Abstract
Appreciation is growing for how chromosomes are organized in three-dimensional space at interphase. Microscopic and high throughput sequence-based studies have established that the mammalian inactive X chromosome (Xi) adopts an alternate conformation relative to the active X chromosome. The Xi is organized into several multi-megabase chromatin loops called superloops. At the base of these loops are superloop anchors, and in humans three of these anchors are composed of large tandem repeat DNA that include DXZ4, Functional Intergenic Repeating RNA Element, and Inactive-X CTCF-binding Contact Element (ICCE). Each repeat contains a high density of binding sites for the architectural organization protein CCCTC-binding factor (CTCF) which exclusively associates with the Xi allele in normal cells. Removal of DXZ4 from the Xi compromises proper folding of the chromosome. In this study, we report the characterization of the ICCE tandem repeat, for which very little is known. ICCE is embedded within an intron of the Nobody (NBDY) gene locus at Xp11.21. We find that primary DNA sequence conservation of ICCE is only retained in higher primates, but that ICCE orthologs exist beyond the primate lineage. Like DXZ4, what is conserved is organization of the underlying DNA into a large tandem repeat, physical location within the NBDY locus and conservation of short DNA sequences corresponding to specific CTCF and Yin Yang 1 binding motifs that correlate with female-specific DNA hypomethylation. Unlike DXZ4, ICCE is not common to all eutherian mammals. Analysis of certain ICCE CTCF motifs reveal striking similarity with the DXZ4 motif and support an evolutionary relationship between DXZ4 and ICCE.
Keywords: X chromosome inactivation, tandem repeat, NBDY, Thp5, superloop anchor, inactive X chromosome, YY1
Introduction
Developmentally regulated gene silencing is achieved by repackaging DNA into facultative heterochromatin, the most extensive example of which is the inactive X chromosome (Xi) (Wutz 2011). The Xi is the product of dosage compensation in mammals (Lee 2011) whereby gene silencing from all but one X chromosome per cell is largely silenced (Balaton and Brown 2016). Silencing is established early during development to ensure balance of X-linked gene expression between the sexes (Lyon 1961), and is brought about in part through the acquisition of repressive covalent DNA (Mohandas et al. 1981) and histone modifications (Boggs et al. 2002; Peters et al. 2002; Plath et al. 2003; Silva et al. 2003). Once established, the Xi can be readily detected as a densely staining mass at the nuclear periphery or in proximity to nucleoli (Barr and Bertram 1949), possibly due to adopting an alternate three-dimensional (3D) conformation relative to the active X chromosome (Xa) (Rego et al. 2008; Naughton et al. 2010; Teller et al. 2011).
While most regions of the human Xi acquire repressive features, several regions adopt a euchromatic configuration (Boggs et al. 2002; Chadwick and Willard 2002) that reside at the interface between alternating multi-megabase (Mb) bands of differentially methylated forms of histone H3 (Chadwick and Willard 2004). The DNA sequence underlying three of the most extensive euchromatin signals correspond to large tandem repeat DNA including the macrosatellite DXZ4 (Chadwick 2008), the Functional Intergenic Repeating RNA Element (FIRRE) (Hacisuleyman et al. 2014), formerly referred to as X130 based on the chromosomal coordinates (Horakova, Moseley et al. 2012), and the Inactive-X CTCF-binding Contact Element (ICCE) (Darrow et al. 2016), formerly referred to as X56 (Horakova, Moseley et al. 2012). Each of these three repeat elements are bound exclusively on the Xi allele by the architectural protein CCCTC-binding factor (CTCF) (Chadwick 2008; Horakova, Moseley et al. 2012) and despite residing between ∼15 and 75 Mb apart (fig. 1A), the three elements make frequent Xi-specific contact with one another (Horakova, Moseley et al. 2012; Rao et al. 2014; Tang et al. 2015) organizing the Xi into several massive chromatin superloops. The macrosatellite DXZ4 divides the Xi into two superdomains (Rao et al. 2014; Deng et al. 2015; Giorgetti et al. 2016) and contributes to the proper 3D organization of the Xi (Darrow et al. 2016; Giorgetti et al. 2016; Bonora et al. 2018). While DXZ4 (Giacalone et al. 1992; Chadwick 2008; McLaughlin and Chadwick 2011; Tremblay et al. 2011; Horakova, Calabrese et al. 2012; Horakova, Moseley et al. 2012) and FIRRE (Horakova, Moseley et al. 2012; Hacisuleyman et al. 2014; Yang et al. 2015) have been characterized to some extent, the ICCE repeat remains largely unexplored (Horakova, Moseley et al. 2012). Here we further characterize the ICCE locus and reveal that like DXZ4 (McLaughlin and Chadwick 2011; Horakova, Calabrese et al. 2012), primary DNA sequence conservation of ICCE drops off rapidly beyond higher primates with the notable exception of the CTCF binding sites as well as tandem repeat organization and location in a syntenic region of the X chromosome. However, unlike DXZ4 and FIRRE, ICCE is not universal to eutherian mammals and DNA sequence analysis reveals an evolutionary relationship between the DXZ4 and ICCE repeats.
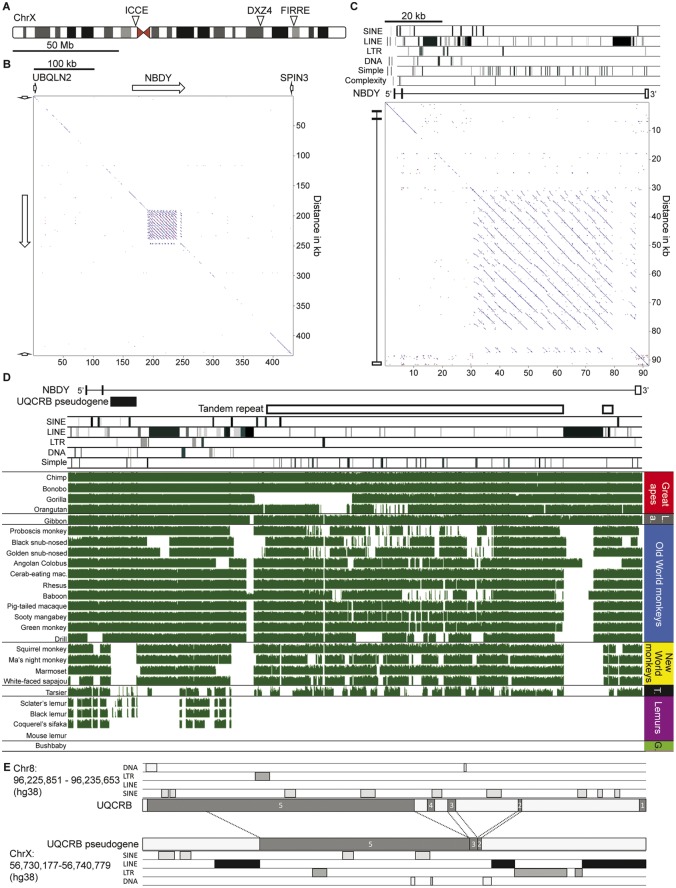
Fig. 1.
—Characterization of the ICCE tandem repeat in primates. (A) Ideogram of the human X chromosome showing the relative locations of the ICCE, DXZ4 and FIRRE tandem repeats. (B) Pairwise alignment of the UBQLN2–SPIN3 interval revealing the presence of the ICCE tandem repeat embedded within the NBDY gene. White arrows indicate gene locations and genomic span. Interval corresponds to hg38 chrX: 56, 566, 788-57, 000, 444. (C) Pairwise alignment of the human NBDY gene locus. Location of various repeat elements is shown at the top, under which are annotated exons 1–3 of NBDY. Interval corresponds to hg19 chrX: 56, 754, 911-56, 846, 086. (D) Sequence conservation at the NBDY locus in primates. The location of NBDY exons 1–3 are indicated as is the ICCE tandem repeat and UQCRB pseudogene. The location of various repeat elements is indicated. Below this are various primate NBDY loci organized into the groupings indicated to the right (LA, Lesser ape; T, Tarsier; G, Galago). Green blocks represent conserved DNA sequence whereas gaps represent a lack of DNA sequence conservation or gaps. Interval corresponds to hg38 chrX: 56, 726, 572-56, 818, 568. (E) Comparison of the UQCRB gene locus on chromosome 8 to the processed pseudogene copy embedded within the NBDY locus. UQCRB exons are indicated as are repeat elements.
Materials and Methods
All DNA sequences used for generation of phylogenetic trees, multiple sequence alignments, and Sequence Logos can be found in Supplementary Material online.
DNA Sequence Analysis—Pairwise Aliments
Pairwise alignments were achieved using the default settings of the YASS genomic similarity search tool (Noe and Kucherov 2005) (http://bioinfo.lifl.fr/yass/index.php; last accessed August 17, 2018). DNA sequences were extracted from the UCSC Genome Browser (Kent et al. 2002) (https://genome.ucsc.edu; last accessed August 17, 2018) using the build and coordinates indicated in the figure legends. In each case, the sequence used for pairwise alignment is extracted both with and without repetitive elements masked as “N”. Output images were edited according to the website instructions to remove histograms and colors were edited from red-green to blue-red to facilitate visualization by all readers.
DNA and Protein Sequence Analysis—Sequence Logos
DNA or protein sequences were converted to FASTA format before pasting into the sequence alignment window of ClustalW (version 1.83) (www.genome.jp/tools-bin/clustalw; last accessed August 17, 2018). Multiple sequence alignments were executed with DNA or protein selection using default settings. Rooted phylogenetic trees with branch length were generated using unweighted pair group method with arithmetic mean (UPGMA). The .aln output file was copied and pasted into the BoxShade server (version 3.21) (https://embnet.vital-it.ch/software/BOX_form.html; last accessed August 17, 2018) with ALN format selected, or into WebLogo 3 (Crooks et al. 2004) (http://weblogo.threeplusone.com; last accessed August 17, 2018). For WebLogo 3, output was set as PDF vector format, sequence type was selected as DNA or Protein accordingly.
DNA Sequence Analysis—CTCF and YY1 Motif Search
The identification of CTCF motifs was achieved by extracting from the UCSC Genome Browser (Kent et al. 2002) (https://genome.ucsc.edu; last accessed August 17, 2018) the DNA sequence under CTCF ChIP-Seq peaks at the human NBDY locus. The sequence was then used to identify CTCF and YY1 motifs using default settings for JASPAR (Khan et al. 2018) (http://jaspar.genereg.net; last accessed August 17, 2018) based on genome wide human CTCF and YY1 sites (ID MA0139.1 for CTCF; MA0095.1 and MA0095.2 for YY1).
DNA Sequence Analysis—DNA Methylation
DNA methylation at the FIRRE and ICCE loci was assessed using publicly available NIH Roadmap Epigenomics Program data (Bernstein et al. 2010) for MethylC-Seq DNA sequencing (Lister et al. 2009).
Results and Discussion
Characterization of the Primate ICCE Repeat
In humans, the ICCE repeat is located between 56 and 57 Mb on the X chromosome (fig. 1A) and is actually embedded within the Negative Regulator of P-Body Association (NBDY) gene, between Ubiquilin 2 (UBQLN2) on the distal side and Spindilin Family Member 3 (SPIN3) gene on the proximal side (fig. 1B). The easiest way to expose the ICCE tandem repeat is via a pairwise alignment of the DNA sequence from the genomic interval, resulting in the characteristic multiple parallel line pattern off the diagonal due to translocating immediately adjacent sequence alignments (fig. 1B).
NBDY was originally annotated as a long noncoding RNA (LOC550643), but more recently it was found to encode a micropeptide that is involved in P-body formation and nonsense mediated decay (D'Lima et al. 2017). Close examination reveals that the ICCE repeat is embedded within intron-2 of NBDY and that with the exception of numerous simple repeats, it is itself largely devoid of long interspersed elements (LINE), short interspersed elements (SINE) and long terminal repeats (fig. 1C). The notable exception is disruption of the tandem repeat at the proximal edge by the insertion of an apparent full-length 6.4 kb nonintact L1PA7 LINE element (Penzkofer et al. 2017), whereas the distal edge is characterized by numerous broken LINE fragments.
Examination of DNA sequence conservation of the NBDY locus in the great and lesser apes reveals the interval to be highly conserved, with most variation occurring in the tandem repeat itself (fig. 1D), which likely reflects repeat unit copy number variation typical of large tandem repeats (Tremblay et al. 2010; Schaap et al. 2013) and has been observed for DXZ4 in the great apes (McLaughlin and Chadwick 2011). As expected, the NBDY locus shows most variation as one moves through the primate tree. Outside of the great and lesser apes, the large LINE element that disrupts the proximal edge of the array is absent, suggesting that it was either lost, or more likely gained after divergence of the apes. Likewise, the closest LINE fragment to the distal edge of the tandem repeat is largely absent outside of the apes. The ICCE repeat itself is relatively well conserved within the Old and New World monkeys with noticeable sequence conservation loss in tarsier and lack of conservation in lemurs and galago, which is consistent with observations made for DXZ4 (Tremblay et al. 2010; Schaap et al. 2013). Distal to ICCE and close to exon-2 of NBDY is a pseudogene copy of Ubiquinol-Cytochrome C Reductase Binding Protein (UQCRB) that appears to have been acquired after the split between Catarrhines and Platyrrhines (fig. 1D). The pseudogene corresponds to exons 2, 3, 4, and most of exon-5 of the spliced gene that is located on chromosome 8 (fig. 1E) and is flanked by a DNA repeat and LINE elements at the NBDY locus.
Characterization of the ICCE Locus Outside Primates
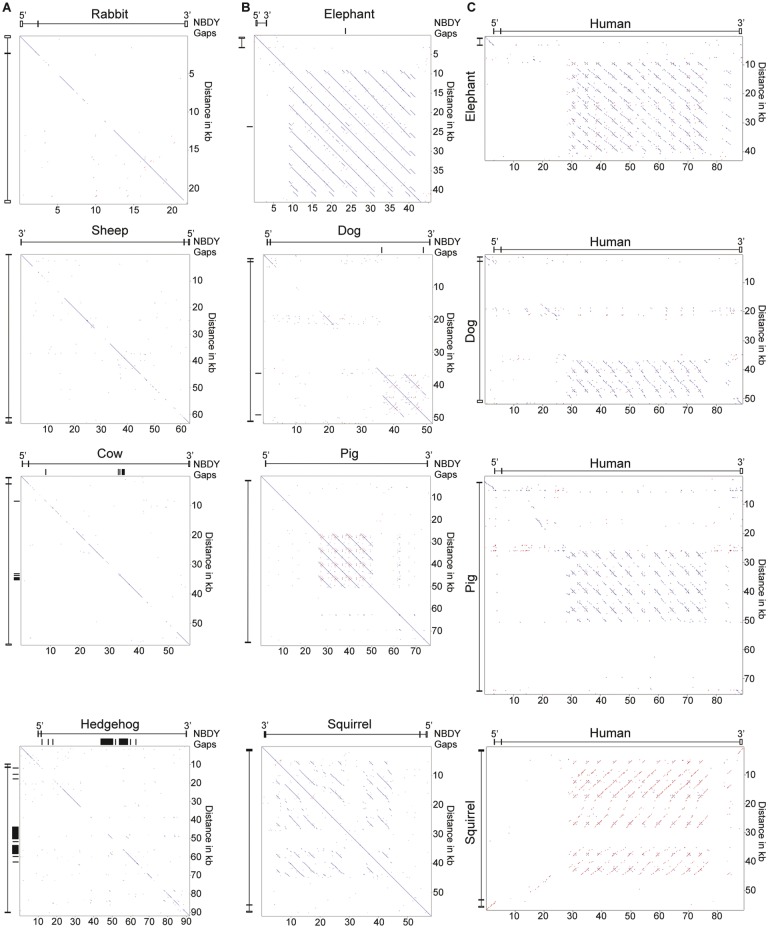
The rapid divergence of the NBDY locus in Strepsirrhini suggests that ICCE may be specific to the suborder Haplorhini. However, exploration of the DXZ4 tandem repeat showed that it too rapidly diverged in primary DNA sequence content through primates, but what is conserved outside the primate lineage is tandem repeat organization, presence of the CTCF binding motif and retention of its physical location adjacent and downstream of the Plastin 3 (PLS3) gene (McLaughlin and Chadwick 2011; Horakova, Calabrese et al. 2012). To assess if this is also the case for ICCE, we looked for the presence of a large tandem repeat in the vicinity of the NBDY ortholog in a large number of eutherian mammals. In most mammals, a tandem repeat could not be detected within the NBDY locus (fig. 2A), nor the more extensive surrounding interval (data not shown). However, a large tandem repeat was detected within the NBDY locus in elephant, dog, pig, and squirrel (fig. 2B). Furthermore, the tandem repeat shared weak but detectable sequence similarity with the human tandem repeat (fig. 2C) supporting these as orthologous ICCE elements.
Fig. 2.
—Presence or absence of a tandem repeat embedded within the NBDY gene locus in various mammals. (A) Pairwise alignments of the NBDY gene locus in representative mammals lacking evidence of an ICCE ortholog, and (B) those with an ICCE ortholog. The location of the NBDY exons are indicated as are any gaps in the genome assembly. (C) Pairwise alignment between the NBDY locus of mammals carrying a tandem repeat and the human NBDY locus. Coordinates of interval represented and build are; Rabbit oryCun2, chrX: 37, 710, 438-37, 732, 512; Sheep oviAri3, chrX: 46, 677, 179-46, 740, 585; Cow bosTau8 chrX: 99, 188, 225-99, 245, 611; Hedgehog eriEur2 JH836267: 547, 376-639, 175; Elephant loxAfr3 scaffold_32: 15, 307, 624-15, 604, 333; Dog canFam3 chrX: 47, 935, 333-47, 987, 627; Pig susScr11 chrX: 49, 155, 577-49, 232, 077; Squirrel speTri2 JH393629: 314, 049-372, 146; Human hg19 chrX: 56, 755, 692-56, 844, 806.
The ICCE Locus Is Largely Absent from Rodents
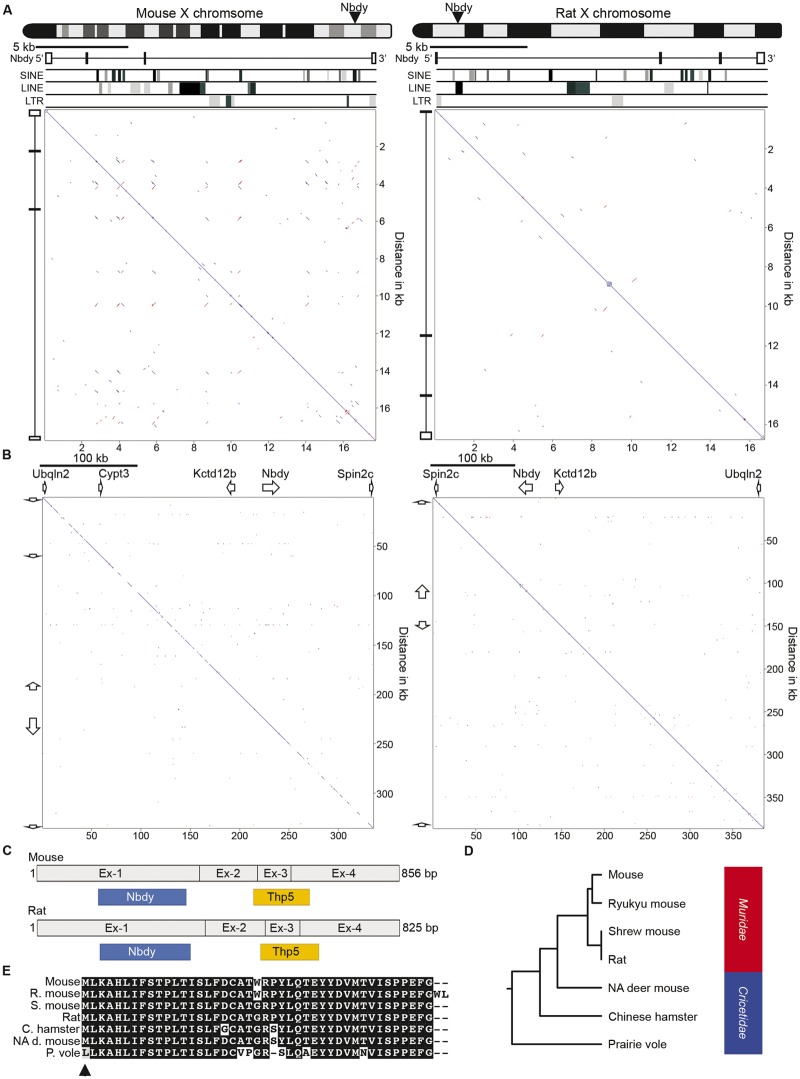
Among the placental mammals, Rodentia is the largest and most diverse order (Macdonald and Norris 2001). The detection of ICCE in squirrel (fig. 2C) indicated the possible presence of the ICCE locus in rodents. Therefore, we examined the rat and lab mouse genomes to see if an ICCE ortholog could be detected. As expected, the Nbdy gene maps to the X chromosome in mouse and rat, but pairwise alignment of the Nbdy locus (both of which span a shorter genomic interval than in most mammals) did not show any evidence of a tandem repeat (fig. 3A), nor was a tandem repeat detected when the pairwise comparison was extended substantially beyond the Nbdy locus, despite shared gene order with the human interval (fig. 3B).
Fig. 3.
—Characterization of the NDBY locus in rodents. (A) Ideograms of the mouse (left) and rat (right) X chromosomes indicating the location of the Nbdy gene locus. Immediately below are pairwise alignments of the Nbdy locus with Nbdy exons and repeat content indicated. The mouse region corresponds to mm10 chrX: 153, 723, 554-153, 741, 296, whereas the rat region is rn5 chrX19, 212, 132-19, 228, 883. (B) Pairwise alignments of an expanded syntenic interval centered on the Nbdy locus in mouse (left, mm9 chrX: 149, 934, 536-150, 270, 027) and rat (right, rn5 chrX: 19, 112, 466-19, 491, 745). White arrows represent gene orientation and approximate span. (C) Comparison of the mouse and rat Nbdy cDNA highlighting exons 1–4 and the ORFs for Nbdy and Thp5. (D) Rooted phylogenetic tree with branch lengths derived using the Thp5 amino acid sequence and the unweighted pair group method with arithmetic mean (UPGMA). Family Muridae and Cricetidae are indicated to the right. (E) Boxshade alignment of the putative Thp5 coding sequence in the rodents indicated (R. mouse, mouse ryukyu; S. mouse, shrewmouse; C. hamster, Chinese hamster; NA d. mouse, North American deer mouse; P. vole, prairie vole). Identical amino acids in all rodents are represented as white letters on a black background whereas differing amino acids are black letters on a white background. Dashed lines represent no amino acid at this position. Black arrowhead indicates initiation codon.
Notably, both mouse and rat Nbdy is composed of four main exons compared with three (or sometimes 2) in other mammals (fig. 3A). Furthermore, in addition to the open reading frame (ORF) for Nbdy, a second ORF has been reported in mouse that codes for another micropeptide called Thp5, a 43 amino acid peptide that is involved in CD4+ T-cell activity (Khan et al. 2012). Therefore, at least in mouse, the Nbdy mRNA encodes two different micropeptides with distinct functions. Consistent with all NBDY orthologs examined, the Nbdy ORF is entirely contained within exon-1, whereas in mouse and conserved in rat, the Thp5 ORF spans exons 2–4 (fig. 3C). Comparison of the complementary DNA (cDNA) sequence of human NBDY (Acc. No. BC062451, 722 bp) with mouse Nbdy (Acc. No. NM_027327, 856 bp) using the nucleotide basic local alignment search tool (BlastN) revealed 84% nucleotide identity between human 1–433 bp and mouse 80–512 bp, as well as 76% nucleotide identity between human 441–690 bp and mouse 598–849 bp. Mouse 513–597 bp (corresponding to mouse exon-3) has no homology in human cDNA. Given that this interval contains most of the Thp5 ORF and is absent from the mature human mRNA, these data strongly support no Thp5 ortholog in humans. Consistent with this, comparison of the sequence of mouse Nbdy exon-3 (78 bp) against the human genome reveals 82.5% sequence identity across the first 40 bp to a LINE element distal to the ICCE tandem repeat, and close to the UQCRBP1 pseudogene. However, the human match lacks the canonical splice acceptor AG sequence where the exon–intron boundary is in mouse, most likely accounting for why it is not retained in the spliced mature message. The remaining 3′ 38 bp of mouse exon-3 has no match in man.
A more extensive search for sequence similarity to the Thp5 ORF revealed that this coding sequence is only conserved in closely related rodents (Family Muridae and Cricetidae), and no coding potential was detected in genomes of Rodentia Family Heteromyidae (kangaroo rat), Heterocephalidae (naked mole-rat), Caviidae (guinea pig), or Sciuridae (squirrel), nor in the lagomorphs (rabbit and pika) of Clade Glires (lagomorphs and rodents) (fig. 3D). Conservation of Thp5 DNA sequence in Muridae and Cricetidae ranged from 92% to 99% DNA sequence identity suggesting that Thp5 may exist in these closely related rodents. However, the start codon is prairie vole is mutated (ATG to TTG) and therefore Thp5 coding potential in this rodent is unlikely (fig. 3E).
Characterization of NBDY
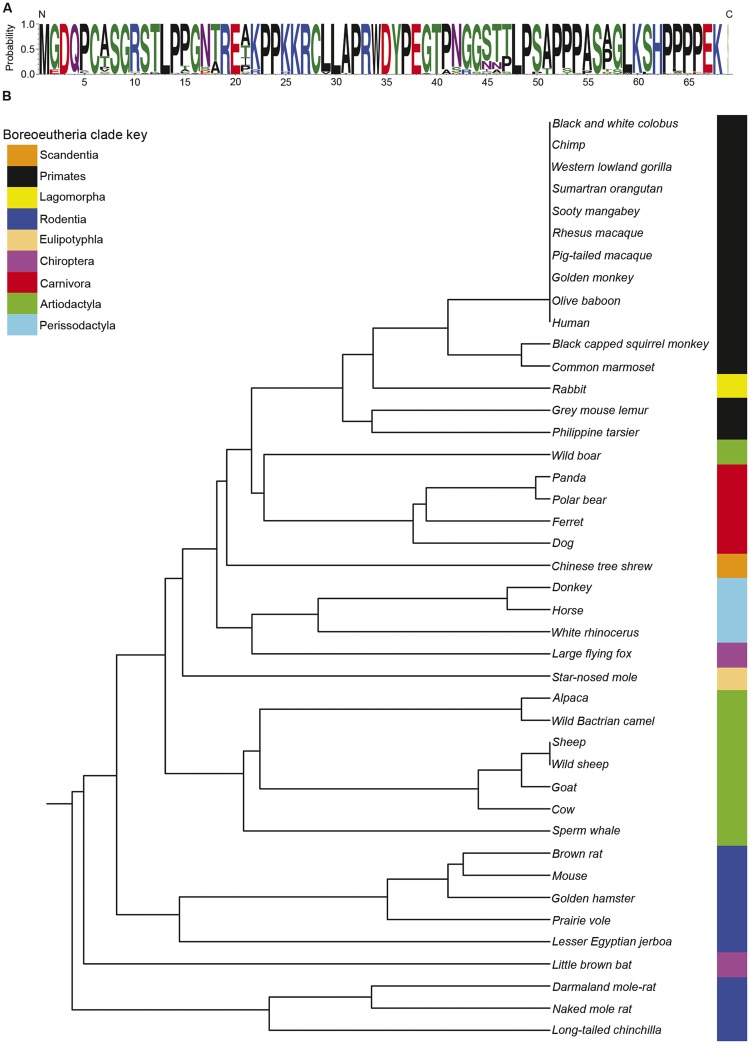
In sharp contrast to Thp5, the NBDY ORF is highly conserved as a short 67–69 amino acid micropeptide (fig. 4A) encoded from the first exon of the transcript in all eutherian mammals (D'Lima et al. 2017). A comparison of the underlying coding sequence of NBDY between a broad range of eutherian mammals produces a phylogenetic tree that closely reflects expectations with the notable exception of rabbit, in which the NBDY coding sequence is more similar to primates than other rodents (fig. 4B). Interestingly, in addition to the X-linked copy, a second identical copy of NBDY ORF exists on rabbit chromosome 15 (2009 build oryCun2) embedded within and antisense to the Interleukin 15 (IL15) gene. Similarly, dog carries a second copy of NBDY ORF embedded within and antisense to the AF4/FMR2 Family Member 3 (AFF3) gene on dog chromosome 10, and microbat has a NBDY ORF embedded within the FK506 Binding Protein 14 (FKBP14) gene. Given their locations outside regions of synteny and orientation relative to the gene in which they are located, each is likely processed pseudogene copies.
Fig. 4.
—The NBDY micropeptide is highly conserved in eutherian mammals. (A) Sequence logo of the primary structure of the NBDY micropeptide using the single letter amino acid code. The amino (N) and carboxy (C) terminus are indicated as is the probability of a particular amino acid at each position. The appearance of a single letter indicates that this amino acid is present at this location in all mammals examined, whereas multiple letters are sized based on their residue frequency. Color of amino acids is based on chemical properties (polar, green; neutral, purple; basic, blue; acidic, red; hydrophobic, black). (B) Rooted phylogenetic tree with branch lengths derived using the NBDY coding DNA sequence and UPGMA. Clustering of members of Orders within the Boreoeutheria clade are highlighted to the right and indicated by the key.
CTCF and the Human ICCE Locus
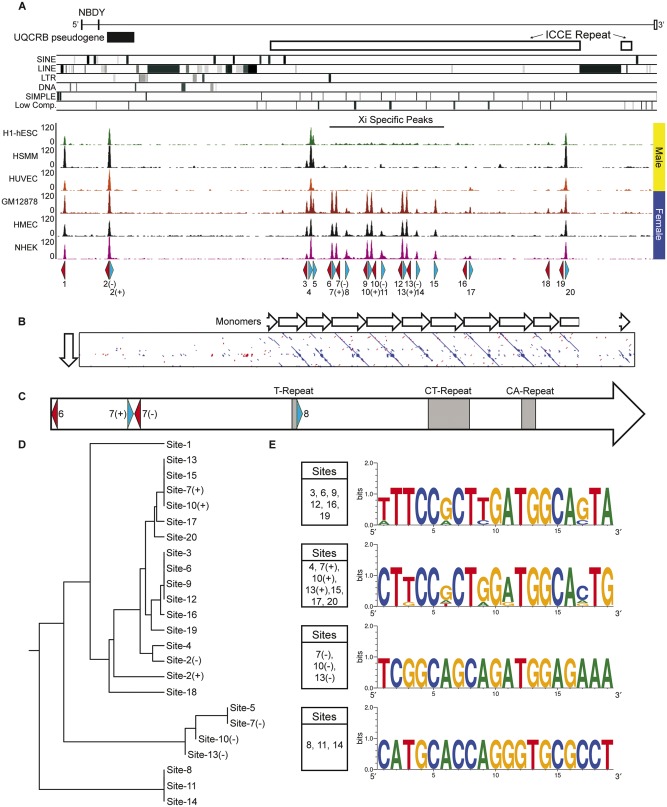
Human DXZ4, FIRRE, and ICCE are all characterized by high-density clustering of CTCF binding sites (Chadwick 2008; Horakova, Moseley et al. 2012; Hacisuleyman et al. 2014). At the human ICCE locus there are 20 CTCF ChIP-Seq peaks in common to several different cell types, located just upstream of the NBDY promoter and into the ICCE repeat, which has the highest density of peaks (fig. 5A). Some are common to males and females and a logical interpretation of this is that these peaks represent binding of CTCF to the male X and female Xa, whereas other peaks are female specific and as such are inferred to correspond to binding to the Xi. These sites are likely contributors to the long-range Xi-specific contacts made between the three repeat elements (Horakova, Moseley et al. 2012) that serve as superdomain anchors (Rao et al. 2014). Using JASPAR (Khan et al. 2018), a single high-quality match for a CTCF binding site was identified under each ChIP-Seq peak, with the exception of sites 2, 7, 10, and 13 that each had a CTCF motif match on each strand (labeled – and +) (fig. 5A). Site-2 (−/+) is located within the UQCRB pseudogene and also corresponds to a ChIP-Seq peak on chromosome 8. The sequence under the peak is >99% identical between the two chromosomes and therefore, whether CTCF is bound at one or both is unclear.
Fig. 5.
—Characterization of CTCF sites within the human NBDY locus. (A) Schematic map of the human NBDY locus showing the location of NBDY exons, the UQCRB pseudogene, the ICCE tandem repeat and the location of various repeat elements at hg19 chrX: 56, 752, 065-56, 844, 234. Beneath this are human CTCF ChIP-Seq profiles for three male samples (H1-hESC, human embryonic stem cell line H1; HSMM, skeletal muscle myoblasts; HUVEC, umbilical vein endothelial cells) and three female samples (GM12878, EBV transformed B-lymphocytes; HMEC, mammary epithelial cells; NHEK, epidermal keratinocytes). Beneath the ChIP-Seq profile are annotated the location of predicted motifs 1–20 and their orientation (blue, forward; red, reverse). Two predicted motifs under a single peak are represented as (+) and (−) for forward and reverse. (B) Pairwise alignment of a single ICCE repeat monomer with the NBDY locus. White arrows represent adjacent monomers. (C) Schematic map of the single monomer from part-B above (corresponding to hg19 chrX: 56, 799, 632-56, 805, 033), showing location of CTCF motifs and microsatellite repeat elements. (D) Rooted phylogenetic tree with branch lengths derived using the DNA sequence of CTCF motifs from the interval by UPGMA. (E) Sequence logos of related CTCF motifs from the NBDY locus. The presence of an identical base at a particular location in a motif is represented by a single base, whereas variation between sites at a particular position in the motif shows two or more bases with relative frequency represented by the size of the height of the base.
The regular periodicity of the CTCF peaks within ICCE reflects the tandem repeat arrangement, which can be more readily observed if the DNA sequence of a single ∼5 kb repeat unit is aligned with the NBDY locus (fig. 5B). The individual repeat units that make up the ICCE array are not as well conserved when compared with one another as adjacent DXZ4 repeat units. Human DXZ4 repeat units share >99% DNA sequence identity with minimal repeat unit length variation (all approximately 3 kb in length) resulting in a highly homogenous array (Tremblay et al. 2011). Within ICCE, the breaks in the diagonal lines in the pairwise alignment as well as substantial length variation clearly illustrate the reduced sequence conservation of repeat units that make up the array (fig. 5B). Up to four CTCF motifs exist within a single ICCE repeat unit; two on the bottom strand and two on the top strand. Additionally, with the exception of three polymorphic simple microsatellite repeats, the monomer sequences are unique to Xp11.21 (fig. 5C). Given that ICCE is composed of approximately ten repeat monomers/fragments that share DNA sequence identity, it is not surprising therefore that CTCF motifs in adjacent repeats are related. This can be seen more clearly when all motifs are compared, resulting in clustering of related CTCF binding sites (fig. 5D). Clustering indicates that most CTCF motifs fall into one of the four related ancestral groups, the consensus of which can be represented graphically as sequence logos (fig. 5E). The sequence logos clearly demonstrate how similar the members of a group are and that consensus sequences differ substantially. Site-1 and Site-2 reside outside of the ICCE interval and have the greatest sequence divergence from the four groups, as does Site-18, represented by a weak CTCF ChIP-Seq peak in two of the six cell types and Site-5, also missing a consistent peak.
Conservation of ICCE Associated CTCF Motifs in Mammals
To gain insight into which sites might be the most important for ICCE function, DNA sequences of the ICCE-embedded CTCF peaks were compared with primate genomes. Given the relatedness of the motifs within clusters, henceforth these will be referred to as either Site-6 (Sites 3, 6, 9, 12, 16, and 19), Site-7 (Sites 4, 7+, 10+, 13+, 15, 17, and 20), Site-7− (Sites 7−, 10−, and 13−), and Site-8 (Sites 8, 11, and 14).
Using BLAST-Like Alignment Tool (BLAT), the Site-8 cluster was found at the ICCE locus in the great apes, lesser apes and Old World monkeys but could not be detected beyond these, suggesting it may have a function restricted to higher primates. The Site-7− cluster was even more restricted and could only be detected in chimpanzee and bonobo. This suggests that the predicted Site-7− motif is unlikely to be an actual CTCF bound site in vivo and that Site-7+ alone is responsible for the experimentally generated ChIP-Seq peak. In contrast, Site-6 and Site-7 were detected at the ICCE locus in all great and lesser apes and all Old and New World monkeys examined. Only matches at ICCE for Site-6 were detected in tarsier. Outside of primates, Site-7 was detected at the ICCE ortholog in pig and dog, whereas Site-6 could be detected at the ICCE ortholog in pig, dog, elephant and squirrel.
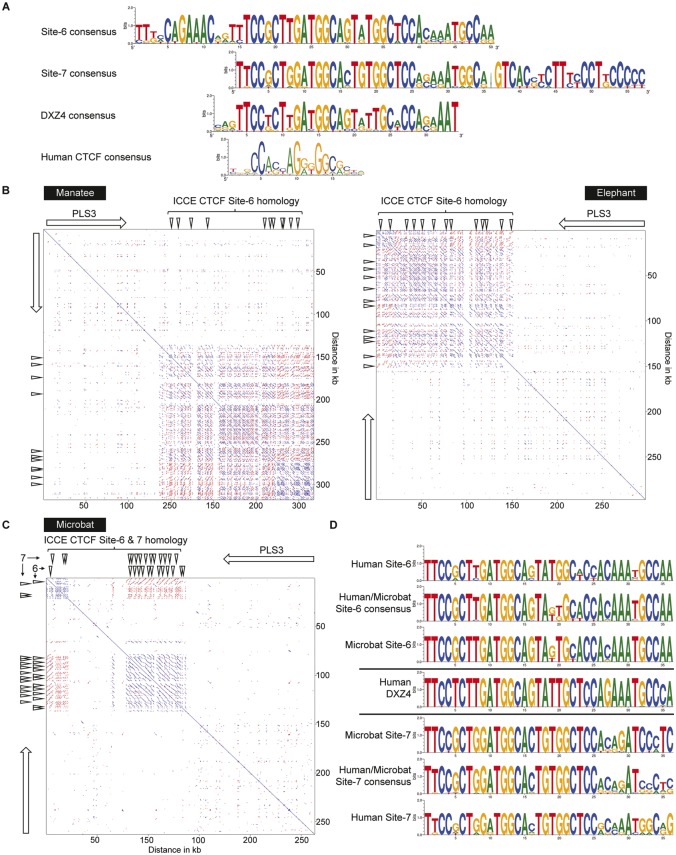
These data support Site-6 and Site-7 as under positive selective pressure and therefore most likely to have an important conserved role in ICCE function. Furthermore, like the highly conserved DXZ4 CTCF motif in mammals (Horakova, Calabrese et al. 2012), sequence conservation extended beyond the core 23 bp predicted CTCF motif to 50 bp for Site-6 and 57 bp for Site-7, both of which are longer than the 34 bp conserved DXZ4 sequence. Generation of sequence logos for mammalian Site-6, Site-7, and DXZ4 and their alignment relative to a genome-wide human CTCF sequence logo reveals that the core of Site-6, Site-7, and DXZ4 are similar and more related to one another than the genome-wide human CTCF consensus (fig. 6A). Notably, Site-6 has unique additional sequence conservation upstream of the CTCF core region, whereas Site-7 has strong conservation downstream of the CTCF core. Besides some predominant C, A, and G residues in the genome-wide consensus, other positions are far more variable, indicating enormous versatility of CTCF binding options throughout the genome (Kim et al. 2007). The ICCE and DXZ4 CTCF motifs are far more rigid in their composition. It is possible that highly related CTCF binding sequences could provide similar functions that differ from other highly related binding sequences. In this case, both DXZ4 and ICCE are Xi-specific superdomain anchors. Perhaps the highly conserved sequence reflects this functional need. It is also conceivable that the conserved sequences up and downstream of the CTCF binding core could reflect binding of other key DNA binding proteins necessary for superdomain anchor function, accounting for why these sequences are under similar positive selective pressure. Regardless, the high sequence similarity between the conserved ICCE CTCF Site-6 and Site-7 with DXZ4 suggest the possibility of an evolutionary relationship between the two tandem repeats whereby ICCE may have originated from duplication, translocation into the NBDY locus and subsequent divergence from DXZ4. Given that not all eutherian mammals appear to have ICCE could reflect its independent loss in some mammals but retention in others.
Fig. 6.
—Conservation of CTCF sites 6 and 7 in mammals with an ICCE ortholog and relationship to the CTCF motif at the DXZ4 tandem repeat. (A) Sequence logos of DNA sequence at and flanking the core predicted CTCF motif at sites 6 and 7 relative to the DXZ4 CTCF sequence logo and human CTCF consensus. Site-6 is shown as the reverse complement and was built from human, pig, dog, elephant, squirrel and tarsier. Site-7 is built from human, pig and dog. The DXZ4 sequence logo was built from previous alignments (Horakova, Calabrese et al. 2012). The human CTCF consensus was adapted from the sequence logo from JASPAR (Khan et al. 2018). (B) Pairwise alignment of the PLS3 locus and downstream region from manatee (left, triMan1 JH594674: 9, 742, 766-10, 060479) and elephant (right, loxAfr3 scaffold_32: 15, 307, 624-15, 604, 333). The white-arrow shows the location, orientation and span of the PLS3 gene and downward facing white arrow-heads indicate sites of sequence homology with human ICCE CTCF Site-6. (C) Pairwise alignment of the microbat PLS3 locus and downstream region (myoLuc2 GL429769: 20, 409, 038-20, 672, 161). The white-arrow shows the location, orientation and span of the PLS3 gene and downward facing white arrow-heads indicate sites of sequence homology with human ICCE CTCF Site-6 and Site-7. (D) Sequence logo of human CTCF Site-6 and -7 compared with microbat CTCF sites 6 and 7, as well as a human-microbat consensus sequence for each aligned with the core human DXZ4 CTCF consensus.
Consistent with the possible evolutionary relationship between DXZ4 and ICCE, BLAT searches using the Site-6 consensus sequence readily detect the DXZ4 tandem repeat downstream of PLS3 in the manatee and elephant genomes (fig. 6B). Notably, in shrew and manatee an ICCE ortholog was not detected in the vicinity of NBDY.
The most intriguing and strongest evidence to support the evolutionary relationship between DXZ4 and ICCE was found in microbat. Like many mammals, microbat does not have a tandem repeat embedded within the NBDY ortholog nor within the interval between UBQLN2 and SPIN3 genes (data not shown), supporting the lack of ICCE in microbat. However, comparison of the ICCE Site-6 and ICCE Site-7 consensus against the microbat genome revealed twenty-seven alternating spatially separate matches to Site-6 and Site-7 (fig. 6C), each located ∼1.5 kb apart and all immediately downstream of PLS3 where DXZ4 has been detected in every mammal examined (Horakova, Calabrese et al. 2012). Microbat was the only mammal in which we found distinct matches to Site-6 and Site-7, indicating that microbat DXZ4 contains two different CTCF motifs arranged in a tandem array similar to what is seen for ICCE. Finally, examination of the sequence of the human and microbat consensus sequences for Site-6 and Site-7 show that despite residing where the DXZ4 ortholog is found in mammals, microbat Site-6 more closely resembles human ICCE Site-6 than the DXZ4 consensus, and microbat Site-7 more closely resembles human ICCE Site-7 than the DXZ4 consensus (fig. 6D).
DNA Underlying CTCF Motifs Is Hypomethylated at the Female FIRRE and ICCE Loci
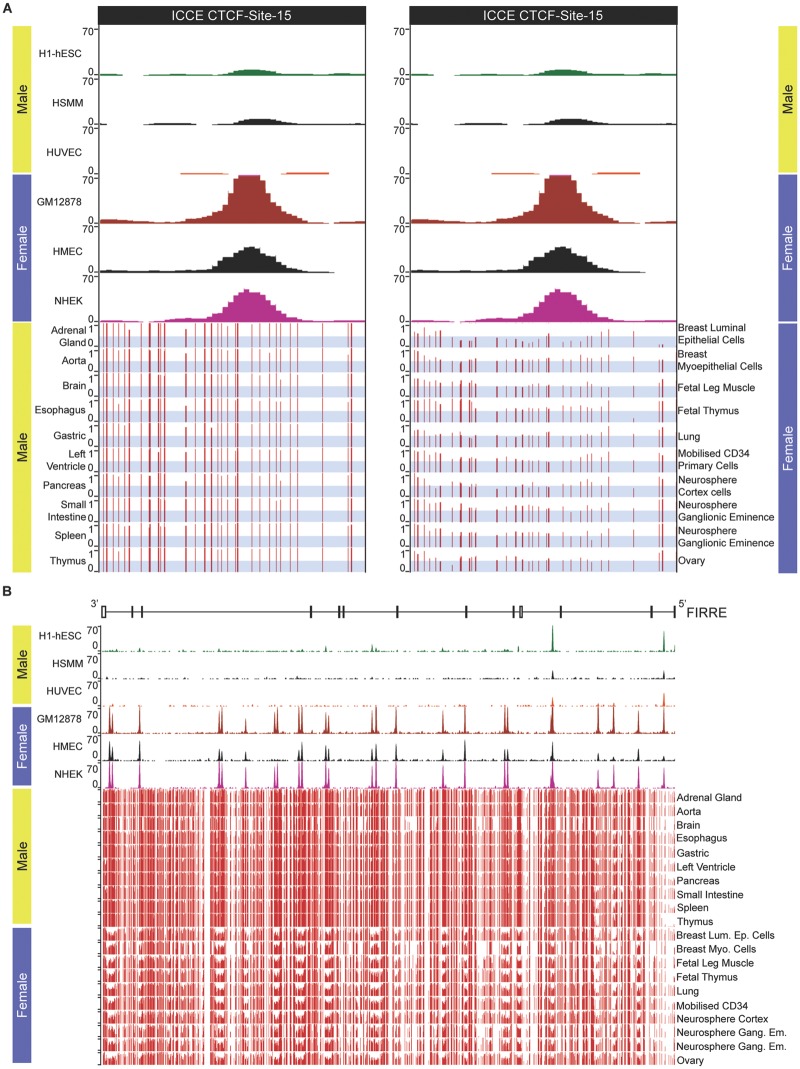
DNA sequences at and surrounding the CTCF binding sites at DXZ4/Dxz4 are hypomethylated in females but not males (Giacalone et al. 1992; Chadwick 2008; Horakova, Calabrese et al. 2012). Therefore, we sought to determine if female-specific CpG hypomethylation is a feature of the ICCE locus. Like all large tandem repeats for which adjacent repeat units are relatively conserved, mapping of epigenomic features is challenging and often excluded from display tracks in genome browsers. This is indeed the case for whole genome massively parallel bisulfite sequencing data at the human DXZ4 and ICCE loci (data not shown). However, sequence flanking CTCF Site-15 (a member of the Site-7 homologous group; fig. 5D) which resides at the proximal edge of ICCE (fig. 5A) is sufficiently different from the more homogenous distal region of the tandem array that unique bisulfite reads could be mapped. Comparison of DNA methylation profile at and immediately around CTCF Site-15 reveals that the interval is predominantly methylated in males, but substantially hypomethylated in females (fig. 7A) consistent with the general inverse relationship between CTCF occupancy and DNA methylation (Wang et al. 2012). Our interpretation of these female-specific data are that like DXZ4/Dxz4 (Giacalone et al. 1992; Chadwick 2008; Horakova, Calabrese et al. 2012), DNA sequence at and flanking CTCF binding sites on the Xi is hypomethylated whereas DNA at the Xa resembles that seen in males and is predominantly methylated.
Fig. 7.
—DNA underlying CTCF motifs is hypomethylated in females at the ICCE and FIRRE loci. (A) Top left and right panels show human CTCF ChIP-Seq for CTCF Site-15 within the proximal edge of the ICCE repeat embedded in the NBDY locus corresponding to hg19 chrX: 56, 809, 447-56, 810, 699. As indicated at the top are three male samples (H1-hESC, human embryonic stem cell line H1; HSMM, skeletal muscle myoblasts; HUVEC, umbilical vein endothelial cells) below which are three female samples (GM12878, EBV transformed B-lymphocytes; HMEC, mammary epithelial cells; NHEK, epidermal keratinocytes). Beneath the ChIP-Seq data are bisulfite-sequence profiles for ten male tissues (left) and ten female tissues or primary cell types (right). The images show read density data wiggle plots. The y axis represents the degree of methylation from no methylation (0) to fully methylated (1). The light gray-blue horizontal bars have been added to indicate the range 0–0.5 to more readily interpret the profiles. (B) CTCF ChIP-Seq and bisulfite sequencing profiles covering the entire FIRRE locus corresponding to hg19 ChrX: 130, 836, 678-130, 964, 671. The schematic at the top represents the extent of the FIRRE gene that is transcribed from right to left 5′–3′. Vertical bars represent exons whereas horizontal lines indicate introns.
To extend this observation further, we examined the relationship between DNA methylation and CTCF occupancy at the FIRRE locus. DNA sequence homology between adjacent repeat units defined by pair-wise alignments is least conserved at FIRRE relative to that of DXZ4 or ICCE (Horakova, Moseley et al. 2012). Consequently, massively parallel bisulfite sequence reads can be assigned throughout FIRRE. As previously reported, most CTCF ChIP-Seq peaks are female specific across the FIRRE locus (Horakova, Moseley et al. 2012; Hacisuleyman et al. 2014; Yang et al. 2015). Comparing the male versus female CTCF ChIP-Seq and DNA methylation profiles reveals that DNA methylation across the region is noticeably lower in females and that regions that are most obviously hypomethylated correlate with CTCF ChIP-Seq peaks (fig. 7B). Therefore, we conclude that the Xi alleles of DXZ4, FIRRE and ICCE are all hypomethylated which is consistent with the presence of female-specific euchromatic histone methylation at all three (Horakova, Moseley et al. 2012).
Conserved YY1 Binding Motifs Adjacent to CTCF Sites at ICCE
Like CTCF, the versatile zinc-finger protein Yin Yang 1 (YY1) is frequently observed at the base of chromatin loops (Beagan et al. 2017) and colocalization of the two proteins is common throughout the genome (Kang et al. 2009; Schwalie et al. 2013). Clustering of YY1 and CTCF binding sites is a feature at regions subject to genomic imprinting (Kim 2008) and within the X-inactivation center (Donohoe et al. 2007; Jeon and Lee 2011). YY1 binding adjacent to CTCF has also been reported at DXZ4 (Moseley et al. 2012; Chen et al. 2016) and the FIRRE locus (Yang et al. 2015; Chen et al. 2016; Hacisuleyman et al. 2016). Therefore, we looked to see if conserved YY1 binding motifs exist in the vicinity of CTCF at ICCE.
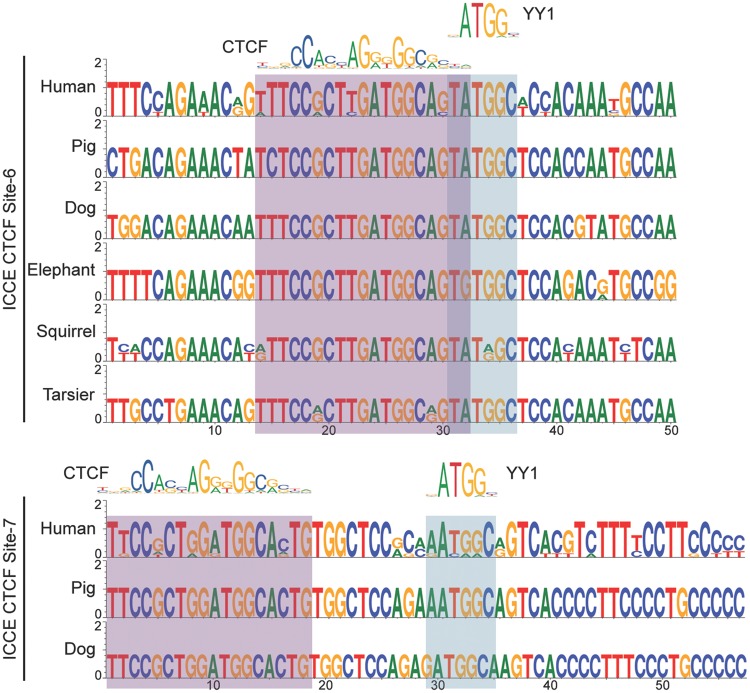
High confidence matches for YY1 binding motifs (Khan et al. 2018) were found immediately abutting the CTCF binding motif at ICCE CTCF Site-6 group and within ten base pairs of the CTCF motif at ICCE CTCF Site-7 group (fig. 8). Examination of this sequence in tarsiers and nonprimate mammals in which we have identified a putative ICCE ortholog reveals that like CTCF, the YY1 binding motifs are conserved. The notable exception was in elephant Site-6 where an A to G mutation disrupts the YY1 motif. Given that DNA methylation negatively impacts CTCF binding (Bell and Felsenfeld 2000; Hark et al. 2000; Kanduri et al. 2000; Filippova et al. 2001) and YY1 (Gaston and Fried 1995; Kim et al. 2003), if these YY1 motifs are occupied, we would predict that this would be Xi-specific. However, DNA methylation did not impact CTCF or YY1 binding to DXZ4 in vitro (Chadwick 2008; Moseley et al. 2012). Nevertheless, it is possible that CTCF and YY1 work together to ensure Xi-specific superloop formation.
Fig. 8.
—A putative YY1 binding motif resides next to the CTCF motif at Sites-6 and Site-7. Sequence logos of ICCE CTCF Site-6 (top) and Site-7 (bottom) for different mammalian ICCE orthologs. The presence of an identical base at a particular location in a motif is represented by a single base, whereas variation between sites at a particular position in the motif shows two or more bases with relative frequency represented by the size of the height of the base. The human CTCF and YY1 consensus sequences were adapted from the sequence logo obtained from JASPAR (Khan et al. 2018). The corresponding bases for each binding motif are highlighted immediately beneath the logos by the red–gray (CTCF) or blue–gray (YY1) shading.
In summary, this inaugural investigation into the ICCE locus has revealed that like DXZ4, the primary DNA sequence of the ICCE superdomain anchor diverges rapidly through the primate lineage but what is conserved are tandem repeat organization, location within the NBDY locus and retention of specific CTCF and YY1 binding motifs. Like DXZ4 and FIRRE, sequences surrounding CTCF occupancy show female-specific DNA hypomethylation, which we interpret as being attributed to the Xi. Unlike DXZ4, ICCE does not appear to be present in all mammals and data support the hypothesis that ICCE originated from DXZ4. If, like DXZ4 (Darrow et al. 2016; Giorgetti et al. 2016; Bonora et al. 2018), ICCE contributes to the proper 3D organization of the Xi remains to be determined.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of General Medical Sciences at the National Institutes of Health [grant number GM117003 to B.P.C.]
Literature Cited
- Balaton BP, Brown CJ.. 2016. Escape artists of the X chromosome. Trends Genet. 32(6):348–359. [DOI] [PubMed] [Google Scholar]
- Barr ML, Bertram EG.. 1949. A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature 163(4148):676–677. [DOI] [PubMed] [Google Scholar]
- Beagan JA, et al. 2017. YY1 and CTCF orchestrate a 3D chromatin looping switch during early neural lineage commitment. Genome Res. 27(7):1139–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G.. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405(6785):482–485. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, et al. 2010. The NIH roadmap epigenomics mapping consortium. Nat Biotechnol. 28(10):1045–1048. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs BA, et al. 2002. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat Genet. 30(1):73–76. [DOI] [PubMed] [Google Scholar]
- Bonora G, et al. 2018. Orientation-dependent Dxz4 contacts shape the 3D structure of the inactive X chromosome. Nat Commun. 9(1):1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick BP. 2008. DXZ4 chromatin adopts an opposing conformation to that of the surrounding chromosome and acquires a novel inactive X-specific role involving CTCF and antisense transcripts. Genome Res. 18(8):1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick BP, Willard HF.. 2002. Cell cycle-dependent localization of macroH2A in chromatin of the inactive X chromosome. J. Cell Biol. 157(7):1113–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick BP, Willard HF.. 2004. Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc Natl Acad Sci U S A. 101(50):17450–17455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, et al. 2016. YY1 binding association with sex-biased transcription revealed through X-linked transcript levels and allelic binding analyses. Sci Rep. 6(1):37324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE.. 2004. WebLogo: a sequence logo generator. Genome Res. 14(6):1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Lima NG, et al. 2017. A human microprotein that interacts with the mRNA decapping complex. Nat Chem Biol. 13(2):174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrow EM, et al. 2016. Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc Natl Acad Sci U S A. 113(31):E4504–E4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, et al. 2015. Bipartite structure of the inactive mouse X chromosome. Genome Biol. 16(1):152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe ME, Zhang LF, Xu N, Shi Y, Lee JT.. 2007. Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Mol Cell 25(1):43–56. [pii] [DOI] [PubMed] [Google Scholar]
- Filippova GN, et al. 2001. CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat Genet. 28(4):335–343. [DOI] [PubMed] [Google Scholar]
- Gaston K, Fried M.. 1995. CpG methylation has differential effects on the binding of YY1 and ETS proteins to the bi-directional promoter of the Surf-1 and Surf-2 genes. Nucleic Acids Res. 23(6):901–909. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacalone J, Friedes J, Francke U.. 1992. A novel GC-rich human macrosatellite VNTR in Xq24 is differentially methylated on active and inactive X chromosomes. Nat Genet. 1(2):137–143. [DOI] [PubMed] [Google Scholar]
- Giorgetti L, et al. 2016. Structural organization of the inactive X chromosome in the mouse. Nature 535(7613):575–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacisuleyman E, et al. 2014. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol. 21(2):198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacisuleyman E, Shukla CJ, Weiner CL, Rinn JL.. 2016. Function and evolution of local repeats in the Firre locus. Nat Commun. 7:11021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hark AT, et al. 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405(6785):486–489. [DOI] [PubMed] [Google Scholar]
- Horakova AH, Calabrese JM. et al. 2012. The mouse DXZ4 homolog retains Ctcf binding and proximity to Pls3 despite substantial organizational differences compared to the primate macrosatellite. Genome Biol. 13(8):R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horakova AH, Moseley SC, McLaughlin CR, Tremblay DC, Chadwick BP.. 2012. The macrosatellite DXZ4 mediates CTCF-dependent long-range intrachromosomal interactions on the human inactive X chromosome. Hum Mol Genet., 21(20):4367–4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y, Lee JT.. 2011. YY1 tethers Xist RNA to the inactive X nucleation center. Cell 146(1):119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanduri C, et al. 2000. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol. 10(14):853–856. [DOI] [PubMed] [Google Scholar]
- Kang K, Chung JH, Kim J.. 2009. Evolutionary Conserved Motif Finder (ECMFinder) for genome-wide identification of clustered YY1- and CTCF-binding sites. Nucleic Acids Res. 37(6):2003–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, et al. 2002. The human genome browser at UCSC. Genome Res 12(6):996–1006. Article published online before print in May 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, et al. 2018. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 46(D1):D1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MM, et al. 2012. CD4+ T cell-derived novel peptide Thp5 induces interleukin-4 production in CD4+ T cells to direct T helper 2 cell differentiation. J Biol Chem. 287(4):2830–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. 2008. Multiple YY1 and CTCF binding sites in imprinting control regions. Epigenetics 3(3):115–118. [pii] [DOI] [PubMed] [Google Scholar]
- Kim J, Kollhoff A, Bergmann A, Stubbs L.. 2003. Methylation-sensitive binding of transcription factor YY1 to an insulator sequence within the paternally expressed imprinted gene, Peg3. Hum Mol Genet. 12(3):233–245. [DOI] [PubMed] [Google Scholar]
- Kim TH, et al. 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128(6):1231–1245. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT. 2011. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat Rev Mol Cell Biol. 12(12):815–826. [pii] [DOI] [PubMed] [Google Scholar]
- Lister R, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462(7271):315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF. 1961. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190:372–373. [DOI] [PubMed] [Google Scholar]
- Macdonald DW, Norris S.. 2001. The new encyclopedia of mammals. Oxford, New York (NY: ): Oxford University Press. [Google Scholar]
- McLaughlin CR, Chadwick BP.. 2011. Characterization of DXZ4 conservation in primates implies important functional roles for CTCF binding, array expression and tandem repeat organization on the X chromosome. Genome Biol. 12(4):R37.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohandas T, Sparkes RS, Shapiro LJ.. 1981. Reactivation of an inactive human X chromosome: evidence for X inactivation by DNA methylation. Science 211(4480):393–396. [DOI] [PubMed] [Google Scholar]
- Moseley SC, et al. 2012. YY1 associates with the macrosatellite DXZ4 on the inactive X chromosome and binds with CTCF to a hypomethylated form in some male carcinomas. Nucleic Acids Res. 40(4):1596–1608. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton C, Sproul D, Hamilton C, Gilbert N.. 2010. Analysis of active and inactive X chromosome architecture reveals the independent organization of 30 nm and large-scale chromatin structures. Mol Cell 40(3):397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noe L, Kucherov G.. 2005. YASS: enhancing the sensitivity of DNA similarity search. Nucleic Acids Res 33(Web Server issue):W540–W543. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzkofer T, et al. 2017. L1Base 2: more retrotransposition-active LINE-1s, more mammalian genomes. Nucleic Acids Res. 45(D1):D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, et al. 2002. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat. Genet 30(1):77–80. [DOI] [PubMed] [Google Scholar]
- Plath K, et al. 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300(5616):131–135. [DOI] [PubMed] [Google Scholar]
- Rao SS, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159(7):1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rego A, Sinclair PB, Tao W, Kireev I, Belmont AS.. 2008. The facultative heterochromatin of the inactive X chromosome has a distinctive condensed ultrastructure. J Cell Sci. 121(7):1119–1127. [pii] [DOI] [PubMed] [Google Scholar]
- Schaap M, et al. 2013. Genome-wide analysis of macrosatellite repeat copy number variation in worldwide populations: evidence for differences and commonalities in size distributions and size restrictions. BMC Genomics 14(1):143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalie PC, et al. 2013. Co-binding by YY1 identifies the transcriptionally active, highly conserved set of CTCF-bound regions in primate genomes. Genome Biol. 14(12):R148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, et al. 2003. Establishment of histone h3 methylation on the inactive x chromosome requires transient recruitment of eed-enx1 polycomb group complexes. Dev Cell 4(4):481–495. [DOI] [PubMed] [Google Scholar]
- Tang Z, et al. 2015. CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell 163(7):1611–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teller K, et al. 2011. A top-down analysis of Xa- and Xi-territories reveals differences of higher order structure at >/= 20 Mb genomic length scales. Nucleus 2(5):465–477. [pii] [DOI] [PubMed] [Google Scholar]
- Tremblay DC, Alexander G Jr, Moseley S, Chadwick BP.. 2010. Expression, tandem repeat copy number variation and stability of four macrosatellite arrays in the human genome. BMC Genomics 11(1):632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay DC, Moseley S, Chadwick BP.. 2011. Variation in array size, monomer composition and expression of the macrosatellite DXZ4. PLoS ONE 6(4):e18969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, et al. 2012. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 22(9):1680–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A. 2011. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat Rev Genet. 12(8):542–553. [pii] [DOI] [PubMed] [Google Scholar]
- Yang F, et al. 2015. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 16(1):52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.