Abstract
Recent strategies targeting the interaction of the programmed cell death ligand-1 (PD-L1, B7-H1, CD274) with its receptor, PD-1, resulted in promising activity in early phase clinical trials. In this study, we used various antibodies and in situ mRNA hybridization to measure PD-L1 in non-small cell lung cancer (NSCLC) using a quantitative fluorescence (QIF) approach to determine the frequency of expression and prognostic value in two independent populations. A control tissue microarray (TMA) was constructed using PD-L1-transfected cells, normal human placenta and known PD-L1-positive NSCLC cases. Only one of four antibodies against PD-L1 (5H1) validated for specificity on this TMA. In situ PD-L1 mRNA using the RNAscope method was similarly validated. Two cohorts of NSCLC cases in TMAs including 340 cases from hospitals in Greece and 204 cases from Yale University were assessed. Tumors showed PD-L1 protein expression in 36% (Greek) and 25% (Yale) of the cases. PD-L1 expression was significantly associated with tumor-infiltrating lymphocytes in both cohorts. Patients with PD-L1 (both protein and mRNA) expression above the detection threshold showed statistically significant better outcome in both series (log-rank P=0.036 and P=0.027). Multivariate analysis showed that PD-L1 expression was significantly associated with better outcome independent of histology. Measurement of PD-L1 requires specific conditions and some commercial antibodies show lack of specificity. Expression of PD-L1 protein or mRNA is associated with better outcome. Further studies are required to determine the value of this marker in prognosis and prediction of response to treatments targeting this pathway.
Non-small cell lung cancer (NSCLC) is the leading cause of cancer-related death in the United States. The overall survival (OS) for metastatic NSCLC is dismal with 5-year survival of <5% and for patients with early stage NSCLC the 5-year survival is <50%.1 Over the past decade, identification of several oncogenic driver mutations have helped improve the outcomes in certain subtypes of patients with NSCLC.2 However, a majority of the patients with lung cancer do not have an actionable molecular aberration. Other treatment approaches, such as immune therapies, are being investigated in clinical trials. Programmed cell death-1 (PD-1) pathway is a major immune checkpoint by which tumors suppress lymphocyte function within the tumor microenvironment, and antibody blockade of PD-1 with its ligands (B7-H1/PD-L1 and B7-DC/PD-L2) showed promising activity in several malignancies.3 In particular, blocking antibodies against PD-1 and PD-L1 have shown clinical activity in NSCLC.4
Preliminary data suggest that tumor PD-L1 protein expression on human cancers using chromogenic-based immunohistochemistry (IHC) in formalin-fixed paraffin-embedded tissue samples (FFPE) may predict clinical response to PD-1/PD-L1 directed therapy.4, 5 There are limited data on the prevalence and the prognostic role of PD-L1 expression in NSCLC. Data from small previously published studies is mixed with some showing poor prognosis and others with no prognostic significance.6, 7, 8, 9 Evaluation of PD-L1 positivity by conventional IHC is not well defined and subject to antibody and assay variability and interpretative subjectivity. In addition, the specificity and reproducibility of the commercially available antibodies has not been thoroughly assessed. Here we report a reproducible assay for tissue PD-L1 protein measurement using quantitative immunofluorescence (AQUA®) with a validated monoclonal antibody and a novel method of in situ measurement of mRNA.
MATERIALS AND METHODS
Patient Cohorts and Tissue Microarrays (TMAs)
The Yale University lung cohort was previously described 10 and included retrospectively collected FFPE from 204 lung carcinomas between December 1988 and October 2003. Detailed cohort description including cancer histotypes is shown in Table 1. An independent retrospective Greek cohort was also used and included FFPE samples from 340 NSCLC diagnosed between 1991 and 2001 at Sotiria General Hospital and Patras University General Hospital in Greece. Tissue specimens were included in a 0.6-mm TMA format as described.10 A custom designed ‘index’ TMA was constructed for reagent titration, assay validation and reproducibility assessment. This index TMA contained samples from FFPE tissue blocks of term human placenta and tonsil as positive controls for endogenous PD-L1 and cores from FFPE prepared, parental Mel624 cells that do not express PD-L1 and Mel624 transfected with PD-L1 with proven overexpression.11,12 Finally, a small series of NSCLC cases with previously measured high, low and intermediate PD-L1 protein levels were also included for assessment of reproducibility on the index preparation. Culture conditions and cell-line TMA construction have been published in detail elsewhere.13
Table 1.
Clinicopathological characteristics of Greek and Yale NSCLC cohorts according to PD-L1 protein status
| Greek NSCLC cohort |
Yale NSCLC cohort |
|||||
|---|---|---|---|---|---|---|
| PD-L1 low | PD-L1 high | P-value | PD-L1 low | PD-L1 high | P-value | |
| Parameter | ||||||
| All patients | N = 228 (75.2%) | N = 75 (24.8%) | N =99 (63.9%) | N = 56 (36.1%) | ||
| Age (years) | P = 0.87 | P = 0.38 | ||||
| ≥70 | 175 | 57 | 64 | 31 | ||
| >70 | 52 | 18 | 33 | 22 | ||
| Unknown | 1 | 0 | 2 | 3 | ||
| Gender | P = 0.42 | P = 0.397 | ||||
| Female | 31 | 7 | 43 | 28 | ||
| Male | 197 | 68 | 55 | 26 | ||
| Unknown | 0 | 0 | 1 | 2 | ||
| Histology | P = 0.88 | P=0.004 | ||||
| Adenocarcinoma | 96 | 28 | 74 | 28 | ||
| Squamous cell | 115 | 37 | 13 | 17 | ||
| Other | 17 | 10 | 10 | 8 | ||
| Unknown | 0 | 0 | 2 | 3 | ||
| Smoker | P = 0.45 | |||||
| Yes | 190 | 56 | NA | NA | ||
| No | 17 | 7 | NA | NA | ||
| Unknown | 21 | 12 | ||||
| Stage | P = 0.011 | P = 0.57 | ||||
| 1 | 70 | 25 | 44 | 30 | ||
| 2 | 52 | 28 | 15 | 7 | ||
| 3 | 78 | 16 | 24 | 11 | ||
| 4 | 27 | 6 | 7 | 4 | ||
| Unknown | 1 | 0 | 9 | 4 | ||
| Inflammation | P = 0.03 | P<0.001 | ||||
| 0–1 | 143 | 36 | 64 | 19 | ||
| 2–3 | 84 | 39 | 35 | 37 | ||
| Unknown | 1 | 0 | 0 | 0 | ||
P-values displayed in bold text reflect the presence of statistical significance (eg, P<0.05).
Antibodies and IHC/Immunofluorescence
For detecting PD-L1 using automated QIF analysis using the AQUA method we screened one rabbit polyclonal antibody from ABCAM (cat # ab58810), and three mouse mAbs, one from Ebiosciences (clone MIH1), one from Biolegend (clone 29E.2A3) and a non-commercial mAb made in Dr Lieping Chen’s lab (clone 5H1) using the index array described above. The specificity of the antibody was evaluated by the staining of control tissues (human placenta and tonsil) and cell line transfectants. A validated antibody showed expression in the transfected Mel624 cells and no expression in parental cells; as well as membranous staining of the syncytotrophoblast layer of the placenta, while showing absence of staining in the stromal and vascular regions. Of the tested antibodies, only the 5H1 antibody met these standards and was used for evaluation of our lung cohorts.
Fresh TMA cuts were deparaffinized and subjected to a range of conventional antigen retrieval conditions, which showed limited and non-reproducible staining. Only the unusual antigen retrieval method using tris-EDTA buffer (Sigma-Aldrich, St Louis, MO, USA) with 0.05% Tween (pH=9.0) and boiling for 20 min at 102 °C in a pressure-boiling container (PT module, Lab Vision) showed consistent results. Slides were then incubated with 1% BlockAce (cat # BUF029, AbDSerotec, Oxford, UK) for 10 min at room temperature and incubated overnight at 4 °C with a solution containing primary PD-L1 antibody at 1:500 dilution (mouse monoclonal, clone 5H1) and a rabbit monoclonal anti-human pancytokeratin antibody at 1:50 (clone AE1/AE3, M3515; Dako Corp., Carpinteria, CA, USA). Sections were incubated for 1 h at room temperature with Alexa 546-conjugated goat anti-rabbit secondary antibody (A11003; Molecular Probes, Eugene, OR, USA) diluted 1:100 in mouse EnVision amplification reagent (K4003, Dako). Cyanine 5 (Cy5) directly conjugated to tyramide (FP1117; Perkin-Elmer) at a 1:50 dilution was used for target antibody detection. Prolong mounting medium (ProLong Gold, P36931; Molecular Probes) with 4′,6-diamidino-2-phenylindole (DAPI) was used to stain nuclei in the histospot (TMA spot). Serial sections from controls slides were run alongside each cohort slide staining.
At least two TMA histospots were evaluated in each case. To address run-to-run variability, slides were stained and analyzed on different days using the same protocol. Linear regression coefficients (R2) between independent runs were high (>0.7, Supplementary Figure S1). Heterogeneity of expression is illustrated by the regression coefficient for each graph and the distance of the heterogeneity is appreciated by comparison of serial sections vs cores from different regions in the blot as seen comparing build 1 vs build 2 of each array (Figure S1 and Figure 2 insets). The specificity of the antibody was demonstrated by the staining of control tissues and cell line transfectants as described and shown in Figure 1.
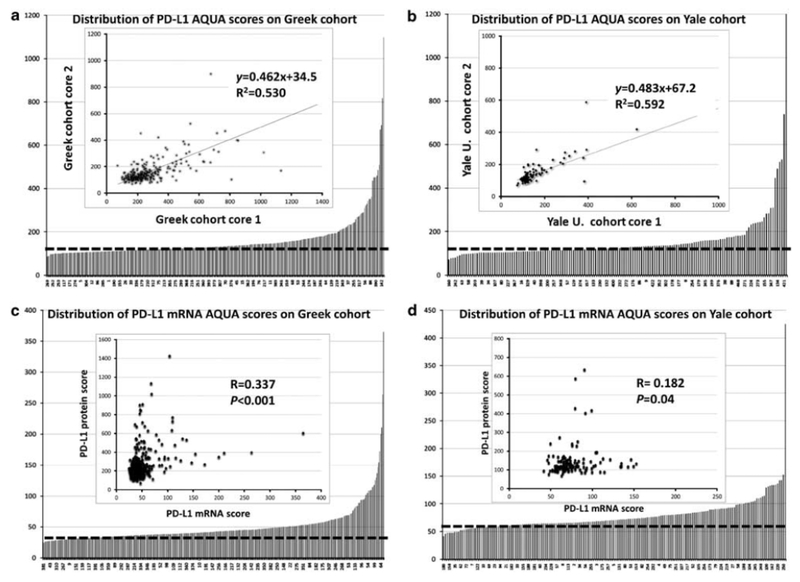
Figure 2. Distribution of PD-L1 protein and mRNA scores in the lung cohorts and assay reproducibility.
(a, b) Distribution of PD-L1 protein scores in cases from the Greek (a) and Yale (b) lung cohorts. The dashed line indicates the cutoff for positive/negative designation as described in the Materials and methods section. Inset shows reproducibility of PD-L1 protein measurements in two different cores (core 1 and core 2) obtained from the same case and paraffin block. R2 represents the linear regression coefficient and y represents the slope, (c, d) Distribution of PD-L1 mRNA scores in cases from the Greek (c) and Yale (d) lung cohorts. The dashed line indicates the cutoff for positive/negative designation as per negative control DapB scores. Inset shows relationship between PD-L1 protein and mRNA in both cohorts. R represents the Spearman correlation coefficient.
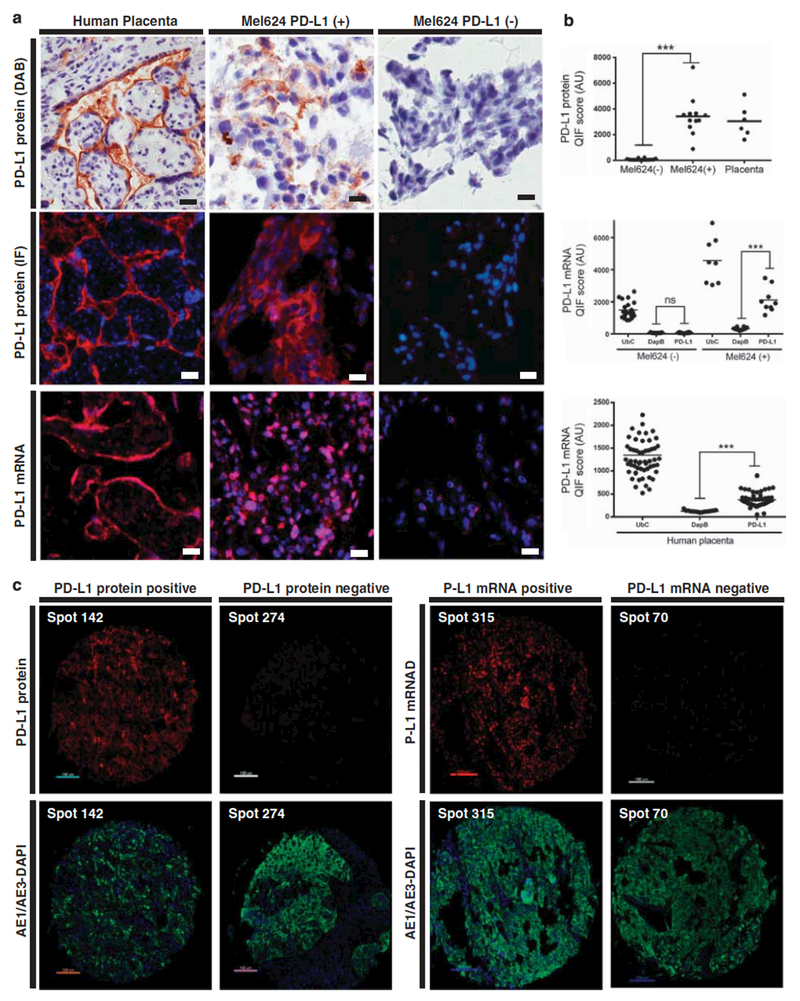
Figure 1. PD-L1 protein and mRNA detection using chromogenic immunohistochemistry and quantitative fluorescence in cell lines and human tissues.
(a) Representative microphotographs showing PD-L1 protein expression using diaminobenzidine (brown chromogen, upper panels), immunofluorescence (red channel, middle panels) and fluorescent in situ PD-L1 mRNA (red channel, bottom panels) in human placenta, PD-L1 expressing Mel624 cells and parental Mel624 cells, × 400 magnification. Bar=60 μm. (b) PD-L1 protein and mRNA fluorescence measurements in several areas of FFPE preparations containing parental Mel624 (Mel624 (−)), PD-L1 Mel624 transfectants (Mel624 (+)) or human placenta. Each dot shows the fluorescence score within the mask in an independent field of view and scores are expressed as arbitrary units (AU). ***P<0.001, NS, not significant, (c) Representative fluorescence microphotographs of PD-L1 protein (left panels) staining in a positive (histospot 142) and negative NSCLC sample (histospot 274). Right panels show representative in situ PD-L1 mRNA staining in a positive (histospot 315) and a negative (histospot 70) NSCLC case. Nuclei were stained with DAPI. × 400 magnification. Bar=100 μm.
In Situ mRNA Hybridization
In situ detection of PD-L1 transcripts in FFPE TMA samples was performed using the RNAscope assay (Advanced Cell Diagnostics, CA, USA) coupled to fluorescence detection as recently described.4 Briefly, 5 μm sections were deparaffinized, boiled with pre-amplification reagent for 15 min and submitted to protease digestion followed by hybridization for 2 h with a mixture containing target probes to either human PD-L1, ubiquitin C (UbC) as a positive control or the bacterial gene DapB as a negative control. Hybridization signals were detected with Cy5-tyramide. Preparations were then incubated with a wide-spectrum rabbit anti-cow cytokeratin antibody (Z0622 1:100, Dako) in BSA/tris-buffered saline for 1 h at room temperature followed by detection with a secondary Alexa-546-conjugated goat anti-rabbit antibody (1:100, Molecular Probes). Slides were then mounted using ProlongGold plus DAPI to define nuclei.
Automated QIF
Automated QIF analysis using the AQUA method enables objective measurement of protein concentration within user defined cellular compartments, as described in detail elsewhere.13, 14, 15 Briefly, the AQUA score of PD-L1 in the tumor compartment was calculated by dividing the PD-L1 compartment pixel intensities by the area of the compartment within which they were measured. AQUA scores were normalized to the exposure time and bit depth at which the images were captured, allowing scores collected at different exposure times to be comparable. All acquired histospots were visually evaluated by the experimenters and cases with staining artifacts and <2% tumor (cytokeratin staining) were excluded from the analysis.
Evaluation of Tumor-Infiltrating Lymphocytes
The degree of tumor-infiltrating lymphocytes (TILs) was scored on the hematoxylin and eosin-stained TMA preparations by two pathologists independently. The scoring was performed as described16 using a four-tiered scale.
Determination of PD-L1 Protein and mRNA Expression
PD-L1 protein cutoff for expression in our study was defined as the AQUA score of first signal detection beyond the signal intensity in FFPE samples from normal lung and negative controls. We also checked visually for the staining pattern in each case to confirm that those cases below the noise range showed nonspecific signal (eg, signal detected equally in normal and tumor areas; and without defined compartment distribution). Those above the noise range showed a more specific staining pattern.
The cutoff for PD-L1 mRNA expression was determined using the average AQUA score of the negative control ISH with DapB, a bacterial mRNA. Cases with PD-L1 mRNA signal above the DapB score in the same TMA and run were considered as having high expression and cases with scores equal or lower were considered as low PD-L1 signal. UbC is a tissue quality control as it is expressed in all tissues. Cases with UbC scores below 100 were excluded from further analysis as this suggests degraded mRNA and poor quality specimens.
Statistical Analysis
Patient characteristics were compared between the two cohorts using t-test for continuous variables and Χ2 test for categorical variables. OS functions between patients with high and low PD-L1 were compared using log-rank test. Multivariate Cox proportional models were built to examine the effect of PD-L1 on OS adjusted by the effect of stage, histology and TILs. SPSS version 19 statistical software was used.
RESULTS
PD-L1 Protein and mRNA Assay Validation
To validate the QIF-based method for PD-L1 protein measurements in FFPE tumors each antibody selected was tested on a set of endogenous controls and cell line transfectants. As shown in Figure 1, PD-L1 protein was detected using the 5H1 antibody in both IHC and QIF in the trophoblastic cells of mature human placenta but was absent in the mesenchymal stromal cells and vessels of the chorionic villi (Figure 1a). In addition, PD-L1 signal was detected with a predominantly membranous/perinuclear pattern in melanoma Mel624 cells stably transfected with human PD-L1 cDNA but not in parental Mel624 cells (Figures 1a and b). Other antibodies tested, including clone 29E.2A3, clone MIH1 and the rabbit polyclonal antibody from Abeam failed to show this pattern. Although PD-L1 AQUA scores in preparations from human placenta and Mel624 transfectants showed focal variation (eg, heterogeneity), the average protein levels were comparably higher than in parental Mel624 cells (Figure 1b, upper panel). In NSCLC samples, PD-L1 protein also showed a predominantly membranous staining pattern and was observed frequently within the tumor compartment (Figure 1c). A proportion of NSCLC cases showed stromal PD-L1 protein immunoreactivity and a fraction showed absence of PD-L1 signal (Figure 1c).
Using the previously described RNAscope method for in situ mRNA detection in TMAs, we measured the in situ PD-L1 mRNA in the two lung cohorts.15 Similar to PD-L1 protein, PD-L1 mRNA was detected predominantly in the trophoblastic (cytokeratin positive) compartment of the placenta and only in Mel624 transfectants (Figure 1a, bottom). PD-L1 transcripts were identified as multiple small dots distributed throughout the cells (Figures 1a and c). As shown in Figure 1b, elevated PD-L1 mRNA levels (above DapB) were detected only in Mel624 transfectants and human placenta, but not in parental Mel624 cells. High UbC mRNA was recognized in all studied preparations indicating mRNA preservation and bacterial DapB mRNA transcripts were comparably low. In NSCLC samples, PD-L1 mRNA showed a similar staining pattern and was located predominantly in the tumor (cytokeratin positive) compartment (Figure 1c, right).
PD-L1 protein levels were measured in 303 (86%) and 155 (76%) specimens of the Greek cohort and Yale University cohort, respectively. Failures were due to insufficient tumor in the histospots. In both cohorts, PD-L1 protein levels showed a wide range of scores (Figures 2a and b). The cutoff for PD-L1 protein signal was comparable in the two cohorts and 25% and 36% of the cases showed signal above the detection threshold. The reproducibility of PD-L1 protein assay was determined by the correlation of measurements in serial sections from the Greek cohort and the index lung array (R2=0.9 and 0.88, respectively, Supplementary Figure S1), confirming the robustness of the assay. To assess tumor heterogeneity, regressions were calculated in the two cohorts by comparison of histospots from different TMA cores taken at least 3 mm apart. The linear regression coefficients were 0.53 and 0.59, reflecting some degree of heterogeneity in PD-L1 expression (Figures 2a and b, insets).
PD-L1 mRNA levels above the noise threshold were detected in 53% and 51% of the Greek and Yale cohort, respectively (Figures 2c and d). Reproducibility of in situ PD-L1 mRNA measurements was high with correlation coefficients of 0.79 between serial sections and 0.48 between different cores of the same tumor (Supplementary Figure S1). Interestingly, PD-L1 protein and mRNA levels showed a complex non-linear positive association in the two cohorts with a Spearman’s correlation coefficient of 0.33 in the Greek and 0.18 in the Yale cohort (P<0.001 and P=0.04, Figures 2c and d, insets). In the Greek cohort, PD-L1 protein expression was seen in 75 cases, including 28 adenocarcinomas (ADCs, 23.4%) and 37 squamous cell carcinomas (SCCs, 24.3%). In the Yale University cohort, PD-L1 protein signal was seen in 56 patients including 28 ADCs (27.5%) and 17 SCCs (56.7%; Table 1). Elevated PD-L1 protein levels were positively correlated with the presence of lymphocytic infiltrate in both cohorts (P=0.03 and <0.001, respectively). In the Yale cohort, SCC showed higher PD-L1 levels than ADC (56.7% vs 27.5%; P=0.009) despite having comparable lymphocytic infiltrates (Table 1). Tumor PD-L1 protein expression was not associated with age, gender, race or smoking status (Table 1).
In the Greek collection, PD-L1 mRNA expression was found in 167 cases, 53 ADCs (31.7%) and 83 SCCs (49.7%) (Table 2). In the Yale cohort, 88 cases showed high PD-L1 mRNA signal including 53 ADCs (60%) and 17 SCCs (19.3%). As for PD-L1 protein, mRNA expression was significantly associated with lymphocytic infiltration in the Greek cohort (P=0.0002) and the Yale cohort (P=0.001).
Table 2.
Clinicopathological characteristics of Greek and Yale NSCLC cohorts according to PD-L1 mRNA status
| Greek NSCLC cohort |
Yale NSCLC cohort |
|||||
|---|---|---|---|---|---|---|
| PD-L1 mRNA low | PD-L1 mRNA high | P-value | PD-L1 mRNA low | PD-L1 mRNA high | P-value | |
| Parameter | ||||||
| All patients | N =147 (46.8%) | N =167 (53.2%) | N=85 (49.2%) | N = 88 (50.8%) | ||
| Age (years) | P=0.1602 | P=0.1523 | ||||
| <70 | 101 | 126 | 51 | 45 | ||
| ≥70 | 38 | 31 | 24 | 36 | ||
| Unknown | 8 | 10 | 10 | 7 | ||
| Gender | P=0.9113 | P=0.3424 | ||||
| Female | 17 | 20 | 35 | 45 | ||
| Male | 122 | 138 | 42 | 38 | ||
| Unknown | 8 | 9 | 8 | 5 | ||
| Histology | P=0.2103 | P=0.3756 | ||||
| Adenocarcinoma | 58 | 53 | 55 | 53 | ||
| Squamous cell | 65 | 84 | 11 | 17 | ||
| Other | 16 | 20 | 9 | 11 | ||
| Unknown | 8 | 10 | 11 | 14 | ||
| Smoker | P=0.7697 | |||||
| Yes | 119 | 125 | NA | NA | ||
| No | 9 | 12 | NA | NA | ||
| Unknown | 19 | 30 | ||||
| Stage | P=0.2196 | P=0.6481 | ||||
| 1 | 44 | 50 | 36 | 40 | ||
| 2 | 32 | 48 | 11 | 14 | ||
| 3 | 45 | 45 | 19 | 13 | ||
| 4 | 16 | 12 | 5 | 9 | ||
| Unknown | 10 | 12 | 14 | 12 | ||
| Inflammation | P= 0.0002 | P= 0.001 | ||||
| 0–1 | 101 | 79 | 66 | 19 | ||
| 2–3 | 46 | 88 | 34 | 49 | ||
| Unknown | 0 | 0 | 0 | 5 | ||
P-values displayed in bold text reflect the presence of statistical significance (eg, P<0.05).
Association of PD-L1 with Survival in Patients with NSCLC
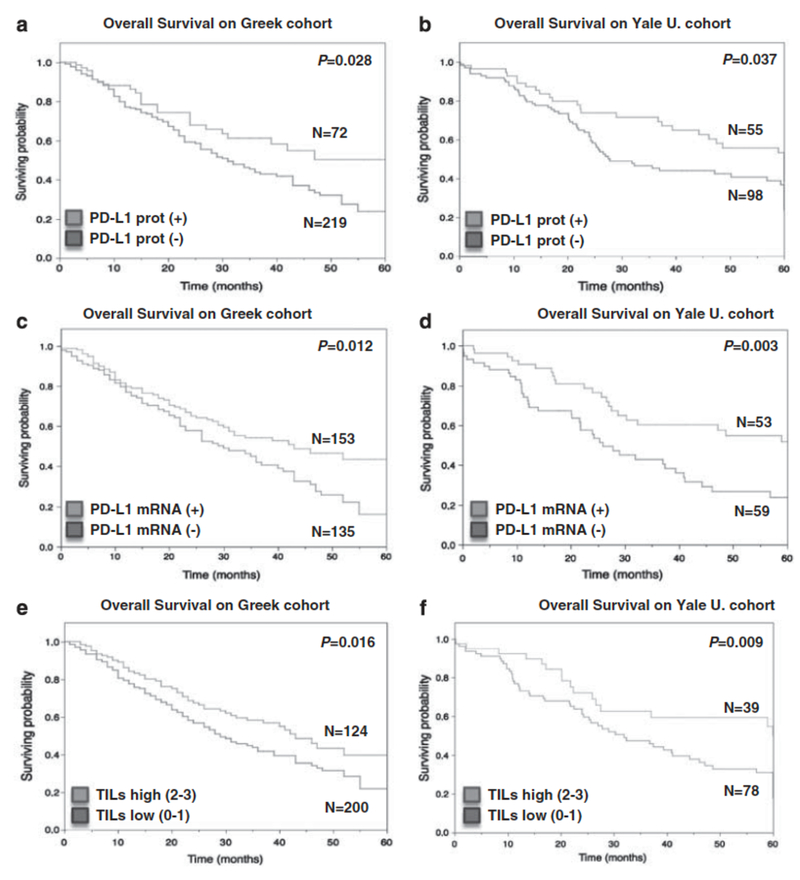
In the Greek cohort, PD-L1 protein expression was associated with longer survival (median survival not reached vs 31 months; log-rank P=0.028; Figure 3a). High PD-L1 protein resulted in 39% risk reduction in NSCLC patients at Cox proportional univariate analysis (HR=0.61 CI (0.39–0.95), P=0.031). Similarly, analysis of the Yale cohort showed that elevated PD-L1 protein expression was associated with better outcome (median survival 60 months vs 27.8 months; log-rank P=0.037; Figure 3b). High PD-L1 protein signal in the Yale cohort resulted in 37% risk reduction (HR=0.63 CI (0.40–0.98), P=0.043).
Figure 3. PD-L1 positivity and increased TILs is associated with better outcome in lung carcinomas.
(a, b) Kaplan-Meier graphical analysis of 5-year survival in patients with lung carcinomas in the Greek cohort (a) and the Yale University cohort (b) according to PD-L1 protein status. In both cohorts PD-L1-positive patients (black line) showed longer survival compared with negative cases (gray lines, log-rank P=0.0276 for Greek cohort and P=0.0369 for the Yale University cohort). (c, d) 5-Year survival in patients with lung carcinomas in the Greek cohort (c) and the Yale University cohort (d) according to PD-L1 mRNA status. PD-L1 mRNA-positive patients (black line) showed longer survival compared with negative cases (gray lines, log-rank P=0.012 for Greek cohort and P=0.003 for the Yale University cohort). (e, f) 5-Year survival in patients with lung carcinomas in the Greek cohort (e) and the Yale University cohort (f) according to the lymphocytic infiltrate. Cases with high lymphocytic infiltrate (black line, scores 2 and 3) showed longer survival compared with cases with low TILs (gray lines, scores 0 and 1) in both cohorts (log-rank P=0.0158 and 0.009, respectively). The number of cases in each group is indicated within the chart.
Similar to protein measurements, PD-L1 mRNA expression was significantly associated with longer OS in both studied cohorts (log-rank P=0.012 in the Greek and P=0.003 in the Yale cohort, Figures 3c and d). In Cox proportional univariate analysis of Greek and Yale cohorts, patients with high PD-L1 mRNA had 36% and 50% risk reduction respectively (HR=0.64 CI (0.46–0.90), P=0.011 in the Greek cohort and HR=0.50 CI (0.30–0.82), P=0.006, in the Yale cohort). In addition, increased TILs was associated with better outcome in the Greek cohort (log-rank P=0.015, Figure 3e) and the Yale cohort (log-rank P=0.009, Figure 3f). In multivariate analysis, PD-L1 protein appears to be nearly independent of stage and TILs (P=0.05 and 0.06, Supplementary Table S1). PD-L1 mRNA was independent of stage, histology and TILs in the Yale cohort (P<0.001) but not in the Greek cohort (P=0.214; Supplementary Table S1).
DISCUSSION
Strategies targeting the PD-1/PD-L1 axis are currently under investigation and have shown evidence of antitumor activity.4, 5, 17 PD-L1 protein expression in the tumor cells has been suggested as a predictive tool. Here, we describe a reproducible QIF-based assay for PD-L1 protein and mRNA determination in FFPE tumor samples.
As for most cancer biomarkers, determination of PD-L1 levels in FFPE samples has been generally performed using chromogenic IHC using various antibodies with various levels of validation. Thresholds for positivity have not been clearly defined and reproducibility has not been formally assessed. Most studies have used subjective visual PD-L1 levels estimation as indicator of positivity. In addition, a 5% proportion of membrane-positive tumor cells have been used as cutoff for tumor positivity2, 17 as this cut-point is reported to be associated with clinical response to anti-PD-1 therapy.17
Our data show that both tumor PD-L1 protein and mRNA expression is consistently associated with increased local lymphocytic infiltrate and longer OS, independent of age, stage and histotype in two independent NSCLC cohorts. In contrast to this study, in some of the previous reports, PD-L1 protein expression was associated with worse outcome in several neoplasms, including NSCLC.7, 8, 9, 18, 19, 20, 21 However, these studies are limited by the use of relatively small cohorts (n=14–214) and more importantly, by the absence of validated PD-L1 determination methods including key parameters such as antibody specificity, assay reproducibility and heterogeneity of marker expression.
Our observations are consistent with two recent studies in metastatic melanomas and Merkel-cell carcinomas using the validated monoclonal antibody clone 5H1 and showing association of PD-L1 protein expression with increased TILs and longer survival.12, 22 Moreover, a recent study in a large colorectal cancer cohort found positive association between PD-L1 protein positivity, tumor infiltration by CD8+ TILs and OS using two different commercially available antibodies.23 Although the ultimate effects of PD-L1 signaling in the tumor are not completely understood, they are likely affected by the presence of specific immune regulatory cells and co-activation of additional immune checkpoint pathways. In support of this notion, tumor infiltration by CD8+ T lymphocytes was recently shown to be associated with increased FOXP3+ regulatory T cells, PD-L1 protein/mRNA and IDO expression in human metastatic melanomas.24 Of note, induction of these co-inhibitory pathways in the tumor microenvironment required the presence of CD8+ T cells and IFN-γ expression in a murine melanoma model.24 Therefore, rather than an indication of tumor immune-evasion, expression of PD-L1 by tumor cells might reflect the presence of (ineffective) antitumor immune pressure mediated by TILs. Further studies will be required to clarify this in other solid tumor models and carefully weigh the effect of additional active co-stimulatory pathways.
Finally, though yet unpublished, preliminary data from our group also showed a positive association of PD-L1 protein expression and survival in glioblastoma, breast cancer, and head and neck tumors. Other correlations with PD-L1 expression included squamous cell histology, giant cell histology and earlier stage of disease showed higher PD-L1 levels. The biological determinants and potential clinical implications of these observations are unknown and require further studies.
This work should be considered in light of a number of limitations. One major weakness of our study is that it includes only retrospectively collected samples and information. The retrospective nature of the collection is associated with potential bias from variable treatment. However, the fact that two cohorts from different continents show concordant results suggests the observation may be treatment independent. A second limitation is that the work was performed on TMAs. TMAs may under-represent heterogeneity, but this issue appears to be less significant than anticipated. However, more significantly, TMAs are not used as standard diagnostic samples. Therefore, future studies testing PD-L1 in whole tissue sections, including measurement of field to field heterogeneity are underway. These studies may define the number of fields of view required to represent PD-L1 that are associated with outcome, or more importantly, response to targeted therapy.
In summary, the measurement of PD-L1 has the potential to identify subsets of NSCLC patients with an inflammatory tumor microenvironment that predicts for better outcome and also may predict for response to PD-1 pathway blockade. Our work describes new tools with the potential for more accurate and reproducible measurement of PD-L1. Furthermore, we describe the population distribution of PD-L1 protein and mRNA expression in NSCLC by examination of two independent cohorts. The prognostic value of each will need to be considered in analysis of biomarker-associated clinical trials. Future studies will test the capacity of tumor PD-L1 expression in predicting response to targeted immunotherapy in lung cancer patients.
Supplementary Material
Acknowledgements
This work was supported by grants from the Breast Cancer Research Foundation to David L Rimm and the ASCO Conquer Cancer Foundation through the Young Investigator Award for Vamsidhar Velcheti, MD.
Footnotes
Competing interests
The authors declare no conflict of interest.
Contributor Information
Vamsidhar Velcheti, Solid Tumor, Oncology, Taussig Cancer Institute, Cleveland Clinic, Cleveland, OH, USA.
Kurt A Schalper, Department of Pathology, Yale School of Medicine, New Haven, CT, USA.
Daniel E Carvajal, Department of Pathology, Yale School of Medicine, New Haven, CT, USA.
David L Rimm, Department of Pathology, Yale School of Medicine, New Haven, CT, USA.
Valsamo K Anagnostou, Department of Medical Oncology, Yale School of Medicine, New Haven, CT, USA.
Konstantinos N Syrigos, Department of Medical Oncology, Yale School of Medicine, New Haven, CT, USA.
Mario Sznol, Department of Medical Oncology, Yale School of Medicine, New Haven, CT, USA.
Roy S Herbst, Department of Medical Oncology, Yale School of Medicine, New Haven, CT, USA.
Scott N Gettinger, Department of Medical Oncology, Yale School of Medicine, New Haven, CT, USA.
Lieping Chen, Department of Medical Oncology, Yale School of Medicine, New Haven, CT, USA; Department of Immunobiology, Yale School of Medicine, New Haven, CT, USA.
Konstantinos N Syrigos, Oncology Unit, Department of Medicine, University of Athens, Sotiria General Hospital, Athens, Greece.
References
- 1.Detterbeck FC, Boffa DJ, Tanoue LT. The new lung cancer staging system. Chest 2009;136:260–271. [DOI] [PubMed] [Google Scholar]
- 2.Petrelli NJ, Winer EP, Brahmer J et al. Clinical cancer advances 2009: major research advances in cancer treatment, prevention, and screening—a report from the American Society of Clinical Oncology. J Clin Oncol 2009;27:6052–6069. [DOI] [PubMed] [Google Scholar]
- 3.Chen L Co-inhibitory molecules of the B7-CD28 family in the control of T-cell immunity. Nat Rev Immunol 2004;4:336–347. [DOI] [PubMed] [Google Scholar]
- 4.Sznol M, Chen L. Antagonist antibodies to PD-1 and B7-H1 (PD-L1) in the treatment of advanced human cancer. Clin Cancer Res 2013;19:1021–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brahmer JR, Tykodi SS, Chow LQ et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. New Eng J Med 2012;366:2455–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mu C-Y, Huang J-A, Chen Y et al. High expression of PD-L1 in lung cancer may contribute to poor prognosis and tumor cells immune escape through suppressing tumor infiltrating dendritic cells maturation. Med Oncol 2011. ;28:682–688. [DOI] [PubMed] [Google Scholar]
- 7.Konishi J, Yamazaki K, Azuma M et al. B7-H1 expression on non-small cell lung cancer cells and its relationship with tumor-infiltrating lymphocytes and their PD-1 expression. Clin Cancer Res 2004;10:5094–5100. [DOI] [PubMed] [Google Scholar]
- 8.Chen YB, Mu CY, Huang JA. Clinical significance of programmed death-1 ligand-1 expression in patients with non-small cell lung cancer: a 5-year-follow-up study. Tumori 2012;98:751–755. [DOI] [PubMed] [Google Scholar]
- 9.Boland JM, Kwon ED, Harrington SM et al. Tumor B7-H1 and B7-H3 expression in squamous cell carcinoma of the lung. Clin Lung Cancer 2013;14:157–163. [DOI] [PubMed] [Google Scholar]
- 10.Anagnostou VK, Syrigos KN, Bepler G et al. Thyroid transcription factor 1 is an independent prognostic factor for patients with stage I lung adenocarcinoma. J Clin Oncol 2009;27:271–278. [DOI] [PubMed] [Google Scholar]
- 11.Dong H, Strome SE, Salomao DR et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med 2002;8:793–800. [DOI] [PubMed] [Google Scholar]
- 12.Taube JM, Anders RA, Young GD et al. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med 2012;4:127ra37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCabe A, Dolled-Filhart M, Camp RL et al. Automated quantitative analysis (AQUA) of in situ protein expression, antibody concentration, and prognosis. J Natl Cancer Inst 2005;97:1808–1815. [DOI] [PubMed] [Google Scholar]
- 14.Bordeaux JM, Cheng H, Welsh AW et al. Quantitative in situ measurement of estrogen receptor mRNA predicts response to tamoxifen. PloS One 2012; e36559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med 2002;8:1323–1327. [DOI] [PubMed] [Google Scholar]
- 16.Klintrup K, Mäkinen JM, Kauppila S et al. Inflammation and prognosis in colorectal cancer. Eur J Cancer 2005;41:2645–2654. [DOI] [PubMed] [Google Scholar]
- 17.Topalian SL, Hodi FS, Brahmer JR et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. New Engl J Med 2012;366:2443–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamanishi J, Mandai M, Iwasaki M et al. Programmed cell death 1 ligand 1 and tumor-infiltrating CD8+ T lymphocytes are prognostic factors of human ovarian cancer. Proc Natl Acadl Sci USA 2007;104:3360–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mu CY, Huang JA, Chen Y et al. High expression of PD-L1 in lung cancer may contribute to poor prognosis and tumor cells immune escape through suppressing tumor infiltrating dendritic cells maturation. Med Oncol 2011. ;28:682–688. [DOI] [PubMed] [Google Scholar]
- 20.Nomi T, Sho M, Akahori T et al. Clinical significance and therapeutic potential of the programmed death-1 ligand/programmed death-1 pathway in human pancreatic cancer. Clin Cancer Res 2007;13:2151–2157. [DOI] [PubMed] [Google Scholar]
- 21.Thompson RH, Kuntz SM, Leibovich BC et al. Tumor B7-H1 is associated with poor prognosis in renal cell carcinoma patients with long-term follow-up. Cancer Res 2006;66:3381–3385. [DOI] [PubMed] [Google Scholar]
- 22.Lipson EJ, Vincent JG, Loyo M et al. PD-L1 expression in the Merkel cell carcinoma microenvironment: association with inflammation, Merkel cell polyomavirus, and overall survival. Cancer Immunol Res 2013;1:54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Droeser RA, Hirt C, Viehl CT et al. Clinical impact of programmed cell death ligand 1 expression in colorectal cancer. Eur J Cancer 2013, pii S0959-8049:00123–00128. [DOI] [PubMed] [Google Scholar]
- 24.Spranger S, Spaapen RM, Zha Y et al. Up-regulation of PD-L1, IDO, and Tregs in the melanoma tumor microenvironment is driven by CD8+ T cells. Sci Transl Med 2013;5:200ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.