Abstract
Staphylococcus aureus is an opportunistic pathogen that causes a range of serious infections associated with significant morbidity, by strains increasingly resistant to antibiotics. However, to date all candidate vaccines have failed to induce protective immune responses in humans. We need a more comprehensive understanding of the antigenic targets important in the context of human infection. To investigate infection-associated immune responses, patients were sampled at initial presentation and during convalescence from three types of clinical infection; skin and soft tissue infection (SSTI), prosthetic joint infection (PJI) and pediatric hematogenous osteomyelitis (PHO). Reactivity of serum IgG was tested with an array of recombinant proteins, representing over 2,652 in-vitro-translated open reading frames (ORFs) from a community-acquired methicillin-resistant S. aureus USA300 strain. High-level reactivity was demonstrated for 104 proteins with serum IgG in all patient samples. Overall, high-level IgG-reactivity was most commonly directed against a subset of secreted proteins. Although based on limited surveys, we found subsets of S. aureus proteins with differential reactivity with serum samples from patients with different clinical syndromes. Together, our studies have revealed a hierarchy within the diverse proteins of the S. aureus “immunome”, which will help to advance efforts to develop protective immunotherapeutic agents.
Introduction
Staphylococcus aureus is an opportunistic bacterial pathogen that causes a range of serious infections associated with significant morbidity, with 10,000 deaths per year in the US alone1–3. S. aureus is also a common commensal microorganism, chronically colonizing approximately 30 percent of adults, with the remainder intermittently colonized4,5. Antibiotic resistant strains, including methicillin-resistant S. aureus (MRSA), have become more prevalent, especially in community settings6, and increasing resistance to other commonly used antibiotic treatments has also been documented7,8. Mobile genetic elements enable efficient horizontal transfer of antibiotic resistance genes and other virulence factors9,10, resulting in the rapid genetic diversification of S. aureus strains11. Due to this genomic plasticity12, coupled with the slow development of new antibiotics for clinical use, there has been greatly increased interest in the development of vaccines and therapeutic immunotherapies directed against S. aureus.
The need for an effective vaccine is well recognized, and thus a range of S. aureus surface antigens, as well as whole organism preparations, have been evaluated13–15. Yet despite extensive efforts to develop vaccines against S. aureus, a broadly protective clinical vaccine has not been validated in controlled clinical trials16, as none has imparted protective immunity from serious infections, while in some cases vaccination has led to worse outcomes17. In part, this may be due to our current incomplete understanding of host-commensal/pathogen relationships and the range of S. aureus products that can represent antigenic targets for host immune defenses.
Candidate vaccines for S. aureus are generally first validated in mouse models, before a clinical trial is even considered. Yet murine models of infection display important differences from human infections, and therefore may have inherent issues for the identification of key features of human immunity18. Indeed, the set of immunodominant antigens recognized in mouse infection may not accurately identify antigens critical for controlling human infections19. Interpretation of animal model data is further complicated by evidence that chronic carrier states and recurrent S. aureus infections are common in humans, and these exposures do not induce immune responses sufficient to protect from subsequent infections20. Furthermore, protection from specific types of clinical infection syndromes may require antibody responses to different sets of staphylococcal antigens. Thus, it is essential that we perform more complete interrogations of immune responses in patients with different clinical syndromes.
To help guide the development of effective immune protective and therapeutic agents, we sought to perform unbiased surveys of human immune responsiveness to all potential protein antigens encoded by the genome of epidemic community-acquired MRSA (CA-MRSA) strain USA300. Our goal was to better understand which S. aureus proteins are recognized during human infection, as well as those which are rarely or never recognized by our immune systems. We postulated that the results from these simple surveys would in part provide an essential step in the assembly of an effective combinatorial vaccine.
S. aureus can cause different clinical infection syndromes, which in part may result from expression of different virulence factors by the infecting strain21–23. To identify whether infections commonly lead to antibody responses to different sets of S. aureus proteins, we used serum samples, collected at acute and convalescent time points, from representative patients with: adult skin and soft tissue infection (SSTI), adult prosthetic joint infection (PJI), and pediatric hematogenous osteomyelitis (PHO). To investigate which open reading frames (ORFs) can encode for immunogenic proteins, we used solid-phase arrays printed with in vitro-translated recombinant proteins from every ORF from a prototypic clinical MRSA strain. This view of the reactivity across all ORFs is enhanced by the use of longitudinal samples from human patients with a variety of clinical syndromes, providing a novel and comprehensive view of what the immune system is recognizing from S. aureus. From these unbiased investigations, we identified a hierarchy of antigenic proteins from S. aureus, which are recognized by the immune systems of individuals recovering from S. aureus infections.
Results
Selection of representative patients with serum antibody responses against common S. aureus antigens
To investigate patterns of immune responsiveness to the S. aureus immunome, we recruited a total of 95 patients with S. aureus infection from three cohorts; adults with SSTI (n = 55) or PJI (n = 12), and a pediatric cohort (n = 28) with hematogenous osteomyelitis. For each patient in the study, we also recovered the infecting S. aureus isolate, and screened for colonization of the nares and groin.
For the initial characterization of patient immune responses, serum samples from initial clinical presentation and follow-up visits were used to quantitate IgG reactivity with 46 recombinant antigens, including S. aureus proteins and control antigens (Supplementary Table S1). We prioritized male patients from each of the three clinical cohorts for further study based on detection of increased IgG-reactivity with three or more S. aureus antigens in the short-term follow-up blood sample compared to the baseline sample (Supplementary Fig. S1). These representative patients ranged from 9 to 63 years of age (median 39 years). Of the seven selected patients, four were from the SSTI cohort, one from the PJI cohort, and two from the PHO cohort (Table 1).
Table 1.
Patients selected for S. aureus protein array analysis.
| Patient ID | Age | Gender | S. aureus cohort | Other medical conditions | Prior S. aureus infection^ | Prior antibiotic course | Days Infected Prior to Visit 1* | Days from Visit 1 to Visit 2 | Days from Visit 1 to Visit 3 |
|---|---|---|---|---|---|---|---|---|---|
| SSTI 1 | 54 | Male | SSTI | Seizures, HCV | No | No | 3 | 37 | 178 |
| SSTI 2 | 25 | Male | SSTI | None | No | Yes | 5 | 48 | 195 |
| SSTI 3 | 61 | Male | SSTI | HCV, pulmonary fibrosis, hypothyroidism | No | No | 3 | 39 | 207 |
| SSTI 4 | 50 | Male | SSTI | None | No | No | 4 | 41 | 173 |
| PJI 1 | 63 | Male | Prosthetic Joint (PJI) | Peripheral vascular disease, chronic liver failure, ulcerative colitis, alcoholic cirrhosis, recurrent C. difficile | N/A | N/A | N/A | 45 | 183 |
| PHO 1 | 9 | Male | Septic Arthritis (PHO) | Eczema | N/A | N/A | N/A | 27 | 190 |
| PHO 2 | 11 | Male | Osteomylitis (PHO) | None | N/A | N/A | N/A | 41 | 223 |
Patient identification numbers, as well as their age, gender, S. aureus proven syndrome, any co-morbid medical conditions are included for all patients selected for the protein array, in addition to the number of days between acute clinical presentation and both the short-term and long-term follow-up visits. Patients in the SSTI cohort provided additional information such as prior and subsequent S. aureus infections or antibiotic courses prior to enrollment, as well as patient-reported duration of symptoms that preceded the day of enrollment. N/A: Data not available. ^This designation was assigned based on the patient’s statement that there was no history provided of an infection requiring attention from a healthcare professional, and a course of antibiotics. *Patient reported, days with symptoms prior to hospital visit.
Differential serum IgG reactivity within the S. aureus proteome
We next sought to characterize each of the patient serum samples for the presence of IgG-antibodies against S. aureus antigens in our solid-phase printed immunome arrays. Each array included 2,652 ORF-encoded recombinant proteins from epidemic CA-MRSA strain USA300, and each ORF product was individually ranked based on mean reactivity with IgG across a total of 21 patient visit serum samples (Supplementary Fig. S2). An ORF-encoded protein was classified as immune reactive if the associated IgG signal was at least two-fold above the background level of control spot fluorescence on an array, as described24,25. Whereas most ORF protein products were devoid of significant IgG-reactivity with any of the samples tested (Supplementary Table S2), 1,086 of these 2,652 polypeptides instead displayed IgG-reactivity with one or more serum sample (Supplementary Table S3).
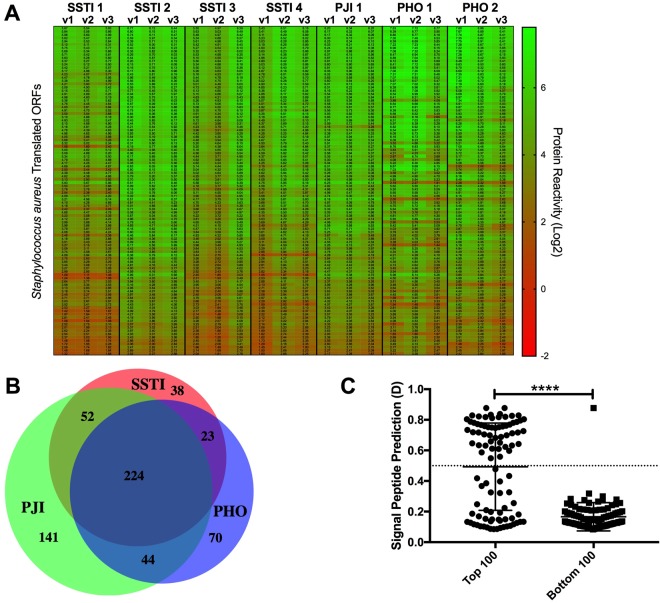
To identify the most immune-reactive S. aureus proteins, we ranked all of the ORF products by their relative IgG reactivity with all of the serum samples, with reactivity shown for the top 100 shown in Fig. 1A and listed in Supplementary Table S4, and the top 50 most IgG-reactive proteins (Table 2). Although there was some variability in reactivity between patients, we found more limited differences in samples obtained at different time points from in an individual (Fig. 1A). We thereby found an overall hierarchy amongst these ORFS, with a defined subset that were IgG-reactive across each of the different subjects from the three infection types that were evaluated.
Figure 1.
Hierarchy of reactivity for human serum IgG to antigens from S. aureus USA300 MRSA in different clinical infection types. Relative IgG binding reactivity was measured using a chip-based method for 2,652 ORFs in the S. aureus FPR3757 reference genome for seven patients, with SSTI (n = 4), PJI (n = 1), and PHO (n = 2) at three different time points of infection (acute, short term follow-up, and long-term follow-up). (A) Proteins were ranked based on overall mean IgG-antibody reactivity within patient serum samples. The 100 proteins with the top IgG reactivity proteins with each of the 21 serum samples are depicted. (B) The mean antibody reactivity value for each infection group (SSTI, PJI and PHO) across all the samples was determined, and antigens with mean reactivity of 1.0 or greater (base 2) were included in the analysis. (C) ORF sequences from the top and bottom 100 antigens identified in the protein array were analyzed with the SignalP Server and the Signal Peptide Prediction value, termed D, was obtained. A cutoff of D = 0.5 or greater predicted there was a signal peptide associated with the particular ORF. The top and bottom 100 IgG-reactive ORF-encoded antigens displayed significant differences (p < 0.0001) in the representation of predicted signal peptides.
Table 2.
50 most IgG-reactive S. aureus antigen by protein immunoarray.
| No. | Locus Tag | Protein Name/Description | Signal Peptide | Transmembrane |
|---|---|---|---|---|
| 1 | SAUSA300-0113 | immunoglobulin G binding protein A (SpA) | + | + |
| 2 | SAUSA300-0398 | superantigen-like protein SSL4 | + | − |
| 3 | SAUSA300-0403 | superantigen-like protein SSL9 | + | − |
| 4 | SAUSA300-2366 | gamma-hemolysin component C (hlgC) | + | − |
| 5 | SAUSA300-2579 | N-acetylmuramoyl-L-alanine amidase domain protein | + | − |
| 6 | SAUSA300-0883 | MAP domain-containing extracellular adherence protein Eap | + | − |
| 7 | SAUSA300-2364 | IgG-binding protein SBI (sbi) | + | − |
| 8 | SAUSA300-1029 | Iron-regulated surface determinant Protein A (IsdA) | + | + |
| 9 | SAUSA300-1028 | Iron-regulated surface determinant Protein B (IsdB) | + | + |
| 10 | SAUSA300-1920 | chemotaxis-inhibiting protein CHIPS (chs) | + | − |
| 11 | SAUSA300-1382 | Panton-Valentine leukocidin, LukS-PV (lukS-PV) | + | − |
| 12 | SAUSA300-2367 | gamma-hemolysin component B (hlgB) | + | − |
| 13 | SAUSA300-1768 | leukotoxin LukD (lukD) | + | − |
| 14 | SAUSA300-1917 | map protein, programmed frameshift (map) | + | − |
| 15 | SAUSA300-0651 | peptidase M23 | + | − |
| 16 | SAUSA300-0951 | V8 protease (sspA) [3.4.21.19] | + | − |
| 17 | SAUSA300-0408 | heme uptake protein | + | − |
| 18 | SAUSA300-0438 | CHAP domain family | + | − |
| 19 | SAUSA300-0409 | peroxidase inhibitor | + | + |
| 20 | SAUSA300-0693 | electron transfer DM13 | + | − |
| 21 | SAUSA300-0214 | Sugar phosphate isomerase/epimerase PFAM family PF07582 | − | − |
| 22 | SAUSA300-2506 | immunodominant staphylococcal antigen A precursor (isaA) | + | − |
| 23 | SAUSA300-1058 | alpha-hemolysin precursor (HLA) | + | − |
| 24 | SAUSA300-0548-s2 | sdrE protein (sdrE) | + | + |
| 25 | SAUSA300-0776 | thermonuclease precursor (nuc) [3.1.31.1] | + | + |
| 26 | SAUSA300-0547-s2 | sdrD protein (sdrD) | + | + |
| 27 | SAUSA300-2572 | zinc metalloproteinase aureolysin (aur) [3.4.24.29] | + | − |
| 28 | SAUSA300-0370 | SEP staphylococcal enterotoxin | − | − |
| 29 | SAUSA300-1030 | iron transport associated domain protein | + | + |
| 30 | SAUSA300-1476 | acetyl-CoA carboxylase, biotin carboxyl carrier protein (accB) | − | − |
| 31 | SAUSA300-1922 | staphylokinase precursor (sak) | + | + |
| 32 | SAUSA300-0862 | glycerophosphoryl diester phosphodiesterase (glpQ) [3.1.4.46] | + | − |
| 33 | SAUSA300-1055 | fibrinogen-binding protein (efb) | + | − |
| 34 | SAUSA300-1481 | hypothetical mating channel protein | + | + |
| 35 | SAUSA300-0955-s1 | autolysin (atl) [3.5.1.28] | + | − |
| 36 | SAUSA300-0774 | secretory extracellular matrix, plasma binding protein (empbp) | + | − |
| 37 | SAUSA300-0950 | cysteine protease precursor (sspB) [3.4.22.48] | + | + |
| 38 | SAUSA300-1381 | Panton-Valentine leukocidin, LukF-PV (lukF-PV) | + | − |
| 39 | SAUSA300-2441 | fibronectin binding protein A (fnbA) | + | − |
| 40 | SAUSA300-2136 | iron compound ABC transporter, iron compound-binding protein | + | − |
| 41 | SAUSA300-2440 | fibronectin binding protein B (fnbB) | + | − |
| 42 | SAUSA300-0242 | sorbitol dehydrogenase (gutB) [1.1.1.14] | − | − |
| 43 | SAUSA300-1985 | serine-aspartate repeat family protein, SdrH (sdrH) | + | + |
| 44 | SAUSA300-0279 | membrane protein YhgE, type VII secretion protein EsaA) | − | + |
| 45 | SAUSA300-0708 | histidinol-phosphate aminotransferase (hisC) [2.6.1.9] | − | − |
| 46 | SAUSA300-0399 | superantigen-like protein SSL5 | + | − |
| 47 | SAUSA300-0955-s2 | autolysin (atl) [3.5.1.28] | + | − |
| 48 | SAUSA300-0395 | superantigen-like protein SSL1 | + | − |
| 49 | SAUSA300-1031 | heme ABC transporter permease | − | + |
| 50 | SAUSA300-1512 | penicillin-binding protein 3 (pbp3) | − | + |
IgG reactivity for each antigen was determined across all patients and samples, with ranking determined by overall mean reactivity, in addition to the presence of reactivity in each patient at each time point (see Methods). Numbers in brackets [] represent the enzyme commission designation (see www.genome.jp/kegg/).
IgG reactivity correlated in part with structural domain predictions
For each infection group, we calculated the average reactivity for each individual at each time point compared to the other infection groups. An average reactivity of two-fold over background was considered a positive immune response. The specific proteins associated with each infection group were identified, and the average reactivity was estimated (Supplementary Table S5). A subset of antigens was reactive in individuals from all the three infection groups (n = 224), whereas other antigens (n = 253) were reactive with sera from only one of the three infection groups (Fig. 1B). Subsets of proteins which were the most highly IgG-reactive were identified for each of the clinical infection syndromes. Although only a limited number of patients per clinical group were studied, amongst the most highly IgG-reactive proteins we identified subsets of proteins that uniquely distributed with each of the different infection groups (Supplementary Table S5).
Although different proteins were associated with different clinical syndromes, secreted virulence factors were generally highly represented at the top of these hierarchies (Table 2). Notably, SSTI syndrome alone was associated with reactivity to a specific cell surface protein26, encoded by SAUSA300-1327 (Supplementary Table S5). This gene encodes for extracellular matrix-binding protein or ebh27, which contains many FIVAR (Found In Various Architectures) protein domains that are postulated to bind fibronectin and N-acetyl glucosamine in the host extracellular matrix28. Notably, due to the large size of the predicted ORF, 16 segments were separately printed on the array, and we found that seven of these ebh polypeptides were recognized by antibodies in the serum of one or more of the SSTI patients (Supplementary Table S5).
We next analyzed the genotypes of the infecting isolates from the representative patients chosen for this study. Five out of the seven patients were infected with MRSA isolates (Supplementary Table S6), while the two PHO patients were instead infected with MSSA. We also examined the phylogenetic relationship between the patient infecting S. aureus isolates and the reference strain, FPR3575, used in the array (Supplementary Fig. S3). Curiously, based on our genome analyses the isolates from each infection group did not cluster together. We then compared the ORF sequence homology of the 100 top IgG-reactive proteins in each of the 7 infecting strains and the reference USA300 strain used in the array by BLAST identity (Supplementary Fig. S4). The results indicate that the isolates are polymorphic, and among the top 100 proteins there are many cases in which an isolate either does not contain a homologous protein or contain a variant of the protein with differences in sequence. Thus, we detected sequence variations between isolates and in synonymous ORFs that could affect recognition of associated epitopes.
Among USA300 antigens that were highly reactive with sera from all 7 patients, we screened antigen sequences for the presence of a secretion signal peptide or transmembrane domains. As expected, ORFs with signal peptides were more common amongst the top 100 IgG-reactive proteins (Table 2) compared to the 100 with lowest reactivity levels (p < 0.0001, Fig. 1C). In contrast, we found that ORFs predicted to contain transmembrane domains were not differentially represented in the top and bottom IgG-reactive sets of potential antigens (Supplementary Fig. S5). The representation of motifs predictive of transmembrane domains therefore did not correlate with the level of IgG-reactivity. Furthermore, many of the genes for proteins that were non-reactive with circulating IgG (Supplementary Table S2), were associated with intracellular bacterial functions, such as ribosome-binding factor A (rbfA – SAUSA300-1163) or 30S ribosomal protein S14 (rpsN – SAUSA300-2191). Collectively, these observations suggest that antibody reactivity was primarily against secreted antigens that likely become accessible for immune recognition by the host.
Amongst the top antibody-reactive proteins, the IgG-binding proteins, Staphylococcal Protein A (SpA) and Staphylococcal binding immunoglobulin protein (Sbi) ranked first and seventh for overall reactivity, respectively. Yet due to the non-immune nature of the binding specificities of these virulence factors29,30 the results for these microbial products must be deemed as not informative for investigations of host immune responses.
Concordance of IgG-antibody reactivity in solid-phase printed protein and bead-based assays
As immunoreactivity may in part be affected by technical features of the assay, we also assessed whether the level of IgG reactivity for a given S. aureus antigen in the chip-based approach correlated with the level of S. aureus expressed antigen reactivity in a multiplex bead-based assay system. Notably, the bead-based assay detected IgG reactivity in a generally broader dynamic range, and for many Leukocidin family members this enabled the detection of longitudinal increases in IgG antibody responses in the sequential samples from each of these selected patients (Supplementary Fig. S1). In general, although specific reactivity values differed, we found that secreted toxins, especially members of the Leukocidin family, were amongst the most immunogenic S. aureus antigens by both approaches (Table 2).
Finally, we compared our results to available data on the level of S. aureus gene expression during clinical infection that is no doubt an important determinant of the potential immunogenicity of individual S. aureus genes. We used data reported for S. aureus gene expression levels determined from ex vivo analyses of mRNA isolated from active clinical cutaneous abscesses, as reported by Date et al.31. Gene expression data from this study was normalized with levels detected in in vitro cultures of the same S. aureus strain (USA300) that was used as the source of proteins in our microarray. Our analyses found that during active infection 26 of our top 50 immune-reactive proteins also displayed two-fold or greater increases in gene-specific transcript expression (Supplementary Fig. S6). Interestingly, a prominent member of the Leukocidin family, LukF-PV, displayed an 89-fold increase in RNA levels in skin abscesses compared to laboratory culture31. Thus, antigen reactivity in our system correlated with in vivo bacterial gene expression during clinical infection, which strongly supported the relevance of our methodological approach for the identification of S. aureus proteins involved in host immune responses. In summary, based on recognition by host serum IgG we have identified a hierarchical set of staphylococcal antigens, and found that a limited group of these microbial products appear to be immunologically targeted in patients with a range of clinical infection syndromes (Fig. 1B).
Discussion
Herein, we report findings from an unbiased approach to identify antigens associated with different types of S. aureus clinical infections. Using an antigen array of all ORFs in a USA300 CA-MRSA clinical strain, we measured serologic IgG reactivity against every potential staphylococcal protein and found that only a small subset of total ORFs encoded for proteins that are immunologically-recognized across all infection types. In addition, we detected IgG-binding reactivity in one or more serum sample with nearly half of the S. aureus ORF protein products; 1,028 out of the 2,652 tested, which at first glance represents an overwhelming number of potential antigenic targets (Supplementary Table S3).
Among the top IgG-reactive antigens were the subunits of Panton-Valentine Leukocidin (i.e., LukS/F-PV). The heterodimeric S. aureus antigen LukS/F has been reported to be highly associated with SSTI21,32. In our comparisons in disease syndromes, the genes for LukS and LukF were amongst the most up-regulated31 and these also appeared to be amongst the most immunogenic targets in patients with skin abscess (Supplementary Fig. S6). Notably, although IgG responses to LukS and LukF were highly up-regulated in all patients studied, only isolates from SSTI patients contained genes for the LukS/F genes (Supplementary Table S6). This could reflect persistent responses from prior exposure to a strain that produced these gene products. Alternatively, this could also reflect the IgG cross-reactivity with epitopes expressed by different members of the Leukocidin family, which reiterates recently reported findings from more focused studies of a larger number of patients33.
In several important instances, reactivity documented in the printed-arrays also correlated with post infection increases in antigen-reactive IgG titers demonstrated in our multiplex bead-based analysis. In particular, LukS-PV ranked at number 11 and LukF-PV, the partner subunit in the heterodimeric protein, ranked at 38 in overall IgG reactivity in these array studies (Table 2), and very similar patterns of increases in antibody reactivity over time were demonstrated in our bead-based assay (Supplementary Fig. S1). In a recent study we showed that infection can induce raised antibody responses to a number of S. aureus gene protein products, but such enhanced immune responses to infection do not generally persist long-term33. The technical approach used in the current report enables more complete antigenic surveys with more sensitive detection of IgG binding. By including sera from a follow-up visit at about 6-weeks that was after a course of antibiotics, we ensured the best detection of even transiently enhanced responses to staphylococcal proteins. Furthermore, proteins with the greatest IgG reactivity were amongst the most highly expressed during cutaneous infection based on gene expression data (Supplementary Fig. S6). Cumulatively these findings suggest that our immune systems often have exuberant, yet transient, responses to specific virulence factors that are associated with highly up-regulated expression during S. aureus infection of the human host.
Among the most reactive recombinant proteins identified were SpA and Sbi, which are IgG-binding proteins implicated as countermeasures that actively subvert host defenses30,34,35. SpA, a virulence factor that is both expressed on the bacterial surface and secreted36, promotes immune evasion due to its ability to bind the Fc portions of IgG antibodies, interfering with opsonophagocytic clearance. SpA also has a separate binding site for Fab encoded by VH3-family genes that convey the properties of a VH-targeted B-cell superantigen37,38. In our study, SpA, which was identified as the most IgG-reactive recombinant S. aureus protein in all serum samples (Table 2). Although it is unclear from these surveys whether the high reactivity comes from the binding of the Fab or Fc portions of the IgG antibodies, it provided a validating technical control for detection of IgG reactivity. Notably, Iron-regulated surface determinant Protein B (IsdB) was also identified within the top 10 IgG reactive proteins as well as the related IsdA (Table 2). In a recent report, NEAT1 and NEAT2 domains of IsdB were associated with a different type of non-immune Fab-binding interaction specific for germ-line VH gene segment CDR2 motifs that may reflect co-evolution of the human immune system with this opportunistic microbial pathogen39.
Many other S. aureus genes, including several in our top 100 list for IgG-reactivity, are currently annotated as hypothetical proteins of unknown structure (Table 2, Supplementary Table S3) due to current uncertainty regarding their biologic roles. However, the detection of IgG reactivity may suggest these proteins are expressed in the context of infection. Further investigations of the structure and function of “hypothetical” proteins with prominent IgG reactivity may illuminate currently obscure facets of the pathogenesis of S. aureus infection. Notably, SSTI syndrome alone was associated with reactivity for an extracellular matrix-binding protein, ebh, encoded by SAUSA300-1327 (Supplementary Table S5), which should be further considered as a candidate antigen in an experimental vaccine.
A previous report of the immunome associated with S. aureus infection assessed reactivity of serum antibodies against staphylococcal antigens using 2-D immunoproteomics and mass spectrometry, which by comparison are time and labor-intensive approaches40. In one recent paper, a printed array-based strategy similar to ours was used but only 44 antigens examined41. Other studies have used ELISA or bead-based arrays to assess responses to much more limited sets of selected S. aureus antigens15,42–44. Whereas Etz et al. described surveys of the S. aureus genome that assessed binding to short S. aureus gene product fragments displayed on bacterial cells, few potential antigens, including a coagulase, as well as SpA, were identified45. By comparison, our studies used a much more comprehensive array of in vitro-translated antigens, representing products of the entire genome of a clinical isolate of S. aureus, with a method amenable to high-throughput analyses. We thereby identified antigens that were recognized immunologically across several groups of patients with different types of clinical infection. We have also identified the large set of staphylococcal antigens that are not reactive with human serum IgG antibodies. Taken together, we have provided a comprehensive overview of the immunome during the acute phase and during the resolution of S. aureus infection.
The design of this methodological approach also involved a level of compromise as there are several inherent technical limitations. First, although our microarray studies were unbiased and comprehensive, these are based on a single reference strain (USA300, FPR3757) and therefore could not detect antigens not represented in this strain. There is such large diversity of gene composition between S. aureus strains that it would be very difficult to generate a protein array containing all potential ORFs found in any and all S. aureus strain, therefore this single reference genome was chosen. Likewise, in some cases the microarray may not have been able to detect antibody binding to antigen variants unique to other infecting strains, which may be polymorphic from the microarray reference strain (Supplementary Fig. S4). Second, in these studies of in vitro translated S. aureus ORFs printed onto the microarray we could not control for altered protein folding and conformation, or post-translational modifications that could affect the range of epitope expression. Third, compared to results from our bead-based multiplex assay, we found that there was a more limited dynamic range for antibody reactivity in the printed protein array studies, and binding levels were more quickly saturated for recombinant proteins encoded by the same gene sequence33. Therefore, we chose to use a ranking based hierarchy method, instead of using values from a semi-quantitative method that we felt would have less inherent utility. Using a ranking based method, we are able to normalize for differences between patients from overall average MFI for IgG binding. For practical reasons, for each patient and time point we used a single dilution of serum (1:2) and based on the absence of truly naïve human serum we could not otherwise normalize serum concentrations across the time points of patients and between patients. Inherent to the technical approach of the printed protein microarray, non-protein antigens were outside of the scope of our study, as this report was meant to test the potential utility of this printed array technical approach to aid in prioritizing potential protein antigens. In the future, these studies will be extended to larger patient groups and studies of ORF products from multiple strains.
We emphasize that S. aureus is an opportunistic pathogen, and a common commensal colonizer of the nares and cutaneous sites, with evidence of near universal immune exposure after infancy. We therefore focused on studies of patients with infection-associated increases in antibody responses during subsequent visits. In fact, all of the adult patients we investigated had significant IgG anti-Staphylococcal antibody reactivity at the time of recruitment, and it is therefore unknown whether such responses are from the current infection or from prior exposures. Furthermore, structurally homologous toxins expressed during the active infection could induce cross-reactive antibodies to antigens that themselves are not actively being expressed, such as the Panton-Valentine (i.e., LukS/F) exotoxin mentioned previously, which had high IgG-reactivity in the immunoassay even though we found that the gene was only present in 4 out of the 7 patients tested (Supplementary Fig. S4, Supplementary Table S6)33. In addition to the Leukocidin toxins, cross-reactivity has been reported within other antigenic groups like the S. aureus superantigen family46. While our studies focused on elucidating the reactivity of IgG antibodies with the S. aureus immunome, we are aware that other isotypes may also be important, and especially IgA antibodies that play essential roles at mucosal surfaces. Despite these caveats, our investigations were informative and shed light on the immunoreactivity patterns that were shared, or which varied, between different subjects, and in those with different clinical syndromes.
In part, we initiated these studies as past vaccine candidates have not met their primary endpoints47,48. The failure of past vaccine attempts could be due to the S. aureus antigens chosen for these vaccines, and that in many cases only single proteins were used. We therefore initiated the current studies because we believed it was time to take a step back and perform unbiased surveys of a more complete set of S. aureus antigens that may be recognized by serum antibodies from well-characterized patients recovering from different clinical infection syndromes.
Our studies have provided an overview of the immunologic potential of individual antigens within the S. aureus genome, which we hope will contribute to the advancement of vaccine research for this pathogen. From our findings, we postulate that the proteins associated with strong antibody immunoreactivity in all convalescent patients may be relevant to the development of a broad-reaching vaccine, whereas proteins with immunoreactivity associated with only certain infection groups may reflect features deriving from mechanisms of pathogenicity of the associated infection syndrome that at times may be linked to specific infecting strains of S. aureus. As S. aureus genomes shows great genetic diversity and plasticity, efforts such as ours to objectively characterize levels of antigenicity of the broad range of staphylococcal proteins, in the context of human infection, are critical for the development of effective clinical vaccines.
Methods
Patient cohort
Patients were enrolled and informed consent was obtained following IRB approval under institutional supervision at three university medical centers: SSTI and uninfected adult controls at Bellevue Hospital and NYU Tisch Hospital; Prosthetic Joint Infection (PJI) patients and uninfected controls at the Hospital for Special Surgery (HSS); and pediatric patients with hematogenous osteomyelitis (PHO) at Vanderbilt Medical Center. Blood samples and cultures were obtained from colonizing (nares, groin) and infecting sites in patients. Eligibility was confirmed by culture-based documentation of S. aureus infection by the clinical microbiology laboratory in accordance with IRB protocols. All S. aureus isolates were preserved for genomic characterization. Genotyping involved a combination of standard molecular methods including spa (staphylococcal protein A) and SCCmec typing and screening for presence of pvl (Supplementary Table S6)49. For analysis of antigen polymorphism, DNA from S. aureus isolates was extracted as described50,51. DNA from each patient isolate was then sequenced to a depth of approximately 150X with 150 base paired-end reads on the Illumina HiSeq4000. Adapters and low-quality bases were trimmed from the ends of reads using Trimmomatic v 0.3352. Draft genomes were assembled from the sequence data for each isolate using SPAdes v 3.9.053. All genome assemblies have N50 > 100 Kb and (sum of contigs) genome length from 2.8 to 3.1 Mb.
At initial presentation to the hospital with symptoms patient blood samples and complete blood counts were obtained, then patients received a course of antibiotics as determined by their physicians. Further blood samples were recovered at short-term (after 27–48 days), and long-term follow-up (after 173–223 days). Among the data collected were patient reports of past exposure to S. aureus and infections, co-morbidities, and past antibiotic usage. Seven S. aureus patients, chosen from a larger cohort (n = 95) for analysis of serum antibody responses, based on evidence of induction of increased serum IgG responses to S. aureus antigens during convalescence33 (Supplementary Fig. S1). Four of the patients had SSTI, one had PJI, and two had PHO. Three serum samples from each of the seven S. aureus infected patients (obtained at initial presentation, short-term follow-up, and long-term follow-up) were used in the analysis.
Bead-based multiplex immunoassays
To evaluate antigen-specific IgG-antibody reactivity, immunoassays were performed with cocktails of beads, each identified via internal fluorophores (MagPlex microspheres, Luminex). Each bead was coupled to an individual recombinant S. aureus protein, expressed in S. aureus, or control protein/ analyte (Supplementary Table S1), adapting previously described methods54. Assays were performed on serum sample at 1:100, 1:1000, 1:10,000 and 1:100,000 dilutions, with IgG binding detected with a PE-labeled goat anti-Human IgG Fc secondary antibody (eBioscienceTM, CAT:12-4998-82), using the MagPix xMap (Luminex) platform. Custom software was used to generate binding curves for each antigen, from which comparisons to the acute visit baseline sample were presented as fold-change in IgG titer.
Solid-phase recombinant protein printed microarrays
Following their previously developed standard protocol (Antigen Discovery Inc.). 2,652 open reading frames (ORFs) from the USA300 genotype MRSA strain, FPR3757, were expressed in vitro, and printed onto nitrocellulose coated slides (for strain details, see www.atcc.org/Products/All/BAA-1556). 34 ORFs were split into a varying number of segments based on the length of the ORF. These are annotated SAUSA300-0000-s1 as an example for segment one of an ORF. Individual replicate slides were then incubated with blocking buffer. The patient samples were each diluted 1:100 in blocking buffer and incubated with a replicate slide, which were washed, and secondary PE-conjugated goat anti-human IgG (Fc gamma-specific) was added. After washing, slides were then scanned using Genepix 4300 scanner (Molecular Devices), with data analyzed using Genepix software with a validated protocol. IgG-binding data were normalized against control negative regions on the slide and log2 transformed, with the normalized value of 0 indicating no difference compared to background, and 1.0 indicating twice the signal compared to background.
Data analysis
Normalized data were used to rank immune reactivity for each antigen, with further analysis using MATLAB® R2016b (MathWorks) and GraphPad Prism7® (Instat). Signal-peptide containing ORFs were identified with the CBS Prediction Server SignalP 4.1 (www.cbs.dtu.dk/services/SignalP/), by assignment of a Signal Peptide Prediction value (D), and a cutoff value of D = 0.5 or greater was used to identify the ORFs with signal sequences as previously described55,56. To identify transmembrane ORFs, we used the CBS Prediction Server TMHMM Server v 2.0 (www.cbs.dtu.dk/services/TMHMM/) to predict the number of transmembrane helices as previously described57,58. Significance between signal peptide containing proteins within the top 100 reactive with serum IgG (Fig. 1C), as well as for transmembrane containing proteins (Supplementary Fig. S5), was determined using a two-tailed Mann-Whitney test. Venn Diagram was made using BioVenn59.
Gene content was assayed by translated BLAST search of the draft genome contigs for each isolate, using as queries, protein sequences from the annotated genome of strain USA300 FPR-3757. The draft genome contigs from each isolate were built into individual BLAST databases then searched with each of the query proteins. The percent amino acid identity of the best match for each protein to each genome is reported. Proteins that match the draft genome with less than 50 percent amino acid identity are clearly absent (the best match is to a distantly related protein). Matches with greater than 90 percent identity are clearly present as closely related isoforms. Matches in the 50–90 percent range represent either distant gene isoforms or closely related members of a multi-gene family.
The phylogenetic tree was calculated using the shared kmer distance method as implemented in the AAF software v 20171001 with kmer size of 25. Assembled draft genome contigs were used as genome data for each isolate and the GenBank draft genome assembly for strain USA300 FPR-3757 (GCA-000013465) was used as a reference60. Phylogenetic tree drawings were made using FigTree v 1.4.3.
Electronic supplementary material
Acknowledgements
We thank Kayan Tam, Alexis Sommerfield and William Sause in the Torres lab for cloning, purifying and providing several of the S. aureus recombinant antigens, Adriana Heguy and the NYU Genome Technology Center, and Krista Trappl, Joe Campo and Angela Yee from Antigen Technology Inc. This work was supported through the NIH/NIAID contract HHS N272201400019C, “B Cell Epitope Discovery and Mechanisms of Antibody Protection in Staphylococcus aureus”, and T32 GM066704 (E.E.R.).
Author Contributions
G.J.S. conceived and designed the experiment; I.P.T., W.K.C. and A.O.M assisted in patient recruitment; E.E.R., G.J.S. and S.M.B. analyzed the results; E.E.R. wrote the manuscript; and G.J.S, A.J.P., B.S., S.M.B., I.P.T., Y.F., D.N.H. and V.J.T. reviewed the manuscript.
Data Availability
The datasets generated and analyzed in the current report are available from the corresponding author on reasonable request.
Competing Interests
V.J.T. is an inventor on patents and patent applications filed by New York University. I.P.T. serves as an investigator on studies funded by GlaxoSmithKline and Horizon Pharma. None of these studies conflict with the contents of this manuscript.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-31424-3.
References
- 1.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Klein E, Smith DL, Laxminarayan R. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999-2005. Emerg Infect Dis. 2007;13:1840–1846. doi: 10.3201/eid1312.070629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG., Jr. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kluytmans J, van Belkum A, Verbrugh H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev. 1997;10:505–520. doi: 10.1128/cmr.10.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wertheim HF, et al. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005;5:751–762. doi: 10.1016/S1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- 6.David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev. 2010;23:616–687. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 2009;7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pantosti A, Sanchini A, Monaco M. Mechanisms of antibiotic resistance in Staphylococcus aureus. Future Microbiol. 2007;2:323–334. doi: 10.2217/17460913.2.3.323. [DOI] [PubMed] [Google Scholar]
- 9.Winston, L. G. & Chambers, H. F. In Antimicrobial Drug Resistance 735–748 (Humana Press, 2009).
- 10.McCarthy AJ, et al. Extensive horizontal gene transfer during Staphylococcus aureus co-colonization in vivo. Genome Biol Evol. 2014;6:2697–2708. doi: 10.1093/gbe/evu214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jamrozy D, et al. Evolution of mobile genetic element composition in an epidemic methicillin-resistant Staphylococcus aureus: temporal changes correlated with frequent loss and gain events. BMC Genomics. 2017;18:684. doi: 10.1186/s12864-017-4065-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Copin R, Shopsin B, Torres VJ. After the deluge: mining Staphylococcus aureus genomic data for clinical associations and host-pathogen interactions. Curr Opin Microbiol. 2018;41:43–50. doi: 10.1016/j.mib.2017.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Missiakas D, Schneewind O. Staphylococcus aureus vaccines: Deviating from the carol. J Exp Med. 2016;213:1645–1653. doi: 10.1084/jem.20160569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Middleton JR. Staphylococcus aureus antigens and challenges in vaccine development. Expert Rev Vaccines. 2008;7:805–815. doi: 10.1586/14760584.7.6.805. [DOI] [PubMed] [Google Scholar]
- 15.Glowalla E, Tosetti B, Kronke M, Krut O. Proteomics-based identification of anchorless cell wall proteins as vaccine candidates against Staphylococcus aureus. Infect Immun. 2009;77:2719–2729. doi: 10.1128/IAI.00617-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giersing BK, Dastgheyb SS, Modjarrad K, Moorthy V. Status of vaccine research and development of vaccines for Staphylococcus aureus. Vaccine. 2016;34:2962–2966. doi: 10.1016/j.vaccine.2016.03.110. [DOI] [PubMed] [Google Scholar]
- 17.Fowler VG, et al. Effect of an investigational vaccine for preventing Staphylococcus aureus infections after cardiothoracic surgery: a randomized trial. JAMA. 2013;309:1368–1378. doi: 10.1001/jama.2013.3010. [DOI] [PubMed] [Google Scholar]
- 18.Parker D. Humanized Mouse Models of Staphylococcus aureus Infection. Front Immunol. 2017;8:512. doi: 10.3389/fimmu.2017.00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bagnoli F, Bertholet S, Grandi G. Inferring reasons for the failure of Staphylococcus aureus vaccines in clinical trials. Front Cell Infect Microbiol. 2012;2:16. doi: 10.3389/fcimb.2012.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown AF, Leech JM, Rogers TR, McLoughlin RM. Staphylococcus aureus Colonization: Modulation of Host Immune Response and Impact on Human Vaccine Design. Front Immunol. 2014;4:507. doi: 10.3389/fimmu.2013.00507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lina G, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29:1128–1132. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 22.Vandenesch F, et al. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg Infect Dis. 2003;9:978–984. doi: 10.3201/eid0908.030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otto M. Staphylococcus aureus toxins. Curr Opin Microbiol. 2014;17:32–37. doi: 10.1016/j.mib.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molina, D. M., Morrow, W. J. W. & Liang, X. In Drug Discovery, Development, and Manufacturing Vol. 17 (John Wiley & Sons, Inc., 2010).
- 25.Noroozi Z, et al. A multiplexed immunoassay system based upon reciprocating centrifugal microfluidics. Rev Sci Instrum. 2011;82:064303. doi: 10.1063/1.3597578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diep BA, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 27.Clarke SR, Harris LG, Richards RG, Foster SJ. Analysis of Ebh, a 1.1-megadalton cell wall-associated fibronectin-binding protein of Staphylococcus aureus. Infect Immun. 2002;70:6680–6687. doi: 10.1128/IAI.70.12.6680-6687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams RJ, Henderson B, Nair SP. Staphylococcus aureus fibronectin binding proteins A and B possess a second fibronectin binding region that may have biological relevance to bone tissues. Calcif Tissue Int. 2002;70:416–421. doi: 10.1007/s00223-001-2073-z. [DOI] [PubMed] [Google Scholar]
- 29.Sjodahl J. Repetitive sequences in protein A from Staphylococcus aureus. Arrangement of five regions within the protein, four being highly homologous and Fc-binding. Eur J Biochem. 1977;73:343–351. doi: 10.1111/j.1432-1033.1977.tb11324.x. [DOI] [PubMed] [Google Scholar]
- 30.Zhang L, Jacobsson K, Vasi J, Lindberg M, Frykberg L. A second IgG-binding protein in Staphylococcus aureus. Microbiology. 1998;144(Pt 4):985–991. doi: 10.1099/00221287-144-4-985. [DOI] [PubMed] [Google Scholar]
- 31.Date SV, et al. Global gene expression of methicillin-resistant Staphylococcus aureus USA300 during human and mouse infection. J Infect Dis. 2014;209:1542–1550. doi: 10.1093/infdis/jit668. [DOI] [PubMed] [Google Scholar]
- 32.Couppie P, Cribier B, Prevost G. Leukocidin from Staphylococcus aureus and cutaneous infections: an epidemiologic study. Arch Dermatol. 1994;130:1208–1209. doi: 10.1001/archderm.130.9.1208. [DOI] [PubMed] [Google Scholar]
- 33.Pelzek, A. J. et al. Human Memory B Cells Targeting Staphylococcus aureus Exotoxins Are Prevalent with Skin and Soft Tissue Infection. MBio9, 10.1128/mBio.02125-17 (2018). [DOI] [PMC free article] [PubMed]
- 34.Uhlen M, et al. Complete sequence of the staphylococcal gene encoding protein A. A gene evolved through multiple duplications. J Biol Chem. 1984;259:1695–1702. [PubMed] [Google Scholar]
- 35.Zhao, F., Chong, A. S. & Montgomery, C. P. Importance of B Lymphocytes and the IgG-Binding Protein Sbi in Staphylococcus aureus Skin Infection. Pathogens5, 10.3390/pathogens5010012 (2016). [DOI] [PMC free article] [PubMed]
- 36.Schneewind O, Model P, Fischetti VA. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-H. [DOI] [PubMed] [Google Scholar]
- 37.Falugi F, Kim HK, Missiakas DM, Schneewind O. Role of protein A in the evasion of host adaptive immune responses by Staphylococcus aureus. MBio. 2013;4:e00575–00513. doi: 10.1128/mBio.00575-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silverman GJ, Goodyear CS. Confounding B-cell defences: lessons from a staphylococcal superantigen. Nat Rev Immunol. 2006;6:465–475. doi: 10.1038/nri1853. [DOI] [PubMed] [Google Scholar]
- 39.Yeung YA, et al. Germline-encoded neutralization of a Staphylococcus aureus virulence factor by the human antibody repertoire. Nat Commun. 2016;7:13376. doi: 10.1038/ncomms13376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holtfreter S, Kolata J, Broker BM. Towards the immune proteome of Staphylococcus aureus - The anti-S. aureus antibody response. Int J Med Microbiol. 2010;300:176–192. doi: 10.1016/j.ijmm.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 41.Kloppot P, et al. Microarray-based identification of human antibodies against Staphylococcus aureus antigens. Proteomics Clin Appl. 2015;9:1003–1011. doi: 10.1002/prca.201400123. [DOI] [PubMed] [Google Scholar]
- 42.Dryla A, et al. Comparison of antibody repertoires against Staphylococcus aureus in healthy individuals and in acutely infected patients. Clin Diagn Lab Immunol. 2005;12:387–398. doi: 10.1128/CDLI.12.3.387-398.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.den Reijer, P. M. et al. Combining in vitro protein detection and in vivo antibody detection identifies potential vaccine targets against Staphylococcus aureus during osteomyelitis. Med Microbiol Immunol, 11–22 (2016). [DOI] [PMC free article] [PubMed]
- 44.Fritz SA, et al. A serologic correlate of protective immunity against community-onset Staphylococcus aureus infection. Clin Infect Dis. 2013;56:1554–1561. doi: 10.1093/cid/cit123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Etz H, et al. Identification of in vivo expressed vaccine candidate antigens from Staphylococcus aureus. Proc Natl Acad Sci USA. 2002;99:6573–6578. doi: 10.1073/pnas.092569199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bavari S, Ulrich RG, LeClaire RD. Cross-reactive antibodies prevent the lethal effects of Staphylococcus aureus superantigens. J Infect Dis. 1999;180:1365–1369. doi: 10.1086/314977. [DOI] [PubMed] [Google Scholar]
- 47.Salgado-Pabon W, Schlievert PM. Models matter: the search for an effective Staphylococcus aureus vaccine. Nat Rev Microbiol. 2014;12:585–591. doi: 10.1038/nrmicro3308. [DOI] [PubMed] [Google Scholar]
- 48.Verkaik NJ, van Wamel WJ, van Belkum A. Immunotherapeutic approaches against Staphylococcus aureus. Immunotherapy. 2011;3:1063–1073. doi: 10.2217/imt.11.84. [DOI] [PubMed] [Google Scholar]
- 49.Mendes RE, et al. Characterization of methicillin-resistant Staphylococcus aureus strains recovered from a phase IV clinical trial for linezolid versus vancomycin for treatment of nosocomial pneumonia. J Clin Microbiol. 2012;50:3694–3702. doi: 10.1128/JCM.02024-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altman DR, et al. Transmission of methicillin-resistant Staphylococcus aureus via deceased donor liver transplantation confirmed by whole genome sequencing. Am J Transplant. 2014;14:2640–2644. doi: 10.1111/ajt.12897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benson MA, et al. Evolution of hypervirulence by a MRSA clone through acquisition of a transposable element. Mol Microbiol. 2014;93:664–681. doi: 10.1111/mmi.12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pelzek AJ, et al. Persistence of disease-associated anti-citrullinated protein antibody-expressing memory B cells in Rheumatoid Arthritis in clinical remission. Arthritis Rheumatol. 2017;69:1176–1186. doi: 10.1002/art.40053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nielsen H. Predicting Secretory Proteins with SignalP. Methods Mol Biol. 2017;1611:59–73. doi: 10.1007/978-1-4939-7015-5_6. [DOI] [PubMed] [Google Scholar]
- 56.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 57.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 58.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 59.Hulsen T, de Vlieg J, Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fan H, Ives AR, Surget-Groba Y, Cannon CH. An assembly and alignment-free method of phylogeny reconstruction from next-generation sequencing data. BMC Genomics. 2015;16:522. doi: 10.1186/s12864-015-1647-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed in the current report are available from the corresponding author on reasonable request.