecular properties representing the intra- and interpatient heterogeneity characterizing castrate-resistant prostate cancer (CRPC). Yet, translating this information to basic biological principles and therapeutic vulnerabilities has been constrained by the limited numbers of tractable prostate cancer models. CRPC patient-derived xenografts (PDXs) recapitulate the genetic and phenotypic diversity of the disease, but the full utility of these models for high-throughput and mechanistic analyses has not been realized due to their lack of robust growth in vitro. Here we describe the derivation, the molecular and phenotypic characterization, and the utility for drug testing of a platform of LuCaP CRPC PDX-derived and patient biopsy-derived organoids that markedly expands the number and diversity of in vitro CRPC models. There are immediate implications for therapeutic screening as well as for long-term potential to advance the understanding of fundamental mechanisms underlying disease pathogenesis and resistance to therapy.
Acknowledgments
Financial support
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. A.G.S. is a recipient of a Prostate Cancer Foundation Young Investigator Award and Department of Defense Prostate Cancer Research Program, W81XWH-16-1-0433. Support for LuCaP PDX generation and maintenance: The Prostate Cancer Foundation, NIH PO1 CA163227, the PNW Prostate Cancer SPORE NIH P50 CA097186 and the Department of Defense Prostate Cancer Research Program W81XWH-14-2-0183.
The authors wish to express their gratitude to the patients and the families of the patients who contributed to this study. We would like to thank Drs. Peter Nelson and Ilsa Coleman (Fred Hutchinson Cancer Research Center) for generously sharing the LuCaP PDX RNA-seq data. Additionally, we would like to thank Ross Lake and the LGCP microscopy core at the NCI/CCR.
Footnotes
Conflicts of interest: The authors declare no potential conflicts of interest.
References
- 1.Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer. 2015;15(12):701–11. doi: 10.1038/nrc4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22(3):298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network. Electronic address scmo, Cancer Genome Atlas Research N. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163(4):1011–25. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar A, Coleman I, Morrissey C, Zhang X, True LD, Gulati R, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med. 2016;22(4):369–78. doi: 10.1038/nm.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161(5):1215–28. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boutros PC, Fraser M, Harding NJ, de Borja R, Trudel D, Lalonde E, et al. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat Genet. 2015;47(7):736–45. doi: 10.1038/ng.3315. [DOI] [PubMed] [Google Scholar]
- 7.Sobel RE, Sadar MD. Cell lines used in prostate cancer research: a compendium of old and new lines--part 1. J Urol. 2005;173(2):342–59. doi: 10.1097/01.ju.0000141580.30910.57. [DOI] [PubMed] [Google Scholar]
- 8.Sobel RE, Sadar MD. Cell lines used in prostate cancer research: a compendium of old and new lines--part 2. J Urol. 2005;173(2):360–72. doi: 10.1097/01.ju.0000149989.01263.dc. [DOI] [PubMed] [Google Scholar]
- 9.Parisotto M, Metzger D. Genetically engineered mouse models of prostate cancer. Mol Oncol. 2013;7(2):190–205. doi: 10.1016/j.molonc.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang M, Lin YH, Sun YJ, Zhu S, Zheng J, Liu K, et al. Pharmacological Reprogramming of Fibroblasts into Neural Stem Cells by Signaling-Directed Transcriptional Activation. Cell Stem Cell. 2016;18(5):653–67. doi: 10.1016/j.stem.2016.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aparicio S, Hidalgo M, Kung AL. Examining the utility of patient-derived xenograft mouse models. Nat Rev Cancer. 2015;15(5):311–6. doi: 10.1038/nrc3944. [DOI] [PubMed] [Google Scholar]
- 12.Sachs N, Clevers H. Organoid cultures for the analysis of cancer phenotypes. Curr Opin Genet Dev. 2014;24:68–73. doi: 10.1016/j.gde.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 13.Gao H, Korn JM, Ferretti S, Monahan JE, Wang Y, Singh M, et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med. 2015;21(11):1318–25. doi: 10.1038/nm.3954. [DOI] [PubMed] [Google Scholar]
- 14.Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, Pitts TM, et al. Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol. 2012;9(6):338–50. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen HM, Vessella RL, Morrissey C, Brown LG, Coleman IM, Higano CS, et al. LuCaP Prostate Cancer Patient-Derived Xenografts Reflect the Molecular Heterogeneity of Advanced Disease an--d Serve as Models for Evaluating Cancer Therapeutics. Prostate. 2017 doi: 10.1002/pros.23313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar A, White TA, MacKenzie AP, Clegg N, Lee C, Dumpit RF, et al. Exome sequencing identifies a spectrum of mutation frequencies in advanced and lethal prostate cancers. Proc Natl Acad Sci U S A. 2011;108(41):17087–92. doi: 10.1073/pnas.1108745108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159(1):176–87. doi: 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karthaus WR, Iaquinta PJ, Drost J, Gracanin A, van Boxtel R, Wongvipat J, et al. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 2014;159(1):163–75. doi: 10.1016/j.cell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–5. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 20.van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161(4):933–45. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broutier L, Mastrogiovanni G, Verstegen MM, Francies HE, Gavarro LM, Bradshaw CR, et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med. 2017;23(12):1424–35. doi: 10.1038/nm.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sachs N, de Ligt J, Kopper O, Gogola E, Bounova G, Weeber F, et al. A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity. Cell. 2018;172(1–2):373–86. e10. doi: 10.1016/j.cell.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 23.Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160(1–2):324–38. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drost J, Karthaus WR, Gao D, Driehuis E, Sawyers CL, Chen Y, et al. Organoid culture systems for prostate epithelial and cancer tissue. Nat Protoc. 2016;11(2):347–58. doi: 10.1038/nprot.2016.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526(7574):525–30. doi: 10.1038/nature15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Onnis B, Fer N, Rapisarda A, Perez VS, Melillo G. Autocrine production of IL-11 mediates tumorigenicity in hypoxic cancer cells. J Clin Invest. 2013;123(4):1615–29. doi: 10.1172/JCI59623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bikkavilli RK, Feigin ME, Malbon CC. p38 mitogen-activated protein kinase regulates canonical Wnt-beta-catenin signaling by inactivation of GSK3beta. J Cell Sci. 2008;121(Pt 21):3598–607. doi: 10.1242/jcs.032854. [DOI] [PubMed] [Google Scholar]
- 29.Thornton GP-A Tina M, Deng Bin, Wood C David, Alexander Aronshtam JLC, Sabio Guadalupe, Davis Roger J, Matthews BD Dwight E, Rincon Mercedes. Phosphorylation by p38 MAPK as an Alternative Pathway for GSK3b Inactivation. Science. 2008;320(5876):667–70. doi: 10.1126/science.1156037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sutherland C. What Are the bona fide GSK3 Substrates? Int J Alzheimers Dis. 2011;2011:505607. doi: 10.4061/2011/505607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar S, Boehm J, Lee JC. p38 MAP kinases: key signalling molecules as therapeutic targets for inflammatory diseases. Nat Rev Drug Discov. 2003;2(9):717–26. doi: 10.1038/nrd1177. [DOI] [PubMed] [Google Scholar]
- 32.Luo J. Glycogen synthase kinase 3beta (GSK3beta) in tumorigenesis and cancer chemotherapy. Cancer Lett. 2009;273(2):194–200. doi: 10.1016/j.canlet.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pargellis C, Tong L, Churchill L, Cirillo PF, Gilmore T, Graham AG, et al. Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site. Nat Struct Biol. 2002;9(4):268–72. doi: 10.1038/nsb770. [DOI] [PubMed] [Google Scholar]
- 34.Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, et al. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408(3):297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fabian MA, Biggs WH, 3rd, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23(3):329–36. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 36.Zoubeidi A, Zardan A, Beraldi E, Fazli L, Sowery R, Rennie P, et al. Cooperative interactions between androgen receptor (AR) and heat-shock protein 27 facilitate AR transcriptional activity. Cancer Res. 2007;67(21):10455–65. doi: 10.1158/0008-5472.CAN-07-2057. [DOI] [PubMed] [Google Scholar]
- 37.Roudier MP, True LD, Higano CS, Vesselle H, Ellis W, Lange P, et al. Phenotypic heterogeneity of end-stage prostate carcinoma metastatic to bone. Hum Pathol. 2003;34(7):646–53. doi: 10.1016/s0046-8177(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 38.Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez R, et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Engl J Med. 2015;373(18):1697–708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pomerantz MM, Spisak S, Jia L, Cronin AM, Csabai I, Ledet E, et al. The association between germline BRCA2 variants and sensitivity to platinum-based chemotherapy among men with metastatic prostate cancer. Cancer. 2017;123(18):3532–9. doi: 10.1002/cncr.30808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konstantinopoulos PA, Ceccaldi R, Shapiro GI, D'Andrea AD. Homologous Recombination Deficiency: Exploiting the Fundamental Vulnerability of Ovarian Cancer. Cancer Discov. 2015;5(11):1137–54. doi: 10.1158/2159-8290.CD-15-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ben-David U, Ha G, Tseng YY, Greenwald NF, Oh C, Shih J, et al. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat Genet. 2017;49(11):1567–75. doi: 10.1038/ng.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holmstrom KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat Rev Mol Cell Biol. 2014;15(6):411–21. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 43.McGuire VA, Gray A, Monk CE, Santos SG, Lee K, Aubareda A, et al. Cross talk between the Akt and p38alpha pathways in macrophages downstream of Toll-like receptor signaling. Mol Cell Biol. 2013;33(21):4152–65. doi: 10.1128/MCB.01691-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schewe DM, Aguirre-Ghiso JA. ATF6alpha-Rheb-mTOR signaling promotes survival of dormant tumor cells in vivo. Proc Natl Acad Sci U S A. 2008;105(30):10519–24. doi: 10.1073/pnas.0800939105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gioeli D, Black BE, Gordon V, Spencer A, Kesler CT, Eblen ST, et al. Stress kinase signaling regulates androgen receptor phosphorylation, transcription, and localization. Mol Endocrinol. 2006;20(3):503–15. doi: 10.1210/me.2005-0351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.