Abstract
Clinical application of antigen-specific T regulatory cells (Tregs) offers promise for the treatment of undesirable immune diseases. To achieve this goal, long-term expansion of Tregs is required to obtain sufficient numbers of cells. However, human Tregs are not stable ex vivo. Therefore, we previously developed an innovative Treg expansion protocol using phosphorothioated random oligonucleotides (ODNps25). The addition of ODNps25 successfully resulted in the stabilization of engineered antigen-specific Tregs; however, the mechanism is not fully characterized. We first identified SAMHD1 (sterile alpha motif and HD domain 1) as an ODNps25 binding protein using a UV-crosslinking pull-down strategy. SAMHD1 physically interacted with the 3’ untranslated region (UTR) of Foxp3 mRNA and was translocated from nucleus to cytoplasm after ODNps25 treatment. Importantly, addition of ODNps25 enhanced the interaction of SAMHD1 and Foxp3 mRNA significantly, and this interaction was increased by TCR stimulation. Since ODNps25 binds to the nuclease (HD) domain of SAMHD1, we then established that overexpression of a dNTPase-deficient mutant (D137N) in Tregs significantly stabilized the expression level of Foxp3 protein. Furthermore, we also found that TCR stimulation up-regulates phosphorylation of the threonine residue (Thr592), which is a regulatory site to control SAMHD1 activity, and phosphorylation of Thr592 is critical to control SAMHD1 activity to stabilize the expression of Foxp3 and Helios in Tregs. Taken together, we suggest that the interaction of ODNPs25 in HD or phosphorylation of Thr592 by TCR stimulation interferes with nuclease activity of SAMHD1, thereby stabilizing 3’ UTR of Foxp3 and Helios mRNAs in long-term culture.
Introduction
Regulatory T cells (Tregs) expressing the transcription factor Foxp3 are a distinct subset of CD4+ T cells that regulate immune homeostasis and suppress harmful autoimmunity(1). Cellular therapy with Tregs that have been expanded in vitro has emerged as a potential therapy of autoimmune disease. Foxp3+ Tregs isolated from peripheral blood or umbilical cord are the major sources for the in vitro production of human Tregs for therapeutic purposes. Unfortunately, the in vitro expansion of Tregs is frequently complicated by the loss of Foxp3 expression and loss of Treg suppressive function.
We have previously demonstrated that Foxp3+ Tregs co-expressing the transcription factor Helios (a marker of thymus-derived Tregs) can be expanded in culture by the addition of random sequence phophorothiate backbone 25 base-pair oligodeoxynucleotides (ODNps25). Tregs expanded in the presence of the ODNps25 are epigenetically stable and immunosuppressive in vitro(2). Furthermore, the addition of ODNps25 also stabilized human Factor VIII (FVIII)-specific engineered Treg during the expansion of Tregs that had been retrovirally transduced with a FVIII specific TCR. These engineered, ODNps25-stabilized, Tregs suppressed proliferation and cytokine production by FVIII-specific T effector cells as well as the ex vivo production of FVIII-specific antibody from FVIII-immunized HLA DR1 mice(3).
The stabilization of Foxp3 expression by ODNps25 was not mediated by cytosolic Toll-like receptor (TLR) signaling. The accumulation of the ODNps25 in the cytoplasm of Treg raised the possibility that it was mediating the stabilization of Foxp3 expression by binding to a cytoplasmic receptor. Here we demonstrate that the receptor for the ODNps25 is SAMHD1 (sterile alpha motif histidine-aspartate-domain containing protein 1). SAMHD1 is ubiquitously expressed in cells of the immune system and functions as a triphosphohydrolase enzyme that degrades deoxyribonucleoside triphosphosphatases (dNTPS) (4, 5). SAMHD1 has been best characterized as a virus restriction factor, particularly HIV-1(6–11). Mutations in SAMHD1 are associated with the Aicardi-Goutieres Syndrome (AGS), a systemic autoimmune disease with a hyperactive innate immune response(12). Here, we demonstrate that SAMHD1 plays a critical role in the post-transcriptional processing of Foxp3 and Helios mRNAs in the cytoplasm by binding to their 3’UTRs leading to their degradation. The dNTPase activity of SAMHD1 can be inhibited by TCR induced activation of CDK2 leading to Thr592 phosphorylation of SAMHD1 or by inhibiting the binding of SAMHD1 to the 3’UTRs by ODNps25.
Materials and Methods
General reagents, human peripheral blood
Recombinant human IL-2 was from the NCI Biological Resources Branch Preclinical Repository (NCI, Frederick, MD). Synthesis of phosphorothioate-backboned random deoxyoligonucleotide (ODNps25) followed the previous method(2). Buffy-coat fractions (20–70 year-old healthy men and women) were provided by Department of Transfusion Medicine at the National Institutes of Health (NIH, Bethesda, MD) or purchased from the American Red Cross (Rockville, MD). All procedures were approved by the Uniformed Services University of the Health Sciences Institutional Review Board. All blood donors provided written informed consent in accordance with the Declaration of Helsinki.
ODN/ORN Interference assay
Fluorescein (FITC)-conjugated ODNps10 (ODNps10-FITC, 2 µM) and ODNpe25 (ODNpe25-FITC, 2 µM) were co-added with phycoerythrin (PE)-conjugated ODNps25 (ODNps25-PE, 2 µM) in 3 wk-expanded Tregs, followed by 24 hr-incubation in rIL-2 (200 IU/ml). Incubated cells were extracellular-stained with human CD4 and viability dye and fixed for the FACS analysis. For ODN/ORN competition experiment, indicated dose of biotin-conjugated ODNps25 (Biotin-ODNps25) and ODNps10, or ORN (oligoribonucleotide) ps25 were co-added and incubated as above, respectively. Extracellular CD4-stained cells were fixed, permeabilized, and intracellular-stained with Streptavidin-PE for flow cytometry. Conjugation of fluorochromes or biotin at 3’ end of oligonucleotides was obtained from Integrated DNA Technologies (IDT).
Preparation and expansion of polyclonal T effectors, polyclonal Tregs, and 17195 Tregs
For the preparation of naive T cells (CD4+CD25−CD127+CD45RA+) and Tregs (CD4+CD25hiCD127lo), peripheral blood mononuclear cells (PBMCs) were separated using FACS sorter (FACSAria II, Becton Dickinson) from positively MACS-enriched human CD4+ T cells (Miltenyi Biotech). Native T cells and Tregs were stored in liquid nitrogen until use. Naïve T cells or Tregs (1×106) were pre-stimulated for 48 hrs with anti-CD3ε (5 µg/ml) and anti-CD28 (1 µg/ml) antibodies in RPMI1640 media complemented with 10% Fetal bovine serum (FBS) and recombinant IL-2 (rIL-2, 200 IU/ml). ODNps25 (2 µM) were added in a case of required condition. To generate 17195 Tregs, Tregs were spin-fected (1500 × g, 2 hr, 32° C) with retroviral 17195 TCR in retronectin (10 µg/ml, Clontech) plate. The transduced cells were further expanded with rIL-2 for 9 to14 days.
Pull-down assay of Biotin-ODN/protein complexes after UV-exposed crosslinking
Expanded Tregs (4×107) were co-incubated for 36 hrs with Biotin-ODNps25 (2.5 µM) and ODNps6 (10 µM) together in culture media with rIL-2 (200 IU/ml). Ultraviolet (UV) crosslinking of proteins to ODNs in living cells were performed with one-time UV exposure (150 mJ/cm2) at 254 nm using stratalinker (Stratagene). Cross-linked cells were transferred into two tubes with equal cell number (A and B in Fig 2a), and further crosslinking was performed in B tube with cell permeable disuccinimidyl suberate (DSS, 5 mM) for 30 min at room temperature. Cross-linked Proteins in both A and B tube were extracted by using IP buffer (Invitrogen), and immunoprecipitated by adding streptavidin-conjugated magnetic pull-down beads (50 µl, Dynabeads M-280 Streptavidin, Invitrogen).
Figure 2. SAMHD1 is a cytoplasmic ODNps25 receptor to stabilize co-expression of Foxp3 and Helios in human Tregs.
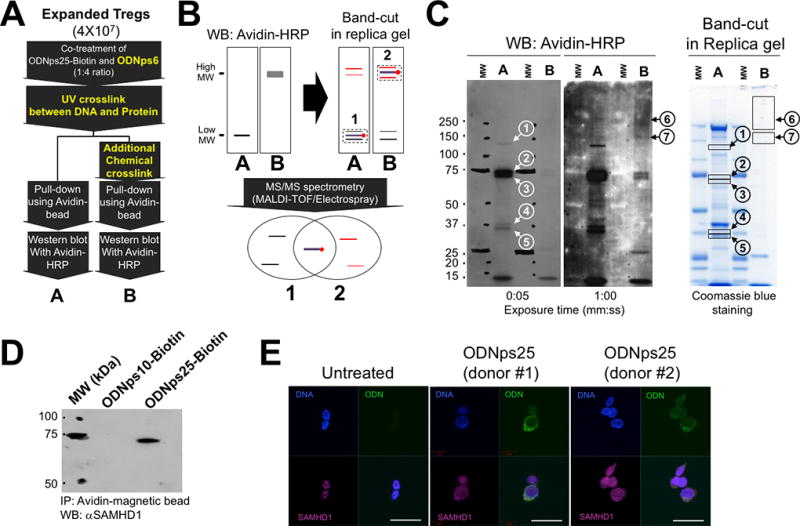
(A) Two-step cross-link strategy to maximize pull-down efficacy of ODNps25-Biotin-interacting proteins. To crosslink ODNps25-biotin and its interacting protein, cells were exposed by UV (150 mJ/cm2), then incubated with disuccinimidyl suberate (DSS, 5 mM) for 30 min at room temperature. ‘A’ and ‘B' represent avidin-dependent pull-down proteins from single UV-crosslinked Tregs and two step cross linked Tregs by UV and DSS, respectively. (B) The principle to Identify proteins crosslinked with ODNps25-biotin covalently by comparing two different pull-down samples (A and B in Fig. 2a). Briefly, the gel positions of ODNps25-biotin and protein covalent complex would be determined by western blot of A and B samples with avidin-HRP, and the protein bands in sliced gel bands from replica Coomassie blue stained gel will be identified by MS/MS spectrometry. Overlapped protein IDs from the list of A and B band-cuts (1 and 2) would be considered as an ODNps25 receptor candidates, and the rest IDs in the lists will be excluded as a non-specific gel slice contaminants. Black thin line and red thin line represent gel contaminant with low molecular weight and with high molecular weight, respectively. (C) Western blot of A and B pull-down samples with avidin-HRP (left two membranes) and band-cut from replica SDS-PAGE gel (right Coomassie blue-stained gel). The numbers in the circle indicate specifically detected bands in A samples (1–5) and B samples (6 and 7) after blotting with avidin-HRP (1:10,000). The closed squares indicate the gel-cut positions corresponding to western bands in replica gel. pull-down samples were prepared as same as in Fig.2C. (D) Confirmation of SAMHD1 binding to ODNps25-biotin using avidin-dependent pull-down and western blot with anti-hSAMHD1 antibody (1:2,000). Data shown is one of two independent experiments with different donors. (E) Confocal microscopy of endogenous SAMHD1 (red) in ODNps25-FITC (green) treated expanded Tregs. Nuclei were counterstained with DAPI (blue). Scale bars, 10 µm. Data shown is one of three independent experiments with different donors.
RNA-protein complex immunoprecipitation
RNA-protein complexes were performed with an RNA ChIP-IT kit (Active Motif) using anti-SAMHD1 antibody (Novus Biologicals) according to the manufacturer’s instructions. The RNAs recovered from the complexes were subjected to RT-PCR and qRT-PCR using primers 5’-CCT CCC CCA TCA TAT CCT TT-3’ and 5’-TTG GGG TTT GTG TTG AGT GA-3’ for Foxp3 3’-UTR; and 5’-GAT TTG GAG ACC TCC GCT AA-3’ and 5’-CAG ACA CAC ACT CAC ACA TGC-3’ for Helios 3’-UTR.
Immunofluorescence staining
Expanded Tregs were incubate with the ODNps25-FITC (2 µM) in rIL-2 (200 IU/ml) for 24 hrs. 17195 TCR transduced Tregs were re-stimulated with anti-CD3ε antibody or FVIII C2 peptide for 36hrs, followed by fixation for 20 min in 4% paraformaldehyde. Fixed cells were stained with anti-SAMHD1 antibody (1:200, Abcam) and Fluor 546-conjugated secondary antibody subsequently (Invitrogen) (1:500). For confocal microscopic analyses, fixed and stained cells were mounted with Vectashield mounting medium (Vector Laboratories, Burlingame, CA). Images were taken by LSM 710 Laser Scanning Microscope (Carl Zeiss, Germany) and processed with ZEN 2009 software.
SDS-PAGE and western blot analysis
Protein samples (5 µg) or pull-down pellets was boiled in reduced sample buffer, and electrophoresed on 4 to 12% Tris-glycine gel (NuPAGE, Invitrogen). To cut the gel slice, gel was stained in coomassie blue solution (SimplyBlue Safestain, Invitrogen). For western blot analysis, protein bands separated is blotted in PVDF membrane, and subsequently probed in primary antibodies against human SAMHD1(1:500, Novus Biologicals), Phospho-SAMHD1 (Thr592) (1:500, Cell Signaling Technology) and Avidin-HRP (1:2000, Cell Signaling Technology), followed by secondary antibody: anti-mouse IgG-HRP (1:2000) and anti-Rabbit IgG-HRP (1:2000). specific band were detected in autoradiography using enhanced chemiluminescence (ECL, Thermo Scientific). For housekeeping control, anti-actin-HRP (1:10,000, Santa Cruz Biotechnology) was used.
RNA preparation, RT-PCR, and qRT-PCR
Total RNA was isolated with the RNeasy Mini kit (QIAGEN) and reverse transcribed by the SuperScript III First-Strand Synthesis System (Invitrogen) with random hexamers. qRT-PCR was performed in triplicate with FastStart Universal SYBR Green Master Premix (Roche) on a 7500 Real-Time PCR System (Applied Biosystems). Relative differences in RNA expression levels were calculated according to the 2−ddCt method and normalized against GAPDH mRNA. The PCR primers used were 5’-TTC TGT CAG TCC ACT TCA CCA-3’ and 5’-AGG TCT GAG GCT TTG GGT G-3’ for Foxp3; 5’-GCT GAG ATC TCC CGA CAG AG-3’ and 5’-CAG GGG TTT CCT GTC ACA CT-3’ for Helios; and 5’-CCA TGG AGA AGG CTG GGG-3’ and 5’-CAA AGT TGT CAT GGA TGA CC-3’ for GAPDH mRNA.
Constructions of retroviral wild-type and mutant SAMHD1s
Dr. Sarah Welbourn and Dr. Klaus Strebel (NIAID, bethesda) kindly provided mammalian expression vectors of Wild type SAMHD1 (SAMWT) and SAMHD1 D137N (SAM137N), and nucleotide sequences for direct point mutagenesis of SAMHD1 T592A (SAMT592A) (sense, 5’-gttatagccccactcatagcacctcaaaaaaagg-3’; antisense, 5’-cctttttttgaggtgctatgagtggggctataac-3’) and SAMHD1 T592E (SAMT592E) (sense, 5’-gttatagccccactcatagaacctcaaaaaaagg-3’; antisense, 5’- cctttttttgaggttctatgagtggggctataac-3’)1. To construct retroviral expression of SAMWT and SAM137N, Inserts of SAMWT and SAMl137N were amplified by polymerase chain reaction (PCR), and transferred into retroviral expression vector (pRetro-X-IRES-Zsgreen, Clontech). For the construction of SAMT592A and SAMT592E, mutant strands were synthesized by hybridization of sense, antisense nucleotides, and pRetro-X-SAMWT-IRES-Zsgreen vector, and then followed by thermal cycling amplification (QuikChange II XL Site-Directed Mutagenesis Kit, Stratagen). The protocol in detail is followed by manufacturer’s instruction.
Flow cytometry
For intracellular staining of Foxp3 and Helios, cells were extracellular stained with anti-CD4-PECy7 (RPA-T4, TONBO Biosciences) and Fixable Viability Dye eFluor780 (eBioscience) in FACS buffer, followed by fixation with 2% paraformaldehyde solution for 10 min at 37°C. After permeabilization in 0.1% Triton X-100 overnight, cells were further stained intracellularly with antibodies for Foxp3-APC (236A/E7, eBioscience) and Helios-PE (22F6, Biolegend). All FACS data were collected by FACSDiva software (Becton Dickinson) using instrument BD LSRII (Becton Dickinson), and analyzed using FlowJo software (Flowjo).
Statistics
Statistical number were calculated using Prism (GraphPad Software) and Excel (Microsoft). Normality was confirmed using one-way ANOVA. Students’ t test was used to determine significant differences between groups. *, p<0.05; **, p<0.01; ***, p<0.001. The details of statistical analyses are described in the figure legends.
Results
Identification of an intracellular receptor for ODNps25 with an ODN/ oligoribonucleotide (ORN) interference assay
In our previous study, we demonstrated that the addition of ODNps25 resulted in the maintenance of suppressive human Foxp3+Helios+ Tregs ex vivo. The accumulation of the ODNps25 in the cytoplasm of Treg suggested that it might be binding to a cytoplasmic receptor. However, the existence of non-specific cytoplasmic and nuclear nucleotide-binding proteins renders the identification of an ODNps25-specific receptor in Tregs difficult. To address this problem, polyclonal human Tregs were expanded by stimulation with anti-CD3ε/CD28 antibodies in the presence of rIL-2 for 3 weeks. PE-conjugated ODNps25 (ODNps25-PE), ODNps10-FITC, or ODNpe25-FITC were then added and the accumulation of all three molecules was analyzed after 24 hours. Only ODNps25 showed specific accumulation in Treg cells and neither the short-length ODN (ODNps10) nor the phosphodiester backboned ODN (ODNpe25) accumulated to a significant extent (Fig. 1A). Furthermore, we established an ODN/ORN interference assay in which different sizes of ODNs or ORN were added as competitors to block the binding of biotinylated ODNps25 (Biotin-ODNps25). The relative binding interference was measured by intracellular flow cytometry of biotinylated-ODNps25 24 hrs later. Receptor binding of biotin-ODNps25 was proportionally diminished by increasing concentrations of ODNps25, but not by ODNps10 or by phosphorothioated oligoribonucleotide (ORNps25) (Fig. 1BC). The results clearly indicate that an intracellular receptor (s) in Treg cells preferentially binds to ODNps25 rather than ORNs or short length ODNs.
Figure 1. ODNps25 receptor specifically binds to phosphothioate backboned deoxyoligonucleotides.
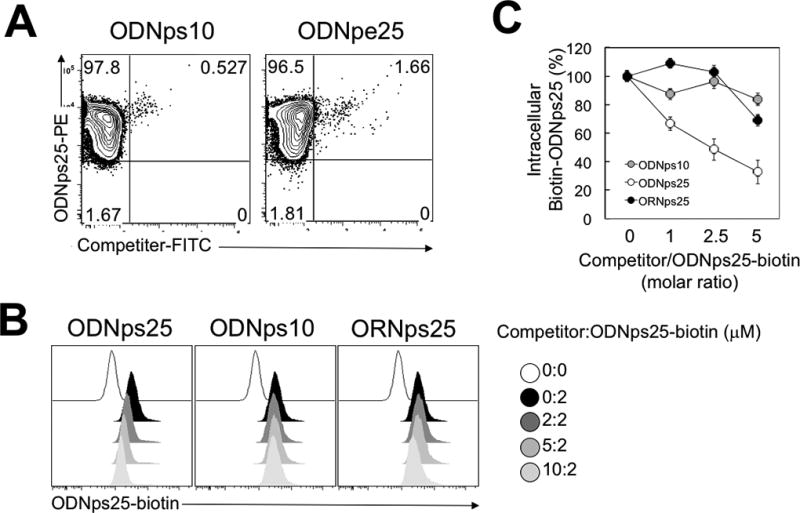
(A) Intracellular uptake analysis of competitor-FITC and ODNps25-PE in co-treated human Tregs. FACS analysis was performed at 24 hrs after co-treatment of competitor-FITC and ODNps25-PE. ODNpe25, 25 base-pair length phosphorodiester backboned deoxynucleotide. Data shown is one of three independent experiments with different donors. (B) Competitive interaction between ODNps25 receptor and Biotin-ODNps25 in the presence of interfering oligonucleotides. Uptake of Biotin-ODNps25 was measured by intracellular staining with PE-conjugated streptavidin. ODNps25, 25 base-pair length phosphorothioate backboned deoxynucleotide; ODNps10, 10 base-pair length phosphorothioate backboned deoxynucleotide; ORNps25, 25 base-pair length ribonucleotide; ODNps25-biotin, ODNps25 conjugated with biotin at 5’-end. Data shown is one of three independent experiments with different donors. (C) Summarized graph of histogram shown in (B). Intracellular Biotin-ODNps25 (%) in y axis is calculated by [MFI (indicated ratio)−MFI (0:0)]/ [MFI (0:2)−MFI (0:0)] ×100. Dots indicate mean ± SD in the triplicate.
Isolation of the ODNps25 receptor using pull-down technology
We next attempted to identify the ODNps25 receptor using an avidin-mediated pull-down method after addition of biotin-ODNps25 to Tregs. Treg cells were first expanded for two weeks by stimulation with anti-CD3ε/CD28 and IL-2 in the presence of ODNps25. The expanded Tregs were then cultured for 36 hours in IL-2 alone in the presence of Biotin-ODNps25 prior to the pull-down assay. To minimize potential non-specific binding of Biotin-ODNps25, a four-fold molar excess of ODNps6 was added together with biotin-ODNps25 (Step 1 in Fig. 2A). The biotin-ODNps25 was covalently cross-linked with potential receptor proteins by UV exposure, which enabled photoreaction-mediated covalent crosslinking of thymines of the ODN DNA to cysteine and lysine in binding proteins (Step 2 in Fig. 2A). Complexes of the biotin-ODNps25 and its receptor were then pulled down with avidin-magnetic beads (A in Fig. 2A and Fig. 2B), followed by analysis on Western blotting with avidin-HRP. Five distinct bands were detected (bands 1–5 in A of Fig. 2C). To eliminate non-specific protein contamination in cut gel slices, the bands detected in lane A were “upshifted” by incubation with additional cell-permeable chemical cross-linker and further exposure to UV (B in Fig. 2A and Fig. 2B). In the upshifted lane, the five bands detected in Lane A were no longer detected and two upshifted high molecular weight bands were seen (6 and 7 in B of Fig. 2C). The seven bands in lane A and B were sliced from replica Coomassie stained gels, and their protein identities (IDs) were analyzed by MS-to-MS spectrometry analysis. Protein IDs specific to ODNps25 were re-sorted by overlapping lists in lane A slices and in lane B. Final selected binding candidates for specific binding to ODNps25 are summarized in Table 1. Identified candidates were grouped in four categories based on their characteristic functions: mitochondrial enzymes with biotin cofactor, photo damage-related proteins, heterogeneous ribonucleoproteins, and RNA-processing proteins. We selected SAMHD1 for further study based on its nuclease function and potential to control transcriptomes in Tregs.
Table 1.
Summarized pull-down proteins commonly identified in both UV cross-linked bands (1–5) and upshifted extra cross-linked bands (6, 7)
| Category | Accession | Score | Descriptions |
|---|---|---|---|
|
| |||
| Mitochondrial enzymes with Biotin cofactor | MCCB_HUMAN | 348.2093251 | (Q9HCC0) Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial |
| Mitochondrial enzymes with Biotin cofactor | MCCA_HUMAN | 249.9351293 | (Q96RQ3) Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial |
| Mitochondrial enzymes with Biotin cofactor | PCCA_HUMAN | 93.580363 | (P05165) Propionyl-CoA carboxylase alpha chain, mitochondrial |
| Mitochondrial enzymes with Biotin cofactor | DLDH_HUMAN | 165.104467 | (P09622) Dihydrolipoyldehydrogenase, mitochondrial |
| Photo damage-related proteins | ARGI1_HUMAN | 349.6449536 | (P05089) Arginase-1 |
| Photo damage-related proteins | CASPE_HUMAN | 198.6111486 | (P31944) Caspase-14 |
| Heterogeneous ribonucleoproteins | ROA2_HUMAN | 1363.407797 | (P22626) Heterogeneous nuclear ribonucleoproteinsA2/B1 |
| Heterogeneous ribonucleoproteins | ROA1_HUMAN | 579.4896169 | (P09651) Heterogeneous nuclear ribonucleoproteinA1 |
| Heterogeneous ribonucleoproteins | HNRPU_HUMAN | 153.036719 | (Q00839) Heterogeneous nuclear ribonucleoproteinU |
| Heterogeneous ribonucleoproteins | ROA0_HUMAN | 122.6228502 | (Q13151) Heterogeneous nuclear ribonucleoproteinA0 |
| Heterogeneous ribonucleoproteins | HNRDL_HUMAN | 80.05434487 | (O14979) Heterogeneous nuclear ribonucleoproteinD-like |
| RNA-processing proteins | DHX9_HUMAN | 113.3291351 | (Q08211) ATP-dependent RNA helicase A |
| RNA-processing proteins | EF1A1_HUMAN | 82.56966189 | (P68104) Elongation factor 1-alpha 1 |
| Stress-Related proteins | CH60_HUMAN | 470.962906 | (P10809) 60 kDaheat shock protein, mitochondrial |
| Stress-Related proteins | GRP75_HUMAN | 149.0790893 | (P38646) Stress-70 protein, mitochondrial |
| SAMH1_HUMAN | 349.0241875 | (Q9Y3Z3) SAM domain and HD domain-containing protein 1 | |
SAMHD1 binds ODNps25 in the cytoplasm
SAMHD1 has been characterized as a nuclease resident in the nucleus, but its cellular function remains poorly characterized. To address the potential functional role of SAMHD1 in Tregs, we first confirmed physical binding of ODNps25 with SAMHD1 by probing a Western blot with anti-SAMHD1 antibody after pull-down of UV cross-linked short biotin-ODNps10 or biotin-ODNps25 in Tregs. Physical interaction was specifically detected only in biotin-ODNps25 pull-down samples, but not in biotin-ODNps10 samples (Fig. 2D). Because we have previously shown that ODNps25 specifically accumulates in the cytoplasm of Tregs after treatment(2), we tested whether SAMHD1, which is normally found only in the nucleus, can be co-localized with ODNps25 in the cytoplasm. FITC-conjugated ODNps25 (FITC-ODNps25) was added to Tregs, followed by immunostaining of SAMHD1. In the absence of ODNps25, endogenous SAMHD1 was predominantly detected in the nucleus of Tregs (untreated in Fig. 2E). However, in the presence of FITC-ODNps25, SAMHD1 was localized in the cytoplasm and co-localized with FITC-ODNps25 (Fig. 2E). Taken together, we concluded that SAMHD1 physically interacts with ODNps25 in cytoplasm of human Tregs.
ODNps25 stabilize Foxp3 and Helios expression by inhibiting SAMHD1 dNTPase function
As the primary function of SAMHD1 is secondary to its dNTPase activity that is regulated allosterically following dGTP binding, we first examined if the lack of stability of expression of Foxp3 and Helios during Treg expansion required the allosteric regulation of SAMHD1. We prepared retroviral GFP-reporter constructs encoding wild-type SAMHD1 (SAMWT) and a dNTP-deficient SAMHD1 mutant (SAM137N) which is not subject to allosteric regulation (Fig. 3A). Pre-stimulated polyclonal Tregs were then transduced with the retroviral vector alone (Mock), SAMWT vector, SAM137N vector, or with ODNps25 and expanded for 3 weeks in the presence of anti-CD3ε antibody and rIL-2. Intracellular Foxp3 and Helios were analyzed in transduced and untransduced cells, respectively (GFP+ and GFP− cells in Fig. 3B). The Mock Treg group showed significant loss of Foxp3+Helios+ cells and this reduction was prevented by addition of ODNps25. Importantly, the reduction was minimally reversed by overexpression of SAMWT, but Tregs transduced with the dNTPase-deficient SAM137N construct retained high percentages of Foxp3+Helios+ cells (Fig. 3E and top panels in Fig. 3C) whereas untransduced GFP− Tregs showed no significant difference in each group (Fig. 3D). Taken together, these results strongly suggest that the dNTPase activity of SAMHD1 is responsible for the instability of Foxp3 and Helios expression in Treg.
Figure 3. The blockage of dNTPase activity of SAMHD1 retains co-expression of Foxp3 and Helios in human Tregs.
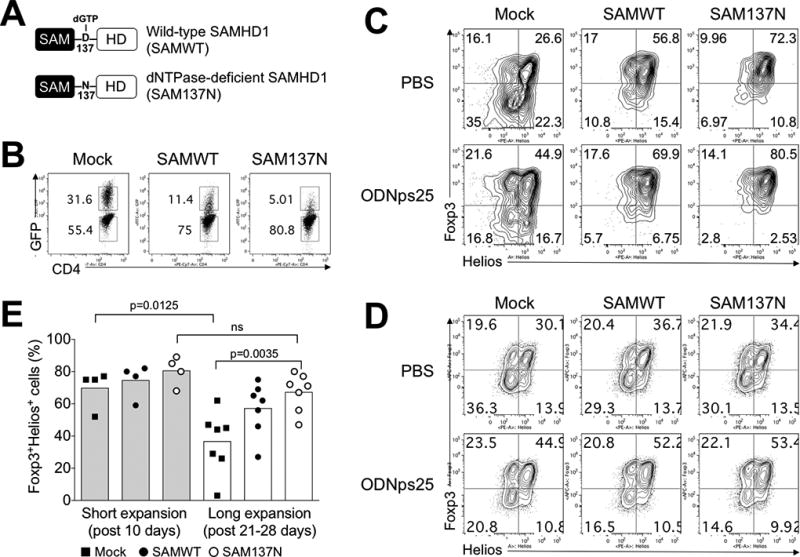
(A) Expression constructs of Wild-type SAMHD1 (SAMWT) and dNTPase-deficient SAMHD1 (SAM137N). To construct retroviral SAMWT and SAM137N, HA-SAMHD1 and HA-SAMHD1 D137N (kindly gifted from Dr. Welbourn, NIAID, NIH) was re-cloned in retroviral expression vector (pRetro-X-IRES-Zsgreen1), and produced viral particles for amphotropic transduction in Tregs. (B) Transduction efficiency of in SAMWT and SAM137N. The preparation and expansion protocol were described in Methods. Briefly, FACS-sorted Tregs from healthy donors (n=4 for short expansion; n=7 for long expansion) were stimulated with anti-CD3ε/CD28 antibodies, and transduced with retroviral Mock vector (Mock), SAMWT, and SAM137N. GFP expression was measured at day 19 after transduction. The number in the plot indicates the frequency of GFP+ and GFP− Tregs, respectively. (C) Stabilization of Foxp3 and Helios expression in GFP+ SAMWT and SAM137N Tregs. Foxp3 and Helios were measured by intracellular FACS staining after short- and long-term expansion periods, respectively. Data are presented as the mean of MFI in each donors. The p values were calculated by two-tailed Student’s t-test. (D) The expression of Foxp3 and Helios in GFP− Tregs. (E) ODNps25 effect to stabilize Foxp3 and Helios expressions in SAMHD1 construct-overexpressed Tregs. Transduction and expansion culture of Tregs was same as described in Fig. 3C in the presence of ODNps25 (2 µM) or not. FACS analysis is performed by gating of intercellular stained GFP+ transduced cells. Percent of Foxp3 and Helios expression is indicated in quadrant gate. Data in C and D is one of seven independent experiments with different donors.
SAMHD1 directly interacts with Foxp3 and Helios mRNAs
To directly address the role of SAMHD1 in the post-transcriptional regulation of Foxp3 and Helios mRNAs, we determined whether SAMHD1 bound either of the mRNAs using a qPCR specific for Foxp3 and Helios transcripts after RNA immunoprecipitation (RIP-qPCR) with anti-SAMHD1 antibody in ODNps25-treated Tregs. To map the potential SAMHD1 binding site in Foxp3 and Helios transcripts, the qPCR was accessed with probes specific for the upstream 5'-coding regions of mRNAs (P1 and P2 in Fig. S1A) and the 3'-untranslated region (UTR) (P3 and P4 in Fig. S1A) of Foxp3 and Helios, respectively. The RIP-qPCR indicated that SAMHD1 does not interact with 5’-coding regions, but specifically binds to 3'-UTRs in both Foxp3 and Helios transcripts. Importantly, binding of SAMHD1 to 3-UTRs was markedly enhanced by ODNps25 treatment (Fig. S1C, Fig S1D, Fig. 4B, and Fig 4C). No change in protein level of SAMHD1 in the presence of ODNps25 could be detected by Western blot. (Fig. S1E).
Figure 4. SAMHD1 specifically binds to 3’-UTR of Foxp3 and Helios, and stabilize their post-transcription by TCR stimulation as well as ODNps25 addition.
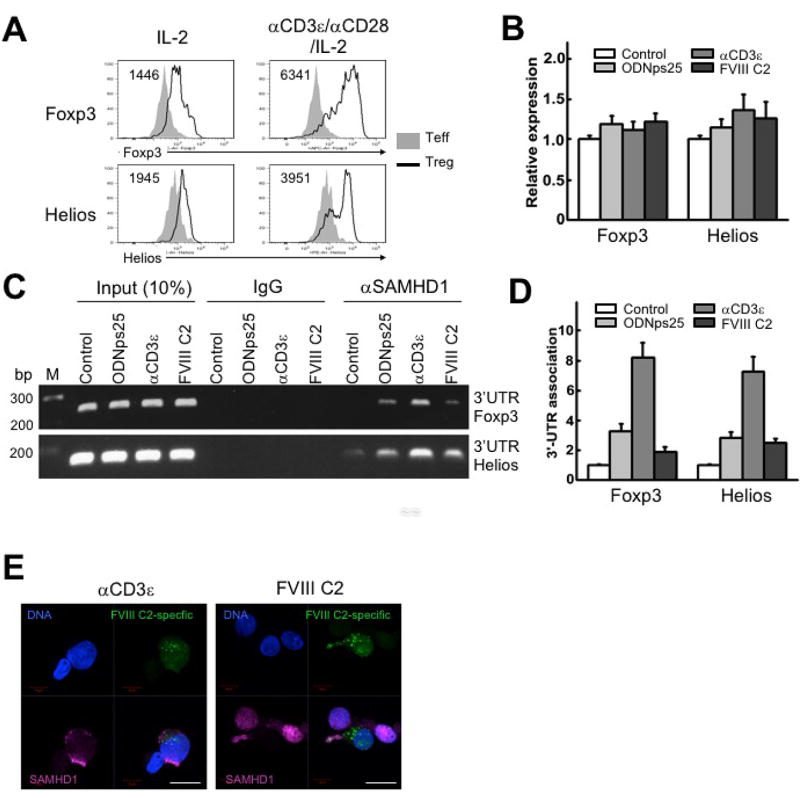
(a) Relative induction of Foxp3 and Helios proteins by in vitro CD3/CD28-stimulation in Tregs. Rested polyclonal Tregs, expanded as in Methods, were re-stimulated by adding anti-CD3ε (0.5 µg/ml), CD28 antibodies (0.2 µg/ml) and recombinant IL-2 (200 IU/ml) together with -irradiated PBMCs for 36 hrs. The number positioned in upper left corner in the histogram plots indicates median of MFI of Foxp3 or Helios, respectively. The plots shown is one of three independent experiments with different donors. (B) Relative expression of total mRNA of Foxp3 and Helios in 17195TCR-transduced Tregs stimulated with ODNps25, anti-CD3ε antibody, or FVIII C2 peptide. Preparation and in vitro expansion of 17195 TCR-transduced Tregs (17195 Tregs) is followed as shown in Rf. Expanded 17195 Tregs were rested in IL-2 deprived culture media overnight, and re-stimulated with ODNps25 (2 µM), anti-CD3ε antibody (0.5 µg/ml), or FVIII C2 peptide (1 µg/ml) for 36 hrs. Quantitative RT-PCR (qRT-PCR) of Foxp3 and Helios was performed with primers specific to 5’encoding region of Foxp3 and Helios (P1 and P2 for Foxp3 shown in Supplemental Fig.1). Data was normalized with external HPRT control. (C) RNA immunoprecipitation (RIP)-qPCR against SAMHD1 to detect 3’UTR of Foxp3 and Helios. RIP analysis was performed from the whole extract of re-stimulated 17195Tregs in (A). Amplification was performed by qPCR with primer specific to 3’UTR of Foxp3 and Helios (P3 and P4 for Foxp3 shown in Supplemental Fig.1). The experiments of B, C, and D are one of two independent experiments with different donors. (D) Relative quantification was measured by SYBR green-qPCR. (E) Confocal microscopy of SAMHD1 (red) in re-stimulated 17195 TCR transduced (green) Tregs. Cells were re-stimulated by adding anti-CD3ε antibody (0.5 µg/ml) or FVIII C2 peptide (1 µg/ml) for 36 hrs. FVIII-specific TCR expressed cells are recognized by the expression of green fluorescent protein (GFP). Nuclei were counterstained with DAPI (blue). Scale bars, 10 µm.
Effect of TCR signaling on the binding of SAMHD1 to Foxp3 and Helios mRNAs
Activation of Tregs via the TCR results in enhancement of the levels of expression of both Foxp3 and Helios proteins (1446 to 6341 and 1945 to 3951in Mean Fluorescence Intensity in Fig. 4A). To determine if TCR signaling also enhanced the physical interaction of SAMHD1 with Foxp3 and Helios mRNAs, we expanded polyclonal or FVIII C2-specific Tregs (17195 Tregs)(3) with anti-CD3ε antibody or FVIII C2 (2191–2220) peptide in the presence of rIL-2 for 36 hours, followed by qRT-PCR for Foxp3 and Helios and RIP-qPCR with anti-SAMHD1 antibody. While total Foxp3 and Helios mRNAs in polyclonal Tregs or 17195 Tregs were not significantly changed by stimulation with anti-CD3ε antibody or FVIII C2 (2191–2220) peptide (Fig. 4B), RIP-qPCR showed strong induction of the interaction of SAMHD1 and Foxp3/Helios 3’UTRs (Fig. 4CD). Addition of ODNps25 in the absence of TCR stimulation also enhanced the interaction of SAMHD1 with Foxp3 and Helios mRNAs, but to a much lesser extent than that seen after TCR stimulation. These results suggest that the TCR-mediated induction of SAMHD1–3'UTR binding correlates with the translation of Foxp3/Helios, as SAMHD1 accumulated in the cytoplasm in manner similar to that seen with ODNps25 (compare Fig. 4E and 2E).
TCR signaling modulates SAMHD1 function by phosphorylation of Thr592
Previous studies have shown that phosphorylation of SAMHD1 at Thr592 deactivates its dNTPase activity and reverses HIV restriction in U927 and primary myeloid cells(13, 14). Thr592 of SAMHD1 can also be phosphorylated by TCR stimulation in mouse and human CD4+ T cells(14, 15). We then determined if the TCR-mediated effects on stabilization of Foxp3 and Helios mRNAs were also secondary to reduction of the enzymatic activity of SAMHD1 via Thr592 phosphorylation in CD4+ T conventional (Tconv) cells and Tregs. While both Tconv cells and Tregs expressed pThr592 in the absence of stimulation, the level of pThr592 was further enhanced following anti-CD3ε/CD28 stimulation in both cell types with a greater degree of enhancement in Tregs (Fig. 5A). To determine if phosphorylation of Thr592 is related to the stabilization of Foxp3 and Helios during long-term culture of Tregs, we transfected Tregs with either phospho-deficient (SAM592A) or phosphomimetic (SAM592E) SAMHD1 mutants (Fig. 5B), and the expression of Foxp3 and Helios was monitored subsequently during the expansion cultures. High percentages (76–79%) of Foxp3+Helios+ cells were maintained in cultures of Tregs transfected with SAMWT, SAM592A, or SAM592E at initial expansion stage (Post 5 days in Fig. 5C). Over 29 days of culture, the percentages of Foxp3+Helios+ cells were decreased up to 44.1% in Mock Treg culture and the frequency of double positive cells was relatively stabilized by SAMWT overexpression (61.3%) and SAM592E overexpression (51.6%) (Fig. 5C), but not in cells transduced with SAM592A (39.8%). Thus, SAMHD1 must be phosphorylated at Thr592 for inhibition of dNTPase activity and stabilization of Foxp3 and Helios during long-term Treg culture.
Figure 5. Thr592 phosphorylation of SAMHD1 due to TCR stimulation stabilizes the expression of Foxp3 and Helios in Tregs.
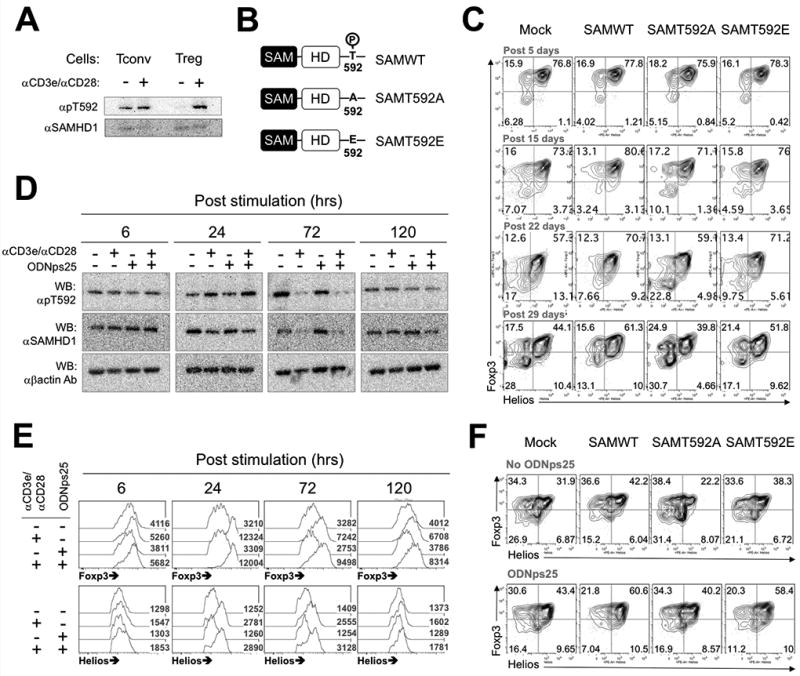
(A) The592 Phosphorylation of SAMHD1 by TCR stimulation. 2 wk-expanded conventional T cells (Tconv) or Tregs were re-stimulated with plate-coated anti-CD3ε /CD28 antibodies and IL-2 or IL-2 alone condition for 24 hrs. Western blots were performed with total proteins extracted in RIPA. Data shown is one of three independent experiments with different donors. (B) Expression constructs of phospho-deficient (SAMT592A) and phosphomimetic (SAMT592E) SAMHD1. (C) Stabilization of Foxp3 and Helios expressions in SAMWT, SAMT592A, or SAMT592E Tregs. The protocols for Transduction, expansion and FACS analysis is same as in Fig 3H. The day displayed above panels indicates post-time after primary stimulation. The plots shown is one of three independent experiments with different donors. (D) Kinetics of Thr592 phosphorylation of SAMHD1 in Tregs during CD3 stimulation with ODNps25. 2 wk-expanded Tregs (1×106) were restimulated with indicated stimuli for given times. CD3 stimulation was applied by incubating the cells in plate-coated anti-CD3ε Ab. After incubation, the cells were harvested, and transferred to two tubes in an equal cell number. One was used to total protein extraction for the western blot, and the cells in the other tubes were stained for intracellular FACS of the Foxp3 and Helios. The number in histogram plots (numbers in the plot) represents mean fluorescence intensity (MFI) of histogram (E). (F) Stabilization of Foxp3 and Helios expressions in GFP+ SAMWT, SAMT592A, and SAMT592E Tregs in the addition of ODNps25 or not. The protocols for Transduction, expansion and FACS analysis is same as in Fig 3H except for the ODNps25 treatment (2 µM). Intracellular Foxp3 and Helios is measured at day 20 after initial stimulation. Data shown is one of two experiments with different donors.
To further explore the relationship between Thr592 phosphorylation and the expression of Foxp3/Helios during TCR stimulation, kinetic analyses were performed to determine the SAMHD1 phosphorylation status and intracellular levels of Foxp3 and Helios. By 6 hrs after CD3 stimulation or ODNps25 addition, there were no significant changes in the levels of Thr592 phosphorylation and Foxp3/Helios expressions (6 in Fig. 5D). After 24 hrs, Thr592 phosphorylation was increased in TCR-activated Tregs. The enhancement of Thr592 phosphorylation by TCR stimulation correlated with the enhancement of the levels of Foxp3 and Helios proteins as determined by FACS (24 in Fig. 5D). The addition of ODNps25 in the presence or absence of TCR stimulation had no effects on the level of Thr592 phosphorylation. Interestingly, the levels of SAMHD1 protein were significantly reduced after 72 hrs of culture, but the levels of Foxp3 and Helios remained elevated at this time point (72 hr in Fig. 5D). ODNps25 did not contribute to the downregulation of SAMHD1 protein in either control or TCR-stimulated cultures. After 120 hours of culture, total SAMHD1 protein levels, the levels of Thr592 phosphorylation, and the levels of Foxp3 were similar to those of unstimulated cells (120 in Fig. 5D).
Finally, to confirm that the stabilization of Foxp3 and Helios expression by ODNps25 is unrelated to the TCR induced phosphorylation of SAMHD1 Thr592, we transduced Tregs with SAM592E or SAM592A (Fig. 5E). Addition of ODNps25 stabilized the expression of Foxp3+Helios+ cells in expanded cultures to the same extent in the presence of SAMWT, SAM592A, SAM592E (Fig. 5F). Taken together, these results demonstrate that Tregs regulate intracellular Foxp3 and Helios expressions via dephosphorylation of SAMHD1 Thr592 and downregulation of SAMHD1 protein itself, and that ODNps25 controls the activity of SAMHD1 via direct block of the HD domain rather than dephosphorylation of SAMHD1 Thr592 and that ODN and TCR stimulation stabilize the expression of Foxp3 and Helios via different mechanisms. It is still unknown how TCR stimulation in Tregs controls the phosphorylation and downregulation of SAMHD1 sequentially, which will be a subject of further investigation.
Discussion
We had previously observed that the addition of ODNps25 to long-term expansion cultures of hTreg resulted in stabilization of the expression of two transcription factors critical for Treg function, Foxp3 and Helios. While the role of Helios remains poorly defined, its expression in hTreg distinguishes bona fide Treg from Tconv cells that have upregulated Foxp3 expression secondary to T cell activation(16–19). As the latter population may readily lose Foxp3 expression and revert to Tconv cells, we believe the optimal preparation of hTregs for cellular therapy should contain a high percentage of Foxp3+Helios+ T cells. ODNps25-mediated stabilization has been shown to be critical for genetic engineering of human Tregs without the loss of suppressive function(3). The goals of the present experiments were to identify the cellular receptor for ODNps25 and to determine how it stabilized the expression of these two transcription factors. We used a pull-down approach by adding biotin-ODNps25, covalently cross-linking potential targets by UV exposure, and precipitated potential complexes with avidin-magnetic beads. When the complexes were separated by gel electrophoresis and then analyzed by mass spectroscopy the most prominent candidate identified was SAMHD1, a dNTPase. We propose a model (fig. 6) in which in the absence of the ODNps25 or TCR stimulation, dephosphorylated SAMHD1 binds to the 3’UTRs of both Foxp3 and Helios in the nucleus and following transfer of the complex to the cytoplasm results in processing of the 3’UTRs and a decrease in Foxp3 and Helios mRNA. The ODNps25 functions by binding to the SAMHD1–3’UTR complex in the cytoplasm, inhibiting its dNTPase activity resulting in enhancement of the stability of Foxp3 and Helios mRNA. TCR stimulation functions by a separate, but parallel, mechanism by activating CDK2-mediated phosphorylation of SAMHD1 T592(13–15, 20) resulting in inhibition of the enzymatic activity of SAMHD1 again resulting in stabilization of Foxp3 and Helios mRNAs. While it remains possible that the enhanced expression of Foxp3 and Helios by ODNps25 is secondary to an effect on the ODN on non-Treg that contaminate our sorted Treg populations, we believe that this possibility is unlikely. First, we have never observed the induction of Helios expression by non-Treg following TCR stimulation alone or even in the presence of added TGF-β. Secondly, we have intentionally contaminated our sorted Treg population with non-Tregs and did not observe any enhancement of Foxp3 and Helios expression above that seen with Tregs cultured in the absence of added non-Treg. Lastly, we intentionally induced Foxp3 expression in purified non-Treg by TCR stimulation in the presence of TGF-β and then added ODNps25 to the cultures. We failed to observe stabilization of Foxp3 expression or the induction of Helios expression. Thus, we conclude that the effects of ODNps25 are specific for thymus-derived Treg.
Figure 6. SAMHD1-mediated stability regulation of Foxp3 and Helios mRNAs in Tregs.
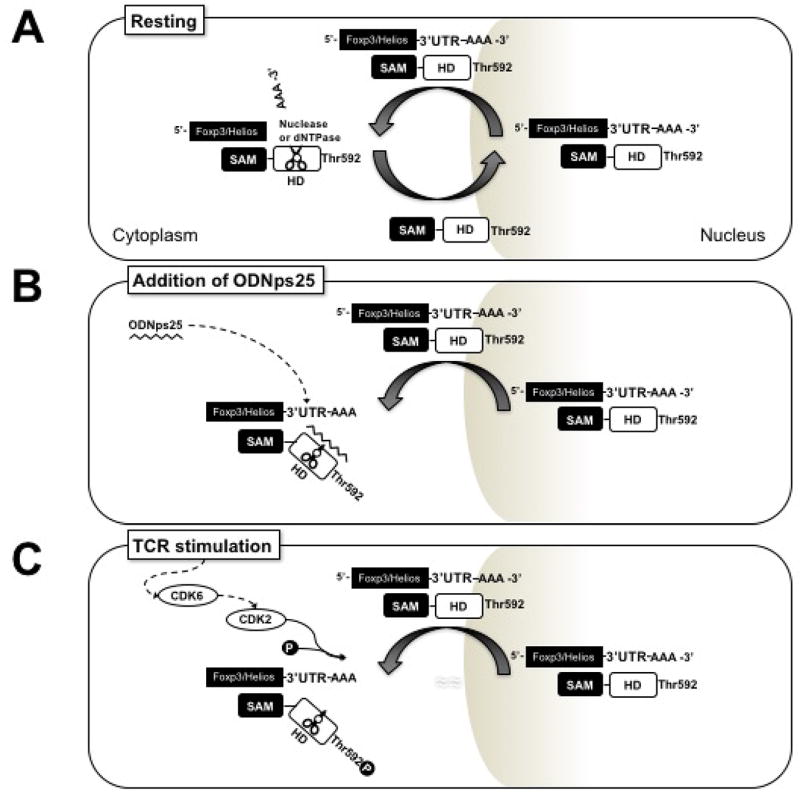
(A) Upon resting status, dephosphorylated SAMHD1 binds to 3’ UTR of Foxp3 and Helios mRNAs in nucleus, and then transfer those to cytoplasm to process 3’ UTR of mRNAs. After the processing, SAMHD1 is shuttled back to nucleus. (B) However, if ODNps25 is added in the cells, ODNps25 preferentially binds to SAMHD1–3’UTR complex in cytoplasm. The binding inhibits dNTPase activity of SAMHD1 and subsequent processing of 3’ UTRs, resulting of accumulation of the complex and enhancement of stability of Foxp3 and Helios mRNAs. (C) In TCR stimulation, CDK2-mediated Thr592 phosphorylation of SAMHD1 inhibits dNTPase activity, resulting of the induction of Foxp3 and Helios expression as well.
As shown in Table 1, several RNA-binding/stabilizing proteins (e.g., heterogeneous ribonucleoproteins) as well as SAMHD1 were positively identified as an ODNps25-interacting protein. Among them, we confirmed that hnRNPA2/B1 proteins bind directly to ODNps25 and FoxP3 mRNA, but the binding pattern to Foxp3 mRNA in the presence of ODNps25 was different from that observed with SAMHD1. No interactions between hnRNPA2/B1 and SAMHD1 were seen in immunoprecipitation and RPA studies (data not shown), Further investigation of the function of hnRNPA2/B1 will be addressed in future studies.
SAMHD1 was originally described in 2000 as an IFNγ-induced protein in dendritic cells(21) and later shown to play a critical role as a host restriction factor inhibiting HIV-1 infection. Mutations in SAMHD1 are responsible for one form of the Aicardi-Goutieres Syndrome (AGS5), a type I interferonopathy. Interferonopathies are caused either by an increase in the burden of nucleic acids as in AGS or by constitutive activation of nucleic acid receptors (e.g., Sting associated vasculopathy, Singleton-Merten Syndrome). While the role of SAMHD1 in HIV infection has been extensively studied, very little is known about the function of SAMHD1 in normal lymphocyte physiology. Of note, the cytokines IL-2 and IL-7 have been shown to induce SAMHD1 T592 phosphorylation in HIV infected cells abrogating its antiviral activity(22). The effects of IL-7 were more prominent than those of IL-2. We have not examined the effects of either IL-2 or IL-7 on SAMHD1 phosphorylation in Treg expansion cultures, but the routine addition of high concentrations of IL-2 during Treg expansion is not sufficient to prevent downregulation of Foxp3 or Helios.
We were unsuccessful in our attempts to knock down SAMHD1 in hTregs using shRNAs that have been reported to be useful in other cell types including monocyte/macrophage cell lines(6, 7) and primary CD4 T cells(11). To further define the role of SAMHD1 in Treg function, we have used a series of gain/loss of function mutants (SAMWT, SAM137N, SAM592A, and SAM592E) in overexpression studies. Most importantly, overexpression of SAM137N which results in a loss of dGTP binding, oligomeric activation, and dNTPase activity clearly defined the role of SAMHD1 in the processing of Foxp3 and Helios 3’UTRs. We have not yet defined the specific binding site for the blocking ODNs, but it is likely that they preferentially interact with the HD domain in SAMHD1 which has been postulated to regulate SAMHD1 nuclease activity(23). In figure 3C and 3E, cells transduced with SAMWT maintained a high level of FoxP3 and Helios expression. We have no direct data to explain this observation, but hypothesize that when wild-type SAMHD1 is overexpressed in the presence of endogenous SAMHD1, an excess amount of SAMHD1 is present and remained without occupying FoxP3 and Helios transcripts in the cells. Consequently, phosphorylation of SAMHD1 complexed with transcripts could be neutralized by excess unoccupied SAMHD1 resulting in stabilization of the FoxP3 and Helios transcripts. As indirect evidence of this hypothesis, overexpression of SAM137N showed extreme stabilization (Figure 3C and 3E).
In addition to SAMHD1, in our analysis of the mass spectrometry data (Table 1), we identified several other groups of proteins as possible ODNps25 receptors. One of the groups identified included components of heterogeneous ribonucleoprotein particles (hnRNPs) that respond to mRNA transcription in the nucleus and modulate post-transcriptional activity in the cytoplasm(24–26). Considering the cytoplasmic accumulation of ODNps25-SAMHD1 complex, it remains possible that hnRNPs cooperate with SAMHD1 during post-transcriptional control of Foxp3 and Helios in Tregs. This requires further investigation.
We originally identified ODNps25 as a stabilizer of Foxp3/Helios expression during an analysis of the potential role of the effects of TLR9 ligands on the expression of Foxp3. While the addition of a TLR9 agonist resulted in stabilization of Foxp3 expression, a TLR antagonist was equally effective. Subsequent studies demonstrated that any random sequence ODN was effective and that the optimal size was a 25 mer. The major finding in this report is that the ODNs mediate their effects by inhibiting the activity of SAMHD1, a dNTP hydrolase that is widely expressed in different cell types. Studies of the function of SAMHD1 have focused on its role in innate immune cells or T lymphocytes in processing nucleic acids derived from exogenous infectious agents or produced endogenously. Our studies suggest that SAMHD1 also modulates the post-transcriptional activity of two key transcription factors that are critical for Treg function. It remains to be determined what other normal cellular pathways (mRNAs) are targeted by SAMHD1. Although SAMHD1 is a universal enzyme that is expressed in both conventional T cells and Tregs, SAMHD1 showed selective stabilization activity of particular key gene products in Tregs. To examine this further, we analyzed SAM domain-specific binding secondary structures and consensus sequences in mRNAs of Foxp3 and Helios, but there were no binding motifs or sequences for SAM. One possible hypothesis is an existence of Treg-specific linker protein, by which signature mRNAs indirectly interact with SAM domain of SAMHD1 to process 3’UTRs.
Drug-induced modulation of SAMHD1 activity in malignant cells has been aimed at increasing its enzymatic activity(27) as down-regulation of SAMHD1 activity may promote cancer cell proliferation. The use of agents that downregulate SAMHD1 activity in vivo to promote Treg function must be approached with caution as such a therapeutic approach might increase the intracellular nucleic acid pool and thereby promote inflammation.
Supplementary Material
Acknowledgments
Supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, NIH.
We thank Drs. Sarah Welbourn and Klaus Strebel (NIAID, NIH) for expression vector constructs and sequences of SAMHD1 137N and SAMHD1 592A mutants. Y.C.K. designed and conducted overall experiments and wrote the manuscript; K.K.K. designed and conducted biotin/avidin-mediated pull-down, qRT-PCR, RNA precipitation, and other biochemical analyses; J.Y. conducted western blot analyses and immunofluorescence staining; E.M.S and D.W.S. supervised the research and edited the manuscript.
Abbreviations used in this article
- Tregs
regulatory T cells
- ODNps25
25mer-phosphorothioated random oligonucleotides
- hTregs
human Tregs
- SAMWT
Wild-type SAMHD1
- Tconv
conventional CD4+ T cell
Footnotes
There is no conflict of interests among the authors.
References
- 1.Shevach EM. Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity. 2009;30:636–645. doi: 10.1016/j.immuni.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 2.Kim YC, Bhairavabhotla R, Yoon J, Golding A, Thornton AM, Tran DQ, Shevach EM. Oligodeoxynucleotides stabilize Helios-expressing Foxp3+ human T regulatory cells during in vitro expansion. Blood. 2012;119:2810–2818. doi: 10.1182/blood-2011-09-377895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim YC, Zhang AH, Su Y, Rieder SA, Rossi RJ, Ettinger RA, Pratt KP, Shevach EM, Scott DW. Engineered antigen-specific human regulatory T cells: immunosuppression of FVIII-specific T- and B-cell responses. Blood. 2015;125:1107–1115. doi: 10.1182/blood-2014-04-566786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, de Carvalho LP, Stoye JP, Crow YJ, Taylor IA, Webb M. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480:379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- 5.Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, Bloch N, Maudet C, Bertrand M, Gramberg T, Pancino G, Priet S, Canard B, Laguette N, Benkirane M, Transy C, Landau NR, Kim B, Margottin-Goguet F. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat Immunol. 2012;13:223–228. doi: 10.1038/ni.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim ES, Emerman M. HIV: Going for the watchman. Nature. 2011;474:587–588. doi: 10.1038/474587a. [DOI] [PubMed] [Google Scholar]
- 9.Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, Schenkova K, Ambiel I, Wabnitz G, Gramberg T, Panitz S, Flory E, Landau NR, Sertel S, Rutsch F, Lasitschka F, Kim B, Konig R, Fackler OT, Keppler OT. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat Med. 2012;18:1682–1687. doi: 10.1038/nm.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beloglazova N, Flick R, Tchigvintsev A, Brown G, Popovic A, Nocek B, Yakunin AF. Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutieres syndrome and HIV-1 restriction. J Biol Chem. 2013;288:8101–8110. doi: 10.1074/jbc.M112.431148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryoo J, Choi J, Oh C, Kim S, Seo M, Kim SY, Seo D, Kim J, White TE, Brandariz-Nunez A, Diaz-Griffero F, Yun CH, Hollenbaugh JA, Kim B, Baek D, Ahn K. The ribonuclease activity of SAMHD1 is required for HIV-1 restriction. Nat Med. 2014;20:936–941. doi: 10.1038/nm.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, Fuller JC, Jackson RM, Lamb T, Briggs TA, Ali M, Gornall H, Couthard LR, Aeby A, Attard-Montalto SP, Bertini E, Bodemer C, Brockmann K, Brueton LA, Corry PC, Desguerre I, Fazzi E, Cazorla AG, Gener B, Hamel BC, Heiberg A, Hunter M, van der Knaap MS, Kumar R, Lagae L, Landrieu PG, Lourenco CM, Marom D, McDermott MF, van der Merwe W, Orcesi S, Prendiville JS, Rasmussen M, Shalev SA, Soler DM, Shinawi M, Spiegel R, Tan TY, Vanderver A, Wakeling EL, Wassmer E, Whittaker E, Lebon P, Stetson DB, Bonthron DT, Crow YJ. Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet. 2009;41:829–832. doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.St Gelais C, de Silva S, Hach JC, White TE, Diaz-Griffero F, Yount JS, Wu L. Identification of cellular proteins interacting with the retroviral restriction factor SAMHD1. J Virol. 2014;88:5834–5844. doi: 10.1128/JVI.00155-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cribier A, Descours B, Valadao AL, Laguette N, Benkirane M. Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep. 2013;3:1036–1043. doi: 10.1016/j.celrep.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 15.Pauls E, Ruiz A, Badia R, Permanyer M, Gubern A, Riveira-Munoz E, Torres-Torronteras J, Alvarez M, Mothe B, Brander C, Crespo M, Menendez-Arias L, Clotet B, Keppler OT, Marti R, Posas F, Ballana E, Este JA. Cell cycle control and HIV-1 susceptibility are linked by CDK6-dependent CDK2 phosphorylation of SAMHD1 in myeloid and lymphoid cells. J Immunol. 2014;193:1988–1997. doi: 10.4049/jimmunol.1400873. [DOI] [PubMed] [Google Scholar]
- 16.Thornton AM, Korty PE, Tran DQ, Wohlfert EA, Murray PE, Belkaid Y, Shevach EM. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol. 2010;184:3433–3441. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sebastian M, Lopez-Ocasio M, Metidji A, Rieder SA, Shevach EM, Thornton AM. Helios Controls a Limited Subset of Regulatory T Cell Functions. J Immunol. 2016;196:144–155. doi: 10.4049/jimmunol.1501704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Getnet D, Grosso JF, Goldberg MV, Harris TJ, Yen HR, Bruno TC, Durham NM, Hipkiss EL, Pyle KJ, Wada S, Pan F, Pardoll DM, Drake CG. A role for the transcription factor Helios in human CD4(+)CD25(+) regulatory T cells. Mol Immunol. 2010;47:1595–1600. doi: 10.1016/j.molimm.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim HJ, Barnitz RA, Kreslavsky T, Brown FD, Moffett H, Lemieux ME, Kaygusuz Y, Meissner T, Holderried TA, Chan S, Kastner P, Haining WN, Cantor H. Stable inhibitory activity of regulatory T cells requires the transcription factor Helios. Science. 2015;350:334–339. doi: 10.1126/science.aad0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Welbourn S, Dutta SM, Semmes OJ, Strebel K. Restriction of virus infection but not catalytic dNTPase activity is regulated by phosphorylation of SAMHD1. J Virol. 2013;87:11516–11524. doi: 10.1128/JVI.01642-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li N, Zhang W, Cao X. Identification of human homologue of mouse IFN-gamma induced protein from human dendritic cells. Immunol Lett. 2000;74:221–224. doi: 10.1016/s0165-2478(00)00276-5. [DOI] [PubMed] [Google Scholar]
- 22.Coiras M, Bermejo M, Descours B, Mateos E, Garcia-Perez J, Lopez-Huertas MR, Lederman MM, Benkirane M, Alcami J. IL-7 Induces SAMHD1 Phosphorylation in CD4+ T Lymphocytes, Improving Early Steps of HIV-1 Life Cycle. Cell Rep. 2016;14:2100–2107. doi: 10.1016/j.celrep.2016.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goncalves A, Karayel E, Rice GI, Bennett KL, Crow YJ, Superti-Furga G, Burckstummer T. SAMHD1 is a nucleic-acid binding protein that is mislocalized due to aicardi-goutieres syndrome-associated mutations. Hum Mutat. 2012;33:1116–1122. doi: 10.1002/humu.22087. [DOI] [PubMed] [Google Scholar]
- 24.Pinol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 25.Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 26.Han SP, Tang YH, Smith R. Functional diversity of the hnRNPs: past, present and perspectives. Biochem J. 2010;430:379–392. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- 27.Rampazzo C, Tozzi MG, Dumontet C, Jordheim LP. The druggability of intracellular nucleotide-degrading enzymes. Cancer Chemother Pharmacol. 2016;77:883–893. doi: 10.1007/s00280-015-2921-6. [DOI] [PubMed] [Google Scholar]
- 28.Welbourn S, Strebel K. Low dNTP levels are necessary but may not be sufficient for lentiviral restriction by SAMHD1. Virology. 2016;488:271–277. doi: 10.1016/j.virol.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


