Abstract
Polycystic ovary syndrome (PCOS) is the most common endocrinopathy in women of reproductive age, with a prevalence of 6–8%. Although the etiology of PCOS has been investigated extensively, the association between genetic predisposition and PCOS risk is largely unknown. In this study, we genotyped 63 SNPs in 10 genes among 361 PCOS patients and 331 healthy controls in a Chinese Han population. The following variant alleles were significantly associated with decreased PCOS risk: ESR1 rs9340799 (P = 0.000), PPARG rs709154 (P = 0.013), and rs1151996 (P = 0.013), HMGA2 rs2272046 (P = 0.000), MTHFR rs1801133 (P = 0.000). Accordingly, the following genotypes at various loci were associated with reduced PCOS risk: GA genotype at rs9340799 (P < 0.0001) in ESR1, TA genotype at rs709154(P < 0.0001) in PPARG and CA genotype at rs2272046 (P < 0.0001) in HMGA2. Moreover, GA genotype at rs1999805 (P = 0.013) in ESR1 and TT genotype at rs1801133 in MTHFR (P < 0.0001) correlated with elevated PCOS risk. Furthermore, haplotype analysis revealed significant differences in haplotype distributions of CYP11A1, ESR2 and PPARG gene between cases and controls. In addition to confirming that ESR1 rs9340799, HMGA2 rs2272046 and MTHFR rs1801133 are related to the risk of PCOS, these findings also provide the first evidence that PPARG rs709154 and ESR1 rs1999805 are significantly associated with PCOS risk in a Chinese population. Further functional studies are warranted to elucidate the underlying biological mechanisms.
Keywords: polycystic ovary syndrome, single-nucleotide polymorphism, infertility, endocrinopathy, variant alleles
Introduction
Polycystic ovary syndrome (PCOS) is the most common endocrinopathy in women of reproductive age, with a prevalence of 6–8% (1). This syndrome is characterized by hyperandrogenism, amenorrhea or oligomenorrhea and polycystic ovaries (2, 3). Women with PCOS are potentially at elevated risk of multiple diseases and disorders, including infertility (4), insulin resistance (5), obesity (6), premature carotid arteriosclerosis (7), type 2 diabetes mellitus (8), and mood disorders, such as depression and anxiety (9, 10). Epidemiological studies suggest that androgen excess, lifestyle (11), ovulatory dysfunction, alteration in intrauterine environment (12), adipose tissue dysfunction and gonadotropin abnormalities can contribute to PCOS risk (13). The etiology of PCOS is complicated and not well elucidated.
A substantial body of evidence has implicated various genetic factors in PCOS development (14). Related studies have been conducted on at least 70 genes, which roles are involved in the main process of this disease, such as steroid synthesis (CYP11A1 and CYP19A1) (15), steroid action (ESR1, ESR2, and PGR) (16), lipid metabolism (PPARG and MTHFR) (17–19), insulin action (HMGA2) (20) and embryonic development (SUMO1P1) (21, 22) have been implicated in PCOS. However, the heterogeneity and generalizability of these genetic associations are not enough to explain clearly the considerable genetic susceptibility for this endocrine-metabolic disorder. For example, FSHR rs6166 was associated with increased PCOS risk in a study of 377 Chinese PCOS patients and 388 age-matched healthy controls (23), but not in a study in a different ethnic population (24). ESR1 rs9340799 has been associated with increased PCOS risk in a Pakistani population (25), but not in a Brazilian population (26).
Therefore, finding as many single-nucleotide polymorphisms (SNPs) as possible through population studies will provide valuable assistance to clinicians and patients. Except for the previous reported genes which are involved in the pathogenesis of PCOS, morphological changes in the ovary is a key part in the development of PCOS. Related studies have proved that LAMC1 can promote the development of follicles (27) and some SNP polymorphisms were significantly associated with premature ovarian failure (POF) (28). Because of the role of follicular development in the development of PCOS, the association between LAMC1 polymorphism and PCOS risk deserved studying.
Considering the important role of genetic factors in this pathogenesis of PCOS, the aim of the study was to discover some genetic variations associated with these abnormal pathological mechanisms. So we have chosen several sites in CYP11A1, CYP19A1, ESR1, ESR2, PGR, PPARG, LAMC1, HMGA2, MTHFR, and SUMO1P1 as our research object according to the functional description in the literature. We genotyped for 63 SNPs including 40 which had never been assessed for their potential association with PCOS in 10 known pathogenicity-causing genes by a case-control study of 361 PCOS cases and 331 controls in a Chinese population.
Materials and methods
Sample collection
A total of 361 PCOS patients and 331 controls, all of Han Chinese ethnicity, were included in this multi-center study. Subjects were recruited between January 2015 and February 2017 at the Reproductive Hospital Affiliated to Shandong University, at the Women's Hospital of the School of Medicine of Zhejiang University and at the Renji Hospital Affiliated to Shanghai Jiao Tong University. PCOS was diagnosed based on the NIH criteria [National Institutes of Health/National Institute of Child Health and Human Development (NIH/NICHD) in April 1990], including biochemical and/or clinical hyperandrogenism and ovulatory dysfunction, after the exclusion of related or other disorders (1). This study was approved by the Shanghai Xinhua Hospital Research Ethics Board and all subjects gave their informed consent for inclusion before they participated in the study. Women were excluded if they had been diagnosed with 21-hydroxylase-deficiency, androgen-producing tumors, hyperprolactinemia, non-classical adrenal hyperplasia, Cushing's syndrome, and active thyroid disease, since these conditions likely affect reproductive physiology (19, 29). All the controls were defined as women with normal hormonal status and regular menstrual cycles at intervals of 28–35 days. Information on participants was collected from medical records.
Biochemical and hormonal analyses
Peripheral blood samples were taken from participants in a fasting state on days 3–5 of the menstrual cycle. Blood samples were collected into EDTA anticoagulant tubes, centrifuged, and stored at −80°C. Levels of luteinizing hormone (LH), follicle-stimulating hormone (FSH), testosterone (T), and estradiol (E2) were measured using a chemiluminescent analyzer (Beckman Coulter, Fullerton, CA, USA). The limit of detection (LOD) of LH, FSH, T and E2 was 0.1 IU/L, 0.1 IU/L, 0.1 nmol/L and 0.1 pg/ml respectively. They intra- and inter-assay coefficients of variations (CV) were < 6% and < 10%, respectively.
SNP selection and genotyping
A total of 63 SNPs were selected in the following 10 genes, mainly based on literature review: CYP11A1, CYP19A1, ESR1, ESR2, PGR, PPARG, MTHFR, HMGA2, LAMC1, and SUMP1O1 (21). Five of the SNPs were 3′-flanking variants; 57 SNPs, intronic variants; 3 SNPs in exons, non-synonymous variants; 3 SNPs in exons, synonymous variants; and 2 SNPs near 5′-flanking regions. Genomic DNA was extracted from the buffy coat using DNA isolation kits (TIANGEN, China) according to the manufacturer's protocol. Genotyping was carried out using polymerase chain reaction ligation detection reaction (PCR-LDR). Genotyping was performed by technicians blinded to case or control status. In addition, 5% of samples were re-genotyped, and the results agreed 100% with the first results. PCRs were carried out on an ABI 7300 (Applied Biosystem, Foster City, CA, USA) in a total volume of 20 μl including 20 ng genomic DNA, 2.5 mM MgCl2, 0.2 mM dNTPs, 0.5 μM each primer, 1 × PCR Buffer and 1 U Hot-Start Taq DNA polymerase (QIAgen). Cycling parameters were set as follows: 95°C for 10 min, 38 cycles at 94°C for 15 s, 60°C for 1.5 min, 72°C for 60 s, followed by 72°C for 10 min. The ligation reaction was carried out in a final volume of 20 μl containing 2 μl template DNA, 1 μl 10 × ligation buffer, 20 U Taq DNA ligase and 1 pmol of each discriminating probe (New England Biolabs, USA). The LDR parameters were as follows: 94°C for 1.5 min, 35 cycles at 94°C for 15 s and 58°C for 1.5 min. Following the LDR reaction, 1 μl LDR reaction product was mixed with 1 μl loading buffer as well as 1 μl ROX. The mixture was then analyzed using the ABI 7900HT (Applied Biosystems).
Statistical analysis
Hormone levels, differences in age and body mass index (BMI) between controls and cases were evaluated using Student's t-test. The fit of results for each SNP in controls was assessed against expectations based on Hardy-Weinberg equilibrium (HWE) using the χ2-test. Associations between selected polymorphisms and PCOS were assessed using unconditional logistic regression with adjustment for BMI and age.
Pairwise linkage disequilibrium (LD) among the selected SNPs was examined using Lewontin's standardized coefficient D' and LD coefficient r2 (30). Haplotype blocks were defined by the method of (31) in the publicly available Haploview software (www.broadinstitute.org/haploview/downloads) using default settings: the confidence interval for a strong LD was minimal for upper 0.98 and low 0.7 and maximal for a strong recombination of 0.9, and a fraction of strong LD in informative comparisons was at least 0.95. Haplotype analysis was performed using the SNP stats tool (www.snpstats.net/snpstats/start.htm?q=snpstats/start.htm), with adjustment for age and BMI where appropriate. Multiple test corrections were performed for each gene using Bonferroni correction. All statistical analyses were performed using SAS 9.4 software and R. P < 0.05 was considered statistically significant.
Results
Characteristics of the participants
The characteristics of the participants are shown in Table 1. The age of cases ranged from 19 to 40 years (mean ± SD, 28.1 ± 3.7); the age of controls, from 22 to 45 years (28.4 ± 4.2, P = 0.041). As expected, levels of LH, testosterone and BMI in the PCOS group were significantly higher than that in healthy controls.
Table 1.
Characteristics of participants.
| Characteristic | Controls (n = 331) | Cases (n = 361) | Pa |
|---|---|---|---|
| Age (years) | 28.4 ± 4.2 | 28.1 ± 3.7 | 0.364 |
| BMI (kg/m2) | 21.5 ± 3.4 | 24.9 ± 4.3 | <0.0001 |
| < 24 | 282 (85.2%) | 169 (46.8) | |
| ≥24 | 49 (14.8%) | 192 (53.2) | |
| LH (IU/L) | 4.1 ± 4.7 | 7.0 ± 5.8 | <0.0001 |
| FSH (IU/L) | 4.1 ± 3.6 | 5.4 ± 2.6 | 0.0002 |
| LH/FSH | 0.6 ± 0.7 | 1.2 ± 1.1 | <0.0001 |
| T (nmol/L) | 2.8 ± 3.1 | 23.7 ± 23.4 | <0.0001 |
| E2 (pg/ml) | 88.2 ± 75.3 | 104.7 ± 165.8 | 0.775 |
Continuous data were expressed as mean ± SD; BMI, body mass index; LH, luteinizing hormone; FSH, follicle-stimulating hormone; T, testosterone; E2, estradiol.
Boldface indicates P < 0.05.
Association between individual SNPs and PCOS risk
Results on minor allele frequencies and HWE at selected SNPs are shown in Table 2. Among the 63 polymorphisms, five deviated from HWE (P < 0.05): rs1801132, rs1884051, rs722208, rs932477, and rs728524. Minor allele frequencies of 0 were observed for the following five SNPs: rs1913474, rs1042839, rs480851, rs1805192, and rs709150. These 10 SNPs were excluded from subsequent analysis. Among 53 polymorphic locus analysis, we found that the following variant alleles were significantly associated with decreased PCOS risk: The A>G allele distributions at rs9340799 in ESR1 (P = 0.000), the A>T at rs709154 (P = 0.013), and A>C at rs1151996 (P = 0.013) in PPARG. the A>C at rs2272046 in HMGA2 (P = 0.000), and the T>C at rs1801133 in MTHFR (P = 0.000, Table 3). Further we detected the following genotype distribution were associated with decreased PCOS risk: GA genotype at rs9340799 (P < 0.0001) in ESR1, TA genotype at rs709154 (P < 0.0001) in PPARG, CA genotype at rs2272046 (P < 0.0001) in HMGA2. Moreover, GA genotype at rs1999805 (P = 0.013) in ESR1 and TT genotype at rs1801133 in MTHFR (P < 0.0001) correlated with elevated PCOS risk (Table 4). We conducted a stratified analysis of risk-related loci by BMI (BMI < 24, BMI ≥ 24; Supplementary Table 1). Finally we found that BMI may not affect the risk of rs9340799 and rs1999805 in ESR1, rs2272046 in HMGA2, rs1801133 in MTHFR, rs709154 in PPARG gene and PCOS.
Table 2.
Information about minor allele frequency and Hardy-Weinberg equilibrium at selected SNPs.
| Gene | SNP | Genomic position | Position in gene | MAF | HWE | Genotyping |
|---|---|---|---|---|---|---|
| Rate (%) | ||||||
| CYP11A1 | rs9806234 | 74663033 | Intron | 0.35 | 0.19 | 96.3 |
| rs11632698 | 74637867 | Intron | 0.25 | 0.47 | 99.2 | |
| rs11638442 | 74634899 | Intron | 0.25 | 0.62 | 99.6 | |
| rs16968478 | 74662811 | Intron | 0.45 | 0.12 | 97.2 | |
| rs1843090 | 74643034 | Intron | 0.41 | 0.69 | 96.5 | |
| rs2073475 | 74661894 | 5′ near gene | 0.41 | 0.56 | 97.8 | |
| rs2279357 | 74630623 | Intron | 0.41 | 0.56 | 97.4 | |
| rs4887139 | 74661945 | 5′ near gene | 0.1 | 1.00 | 97.2 | |
| rs7173655 | 74632362 | Intron | 0.4 | 0.43 | 97.9 | |
| CYP19A1 | rs2414096 | 51529779 | Intron | 0.47 | 0.70 | 96.5 |
| rs700518 | 51529112 | Synonymous exon | 0.47 | 1.00 | 96.7 | |
| ESR1 | rs1709183 | 152193996 | Intron | 0.41 | 0.56 | 98.4 |
| rs1801132 | 152265522 | Synonymous exon | 0.48 | 0.00b | 98.4 | |
| rs1884051 | 152283279 | Intron | 0.47 | 0.01b | 98.2 | |
| rs2228480 | 152420095 | Intron | 0.26 | 0.06 | 97.9 | |
| rs2234693 | 152163335 | Intron | 0.41 | 0.69 | 98.9 | |
| rs3020314 | 152270672 | Intron | 0.14 | 0.07 | 100.0 | |
| rs3778082 | 152387664 | Intron | 0.33 | 0.52 | 97.4 | |
| rs3778099 | 152418575 | Intron | 0.37 | 0.22 | 97.2 | |
| rs3798573 | 152389362 | Intron | 0.34 | 0.40 | 96.9 | |
| rs3798577 | 152421130 | Intron | 0.46 | 0.33 | 98.4 | |
| rs722208 | 152322885 | Intron | 0.49 | 0.01b | 97.8 | |
| rs851982 | 152024985 | Intron | 0.13 | 0.60 | 96.7 | |
| rs9322331 | 152162317 | Intron | 0.03 | 1.00 | 98.8 | |
| rs932477 | 152304596 | Intron | 0.39 | 0.01b | 99.4 | |
| rs9340799 | 152163381 | Intron | 0.19 | 0.61 | 96.9 | |
| rs1913474 | 152208722 | Intron | 0.00a | 1.00 | 23.4 | |
| rs1999805 | 152068364 | Intron | 0.33 | 0.23 | 97.4 | |
| rs728524 | 152303437 | Intron | 0.21 | 0.02b | 97.9 | |
| ESR2 | rs1255998 | 64693871 | 3′ utr | 0.48 | 0.55 | 96.3 |
| rs928554 | 64694195 | 3′ utr | 0.66 | 0.31 | 97.9 | |
| rs960070 | 64745179 | Intron | 0.25 | 1.00 | 94.3 | |
| rs1152579 | 64695087 | Intron | 0.3 | 0.83 | 96.7 | |
| rs944459 | 64699358 | Intron | 0.27 | 0.10 | 97.9 | |
| PGR | rs1042839 | 100922202 | Synonymous exon | 0.00a | 1.00 | 100 |
| rs471767 | 100905297 | 3′ utr | 0.05 | 1.00 | 99.8 | |
| rs484389 | 100909809 | 3′ utr | 0.16 | 0.26 | 96.7 | |
| rs508653 | 100962200 | Intron | 0.15 | 0.06 | 98.9 | |
| rs547378 | 100922939 | Intron | 0.16 | 0.26 | 99.4 | |
| rs561650 | 100915894 | Intron | 0.14 | 0.69 | 98.4 | |
| rs566351 | 100985014 | Intron | 0.15 | 0.42 | 98.2 | |
| rs572483 | 100967572 | Intron | 0.15 | 0.07 | 97.6 | |
| rs613120 | 100974278 | Intron | 0.15 | 0.06 | 98.6 | |
| rs480851 | 100976191 | Intron | 0.00a | 1.00 | 98.9 | |
| rs500760 | 100909991 | Synonymous exon | 0.16 | 0.26 | 97.3 | |
| rs608995 | 100905733 | 3′ utr | 0.16 | 0.26 | 96.7 | |
| PPARG | rs1801282 | 12393125 | Intron | 0.06 | 1.00 | 99.1 |
| rs1805192 | 12421238 | Non-synonymous exon | 0.00a | 1.00 | 100 | |
| rs709149 | 12450354 | Intron | 0.43 | 0.51 | 95.5 | |
| rs709150 | 12451337 | Intron | 0.00a | 1.00 | 28.2 | |
| rs709151 | 12454999 | Intron | 0.39 | 0.18 | 98.2 | |
| rs709154 | 12456834 | Intron | 0.36 | 0.25 | 98.6 | |
| rs1151996 | 12445807 | Intron | 0.47 | 1.00 | 98.2 | |
| LAMC1 | rs1413390 | 183096634 | Intron | 0.45 | 0.55 | 95.9 |
| rs1537520 | 183084229 | Intron | 0.45 | 0.43 | 98.2 | |
| rs20558 | 183094547 | Non-synonymous exon | 0.47 | 0.24 | 98.6 | |
| rs2296293 | 183095477 | Intron | 0.45 | 0.16 | 96.5 | |
| rs2296300 | 183099701 | Intron | 0.45 | 0.33 | 98.8 | |
| rs6424888 | 183085696 | Intron | 0.45 | 0.33 | 98.8 | |
| rs6672093 | 183079853 | Intron | 0.47 | 0.43 | 98.6 | |
| HMGA2 | rs2272046 | 66224461 | Intron | 0.07 | 0.21 | 99.8 |
| MTHFR | rs1801133 | 11856378 | Non-synonymous exon | 0.47 | 0.42 | 92.7 |
| SUMO1P1 | rs6022786 | 52447303 | Intron | 0.33 | 0.54 | 96.7 |
Boldface is used for minor allele frequencies of 0.
Boldface for deviation from HWE.
SNP, single-nucleotide polymorphism; MAF, minor allele frequency; HWE, Hardy-Weinberg equilibrium, HWE P value in the control group.
Table 3.
Allele frequencies of the selected genetic variants analyzed in PCOS patients and controls.
| No. | Gene | SNP | Base | OR (95%CI) | Pa | Pajusted |
|---|---|---|---|---|---|---|
| Change | ||||||
| 1 | CYP11A1 | rs9806234 | G>A | 1.15(0.88,1.50) | 0.299 | 0.474 |
| 2 | rs11632698 | A>G | 0.87(0.66,1.14) | 0.311 | 0.479 | |
| 3 | rs11638442 | C>G | 0.89(0.67,1.17) | 0.393 | 0.593 | |
| 4 | rs16968478 | A>G | 0.97(0.76,1.22) | 0.779 | 0.874 | |
| 5 | rs1843090 | G>A | 0.91(0.71,1.17) | 0.475 | 0.684 | |
| 6 | rs2073475 | C>T | 0.77(0.60,0.99) | 0.042 | 0.150 | |
| 7 | rs2279357 | C>T | 0.91(0.71,1.17) | 0.480 | 0.685 | |
| 8 | rs4887139 | A>G | 2.10(1.36,3.26) | 0.001 | 0.068 | |
| 9 | rs7173655 | C>T | 0.97(0.75,1.26) | 0.832 | 0.908 | |
| 10 | CYP19A1 | rs2414096 | G>A | 1.15(0.91,1.45) | 0.251 | 0.419 |
| 11 | rs700518 | T>C | 1.06(0.83,1.34) | 0.664 | 0.783 | |
| 12 | ESR1 | rs1709183 | C>T | 1.02(0.79,1.31) | 0.900 | 0.955 |
| 13 | rs2228480 | G>A | 1.46(1.08,1.98) | 0.013 | 0.079 | |
| 14 | rs2234693 | T>C | 1.03(0.80,1.32) | 0.840 | 0.910 | |
| 15 | rs3020314 | C>T | 0.71(0.49,1.04) | 0.081 | 0.218 | |
| 16 | rs3778082 | G>A | 0.76(0.58,1.00) | 0.053 | 0.166 | |
| 17 | rs3778099 | T>C | 0.92(0.68,1.25) | 0.594 | 0.740 | |
| 18 | rs3798573 | A>G | 0.74(0.57,0.97) | 0.027 | 0.121 | |
| 19 | rs3798577 | T>C | 0.67(0.51,0.87) | 0.003 | 0.124 | |
| 20 | rs851982 | T>C | 1.68(1.14,2.47) | 0.008 | 0.218 | |
| 21 | rs9322331 | C>T | 1.46(0.70,3.04) | 0.314 | 0.479 | |
| 22 | rs9340799 | A>G | 0.50(0.37,0.67) | 0.000 | 0.000 | |
| 23 | rs1999805 | G>A | 1.22(0.95,1.57) | 0.123 | 0.280 | |
| 24 | ESR2 | rs1255998 | C>G | 1.05(0.83,1.33) | 0.656 | 0.783 |
| 25 | rs928554 | T>C | 0.83(0.63,1.08) | 0.159 | 0.324 | |
| 26 | rs960070 | G>C | 1.36(1.01,1.84) | 0.042 | 0.150 | |
| 27 | rs1152579 | G>A | 0.75(0.58,0.98) | 0.036 | 0.135 | |
| 28 | rs944459 | C>T | 0.90(0.69,1.17) | 0.430 | 0.643 | |
| 29 | PGR | rs471767 | A>G | 1.19(0.68,2.09) | 0.538 | 0.716 |
| 30 | rs484389 | A>G | 1.46(1.04,2.06) | 0.030 | 0.124 | |
| 31 | rs508653 | C>A | 1.35(0.96,1.91) | 0.085 | 0.222 | |
| 32 | rs547378 | G>A | 1.49(1.06,2.09) | 0.022 | 0.102 | |
| 33 | rs561650 | T>C | 1.50(1.04,2.17) | 0.031 | 0.125 | |
| 34 | rs566351 | C>T | 1.42(0.99,2.01) | 0.053 | 0.166 | |
| 35 | rs572483 | T>C | 1.31(0.93,1.84) | 0.117 | 0.274 | |
| 36 | rs613120 | A>G | 1.29(0.92,1.81) | 0.143 | 0.299 | |
| 37 | rs500760 | T>C | 1.45(1.03,2.04) | 0.032 | 0.126 | |
| 38 | rs608995 | A>T | 1.50(1.07,2.11) | 0.019 | 0.095 | |
| 39 | PPARG | rs1801282 | C>G | 0.56(0.35,0.91) | 0.020 | 0.095 |
| 40 | rs709149 | G>A | 0.87(0.66,1.14) | 0.311 | 0.479 | |
| 41 | rs709151 | C>T | 0.72(0.56,0.94) | 0.017 | 0.092 | |
| 42 | rs709154 | A>T | 0.65(0.50,0.85) | 0.001 | 0.013 | |
| 43 | rs1151996 | A>C | 0.66(0.51,0.85) | 0.001 | 0.013 | |
| 44 | LAMC1 | rs1413390 | A>G | 1.19(0.92,1.53) | 0.191 | 0.346 |
| 45 | rs1537520 | T>C | 1.19(0.92,1.55) | 0.180 | 0.340 | |
| 46 | rs20558 | C>T | 1.18(0.91,1.53) | 0.209 | 0.361 | |
| 47 | rs2296293 | A>G | 1.26(0.97,1.65) | 0.082 | 0.218 | |
| 48 | rs2296300 | A>G | 1.18(0.91,1.53) | 0.213 | 0.363 | |
| 49 | rs6424888 | G>A | 1.18(0.91,1.53) | 0.204 | 0.356 | |
| 50 | rs6672093 | C>T | 1.15(0.89,1.49) | 0.274 | 0.443 | |
| 51 | HMGA2 | rs2272046 | A>C | 0.28(0.18,0.43) | 0.000 | 0.000 |
| 52 | MTHFR | rs1801133 | T>C | 0.62(0.49,0.80) | 0.000 | 0.000 |
| 53 | SUMO1P1 | rs6022786 | G>A | 0.74(0.57,0.95) | 0.019 | 0.095 |
Boldface indicates P < 0.05; CI, confidence interval; OR, odds ratio; SNP, single-nucleotide polymorphism; PCOS, polycystic ovarian syndrome.
P-value from unconditional logistic regression with adjustment for BMI and age.
Pajusted were performed for each gene using Bonferroni correction.
Table 4.
Genotype frequencies of selected polymorphisms analyzed in PCOS patients and controls.
| Gene | SNP | Controls | Cases | OR (95%CI) | Pa | Pajusted |
|---|---|---|---|---|---|---|
| (n = 331) | (n = 361) | |||||
| CYP11A1 | rs9806234 | |||||
| G/G | 124 (40.0) | 139 (40.1) | Reference | |||
| G/A | 156(50.3) | 167 (48.1) | 1.02(0.71–1.46) | 0.926 | 0.974 | |
| A/A | 30 (9.7) | 41 (11.8) | 1.50(0.83–2.69) | 0.178 | 0.340 | |
| rs11632698 | ||||||
| A/A | 160 (50.2) | 192 (53.6) | Reference | |||
| A/G | 135 (42.3) | 152 (42.5) | 0.96 (0.68–1.36) | 0.830 | 0.908 | |
| G/G | 24 (7.5) | 14 (3.9) | 0.60 (0.28–1.25) | 0.170 | 0.340 | |
| rs11638442 | ||||||
| C/C | 160 (50.8) | 194 (53.7) | Reference | |||
| C/G | 131 (41.6) | 153 (42.4) | 1.01 (0.71–1.44) | 0.943 | 0.974 | |
| G/G | 24 (7.6) | 14 (3.9) | 0.59 (0.28–1.23) | 0.158 | 0.324 | |
| rs16968478 | ||||||
| A/A | 67 (20.6) | 104 (29.7) | Reference | |||
| G/A | 202 (62.0) | 190 (54.3) | 0.59 (0.39–0.88) | 0.010 | 0.068 | |
| G/G | 57 (17.5) | 56 (16.0) | 0.55 (0.32–0.94) | 0.028 | 0.122 | |
| rs1843090 | ||||||
| G/G | 102 (32.5) | 125 (35.7) | Reference | |||
| G/A | 162 (51.6) | 172 (49.1) | 0.81 (0.56–1.18) | 0.266 | 0.435 | |
| A/A | 50 (15.9) | 53 (15.1) | 0.89 (0.53–1.48) | 0.652 | 0.783 | |
| rs2073475 | ||||||
| C/C | 92 (28.3) | 122 (34.7) | Reference | |||
| C/T | 181 (55.7) | 182 (51.7) | 0.76 (0.52–1.10) | 0.142 | 0.299 | |
| T/T | 52 (16.0) | 48 (13.6) | 0.60 (0.35–1.01) | 0.055 | 0.169 | |
| rs2279357 | ||||||
| C/C | 102 (30.8) | 119 (34.1) | Reference | |||
| T/C | 179 (54.1) | 179 (51.3) | 0.80 (0.55–1.16) | 0.243 | 0.410 | |
| T/T | 50 (15.1) | 51 (14.6) | 0.89 (0.53–1.50) | 0.675 | 0.785 | |
| rs4887139 | ||||||
| A/A | 280 (87.2) | 278 (79.0) | Reference | |||
| G/A | 41 (12.8) | 69 (19.6) | 1.91 (1.21–3.04) | 0.006 | 0.055 | |
| G/G | 0 (0.0) | 5 (1.4) | NA | 0.977 | 0.980 | |
| rs7173655 | ||||||
| C/C | 101 (30.6) | 113 (32.1) | Reference | |||
| C/T | 186 (56.4) | 189 (53.7) | 0.95 (0.65–1.38) | 0.777 | 0.874 | |
| T/T | 43 (13.0) | 50 (14.2) | 0.96 (0.56–1.65) | 0.888 | 0.948 | |
| CYP19A1 | rs2414096 | |||||
| G/G | 106 (34.0) | 99 (28.3) | Reference | |||
| G/A | 131 (42.0) | 174 (49.7) | 1.40 (0.94–2.08) | 0.100 | 0.249 | |
| A/A | 75 (24.0) | 77 (22.0) | 1.28 (0.80–2.04) | 0.304 | 0.477 | |
| rs700518 | ||||||
| T/T | 90 (29.9) | 100 (28.4) | Reference | |||
| T/C | 143 (47.5) | 177 (50.3) | 1.05 (0.70–1.58) | 0.801 | 0.892 | |
| C/C | 68 (22.6) | 75 (21.3) | 1.11 (0.68–1.81) | 0.668 | 0.783 | |
| ESR1 | rs1709183 | |||||
| C/C | 97 (29.5) | 121 (34.1) | Reference | |||
| T/C | 189 (57.4) | 178 (50.1) | 0.75 (0.52–1.09) | 0.133 | 0.294 | |
| T/T | 43 (13.1) | 56 (15.8) | 1.22 (0.72–2.08) | 0.460 | 0.675 | |
| rs2228480 | ||||||
| G/G | 194 (60.8) | 186 (52.4) | Reference | |||
| G/A | 116 (36.4) | 153 (43.1) | 1.46 (1.03–2.07) | 0.034 | 0.130 | |
| A/A | 9 (2.8) | 16 (4.5) | 2.18 (0.86–5.55) | 0.102 | 0.250 | |
| rs2234693 | ||||||
| T/T | 102 (32.5) | 111 (30.9) | Reference | |||
| T/C | 158 (50.3) | 198 (55.2) | 1.11 (0.76–1.63) | 0.576 | 0.735 | |
| C/C | 54 (17.2) | 50 (13.9) | 1.02 (0.61–1.71) | 0.941 | 0.974 | |
| rs3020314 | ||||||
| C/C | 235 (71.0) | 271 (75.1) | Reference | |||
| T/C | 96 (29.0) | 90 (24.9) | 0.72 (0.49–1.04) | 0.081 | 0.218 | |
| rs3778082 | ||||||
| G/G | 129 (42.4) | 164 (46.2) | Reference | |||
| G/A | 146 (48.0) | 158 (44.5) | 0.70 (0.48–1.02) | 0.061 | 0.184 | |
| A/A | 29 (9.5) | 33 (9.3) | 0.65 (0.34–1.22) | 0.176 | 0.340 | |
| rs3778099 | ||||||
| T/T | 37 (34.9) | 137 (36.9) | Reference | |||
| C/T | 57 (53.8) | 195 (52.6) | 0.84 (0.51–1.49) | 0.511 | 0.710 | |
| C/C | 12 (11.3) | 39 (10.5) | 0.84 (0.37–1.86) | 0.659 | 0.783 | |
| rs3798573 | ||||||
| A/A | 126 (40.5) | 157 (44.9) | Reference | |||
| A/G | 154 (49.5) | 160 (45.7) | 0.70 (0.49–1.00) | 0.051 | 0.166 | |
| G/G | 31 (10.0) | 33 (9.4) | 0.58 (0.31–1.08) | 0.089 | 0.229 | |
| rs3798577 | ||||||
| C/C | 72 (22.4) | 52 (14.5) | Reference | |||
| T/C | 184 (57.3) | 206 (57.5) | 1.48 (0.94–2.33) | 0.092 | 0.233 | |
| T/T | 65 (20.2) | 100 (27.9) | 2.23 (1.32–3.78) | 0.003 | 0.124 | |
| rs851982 | ||||||
| T/T | 266 (81.6) | 258 (74.1) | Reference | |||
| T/C | 60 (18.4) | 83 (23.9) | 1.51 (1.00–2.27) | 0.047 | 0.164 | |
| C/C | 0 (0.0) | 7 (2.0) | NA | 0.973 | 0.980 | |
| rs9322331 | ||||||
| C/C | 315 (96.0) | 336 (94.1) | Reference | |||
| T/C | 13 (4.0) | 20 (5.6) | 1.31 (0.60–2.85) | 0.503 | 0.705 | |
| T/T | 0 (0.0) | 1 (0.3) | NA | 0.980 | 0.980 | |
| rs9340799 | ||||||
| A/A | 159 (48.8) | 243 (69.6) | Reference | |||
| G/A | 144 (44.2) | 93 (26.6) | 0.40 (0.28–0.58) | < 0.0001 | <0.0001 | |
| G/G | 23 (7.1) | 13 (3.7) | 0.45 (0.20–1.00) | 0.049 | 0.164 | |
| rs1999805 | ||||||
| G/G | 167 (53.9) | 140 (39.7) | Reference | |||
| G/A | 103 (33.2) | 178 (50.4) | 1.91 (1.32–2.75) | 0.001 | 0.013 | |
| A/A | 40 (12.9) | 35 (9.9) | 0.98 (0.56–1.72) | 0.953 | 0.978 | |
| ESR2 | rs1255998 | |||||
| C/C | 90 (29.7) | 104 (29.5) | Reference | |||
| C/G | 146 (48.2) | 161 (45.7) | 0.99 (0.66–1.48) | 0.959 | 0.978 | |
| G/G | 67 (22.1) | 87 (24.7) | 1.12 (0.70–1.80) | 0.630 | 0.773 | |
| rs928554 | ||||||
| T/T | 128 (41.7) | 179 (50.0) | Reference | |||
| C/T | 148 (48.2) | 152 (42.5) | 0.89 (0.63–1.27) | 0.523 | 0.716 | |
| C/C | 31 (10.1) | 27 (7.5) | 0.60 (0.32–1.15) | 0.124 | 0.280 | |
| rs960070 | ||||||
| G/G | 194 (63.8) | 179 (52.5) | Reference | |||
| G/C | 99 (32.6) | 141 (41.3) | 1.33 (0.92–1.91) | 0.125 | 0.280 | |
| C/C | 11 (3.6) | 21 (6.2) | 2.03 (0.85–4.85) | 0.110 | 0.262 | |
| rs1152579 | ||||||
| G/G | 128 (40.6) | 184 (52.4) | Reference | |||
| G/A | 154 (48.9) | 141 (40.2) | 0.77 (0.54–1.09) | 0.141 | 0.299 | |
| A/A | 33 (10.5) | 26 (7.4) | 0.55 (0.29–1.04) | 0.065 | 0.189 | |
| rs944459 | ||||||
| C/C | 156 (49.4) | 201 (56.3) | Reference | |||
| C/T | 139 (44.0) | 129 (36.1) | 0.77 (0.54–1.09) | 0.136 | 0.297 | |
| T/T | 21 (6.6) | 27 (7.6) | 1.06 (0.55–2.04) | 0.868 | 0.933 | |
| PGR | rs471767 | |||||
| A/A | 299 (90.3) | 325 (90.3) | Reference | |||
| A/G | 32 (9.7) | 35 (9.7) | 1.19 (0.68–2.09) | 0.534 | 0.716 | |
| rs484389 | ||||||
| A/A | 262 (79.2) | 248 (70.9) | Reference | |||
| A/G | 61 (18.4) | 95 (27.1) | 1.67 (1.12–2.49) | 0.013 | 0.079 | |
| G/G | 8 (2.4) | 7 (2.0) | 1.14 (0.34–3.76) | 0.833 | 0.908 | |
| rs508653 | ||||||
| C/C | 267 (81.2) | 264 (73.7) | Reference | |||
| C/A | 53 (16.1) | 84 (23.5) | 1.84 (1.20–2.81) | 0.005 | 0.051 | |
| A/A | 9 (2.7) | 10 (2.8) | 0.68 (0.25–1.88) | 0.457 | 0.684 | |
| rs547378 | ||||||
| G/G | 262 (79.2) | 253 (70.7) | Reference | |||
| G/A | 61 (18.4) | 97 (27.1) | 1.69 (1.14–2.53) | 0.010 | 0.068 | |
| A/A | 8 (2.4) | 8 (2.2) | 1.22 (0.38–3.87) | 0.735 | 0.836 | |
| rs561650 | ||||||
| T/T | 265 (80.1) | 262 (74.0) | Reference | |||
| T/C | 64 (19.3) | 85 (24.0) | 1.44 (0.96–2.15) | 0.077 | 0.216 | |
| C/C | 2 (0.6) | 7 (2.0) | 3.33 (0.59–18.9) | 0.174 | 0.340 | |
| rs566351 | ||||||
| C/C | 260 (79.5) | 256 (72.1) | Reference | |||
| C/T | 62 (19.0) | 89 (25.1) | 1.51 (1.00–2.27) | 0.049 | 0.164 | |
| T/T | 5 (1.5) | 10 (2.8) | 1.46 (0.44–4.85) | 0.538 | 0.716 | |
| rs572483 | ||||||
| T/T | 263 (79.9) | 259 (73.4) | Reference | |||
| C/T | 57 (17.3) | 83 (23.5) | 1.70 (1.12–2.58) | 0.013 | 0.079 | |
| C/C | 9 (2.7) | 11 (3.1) | 0.76 (0.28–2.05) | 0.588 | 0.739 | |
| rs613120 | ||||||
| A/A | 265 (80.1) | 262 (73.8) | Reference | |||
| A/G | 57 (17.2) | 82 (23.1) | 1.66 (1.09–2.52) | 0.017 | 0.092 | |
| G/G | 9 (2.7) | 11 (3.1) | 0.75 (0.28–2.01) | 0.561 | 0.734 | |
| rs500760 | ||||||
| T/T | 262 (79.2) | 247 (70.6) | Reference | |||
| C/T | 61 (18.4) | 95 (27.1) | 1.63 (1.09–2.44) | 0.017 | 0.092 | |
| C/C | 8 (2.4) | 8 (2.3) | 1.25 (0.39–3.99) | 0.702 | 0.810 | |
| rs608995 | ||||||
| A/A | 258 (78.9) | 244 (69.7) | Reference | |||
| T/A | 61 (18.7) | 98 (28.0) | 1.71 (1.15–2.56) | 0.008 | 0.063 | |
| T/T | 8 (2.4) | 8 (2.3) | 1.24 (0.39–3.93) | 0.711 | 0.815 | |
| PPARG | rs1801282 | |||||
| C/C | 271 (83.4) | 321 (89.2) | Reference | |||
| C/G | 54 (16.6) | 39 (10.8) | 0.56 (0.35–0.91) | 0.020 | 0.095 | |
| rs709149 | ||||||
| G/G | 95 (30.0) | 132 (37.9) | Reference | |||
| G/A | 177 (55.8) | 176 (50.6) | 0.73 (0.50–1.07) | 0.108 | 0.261 | |
| A/A | 45 (14.2) | 40 (11.5) | 0.86 (0.48–1.54) | 0.621 | 0.768 | |
| rs709151 | ||||||
| C/C | 85 (26.2) | 133 (37.6) | Reference | |||
| T/C | 189 (58.3) | 182 (51.4) | 0.61 (0.42–0.89) | 0.009 | 0.067 | |
| T/T | 50 (15.4) | 39 (11.0) | 0.60 (0.34–1.04) | 0.069 | 0.197 | |
| rs709154 | ||||||
| A/A | 90 (27.5) | 156 (43.9) | Reference | |||
| T/A | 187 (57.2) | 160 (45.1) | 0.47 (0.33–0.69) | < 0.0001 | <0.0001 | |
| T/T | 50 (15.3) | 39 (11.0) | 0.54 (0.31–0.94) | 0.030 | 0.124 | |
| rs1151996 | ||||||
| A/A | 88 (27.2) | 125 (35.9) | Reference | |||
| C/A | 154 (47.7) | 182 (52.3) | 0.81 (0.55–1.19) | 0.285 | 0.457 | |
| C/C | 81 (25.1) | 41 (11.8) | 0.40 (0.24–0.68) | 0.010 | 0.068 | |
| LAMC1 | rs1413390 | |||||
| A/A | 97 (30.5) | 88 (25.4) | Reference | |||
| G/A | 174 (54.7) | 195 (56.4) | 1.14 (0.77–1.70) | 0.501 | 0.705 | |
| G/G | 47 (14.8) | 63 (18.2) | 1.43 (0.85–2.40) | 0.184 | 0.344 | |
| rs1537520 | ||||||
| T/T | 97 (29.5) | 88 (24.8) | Reference | |||
| C/T | 185 (56.2) | 206 (58.0) | 1.15 (0.78–1.71) | 0.473 | 0.684 | |
| C/C | 47 (14.3) | 61 (17.2) | 1.44 (0.85–2.44) | 0.174 | 0.34 | |
| rs20558 | ||||||
| C/C | 96 (29.2) | 88 (24.6) | Reference | |||
| T/C | 187 (56.8) | 208 (58.3) | 1.12 (0.75–1.65) | 0.585 | 0.739 | |
| T/T | 46 (14.0) | 61 (17.1) | 1.42 (0.84–2.41) | 0.192 | 0.346 | |
| rs2296293 | ||||||
| A/A | 97 (30.1) | 87 (25.0) | Reference | |||
| A/G | 183 (56.8) | 201 (57.8) | 1.13 (0.76–1.69) | 0.536 | 0.716 | |
| G/G | 42 (13.0) | 60 (17.2) | 1.67 (0.97–2.86) | 0.065 | 0.189 | |
| rs2296300 | ||||||
| A/A | 97 (29.3) | 89 (25.0) | Reference | |||
| A/G | 187 (56.5) | 206 (57.9) | 1.12 (0.76–1.66) | 0.570 | 0.735 | |
| G/G | 47 (14.2) | 61 (17.1) | 1.41 (0.83–2.39) | 0.199 | 0.351 | |
| rs6424888 | ||||||
| G/G | 97 (29.3) | 88 (24.6) | Reference | |||
| G/A | 187 (56.5) | 207 (58.0) | 1.12 (0.76–1.66) | 0.559 | 0.734 | |
| A/A | 47 (14.2) | 62 (17.4) | 1.42 (0.84–2.40) | 0.190 | 0.346 | |
| rs6672093 | ||||||
| C/C | 90 (27.9) | 83 (23.2) | Reference | |||
| T/C | 181 (56.0) | 205 (57.4) | 1.09 (0.73–1.64) | 0.661 | 0.783 | |
| T/T | 52 (16.1) | 69 (19.3) | 1.35 (0.80–2.26) | 0.261 | 0.431 | |
| HMGA2 | rs2272046 | |||||
| A/A | 227 (68.6) | 320 (88.9) | Reference | |||
| C/A | 104 (31.4) | 40 (11.1) | 0.28 (0.18–0.43) | < 0.0001 | <0.0001 | |
| MTHFR | rs1801133 | |||||
| C/C | 96 (31.3) | 52 (15.5) | Reference | |||
| T/C | 139 (45.3) | 162 (48.2) | 1.88 (1.19–2.96) | 0.007 | 0.061 | |
| T/T | 72 (23.5) | 122 (36.3) | 2.64 (1.60–4.34) | < 0.0001 | <0.0001 | |
| SUMO1P1 | rs6022786 | |||||
| G/G | 112 (35.0) | 167 (47.7) | Reference | |||
| G/A | 169 (52.8) | 143 (40.9) | 0.58 (0.41–0.84) | 0.046 | 0.160 | |
| A/A | 39 (12.2) | 40 (11.4) | 0.69 (0.39–1.21) | 0.194 | 0.346 |
Boldface indicates P < 0.05; CI, confidence interval; NA, not available; OR, odds ratio; PCOS, polycystic ovarian syndrome; SNP, single-nucleotide polymorphism.
P value was from unconditional logistic regression with adjustment for BMI and age.
Pajusted were performed for each gene using Bonferroni correction.
Haplotype block structure and haplotype analysis
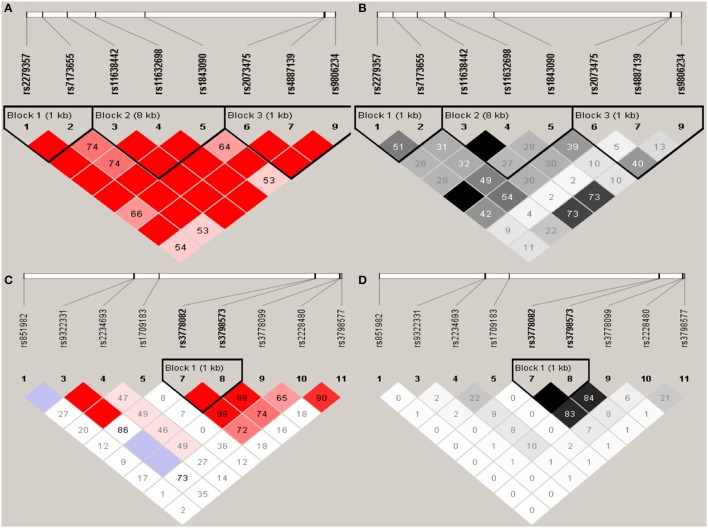
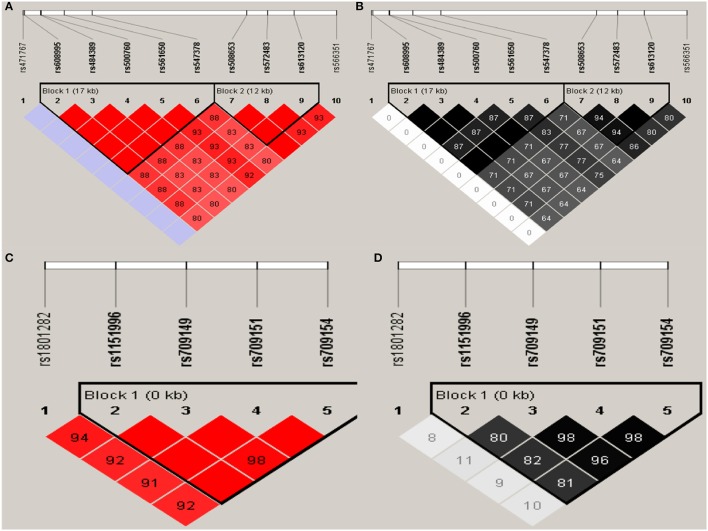
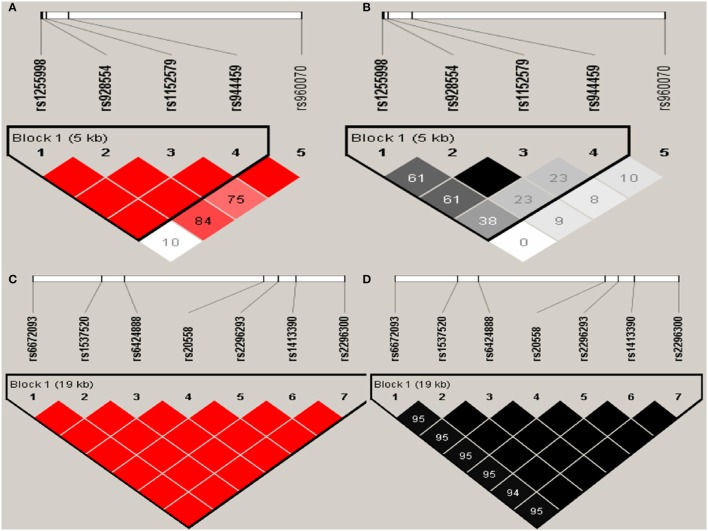
Haploview analysis indicated strong LD among SNPs in the genes CYP11A1, ESR1, ESR2, LAMC1, PGR, and PPARG (Figures 1–3). In CYP11A1, three haplotype blocks were defined using 8 genotyped SNPs. In PGR, two haplotype blocks were formed: the first spanned 17 kb and contained five tested SNPs, and the second contained three tested SNPs. In addition, ESR1, ESR2, LAMC1, and PPARG were constructed respectively with one block. Table 5 summarizes the associations between genetic haplotypes and PCOS risk. After adjusting for age and BMI, the risk of PCOS was significantly increased among individuals carrying the haplotype “CGA” in Block3 of CYP11A1 (OR = 2.30, 95% CI = 1.45–3.66, Pajusted = 0.013), compared with those carrying the most common haplotype “TAG.” Similarly, the risk was also increased among individuals carrying the haplotype “GTGC” in ESR2 (OR = 2.01, 95% CI = 1.33–3.02, Pajusted = 0.020), compared with those carrying the haplotype “GCAC.” Furthermore, one highly protective haplotype “CGCA” in PPARG with 87% reduction in risk of developing PCOS comparing with those carrying the haplotype “AGCA” (OR = 0.13 and 95% CI = 0.04–0.40, Pajusted = 0.010).
Figure 1.
Graphical representations of the SNP locations and LD structure of CYP11A (A,B) and ESR1 (C,D) using tested genotyped SNPs in 331 controls.
Figure 3.
Graphical representations of the SNP locations and LD structure of PGR (A,B) and PPARG (C,D) using tested genotyped SNPs in 331 controls.
Table 5.
Association between CYP11A1, ESR1, ESR2, LAMC, PGR, and PPARG genetic haplotypes and PCOS risk.
| Gene | Haplotypea | Frequencies | Logistic regression | |||
|---|---|---|---|---|---|---|
| Controls | Cases | OR (95% CI)b | P | Pajusted | ||
| CYP1A11 | ||||||
| Block1 | CT | 0.412 | 0.412 | 1.00 (reference) | – | – |
| TC | 0.421 | 0.403 | 0.96 (0.73–1.27) | 0.779 | 0.874 | |
| CC | 0.167 | 0.184 | 1.15 (0.81–1.64) | 0.430 | 0.643 | |
| Block2 | ||||||
| CAA | 0.415 | 0.395 | 1.00 (reference) | – | – | |
| CAG | 0.298 | 0.354 | 1.22 (0.92–1.62) | 0.159 | 0.324 | |
| GGG | 0.287 | 0.251 | 0.95 (0.70–1.28) | 0.702 | 0.810 | |
| Block3 | ||||||
| TAG | 0.437 | 0.394 | 1.00 (reference) | – | – | |
| CAA | 0.281 | 0.242 | 1.06 (0.77–1.45) | 0.702 | 0.810 | |
| CAG | 0.219 | 0.25 | 1.29 (0.94–1.79) | 0.191 | 0.346 | |
| CGA | 0.063 | 0.113 | 2.30 (1.45–3.66) | 5*104 | 0.013 | |
| ESR1 | GA | 0.654 | 0.684 | 1.00 (reference) | – | – |
| AG | 0.346 | 0.317 | 0.72 (0.55–0.93) | 0.013 | 0.079 | |
| ESR2 | GCAC | 0.356 | 0.278 | 1.00 (reference) | – | – |
| CTGT | 0.291 | 0.256 | 1.15 (0.84–1.57) | 0.380 | 0.593 | |
| CTGC | 0.234 | 0.26 | 1.34 (0.97–1.86) | 0.074 | 0.214 | |
| GTGC | 0.119 | 0.199 | 2.01 (1.33–3.02) | 9*104 | 0.020 | |
| LAMC1 | CTGCAAA | 0.563 | 0.519 | 1.00 (reference) | – | – |
| TCATGGG | 0.425 | 0.462 | 1.20 (0.93–1.56) | 0.170 | 0.340 | |
| TTGCAAA | 0.013 | 0.015 | 1.07 (0.38–3.03) | 0.900 | 0.955 | |
| PGR | ||||||
| Block1 | AATTG | 0.884 | 0.838 | 1.00 (reference) | – | – |
| TGCCA | 0.103 | 0.142 | 1.52 (1.06–2.20) | 0.025 | 0.101 | |
| TGCTA | 0.014 | 0.015 | 1.12 (0.41–3.04) | 0.830 | 0.908 | |
| Block2 | CTA | 0.887 | 0.85 | 1.00 (reference) | – | – |
| ACG | 0.107 | 0.144 | 1.35 (0.95–1.90) | 0.090 | 0.233 | |
| PPARG | ||||||
| AGCA | 0.515 | 0.616 | 1.00 (reference) | – | – | |
| CATT | 0.429 | 0.325 | 0.77 (0.59–1.00) | 0.053 | 0.166 | |
| CGCA | 0.048 | 0.006 | 0.13 (0.04–0.40) | 4*104 | 0.010 | |
| CATA | 0.003 | 0.034 | 10.89 (2.32–51.19) | 0.003 | 0.124 | |
Boldface indicates P < 0.05, CI, confidence interval; OR, odds ratio.
Polymorphic bases are listed in 5′-3′ order as in Figure 1.
Adjusted for age and BMI. Pajusted were performed for each gene using Bonferroni correction.
Figure 2.
Graphical representations of the SNP locations and LD structure of ESR2 (A,B) and LAMC1 (C,D) using tested genotyped SNPs in 331 controls.
In CYP11A1, three haplotype blocks were defined using 8 genotyped SNPs. In ESR1, one haplotype block was defined in each gene using the Haploview program with default settings. The confidence interval minima were upper 0.98 and low 0.7 in the case of strong LD; the upper confidence interval maximum was 0.9 for strong recombination; and the fraction of strong LD in informative comparisons had to be at least 0.95. SNPs are indicated by rs number (top: from left to right). (A) The numbers in squares are D′ values (|D′| × 100). The measure of LD (D′) among all possible pairs of SNPs is shown graphically according to the shade of red, where white represents very low D′ and dark red represents very high D′. (B) The measure of LD (r2) among all possible pairs of SNPs is shown graphically according to the shade of gray, where white represents very low r2 and black represents very high r2.
In ESR2 and LAMC1, one haplotype block was defined in each gene using the Haploview program with default settings.
In PGR, two haplotype blocks were defined using 10 genotyped SNPs. In PPARG, one haplotype block was defined in each gene using the Haploview program with default settings.
Discussion
In this PCOS case-control study in a Chinese Han population, we found the following genotypes were associated with a lower risk of developing PCOS: GA genotype of rs9340799 in ESR1, TA genotype of rs709154 in PPARG, and CA genotype of rs2272046 in HMGA. Conversely, the GA genotype of rs1999805 in ESR1, and the TT genotype of rs1801133 in MTHFR were significantly associated with increased risk of PCOS. Associations of ESR1 rs9340799 (25), HMGA2 rs2272046 (21), and MTHFR rs1801133 (32) with PCOS risk have been reported in other studies. In contrast, our results appear to provide the first evidence linking rs1999805 in ESR1 and rs709154 in PPARG with PCOS risk in a Chinese population. These findings provide evidence that polymorphism in genes involved in hormonal action, lipid metabolism and insulin action may modify PCOS risk.
Polymorphism in the ESR1 gene may influence PCOS risk because the protein is necessary for the proper functioning of the hypothalamic-pituitary-ovarian axis. The protein is up-regulated in theca cells of polycystic ovaries, such that the ratio of ESR1 to ESR2 expression is elevated in PCOS, which may contribute to abnormal follicular development (33). Whatever the underlying mechanism, the association between ESR1 polymorphism and risk of PCOS appears to depend in complex ways on ethnicity. A Pakistani study linked the CC genotype of ESR1 rs2234693, the GG genotype of rs9340799, and the CT genotype of rs8179176 with elevated disease risk (25). However, studies in Greeks (34) and Caucasians (35) revealed no differences in frequencies of these genotypes between PCOS patients and controls. The present study found that the GA genotype of rs9340799 protected Chinese women from PCOS. Further study should clarify to what extent these divergent findings reflect ethnicity.
Our results raise the possibility of a correlation between polymorphism at rs1999805 in influencing PCOS risk. We found that the GA genotype of rs1999805 increased that risk in a Chinese population. Since rs1999805 lies in an ESR1 intron, the question arises whether the polymorphism contributes to disease or is simply a marker in LD with other untyped functional variants. Our LD analysis does not predict other disease-relevant sites in this gene. Therefore, the pathway(s) connecting rs1999805 polymorphism with PCOS warrant further exploration.
Our finding of a link between rs709154 and rs1151996 in PPARG, which lies in an intron of the gene, and risk of PCOS may be explained by the fact that the encoded protein functions as a nuclear hormone receptor to play a crucial role in lipogenesis, cell differentiation, inflammatory cytokine production, glucose homeostasis and insulin sensitization (36). We found evidence that PPARG rs709154 and rs1151996 acts as a protective factor against PCOS. Similarly, a meta-analysis has concluded that PPARG rs1801282 C>G polymorphism is associated with decreased PCOS risk. Conversely, the PPARG rs1801282 (Pro12Ala) polymorphism increased PCOS risk in a study comparing 100 African PCOS patients and 120 healthy controls (37). We found PPARG rs709154 to be in strong LD with rs709149 and rs709151, but we are unaware of studies investigating possible correlation among these three polymorphisms in PCOS risk. Future work should examine this question.
Our results with rs2272046 in HMGA2 are consistent with those of a previous study of 744 Han Chinese PCOS patients and 895 healthy controls linking the SNP with decreased PCOS risk (21). The polymorphism rs2272046 is located in an HMGA2 intron, raising the possibility that it affects risk of PCOS by altering degradation or translation of the HMGA2 mRNA (38). The SNP rs2272046 is in complete LD with rs74980477 (D′ = 1, r2 = 1), and the latter is predicted to alter the function of the myc gene. Normal expression of myc oncoprotein plays a critical role in initial oocyte growth and autonomous growth of granulosa cells, so dysregulation may affect follicular development (39). However, This study didn't revealed that SUMO1P1 was associated with PCOS risk, and the result was inconsistent with the GAWS studies (21). The population geographic differences, our limited power and clinical features differences in these subjects may partly explain the inconsistent results.
The results with rs1801133 in MTHFR are consistent with an Iranian study linking the CC genotype at this locus with decreased PCOS risk (32). The SNP rs1801133 occurs in an MTHFR exon that changes an Ala to Val, based on South Han Chinese data from the 1000 Genomes Project (40). This polymorphism is associated with acetylation of lysine 27 of the H3 histone protein, and this H3K27Ac mark is believed to enhance transcription, possibly by blocking the spread of the repressive histone mark H3K27Me3, which is usually found near regulatory elements and is considered to enhance transcription (41). In this way, polymorphism at rs1801133 may affect MTHFR expression and thereby affect risk of PCOS.
This study has some limitations. Our relatively small sample size limited the statistical power of our findings. We recruited only Han Chinese participants, preventing the analysis of how our SNP results depend on ethnicity. The fact that five of our 63 SNPs deviated from HWE and 5 had a minor allele frequency of 0 greatly reduced the rate of locus detection. Since our study lacked a validation group, further work is needed to confirm our observed associations between SNPs and PCOS risk.
In summary, we found, the first evidence that polymorphism at ESR1 rs1999805 and PPARG rs709154 were significantly associated with PCOS risk in Han Chinese. GA genotype at rs9340799 in ESR1, TA genotype at rs709154 in PPARG, CA genotype at rs2272046 in HMGA2, and GA genotype at rs6022786 in SUMO1P1 may be associated with a lower risk of developing PCOS. Conversely, GA genotype at rs1999805 in ESR1, TT genotype at rs1801133 in MTHFR correlated with elevated PCOS risk. Our findings may help clarify the genetics of PCOS and generate leads for further functional research on the disease. Further investigations with larger, multi-ethnic samples are required to confirm our results.
Author contributions
XJ contributed significantly to analysis and wrote the manuscript. JZ and WW conceived and designed the experiments. JS and DC helped perform the analysis with constructive discussions. WC, YS, and WZ helped collect data and test specimens. XY designed the experiments and approved the final version.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge Kefu Tang for a critical advising on data analysis of this article.
Footnotes
Funding. This study was supported by National Key Research Program of China (No.2016YFC1305204), the Chinese National Natural Science Foundation (No. 81773411), the National Basic Research Program of China (No. 2014CB943300), Shanghai Municipal Education Commission—Gaofeng Clinical Medicine Grant Support (No. 20152220), and Shanghai Children's Health Services Capacity Program (No. GDEK201708).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2018.00504/full#supplementary-material
References
- 1.Goodarzi MO, Azziz R. Diagnosis, epidemiology, and genetics of the polycystic ovary syndrome. Best Pract Res Clin Endocrinol Metab. (2006) 20:193–205. 10.1016/j.beem.2006.02.005 [DOI] [PubMed] [Google Scholar]
- 2.Sheehan MT. Polycystic ovarian syndrome: diagnosis and management. Clin Med Res. (2004) 2:13–27. 10.3121/cmr.2.1.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apridonidze T, Essah PA, Iuorno MJ, Nestler JE. Prevalence and characteristics of the metabolic syndrome in women with polycystic ovary syndrome. J Clin Endocrinol Metab. (2005) 90:1929–35. 10.1210/jc.2004-1045 [DOI] [PubMed] [Google Scholar]
- 4.Hart R. PCOS and infertility. Panminerva Med. (2008) 50:305–14. [PubMed] [Google Scholar]
- 5.Diamanti-Kandarakis E, Dunaif A. Insulin resistance and the polycystic ovary syndrome revisited: an update on mechanisms and implications. Endocr Rev. (2012) 33:981–1030. 10.1210/er.2011-1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Messinis IE, Messini CI, Anifandis G, Dafopoulos K. Polycystic ovaries and obesity. Best Pract Res Clin Obstet Gynaecol. (2015) 29:479–88. 10.1016/j.bpobgyn.2014.11.001 [DOI] [PubMed] [Google Scholar]
- 7.Vural B, Caliskan E, Turkoz E, Kilic T, Demirci A. Evaluation of metabolic syndrome frequency and premature carotid atherosclerosis in young women with polycystic ovary syndrome. Hum Reprod. (2005) 20:2409–13. 10.1093/humrep/dei100 [DOI] [PubMed] [Google Scholar]
- 8.Orio F, Muscogiuri G, Nese C, Palomba S, Savastano S, Tafuri D, et al. Obesity, type 2 diabetes mellitus and cardiovascular disease risk: an uptodate in the management of polycystic ovary syndrome. Eur J Obstet Gynecol Reprod Biol. (2016) 207:214–9. 10.1016/j.ejogrb.2016.08.026 [DOI] [PubMed] [Google Scholar]
- 9.Hollinrake E, Abreu A, Maifeld M, Van Voorhis BJ, Dokras A. Increased risk of depressive disorders in women with polycystic ovary syndrome. Fertil Steril. (2007) 87:1369–76. 10.1016/j.fertnstert.2006.11.039 [DOI] [PubMed] [Google Scholar]
- 10.Kerchner A, Lester W, Stuart SP, Dokras A. Risk of depression and other mental health disorders in women with polycystic ovary syndrome: a longitudinal study. Fertil Steril. (2009) 91:207–12. 10.1016/j.fertnstert.2007.11.022 [DOI] [PubMed] [Google Scholar]
- 11.Karimzadeh MA, Javedani M. An assessment of lifestyle modification versus medical treatment with clomiphene citrate, metformin, and clomiphene citrate-metformin in patients with polycystic ovary syndrome. Fertil Steril. (2010) 94:216–20. 10.1016/j.fertnstert.2009.02.078 [DOI] [PubMed] [Google Scholar]
- 12.Dumesic DA, Goodarzi MO, Chazenbalk GD, Abbott DH. Intrauterine environment and polycystic ovary syndrome. Semin Reprod Med. (2014) 32:159–65. 10.1055/s-0034-1371087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodarzi MO, Dumesic DA, Chazenbalk G, Azziz R. Polycystic ovary syndrome: etiology, pathogenesis and diagnosis. Nat Rev Endocrinol. (2011) 7:219–31. 10.1038/nrendo.2010.217 [DOI] [PubMed] [Google Scholar]
- 14.Jones MR, Goodarzi MO. Genetic determinants of polycystic ovary syndrome: progress and future directions. Fertil Steril. (2016) 106:25–32. 10.1016/j.fertnstert.2016.04.040 [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Wu X, Cao Y, Yi L, Chen J. A microsatellite polymorphism (tttta)n in the promoter of the CYP11a gene in Chinese women with polycystic ovary syndrome. Fertil Steril. (2006) 86:223–6. 10.1016/j.fertnstert.2005.12.037 [DOI] [PubMed] [Google Scholar]
- 16.Anagnostou E, Drakakis P, Marinopoulos S, Mavrogianni D, Loutradis D. The impact of genetics profile (gene polymorphisms) in obese non-PCOS women entering an IVF/ICSI program. Curr Drug Targets (2013) 14:850–5. 10.2174/1389450111314080004 [DOI] [PubMed] [Google Scholar]
- 17.Sudo S, Kudo M, Wada S, Sato O, Hsueh AJ, Fujimoto S. Genetic and functional analyses of polymorphisms in the human FSH receptor gene. Mol Hum Reprod. (2002) 8:893–9. 10.1093/molehr/8.10.893 [DOI] [PubMed] [Google Scholar]
- 18.Wu JB, Zhai JF, Yang J. Role of methylenetetrahydrofolate reductase C677T and A1298C polymorphisms in polycystic ovary syndrome risk. Genet Mol Res. (2016) 15. 10.4238/gmr15048570 [DOI] [PubMed] [Google Scholar]
- 19.Thangavelu M, Godla UR, Paul Solomon FD, Maddaly R. Single-nucleotide polymorphism of INS, INSR, IRS1, IRS2, PPAR-G and CAPN10 genes in the pathogenesis of polycystic ovary syndrome. J Genet. (2017) 96:87–96. 10.1007/s12041-017-0749-z [DOI] [PubMed] [Google Scholar]
- 20.Day FR, Hinds DA, Tung JY, Stolk L, Styrkarsdottir U, Saxena R, et al. Causal mechanisms and balancing selection inferred from genetic associations with polycystic ovary syndrome. Nat commun. (2015) 6:8464. 10.1038/ncomms9464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi Y, Zhao H, Shi Y, Cao Y, Yang D, Li Z, et al. Genome-wide association study identifies eight new risk loci for polycystic ovary syndrome. Nat Genet. (2012) 44:1020–5. 10.1038/ng.2384 [DOI] [PubMed] [Google Scholar]
- 22.Chen ZJ, Zhao H, He L, Shi Y, Qin Y, Shi Y, et al. Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat Genet. (2011) 43:55–9. 10.1038/ng.732 [DOI] [PubMed] [Google Scholar]
- 23.Kim JJ, Choi YM, Hong MA, Chae SJ, Hwang K, Yoon SH, et al. FSH receptor gene p. Thr307Ala and p. Asn680Ser polymorphisms are associated with the risk of polycystic ovary syndrome. J Assist Reprod Genet. (2017) 34:1087–93. 10.1007/s10815-017-0953-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du J, Zhang W, Guo L, Zhang Z, Shi H, Wang J, et al. Two FSHR variants, haplotypes and meta-analysis in Chinese women with premature ovarian failure and polycystic ovary syndrome. Mol Genet Metab. (2010) 100:292–5. 10.1016/j.ymgme.2010.03.018 [DOI] [PubMed] [Google Scholar]
- 25.Liaqat I, Jahan N, Krikun G, Taylor HS. Genetic polymorphisms in Pakistani women with polycystic ovary syndrome. Reprod Sci. (2015) 22:347–57. 10.1177/1933719114542015 [DOI] [PubMed] [Google Scholar]
- 26.Silva FS, Soter MO, Sales MF, Candido AL, Reis FM, Silva IF, et al. Estrogen receptor alphalpha gene (ESR1) PvuII and XbaI polymorphisms are associated to metabolic and proinflammatory factors in polycystic ovary syndrome. Gene (2015) 560:44–9. 10.1016/j.gene.2015.01.037 [DOI] [PubMed] [Google Scholar]
- 27.Irving-Rodgers HF, Rodgers RJ. Extracellular matrix of the developing ovarian follicle. Semin Reprod Med. (2006) 24:195–203. 10.1055/s-2006-948549 [DOI] [PubMed] [Google Scholar]
- 28.Pyun JA, Cha DH, Kwack K. LAMC1 gene is associated with premature ovarian failure. Maturitas (2012) 71:402–6. 10.1016/j.maturitas.2012.01.011 [DOI] [PubMed] [Google Scholar]
- 29.Ben Salem A, Megdich F, Kacem O, Souayeh M, Hachani Ben Ali F, Hizem S, et al. Vascular endothelial growth factor (VEGFA) gene variation in polycystic ovary syndrome in a Tunisian women population. BMC Genomics (2016) 17(Suppl. 9):748. 10.1186/s12864-016-3092-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewontin RC. On measures of gametic disequilibrium. Genetics (1988) 120:849–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science (2002) 296:2225–9. 10.1126/science.1069424 [DOI] [PubMed] [Google Scholar]
- 32.Naghavi A, Mozdarani H, Garshasbi M, Yaghmaei M. Prevalence of methylenetetrahydrofolate reductase C677T polymorphism in women with polycystic ovary syndrome in southeast of Iran. J Med Life (2015) 8:229–32. [PMC free article] [PubMed] [Google Scholar]
- 33.Hegele-Hartung C, Siebel P, Peters O, Kosemund D, Muller G, Hillisch A, et al. Impact of isotype-selective estrogen receptor agonists on ovarian function. Proc Natl Acad Sci USA. (2004) 101:5129–34. 10.1073/pnas.0306720101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nectaria X, Leandros L, Ioannis G, Agathocles T. The importance of ERalpha and ERbeta gene polymorphisms in PCOS. Gynecol Endocrinol. (2012) 28:505–8. 10.3109/09513590.2011.649811 [DOI] [PubMed] [Google Scholar]
- 35.Valkenburg O, Uitterlinden AG, Themmen AP, de Jong FH, Hofman A, Fauser BC, et al. Genetic polymorphisms of the glucocorticoid receptor may affect the phenotype of women with anovulatory polycystic ovary syndrome. Hum Reprod. (2011) 26:2902–11. 10.1093/humrep/der222 [DOI] [PubMed] [Google Scholar]
- 36.He W. PPARgamma2 polymorphism and human health. PPAR Res. (2009) 2009:849538. 10.1155/2009/849538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zaki M, Hassan N, El-Bassyouni HT, Kamal S, Basha W, Azmy O, et al. Association of the Pro12Ala polymorphism with the metabolic parameters in women with polycystic ovary syndrome. Open Access Maced J Med Sci. (2017) 5:275–80. 10.3889/oamjms.2017.088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chorev M, Carmel L. The function of introns. Front Genet. (2012) 3:55. 10.3389/fgene.2012.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li S, Maruo T, Ladines-Llave CA, Kondo H, Mochizuki M. Stage-limited expression of myc oncoprotein in the human ovary during follicular growth, regression and atresia. Endocrine J. (1994) 41:83–92. 10.1507/endocrj.41.83 [DOI] [PubMed] [Google Scholar]
- 40.Genomes Project C, Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature (2010) 467:1061–73. 10.1038/nature09534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Higgins GA, Allyn-Feuer A, Handelman S, Sadee W, Athey BD. The epigenome, 4D nucleome and next-generation neuropsychiatric pharmacogenomics. Pharmacogenomics (2015) 16:1649–69. 10.2217/pgs.15.111 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.