Abstract
To identify metabolic pathways that were perturbed in pancreatic cancer (PC), we investigated gene‐metabolite networks by integration of metabolomic and transcriptomic. In this research, we undertook the metabolomic study of 43 paired human PC samples, aiming to identify key metabolic alterations in PC. We also carried out in vitro experiments to validate that the key metabolite cytidine and its related gene ENTPD8 played an important role in PC cell proliferation. We screened out 13 metabolites differentially expressed in PC tissue (PCT) by liquid chromatography/mass spectrometry analysis on 34 metabolites, and the partial least square discrimination analysis results revealed that 9 metabolites among them were remarkably altered in PCT compared to adjacent noncancerous tissue (variable importance in projection >1, P < .05). Among the 9 metabolites, 7 might be potential biomarkers. The most significantly enriched metabolic pathway was pyrimidine metabolism. We analyzed 351 differentially expressed genes from The Cancer Genome Atlas and intersected them with Kyoto Encyclopedia of Genes and Genomes metabolic pathways. We found that ENTPD8 had a gene‐metabolite association with cytidine in the CTP dephosphorylation pathway. We verified by in vitro experiments that the CTP dephosphorylation pathway was changed in PCT compared with adjacent noncancerous tissue. ENTPD8 was downregulated in PCT, causing a reduction in cytidine formation and hence weakened CTP dephosphorylation in pyrimidine metabolism.
Keywords: metabolic pathway, metabolite, metabolomic, pancreatic cancer, transcriptomic
Abbreviations
- ANT
adjacent noncancerous tissue
- CMP
cytidine monophosphate
- ENTPD8
ectonucleoside triphosphate diphosphohydrolase 8
- ENTPD
ecto‐NTPDase
- FC
fold change
- FDR
false discovery rate
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LC
liquid chromatography
- MS
mass spectrometry
- NC
negative group
- NTPDase
nucleoside triphosphate diphosphohydrolase
- PC1
primary component 1
- PC2
primary component 2
- PC
pancreatic cancer
- PCT
pancreatic cancer tissue
- PI
propidium iodide
- PLS‐DA
partial least square discrimination analysis
- qRT‐PCR
quantitative RT‐PCR
- Q‐TOF
quadrupole time‐of‐flight
- si‐ENTPD8
siRNA directed against ENTPD8
- si‐NC
negative siRNA
- TOF
time‐of‐flight
- VIP
variable importance in projection
1. INTRODUCTION
High mortality rates have been reported in PC patients, with PC identified as the second leading cause of cancer‐related deaths.1, 2 The low survival rate is closely related to the aggressive growth property of this disease and the absence of early diagnosis.3 Hence, it is urgent to investigate the metabolic characterization of PC and find potential biomarkers for predicting PC risk.
Metabolomic is one of the omics technologies enabling quantification of massive small molecules, termed metabolites, simultaneously.4 Considering that the metabolome is a close molecular representation of tumor phenotype in cancer, metabolomic might be able to determine clinically relevant metabolomic subclasses.5 Untargeted MS analysis is a proven universal analysis method, which shows high intra‐assay precision and linear detection for the majority of compounds tested. However, because of the massive number of compounds, the technique possesses certain drawbacks in specific detection.6 Fortunately, targeted metabolomic specialize in measuring a set of known metabolites with high sensitivity and good reproducibility, which could fill the gap of the untargeted experiment.7 Typically, the detection set was normally hard to define, making the method a dilemma.8 The parallel application of the two analytical techniques could contribute to absolute quantification of specific metabolites and discover more novel compounds that can serve as diagnostic cancer biomarkers. Cho et al9 revealed urinary biomarkers for discriminating obese from normal‐weight adolescents by combining untargeted and targeted metabolomic profiling. Shang et al10 unveiled three significantly abundant metabolites, galactinol, melibiose, and melatonin, in papillary thyroid cancer and validated these potential diagnostic biomarkers using gas chromatography‐TOF‐MS‐based targeted and untargeted metabolomic approaches. Transcriptomic is another omics technology that analyzes all RNA molecules transcribed from DNA in cells, tissues, or organs.11 Combined with transcriptomic, systematic investigation of metabolomic data can be largely enhanced by linking known metabolites and genes through their shared metabolic pathways.4 Recently, researchers have taken advantage of the integration of metabolomic and transcriptomic to study the perturbed metabolic pathways in carcinomas, such as pancreatic ductal adenocarcinoma,12 prostate cancer13, and clear cell renal cell carcinoma.14 With the application of various emerging methods and materials in fields, more specific biomarkers were revealed.
Pyrimidine is an aromatic heterocyclic organic compound similar to pyridine, and pyrimidine biosynthesis occurs both in the body and through organic synthesis. As a pyrimidine component of RNA, cytidine has been found to control neuronal‐glial glutamate cycling, with supplementation decreasing midfrontal/cerebral glutamate/glutamine levels.15 Cytidine triphosphate is a pyrimidine nucleoside triphosphate acting as an inhibitor of the enzyme aspartate carbamoyltransferase, which is used in pyrimidine biosynthesis. Nucleoside triphosphate diphosphohydrolases are omnipresent enzymes for hydrolyzing nucleoside triphosphates and nucleoside diphosphates, and they have an extra plasmic orientation.16 In the ENTPD family, ENTPD8 is the latest member to be discovered and characterized; it is a transmembrane protein that regulates the concentration of the agonists of P1 and P2 receptors at the cell surface.17
In this study, we aimed to identify both metabolic biomarkers and metabolic pathways that had been perturbed in PC by integration of metabolomic and transcriptomic. We screened the differentially expressed metabolite cytidine and its related gene ENTPD8 in PC and we verified in vitro that the CTP dephosphorylation pathway in PC was altered.
2. MATERIALS AND METHODS
2.1. Patients and tissue collection
After acquiring written informed consent and approval, including acknowledging purposes, contents, and risks, 43 matched pancreatic tumor and ANTs were collected from patients with PC after surgery at the Third Affiliated Hospital of Soochow University (Changzhou, China). Detailed information of patients is available in Table 1. Eighty‐six tissue samples consisting of 43 pairs of PCTs and ANT were covered in this study, coming from 23 men and 20 women. The average age of patients was 67.9 years (range, 43.2‐85.7 years). Medical histories of all these patients were recorded, and no diabetic patients were found. There was no chemotherapy or radiotherapy prior to the examination. Tissues for this study were flash frozen after surgery and stored at −80°C before metabolomic characterization. The use of tissue samples was agreed by the ethics committee of the Third Affiliated Hospital of Soochow University. All the experimental procedures were approved by the Third Affiliated Hospital of Soochow University, and they conform with the provisions of the Declaration of Helsinki.
Table 1.
Characteristics of patients with pancreatic cancer (PC)
| Characteristic | Patients (n = 43) |
|---|---|
| Age (years) | 57.8 ± 19.7 |
| Gender, male/female | 21/22 |
| BMI | 24.2 ± 5.3 |
| Disease | |
| Stage I | 2 |
| Stage II | 12 |
| Stage III | 21 |
| Unidentified | 8 |
| Family PC history, n (%) | 5 (11.6%) |
BMI, body mass index.
2.2. Chemicals and reagents
Agilent 1290 Infinity LC and Agilent 6410 Q‐TOF MS/MS were provided by Agilent Technologies (Santa Clara, CA, USA). SpeedVac SPD121P used for centrifuges was provided by Thermo Fisher Scientific (Waltham, MA, USA). GTR16‐2 was provided by BJSDBL (Beijing, China), and XW‐80A was provided by Shanxi (Shanghai, China). IKA T10basic‐S25 was provided by IKA (Staufen, Germany), and double distilled water was provided by Millipore (Milan, Italy). Formic acid (#399388), acetonitrile (#34851), 2‐propanol ethyl carbinol (#1570428), methanol (#34860), saturated fatty acid methyl esters (#ME10‐1KT), xylene substitute (#A5597), ethanol (#443611), hematoxylin (#H9627), hydrochloric acid (32 wt. % in H2O, #W530574), ammonia (28% NH3 in H2O, #338818), and eosin (#E4009) were purchased from Sigma‐Aldrich (Milwaukee, WI, USA).
2.3. Sample collection
For each specimen, 10 mg wet tissue was weighed and placed into a 2‐mL centrifuge tube. A porcelain ball and 300 μL ice‐cold methanol with internal standards were added, followed by homogenization in a frozen mixed ball grinding machine. Next, 0.4 mL acetonitrile solution with 0.2% formic acid was added, and the mixture was incubated at 4°C for 30 minutes, followed by centrifugation for 3 minutes (15 000 g, 4°C). Lipids and hydrophobic metabolites were primarily distributed into the upper layer, and then transferred into a clean centrifuge tube and stored at −80°C. Thirty microliter samples were redissolved at −20°C in 1 mL acetonitrile/isopropanol/water (3:3:2) solvent and centrifuged for 2 minutes (15 000 g). The supernatant containing metabolites was dried in a vacuum centrifuge. Sequentially, 30 μL acetonitrile/water (1:1) solution and 2 μL trimethylsilyl propionic acid were added to the samples for later LC‐MS analysis. The tissues were used for all following assays such as immunohistochemistry and qRT‐PCR.
2.4. Liquid chromatography‐MS analysis
Liquid chromatography‐MS analysis was carried out with a ultra high performance liquid chromatography system (Infinity 1290; Agilent Technologies) connected to a Q‐TOF/MS instrument (Agilent 6410 Q‐TOF MS/MS; Agilent Technologies). Elution with solvent A (0.1% formic acid) and solvent B (acetonitrile) in a linear gradient manner was undertaken at a flow rate of 0.4 mL/min as follows: 5% B (0‐2 minutes), 50% B (2‐10 minutes), 100% B (10‐18 minutes), 100% B (18‐18.5 minutes), 5% B (18.5‐19 minutes), and 5% B (19‐20 minutes). The injection volume was 5 μL, the flow rate was 300 μL/min, and the temperature was 40°C. Electrospray ionization in positive ionization mode was carried out with the following parameters: the capillary voltage and cone voltage were set at 4.5 and 35 kV, respectively, the drying gas temperature was set at 400°C, and the mass range was scanned as m/z 50‐1000. The LC‐MS data were collected and analyzed using MassHunter B.03.01 (Agilent Technologies).
2.5. Quantitative RT‐PCR
Total RNAs extracted by TRIzol reagent (Invitrogen, Carlsbad, CA, USA) were reverse‐transcribed into cDNA with a Transcriptor First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). The primers for qRT‐PCR were synthesized by Sangon Biotech (Shanghai, China) and are shown in Table 2. All reactions were carried out using an Applied Rotor‐Gene 6000 Real‐Time PCR System (Corbett Research, Mortlake, Vic., Australia) in triplicate. Data from real‐time PCR were analyzed using the method.
Table 2.
Primer sequences used in this study
| Primer | Sequence (5′‐3′) |
|---|---|
| ENTPD8 forward | GCCTCACGGCACTCATTCTC |
| ENTPD8 reverse | CGCATCAAACACGATCCCAA |
| β‐Actin forward | GGACTTCGAGCAAGAGATGG |
| β‐Actin reverse | AGCACTGTGTTGGCGTACAG |
2.6. Immunohistochemical analysis
Tissues obtained from the same study cohorts in the metabolites analysis were fixed in neutral‐buffered 10% formalin solution, embedded in paraffin, and sectioned at a thickness of 4 μm. Sections were rehydrated and incubated with 3% H2O2 methanol for 10 minutes, 10% goat serum for 30 minutes at room temperature, 50 μL primary antibodies (ENTPD8 antibodies; Saierbio, Tianjin, China) at 4°C overnight, 40‐50 μL secondary antibodies at room temperature for 1 hour, and then peroxidase‐conjugated streptavidin at 37°C for 30 minutes. Sequentially, DAB used for staining was added for 5‐10 minutes, and then sections were washed in water. Finally, sections were rapidly dehydrated in graded ethanol, cleared in Xylene, and mounted with neutral gum for observation.
2.7. Western blot analysis
The cells were dissolved in RIPA, and the concentration was determined using a BCA Protein Assay Kit (Beyotime Biotechnology, Shanghai, China). Fifty micrograms of protein for each sample was separated on 4%‐12% NuPAGE gel with MOPS SDS running buffer (Thermo Fisher Scientific) and transferred to a PVDF membrane. After being blocked for 30 minutes at room temperature, membranes were probed with primary antibodies (rabbit anti‐human β‐actin, ENTPD8 antibodies; Beyotime Biotechnology) and GAPDH overnight at 4°C and washed four times with 1× TBST. Sequentially, HRP‐conjugated anti‐rabbit secondary antibody (ZSBIO, Beijing, China) was diluted and incubated with membranes for 4 hours at room temperature, and then washed four times with 1× TBST. Finally, rinsed PVDF membranes were placed in a dark room, and electrochemiluminescent was then used for staining. Immunoreactive bands of the protein expression were normalized to the intensity of the corresponding bands for β‐actin.
2.8. Cell culture
Human pancreatic cancer cell lines PANC‐1 and CFPAC‐1 were purchased from the cell bank of the Chinese Academy of Sciences (Shanghai, China). PANC‐1 cells were incubated in DMEM supplemented with 10% FBS and 1.5 g/L NaHCO3. CFPAC‐1 cells were grown in DMEM with 10% FBS. All the cells were cultured at 37°C with 5% CO2.
2.9. Cell transfection
An NC group of scrambled siRNA and an experimental group of si‐ENTPD8 were designed by BLOCK‐iT RNAi Designer Software (Invitrogen), and the sequences are shown in Table 3. Plasmid pcDNA3.1 (+) (#V79020; Invitrogen) was used to construct pcDNA3.1‐ ENTPD8 (1 μg/2 × 105 cells). Cells were transfected using Lipofectamine 2000 (Invitrogen), and the final concentration of transfected cells was 60 nmol/L. The final concentration was stored to support experiments including ENTPD8 overexpression.
Table 3.
Small interfering RNA sequences used in this study
| siRNA | Sequence (5′‐3′) |
|---|---|
| si‐ENTPD8a sense | GGAAUCUCCUCCUACACUU |
| si‐ENTPD8a antisense | AAGUGUAGGAGGAGAUUCC |
| si‐ENTPD8b sense | CCAACUUCUACUACACCUU |
| si‐ENTPD8b antisense | AAGGUGUAGUAGAAGUUGG |
| si‐NC sense | GGACCUCUCCUACAUACUU |
| si‐NC antisense | AAGUAUGUAGGAGAGGUCC |
si‐NC, negative scrambled siRNA.
2.10. Cell Counting Kit‐8 assay
The CCK‐8 assay (Biotechwell, Shanghai, China) detected cell proliferation of PANC‐1 and CFPAC‐1 cells. Logarithmically growing cells were digested with 0.25% trypsin and cultured in RPMI‐1640 medium supplemented with 10% FBS. Transfected cells were seeded into 96‐well plates at 37°C with 5% CO2. After adherence, culture medium was replaced by RPMI‐1640 without FBS 24‐hour incubation. At 0, 24, 36, and 48 hours, optical density was determined by the microplate reader at 450 nm. Each group had six complex holes, and average optical density values were taken as statistics.
2.11. Flow cytometry assay
Cell apoptosis was detected by an Annexin V Apoptosis Detection Kit (Beyotime Biotechnology). At 36 hours after transfection, cells were collected followed by transfection by group. There were three treatment groups: (i) supplemented with PI; (ii) supplemented with annexin V‐FITC; and (iii) no supplementation. Cells were centrifuged and transferred into eppendorf tubes. Each tube contained 106 cells. Next, cells were washed twice with PBS, and then 100 μL binding buffer and 10 μg annexin V (20 μg/mL) marked with FITC were added for 30 minutes at room temperature. After antidark reaction with 5 μL PI (50 μg/mL) for 5 minutes, 400 μL binding buffer was added, and FACScan (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) was used for quantitative detection. The group without Annexin V‐FITC or PI was taken as negative control group. FACS Diva software (Becton, Dickinson and Company) was used for data analysis.
2.12. Cell metabolite detection
Cells were quenched with 12 mL ice‐cold 0.9% (w/v) NaCl for 5 minutes, then the mixture was transferred into a centrifuge tube by pipetting and centrifuged at 15 000 g for 3 minutes at −19°C, and the supernatant was discarded. Five milliliters of 50% methanol was added to the tube and vortex‐mixed for 10 seconds and then transferred into the liquid nitrogen and submerged for 3 minutes. Samples were thawed for 5 minutes at room temperature and then vortex‐mixed for 10 seconds. The procedure was repeated twice, and the samples were centrifuged at 21 000 g for 5 minutes at 4°C, and the supernatant was ready for LC‐MS analysis.
2.13. Statistical analysis
Online software Metaboanalyst 3.0 (http://www.metaboanalyst.ca) was used for data visualization, PLS‐DA construction, and enriched pathway analysis. Data were normalized by median and transformed with generalized logarithm. The variables were selected based on VIP >1 from the normalized peak intensity. Transcriptomic analysis was based on the PC patient mRNA dataset downloaded from the GDC website (https://portal.gdc.cancer.gov/). The sample identifiers used in our experiment are listed in Table S1 and data of PC tissues and adjacent tissues are available on the website https://cancergenome.nih.gov/. The analysis was undertaken in RStudio (https://www.rstudio.com/). Normalization and variation analyses were executed by DESeq2 package (http://bioconductor.org/biocLite.R), and P < .05 and |log2FC| > 1 were considered to indicate significant difference. Genes related to potential metabolic biomarkers were obtained from the KEGG platform (http://www.kegg.jp/kegg/pathway.html), which consists of gene maps and metabolites maps, from which we could determine genic information related to metabolites.
The following tissue and cell experiments analyses were presented with GraphPad Prism 6.0 software (La Jolla, CA, USA).
3. RESULTS
3.1. Thirteen metabolites differentially expressed in PCT according to LC‐MS analysis
Paired PCTs and ANTs from 86 patients were analyzed through metabolomic by LC‐MS. The LC‐MS analysis was based on retention time, molecule weight, and fragment ions. A total of 34 metabolites were quantified and we evaluated the levels of these compounds (Table S2). Among them, 13 kinds of metabolites were differentially expressed in PCT compared to ANT (P < .05; FDR < 0.05; Table 4), including 7 upregulated metabolites, such as 2‐hydroxybutyrate, isoleucine, and taurine, and 6 downregulated metabolites, such as adenosine, 3‐methylhistidine, and cytidine. Among all these metabolites, cytidine showed a significant decrease in PCT according to the logFC value in Table 4. The relative levels of metabolites in PCT and ANT were visualized on a heat map (Figure 1A).
Table 4.
T test of all metabolites expressed in pancreatic cancer tissue (PCT) according to liquid chromatography‐mass spectrometry analysis
| Metabolite | P‐value | FDR | Log2(FC) |
|---|---|---|---|
| 2‐Hydroxybutyrate | 2.7E‐19 | 9.1E‐18a | 0.522 |
| Adenosine | 8.8E‐14 | 1.4E‐12a | −0.437 |
| 3‐Methylhistidine | 1.2E‐13 | 1.4E‐12a | −0.267 |
| Isoleucine | 4.7E‐08 | 4.0E‐07a | 0.230 |
| Cytidine | 1.6E‐07 | 9.2E‐07a | −3.080 |
| Acetylcholine | 1.6E‐07 | 9.2E‐07a | −0.175 |
| Taurine | 6.9E‐06 | 3.4E‐05a | 0.944 |
| Glycerol | 3.0E‐03 | 1.3E‐02a | 0.744 |
| Betaine | 1.0E‐02 | 3.9E‐02a | −0.354 |
| Acetate | 1.2E‐02 | 3.9E‐02a | 0.153 |
| 3‐Hydroxybutyrate | 1.3E‐02 | 3.9E‐02a | 0.125 |
| Leucine | 1.4E‐02 | 3.9E‐02a | 0.142 |
| GPCho | 1.6E‐02 | 4.1E‐02a | −0.239 |
| Lactate | 2.2E‐02 | 5.2E‐02 | 1.314 |
| Choline | 2.8E‐02 | 6.3E‐02 | −0.157 |
| Glutamate | 3.6E‐02 | 7.6E‐02 | 0.292 |
| Valine | 4.9E‐02 | 9.8E‐02 | 0.120 |
| Creatine | 6.3E‐02 | 1.2E‐01 | 0.135 |
| Glutamine | 7.8E‐02 | 1.4E‐01 | −0.077 |
| Ethanolamine | 1.5E‐01 | 2.6E‐01 | 0.071 |
| 1‐Methylhistidine | 2.0E‐01 | 3.2E‐01 | 0.042 |
| Kynurenate | 2.2E‐01 | 3.4E‐01 | −0.013 |
| Citrate | 3.0E‐01 | 4.4E‐01 | −0.107 |
| Threonine | 3.5E‐01 | 5.0E‐01 | −0.083 |
| Adipate | 3.8E‐01 | 5.2E‐01 | 0.129 |
| Lysine | 4.2E‐01 | 5.5E‐01 | −0.051 |
| Ascorbate | 5.3E‐01 | 6.7E‐01 | −0.024 |
| Succinate | 5.7E‐01 | 6.9E‐01 | 0.009 |
| Dimethylglycine | 6.4E‐01 | 7.5E‐01 | 0.003 |
| Glycine | 8.1E‐01 | 9.2E‐01 | 0.046 |
| Inosine | 8.7E‐01 | 9.5E‐01 | 0.038 |
| Aspartate | 9.0E‐01 | 9.6E‐01 | 0.006 |
| PCho | 9.3E‐01 | 9.6E‐01 | −0.006 |
| Glucose | 9.8E‐01 | 9.8E‐01 | 0.003 |
P < .05, statistical significance.
Log2(FC) > 0 indicates that this metabolite is upregulated in PCT. Log2(FC) < 0 indicates that this metabolite is downregulated in PCT.
FC, fold change.
Figure 1.
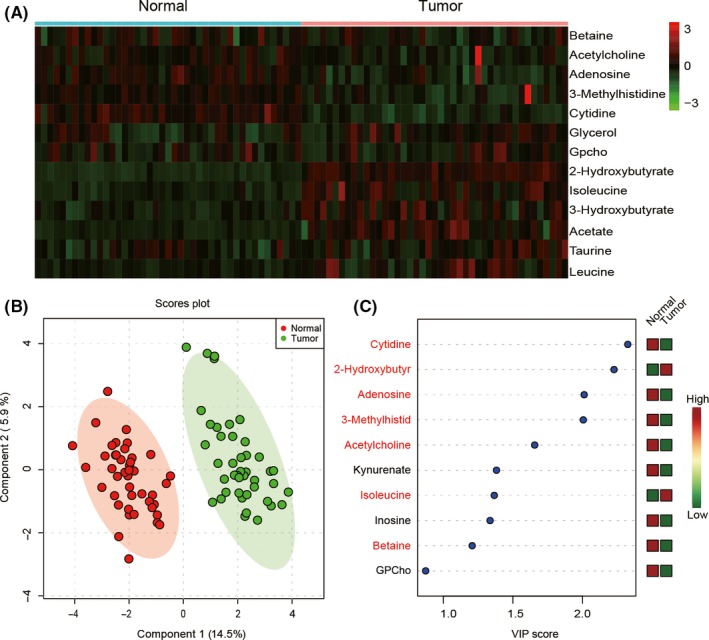
Metabolic biomarker analysis in pancreatic cancer tissue (Tumor) and adjacent nontumor tissue (Normal). A, 13 altered metabolites according to liquid chromatography/mass spectrometry analysis visualized by a heat map. B, Partial least square discrimination analysis (PLS‐DA) model showed that tumor and normal samples were clearly separated in the x‐axis direction. Each point represents an independent sample, and the ellipse represents the 95% confidence interval. C, 10 metabolites with the highest variable importance in projection (VIP) scores of PC1 were screened out by the PLS‐DA model. Seven potential biomarkers were shown in red
3.2. Metabolic biomarkers of PC were confirmed using PLS‐DA model
Partial least square discrimination analysis is a supervised partial least squares analysis mainly used to find potential biomarkers by analyzing metabolic differences from massive data. Data of 34 metabolites were used in PLS‐DA. Primary component 1 and PC2 could explain 20.4% difference. According to the PLS‐DA model, PCTs and ANTs could be clearly separated in the x‐axis direction. The model quality was measured by R 2 and Q 2 value, R 2 = 0.79, Q 2 = 0.71. In Figure 1(B), each data point on the graph represents an independent sample, and the ellipse represents the 95% confidence interval. Variable importance in projection score indicators are mainly used to describe the weight of independent variables in the interpretation of the dependent variable. The greater the VIP score is, the greater the impact on PC1 is. According to the VIP scores, cytidine was supposed to have the largest impact on PC1, which might be a new biomarker. In this study, metabolites with VIP > 1 as well as P < .05 were screened as metabolic biomarkers. Nine metabolic biomarkers with the most contributions to PC1 were screened out (Figure 1C). Combined with the t test, 7 biomarkers were screened out, including cytidine, 2‐hydroxybutyrate, adenosine, 3‐methylhistid, acetylcholine, isoleucine, and betaine.
3.3. Integrated metabolic and transcriptomic analyses suggested major altered pathways in PC
To trace the upstream metabolic pathways of metabolic biomarkers, metabolite sets enrichment analysis was carried out using Metaboanalyst online software. Pathway enrichment analysis depends on the compounds and their concentration, which is based on KEGG and the Human Metabolome Database, and overrepresentation analysis was implemented using the hypergeometric test to evaluate whether a particular metabolite set is represented more than expected by chance within compounds. Among the 17 metabolic pathways with FDR < 0.05, pyrimidine metabolism containing cytidine was screened as the most significantly enriched (Figure 2A). Data from The Cancer Genome Atlas were compared with R project, and 351 mRNAs with P < 0.05 and |log2FC| > 1 were screened by Student's t test and identified as differentially expressed mRNAs. The top 10 upregulated and downregulated mRNAs are shown in Figure 2B. We matched the pyrimidine pathways to the KEGG database, and KEGG pathway maps collected related genes of manually drawn pathways, from which we obtained 290 corresponding mRNAs. By intersection, we found that ENTPD8 existed in both mRNA sets coming from KEGG and TCGA (Figure 2C). Moreover, according to KEGG, there was a correlation between ENTPD8 and metabolic biomarker cytidine in the pyrimidine metabolism (Figure 2D). The remaining genes on this metabolic pathway, such as CTPS1, CTPS2, ENTPD1, ENTPD2, and NT5E, had no statistical difference under the conditions of P < .05, FDR < 0.05. We confirmed that both cytidine and ENTPD8 were downregulated in PCT, and ENTPD8, which catalyzed formation of cytidine, was encoded by ENTPD8 mRNA. Hence, we hypothesized that CTP dephosphorylation in PCT was weakened.
Figure 2.
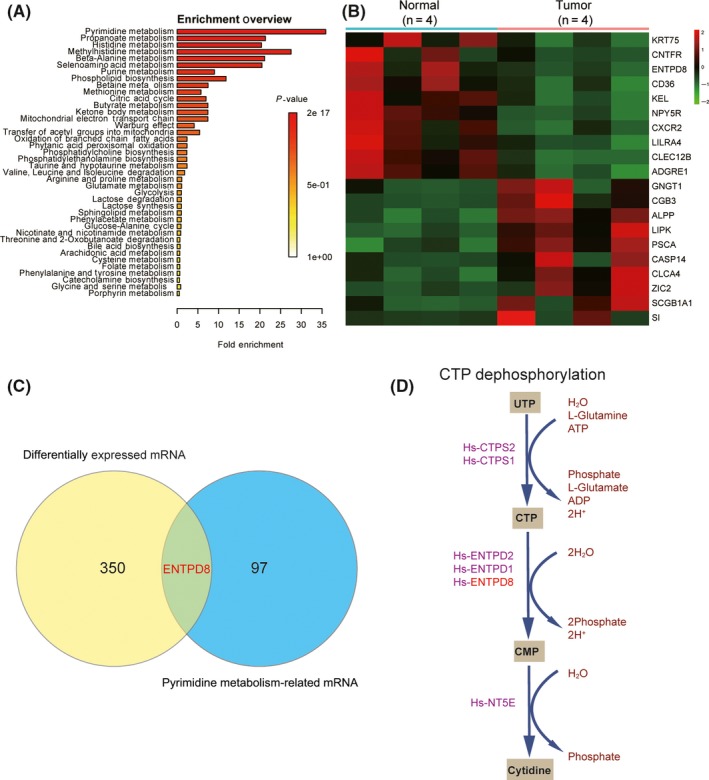
Enriched metabolomic pathway analysis in pancreatic cancer tissue. A, Overview of metabolite sets enrichment. P‐value represents the enrichment level; length of bar represents the amount of metabolites. B, Heat map of the top 10 upregulated and downregulated mRNAs in pancreatic cancer tissue. Data were acquired from The Cancer Genome Atlas. C, Venn diagram of differentially expressed mRNAs and pyrimidine metabolism related mRNAs. D, CTP dephosphorylation in pyrimidine metabolism involves metabolic biomarker cytidine and its related gene ENTPD8
3.4. ENTPD8 downregulated in PCT compared to ANT
To validate our hypothesis, qRT‐PCR was undertaken to measure the expression level of ENTPD8 mRNA. We found that the relative expression of ENTPD8 mRNA in PCT was lower than that in ANT (P < .05; Figure 3A). Furthermore, results of immunohistochemical assay and Western blot analysis revealed that the relative expression of ENTPD protein was also downregulated in PCT compared with ANT (Figure 3B,C). In short, ENTPD8 and its protein had lower expression levels in PCT compared to ANT.
Figure 3.
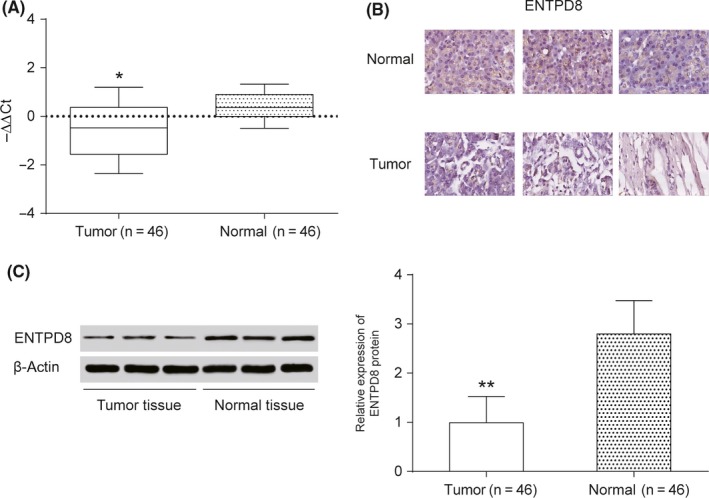
Expression level of ENTPD8 in pancreatic cancer tissue. A, Quantitative RT‐PCR was undertaken to measure the relative expression level of ENTPD8 mRNA in tumor tissues and normal tissues. B, Immunohistochemical assay shows that ENTPD8 appears positive in the normal cell nucleus. C, Western blot analysis to measure ENTPD8 protein expression level. *P < .05; **P < .01, indicates statistical significance compared with negative group
3.5. Overexpression of ENTPD8 suppressed cell viability and promoted cell apoptosis in PCT
The expression level of ENTPD8 was quantified by qRT‐PCR assay after transfection. The expression of ENTPD8 was higher in the overexpression group, and the expression of ENTPD8 in si‐groups was lower than NC (P < .001; Figure 4A). ENTPD8 protein expression was confirmed to be upregulated in the ENTPD8 group and downregulated in si‐ENTPD8 group by Western blot after transfection (Figure 4B). The CCK‐8 assay then showed a strengthened cell viability in the si‐ENTPD8 group at 72 hours but a weakened cell viability in the ENTPD8 group, indicating that ENTPD8 could inhibit PC cell viability (P < .05, Figure 4C). Additionally, flow cytometry results (Figure 5A) revealed a higher apoptosis rate in the ENTPD8 group compared to the NC group (P < .01; Figure 5B), whereas the apoptosis rate was attenuated after transfection with si‐ENTPD8 (P < .05; Figure 5B), indicating that ENTPD8 could promote cell apoptosis in PCT.
Figure 4.
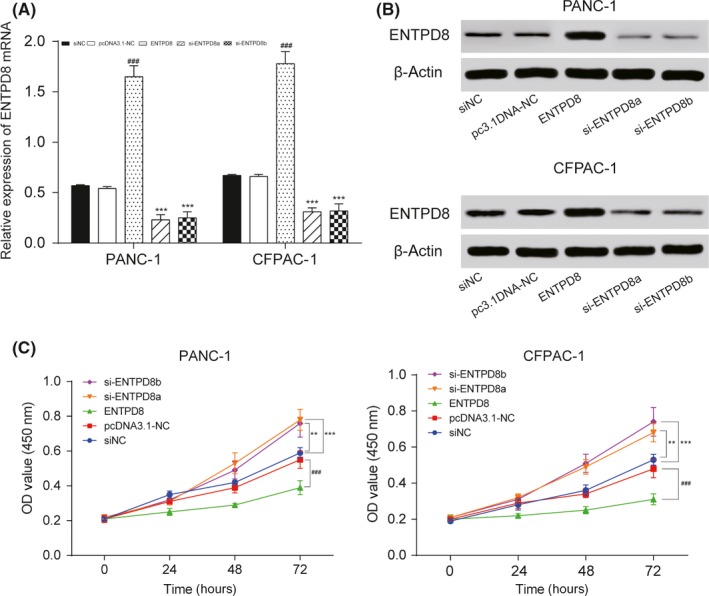
Effects of ENTPD8 on proliferation and protein expression of PANC‐1 and CFPAC‐1 pancreatic cancer cells. A, After transfection, the relative expression level of ENTPD8 was measured by quantitative RT‐PCR. B, After transfection, the relative expression level of ENTPD8 protein was measured by Western blot analysis, and its expression was upregulated in the ENTPD8 group and downregulated in the si‐ENTPD8 group. C, CCK‐8 assay shows that cell viability in the ENTPD8 group was inhibited. **P < .01, ***P < .001 indicated statistical significance compared with the negative (NC) group. ### P < .001 indicates statistical significance compared with pcDNA3.1‐NC group
Figure 5.
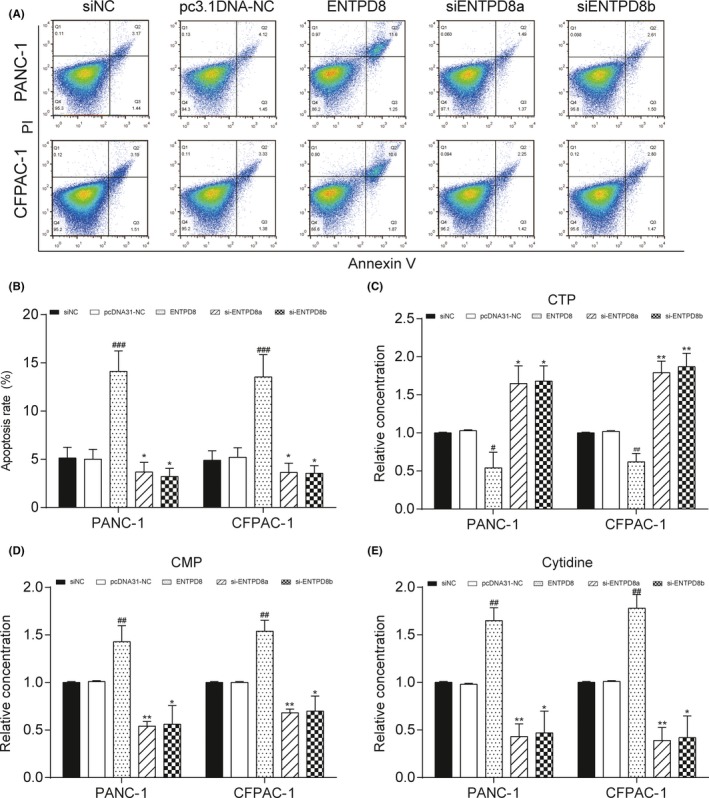
Effect of ENTPD8 on apoptosis and metabolites in two pancreatic cancer cell lines. A,B, Flow cytometry assay shows that ENTPD8 promoted cell apoptosis in both PANC‐1 and CFPAC‐1 cells. C‐E, Relative metabolite concentrations of CTP (C), cytidine monophosphate (CMP) (D) and cytidine (E) in cell lines. *P < .05, **P < .01 indicates statistical significance compared with the negative (NC) group. # P < .05, ## P < .01, ### P < .001 indicates statistical significance compared with pcDNA3.1‐NC group
3.6. ENTPD8 affected the amount of CTP, CMP, and cytidine
The cell metabolites analysis revealed that overexpression of ENTPD8 could downregulate the amount of CTP, while the amount of CMP and cytidine was upregulated. The amount of CTP was upregulated when ENTPD8 was knocked down and the amount of CMP and cytidine was at a lower level (all P < .05; Figure 5C‐E). We supposed that ENTPD8 could catalyze the dephosphorylation of CTP to CMP in the pyrimidine metabolism pathway and the amount of cytidine relied on the amount of CMP because it was dephosphorylated by CMP according to the metabolism pathway. In conclusion, the expression of ENTPD8 can affect the amount of these three metabolites.
4. DISCUSSION
It is well recognized that metabolomic can provide snapshots of physiological conditions in biological samples by acquiring qualitative and quantitative information of low‐molecular‐mass endogenous metabolites,18 and it has been applied to identify potential molecular biomarkers of various cancers.19, 20, 21 A previous study suggested that malignant cancers had markedly altered intermediate metabolites in metabolic pathways compared with nontumor tissue.22 In addition, Budhu et al23 found that metabolic profiles were significantly different among several cancer types, and those distinct metabolites or metabolic pathways in particular cancer types might be helpful to distinguish primary tumors. Fontana et al24 identified metabolites that might be used to predict 1‐year mortality of PC patients by analyzing potential alterations in metabolites in PC serum samples. However, an effective integration of metabolomic with transcriptomic data could better interpret results that could not be obtained from one single metabolomic dataset.25
To date, numerous researchers have unveiled significantly altered pathways and the associated metabolites with high diagnostic accuracy through merging metabolomic and transcriptomic. Chen et al26 showed that the combination of metabolomic and transcriptomic signatures effectively distinguished malignant osteosarcoma with a higher diagnostic accuracy than other techniques. Ren et al13 also observed alterations in certain metabolisms at both metabolic and transcriptional levels to reveal that S1PR2 played a crucial role in prostatic carcinogenesis through the Rac and Rho pathways. Zhang et al took advantage of the integration of metabolomic and transcriptomic to unveil altered lipid metabolic pathways in pancreatic ductal adenocarcinoma.12 Herein, we screened 13 altered metabolites in PCT by LC‐MS metabolomic and the PLS‐DA model, and found that these altered metabolites were mainly enriched in pyrimidine metabolism. By analyzing both metabolomic and transcriptomic data, only one gene, ENTPD8, was found to be not only differentially expressed in PCT but also involved in the altered metabolic pathway. According to our findings, we hypothesized that CTP dephosphorization in pyrimidine metabolism was altered in PCT.
The ENTPD family was hypothesized to associate with inflammation and reactive oxygen species, which are related to cancer to some extent; for instance, ENTPD8, ENPP2, and ENPP3 were downregulated after calving.27 Moreover, ENTPD8 has been identified as a key regulator gene in chicken metabolism with the application of chicken time series microarray data.28 In the present study, we combined metabolomic with transcriptomic to conjointly analyze the altered metabolic pathways in PCT compared with normal tissues, and we investigated the mechanism of the altered metabolite cytidine and its related gene ENTPD8 in PC for the first time.
Ectonucleoside triphosphate diphosphohydrolases were significant ectonucleotidases that hydrolyzed nucleoside triphosphates and nucleoside diphosphates to monophosphate forms.29 Rosemberg et al. revealed that NTPDase members might regulate nucleotide hydrolysis in zebrafish tissues.30 ENTPD8 was also reported to be involved in ATP metabolism,31 and Roszek et al30 found that genes of nucleotidases were significantly overexpressed in osteoblasts compared with growing murine mesenchymal stem cells. Our study first investigated the mechanism of ENTPD8 in particular cancer. We confirmed that ENTPD8 was downregulated in PCT compared with ANT in vitro, and we verified that overexpression of ENTPD8 promoted apoptosis and inhibited viability of PC cells. The potential mechanism of ENTPD8 involved in cell apoptosis and proliferation might be that the low‐level expression of ENTPD8 can inhibit the metabolism of CTP to CMP so that the amount of CTP might be higher. The high level of CTP will metabolize into dCTP, which is involved in DNA synthesis in cancer cells. Overexpressed ENTPD8 might promote the CTP metabolized into cytidine and promoted apoptosis as well as inhibited proliferation.
This study exploited advanced metabolomic and transcriptomic profiling techniques to analyze metabolites and gene transcripts, so results from a single metabolomic dataset were strengthened. However, our limitation lay in the limited source of PC cell lines. Hence, future research could work on verifying the generality of this result in different cell lines or different types of PC.
In summary, our study intended to identify new biomarkers and outlined the mechanism of altered metabolism cytidine as well as its related gene ENTPD8 in PC with integrated analysis of metabolomic and transcriptomic. We found 7 biomarkers and revealed that ENPTD8 engaged in CTP dephosphorization in pyrimidine metabolism was highly deregulated in PCT, and the expression level of ENTPD8 was negatively related to PC tumor formation. Our findings suggested that ENTPD8 and cytidine were closely related to PC tumorigenesis, which could be a target for early detection and effective therapy of PC in the future.\
CONFLICT OF INTEREST
None.
Supporting information
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (No. 81502002) and the Natural Science Foundation of Jiangsu Province (No. BK20150254).
An Y, Cai H, Yang Y, et al. Identification of ENTPD8 and cytidine in pancreatic cancer by metabolomic and transcriptomic conjoint analysis. Cancer Sci. 2018;109:2811–2821. 10.1111/cas.13733
Funding information
National Natural Science Foundation of China, Grant/Award No. 81502002; Natural Science Foundation of Jiangsu Province, Grant/Award No. BK20150254.
Yong An and Huihua Cai contributed equally to this work.
Contributor Information
Donglin Sun, Email: czsdl@sina.com.
Xuemin Chen, Email: tomuer@126.com.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7‐30. [DOI] [PubMed] [Google Scholar]
- 2. Duell EJ, Lucenteforte E, Olson SH, et al. Pancreatitis and pancreatic cancer risk: a pooled analysis in the International Pancreatic Cancer Case–Control Consortium (PanC4). Ann Oncol. 2012;23:2964‐2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang H, Wang Y, Gu X, Zhou J, Yan C. Metabolomic profiling of human plasma in pancreatic cancer using pressurized capillary electrochromatography. Electrophoresis. 2011;32:340‐347. [DOI] [PubMed] [Google Scholar]
- 4. Auslander N, Yizhak K, Weinstock A, et al. A joint analysis of transcriptomic and metabolomic data uncovers enhanced enzyme‐metabolite coupling in breast cancer. Sci Rep. 2016;6:29662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Farshidfar F, Weljie AM, Kopciuk KA, et al. A validated metabolomic signature for colorectal cancer: exploration of the clinical value of metabolomics. Br J Cancer. 2016;115:848‐857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Miller MJ, Kennedy AD, Eckhart AD, et al. Untargeted metabolomic analysis for the clinical screening of inborn errors of metabolism. J Inherit Metab Dis. 2015;38:1029‐1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hu S, Wang J, Ji EH, Christison T, Lopez L, Huang Y. Targeted metabolomic analysis of head and neck cancer cells using high performance ion chromatography coupled with a Q exactive HF mass spectrometer. Anal Chem. 2015;87:6371‐6379. [DOI] [PubMed] [Google Scholar]
- 8. Such‐Sanmartin G, Bache N, Callesen AK, Rogowska‐Wrzesinska A, Jensen ON. Targeted mass spectrometry analysis of the proteins IGF1, IGF2, IBP2, IBP3 and A2GL by blood protein precipitation. J Proteomics. 2015;113:29‐37. [DOI] [PubMed] [Google Scholar]
- 9. Cho K, Moon JS, Kang JH, et al. Combined untargeted and targeted metabolomic profiling reveals urinary biomarkers for discriminating obese from normal‐weight adolescents. Pediatr Obes. 2017;12:93‐101. [DOI] [PubMed] [Google Scholar]
- 10. Shang X, Zhong X, Tian X. Metabolomics of papillary thyroid carcinoma tissues: potential biomarkers for diagnosis and promising targets for therapy. Tumour Biol. 2016;37:11163‐11175. [DOI] [PubMed] [Google Scholar]
- 11. Farid SG, Morris‐Stiff G. “OMICS” technologies and their role in foregut primary malignancies. Curr Probl Surg. 2015;52:409‐441. [DOI] [PubMed] [Google Scholar]
- 12. Zhang G, He P, Tan H, et al. Integration of metabolomics and transcriptomics revealed a fatty acid network exerting growth inhibitory effects in human pancreatic cancer. Clin Cancer Res. 2013;19:4983‐4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ren S, Shao Y, Zhao X, et al. Integration of metabolomics and transcriptomics reveals major metabolic pathways and potential biomarker involved in prostate cancer. Mol Cell Proteomics. 2016;15:154‐163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hakimi AA, Reznik E, Lee CH, et al. An integrated metabolic atlas of clear cell renal cell carcinoma. Cancer Cell. 2016;29:104‐116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Machado‐Vieira R, Salvadore G, DiazGranados N, et al. New therapeutic targets for mood disorders. ScientificWorldJournal. 2010;10:713‐726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chiang WC, Knowles AF. Transmembrane domain interactions affect the stability of the extracellular domain of the human NTPDase 2. Arch Biochem Biophys. 2008;472:89‐99. [DOI] [PubMed] [Google Scholar]
- 17. Pelletier J, Salem M, Lecka J, Fausther M, Bigonnesse F, Sevigny J. Generation and characterization of specific antibodies to the murine and human ectonucleotidase NTPDase8. Front Pharmacol. 2017;8:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tian H, Lam SM, Shui G. Metabolomics, a powerful tool for agricultural research. Int J Mol Sci. 2016;17:pii: E1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang H, Wang L, Hou Z, et al. Metabolomic profiling reveals potential biomarkers in esophageal cancer progression using liquid chromatography‐mass spectrometry platform. Biochem Biophys Res Commun. 2017;491:119‐125. [DOI] [PubMed] [Google Scholar]
- 20. Ohshima M, Sugahara K, Kasahara K, Katakura A. Metabolomic analysis of the saliva of Japanese patients with oral squamous cell carcinoma. Oncol Rep. 2017;37:2727‐2734. [DOI] [PubMed] [Google Scholar]
- 21. Wojakowska A, Chekan M, Widlak P, Pietrowska M. Application of metabolomics in thyroid cancer research. Int J Endocrinol. 2015;2015:258763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Itoi T, Sugimoto M, Umeda J, et al. Serum metabolomic profiles for human pancreatic cancer discrimination. Int J Mol Sci. 2017;18:pii: E767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Budhu A, Terunuma A, Zhang G, Hussain SP, Ambs S, Wang XW. Metabolic profiles are principally different between cancers of the liver, pancreas and breast. Int J Biol Sci. 2014;10:966‐972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fontana A, Copetti M, Di Gangi IM, et al. Development of a metabolites risk score for one‐year mortality risk prediction in pancreatic adenocarcinoma patients. Oncotarget. 2016;7:8968‐8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ortiz‐Villanueva E, Navarro‐Martin L, Jaumot J, et al. Metabolic disruption of zebrafish (Danio rerio) embryos by bisphenol A. An integrated metabolomic and transcriptomic approach. Environ Pollut. 2017;231:22‐36. [DOI] [PubMed] [Google Scholar]
- 26. Chen K, Zhu C, Cai M, et al. Integrative metabolome and transcriptome profiling reveals discordant glycolysis process between osteosarcoma and normal osteoblastic cells. J Cancer Res Clin Oncol. 2014;140:1715‐1721. [DOI] [PubMed] [Google Scholar]
- 27. Seo J, Osorio JS, Schmitt E, et al. Hepatic purinergic signaling gene network expression and its relationship with inflammation and oxidative stress biomarkers in blood from peripartal dairy cattle. J Dairy Sci. 2014;97:861‐873. [DOI] [PubMed] [Google Scholar]
- 28. Ma T, Xu L, Wang H, et al. Mining the key regulatory genes of chicken inosine 5′‐monophosphate metabolism based on time series microarray data. J Anim Sci Biotechnol. 2015;6:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sansom FM, Riedmaier P, Newton HJ, et al. Enzymatic properties of an ecto‐nucleoside triphosphate diphosphohydrolase from Legionella pneumophila: substrate specificity and requirement for virulence. J Biol Chem. 2008;283:12909‐12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rosemberg DB, Rico EP, Langoni AS et al. NTPDase family in zebrafish: nucleotide hydrolysis, molecular identification and gene expression profiles in brain, liver and heart. Comp Biochem Physiol B Biochem Mol Biol. 2010;155:230‐240. [DOI] [PubMed] [Google Scholar]
- 31. Fausther M, Lecka J, Soliman E, et al. Coexpression of ecto‐5′‐nucleotidase/CD73 with specific NTPDases differentially regulates adenosine formation in the rat liver. Am J Physiol Gastrointest Liver Physiol. 2012;302:G447‐G459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


