Abstract
Cell type origin is one of the factors that determine molecular features of tumors, but resources to validate this concept are scarce because of technical difficulties in propagating major cell types of adult organs. Previous attempts to generate such resources to study breast cancer have yielded predominantly basal-type cell lines. We have created a panel of immortalized cell lines from core breast biopsies of ancestry-mapped healthy women that form ductal structures similar to normal breast in 3D cultures and expressed markers of major cell types including the luminal-differentiated cell-enriched ERα-FOXA1-GATA3 transcription factor network. We have also created cell lines from PROCR (CD201)+/EpCAM- cells that are likely the “normal” counterpart of the claudin-low subtype of breast cancers. RNA-seq and PAM50 intrinsic subtype clustering identified these cell lines as the “normal” counterparts of luminal A, basal, and normal-like subtypes and validated via immunostaining with basal-enriched KRT14 and luminal-enriched KRT19. We further characterized these cell lines by flow cytometry for distribution patterns of stem/basal, luminal-progenitor, mature/differentiated, multi-potent PROCR+ cells, and organogenesis-enriched epithelial/mesenchymal hybrid cells using CD44/CD24, CD49f/EpCAM, CD271/EpCAM, CD201/EpCAM, and ALDEFLUOR assays and E-Cadherin/Vimentin double-staining. These cell lines showed inter-individual heterogeneity in stemness/differentiation capabilities and baseline activity of signaling molecules such as NF-κB, AKT2, pERK, and BRD4. These resources can be used to test the emerging concept that genetic variations in regulatory regions contribute to widespread differences in gene expression in ‘normal’ conditions among the general population and can delineate the impact of cell type origin on tumor progression.
Introduction
Normal breast epithelial cells are hierarchically organized broadly into bipotent mammary stem/basal (MaSCs), luminal progenitor, and mature/differentiated luminal cells (1,2). Luminal progenitor cells have been further classified into bipotent and committed progenitor cells based on cell surface marker profiles and expression patterns of keratins (2). While basal cells express keratin 14 (KRT14) and luminal cells express keratin 19 (KRT19), cells expressing both keratins show luminal progenitor phenotype (3). Each of these cell types is associated with distinct transcription factor networks; TP63 and NFIB in basal cells, ELF5 and EHF in luminal progenitors, and ESR1 and FOXA1 in luminal cells (4). Although 11 different cell types have been described, it is acknowledged that current methods of sorting and classifying cell types based on surface markers and keratin expression may underestimate the level of heterogeneity in the normal breast (5). Furthermore, recent studies have identified inter-individual genetic variations in non-coding regions affecting gene expression across tissues, thus supporting the concept of inter-individual variability in the normal breast (6–8). Therefore, a clear understanding of the normal breast heterogeneity and signaling pathway differences is needed for better classification of breast tumors and for assessing tumor heterogeneity.
Breast cancers have been sub-classified into five intrinsic subtypes based on gene expression patterns in tumors (9). These include estrogen receptor alpha (ERα)-positive luminal A and luminal B subtypes, HER2+ subtype, basal-subtype and normal-like subtype. Another relatively rare molecular subtype called the claudin-low has been added subsequently, which is believed to originate from MaSCs (10). It is suggested that bipotent progenitor or luminal progenitors are the cell-type-origin of basal breast cancers (11). HER2+ tumors may arise from late luminal progenitors, whereas luminal A and luminal B breast cancers may originate from differentiated luminal cells (11). Experimental validation of these possibilities is still challenging because most of the prior culturing methods favored the outgrowth of basal-like breast epithelial cells and primary cells need to be directly used for transformation to obtain tumors with luminal and basal-like characteristics (12). Indeed, the most commonly used human mammary epithelial cells (HMECs) and MCF10A cells have basal-like gene expression pattern and transformation of these cells gives rise to squamous cell carcinomas instead of adenocarcinomas (13,14). Only one study has reported a method to generate cells with luminal characteristics and transformed counterpart of these cells giving rise to tumors resembling human breast adenocarcinomas (13). For unknown reasons, this methodology has not been adapted widely.
The majority of “normal” tissue for breast cancer-related studies is derived from reduction mammoplasty or tissues adjacent to normal. However, a recent study that compared normal breast tissue donated by healthy volunteers to Komen Normal Tissue Bank at the Indiana University, reduction mammoplasty, and tumor adjacent normal tissues found significant levels of histologic abnormalities in reduction mammoplasty as well as in tumor-adjacent normal specimens (15). Additionally, “normal” tissue adjacent to tumors undergoes extensive DNA methylation changes, specifically targeting transcription factor binding sites specifying chromatin architecture and stem cell differentiation pathways including Wnt and FGF signaling networks, due to “field effects” attributed to tumors (16). Thus, although several publications in literature have described generation of breast epithelial cell lines using tissues from reduction mammoplasty (12,17–26), these cell lines are less likely suitable for mechanistic studies because of inherent genomic abnormalities and limited luminal cell features. Moreover, a maximum of four cell lines were created in each of these studies, which may not be sufficient to study inter-individual variations in signaling pathways downstream of specific genomic aberrations. The mechanistic studies in breast cancer field, therefore, would benefit from having multiple normal/immortalized cells with luminal characteristics from healthy women to critically evaluate interplay between cell-type-origin, inter-individual differences in normal genome, and cancer-specific genomic aberrations driving tumor growth.
We recently reported a modified epithelial reprogramming growth condition that allowed propagation of breast epithelial cells with stem/basal, luminal progenitor, and differentiated/mature cell characteristics from core breast biopsies of healthy women (27). We reported significant inter-individual variability in terms of specific cell types and an enrichment of PROCR (CD201)+/EpCAM- subpopulation of cells in women of African Ancestry. Here we describe creation of a panel of immortalized breast epithelial cell lines with gene expression pattern closely resembling the intrinsic subtypes of breast cancer and with features of cells prevalent during organogenesis (28). This resource will be made available to research community with the goal to address critical issues related to interplay between cell-type-origin and specific genomic aberration driving breast cancer and to provide diversity in “normal” cell types reflecting inter-individual variation and response to exogenous stimuli.
Materials and Methods
Primary cell culture and immortalization
Primary breast epithelial cells were created from fresh or cryopreserved, de-identified normal breast tissues of healthy Caucasian, Hispanic and African American women donated to the Komen Tissue Bank (KTB) after informed written consent from subjects. All experiments were carried out in accordance with the approved guidelines of the Indiana University Institutional Review Board. International Ethical Guidelines for Biomedical Research Involving Human Subjects were followed. Tissues were dissociated using a 10% collagenase/hyaluronidase mixture (#07919, Stem Cell Technologies) and 10 μM ROCK inhibitor (ALX-270–333-M005, Enzo Life Sciences) in culture media for two hours at 37°C. The dissociated cells were filtered through sterile 70-micron filter, washed in media and centrifuged for 5 minutes at 1000 RPM. The cells were co-cultured on irradiated murine embryonic fibroblasts (Applied StemCell, Inc.) using media described previously (27). Cells were immortalized by human telomerase gene (hTERT) retrovirus using the vector pLXSN-hTERT. Retrovirus was prepared using the amphophoenix cells and primary cells were infected with viral supernatant along with 8 μg/ml polybrene (H9268, Sigma-Aldrich) for 4 hours and immortalized cells were selected by 100 μg/ml G418 (61–234, Corning). Since all cell lines are derived from normal breast tissues and are not established cell lines, authentication is not possible. However, based on karyotyping (Figure S1A), it is clear that these are not contaminating established cell lines. Most of the cell lines used are <20 passages. Cell lines in the laboratory are typically tested for mycoplasma once in two years.
Genetic ancestry mapping and genotype analysis
Leukocyte DNA was obtained from the KTB. Blood was collected using the BD-Vacutainer spray-coated K3EDTA tubes (Becton Dickinson). Tubes were centrifuged for 15 min at 2000 RPM. Once the upper phase (plasma) was removed, the tubes were stored in −80oC. Peripheral blood leukocyte DNA was extracted using AutoGenprep 965 (Autogen, Inc.). Genotyping was performed using the KASP technology (LGC Genomics) and a 41-SNP panel (labeled 41-AIM panel) selected from Nievergelt et al, (29). Genotype analysis using the 41 SNPs panel along with a Bayesian clustering method (Structure Software V2.3.4) was able to discern continental origins including European (Caucasian)/Middle East, East Asia, Central/South Asia, Africa, Americas, and Oceania. A reference set was obtained from the Human Genome Diversity Project (HGDP).
RNA sequencing
The concentration and quality of total RNA samples were first assessed using Agilent 2100 Bioanalyzer. A RIN (RNA Integrity Number) of five or higher was required to pass the quality control. 500 nanograms of RNA per sample were used to prepare dual-indexed strand-specific cDNA library using TruSeq Stranded mRNA Library Prep Kit (Illumina). The resulting libraries were assessed for its quantity and size distribution using Qubit and Agilent 2100 Bioanalyzer. 200 picomolar pooled libraries were utilized per flowcell for clustering amplification on cBot using HiSeq 3000/4000 PE Cluster Kit and sequenced with 2×75bp paired-end configuration on HiSeq4000 (Illumina) using HiSeq 3000/4000 PE SBS Kit. A Phred quality score (Q score) was used to measure the quality of sequencing. More than 90% of the sequencing reads reached Q30 (99.9% base call accuracy).
The sequencing data were first assessed using FastQC (Babraham Bioinformatics) for quality control. All sequenced libraries were mapped to the human genome (UCSC hg19) using STAR RNA-seq aligner with the following parameter: “--outSAMmapqUnique 60”. The reads distribution across the genome was assessed using bamutils (from ngsutils) (30). Uniquely mapped sequencing reads were assigned to hg19 refGene genes using featureCounts (from subread) with the following parameters: “-s 2 –p –Q 10”. Genes with read count per million (CPM) > 0.5 in more than three of the samples were kept. The data were normalized using TMM (trimmed mean of M values) method. Differential expression analysis was performed using edgeR R package (31). False discovery rate (FDR) was computed from p-values using the Benjamini-Hochberg procedure. The multi-dimensional scaling (MDS) plot was drawn using plotMDS function in edgeR to visualize the differences between the expression profiles of different samples in two dimensions. Raw sequencing data have been submitted to GEO (accession number GSE108541).
Predictor Analysis of Microarray (PAM50) classification
Gene expression was averaged from three replicates for each cell line. Based on the nearest PAM50 centroid algorithm, intrinsic breast cancer subtypes were assigned using the Pam50.robust model from genefu R package (32). Luminal cell line MCF7 and basal cell lines human mammary epithelial cells (HMEC) and MCF10A were included as controls. Unsupervised hierarchical clustering of PAM50 gene was performed and shown by using the function of heatmap.2 in gplots R package.
Heatmap of pairwise comparison of cell lines
Differential expression analysis was performed between every two different cell lines. A total of 11,722 genes with FDR < 0.01 and absolute value of fold change > 1 in any pairwise comparison was selected for unsupervised hierarchical clustering.
Flow cytometry, Flow cytometry imaging and Data analysis
Breast epithelial cells were collected by trypsinization, stained using antibodies CD49f-APC (FAB13501A), AXL-APC (FAB154A) (R&D Systems), PROCR (CD201)-PE (130–105-256), EpCAM-PE (130–091-253), EpCAM-APC (130–091-254), EpCAM- VioBlue (130–097-324) (Miltenyi Biotech Inc.), CD271-APC (345108) (Biolegend), CD44-APC (559942), CD24-PE (555428) (BD Pharmingen), Pan-Cytokeratin-FITC (F3418) (Sigma Aldrich) and ALDEFLUOR (01700) (STEMCELL Technologies), and were acquired using a BD LSR II flow cytometer. Data were analyzed using CellQuest or FlowJo software. Forward and side scatter were used to ensure that only live cells were considered in the analysis. Gating was done using appropriate FITC (555573), PE (555749), APC (555576) (BD Pharmingen) and VioBlue (130–094-670) (Miltenyi Biotech) isotype control antibodies and only a representative isotype control for two fluorescent markers are shown.
For imaging flow cytometry, 500,000–1,000,000 cells per sample were stained with the following conjugated antibodies: 1:100 mouse anti-Vimentin Clone V9-Cy3, (C-9080, Sigma-Aldrich), and 1:20 mouse anti-CD324/E-Cadherin Clone 67A4-APC, (A15717, Molecular Probes). The nucleus was stained with 1:5000 Hoechst 33342 (H-3570, Molecular Probes). Cells were fixed and permeabilized using BD Cytofix/Cytoperm solution (51–2090KZ, BD Bioscience) for 20 minutes at 4°C. Blocking was done with 5% goat serum in 1X BD Perm/Wash solution (51–2091KZ, BD Bioscience) for 15 minutes at 4°C. Single color control samples and cocktail sample with all the antibodies were incubated for 30 minutes at 4°C in dark. Cells were washed twice for 5 minutes between each step with 1x BD Perm/Wash solution. The last step of washing was done twice with 1x PBS to eliminate the detergent from the cell suspension. Cells were transferred to the UltraClear boil proof microcentrifuge tubes (Inc-607-GMT, Dot Scientific). The stained cells were stored in 1% formalin in dark at 4°C. 5000 images were analyzed using ImageStream Data Exploration and Analysis Software (IDEAS). In-focus cells were evaluated after gating on live single cells based on an aspect ratio near 1 and a low area of the bright field.
Antibodies and Western blotting analysis
Primary antibody against p16 (ab108349) was purchased from Abcam. An antibody against ERK (sc-94) was purchased from Santa Cruz Biotechnologies. Antibodies against pERK (9101), AKT1 (2967), AKT2 (5239), pAKT1 (9018), pAKT2 (8599), AKT (9272) and BRD4 (13440) were purchased from Cell Signaling Technologies. Antibody against E-cadherin (610181) was purchased from BD Biosciences. Antibody against Vimentin (C9080) and β-actin (A5441) were purchased from Sigma-Aldrich. Horseradish peroxidase (HRP) linked secondary antibodies against mouse, rabbit and goat were purchased from Santa Cruz Biotechnologies. Cell lysates prepared in radioimmunoassay buffer were analyzed by Western blotting as described previously (33).
Mammosphere formation assay and cell proliferation assay
Single cell suspensions were cultured to form mammospheres in ultra-low attachment 6-well plate at a density of 5000 cells/mL in MammoCult basal medium (human) supplemented with MammoCult proliferation supplement (human), Heparin, 1% penicillin/streptomycin (STEMCELL Technologies Inc.), and hydrocortisone. Phase contrast images of mammospheres were taken at day 6. For secondary and tertiary cultures, mammospheres were collected by centrifugation, washed, trypsinized, filtered through 40μm filter and equal number of cells were replated in mammosphere media. Mammospheres were counted using ImageJ. For cell proliferation assay, cells were seeded in 96-well plates followed by treatment with TGFβ1 (1 ng/ml; 7754-BH, R&D Systems) for 3–4 days. At the end of treatment, Bromodeoxyuridine (BrDU) incorporation-ELISA was done using BrDU proliferation assay kit (Calbiochem/Millipore) according to the manufacturer’s instructions.
Karyotyping of immortalized cell lines
Metaphase chromosome spreads from KTB cell lines at passage between 10–20 were prepared by culturing proliferative cells with 0.05 μg/ml colcemid (10295892001, Sigma-Aldrich) for 2–12 hrs depending on the cell proliferation rate followed by resuspension in hypotonic solution (75 mM KCl) for 15 min at 37 °C. Cells were fixed by resuspension in Carnoy’s Fixative (3:1 methanol:acetic acid) onto a clean dry slide and allowed to air dry. Fixed cells were stained with Hoechst 33342 (Molecular Probes H-3570) and analyzed under microscope. More than five metaphase chromosome spreads were analyzed for each cell line.
3D in vitro matrigel, collagen, and hydrogel assays
Eight-chamber cover glass system (155409, Lab-Tek II) was coated with 40 μl of matrigel (354234, Corning) per well and incubated for 30 minutes at 37 °C for the gel to solidify. 6000 cells in 400 μl of overlay media comprising primary cell media with 2% matrigel were added to the matrigel coated wells. The media was changed every 3–4 days and cells were cultured for 10 to 12 days.
Floating Collagen scaffolds were prepared based on a protocol from Linnemann et al. 2015, (34) with some modifications. 5000 cells per well in a 48 well plate were used. The final concentration of the rat-tail collagen I (A10483–01, Corning) used for the scaffold was 1.3 mg/ml. 550 mM of HEPES buffer amounting to 1/10th volume of the collagen was used as a neutralizing solution. The neutralizing solution was first added to the cell suspension, followed by a quick addition of rat-tail collagen. 200 μL of the collagen-cell mixture was immediately plated into 48 well plate kept on ice. The gels were incubated for 1 hour at 37ºC to promote polymerization. 500 μL of Mammary Epithelial Basal Medium (CC-3151, Lonza) with 3 μM Y-27632 and 10 μM forskolin were carefully added to wells. The gels were then detached from the bottom of the well by encircling pipet tip around the edge of the gel. The media was changed every 3 days and cells were cultured for up to 10 days. After day 3, Y-27632 was withdrawn from the medium.
Floating hydrogel scaffold was prepared based on protocol from Miller et al (35). Briefly, 10,000 cells per well were used in a 4-chamber slide. 25x extracellular matrix was made by adding 0.5 mg/ml laminin (23017015, Thermo Fisher), 0.25 mg/ml hyaluronic acid (385908, Millipore), and 0.5 mg/ml fibronectin (33016015, Thermo Fisher) in PBS. The final concentration of rat-tail collagen I was 1.3 mg/ml. To make 1 ml of the hydrogel, collagen I was first neutralized with 12.5% of 0.1N NaOH followed by the addition of ice cold MEGM to make the volume 910 μl. 40 μl of extracellular matrix was quickly added. Lastly, 50 μl of cell suspension was added to the mixture and pipetted up and down 3–4 times to uniformly distribute the cells in the mixture. 200 μl of the hydrogel mixture was added to the 4-well chamber slide kept on ice. The gel was incubated for 1 hour at 37ºC. 1 ml MEGM was added after the polymerization was complete. The gels were then detached from the bottom of the well by encircling a P200 pipet tip around the edge of the gel. All the steps were done on ice. Media was changed twice a week and the cells were cultured for 10–14 days.
RNA extraction and quantitative real-time PCR
Total RNA was isolated using RNeasy Kit (74106, Qiagen) and 1 μg of RNA was used to synthesize cDNA with Bio-RAD iScript cDNA Synthesis Kit (170–8891). Quantitative real-time PCR (qRT-PCR) was performed using Taqman universal PCR mix and predesigned gene expression assays with best coverage from Applied Biosystems. The following assays were used in our study: ACTB (Hs01060665_g1), ESR1 (Hs01046816_m1), GATA3 (Hs00231122_m1), FOXA1 (Hs04187555_m1), FOXC1 (Hs00559473_s1), ZEB1 (Hs01566408_m1), SNAI1 (Hs00195591_m1), SNAI2 (Hs00161904_m1), TWIST1 (Hs00361186_m1), TWIST2 (Hs00382379_m1), EHF (Hs00171917_m1) and TP63 (Hs00978340_m1).
Electrophoretic mobility shift assay (EMSA)
EMSA with whole cell extracts from immortalized cell lines was performed as described previously (33). Oligonucleotides with consensus DNA binding sites for NF-κB (E3292), AP-1 (E3201), and OCT-1 (E3242) were purchased from Promega Corporation.
Immunofluorescence of monolayer culture and 3D collagen structures
Cells were seeded in monolayer glass bottomed microwell dish (P35G-0–14-C, MatTek Corporation) overnight and fixed with 4% paraformaldehyde for 10 minutes at room temperature (RT). Cells were blocked with a blocking buffer comprising 5% goat serum in 1% BSA and 0.1% triton-x-100 in PBS for 1 hour at RT or overnight at 4°C. KRT14 (ab7800, Abcam) and KRT19 (Abcam, ab52625) primary antibodies were diluted to 1:200 in the antibody diluents (S0809, Dako) and added to cells for 1.5 hours at RT. Secondary goat anti-rabbit Alexa Fluor 555 (A21428, Life Technologies) and secondary rabbit anti-mouse Alexa Fluor 488 (A11059, Life Technologies) antibodies were diluted to 1:400 and added to cells for 1 hour in dark at RT. Nuclear staining was done with 1:5000 Hoechst 33342 (H-3570, Molecular Probes) for 5 minutes. Cells were washed 3 times for 2 minute each, between every step. Cells were stored in PBS in dark and images were taken within 48 hours of staining with Olympus FV1000 MPE inverted confocal microscope.
Cells in the collagen and hydrogel scaffolds were rinsed twice for 10 minutes with 0.05% Tween 20 in PBS (PBST) and fixed with 4% paraformaldehyde for 20 minutes, followed by quenching with 0.15 M glycine for 10 minutes at RT. Cells were permeabilized with 0.15% triton-x-100 in PBST at RT overnight and were blocked with 5% goat serum and 5% fetal calf serum in PBST for at least 2 hours. KRT14 and KRT19 primary antibodies were diluted to 1:200 in PBST and cells were incubated overnight at 4°C. Secondary goat anti-rabbit Alexa Fluor 555 and rabbit anti-mouse Alexa Fluor 488 antibodies were diluted to 1:300 and added to cells for 2 hour in dark at RT. The nuclei were stained with 1:2500 Hoechst 33342 for 15 minutes. All steps were carried out on a rocker and cells were washed twice for 10 minutes with PBST between every step. The stained scaffolds were stored in PBS and imaged with Olympus FV1000 MPE inverted confocal microscope by placing the gel on the glass surface of the microwell dish with PBS to help the gel stick to the bottom without floating. The XLUMPLFLN-W 20x long working distance objective was used to enable imaging of cells embedded in the gel and 3D images were processed using Imaris software.
Statistical analyses
All experiments were conducted in three biological replicates. Statistical analyses were performed using Prism software program (version 6.0). qPCR, FACS and cell proliferation assay were analyzed using one-way ANOVA. P values <0.05 were considered statistically significant.
Results
Isolation and immortalization of primary breast epithelial cells:
We generated immortalized cell lines from core biopsies of normal breast by overexpressing human telomerase gene (hTERT) in primary cells isolated and propagated under modified epithelial reprogramming assay (Figures 1A). To evaluate the ability of immortalized cell lines (called KTB cell lines hereafter) to form acini resembling normal breast, we grew cells on matrigel. KTB immortalized cell lines formed acini of varying sizes, similar to the acini formed by the most commonly used “normal” cell line MCF10A (Figures 1B).
Figure 1: Generation of cell lines from core biopsies of ancestry mapped healthy women (KTB cell lines).
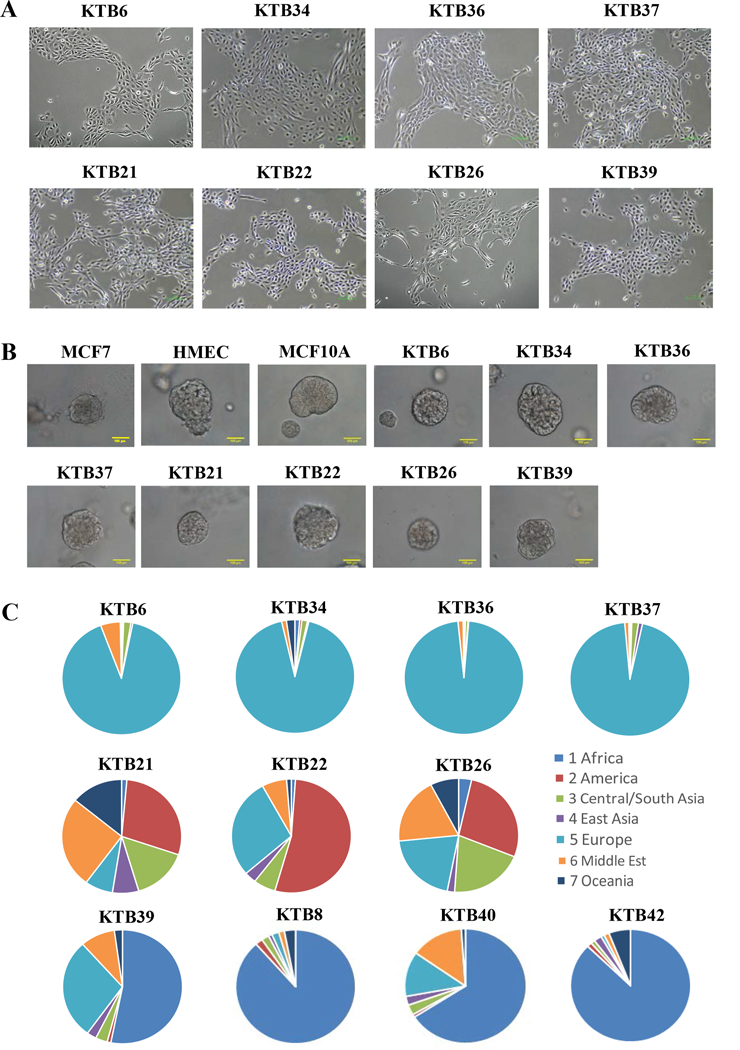
A) Phase contrast images of KTB cell lines. B) KTB cell lines form acini on matrigel. C) Highly discriminative ancestry informative 41-SNP genomic analyses of KTB cell lines show expected heterogeneity in Hispanic women (KTB21, KTB22, and KTB26) compared to Caucasian (KTB6, KTB34, KTB36, and KTB37) and African American Women (KTB8, KTB39, KTB40, and KTB42).
To ensure that KTB cell lines are representative of women of different genetic ancestry, all samples were subjected to highly discriminative ancestry informative 41-SNP (single nucleotide polymorphism) genomic analyses (29). As shown in Figure 1C, while self-reported White women were enriched for European ancestry markers (KTB6, KTB34, KTB36 and KTB37), expectedly, the self-reported Hispanic women (KTB21, KTB22 and KTB26) displayed highly heterogeneous ancestry marker distribution. Four self-reported African American women (KTB8, KTB39, KTB40, and KTB42) had inherited >50% of African ancestry markers. To ensure diploid nature of immortalized cell lines, karyotype analyses were performed after arresting cells at metaphase of the cell cycle by colcemid treatment and, which confirmed 46 chromosomes in all samples (Figure S1A). Several cell lines have undergone >20 passages and remained diploid.
Molecular subtype classification of KTB cell lines:
To determine whether gene expression patterns in immortalized KTB cell lines differ from the commonly used normal cell lines MCF10A and HMECs, we performed RNA-seq analyses. A cell line derived from a BRCA2 mutant carrier 1505–10B was also included in the analyses. Principle component analyses (PCA) revealed distinct differences in gene expression pattern between KTB cell lines compared to HMEC, MCF10A, and the luminal breast tumor cell line MCF7 (Figure 2A). Independent PCA analyses of KTB cell lines showed significant differences between cell lines, further confirming inter-individual variations in characteristics of “normal” breast cell lines (Figure 2B).
Figure 2: Classification of KTB cell lines based on PAM50 subtype predictor.
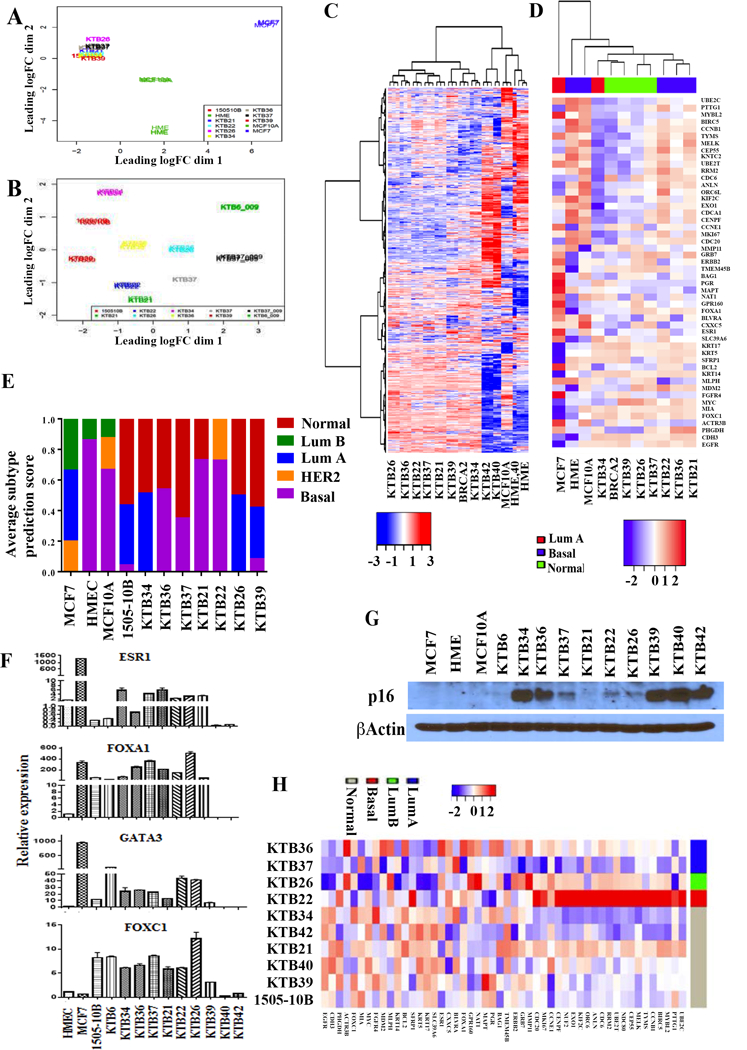
A) PCA analyses of RNA-seq data of all immortalized cell lines including HME, MCF10A and MCF7. 1505–10B is a BRCA2 mutant cell line. B) PCA analyses of RNA-seq data of KTB cell lines show heterogeneity in gene expression. C) Unsupervised clustering of RNA-seq data shows distinct gene expression pattern in KTB cell lines except KTB40 and KTB42 compared to MCF10A and HME. KTB40 and KTB42 clustered with HME and MCF10A. D) Classification of KTB cell lines based on PAM50 subtype classifier. E) Subtype prediction scores of individual cell lines. F) Expression levels of luminal-enriched ESR1, FOXA1, GATA3, and basal-enriched FOXC1 in different KTB cell lines. G) p16(INK4) expression in different cell lines. H) Classification of primary cells prior to immortalization based on PAM50 subtype classifier.
Unsupervised clustering analyses separated KTB cell lines from HMEC and MCF10A with the exception of KTB40 and KTB42, which clustered with HMEC and MCF10A (Figure 2C). Previous studies have shown enrichment of basal cell gene expression pattern in HMEC and MCF10A but the KTB cell lines displayed a distinct gene expression pattern compared to HMEC and MCF10A cell lines (14). We performed ingenuity pathway analyses of 733 genes expressed at higher level in KTB luminal cell lines compared to HMEC, MCF10A, KTB40, and KTB42 (Table S1, Subcluster genes). Canonical pathways typically enriched in luminal cells including ErbB and PAK signaling were elevated in KTB cell lines (11,36) (Table S1, IPA_Pathway).
Encouraged by these results, we next determined whether KTB cell lines represent “normal” counterparts of intrinsic subtypes of breast cancer. Intrinsic subtypes have been predicted to originate from normal counterparts but it has never been demonstrated experimentally (11). Using PAM50 model in genefu, we calculated the subtype probability score for each cell line. KTB34 represented luminal A, KTB21, KTB22, and KTB36 represented basal and KTB26, KTB37, KTB39, and 1505–10B (BRCA2 mutant) represented normal-like subtypes (Figures 2D and 2E). These results document the presence of immortalized cells with gene expression pattern similar to intrinsic subtypes of breast cancer.
To further characterize KTB cell lines for luminal and basal-enriched gene expression, we measured the expression levels of ESR1, FOXA1, GATA3, and FOXC1. While ESR1, FOXA1, and GATA3 are expressed predominantly in luminal cells and form a hormone-responsive lineage-specific transcription factor network, FOXC1 expression identifies cells with basal cell-enriched gene expression (37,38). Indeed, all four of these transcription factors were expressed at variable levels in KTB cell lines and to our knowledge, this is the first report to document immortalized breast epithelial cell lines expressing ESR1, GATA3, and FOXA1 (Figure 2F). We also noted another difference in KTB cell lines and general observation about immortalization of breast epithelial cells. Immortalization is often associated with the loss of cell cycle inhibitor and senescence promoter p16(INK4) (39). However, RNA-seq results showed significant expression of p16(INK4) transcripts in KTB cell lines. Western blotting also confirmed expression of p16(INK4) in several of KTB cell lines (Figure 2G). Thus, immortalized KTB cell lines are unique in their characteristics compared to available cell lines including the expression of luminal-enriched transcription factors and p16(INK4).
To rule out the possibility that intrinsic subtype phenotype is acquired during immortalization, we performed RNA-seq analyses of primary cells prior to immortalization. PAM50 classification identified primary cells with luminal A, luminal B, basal, and normal-like features (Figure 2H). It does appear that immortalization selects for specific subtypes as intrinsic subtype classification prior and after immortalization did not overlap. In particular, KTB40 and KTB42 prior to immortalization clustered as “normal-like” subtype. However, we do note a caveat in our analyses, as RNA-seq of primary and immortalized cells was done at different times due to volume of samples and these comparisons have limitations. Nonetheless, our results clearly show that cells derived from the normal breast prior and after immortalization have gene expression patterns overlapping the gene expression patterns in intrinsic subtypes of breast cancer.
Keratin expression pattern identifies distinct subpopulation of cells within immortalized KTB cell lines:
Keratin 14 (KRT14) is typically expressed at higher levels in basal cells, whereas keratin 19 (KRT19) is expressed in luminal cells. Cells that express both KRT14 and KRT19 (KRT14+/KRT19+) are considered to have luminal progenitor properties (3). We used immunofluorescence to determine expression pattern of these two keratins. Consistent with the results of PAM50 classification, the luminal A cell line KTB34 was strongly positive for KRT19, similar to MCF7, although low level KRT14 expression was seen in this cell line, whereas the basal cell line KTB22 was strongly positive for KRT14 (Figure 3A). The normal-like cell line KTB39 was KRT14+/KRT19+.
Figure 3: Luminal and basal cell-enriched features of KTB cell lines.
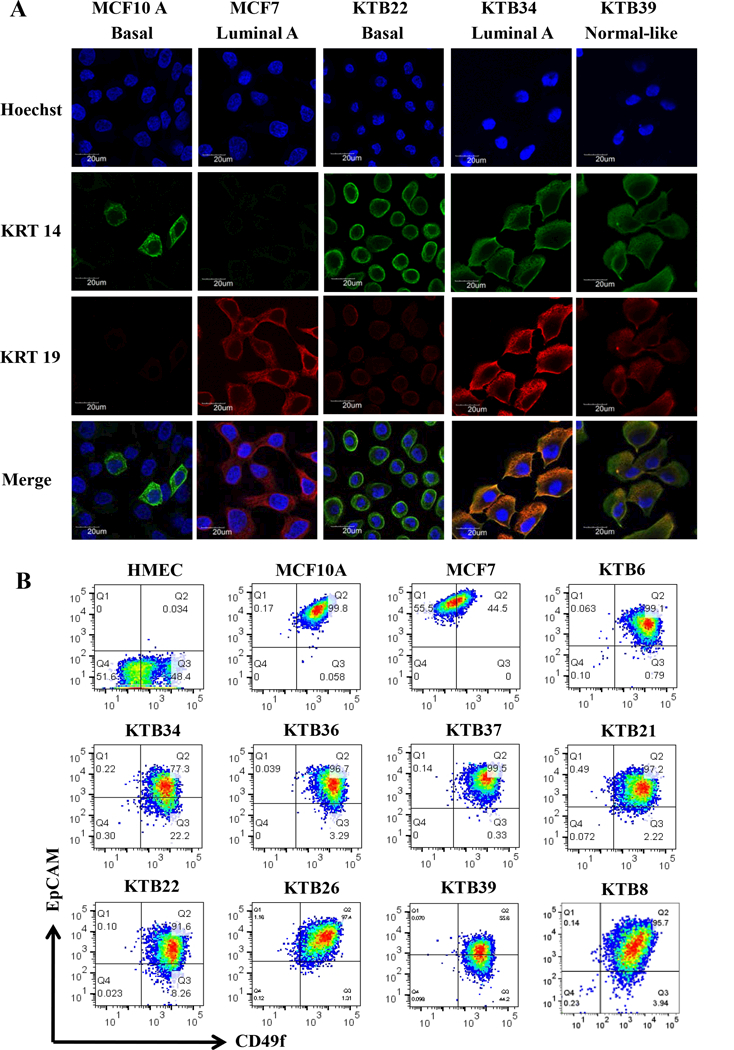
Representative data are presented. A) KRT14 and KRT19 staining patterns of different cell lines. MCF10A and MCF7 were used as controls. B) CD49f and EpCAM staining patterns of cell lines. CD49f+/EpCAM-, CD49f+/EpCAM+ and CD49f-/EpCAM+ cells correspond to stem/basal, luminal-progenitor, and mature-luminal cells, respectively.
Phenotypic heterogeneity in immortalized KTB cell lines:
In order to confirm that the immortalized cell lines show the inter-individual variability as previously shown for the primary breast epithelial cells (27), we subjected KTB cell lines prior to and after immortalization to phenotypic analyses using additional markers that define stem/basal, luminal-progenitor, luminal-differentiated, luminal, and basal cells (1). CD49f+/EpCAM, CD49f+/EpCAM+, and CD49f-/EpCAM+ cells are described as breast stem, luminal progenitor, and mature/differentiated cells, respectively (1). Primary cells prior to immortalization showed inter-individual variation in the levels of all three populations of cells (Figure S1B). CD44/CD24 staining pattern also confirmed inter-individual heterogeneity. After immortalization, this variation decreased considerably with the majority of cell lines displaying luminal progenitor phenotype (CD49f+/EpCAM+) (Figure 3B). However, there was some variation in the intensity of staining as well as the presence of CD49f+/EpCAM- stem/basal subpopulation (Figures 3B). CD49f/EpCAM staining patterns in immortalized KTB cell lines were different from MCF10A, HMEC and MCF7. CD201 (PROCR), which identifies cells with multi-potent stem cell activity in the mouse mammary gland (40), was expressed at variable levels in immortalized KTB cell lines with few cell lines containing a mix of CD201+/EpCAM+ and CD201-/EpCAM+ cells, while the others with only CD201-/EpCAM+ subpopulation (Figure S2). Cell lines showed similar differences in the staining pattern for CD271, which is expressed predominantly in basal cells (41) (Figure S2).
CD44/CD24 and ALDEFLUOR staining identify inter-individual differences in normal counterparts of breast cancer stem cells that transition between epithelial and mesenchymal state:
Recent studies have shown that breast cancer stem cells transition between epithelial and mesenchymal state with proliferative epithelial-like stem cells being positive for aldehyde dehydrogenase (ALDH) and invasive/quiescent mesenchymal stem-like cells being CD44+/CD24- (42). Gene expression pattern in epithelial and mesenchymal cancer stem cells are remarkably similar to luminal and basal cells of the normal breast. We characterized KTB cell lines for the presence of these two subpopulations of cells. While most of the cells in the KTB cell lines were CD44+/CD24+, CD44+/CD24- population varied between cell lines (Figure 4A). As with CD49f/EpCAM staining pattern, immortalized cells compared to cells prior to immortalization showed more homogeneity in CD44/CD24 staining. Significant variations in the levels of ALDEFLUOR+ cells were noted between cell lines (Figures 4B and S3A). AXL, which has recently been shown to drive EMT and confers resistance to PI3K inhibitors (43), was expressed at variable levels in cell lines (Figure S3B). Collectively, these multiple assays document enormous inter-individual heterogeneity in the normal breast, which is at least partially reflected in immortalized cell lines.
Figure 4: KTB cell lines show different levels of CD44+/CD24- and ALDEFLUOR+ stem cells based on culture condition.
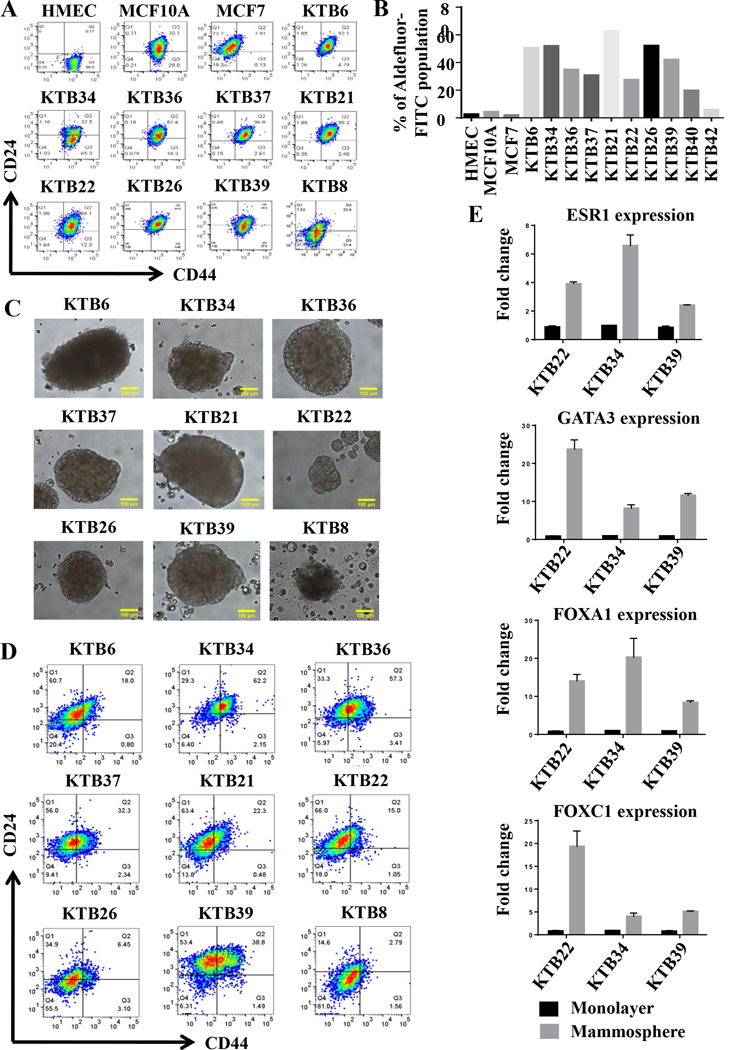
A) CD44 and CD24 staining pattern of KTB cell lines. B) ALDEFLUOR+ stem cells in different cell lines. C) KTB cell lines form mammospheres of variable sizes. D) Cells in mammospheres show different levels of differentiation based on CD44/CD24 staining pattern. E) Cells in mammospheres show elevated levels of stem/basal (FOXC1) and luminal differentiation markers (ESR1, FOXA1, and GATA3).
Stemness properties of immortalized cell lines show inter-individual variability:
To determine whether KTB cell lines show inter-individual variability in stemness, we performed mammosphere assay in all cell lines. KTB cell lines showed variable size of spheres indicating differences in stemness property of each cell line (Figures 4C). Mammospheres were subjected to flow cytometry for CD49f/EpCAM, CD201/EpCAM and CD44/CD24 expression to distinguish stem, luminal progenitor, basal and mature/differentiated cells. This assay suggested inter-individual differences in differentiation capacity (Figures 4D). For example, CD44 and CD24 staining of mammospheres showed inter-individual variability in the ratio between CD44+/CD24−, CD44−/CD24+ and CD44+/CD24+ cells. Notably, more CD44-/CD24+ cells, which were never observed under 2D culture of primary cells before or after immortalization (Figure 4A), (27), were found in mammosphere cultures of most of the cell lines (Figure 4D). Similar increase in CD49f-/EpCAM+ cells, representing mature/differentiated cells, was noted in mammosphere cultures of few cell lines (Figure S4). We further confirmed inter-individual differences in differentiation under mammosphere condition by comparing the expression levels of ESR1, FOXA1, GATA3, and FOXC1 under 2D and mammosphere conditions. The expression levels of ESR1, FOXA1, and GATA3 increased substantially in cells from mammosphere cultures compared to 2D cultures (Figure 4E). Thus, stemness and differentiation capabilities show inter-individual variability and cell lines/assay systems described here are useful for assessment at individual level.
PROCR (CD201)+/EpCAM- cells enriched in African American women correspond to naturally occurring ZEB-1 positive cells with epithelial to mesenchymal transition (EMT) phenotype:
KTB40 and KTB42 cell lines distinguished themselves from the rest of the cell lines with respect to gene expression patterns and cell surface marker profiles. These cell lines were derived from African American women. During our efforts to immortalize breast epithelial cells from African American women, we consistently observed rapid immortalization of cells with EMT features, which overtook colonies with epithelial phenotype. We placed additional efforts to characterize these cell lines. These cells at low density displayed epithelial morphology but acquired fibroblastic phenotype with increasing density (Figure 5A). These cell lines contained heterogeneous population of cells with CD201+/EpCAM-, CD201-/EpCAM-, CD201+/CD90-, CD201+/CD90+, CD201-/CD90+, and CD44+/CD24- cell surface marker profiles (Figure 5B). The majority of cells stained positive for pan-keratin antibody (Figure 5B). We included CD90 in this analysis because CD90 is expressed in MaSC, fibroblasts, adipocytes, and mesenchymal stem cells (44). Unlike other KTB cell lines described above, these cells expressed EMT and stemness-associated transcription factors ZEB1, TWIST1, TWIST2, SNAI1 and SNAI2 (Figures 5C and 5D) (45,46). KTB40 and KTB42 did not demonstrate E-Cadherin positivity in western blotting while the remaining cell lines expressed significant levels of E-cadherin (Figure 5E). Vimentin expression was significantly higher in KTB40 and KTB42 compared to other KTB cell lines.
Figure 5: CD201+/EpCAM- cells express pan-keratin but display features of EMT.
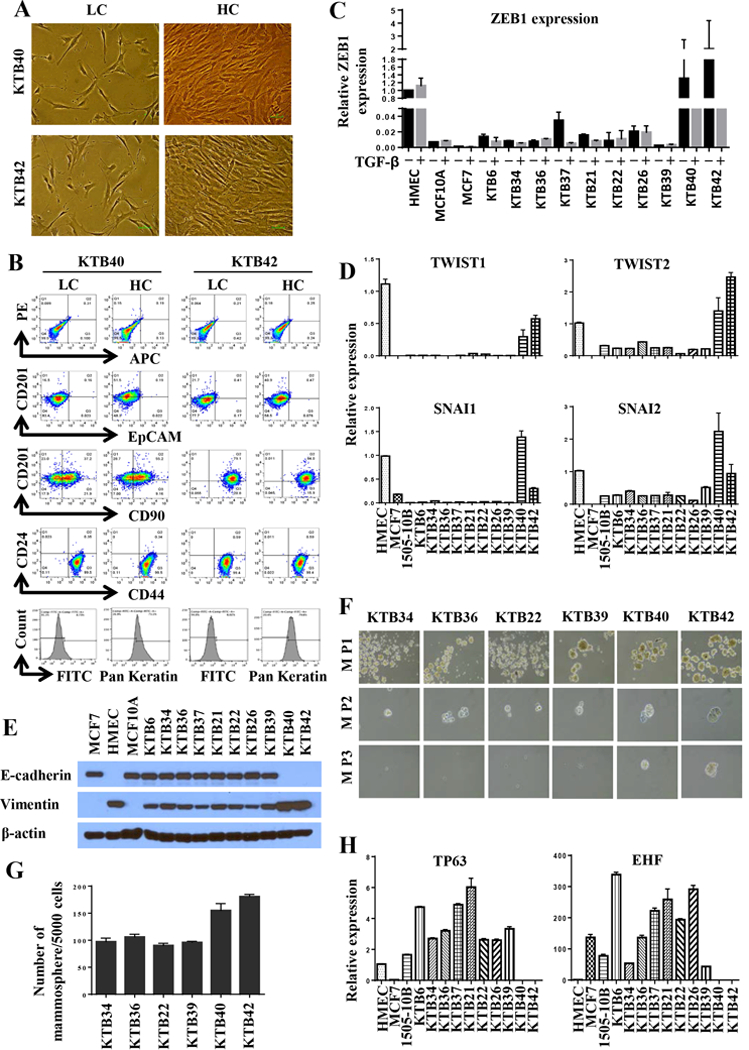
A) Confluence-dependent changes in morphology of CD201+/EpCAM- cells. B) CD24, CD44, CD90, CD201, EpCAM, and pan-keratin staining pattern of cell lines. C) ZEB1 expression levels in various KTB cell lines with and without TGFβ1 treatment. ZEB1 expression in KTB40 and KTB42 was significantly higher than in other KTB cell lines (p<0.0001). D) TWIST1, TWIST2, SNAI1 and SNAI2 expression levels in various KTB cell lines. A TWIST1, TWIST2, SNAI1 and SNAI2 expression in KTB40 and KTB42 was significantly higher than in other KTB cell lines. E. E-cadherin and vimentin expression levels in various KTB cell lines. F) Self-renewal capacity of various cell lines as measured by mammosphere assay. Primary mammospheres of KTB40 and KTB42 but not other KTB cell lines formed tertiary mammospheres. M P1, mammosphere passage 1; M P2 mammosphere passage 2; M P3, mammosphere passage 3. G) Mammosphere forming efficiency of various KTB cell lines. KTB40 and KTB42 cell lines were more efficient than other cell lines in generating mammospheres. H) TP63 and EHF expression levels in various KTB cell lines.
Lack of E-cadherin but elevated vimentin provided further evidence for EMT features of KTB40 and KTB42 cell lines. Since EMT features partially overlap with stemness (47), serial dilution mammosphere assay was performed to determine whether KTB40 and KTB42 differ in stemness relative to other KTB cell lines. KTB40 and KTB42 displayed higher mammosphere forming ability than other KTB cell lines and only these two cell lines were able to form tertiary mammospheres (Figure 5F and 5G).
It is puzzling to detect low levels of vimentin expression in our E-cadherin positive KTB cell lines. This could be due to the presence of distinct subpopulation of cells expressing either E-cadherin or vimentin. Alternatively, there could be a population of cells that express both proteins. Recent studies support the concept of epithelial/mesenchymal or luminal/basal hybrid cells playing a role in organogenesis and tumor progression (28,48). To distinguish between these possibilities, we measured the expression levels of luminal progenitor cell enriched EHF and basal cell enriched TP63 transcription factors (4). While KTB40 and KTB42 did not express these two genes, the remaining KTB lines expressed variable levels of both EHF and TP63 (Figure 5H). The most commonly used HMEC cells expressed only TP63. To confirm the presence of hybrid cells, we double stained cells for E-cadherin and vimentin and single cells were analyzed by imaging cytometry. A significant number of cells in KTB34 cell line were double positive. Interestingly, although western blotting did not detect E-Cadherin expression, a small fraction of KTB42 expressed E-cadherin (Figure S5A).
Since our cell lines were derived from core biopsies of healthy women, there is a possibility that a fraction of cells in our cell lines display myoepithelial features. CD10+/EpCAM- cells are considered myoepithelial cells although a recent study showed CD10 positivity of cancer associated fibroblasts (49,50). While KTB40 and KTB42 were CD10+/EpCAM-, the remaining cell lines showed variable levels of CD10 positivity without any distinct subpopulation with CD10+/EpCAM- phenotype (Figure S5B).
Inter-individual differences in basal activity of select signaling networks:
Since both transcriptome and phenotypic analyses documented inter-individual heterogeneity in cell lines, we next examined whether basal signaling events were also different in these cell lines. We measured phospho-ERK, phospho-AKT1, phospho-AKT2, OCT-1, AP-1 and NF-κB DNA binding activity because intrinsic subtypes are known to differ in activity of signaling molecules (51). Phospho-ERK levels varied between cell lines with KTB40 and KTB42 containing highest levels of both phospho-ERK1 and the remaining cell lines showing variable levels of phospho-ERK2 but very low levels of phospho-ERK1 (Figure S6A). Phospho-AKT2 levels varied between cell lines, while phospho-AKT1 levels were nearly similar between cell lines (Figure S6A). NF-κB DNA binding activity also showed cell line variability with KTB40 and KTB42 showing predominant p65:p50 DNA binding activity (Figure S6B). OCT-1 and AP-1 DNA binding activity was least variable among KTB cell lines with the exception of very little OCT-1 DNA binding activity in KTB40 and KTB42 cell lines (Figures S6C and S6D). These results suggest that few of the reported activation of signaling pathways in cancer could be due to higher baseline activity in cells from which cancer originated instead of acquired changes during transformation.
BET bromodomain (BRD) proteins have recently been identified as major regulators of oncogenic transcription factors and BRD4 among them has been targeted therapeutically (52). Two isoforms of BRD4 with opposing functions in cancer progression have been described; a long isoform with tumor suppressor activity and a short isoform with pro-metastatic functions (53). KTB cell lines expressed different levels of both long and short forms of BRD4 (Figure S6E), which suggest that tumors originating from cells expressing short BRD4 isoform are predisposed for metastatic progression.
The effect of transforming growth factor-β (TGFβ) on cell proliferation of KTB cell lines:
TGFβ regulates various biological processes, including cell proliferation, extracellular matrix (ECM) synthesis, angiogenesis, immune response, apoptosis, and differentiation (54). TGFβ also inhibits proliferation of ERα+ normal breast epithelial cells and it is believed that loss of this TGFβ-mediated growth inhibition is essential for the initiation of ERα+ breast cancer (55). To determine the effect of TGFβ on cell proliferation, BrDU-incorporation-ELISA cell proliferation assay was performed. TGFβ1 inhibited proliferation but response varied between cell lines with highest inhibition of KTB34, KTB36, KTB37, KTB21, KTB26 and moderate inhibition of KTB6, KTB22, and KTB39 (Figure 6A). Note that KTB40 and 42, cell lines derived from CD201+/EpCAM- cells, were least sensitive to TGFβ1-mediated growth inhibition. However, there was no correlation between intrinsic subtype classification of these cell lines and the level of response to TGFβ1. We further examined the effect of TGFβ1 on characteristics of the cells with moderate change in proliferation by analyzing the levels of CD49f, CD201, CD271 and EpCAM markers with and without TGFβ1 treatment. TGFβ1-treated KTB6, KTB21, KTB36, KTB37 and KTB39 cells contained higher proportion of cells with basal/stem cell characteristics compared to untreated cells as evident from higher number of CD49f+/EpCAM- and CD271+/EpCAM− subpopulations (Figures 6B and S7). Also, TGFβ1 had cell line-specific effects on CD49f, CD201, CD271 and EpCAM expression (Figure S7). Overall, these results document inter-individual variability in response to TGFβ1, which includes growth inhibition, dedifferentiation or lack of response.
Figure 6: The effect of TGFβ1 on proliferation and differentiation of KTB cell lines.
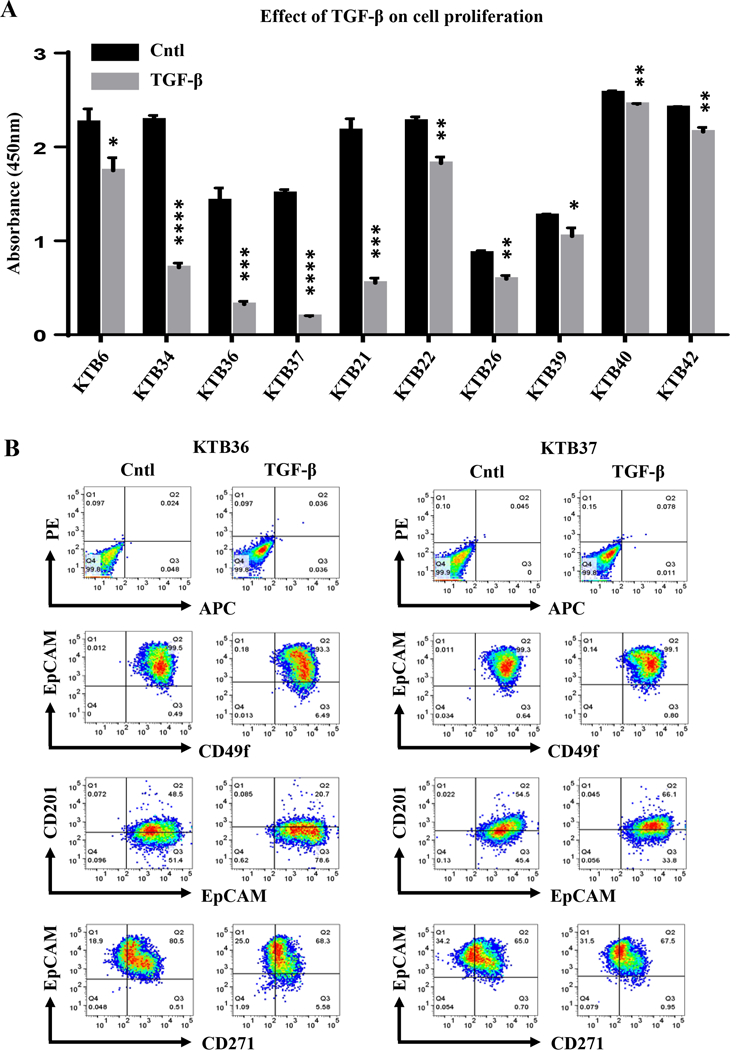
A) KTB cell lines show variable sensitivity to TGFβ1 in cell proliferation assay. The effects indicated by stars are statistically significant but the extent of effects varied between cell lines (*p<0.05, **p<0.004, ***p<0.0007, ****p<0.0001. B) The effect of TGFβ1 on cell surface markers that define stem/progenitor/mature cell status.
KTB cell lines generate ductal-like structure in 3D in vitro breast development assays:
There is growing interest in developing organoid culture system to evaluate growth characteristics and individualized drug screening (56). We used hydrogel and collagen gel 3D culture models to determine whether KTB cell lines could create structures similar to those found in normal breast (34,35). In both hydrogel and collagen scaffolds assays, KTB cell lines formed a complex branched tubular structure when cultured for 10–12 days (Figures 7A and 7B). These ductal structures had some resemblance to the interlobular ducts that form through the process of ductal initiation and branching (Figures 7A and 7B). While the outer layer of these structures contained either KRT14+ or KRT14+/KRT19+ cells, the inner layer contained few cells that stained only for KRT19+ cells (Figures 7C and 7D). We further confirmed that cells in these structures are epithelial by staining for KRT17, which is expressed in all epithelial cell types including mature luminal cells or KRT14+/KRT19+ luminal progenitor cells (57) (Figure 7E). Collectively, resources reported in this study and assays utilized to characterize individual cell lines highlight the need to use a panel of “normal” cells from multiple sources as controls in signaling studies.
Figure 7: KTB cell lines form ductal structures under 3D culture conditions.
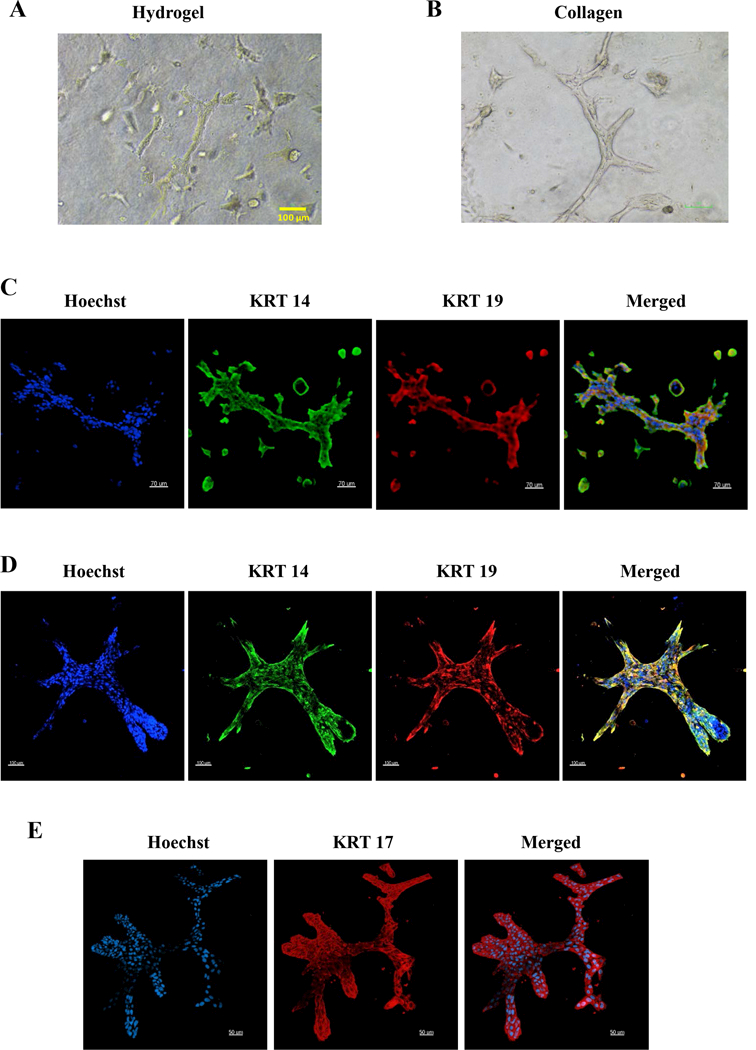
A) Phase contrast images of hydrogel cultures. B) Phase contrast images of collagen cultures. C) KRT14 and KRT19 staining patterns of cells in hydrogel cultures. D) KRT14 and KRT19 staining patterns of collagen cultures. E) KRT17 staining patterns of collagen cultures.
Discussion
Breast cancer research over many decades benefited from the availability of various breast cancer cell lines that maintain genetic and transcriptional features of breast tumors (57). However, modeling features of normal breast epithelial cells in vitro has been exceedingly difficult because culture conditions favored outgrowth of cells with basal cell gene expression patterns with few showing luminal progenitor properties and almost all previously generated breast epithelial cell lines (listed in Table S1, labeled Table comparing similar studies), including those with gene expression patterns overlapping with intrinsic subtypes and luminal progenitor cells, were derived from reduction mammoplasty samples or normal tissues adjacent to tumors with aberrant genomes (13,15,16,57). Moreover, these cell lines have not been characterized for inter-individual heterogeneity because of limited numbers (2–4 immortalized cell lines per study) and have limited luminal characteristics. Thus, limitation in culturing methods and tissue resource severely restricted our ability to mechanistically examine the role of specific genomic aberrations noted in luminal breast cancers, particularly ERα-positive breast cancers (58). For example, PIK3CA mutations are more frequently observed in ERα-positive breast cancers but the mechanistic studies to identify downstream signaling events in non-transformed cells had to use basal-like MCF10A cells that do not express ERα (59). In addition, due to the heterogeneity noted in normal breast as well as the observation that most of the SNPs are located in gene regulatory regions affecting gene expression in cis or in trans (6), there is a need to use multiple isogenic cell lines with and without cancer-specific genome manipulations to identify cancer-specific signaling pathways.
KTB cell lines described in this manuscript fulfill many of the needs of the research community. First, these cell lines are derived from biopsies of healthy women and several of these cell lines express p16(INK), which allows examination of oncogenic process by defined oncogenes with and without p16(INK)-mediated cell cycle and senescence effects. Although most of the in vitro studies have suggested that loss of p16(INK) is necessary for immortalization and consequently for transformation (39), breast tumors in some patients express p16(INK) indicating that its loss of expression is not always needed for transformation. Second, under mammosphere growth conditions, several of KTB cell lines differentiated into cells that express higher levels of luminal restricted transcription factors such as ESR1, FOXA1 and GATA3 as well as CD49f-/EpCAM+ and CD44-/CD24+ phenotypes. Recent genomic analyses of several non-transformed and breast cancer cell lines have revealed non-existence of cell lines corresponding to luminal A breast cancer (57). To our knowledge, immortalized breast epithelial cell lines expressing luminal markers have not been described, but we have for the first time demonstrated that the gene expression pattern in one of the KTB cell lines (KTB34) is comparable to luminal A breast cancer. Third, we have identified cell lines representing basal-like breast cancer. Thus, cell lines with luminal- and basal-enriched gene expression patterns can now be used to generate isogenic cell lines that overexpress amplicons, with deletion of chromosomal regions or mutations unique to specific subtypes of breast cancer. For example, ER+/HER2- tumors are enriched for amplification of CCND1 and FGFR1 and mutations in PIK3CA and ESR1 (60). HER2/ERBB2+ breast cancers have been classified into at least two distinct subtypes; one with luminal-enriched gene expression and other with basal-enriched gene expression including strong co-regulation of hypoxia/EGFR/TNFα/TGFβ/STAT3 pathways (61). KTB cell lines showed differences in response to TGFβ1, baseline DNA activity of transcription factors such as NF-κB and expression levels of pERK, pAKT2 and BRD4. ERBB2 overexpression studies in multiple “normal” cell lines would help to establish the role of cell-type-origin in determining pathways that are co-regulated along with this oncogene. Fifth, in a limited number of cases, we have cells prior to immortalization that have been cryopreserved and gene expression analyzed, which can be used for sequential characterization of genomic/epigenomic changes during immortalization and transformation.
Claudin-low breast cancers and cell lines contain highly motile cells with fibroblastic features that have lost the expression of epithelial markers and at the same time gained gene expression pattern similar to bone-marrow derived mesenchymal stem cells (57). Cell lines derived from CD201+/EpCAM- cells, which are enriched in African American women (27), displayed fibroblastic and stromal features together with high expression level of ZEB1, TWIST1, TWIST2, SNAI1, SNAI2, vimentin and elevated NF-κB and ERK activity. These cell lines also showed enhanced stemness phenotype compared to other KTB cell lines and have partial epithelial characteristics, as E-cadherin expression was detectable in a fraction of cells. ZEB-1 has previously been shown to induce claudin-low mammary tumors and we had demonstrated the ability of NF-κB to induce ZEB1 (62,63). Collectively, these results suggest that CD201+/EpCAM- cells are the cell-type-origin of claudin-low breast cancers with inherent features of EMT and elevated basal NF-κB activity. Triple negative breast cancers (TNBC) have recently been classified into six subgroups with mesenchymal stem-like subgroup showing resemblance to claudin-low breast cancers (64). This TNBC subtype also contains elevated NF-κB activity and show EMT features. African American women develop basal-like and mesenchymal stem-like triple negative breast cancers with higher levels of intra-tumor heterogeneity compared to white women (65) and elevated presence of CD201+/EpCAM- cells in their normal breast may contribute to an increase in mesenchymal stem-like breast cancers.
Overall, the panel of KTB cell lines described in this study would allow us to ask several fundamental questions; 1) to what extent inter-individual differences in baseline signaling activity impact cancer-specific signaling? 2) can we predict the behavior of tumor based on cell type origin, which can be linked back to a specific intrinsic subtype?, and 3) how do we define normal gene expression pattern and expression in how many “normal” cell lines need to be measured as controls for cancer studies? In this respect, Susan G Komen normal tissue bank at Indiana University has cryopreserved >1000 breast biopsies of healthy women with richly annotated demographics and basic health data and we have >90% success rate in generating primary cells from cryopreserved samples. Having demonstrated feasibility of using the cryopreserved tissues to generate cell lines, these resources can be used in future to generate additional cell lines and expand the number of “control” cell lines needed for breast cancer research.
Supplementary Material
Acknowledgements:
We thank Dr. Vimla Band (University of Nebraska) for hTERT vector, flow cytometry and tissue procurement cores at the IU Simon Cancer center and Susan G Komen Tissue Bank for various tissues and reagents. We also thank countless number of women for donating their breast tissue for research purpose as well as volunteers who facilitated tissue collection. This work is supported by DOD-W81XWH-15–1-0707 and R03CA195250–01A1 to H. Nakshatri. Susan G. Komen for the Cure, Breast Cancer Research Foundation and Vera Bradley Foundation for Breast Cancer Research provide funding support to Komen Normal Tissue Bank. Walther Cancer Foundation provided funding support for the Collaborative Core for Cancer Bioinformatics (C3B), which performed RNA-seq data analyses and PAM50 classification.
Footnotes
Conflict of interest: Authors have no conflict of interest to declare
References:
- 1.Visvader JE, Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev 2014;28(11):1143–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raouf A, Zhao Y, To K, Stingl J, Delaney A, Barbara M, et al. Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell Stem Cell 2008;3(1):109–18. [DOI] [PubMed] [Google Scholar]
- 3.Villadsen R, Fridriksdottir AJ, Ronnov-Jessen L, Gudjonsson T, Rank F, LaBarge MA, et al. Evidence for a stem cell hierarchy in the adult human breast. J Cell Biol 2007;177(1):87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pellacani D, Bilenky M, Kannan N, Heravi-Moussavi A, Knapp DJHF, Gakkhar S, Moksa M, Carles A, Moore R, Mungall AJ, Marra MA, Jones SJM, Aparicio S, Hirst M, and Eaves CJ Analysis of normal human mammary epigenomes reveal cell-specfiic active enahcner states and associated transcription factor networks. Cell reports 2016;17:2060–74. [DOI] [PubMed] [Google Scholar]
- 5.Santagata S, Thakkar A, Ergonul A, Wang B, Woo T, Hu R, et al. Taxonomy of breast cancer based on normal cell phenotype predicts outcome. J Clin Invest 2014;124(2):859–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Consortium GT, Laboratory DA, Coordinating Center -Analysis Working G, Statistical Methods groups-Analysis Working G, Enhancing Gg, Fund NIHC, et al. Genetic effects on gene expression across human tissues. Nature 2017;550(7675):204–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lappalainen T, Sammeth M, Friedlander MR, t Hoen PA, Monlong J, Rivas MA, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 2013;501(7468):506–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner JR, Busche S, Ge B, Kwan T, Pastinen T, Blanchette M. The relationship between DNA methylation, genetic and expression inter-individual variation in untransformed human fibroblasts. Genome Biol 2014;15(2):R37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 2001;98(19):10869–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res 2010;12(5):R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prat A, Perou CM. Mammary development meets cancer genomics. Nat Med 2009;15(8):842–4. [DOI] [PubMed] [Google Scholar]
- 12.Keller PJ, Arendt LM, Skibinski A, Logvinenko T, Klebba I, Dong S, et al. Defining the cellular precursors to human breast cancer. Proc Natl Acad Sci U S A 2012;109(8):2772–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ince TA, Richardson AL, Bell GW, Saitoh M, Godar S, Karnoub AE, et al. Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. Cancer Cell 2007;12(2):160–70. [DOI] [PubMed] [Google Scholar]
- 14.Kao J, Salari K, Bocanegra M, Choi YL, Girard L, Gandhi J, et al. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PloS one 2009;4(7):e6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Degnim AC, Visscher DW, Hoskin TL, Frost MH, Vierkant RA, Vachon CM, et al. Histologic findings in normal breast tissues: comparison to reduction mammaplasty and benign breast disease tissues. Breast Cancer Res Treat 2012;133(1):169–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teschendorff AE, Gao Y, Jones A, Ruebner M, Beckmann MW, Wachter DL, et al. DNA methylation outliers in normal breast tissue identify field defects that are enriched in cancer. Nature communications 2016;7:10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fridriksdottir AJ, Villadsen R, Morsing M, Klitgaard MC, Kim J, Petersen OW, et al. Proof of region-specific multipotent progenitors in human breast epithelia. Proc Natl Acad Sci U S A 2017;114(47):E10102–E11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hopkinson BM, Klitgaard MC, Petersen OW, Villadsen R, Ronnov-Jessen L, Kim J. Establishment of a normal-derived estrogen receptor-positive cell line comparable to the prevailing human breast cancer subtype. Oncotarget 2017;8(6):10580–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fridriksdottir AJ, Kim J, Villadsen R, Klitgaard MC, Hopkinson BM, Petersen OW, et al. Propagation of oestrogen receptor-positive and oestrogen-responsive normal human breast cells in culture. Nature communications 2015;6:8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao X, Malhotra GK, Lele SM, Lele MS, West WW, Eudy JD, et al. Telomerase-immortalized human mammary stem/progenitor cells with ability to self-renew and differentiate. Proc Natl Acad Sci U S A 2010;107(32):14146–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arendt LM, Keller PJ, Skibinski A, Goncalves K, Naber SP, Buchsbaum RJ, et al. Anatomical localization of progenitor cells in human breast tissue reveals enrichment of uncommitted cells within immature lobules. Breast Cancer Res 2014;16(5):453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao X, Malhotra GK, Band H, Band V. Derivation of myoepithelial progenitor cells from bipotent mammary stem/progenitor cells. PloS one 2012;7(4):e35338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carter EP, Gopsill JA, Gomm JJ, Jones JL, Grose RP. A 3D in vitro model of the human breast duct: a method to unravel myoepithelial-luminal interactions in the progression of breast cancer. Breast Cancer Res 2017;19(1):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun W, Kang KS, Morita I, Trosko JE, Chang CC. High susceptibility of a human breast epithelial cell type with stem cell characteristics to telomerase activation and immortalization. Cancer Res 1999;59(24):6118–23. [PubMed] [Google Scholar]
- 25.Gudjonsson T, Villadsen R, Nielsen HL, Ronnov-Jessen L, Bissell MJ, Petersen OW. Isolation, immortalization, and characterization of a human breast epithelial cell line with stem cell properties. Genes Dev 2002;16(6):693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chanson L, Brownfield D, Garbe JC, Kuhn I, Stampfer MR, Bissell MJ, et al. Self-organization is a dynamic and lineage-intrinsic property of mammary epithelial cells. Proc Natl Acad Sci U S A 2011;108(8):3264–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakshatri H, Anjanappa M, Bhat-Nakshatri P. Ethnicity-Dependent and -Independent Heterogeneity in Healthy Normal Breast Hierarchy Impacts Tumor Characterization. Scientific reports 2015;5:13526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong J, Hu Y, Fan X, Wu X, Mao Y, Hu B, et al. Single-cell RNA-seq analysis unveils a prevalent epithelial/mesenchymal hybrid state during mouse organogenesis. Genome Biol 2018;19(1):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nievergelt CM, Maihofer AX, Shekhtman T, Libiger O, Wang X, Kidd KK, et al. Inference of human continental origin and admixture proportions using a highly discriminative ancestry informative 41-SNP panel. Investig Genet 2013;4(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breese MR, Liu Y. NGSUtils: a software suite for analyzing and manipulating next-generation sequencing datasets. Bioinformatics 2013;29(4):494–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26(1):139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gendoo DM, Ratanasirigulchai N, Schroder MS, Pare L, Parker JS, Prat A, et al. Genefu: an R/Bioconductor package for computation of gene expression-based signatures in breast cancer. Bioinformatics 2016;32(7):1097–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhat-Nakshatri P, Sweeney CJ, Nakshatri H. Identification of signal transduction pathways involved in constitutive NF-kappaB activation in breast cancer cells. Oncogene 2002;21(13):2066–78. [DOI] [PubMed] [Google Scholar]
- 34.Linnemann JR, Miura H, Meixner LK, Irmler M, Kloos UJ, Hirschi B, et al. Quantification of regenerative potential in primary human mammary epithelial cells. Development 2015;142(18):3239–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller DH, Sokol ES, Gupta PB. 3D Primary Culture Model to Study Human Mammary Development. Methods Mol Biol 2017;1612:139–47. [DOI] [PubMed] [Google Scholar]
- 36.Eswaran J, Li DQ, Shah A, Kumar R. Molecular pathways: targeting p21-activated kinase 1 signaling in cancer--opportunities, challenges, and limitations. Clin Cancer Res 2012;18(14):3743–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eeckhoute J, Keeton EK, Lupien M, Krum SA, Carroll JS, Brown M. Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res 2007;67(13):6477–83. [DOI] [PubMed] [Google Scholar]
- 38.Han B, Bhowmick N, Qu Y, Chung S, Giuliano AE, Cui X. FOXC1: an emerging marker and therapeutic target for cancer. Oncogene 2017;36(28):3957–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garbe JC, Vrba L, Sputova K, Fuchs L, Novak P, Brothman AR, et al. Immortalization of normal human mammary epithelial cells in two steps by direct targeting of senescence barriers does not require gross genomic alterations. Cell Cycle 2014;13(21):3423–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang D, Cai C, Dong X, Yu QC, Zhang XO, Yang L, et al. Identification of multipotent mammary stem cells by protein C receptor expression. Nature 2014;517:81–84. [DOI] [PubMed] [Google Scholar]
- 41.Kim J, Villadsen R, Sorlie T, Fogh L, Gronlund SZ, Fridriksdottir AJ, et al. Tumor initiating but differentiated luminal-like breast cancer cells are highly invasive in the absence of basal-like activity. Proc Natl Acad Sci U S A 2012;109(16):6124–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu S, Cong Y, Wang D, Sun Y, Deng L, Liu Y, et al. Breast Cancer Stem Cells Transition between Epithelial and Mesenchymal States Reflective of their Normal Counterparts. Stem cell reports 2014;2(1):78–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Antony J, Huang RY. AXL-Driven EMT State as a Targetable Conduit in Cancer. Cancer Res 2017;77(14):3725–32. [DOI] [PubMed] [Google Scholar]
- 44.Pan D, Roy S, Gascard P, Zhao J, Chen-Tanyolac C, Tlsty TD. SOX2, OCT3/4 and NANOG expression and cellular plasticity in rare human somatic cells requires CD73. Cell Signal 2016;28(12):1923–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caramel J, Ligier M, Puisieux A. Pleiotropic Roles for ZEB1 in Cancer. Cancer Res 2017. [DOI] [PubMed] [Google Scholar]
- 46.Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell 2016;166(1):21–45. [DOI] [PubMed] [Google Scholar]
- 47.Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Ng Eaton E, et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature 2015;525(7568):256–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pastushenko I, Brisebarre A, Sifrim A, Fioramonti M, Revenco T, Boumahdi S, et al. Identification of the tumour transition states occurring during EMT. Nature 2018;556(7702):463–68. [DOI] [PubMed] [Google Scholar]
- 49.Morel AP, Ginestier C, Pommier RM, Cabaud O, Ruiz E, Wicinski J, et al. A stemness-related ZEB1-MSRB3 axis governs cellular pliancy and breast cancer genome stability. Nat Med 2017;23(5):568–78. [DOI] [PubMed] [Google Scholar]
- 50.Su S, Chen J, Yao H, Liu J, Yu S, Lao L, et al. CD10(+)GPR77(+) Cancer-Associated Fibroblasts Promote Cancer Formation and Chemoresistance by Sustaining Cancer Stemness. Cell 2018;172(4):841–56 e16. [DOI] [PubMed] [Google Scholar]
- 51.Huang KL, Li S, Mertins P, Cao S, Gunawardena HP, Ruggles KV, et al. Proteogenomic integration reveals therapeutic targets in breast cancer xenografts. Nature communications 2017;8:14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shu S, Polyak K. BET Bromodomain Proteins as Cancer Therapeutic Targets. Cold Spring Harb Symp Quant Biol 2016;81:123–29. [DOI] [PubMed] [Google Scholar]
- 53.Alsarraj J, Walker RC, Webster JD, Geiger TR, Crawford NP, Simpson RM, et al. Deletion of the proline-rich region of the murine metastasis susceptibility gene Brd4 promotes epithelial-to-mesenchymal transition- and stem cell-like conversion. Cancer Res 2011;71(8):3121–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heldin CH, Vanlandewijck M, Moustakas A. Regulation of EMT by TGFbeta in cancer. FEBS Lett 2012;586(14):1959–70. [DOI] [PubMed] [Google Scholar]
- 55.Ewan KB, Oketch-Rabah HA, Ravani SA, Shyamala G, Moses HL, Barcellos-Hoff MH. Proliferation of estrogen receptor-alpha-positive mammary epithelial cells is restrained by transforming growth factor-beta1 in adult mice. Am J Pathol 2005;167(2):409–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weeber F, Ooft SN, Dijkstra KK, Voest EE. Tumor Organoids as a Pre-clinical Cancer Model for Drug Discovery. Cell Chem Biol 2017;24(9):1092–100. [DOI] [PubMed] [Google Scholar]
- 57.Prat A, Karginova O, Parker JS, Fan C, He X, Bixby L, et al. Characterization of cell lines derived from breast cancers and normal mammary tissues for the study of the intrinsic molecular subtypes. Breast Cancer Res Treat 2013;142(2):237–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513(7517):202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gustin JP, Karakas B, Weiss MB, Abukhdeir AM, Lauring J, Garay JP, et al. Knockin of mutant PIK3CA activates multiple oncogenic pathways. Proc Natl Acad Sci U S A 2009;106(8):2835–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Turner NC, Neven P, Loibl S, Andre F. Advances in the treatment of advanced oestrogen-receptor-positive breast cancer. Lancet 2017;389(10087):2403–14. [DOI] [PubMed] [Google Scholar]
- 61.Gatza ML, Kung HN, Blackwell KL, Dewhirst MW, Marks JR, Chi JT. Analysis of tumor environmental response and oncogenic pathway activation identifies distinct basal and luminal features in HER2-related breast tumor subtypes. Breast Cancer Res 2011;13(3):R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morel AP, Hinkal GW, Thomas C, Fauvet F, Courtois-Cox S, Wierinckx A, et al. EMT inducers catalyze malignant transformation of mammary epithelial cells and drive tumorigenesis towards claudin-low tumors in transgenic mice. PLoS genetics 2012;8(5):e1002723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chua HL, Bhat-Nakshatri P, Clare SE, Morimiya A, Badve S, Nakshatri H. NF-kappaB represses E-cadherin expression and enhances epithelial to mesenchymal transition of mammary epithelial cells: potential involvement of ZEB-1 and ZEB-2. Oncogene 2007;26(5):711–24. [DOI] [PubMed] [Google Scholar]
- 64.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011;121(7):2750–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Keenan T, Moy B, Mroz EA, Ross K, Niemierko A, Rocco JW, et al. Comparison of the Genomic Landscape Between Primary Breast Cancer in African American Versus White Women and the Association of Racial Differences With Tumor Recurrence. J Clin Oncol 2015;33(31):3621–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


