Abstract
Although the five-year survival rate of chronic lymphocytic leukemia (CLL) patients has risen to >80%, the only potentially curative treatment is allogeneic hematopoietic stem cell transplantation (alloHSCT). To identify possible new monoclonal antibody (mAb) drugs and targets for CLL, we previously developed a phage display–based human mAb platform to mine the antibody repertoire of patients who responded to alloHSCT. We had selected a group of highly homologous post-alloHSCT mAbs that bound to an unknown CLL cell surface antigen. Here we show through next-generation sequencing of cDNAs encoding variable heavy-chain domains that these mAbs had a relative abundance of ~0.1% in the post-alloHSCT antibody repertoire and were enriched ~1,000-fold after three rounds of selection on primary CLL cells. Based on differential RNA-seq and a cell microarray screening technology for discovering human cell surface antigens, we now identify their antigen as Siglec-6. We verified this finding by flow cytometry, ELISA, siRNA knockdown, and surface plasmon resonance. Siglec-6 was broadly expressed in CLL and could be a potential target for antibody-based therapeutic interventions. Our study reaffirms the utility of post-alloHSCT antibody drug and target discovery.
Introduction
Chronic lymphocytic leukemia (CLL) is the most common leukemia in the U.S. with a lifetime risk of ~0.6% and an incidence of ~20,000 cases annually (https://seer.cancer.gov/statfacts/html/clyl.html) (1). Chemoimmunotherapy and small molecule kinase inhibitors have increased the five-year survival rate to >80%. Despite this, allogeneic hematopoietic stem cell transplantation (alloHSCT) has remained the only potentially curative treatment of CLL. Although alloHSCT is not available for most patients and is associated with high morbidity and mortality (2), it underscores the potency of immunotherapy in eradicating CLL and provides an incentive to identify the underlying targets and mechanisms (3). Based on the hypothesis that post-alloHSCT antibodies contribute to the graft-versus-leukemia (GVL) response (4), and the detection of serum antibody titers to CLL cell surface antigens following alloHSCT, we developed a concerted antibody drug and target discovery strategy (5,6). Here we report the identification of Siglec-6 as a target of post-alloHSCT antibodies and confirm its expression in CLL.
Materials and Methods
Cell lines and primary cells
Cell lines MEC-1 (DSMZ #ACC 497) and U937 (ATCC #CRL-1593.2) were newly purchased and used within three passages after thawing without additional cell line authentication and mycoplasma testing. Cell line MEC1-002 was derived from cell line MEC1 by FACS using a BD FACSVantage instrument following staining with chemically biotinylated (BiotinTag Micro Biotinylation Kit; Sigma-Aldrich) JML-1 IgG1 (5) and PE-conjugated streptavidin, cloned by limiting dilution, cryopreserved, and used as described for its parental cell line. Cryopreserved treatment-naïve CLL peripheral blood mononuclear cells (PBMCs) were derived from a natural history study at the NIH Clinical Center registered as NCT00923507 at www.clinicaltrials.gov. The study was approved by the Institutional Review Boards of the NIH Clinical Center and The Scripps Research Institute and written informed consent was obtained in accordance with the Declaration of Helsinki.
Next-generation sequencing
VH-encoding sequences from the unselected and selected post-alloHSCT library (5) in phage display vector pC3C (7) were PCR amplified separately as described (8) and sequenced using an Illumina MiSeq instrument. Next-generation sequencing (NGS) data were analyzed with published and in-house software to trim, cluster, and identify unique sequences. Specifically, the 3′ end adapter sequences and the flanking sequences on read 1 and read 2 from forward and reverse templates were trimmed using CUTADAPT (9). Then each pair of reads was merged into a single sequence using FLASH (10), which required at least 10-bp overlap between the two reads. In order to cluster unique sequences that belong to the same clone, we first sorted these sequences by their frequencies. The most popular sequence was marked as a seed and then compared with a second sequence. If the distance between the two was equal or smaller than the cutoff we set, the second sequence was considered a child of the seed sequence; otherwise, it was marked as another seed. Then the remaining sequences were processed in a similar way by comparing them with all the sequences that had been marked as seed. When a sequence was found to be the child of multiple seed sequences, there was a conflict and the sequence was discarded. Hamming distance was used in the analysis, and only sequences of the same length were compared. The Hamming distance cutoff was set to 2.
Differential RNA-seq
RNA was isolated with a PureLink RNA Mini Kit (Thermo Fisher Scientific). RNA concentration and quality were determined using a NanoDrop spectrophotometer and an Agilent 2100 Bioanalyzer, respectively. Samples with an RNA integrity number ≥9 were sequenced using an Illumina NextSeq 500 system at a targeted depth of 20 million paired-end reads and processed and analyzed with TopHat 2.0.9, HTSeq 0.6.1, and DESeq2 software. Differentially expressed genes encoding transmembrane proteins were then identified.
Retrogenix cell microarray
Primary and confirmatory screens with 20 μg/mL JML-1 IgG1 (5) were carried out exactly as described (8) based on a collection of 4,336 human transmembrane proteins.
Flow cytometry
The binding of 1 μg/mL of chemically biotinylated JML-1 IgG1 and a chemically biotinylated human IgG1 isotype control to MEC1, MEC1-002, and U937 cell lines was detected with PE-conjugated streptavidin (BD Biosciences) by flow cytometry using a BD LSRII instrument. CLL PBMCs were stained using 1 μg/mL of the commercial mouse anti-human Siglec-6 mAb 767329 (R&D Systems) followed by Alexa Fluor 647-conjugated goat anti-mouse IgG pAbs and, separately, 1 μg/mL of chemically biotinylated JML-1 IgG1 followed by PE-conjugated streptavidin. The difference of mean fluorescence intensity (ΔMFI) values from the two separate stainings were plotted to analyze their Pearson correlation coefficient r2 using GraphPad Prism software version 7.
ELISA
The binding of 1 μg/mL of chemically biotinylated JML-1 IgG1 and a chemically biotinylated human IgG1 isotype control to plate-bound human CD22-Fc, CD33-Fc (blue), Siglec-6-Fc (red), and Siglec-8-Fc (purple; all from R&D Systems) was detected using HRP-conjugated streptavidin. HRP-conjugated goat anti-human Fc pAbs (Jackson ImmunoResearch) were used to confirm the presence of the plate-bound Fc fusion proteins.
Surface plasmon resonance
Surface plamson resonance (SPR) studies were carried out on a Biacore X100 instrument as described (8). In brief, SPR sensorgrams were obtained for the binding of JML-1 Fab to Siglec-6-Fc captured on a goat anti-human Fc-coated CM5 chip (GE Healthcare). JML-1 Fab was injected at five different concentrations in the following order: 6.25 nM, 12.5 nM, 25 nM, 50 nM, 100 nM, and again 6.25 nM. The six SPR sensorgrams were used to calculate kon, koff, and Kd.
siRNA knockdown
For knockdown studies, 1 x 106 MEC1-002 cells in a 12-well tissue culture plate were incubated with 1 μM of Accell SIGLEC6-targeting or control siRNAs (both from Dharmacon) for 72 h and then analyzed by flow cytometry using chemically biotinylated JML-1 IgG1 in conjunction with PE-conjugated streptavidin and mouse anti-human Siglec-6 mAb 767329 in conjunction with Alexa Fluor 647-conjugated goat anti-mouse IgG pAbs (Jackson ImmunoResearch).
Statistics
Except for the flow cytometry studies with the 16 CLL PBMCs, which were only analyzed once or twice each due to limited sample availability, at least three independent experiments were carried out for the flow cytometry studies with the cell lines and for the ELISA, SPR, and siRNA knockout studies. Representative experiments are shown for flow cytometry, ELISA (mean ± standard deviation (SD) of technical triplicates), and SPR. The siRNA knockout study is shown as mean ± SD of three independent experiments and analyzed for significance by two-way ANOVA using GraphPad Prism software version 7.
Results and Discussion
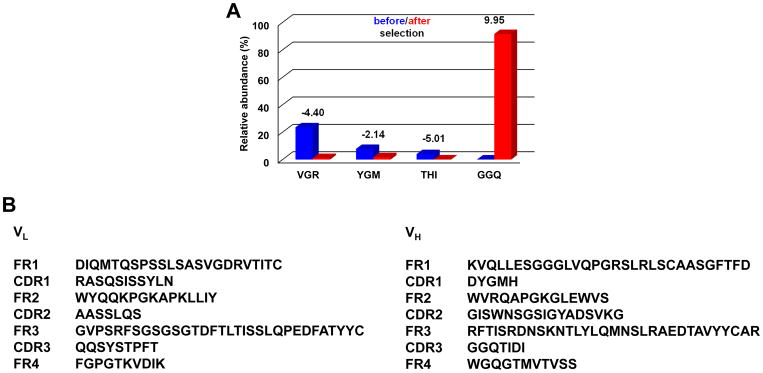
Using reverse transcribed mRNA from cryopreserved PBMCs prepared six months after alloHSCT from a CLL patient, who eventually achieved a complete response, we previously generated a fully human Fab library using our phage display vector pC3C (5). We selected the library by three rounds of whole-cell panning using primary CLL cells from an untreated and unrelated CLL patient and identified a group of highly homologous post-alloHSCT mAbs with identical third complementarity-determining regions of heavy and light chain (HCDR3 and LCDR3, respectively), which indicated a shared CLL cell surface antigen (5). To determine the abundance of these mAbs in the post-alloHSCT antibody repertoire before selection, we sequenced by NGS the cDNA fragments encoding variable heavy chain domain (VH) in the library (Fig. 1A). Of 402,319 sequenced VH with at least one identical repeat, 373 (0.093%) contained the defining HCDR3 amino acid sequence GGQTIDI that we reported previously (5). A relative abundance of ~0.1% on the protein level would correspond to serum antibodies at ~10 μg/mL, a concentration high enough to contribute to the primary CLL cell–directed post-alloHSCT serum antibody titers we detected (5). After selection, the abundance of GGQTIDI increased to 91.5% (1,107,350 of 1,209,577 sequenced VH) (Fig. 1A).
Figure 1. NGS analysis of alloHSCT Fab-phage library selection.
A, shown is the relative abundance of the three most frequent HCDR3 amino acid sequences before selection and the most frequent HCDR3 motif after selection of the alloHSCT Fab-phage library by three rounds of panning on CLL cells. VGR stands for VGRLQYDYYFDS; YGM stands for YGMDV; THI stands for THIVVVIPPGYFDL; GGQ stands for GGQTIDI (the HCDR3 of JML-1). Blue and red columns show the respective relative abundance before and after selection. The values on top of the paired columns give the log2 enrichment. B, amino-acid sequences of the variable light (VL) and heavy chain domains (VH) of selected post-alloHSCT Fab JML-1 (U.S. Patent 8,877,199). FR and CDR denote framework regions and complementarity-determining regions, respectively.
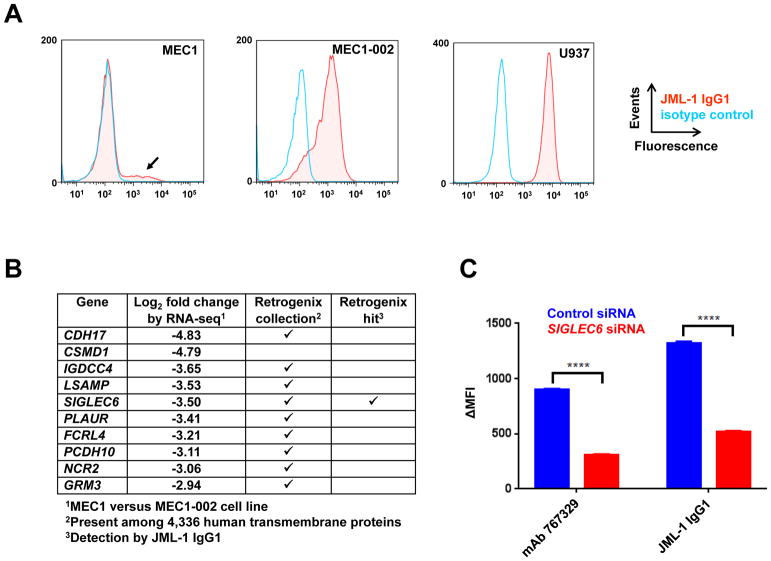
The strongest binder in the panel of post-alloHSCT Fabs was converted to IgG1 and named JML-1 (Fig. 1B) (5). Extensive attempts to identify the antigen of mAb JML-1 by immunoprecipitation and mass spectrometry as well as by cDNA expression cloning failed. These approaches were complicated by the fact that our initial study detected the antigen only on primary CLL cells and not on a large panel of B-cell lines we tested (5). However, we noted that a subpopulation of the CLL cell line MEC1 (11) was stained by mAb JML-1 (Fig. 2A). Using FACS, this subpopulation was enriched and designated MEC1-002 (Fig. 2A). Incubation of MEC1-002 with trypsin, but not with phosphatidylinositol-specific phospholipase C (PI-PLC), revealed loss of mAb JML-1 binding in correlation with incubation time. This suggested that the antigen was a transmembrane protein. A differential RNA-seq analysis of MEC1-002 versus MEC1 cells identified enriched mRNAs encoding transmembrane proteins in MEC1-002 cells (Fig. 2B). To narrow down this list with an independent approach, 4,336 human transmembrane proteins were screened with JML-1 IgG1 using a Retrogenix cell microarray (8). The reproducibly strongest signal in primary and confirmatory screens came from Siglec-6. None of the other transmembrane proteins identified by RNA-seq and present in the cell microarray assay were detected (Fig. 2B). Unlike any human cell line we tested previously, myeloid cell line U937, which is known to express Siglec-6 (12), was strongly bound by mAb JML-1 (Fig. 2A). When MEC1-002 cells were transfected with siRNA targeting SIGLEC6 mRNA, the detection by flow cytometry of both mAb JML-1 and commercial mouse anti-human Siglec-6, mAb 767329, was significantly silenced compared to MEC1-002 cells transfected with control siRNA (Fig. 2C).
Figure 2. Discovery of Siglec-6 as the antigen of mAb JML-1.
A, the binding of biotinylated JML-1 IgG1 (red) and a biotinylated human IgG1 isotype control (light blue) to MEC1 (left), MEC1-002 (middle), and U937 (right) cell lines was detected with PE-conjugated streptavidin by flow cytometry. A representative out of three independent experiments is shown. B, listed are the top ten RNA-seq hits of genes encoding human transmembrane proteins with differential expression in MEC-1 and MEC1-002 cells. Of these, nine were present in the Retrogenix cell microarray of a collection of 4,336 human transmembrane proteins and one (Siglec-6 encoded by SIGLEC6) was detected with JML-1 IgG1 in primary and confirmatory screens. C, MEC1-002 cells were incubated with siRNA targeting SIGLEC6 (red) or a control siRNA (blue) and then analyzed by flow cytometry for binding by either biotinylated JML-1 IgG1 in conjunction with PE-conjugated streptavidin or mouse anti-human Siglec-6 mAb 767329 in conjunction with Alexa Fluor 647-conjugated goat anti-mouse IgG pAbs. Three independent experiments that were carried out are shown as mean ± SD values and analyzed for significance by two-way ANOVA (****, p < 0.0001).
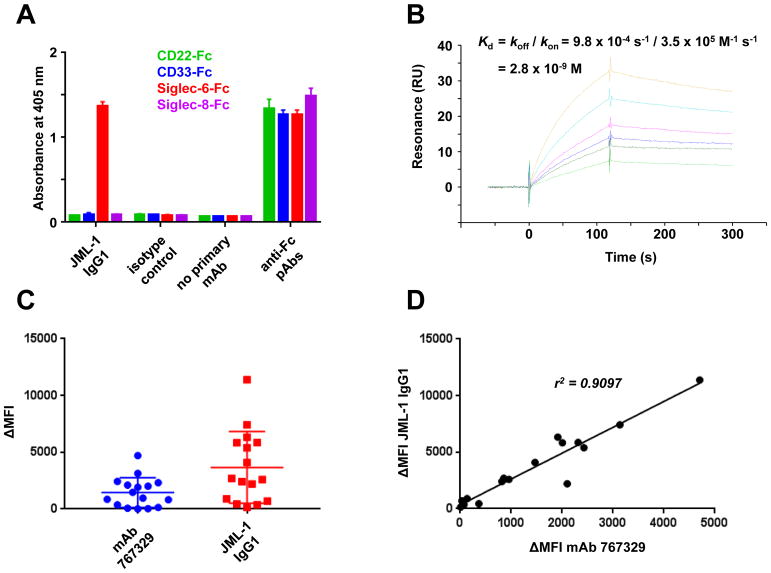
To verify that Siglec-6 is indeed recognized by mAb JML-1, we used commercial Fc fusion proteins of CD22 (a.k.a. Siglec-2), CD33 (a.k.a. Siglec-3), Siglec-6, and Siglec-8. The latter three belong to the Siglec-3 family (13) and share 52–63% amino acid sequence identity in their extracellular domain (12,14). JML-1 IgG1 strongly and exclusively bound to Siglec-6 in ELISA (Fig. 3A). Strong binding (Kd = 2.8 nM) to immobilized Siglec-6 was also found for JML-1 Fab, as measured by SPR (Fig. 3B). In addition, reselection of the alloHSCT Fab-phage library by four rounds of panning on an immobilized recombinant Siglec-6-Fc fusion protein again yielded Fabs with the same HCDR3 and LCDR3 amino-acid sequences as JML-1 (Fig. 1B) as predominant binders.
Figure 3. Validation of Siglec-6 as the antigen of mAb JML-1.
A, the binding of biotinylated JML-1 IgG1 and a biotinylated human IgG1 isotype control to plate-bound human CD22-Fc (green), CD33-Fc (blue), Siglec-6-Fc (red), and Siglec-8-Fc (purple) was detected by ELISA using HRP-conjugated streptavidin. HRP-conjugated goat anti-human Fc pAbs were used to confirm the presence of the plate-bound Fc fusion proteins. Shown are technical triplicates (mean ± SD) of one of three independent experiments. B, SPR sensorgrams obtained on a Biacore X100 instrument for the binding of 6.25 nM (dark green and light green), 12.5 nM (blue), 25 nM (pink), 50 nM (light blue), and 100 nM (light brown) JML-1 Fab to Siglec-6-Fc captured on a goat anti-human Fc-coated CM5 chip. All six SPR sensorgrams were used to calculate kon, koff, and Kd. A representative one of three independent experiments is shown. C, a panel of 16 cryopreserved treatment-naïve CLL PBMCs were stained by flow cytometry using mouse anti-human Siglec-6 mAb 767329 followed by Alexa Fluor 647-conjugated goat anti-mouse IgG pAbs and, separately, biotinylated JML-1 IgG1 followed by PE-conjugated streptavidin. ΔMFI denotes the staining above background staining with corresponding isotype controls. A representative one of the one to two independent experiments for each sample is shown. D, the ΔMFI values from the two separate stainings of each of the 16 primary CLL PBMCs were plotted to analyze their Pearson correlation coefficient r2.
For further validation, we stained a panel of 16 treatment-naïve primary CLL PBMCs with (i) JML-1 IgG1 and (ii) mAb 767329 by flow cytometry (Fig. 3C). As we observed previously for a smaller set of primary CLL cells (5), the MFI revealed a wide range of Siglec-6 cell surface densities on the malignant B cells. The staining of mAbs JML-1 and 767329 correlated strongly (r2 = 0.9097), further corroborating that Siglec-6 was the common antigen (Fig. 3D).
Like other Siglecs, Siglec-6 is characterized by a sialic acid binding site in the extracellular domain and an intracellular ITIM motif (13,15), implying a role in cell adhesion, migration, and signaling, all of which could be attenuated by post-alloHSCT antibodies that contribute to the GVL response. Although the scarcity of post-alloHSCT serum and PBMCs sample collections has precluded us from examining Siglec-6 reactivity in additional CLL patients, our concerted antibody drug and target discovery strategy (i) unveiled Siglec-6 as a broadly expressed CLL cell surface antigen and (ii) delivered fully human anti-human Siglec-6 mAbs. The expression of SIGLEC6 mRNA and Siglec-6 protein in healthy individuals is highly restricted to the placenta, to mast cells, and to exhausted B cells (www.proteinatlas.org/ENSG00000105492-SIGLEC6/tissue) (12, 16). Siglec-6 is not expressed on hematopoietic stem cells (17). Collectively, we found that Siglec-6 is broadly expressed in CLL and warrants interrogation as a candidate target for antibody-based immunotherapeutic interventions. Mining human antibody repertoires by whole-cell panning using phage display or other methods (18) provides a powerful means for antigen-agnostic selection of fully human mAbs. The approaches we used here for antigen identification support concerted antibody drug and target discovery.
Synopsis.
Mining the antibody repertoire of patients responding well to allogeneic hematopoietic stem cell transplantation (alloHST) can unveil targets with therapeutic potential. Through the use of phage display, Siglec-6 was identified as a major antigenic target of such antibodies.
Acknowledgments
We thank Drs. Steven R. Head, Phillip Ordoukhanian, and Shuli Kang from the Next Generation Sequencing and Microarray Core Facility of The Scripps Research Institute in La Jolla, CA for acquiring and analyzing NGS data, and Dr. Pabalu Karunadharma, Wayra Navia, and Nicholas Bild from the Genomics Core of The Scripps Research Institute in Jupiter, FL for acquiring and analyzing differential RNA-seq data. We also thank Marko Modric and Drs. Ivan Samija, Roger R. Beerli, Peter Steinberger, and Christopher S. Hourigan for conducting earlier attempts to discover the antigen of mAb JML-1. This is manuscript 29629 from The Scripps Research Institute.
Grant Support: This study was supported by NIH grant U01 CA174844 and by the Intramural Research Programs of NCI, NIH and NHLBI, NIH.
Footnotes
Disclosure of Potential Conflicts of Interest: The authors declare no competing financial interest. C.R. and S.B. are named inventors on U.S. Patent 8,877,199 claiming the group of post-alloHSCT mAbs that includes JML-1.
References
- 1.Hallek M, Shanafelt TD, Eichhorst B. Chronic lymphocytic leukemia. Lancet. 2018;391:1524–37. doi: 10.1016/S0140-6736(18)30422-7. [DOI] [PubMed] [Google Scholar]
- 2.Boyiadzis M, Foon KA, Pavletic S. Hematopoietic stem cell transplantation for chronic lymphocytic leukemia: potential cure for an incurable disease. Expert Opin Biol Ther. 2007;7:1789–97. doi: 10.1517/14712598.7.12.1789. [DOI] [PubMed] [Google Scholar]
- 3.Mato A, Porter DL. A drive through cellular therapy for CLL in 2015: allogeneic cell transplantation and CARs. Blood. 2015;126:478–85. doi: 10.1182/blood-2015-03-585091. [DOI] [PubMed] [Google Scholar]
- 4.Wu CJ, Ritz J. Induction of tumor immunity following allogeneic stem cell transplantation. Adv Immunol. 2006;90:133–73. doi: 10.1016/S0065-2776(06)90004-2. [DOI] [PubMed] [Google Scholar]
- 5.Baskar S, Suschak JM, Samija I, Srinivasan R, Childs RW, Pavletic SZ, et al. A human monoclonal antibody drug and target discovery platform for B-cell chronic lymphocytic leukemia based on allogeneic hematopoietic stem cell transplantation and phage display. Blood. 2009;114:4494–502. doi: 10.1182/blood-2009-05-222786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byrd JC. Hunting for the Achilles’ heel of CLL. Blood. 2009;114:4324. doi: 10.1182/blood-2009-09-239137. [DOI] [PubMed] [Google Scholar]
- 7.Hofer T, Tangkeangsirisin W, Kennedy MG, Mage RG, Raiker SJ, Venkatesh K, et al. Chimeric rabbit/human Fab and IgG specific for members of the Nogo-66 receptor family selected for species cross-reactivity with an improved phage display vector. J Immunol Methods. 2007;318:75–87. doi: 10.1016/j.jim.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng H, Nerreter T, Chang J, Qi J, Li X, Karunadharma P, et al. Mining naive rabbit antibody repertoires by phage display for monoclonal antibodies of therapeutic utility. J Mol Biol. 2017;429:2954–73. doi: 10.1016/j.jmb.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMB J. 2011;17:10. [Google Scholar]
- 10.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–63. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stacchini A, Aragno M, Vallario A, Alfarano A, Circosta P, Gottardi D, et al. MEC1 and MEC2: two new cell lines derived from B-chronic lymphocytic leukaemia in prolymphocytoid transformation. Leuk Res. 1999;23:127–36. doi: 10.1016/s0145-2126(98)00154-4. [DOI] [PubMed] [Google Scholar]
- 12.Patel N, Brinkman-Van der Linden EC, Altmann SW, Gish K, Balasubramanian S, Timans JC, et al. OB-BP1/Siglec-6. a leptin- and sialic acid-binding protein of the immunoglobulin superfamily. J Biol Chem. 1999;274(32):22729–38. doi: 10.1074/jbc.274.32.22729. [DOI] [PubMed] [Google Scholar]
- 13.Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7:255–66. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 14.Floyd H, Ni J, Cornish AL, Zeng Z, Liu D, Carter KC, et al. Siglec-8. A novel eosinophil-specific member of the immunoglobulin superfamily. J Biol Chem. 2000;275:861–6. doi: 10.1074/jbc.275.2.861. [DOI] [PubMed] [Google Scholar]
- 15.O’Reilly MK, Paulson JC. Siglecs as targets for therapy in immune-cell-mediated disease. Trends Pharmacol Sci. 2009;30:240–8. doi: 10.1016/j.tips.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kardava L, Moir S, Wang W, Ho J, Buckner CM, Posada JG, et al. Attenuation of HIV-associated human B cell exhaustion by siRNA downregulation of inhibitory receptors. J Clin Invest. 2011;121:2614–24. doi: 10.1172/JCI45685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen DH, Ball ED, Varki A. Myeloid precursors and acute myeloid leukemia cells express multiple CD33-related Siglecs. Exp Hematol. 2006;34:728–35. doi: 10.1016/j.exphem.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Beerli RR, Rader C. Mining human antibody repertoires. mAbs. 2010;2:365–78. doi: 10.4161/mabs.2.4.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]