Abstract
Genome reduction is pervasive among maternally inherited bacterial endosymbionts. This genome reduction can eventually lead to serious deterioration of essential metabolic pathways, thus rendering an obligate endosymbiont unable to provide essential nutrients to its host. This loss of essential pathways can lead to either symbiont complementation (sharing of the nutrient production with a novel co-obligate symbiont) or symbiont replacement (complete takeover of nutrient production by the novel symbiont). However, the process by which these two evolutionary events happen remains somewhat enigmatic by the lack of examples of intermediate stages of this process. Cinara aphids (Hemiptera: Aphididae) typically harbor two obligate bacterial symbionts: Buchnera and Serratia symbiotica. However, the latter has been replaced by different bacterial taxa in specific lineages, and thus species within this aphid lineage could provide important clues into the process of symbiont replacement. In the present study, using 16S rRNA high-throughput amplicon sequencing, we determined that the aphid Cinara strobi harbors not two, but three fixed bacterial symbionts: Buchnera aphidicola, a Sodalis sp., and S. symbiotica. Through genome assembly and genome-based metabolic inference, we have found that only the first two symbionts (Buchnera and Sodalis) actually contribute to the hosts’ supply of essential nutrients while S. symbiotica has become unable to contribute towards this task. We found that S. symbiotica has a rather large and highly eroded genome which codes only for a few proteins and displays extensive pseudogenization. Thus, we propose an ongoing symbiont replacement within C. strobi, in which a once “competent” S. symbiotica does no longer contribute towards the beneficial association. These results suggest that in dual symbiotic systems, when a substitute cosymbiont is available, genome deterioration can precede genome reduction and a symbiont can be maintained despite the apparent lack of benefit to its host.
Keywords: symbiont replacement, genome reduction, aphid symbiosis, Serratia symbiotica, Sodalis
Introduction
Many insects with a nutrient restricted diet, depend on vertically inherited obligate nutritional symbionts (Aschner 1934; Nogge 1976, 1981; Ohtaka and Ishikawa 1991; Sacchi et al. 1993; Koga et al. 2003; Hosokawa et al. 2006; Nikoh et al. 2014). These symbionts evolved from once free-living bacterial lineages (Husník et al. 2011; Clayton et al. 2012; Manzano-Marín et al. 2015) and have undergone a series of genomic and phenotypic changes resulting from relaxed selection, continuous bottlenecks, and their metabolic adaptation to the sustained association with their host (Moran 1996; Moran and Plague 2004; Moran et al. 2008; Latorre and Manzano-Marín 2017). These alterations include genome reduction, a simplified metabolism specialized on supplying the host with essential nutrients lacking from its diet, drastic changes in cellular shape, and G + C (uncommon) or A + T-biased genomes.
Aphids (Hemiptera: Aphididae) generally house the obligate vertically transmitted endosymbiotic bacterium Buchnera in specialized cells called bacteriocytes (Buchner 1953; Griffiths and Beck 1975; Munson et al. 1991). This obligate symbiont is capable of producing essential amino acids (hereafter EAAs) and B vitamins (Moran et al. 2005; Akman Gündüz and Douglas 2009; Hansen and Moran 2011; Poliakov et al. 2011; Russell et al. 2013, 2014) that are lacking from the host diet (plant phloem) (Ziegler 1975; Sandstrom and Moran 1999; Akman Gündüz and Douglas 2009), and thus insures the correct development of its host (Mittler 1971; Douglas 1996; Nakabachi and Ishikawa 1999; Akman Gündüz and Douglas 2009). Buchnera underwent a massive genome reduction and established as an obligate symbiont before the diversification of aphids. This is evidenced by its almost-universal presence in aphids (Buchner 1953; Nováková et al. 2013), the high degree of genome synteny displayed among distantly related strains of Buchnera (Tamas et al. 2002; van Ham et al. 2003), and their consistently small genomes. Aphid species from the Lachninae subfamily have been found to harbor Buchnera strains that have ancestrally lost the capacity to synthesize biotin and riboflavin (Pérez-Brocal et al. 2006; Lamelas et al. 2011; Manzano-Marín and Latorre 2014; Manzano-Marín et al. 2016), two essential B vitamins. For the provision of these nutrients, Lachninae aphids and their Buchnera now rely on different co-obligate endosymbionts, most often S. symbiotica (Lamelas et al. 2011; Manzano-Marín and Latorre 2014; Manzano-Marín et al. 2016, 2017). Accordingly, Cinara species (Aphididae: Lachninae) have been consistently found to host an additional bacterial co-obligate symbiont, most commonly S. symbiotica (Lamelas et al. 2008; Burke et al. 2009; Manzano-Marín et al. 2017; Meseguer et al. 2017). An ancestral reconstruction of the symbiotic associations of Cinara with fixed additional symbionts suggests that S. symbiotica was likely the original co-obligate endosymbiont, but has been replaced several times by other bacterial taxa (Meseguer et al. 2017). These new symbionts are phylogenetically affiliated to different lineages, mainly including known aphid facultative symbiotic ones (e.g. Fukatsuia, Sodalis, and Hamiltonella).
Most of our current knowledge from these co-obligate endosymbionts comes from S. symbiotica strains harbored by Lachninae aphids. These symbionts display very different genomic features, ranging from strains holding rather large genomes rich in mobile elements to small genomes rich in A + T and deprived of mobile elements (Manzano-Marín and Latorre 2016). The S. symbiotica strain held by the aphid Cinara tujafilina (hereafter SsCt), shares a considerable genomic similarity to the facultative strain harbored by the pea aphid Acyrthosiphon pisum (hereafter SsAp) (Manzano-Marín and Latorre 2014). This reflects the early stage of genome reduction SsCt is at, which is characterized by a moderately reduced and highly rearranged genome (when compared with free-living relatives), an enrichment of mobile elements (hereafter MEs), and a large-scale pseudogenization (Degnan et al. 2009, 2010; Burke and Moran 2011; Koga and Moran 2014; Manzano-Marín and Latorre 2014; Oakeson et al. 2014). On the other side, the co-obligate S. symbiotica from Tuberolachnus salignus (hereafter SsTs) shows a very small and gene dense genome (Manzano-Marín et al. 2016), similarly to ancient obligate endosymbionts such as Buchnera (Shigenobu et al. 2000; Tamas et al. 2002; van Ham et al. 2003; Pérez-Brocal et al. 2006; Degnan et al. 2011), Blochmannia (Gil et al. 2003; Degnan et al. 2005; Williams and Wernegreen 2015), or Blattabacterium (López-Sánchez et al. 2009; Sabree et al. 2009; Huang et al. 2012; Kambhampati et al. 2013; Patiño-Navarrete et al. 2013; Tokuda et al. 2013). Sitting in between SsCt and SsTs, the S. symbiotica strain housed by Cinara cedri (hereafter SsCc) shows intermediate characteristics between a larger and highly pseudogenized genome and a small and compact one (Lamelas et al. 2011). In Cinara aphids, S. symbiotica has undergone symbiont replacement in different lineages, and thus the endosymbionts’ genomes of species within this genus could provide important clues into reductive genome evolution and the process of symbiont replacement.
Within the aphid Cinara strobi, Jousselin et al. (2016) first reported the presence of Sodalis, Wolbachia, and Serratia bacteria as putative secondary symbionts present in one population of this aphid species using 16S rRNA high-throughput amplicon sequencing. Later, a deeper survey of endosymbionts associated with about 100 Cinara species, using this same technique, showed that only two of these additional symbionts, Sodalis and S. symbiotica, were actually fixed across different populations of C. strobi (Meseguer et al. 2017). The authors found Sodalis to be very abundant in both the amplicon sequencing read set and the whole-genome one. On the other hand, S. symbiotica was found consistently in a lower percentage than Sodalis in all but one sample, and was even found to be almost absent (i.e. represented by very few reads) in one (thus leading to its characterization as a nonfixed symbiont). Further analysis of the riboflavin- and biotin-related biosynthetic genes revealed that Sodalis was able to supplement the previously identified auxotrophies developed by Buchnera strains from Lachninae aphids. This suggested that C. strobi most likely represented a case of co-obligate symbiont replacement, in which the former S. symbiotica was replaced by a younger Sodalis symbiont. However, this results left one unanswered question: What role, if any, is played by the prevalent S. symbiotica strain? We hypothesized that this bacterium could either represent a widely spread facultative lineage (probably resembling SsAp), a transitional state in the symbiont replacement process, or a persistent S. symbiotica strain associated with the ancestor of C. strobi that had established a tripartite mutualistic symbiotic association.
To explore this question, we characterized the symbiotic community of additional populations of C. strobi and defined the fixed bacterial associates of this species. In addition, we assembled the genome of S. symbiotica from this aphid species and evaluated the metabolic capacity of its fixed symbiotic cohort to supply the aphid with EAAs, B vitamins, and other cofactors. Our results suggest that C. strobi houses an ancient, now dispensable, S. symbiotica secondary symbiont along with a co-obligate symbiotic consortium made up of Buchnera and its new partner, Sodalis.
Materials and Methods
Aphid Collection, DNA Extraction, and Sequencing
Cinara strobi individuals were collected in 2015 from five colonies throughout the South Eastern Canada (supplementary table S1, Supplementary Material online) and then kept in 70% ethanol at 6 °C.
For 16S amplicon sequencing, individual aphids from each collected population (3618, 3628, 3629, 3632, and 3682) were washed three times in ultrapure water and total genomic DNA was extracted with the DNEasy Blood & Tissue Kit (Qiagen, Germany), according to the manufacturer’s recommendations. The recovered DNA was then eluted in 70 μl of ultrapure water. We amplified a 251 bp portion of the V4 region of the 16SrRNA gene (Mizrahi-Man et al. 2013), using universal primers, and performed targeted sequencing of indexed bacterial fragments on a MiSeq (Illumina) platform (Kozich et al. 2013), following the protocol described in Jousselin et al. (2016).
For whole-genome sequencing, we prepared DNA samples enriched with bacteria from previously collected colony 3249 following a slightly modified version of the protocol by Charles and Ishikawa (1999) as described in Jousselin et al. (2016). For this filtration protocol 15 aphids for one colony were pooled together. Extracted DNA was used to prepare two custom paired-end libraries in France Génomique. Briefly, 5 ng of genomic DNA were sonicated using the E220 Covaris instrument (Covaris, USA). Fragments were end-repaired, 3′-adenylated, and NEXTflex PCR free barcodes adapters (Bioo Scientific, USA) were added by using NEBNext Ultra II DNA library prep kit for Illumina (New England Biolabs, USA). Ligation products were purified by Ampure XP (Beckman Coulter, USA) and DNA fragments (>200 bp) were PCR-amplified (2 PCR reactions, 12 cycles) using Illumina adapter-specific primers and NEBNext Ultra II Q5 Master Mix (NEB). After library profile analysis by Agilent 2100 Bioanalyser (Agilent Technologies, USA) and qPCR quantification using the KAPA Library Quantification Kit for Illumina Libraries (Kapa Biosystems, USA), the libraries were sequenced using 251 bp paired-end reads chemistry on a HiSeq2500 Illumina sequencer. Additionally, we used reads recovered from paired-end Illumina sequencing of the same colony previously reported in Meseguer et al. (2017).
16S rRNA Amplicon Taxonomic Assignment
We used Mothur v1.3.3 (Schloss and Westcott 2011) to assemble paired-end reads and filter out sequencing errors and chimeras. In brief, the overlapped paired-end reads were assembled with the make.contigs function, and the contigs exceeding 280 bp in length were excluded from further analyses. Remaining unique contigs were then aligned with the V4 portion of reference sequences from the SILVA database v119 (Quast et al. 2013). Sequences that did not align with the V4 fragment were excluded from further analyses. The number of reads resulting from sequencing errors was then reduced by merging rare unique sequences with frequent unique sequences with a mismatch of no more than 2 bp relative to the rare sequences (pre.cluster command in Mothur). We then used the UCHIME program (Edgar et al. 2011) implemented in Mothur to detect chimeric sequences and excluded them from the data set. Following Jousselin et al. (2016), for each sequence, the number of reads per sample was transformed into percentages using an R script and used to compile a frequency table (supplementary table S2, Supplementary Material online). We then removed individual sequences representing less than 1/1,000 of the reads in each sample. Sequences represented by such a small proportion of the reads were generally not arthropod endosymbionts and, in most cases, were not found across PCR replicates of the same sample, suggesting that they could represent contaminants or spurious sequences.
Taxonomic assignation of the remaining sequences was conducted using the RDP classifier (Wang et al. 2007) with the SILVA database v119 and BlastN (Camacho et al. 2009) (only the best hits were reported and when hits with similar scores were found a “multi-affiliation” was reported). Using these assignations and the table of sequence frequencies per sample, we plotted the bacterial composition of each sample. To simplify representation of the results, when different unique sequences were assigned to the same bacterial species (or genus), their frequencies were added.
Genome Assembly and Annotation
Illumina reads were right-tail clipped (using a minimum quality threshold of 20) using FASTX-Toolkit v0.0.14 (http://hannonlab.cshl.edu/fastx_toolkit/, last accessed August 21, 2018). Reads shorted than 75 after the aforementioned clipping were dropped. Additionally, PRINSEQ v0.20.4 (Schmieder and Edwards 2011) was used to remove reads containing undefined nucleotides as well as those left without a pair after the filtering and clipping process. The resulting reads were assembled using SPAdes v3.10.1 (Bankevich et al. 2012) with the options–only-assembler option and k-mer sizes of 33, 55, 77, 99, and 127. From the resulting contigs, those that were shorter than 200 bps were dropped. The remaining contigs were binned using results from a BlastX (Altschul 1997) search (best hit per contig) against a database consisting of the Pea aphid’s proteome and a selection of aphid’s symbiotic bacteria proteomes (supplementary table S3, Supplementary Material online). When no genome was available for a certain lineage, closely related bacteria were used. The assigned contigs were manually screened using the BlastX web server (searching against the nr database) to insure correct assignment. This binning process confirmed the presence of the previously reported putative co-obligate symbionts (Jousselin et al. 2016; Meseguer et al. 2017) (Buchnera aphidicola and a Sodalis sp.) as well as other additional symbionts. One of these additional symbionts was S. symbiotica, for which the first-pass assembly resulted in three contigs assigned to this taxon, two belonging to a putative chromosome with overlapping ends and one much smaller circular contig belonging to a putative plasmid. The resulting contigs were then used as reference for read mapping and individual genome assembly using SPAdes, as described above, with read error correction.
The resulting genomes were annotated using a series of specialized software. First, open reading frame (ORF) prediction was done using prodigal, followed by functional prediction by the BASys web server (Van Domselaar et al. 2005). In order to validate start codons, ribosomal binding sites were predicted using RBSfinder (Suzek et al. 2001). This was followed by noncoding RNA prediction using infernal v1.1.2 (Nawrocki and Eddy 2013) (against the Rfam v12.3 database; Nawrocki et al. 2015), tRNAscan-SE v2.0 (Lowe and Chan 2016), and ARAGORN v1.2.36 (Laslett and Canback 2004). This annotation was followed by manual curation of the genes on UGENE v1.28.1 (Okonechnikov et al. 2012) through on-line BlastX searches of the intergenic regions as well as through BlastP and DELTA-BLAST (Boratyn et al. 2012) searches of the predicted ORFs against NCBI’s nr database. Priority for the BLAST searches was as follows: 1) against Escherichia coli K-12 substrain MG1655; 2) against Yersiniapestis CO92 or Serratia marcescens strain Db11 (for S. symbiotica); and 3) against the whole nr database. The resulting coding sequences (CDSs) were considered to be putatively functional if all essential domains for the function were found or if a literature search supported the truncated version of the protein as functional in a related organism, or if the CDS displayed truncations but retained identifiable domains (details of the literature captured in the annotation file). For S. symbiotica, pseudogenes were also searched based on synteny against available S. symbiotica strains. This prediction was performed using a combination of sequence alignment (with m-coffee; Wallace et al. 2006) and BlastX searches against the NCBI’s nr database (restricted to Serratia taxon ID). This allowed the identification of missed pseudogenes by the previous searches.
Phylogenetic Reconstruction and Rearrangement Analysis
For performing both phylogenetic inferences and analyzing the genetic differences in Serratia from the different aphids, we first ran an orthologous protein clustering analysis using OrthoMCL v2.0.9 (Li et al. 2003; Chen et al. 2007) using a set of S. symbiotica and closely related free-living bacterial strains (supplementary table S4, Supplementary Material online). We then extracted the single copy-core proteins of currently available S. symbiotica genomes and free-living relatives for phylogenetic reconstruction (297 protein groups) and rearrangement analysis (381 protein groups). We then ran MGR v2.03 (Bourque and Pevzner 2002) on the latter set to infer the tree that absolutely minimizes (no heuristics) the number of rearrangements undergone among the strains.
For phylogenetic reconstruction of S. symbiotica, we aligned the single-copy core protein set, gene by gene, using MAFFT v7.220 (Katoh and Standley 2013) (L-INS-i algorithm). We then removed divergent and ambiguously aligned blocks using Gblocks v0.91b (Talavera and Castresana 2007) and concatenated the resulting alignments into a single one (supplementary file S2, Supplementary Material online) for following phylogenetic inference. We used the LG + I + G amino acid substitution model, which incorporates the variability of evolutionary rates across sites in the matrix estimation (Le and Gascuel 2008). Bayesian phylogenetic inference was performed in MrBayes v3.2.5 (Ronquist et al. 2012) running 2 independent analyses with 4 chains each for 300,000 generations and checked for convergence. In order to alleviate long-branch attraction artifacts commonly seen in endosymbionts (Husník et al. 2011; Philippe and Roure 2011), the analysis was also run in Phylobayes v4.1 (Lartillot et al. 2009) under the CAT + GTR + G (four discrete categories) (under eight independent runs) using dayhoff-6-recoded concatenated amino acid alignments. Chains were run and compared using the tracecomp and bpcomp programs, and were considered converged at a maximum discrepancy <0.3 and minimum effective size of 50. None were found to converge even after 30,000 cycles. For further exploration, additional reduced protein data sets were selected for analysis as described above: S. marcescens + S. symbiotica and ribosomal proteins. Phylobayes runs done with a full set of core proteins were not found to converge even after 10,000 cycles. All resulting trees were visualized and exported with FigTree v1.4.1 (http://tree.bio.ed.ac.uk/software/figtree/, last accessed August 21 2018) and edited in Inkscape.
Results
Fixed Symbionts of C. strobi
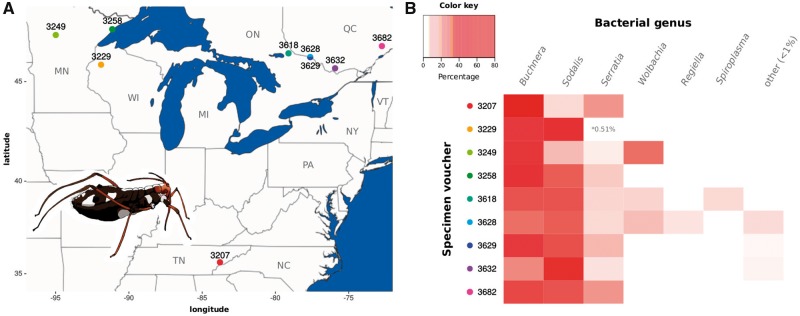
As stated before, C.strobi is distributed throughout eastern North America (Blackman and Eastop 1994). We collected C. strobi individuals from five different populations from the southeast of Canada (3618, 3628, 3629, 3632, and 3682) to complete previous sampling from northeast USA (fig. 1A and supplementary table S1, Supplementary Material online). In order to assess the presence of bacterial associates in geographically distant C. strobi populations, we reanalyzed the 4 C. strobi samples collected in the northeast USA (3229, 3249, 3258, and 3207), and previously included in Meseguer et al. (2017), as well as the newly collected individuals through 16S rRNA high-throughput sequencing (see Materials and Methods: 16S rRNA amplicon taxonomic assignment). Taxonomic assignment of the reads revealed that individuals from all populations harbored not only two symbionts, but three: Buchnera, Sodalis, and S. symbiotica (fig. 1B). It is important to note that sample 3229 showed a very low abundance of S. symbiotica-assigned reads, which prompted Meseguer et al. (2017) to report this symbiont as not being systematically associated with C. strobi. In addition to the three fixed symbionts, we also confirmed the presence of other known aphid facultative symbiont taxa (i.e. Wolbachia, Regiella, and Spiroplasma) in three samples (voucher IDs 3249, 3628, and 3628).
Fig. 1.
—Distribution of sampled C. strobi populations and 16S rRNA high-throughput bacterial symbiont screening. (A) Map showing the north-east USA and south-east Canada regions where the C. strobi samples were collected (coloured points) featuring a cartoon of a C. strobi apterous female adult. (B) Heat map displaying the relative abundance of Illumina reads per taxon per sample. On the top-left, color key for the taxon abundance. On the left, voucher ID for the sampled C. strobi populations with colored dots matching the map in (A).
The genome of S. symbiotica Strain SeCistrobi
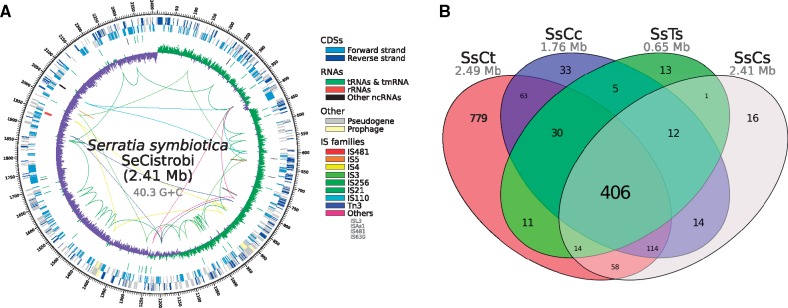
The binning and reassembly process resulted in two assembled circular DNA molecules assigned to S. symbiotica: A chromosome (fig. 2A) and a plasmid, with an average coverage of 83.78× and 32.48×, respectively. The chromosome of S. symbiotica strain SeCistrobi (hereafter SsCs) is 2.41 Mbp and the plasmid is 22.67 kb. Its chromosome has a G + C content of 40.28%, which is slightly lower than that of both the facultative SsAp and the co-obligate SsCt (supplementary table S5, Supplementary Material online). Unlike these two endosymbionts (which possess genomes that are similar in size), SsCs has only 635 protein coding sequences (hereafter CDSs), translating into a staggering low coding density of around 26.3%. This means that around 70% of its genome is noncoding, the highest known for any S. symbiotica. Similarly, its putative plasmid contains only two CDSs (a putative autotransporter β-domain-containing protein and a plasmid replication protein), with the remainder of the molecule containing several pseudogenes mainly belonging to inactivated insertion sequence (hereafter IS) elements. Additionally, the chromosome of SsCs retains two prophage regions, however these do not encode for a single intact protein, but rather show generally highly degraded pseudogenes. Also, unlike SsAp and SsCt, it displays a typical pattern of polarized nucleotide composition in each replichore (G + C skew in fig. 2A and supplementary fig. S1, Supplementary Material online), hinting at a lack of recent chromosome rearrangements. This is consistent with its low number of mobile elements, when compared with SsAp and SsCt, and the complete inactivation of these by pseudogenization and loss of other elements (e.g. inverted repeats in an IS). In regard to ncRNAs, it possesses only one rRNA operon, 38 tRNAs, a tmRNA, and 5 other noncoding RNAs (including the RNase P M1 RNA component and the 4.5S sRNA component of the Signal Recognition Particle).
Fig. 2.
—Genome of S. symbiotica strain SeCistrobi and pangenome of S. symbiotica strains from Lachninae aphids. (A) Genome plot of S. symbiotica strain SeCistrobi. From outermost to innermost, the features on the direct strand, reverse strand, ncRNA features, and G + C skew are represented. For the G + C skew, green=positive; purple=negative. (B) Venn-like diagram displaying the shared (core) and unshared protein-coding genes among currently available S. symbiotica strains.
Regarding its CDS content, it is almost in its entirety a subset of the pan-genome of S. marcescens, except for the two plasmid CDSs (supplementary fig. S2, Supplementary Material online). While the putative autotransporter β-domain-containing protein from SsCs does not cluster with any other proteins, its best five matches in NCBI’s nr database are against other autotransporter β-domain-containing proteins from S. symbiotica strain CWBI-2.3. Therefore it shows as strain specific in our analysis due to the strains chosen for the protein clustering. When compared with co-obligate S. symbiotica strains from Lachninae aphids figure 2B, it shares most of its genetic repertoire with the highly reduced SsCc and SsTs strains. Within the subset of noncore genes, we observed mainly genes retained in degraded pathways, as well as others that reflect differences in pathway retention (such as difference in the metabolism of nucleotides, gluconeogenesis, and sulfur cluster biosynthesis), and genes that code for membrane proteins both involved in the transport of different compounds (such as putrescine import, Sodium/proline symport, and arginine transport) and of unknown function. In terms or DNA repair, SsCs retains mostly the same set of proteins as the most genomically reduced S. symbiotica symbionts (SsCc and SsTs), with the marked exception of SsCc retaining Dam, MutH, MutL, and MutS; thus coding for a mismatch repair system lacking the exonucleases ExoX, XseA, XseB, recJ, and the nonessential HolE protein from the DNA polymerase III.
We reconstructed phylogenetic trees using 297 single-copy CDSs that were shared by all S. symbiotica strains, a selection of free-living Serratia, and Yersinia pestis strain CO92 (as an outgroup). Using MrBayes, we found S. symbiotica as a monophyletic group sister to the S. marcescens clade (supplementary fig. S3A, Supplementary Material online). Given the very long branches leading to the highly reduced SsCs, SsCc, and SsTs; we also ran a phylogenetic reconstruction in Phylobayes with dayhoff-6 recoded alignments and under the CAT + GTR + G (four discrete categories) model. This method is presumably less sensitive to long branch attraction artifacts commonly seen in phylogenies including highly derived endosymbiont lineages (Husník et al. 2011; Philippe and Roure 2011). From all 8 independent chains we ran, only two of them converged, even after 24,000 generations (with some even reaching the 28,000 and 30,000 generations). However, the S. marcescens + S. symbiotica clade, as well as other bipartitions, were lowly supported and/or unresolved (supplementary fig. S3B, Supplementary Material online). Additional reconstructions were performed using subsets of these data with MrBayes and Phylobayes (see Materials and Methods: Phylogenetic Reconstruction and Rearrangement Analysis), finding similar results in the former and alternative topologies for the latter (supplementary file S3, Supplementary Material online). This suggests that additional taxa (e.g. Serratia strains associated with other Cinara species) are probably needed to stabilize the phylogenetic trees. Finally, and like all other currently available S. symbiotica strains, its genome shows many rearrangements (supplementary fig. S3C, Supplementary Material online) when compared with free-living S. marcescens and other S. symbiotica.
Biosynthesis of EAAs and B Vitamins by the Symbiotic Consortium in C. strobi
In previously analyzed co-obligate endosymbiotic systems in Lachninae aphids (Buchnera + secondary symbiont), Buchnera remains as the sole provider of EAAs and the newly acquired symbionts have taken over the role of synthesizing riboflavin (vitamin B2) and biotin (vitamin B7), functions once performed by Buchnera (Lamelas et al. 2011; Manzano-Marín and Latorre 2014; Manzano-Marín et al. 2016; Meseguer et al. 2017). Thus, to infer the role of each fixed symbiont of C. strobi, we searched for the genes involved in the biosynthesis of EAAs (fig. 3), B vitamins, and other cofactors (fig. 4; supplementary fig. S4, Supplementary Material online) in Buchnera, S. symbiotica, and Sodalis from C. strobi and compared them with co-obligate Buchnera + Serratia endosymbiotic systems in Lachninae (using Buchnera-only Aphididae systems as reference).
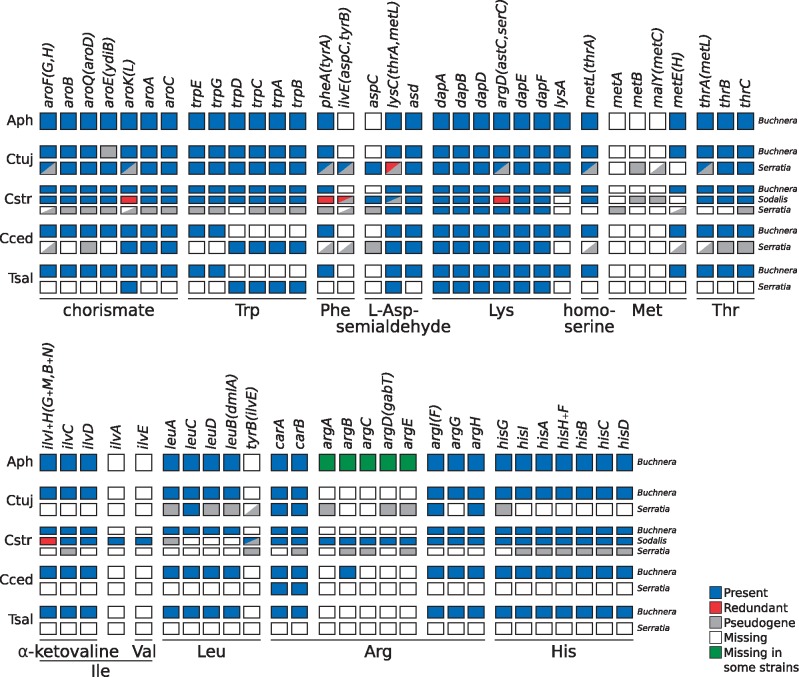
Fig. 3.
—Essential-amino-acid biosynthetic metabolic capabilities of obligate symbiotic consortia of different aphid species. Diagram summarizing the metabolic capabilities of the fixed endosymbiotic consortia of co-obligate symbiotic systems of Lachninae aphids. For comparison, a collapsed representation of Aphididae Buchnera-only systems is used as an outgroup. The names of genes coding for enzymes involved in the biosynthetic pathway are used as column names. Each row’s boxes represent the genes coded by a symbiont’s genome. At the right of each row, the genus for the corresponding symbiont. Abbreviations for the aphids harboring the symbionts is shown at the left of each group rows and goes as follows. Aph=Aphididae; Ctuj=C. tujafilina; Cstr=C. strobi; Cced=C. cedri; Tsal=T. salignus. On the bottom, lines underlining the genes involved in the pathway leading to the compound specified by the name underneath the line. For amino acids, its three letter abbreviation is used.
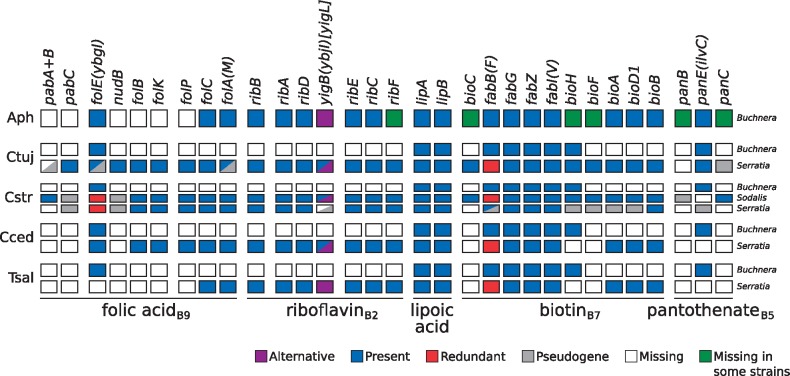
Fig. 4.
—B-vitamin biosynthetic metabolic capabilities of obligate symbiotic consortia of different aphid species. Diagram summarizing the metabolic capabilities of the fixed endosymbiotic consortia of co-obligate symbiotic systems of Lachninae aphids. For comparison, a collapsed representation of Aphididae Buchnera-only systems is used as an outgroup. The names of genes coding for enzymes involved in the biosynthetic pathway are used as column names. Each row’s boxes represent the genes coded by a symbiont’s genome. At the right of each row, the genus for the corresponding symbiont. Abbreviations for the aphids harboring the symbionts is shown at the left of each group rows and goes as follows. Aph=Aphididae; Ctuj=C. tujafilina; Cstr=C. strobi; Cced=C. cedri; Tsal=T. salignus. On the bottom, lines underlining the genes involved in the pathway leading to the compound specified by the name underneath the line.
In terms of EAAs, Buchnera from C. strobi (hereafter BCs), retains the same capabilities as other Buchnera strains. Similarly, Sodalis also retains all genes needed for the biosynthesis of EAAs, except for those of lysine, methionine, and leucine. In the case of SsCs, it has completely lost the potential of de novo synthesizing all EAAs. Nonetheless, it preserves an almost intact route for the synthesis of lysine, resembling the degradation pattern observed for this pathway in other co-obligate S. symbiotica strains.
Regarding B vitamins and other cofactors, we found that BCs is unable to synthesize vitamin B2 and B7, similarly to the other Buchnera from Lachninae aphids. Unlike the Lachninae co-obligate endosymbiotic systems, we determined that SsCs is unable to takeover the role of synthesizing these two vitamins. The vitamin B2 pathway would be interrupted due to the loss of a 5-amino-6-(5-phospho-d-ribitylamino)uracil phosphatase enzyme, preserving only a yigB pseudogene (interrupted by various stop codons and frameshifts). From the genes needed to complement Buchnera’s truncated biotin pathway (bioA, bioD1, and bioB), it preserves only bioB. However, it still retains identifiable pseudogenes for bioA and bioD1. All other pathways for B vitamins and other cofactors are degraded, except for that of lipoic acid. On the other hand, and as previously reported by Meseguer et al. (2017), Sodalis is indeed able to takeover the role as the provider of both riboflavin and biotin, thus being essential for the beneficial symbiosis.
Discussion
Genome degeneration is a common characteristic of vertically inherited mutualistic symbionts of insects (McCutcheon and Moran 2011; Moran and Bennett 2014), and is particularly marked in ancient nutritional mutualistic endosymbionts (Shigenobu et al. 2000; Pérez-Brocal et al. 2006; Rio et al. 2012; Williams and Wernegreen 2015). This genome deterioration can eventually affect pathways involved in the symbiont’s essential functions, such as those involved in essential-amino-acid or B-vitamin biosynthesis. When this occurs, the symbiont is either replaced by a more capable symbiont, or is complemented by a new co-obligate symbiont (Latorre and Manzano-Marín 2017). As members of the Lachninae subfamily, Cinara aphids depend on both Buchnera and an additional symbiont for the supply of essential nutrients, namely EAAs and B-vitamins (Lamelas et al. 2011; Manzano-Marín and Latorre 2014; Meseguer et al. 2017). While S. symbiotica is the most prevalent and putatively ancestral symbiont, it has been replaced by other bacterial taxa in several lineages (Meseguer et al. 2017). Cinara strobi represents such a case, in which the putatively ancient co-obligate S. symbiotica symbiont has been replaced by a Sodalis strain.
Here, we further explored the composition and the role of the fixed symbiotic cohort of the aphid C. strobi. Through the reanalysis of previously reported 16S rRNA NGS amplicon data from geographically distant C. strobi populations plus additional ones, we found that not only Buchnera and Sodalis were fixed, but also S. symbiotica. This third symbiont was previously not deemed as fixed given the low abundance (∼1%) of NGS amplicon reads assigned to this taxon, consistent with the low amount of whole-genome sequence data belonging to S. symbiotica (Meseguer et al. 2017). Thus, the persistent association of this symbiont across populations of C. strobi points towards this being a nonfacultative, hence obligate, symbiotic relationship.
Through whole-genome sequencing of the genome of SsCs, we have provided evidence that SsCs could well be a missing link between the loss of function of a symbiont and the acquisition of a new and more capable one. In spite of SsCc showing a large genome (2.41 Mbp), it displays drastic genome pseudogenization (around 26.3% coding density). This drastically contrast both the “early” co-obligate SsCt (∼2.49 Mbp and 53.4% coding density) and the “modestly” shrunk co-obligate SsCc (1.76 Mbp and 39.0% coding density) (Manzano-Marín and Latorre 2016). This means that the majority of SsCs’ genome is made up of pseudogenes and “genomic wastelands”. This would place the genome in an intermediate state of reduction, before losing bigger chunks of it and thus, evolving a smaller-sized genome. The evolutionary relation SsCs keeps with other S. symbiotica symbionts remains uncertain. Consistent with a previous phylogenetic reconstruction (Manzano-Marín et al. 2016), we found that, through the use of inference methods that alleviate long branch attraction artifacts, the relationships among S. symbiotica lineages is not well resolved. This could be due to the extremely long branches, seen for SsCs, SsCc, and SsTs; when compared with SsAp, SsCt, and other free-living Serratia; which confounds phylogenetic signal (see Philippe and Roure 2011). This makes it difficult to interpret the evolutionary origin and relationships of S. symbiotica endosymbionts solely from phylogenetic data. We expect further large-scale sequencing of these endosymbionts will provide further data to disentangle S. symbiotica phylogenetic relationships.
G + C skew in transitional genomes from some endosymbiotic lineages show an altered pattern, when compared with free-living relatives (Clayton et al. 2012) or long-term highly reduced endosymbionts (Williams and Wernegreen 2015; Manzano-Marín et al. 2016). This perturbation may result from recent chromosome rearrangements likely due to recombination events between repetitive elements, namely ISs (Clayton et al. 2012). The presence of a typical pattern of polarized nucleotide composition in each replichore of SsCs (fig. 2A) points toward long-term genome stability, consistent with the lack of functional mobile elements. This G + C skew pattern is not observed neither in the facultative SsAp nor the co-obligate SsCt (supplementary fig. S1, Supplementary Material online). Therefore, the G + C skew pattern in SsCs, together with its highly degenerated genome and the fixed presence of S. symbiotica in different aphid populations, hints at both a long-term obligate association and a vertical transmission of the symbiont in C. strobi.
When a symbiont replacement occurs, it is expected that the new symbiont will replace the symbiotic functions of the former one. This is seen in different mono- and di-symbiotic systems observed in weevils (Anbutsu et al. 2017), aphids (Vogel and Moran 2013; Meseguer et al. 2017; Chong and Moran 2018), mealybugs (Husnik and McCutcheon 2016; Szabó et al. 2017), and several Auchenorrhyncha (McCutcheon and Moran 2010; Bennett and Moran 2013; Koga and Moran 2014). As observed in all other currently sequenced Buchnera from Lachninae aphids, BCs is unable to provide two essential B vitamins: biotin (B7) and riboflavin (B2). In the case of C. strobi, Meseguer et al. (2017) found that Sodalis was capable of supplementing this deficiencies, thus making this fixed symbiont essential for both Buchnera and the aphid. Here, we have found that these two fixed symbionts indeed are together capable of producing all EAAs and B vitamins for their aphid host and each other. When looking at SsCs, the third fixed symbiont in C. strobi, we found that it is unable to independently synthesize any of the aphid’s essential nutrients. This suggests that this symbiont is no longer contributing to the co-obligate nutritional endosymbiotic consortium in C. strobi but it has persisted in the aphid regardless its metabolic dispensability. The retention of certain enzymes in pathways related to the synthesis of EAAs and B vitamins, can be explained in two ways: 1) The enzymes have not had enough time to accumulate mutations which would render them pseudogenes, and 2) these enzymes participate in other cell-maintenance pathways. Evidence for the former is observed in other S. symbiotica and Sodalis genomes, which display various degraded pathways (theoretically inactive but still coding for several enzymes) (Burke and Moran 2011; Manzano-Marín and Latorre 2014; Oakeson et al. 2014). Relating to the latter, the retention of several genes in the biotin pathway (fabB, fabG, fabZ, fabV, and bioH) is possibly due to their involvement in the pathway leading to cell-membrane biogenesis.
It could be argued that SsCs could be a widespread pathogen. However, the likely ancestral presence of this symbiont as a co-obligate symbiont Meseguer et al. (2017), its universal presence in the sampled populations from the species, the highly reduced genome, the lack of proteins with identifiable eukaryotic-like domains (e.g. ankyrin- or leucine-rich repeats) or diverse secretion systems, does not support this hypothesis.
The genome of SsCs also reveals that a massive genome reduction does not necessarily preclude the symbiont’s replacement. The low amount of intact CDSs that SsCs preserves could be explained by the fixation of Sodalis as a co-obligate symbiont. The long-term association with this new symbiont would thus relax selective pressure on keeping a number of genes, namely those that are redundant. This pattern of gene loss following the acquisition of a companion symbiont can be seen in at least two co-obligate systems: Buchnera + secondary in aphids (Manzano-Marín et al. 2016), and Tremblaya+secondary in mealybugs (Husnik and McCutcheon 2016) (see Latorre and Manzano-Marín 2017). It is worth noting the retention of a mismatch repair system in SsCs, which is involved in the detection of non-Watson–Crick base pairs and strand misalignments arising during DNA replication (Marinus 2012). However, the retention of this system does not, to our knowledge, help explain the retention of a large genome with such a low coding capacity. This retention could rather partly explain the lack of an extreme A + T-biased genome (see Moran et al. 2008), such as the ones held by SsCc and SsTs.
Taken together, the evidence points towards a di-symbiotic co-obligate system in C. strobi, with the two co-obligate partners being Buchnera and Sodalis. Based on an ancestral reconstruction of symbiotic associations in in Cinara (Meseguer et al. 2017), this case would constitute one of secondary co-obligate symbiont replacement. At some point in the lineage of C. strobi, the putative ancient secondary co-obligate S. symbiotica symbiont would have been metabolically replaced by the new and capable Sodalis. Whether the inactivation of the genes involved in the de novo synthesis of both riboflavin and biotin happened before the acquisition of Sodalis (rescue) of after it (takeover) through relaxed selection on the retention of those genes, remains unclear. Following this loss of symbiotic function, S. symbiotica would have continued to thrive within the aphid and be vertically inherited from mother to offspring. The perpetuation of S. symbiotica in C. strobi could hypothetically be a collateral result of a fine-tuned system of symbiont inheritance in the aphid. A similar case could be made for Westeberhardia, the putative ancient endosymbiont of at least some Cardiocondyla ants (Klein et al. 2016). In Cardiocondyla obscurior, the symbiont inhabits the cytoplasm of bacteriocytes and possesses a very small genome (532.68 kb). Its genome lacks intact pathways for the biosynthesis of any EAA or B vitamin, but codes for 4-hydroxyphenylpyruvate. This last can be converted into tyrosine by the ant host, thus the symbiont would hypothetically contribute to cuticle formation during the pupal stage. Interestingly, the authors report on a natural population that has lost this symbiont and seems to thrive in the laboratory (at least under conditions including ad libitum protein provisioning). This reflects Westeberhardia has possibly been retained in other populations despite its apparent dispensability. Thus, the loss of an otherwise long-term symbiont like SsCs would require mutational loss of it and subsequent fixation through drift.
Conclusions
Based on the genome-based metabolic analysis of the pathways involved in the synthesis of EAAs and B-vitamins, we have found that only Buchnera and Sodalis are required for the provision of these nutrients to the aphid. S. symbiotica, the third fixed symbiotic partner, does not seem to be contributing towards the mutualistic consortium, suggesting that it has effectively become a “freeloader” which likely evolved from an ancient co-obligate lineage. Our results reveal that after an obligate symbiont’s metabolic-based replacement, the formerly essential associate can be perpetuated in a consortium despite its dispensability. Also, the genome of SsCs evidences that a long-term symbiont can retain a rather large genome despite its extreme low coding density. We expect the exploration of other Buchnera + S. symbiotica co-obligate systems from closely related lineages to C. strobi will further illuminate the genome reduction process undergone by this symbiont as well as the reasons behind its overstay as a “freeloader” in this aphid species.
Supplementary Material
Acknowledgments
We would like to acknowledge the talented artist/scientist Jorge Mariano Collantes Alegre for the aphid cartoon in figure 1A. This work was supported by the Marie-Curie AgreenSkills+ fellowship program cofunded by the EU’s Seventh Framework Programme (FP7-609398) to A.M.M., the Agropolis foundation/Labex Agro (“Cinara’s microbiome”) to E.J., the France Génomique National Infrastructure, funded as part of the Investissemnt d’Avenir program managed by the Agence Nationale pour la Recherche (ANR-10-INBS-09) to C.O, C.C., and V.B. This publication has been written with the support of the AgreenSkills+ fellowship program that has received funding from the EU’s Seventh Framework Programme under grant agreement N FP7-609398 (AgreenSkills+ contract). We are grateful to the genotoul bioinformatics platform Toulouse Midi-Pyrenees (Bioinfo Genotoul) for providing help and/or computing and/or storage resources. The authors are grateful to the CBGP-HPC computational platform. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Literature Cited
- Akman Gündüz E, Douglas AE.. 2009. Symbiotic bacteria enable insect to use a nutritionally inadequate diet. Proc R Soc Lond B Biol Sci. 276(1658):987–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17):3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anbutsu H, et al. 2017. Small genome symbiont underlies cuticle hardness in beetles. Proc Natl Acad Sci U S A. 114(40):E8382–E8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschner M. 1934. Studies on the symbiosis of the body louse: I. Elimination of the symbionts by centrifugalisation of the eggs. Parasitology 26(03):309–314. [Google Scholar]
- Bankevich A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett GM, Moran NA.. 2013. Small, smaller, smallest: the origins and evolution of ancient dual symbioses in a phloem-feeding insect. Genome Biol Evol. 5(9):1675–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman RL, Eastop VF.. 1994. Aphids on the world’s trees: an identification and information guide. 1st edn Wallingford (UK: ): CAB International, Natural History Museum (London; ). [Google Scholar]
- Boratyn GM, et al. 2012. Domain enhanced lookup time accelerated BLAST. Biol Direct 7: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G, Pevzner PA.. 2002. Genome-scale evolution: reconstructing gene orders in the ancestral species. Genome Res. 12(1):26–36. [PMC free article] [PubMed] [Google Scholar]
- Buchner P. 1953. Endosymbiose der Tiere mit Pflanzlichen Mikroorganismen. Basel: Birkhäuser. [Google Scholar]
- Burke GR, Moran NA.. 2011. Massive genomic decay in Serratia symbiotica, a recently evolved symbiont of aphids. Genome Biol Evol. 3(0):195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke GR, Normark BB, Favret C, Moran NA.. 2009. Evolution and diversity of facultative symbionts from the aphid subfamily Lachninae. Appl Environ Microbiol. 75(16):5328–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles H, Ishikawa H.. 1999. Physical and genetic map of the genome of Buchnera, the primary endosymbiont of the pea aphid Acyrthosiphon pisum. J Mol Evol. 48(2):142–150. [DOI] [PubMed] [Google Scholar]
- Chen F, Mackey AJ, Vermunt JK, Roos DS.. 2007. Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS ONE 2(4):e383.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong RA, Moran NA.. 2018. Evolutionary loss and replacement of Buchnera, the obligate endosymbiont of aphids. ISME J. 12(3): 898–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton AL, et al. 2012. A novel human-infection-derived bacterium provides insights into the evolutionary origins of mutualistic insect-bacterial symbioses. PLoS Genet. 8(11):e1002990.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan PH, Lazarus AB, Wernegreen JJ.. 2005. Genome sequence of Blochmannia pennsylvanicus indicates parallel evolutionary trends among bacterial mutualists of insects. Genome Res. 15(8):1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan PH, et al. 2010. Dynamics of genome evolution in facultative symbionts of aphids. Environ Microbiol. 12(8):2060–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan PH, Ochman H, Moran NA.. 2011. Sequence conservation and functional constraint on intergenic spacers in reduced genomes of the obligate symbiont Buchnera. PLoS Genet. 7(9):e1002252.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan PH, Yu Y, Sisneros N, Wing RA, Moran NA.. 2009. Hamiltonella defensa, genome evolution of protective bacterial endosymbiont from pathogenic ancestors. Proc Natl Acad Sci U S A. 106(22):9063–9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas A. 1996. Reproductive failure and the free amino acid pools in pea aphids (Acyrthosiphon pisum) lacking symbiotic bacteria. J Insect Physiol. 42(3):247–255. [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R.. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27(16):2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil R, et al. 2003. The genome sequence of Blochmannia floridanus: comparative analysis of reduced genomes. Proc Natl Acad Sci U S A. 100(16):9388–9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths GW, Beck SD.. 1975. Ultrastructure of pea aphid mycetocytes: evidence for symbiote secretion. Cell Tissue Res. 159(3):351–367. [DOI] [PubMed] [Google Scholar]
- Hansen AK, Moran NA.. 2011. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc Natl Acad Sci U S A. 108(7):2849–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa T, Kikuchi Y, Nikoh N, Shimada M, Fukatsu T.. 2006. Strict host-symbiont cospeciation and reductive genome evolution in insect gut bacteria. PLoS Biol. 4(10):e337.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Sabree ZL, Moran NA.. 2012. Genome sequence of Blattabacterium sp. strain BGIGA, Endosymbiont of the Blaberus giganteus Cockroach. J Bacteriol. 194(16):4450–4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husník F, Chrudimský T, Hypša V.. 2011. Multiple origins of endosymbiosis within the Enterobacteriaceae (γ-Proteobacteria): convergence of complex phylogenetic approaches. BMC Biol. 9(1):87.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnik F, McCutcheon JP.. 2016. Repeated replacement of an intrabacterial symbiont in the tripartite nested mealybug symbiosis. Proc Natl Acad Sci U S A. 113(37):E5416–E5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousselin E, et al. 2016. Assessment of a 16S rRNA amplicon Illumina sequencing procedure for studying the microbiome of a symbiont-rich aphid genus. Mol Ecol Resour. 16(3):628–640. [DOI] [PubMed] [Google Scholar]
- Kambhampati S, Alleman A, Park Y.. 2013. Complete genome sequence of the endosymbiont Blattabacterium from the cockroach Nauphoeta cinerea (Blattodea: blaberidae). Genomics 102(5-6):479–483. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A, et al. 2016. A novel intracellular mutualistic bacterium in the invasive ant Cardiocondyla obscurior. ISME J. 10(2):376–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga R, Moran NA.. 2014. Swapping symbionts in spittlebugs: evolutionary replacement of a reduced genome symbiont. ISME J. 8(6):1237–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga R, Tsuchida T, Fukatsu T.. 2003. Changing partners in an obligate symbiosis: a facultative endosymbiont can compensate for loss of the essential endosymbiont Buchnera in an aphid. Proc R Soc Lond B Biol Sci. 270(1533):2543–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD.. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina Sequencing Platform. Appl Environ Microbiol. 79(17):5112–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamelas A, et al. 2011. Serratia symbiotica from the aphid Cinara cedri: a missing link from facultative to obligate insect endosymbiont. PLoS Genet. 7(11):e1002357.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamelas A, Gosalbes MJ, Moya A, Latorre A.. 2011. New clues about the evolutionary history of metabolic losses in bacterial endosymbionts, provided by the genome of Buchnera aphidicola from the aphid Cinara tujafilina. Appl Environ Microbiol. 77(13):4446–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamelas A, et al. 2008. Evolution of the secondary symbiont “Candidatus Serratia symbiotica” in Aphid Species of the Subfamily Lachninae. Appl Environ Microbiol. 74(13):4236–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S.. 2009. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25(17):2286–2288. [DOI] [PubMed] [Google Scholar]
- Laslett D, Canback B.. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32(1):11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre A, Manzano-Marín A.. 2017. Dissecting genome reduction and trait loss in insect endosymbionts. Ann N Y Acad Sci. 1389(1):52–75. [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O.. 2008. An improved general amino acid replacement matrix. Mol Biol Evol. 25(7):1307–1320. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS.. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13(9):2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Sánchez MJ, et al. 2009. Evolutionary convergence and nitrogen metabolism in Blattabacterium strain Bge, primary endosymbiont of the cockroach Blattella germanica. PLoS Genet. 5(11):e1000721.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Chan PP.. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano-Marín A, Latorre A.. 2014. Settling Down: the Genome of Serratia symbiotica from the Aphid Cinara tujafilina Zooms in on the process of accommodation to a cooperative intracellular life. Genome Biol Evol. 6(7):1683–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano-Marín A, Latorre A.. 2016. Snapshots of a shrinking partner: genome reduction in Serratia symbiotica. Sci Rep. 6:32590.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano-Marín A, Oceguera-Figueroa A, Latorre A, Jiménez-García LF, Moya A.. 2015. Solving a bloody mess: b-vitamin independent metabolic convergence among gammaproteobacterial obligate endosymbionts from blood-feeding arthropods and the Leech Haementeria officinalis. Genome Biol Evol. 7(10):2871–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano-Marín A, Simon JC, Latorre A.. 2016. Reinventing the wheel and making it round again: evolutionary convergence in Buchnera – Serratia symbiotic Consortia between the distantly related Lachninae Aphids Tuberolachnus salignus and Cinara cedri. Genome Biol Evol. 8(5):1440–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano-Marín A, Szabó G, Simon JC, Horn M, Latorre A.. 2017. Happens in the best of subfamilies: establishment and repeated replacements of co-obligate secondary endosymbionts within Lachninae aphids. Environ Microbiol. 19(1):393–408. [DOI] [PubMed] [Google Scholar]
- Marinus MG. 2012. DNA mismatch repair. EcoSal Plus 5(1). doi:10.1128/ecosalplus.7.2.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA.. 2010. Functional convergence in reduced genomes of bacterial symbionts spanning 200 million years of evolution. Genome Biol Evol. 2:708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA.. 2011. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 10(1):13–26. [DOI] [PubMed] [Google Scholar]
- Meseguer AS, et al. 2017. Buchnera has changed flatmate but the repeated replacement of co-obligate symbionts is not associated with the ecological expansions of their aphid hosts. Mol Ecol. 26(8):2363–2378. [DOI] [PubMed] [Google Scholar]
- Mittler T. 1971. Some effects on the aphid Myzus persicae of ingesting antibiotics incorporated into artificial diets. J Insect Physiol. 17(7):1333–1347. [Google Scholar]
- Mizrahi-Man O, Davenport ER, Gilad Y.. 2013. Taxonomic classification of bacterial 16S rRNA genes using short sequencing reads: evaluation of effective study designs. PLoS One 8(1):e53608.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA. 1996. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc Natl Acad Sci U S A. 93(7):2873–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Bennett GM.. 2014. The tiniest tiny genomes. Annu Rev Microbiol. 68:195–215. [DOI] [PubMed] [Google Scholar]
- Moran NA, Dunbar HE, Wilcox JL.. 2005. Regulation of transcription in a reduced bacterial genome: nutrient-provisioning genes of the obligate symbiont Buchnera aphidicola. J Bacteriol. 187(12):4229–4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, McCutcheon JP, Nakabachi A.. 2008. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 42:165–190. [DOI] [PubMed] [Google Scholar]
- Moran NA, Plague GR.. 2004. Genomic changes following host restriction in bacteria. Curr Opin Genet Dev. 14(6):627–633. [DOI] [PubMed] [Google Scholar]
- Munson MA, Baumann P, Kinsey MG.. 1991. Buchnera gen.nov. and Buchnera aphidicola sp. nov., a Taxon Consisting of the Mycetocyte-Associated, Primary Endosymbionts of Aphids. Int J Syst Bacteriol. 41(4):566–568. [Google Scholar]
- Nakabachi A, Ishikawa H.. 1999. Provision of riboflavin to the host aphid, Acyrthosiphon pisum, by endosymbiotic bacteria, Buchnera. J Insect Physiol. 45(1):1–6. [DOI] [PubMed] [Google Scholar]
- Nawrocki EP, et al. 2015. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 43(Database issue):D130–D137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR.. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29(22):2933–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoh N, et al. 2014. Evolutionary origin of insect-Wolbachia nutritional mutualism. Proc Natl Acad Sci U S A. 111(28):10257–10262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogge G. 1976. Sterility in tsetse flies (Glossina morsitans Westwood) caused by loss of symbionts. Experientia 32(8):995–996. [DOI] [PubMed] [Google Scholar]
- Nogge G. 1981. Significance of symbionts for the maintenance of an optimal nutritional state for successful reproduction in haematophagous arthropods. Parasitology 82(4):101–104. [Google Scholar]
- Nováková E, et al. 2013. Reconstructing the phylogeny of aphids (Hemiptera: aphididae) using DNA of the obligate symbiont Buchnera aphidicola. Mol Phylogenet Evol. 68(1):42–54. [DOI] [PubMed] [Google Scholar]
- Oakeson KF, et al. 2014. Genome degeneration and adaptation in a nascent stage of symbiosis. Genome Biol Evol. 6(1):76–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtaka C, Ishikawa H.. 1991. Effects of heat treatment on the symbiotic system of an aphid mycetocyte. Symbiosis 11:19–30. [Google Scholar]
- Okonechnikov K, Golosova O, Fursov M.. 2012. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28(8):1166–1167. [DOI] [PubMed] [Google Scholar]
- Patiño-Navarrete R, Moya A, Latorre A, Peretó J.. 2013. Comparative genomics of Blattabacterium cuenoti: the Frozen Legacy of an Ancient Endosymbiont Genome. Genome Biol Evol. 5(2):351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Brocal V, et al. 2006. A small microbial genome: the end of a long symbiotic relationship? Science 314(5797):312–313. [DOI] [PubMed] [Google Scholar]
- Philippe H, Roure B.. 2011. Difficult phylogenetic questions: more data, maybe; better methods, certainly. BMC Biol. 9(1):91.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliakov A, et al. 2011. Large-scale label-free quantitative proteomics of the pea aphid-Buchnera symbiosis. Mol Cell Proteomics 10(6):M110.007039.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, et al. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41(Database issue):D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio RVM, et al. 2012. Insight into the transmission biology and species-specific functional capabilities of Tsetse (Diptera: glossinidae) obligate symbiont Wigglesworthia. mBio 3(1):e00240–e00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell CW, Bouvaine S, Newell PD, Douglas AE.. 2013. Shared metabolic pathways in a coevolved insect-bacterial symbiosis. Appl Environ Microbiol. 79(19):6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell CW, et al. 2014. Matching the supply of bacterial nutrients to the nutritional demand of the animal host. Proc R Soc Lond B Biol Sci. 281(1791):20141163.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabree ZL, Kambhampati S, Moran NA.. 2009. Nitrogen recycling and nutritional provisioning by Blattabacterium, the cockroach endosymbiont. Proc Natl Acad Sci U S A. 106(46):19521–19526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacchi L, Grigolo A, Biscaldi G, Laudani U.. 1993. Effects of heat treatment on the symbiotic system of Blattoidea: morphofunctional alterations of bacteriocytes. Boll Zool. 60(3):271–279. [Google Scholar]
- Sandstrom J, Moran N.. 1999. How nutritionally imbalanced is phloem sap for aphids?. Entomol Exp Appl. 91(1):203–210. [Google Scholar]
- Schloss PD, Westcott SL.. 2011. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl Environ Microbiol. 77(10):3219–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R.. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27(6):863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H.. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407(6800):81–86. [DOI] [PubMed] [Google Scholar]
- Suzek BE, Ermolaeva MD, Schreiber M, Salzberg SL.. 2001. A probabilistic method for identifying start codons in bacterial genomes. Bioinformatics 17(12):1123–1130. [DOI] [PubMed] [Google Scholar]
- Szabó G, et al. 2017. Convergent patterns in the evolution of mealybug symbioses involving different intrabacterial symbionts. ISME J. 11(3):715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J.. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577. [DOI] [PubMed] [Google Scholar]
- Tamas I, et al. 2002. 50 million years of genomic stasis in endosymbiotic bacteria. Science 296(5577):2376–2379. [DOI] [PubMed] [Google Scholar]
- Tokuda G, et al. 2013. Maintenance of essential amino acid synthesis pathways in the Blattabacterium cuenoti symbiont of a wood-feeding cockroach. Biol Lett. 9(3):20121153.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Domselaar GH, et al. 2005. BASys: a web server for automated bacterial genome annotation. Nucleic Acids Res. 33(Web Server):W455–W459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ham RCHJ, et al. 2003. Reductive genome evolution in Buchnera aphidicola. Proc Natl Acad Sci U S A. 100(2):581–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel KJ, Moran NA.. 2013. Functional and evolutionary analysis of the genome of an obligate fungal symbiont. Genome Biol Evol. 5(5):891–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace IM, O'Sullivan O, Higgins DG, Notredame C.. 2006. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res. 34(6):1692–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR.. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 73(16):5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LE, Wernegreen JJ.. 2015. Genome evolution in an ancient bacteria-ant symbiosis: parallel gene loss among Blochmannia spanning the origin of the ant tribe Camponotini. PeerJ 3:e881.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler H. 1975. Nature of Transported Substances In: Zimmermann MH, Milburn JA, editors. Encyclopedia of plant physiology. Vol. 1. Transport in Plants I, Chapter 3. Berlin, Heidelberg (Germany: ): Springer; p. 59–100. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.