Mycobacterium abscessus is intrinsically resistant to many antimycobacterial antibiotics, which presents serious problems in therapy. Here, we describe the development of a novel phenotype-based microscopic and computerized imaging drug screening approach.
KEYWORDS: Mycobacterium abscessus, fluorescence assays, in vitro screening
ABSTRACT
Mycobacterium abscessus is intrinsically resistant to many antimycobacterial antibiotics, which presents serious problems in therapy. Here, we describe the development of a novel phenotype-based microscopic and computerized imaging drug screening approach. A pilot screen of 568 compounds from two libraries identified 17 hits. Eleven of these compounds are described for the first time as active against M. abscessus. The impact of growth media on the activity of these compounds was tested, revealing that cation-adjusted Mueller-Hinton broth (MHII) supports better growth of actively replicating M. abscessus and improves the activity of associated compounds.
INTRODUCTION
Mycobacterium abscessus belongs to the family of highly pathogenic nontuberculous mycobacteria (NTM). It is an intracellular pathogen that causes a wide range of diseases (1), from soft tissue infections in trauma patients to chronic respiratory infections in immunosuppressed individuals and cystic fibrosis (CF) patients (2). In CF patients, M. abscessus infections are associated with serious airway damage and lung function decrease (3).
The fast-growing M. abscessus is a chemotherapy-resistant mycobacterium (1), and it is intrinsically resistant to most drugs effective against the related Mycobacterium tuberculosis, including rifampin (4) and isoniazid (5), making treatment options exceedingly challenging (6). Currently, clinical treatment against M. abscessus uses macrolides, such as clarithromycin, amikacin, imipenem, and cefoxitin (7). Yet, therapy requires a combination of three antibiotics that need to be taken for several years, in addition to recommended surgical resection of infected lung tissue (8). This stresses the urgent need for developing new antibiotics tailored specifically to this bacterium. The following study describes our newly developed approach to screen novel compounds against M. abscessus, and a pilot screen of two predefined compound libraries in two different media against this pathogen. This phenotype-based approach is suitable for future high-content screening (HCS).
RESULTS AND DISCUSSION
In vitro fluorescence screening against M. abscessus.
Current screening of compounds against M. abscessus mainly utilizes growth density (optical density at 600 nm [OD600]) (9, 10) measurement or the resazurin/resorufin reaction (11, 12) for quantification of bacterial growth. Although optical density (OD) determination can be used in high-throughput screening (HTS), this method does not distinguish live from dead bacteria and is prone to cross contaminations. In contrast, fluorescence measurement provides higher sensitivity compared to OD- or resazurin-based analysis. Thus, we adapted the rapid, highly specific, phenotype-based automated approach used in other mycobacterial species (13) for M. abscessus drug screening.
We constructed a M. abscessus reporter strain expressing the tomato red fluorescent protein (RFP) from the pTEC27 plasmid (14, 15), which provides a sensitive, consistent, and efficient platform for HCS (16). As the rough morphotype variant of M. abscessus is more prevalent in CF patients (17) we chose it for our screening experiment. The sensitivity and specificity of fluorescence measurements for determination of mycobacterial growth are well documented in the literature (13, 18). Image analysis of the fluorescence signal is used to determine bacterial growth. The use of RFP-labeled M. abscessus can potentially be further developed for intracellular or biofilm HCS assays. Compared to that of the parental strain, the growth of the RFP strain is slightly slower in liquid culture, probably due to microbial fitness associated with the pTEC27 hygromycin selective marker.
We found that absorbance readings at OD600 are consistent with results of RFP-based analysis, as shown in Fig. S1 in the supplemental material. Yet, at subinhibitory concentrations, variations between both readouts were observed. A possible explanation relies on the fact that while OD600 measurement is limited to a single point of measurement at the center of the well, the fluorescence signal intensity from RFP is taken from a large area. Nevertheless, the Z′ score (19) of the RFP assay was determined to be 0.70 (±0.12) on average, similar to that of to assays performed against M. tuberculosis (13), suggesting that these variations are limited.
To address the impact of the assay medium on compound activity, we tested the compounds in both 7H9 broth and cation-adjusted Mueller-Hinton broth (MHII). MHII is recommended by the National Committee for Clinical Laboratory Standards (NCCLS) (20) for rapidly growing mycobacteria, such as M. abscessus, yet several previous publications (9, 10) have used 7H9 broth as the growth medium for M. abscessus drug susceptibility assays.
The RFP assay was validated with reference compounds shown in Table 1. MIC90 values were determined in vitro using the broth microdilution method for both 7H9 and MHII growth media, as both media were used in the past for M. abscessus MIC determinations (9, 10, 21).
TABLE 1.
Activity of reference compounds against M. abscessus pTEC27
Bedaquiline (22), gentamicin (23), and linezolid (24) were used as reference compounds and found to be active against M. abscessus, with very little difference in MICs between the two media (Table 1). Bedaquiline demonstrated outstanding activity in vitro, surpassing the other reference antibiotics (Table 1), which is consistent with data in the literature (10, 25).
A pilot screen of compound libraries against M. abscessus.
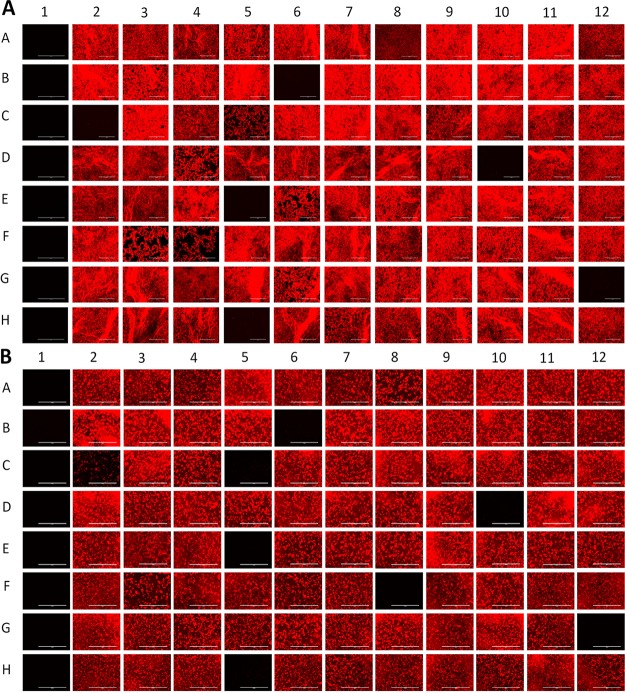
Next, we screened two well-defined compound libraries, the Pathogen Box (http://www.pathogenbox.org/) and GlaxoSmithKline (GSK)'s small-molecule M. tuberculosis leads (26) using our phenotype-based fluorescence assay. Initial screening was performed at single-point concentrations at 10 μM for the Pathogen Box and 25 μM for the GSK leads, using both growth media. Figure 1 shows an example of the fluorescence map obtained for 88 compounds from the Pathogen Box. Growth inhibition corresponds to absence or reduction of RFP signal in wells with active compounds. Compounds with a growth inhibition of >90% were considered hits and can be seen in Fig. 1A (B6, C2, D10, E5, G12, and H5). The impact of the screening medium was also assessed and examples of additional hits in the MHII screen can be seen in Fig. 1B (C5 and F8).
FIG 1.
In vitro screening monitored by RFP fluorescence imaging (A) Pathogen Box plate C in 7H9 (c = 10 μM) compared to (B) Pathogen Box plate C in MHII (c = 10 μM) (G12 contained 175 μM gentamicin, and H12 contained 1% dimethyl sulfoxide [DMSO]).
Analysis of hit compounds.
In total we identified 17 hit compounds out of 568 compounds tested (see Table S1 in the supplemental material). The distribution of hit compounds between the two libraries and their activities in the two media are shown in Fig. 2. The screen of the Pathogen Box at 10 μM resulted in 8 hits in addition to the reference compounds. The screen of GSK small-molecule leads resulted in the identification of 11 hit compounds active against M. abscessus; however, two compounds, MMV688846/GSK1985270A and MMV688845/GSK1055950A, are present in both libraries and showed similar activities in both screens. All hits from the Pathogen Box were active in MHII media; three compounds possessed selective activity in MHII, while five hits were active in both 7H9 broth and MHII. Interestingly, the majority of GSK compounds (with the exception of the two compounds also included in the Pathogen Box), were active either in 7H9 broth or in MHII but not in both. In total, five hit compounds, the entire BRL series hits from GSK, were exclusively active in 7H9 broth; seven compounds possessed selective activity in MHII, while five hits were active in both assays under the conditions tested (Fig. 2).
FIG 2.
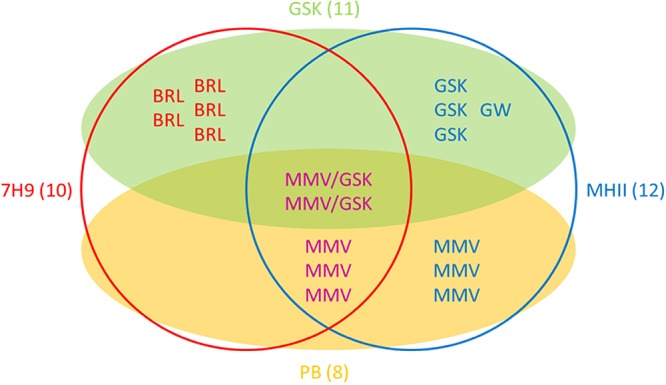
Venn diagram illustrating the source and activity of the 17 identified hit compounds. Compounds are listed by their alphabetic prefixes. Compounds obtained from the GSK series (GSK) are shown within the green shaded oval, while those from the Pathogen Box (PB) are contained within the yellow shaded oval. Two identical compounds belonged to both sets and thus have 2 different identifiers (MMV/GSK). Compounds that are active in 7H9 medium are shown within the red circle, while those active in MHII are contained within the blue circle. Numbers in brackets denote the total number of compounds within the given subset.
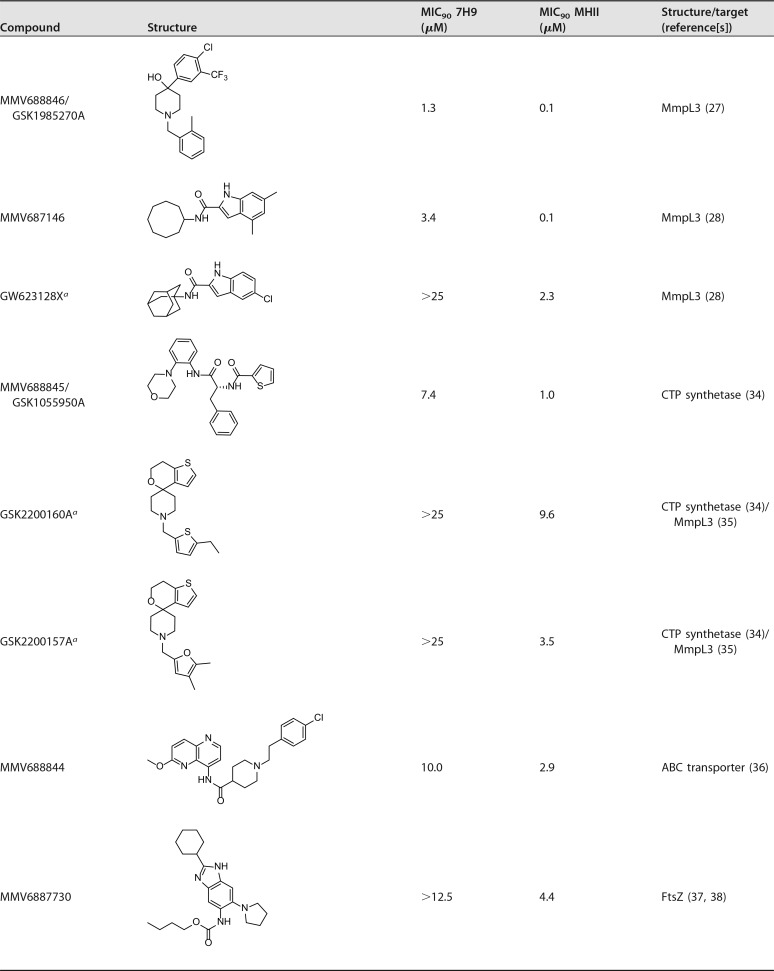
Of the 17 hits, six (MMV688846/GSK1985270A, MMV687146, MMV688845/GSK1055950A, MMV688844, MMV6887730, and MMV688508) were previously shown to be active against M. abscessus (9, 27, 28). However, our screen also identified 11 new hit compounds that are active against this pathogen.
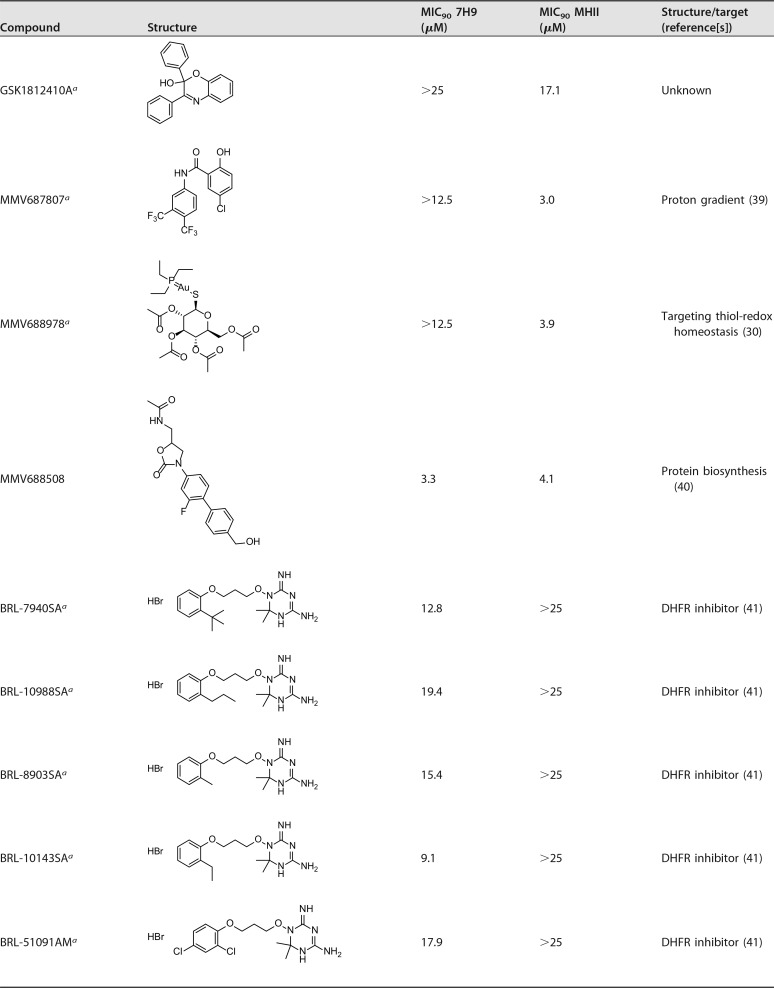
Table 2 shows the results obtained by the screen. Seven of the hits came from the tuberculosis disease set within the Pathogen Box, and one, MMV688978 (Auranofin) is an antirheumatic agent (29) with previously reported antimycobacterial activity (30). We found similar activity for the Pathogen Box in 7H9 medium compared to data published in a recent report by Low et al (9). In their study, however, they identified three additional hit compounds against M. abscessus, most likely because their initial cutoff point concentration was higher (20 μM versus 10 μM in our assay) (9). Interestingly, in our assay, we identified two new hit compounds (MMV688508 and MMV688978) that were active only in MHII media.
TABLE 2.
Activity data of hit compounds
Activity against M. abscessus reported for the first time in this study.
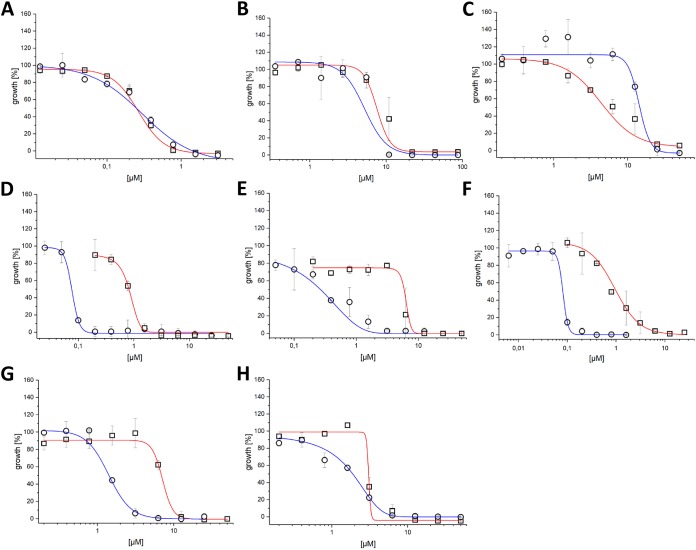
MIC values and dose-dependent killing curves were determined using the broth microdilution method, as shown in Fig. 3 and Fig. S2 in the supplemental material. Due to the high sensitivity of RFP fluorescence, which resulted in large variability in bacterial growth (>10%), killing curves were based on OD600 data and show an average Z′ score (19) of 0.85 (±0.09).
FIG 3.
The effect of growth media on M. abscessus killing curves. Activity against M. abscessus in 7H9 broth (red line, squares) and MHII (blue line, circles) for (A) bedaquiline, (B) gentamicin, (C) linezolid, (D) MMV688846, (E) MMV688845, (F) MMV687146, (G) MMV688844, and (H) MMV688508.
We compared the killing curves for the reference drugs in both media. Bedaquiline killing curves were found to be similar and highly effective in both media (Fig. 3A). Similarly, gentamicin showed a comparable killing curve in both media (Fig. 3B), whereas linezolid had a flatter dose-response slope in 7H9 broth compared to that in MHII (Fig. 3C).
Three inhibitors—MMV688846 (27), MMV687146 (28), and GW623128X (28, 31)—were effective against M. abscessus, with better activity (more than 10-fold lower MIC90) in MHII, as shown in Table 2 and Fig. 3D and F. All three compounds are known to target the Mycobacterial membrane protein Large 3 (MmpL3) (27, 32), an essential protein involved in the transport of mycolic acid and a common target of multiple-drug candidates of diverse chemical scaffolds (33). The phenylalanine derivative MMV688845 (34), a CTP synthetase inhibitor, also showed a lower MIC90 in MHII. The spiropiperidines GSK2200160A and GSK2200157A are also considered to be CTP synthetase inhibitors (34); however, some analogues of this compound class have been shown to target MmpL3 (35). Regardless of the mode of action, both analogues possess selective activity in MHII medium (concentration [c], ≤25 μM).
MMV688844 (36), an ATP-binding cassette (ABC) transporter inhibitor, showed activity against M. abscessus in both media, but with higher potency in MHII. The benzimidazole MMV687730 (37, 38), reported as an inhibitor of the filamenting temperature-sensitive mutant Z (FtsZ) protein, showed selective activity in MHII medium (c, <12.5 μM).
MMV688978 (29, 30) and MMV687807 (39) showed activity against M. abscessus only in MHII medium. The antirheumatic drug MMV688978 has been shown to target thiol-redox homeostasis (30), while MMV687807 is believed to disrupt the mycobacterial membrane proton gradient (39). We have also shown that GSK1812410A (26) is active against M. abscessus specifically in MHII medium. The target of this antimycobacterial compound remains unknown.
The MIC90 values for MMV688508, a synthesis intermediate of radezolid (40), are similar in both media, but the killing curve slope is less steep in MHII compared to that in 7H9 broth.
A set of five compounds sharing a dihydrotriazine moiety in their structure (BRL-7940SA, BRL-10988SA, BRL-8903SA, BRL-10143SA, and BRL-51091AM) were found to be active against M. abscessus exclusively in 7H9 medium, with an MIC90 range between 9.1 to 19.4 μM. Compounds of the BRL series were shown to target dihydrofolate reductase (DHFR) (41), with activity against both Plasmodium falciparum (42) and M. tuberculosis (26) Most likely, excess levels of thymidine present in MHII media allow M. abscessus to tolerate higher levels of these compounds, a phenomenon reported previously for sulfamethoxazole compounds (43).
The results presented above demonstrated a distinct effect of the assay medium. Out of the 17 hit compounds reported, 11 have an increased or selective activity in MHII. With the exception of the five dihydrotriazines mentioned above, similar or decreased MIC90 values were determined in MHII. Since both media are complex, it is challenging to correlate the differential activity to a single ingredient, but there are general differences. MHII broth contains 0.3% beef extract and 1.75% casein hydrolysate, compared to ADS (0.8% sodium chloride, 5.0% bovine serum albumin, and 2.0% dextrose in purified water)-supplemented 7H9 broth, which uses 0.5% albumin and 0.2% glucose as carbon source to support bacterial growth. The growth rate of M. abscessus in MHII is clearly higher than that in 7H9 broth, as shown in Fig. S3 in the supplemental material, which indicates higher metabolic activity in MHII. This observation can be explained, because MHII is a rich broth with excess carbon sources (44). This may induce higher susceptibility of M. abscessus to compounds active against replicating bacteria. On a different note, magnesium and calcium cation concentrations in MHII are adjusted according to NCCLS standards (20).
Phenotypic screening is widely used for antimycobacterial drug discovery; thus, the impact of assay medium is of great interest for the screening process. As such, to avoid missing identification of potential hit compounds, an ideal media should be selected. Currently, based on our results, except for the BRL compounds (as outlined above), screening in MHII is preferable for M. abscessus. Once hit compounds are identified, follow-up ex vivo and in vivo experiments will be used to assess and improve suitability and druggability of potential compounds.
In conclusion, we have shown that our newly developed phenotype-based fluorescence assay is useful for rapid screening of compound libraries against M. abscessus. Our success in identifying hit compounds emphasizes the advantage of testing preselected compound sets with activity against other infective agents, especially those obtained from tuberculosis (TB) drug discovery programs, as an effective approach for discovery of novel active compounds against M. abscessus. The set of hit compounds identified in this study encourages further hit-to-lead development to improve activity and pharmacokinetic properties.
MATERIALS AND METHODS
Equipment.
Equipment consisted of an Evos FL auto cell imaging system fluorescence microscope (Thermo Fisher) and a Victor2 1420 multilabel counter plate reader (PerkinElmer).
Compound libraries.
The Pathogen Box (http://www.pathogenbox.org/), established by the Medicines for Malaria Venture (MMV), is composed of 400 compounds active against various pathogens. Of these molecules, 129 are growth inhibitors of Mycobacterium tuberculosis, while the other 271 compounds show activity against other pathogens.
The small-molecule leads established by GSK (26) are a collection of potent antimycobacterial compounds.
Bacterial cells and culture media.
For transformation with the pTEC27 plasmid, M. abscessus ATCC 19977T R (rough form) was used. The plasmid pTEC27 carried a gene for expression of tomato RFP (15).
For transformation, 7H9 medium supplemented with 10% ADS and 0.05% Tween 80 was inoculated with M. abscessus ATCC 19977T and incubated for 1 to 2 days (OD600, 0.2 to 1.0). M. abscessus electrotransformation was carried out using Gene Pulser (Bio-Rad) at 2.50 kB, 25 μF, and 1,000 Ω.
For isolation of transformed cells, the suspension was plated on 7H10 plates with hygromycin (500 μg/ml). M. abscessus expressing tomato RFP has been used for the activity assays.
Stocks of the bacteria grown in complete 7H9 broth were stored in approximately 15% glycerol at −80°C.
Using an inoculation loop, bacteria were spread on 7H10 plates (containing hygromycin 500 μg/ml) and grown for 5 days in an incubator at 37°C. New plates were prepared every month from frozen stock.
Bacteria were grown in complete 7H9 broth supplemented with 10% ADS and 0.05% Tween 80, respectively, in MHII broth supplemented with 0.05% tyloxapol after scraping one colony of the 7H10 plate. Hygromycin (400 μg/ml) was added for M. abscessus pTEC27 growth. The culture was grown for 3 or 4 days before sub culturing in liquid broth. Subculturing was only done for 1.5 weeks, after those new colonies from the 7H10 plate were used for new cultures. The culture volume was 10 ml in a 50-ml Falcon tube. The tubes were covered to protect the photosensitive hygromycin and shaken in an incubator at 37°C.
Solid cultures were grown on 7H10 medium supplemented with 0.5% glycerol and 10% ADS. For M. abscessus pTEC27, hygromycin (500 μg/ml) was added.
Liquid cultures were grown in 7H9 medium supplemented with 10% ADS and 0.05% Tween 80, respectively, in MHII supplemented with 0.05% tyloxapol. For M. abscessus pTEC27, hygromycin (400 μg/ml) was added.
ADS supplement is a filter-sterilized solution of 0.8% sodium chloride, 5.0% bovine serum albumin, and 2.0% dextrose in purified water.
Single-point in vitro activity determination.
The compound libraries were screened in 96-well flat bottom Corning Costar plates at a concentration of 10 μM (Pathogen Box) or 25 μM (M. tuberculosis leads by GSK) in 7H9 medium supplemented with 10% ADS and 0.05% Tween 80, respectively, in MHII medium supplemented with 0.05% tyloxapol at a final volume of 100 μl. The concentration of the inoculum was 5 × 105 cells/ml (OD600, 0.1 [1 × 108 CFU/ml]). The starting inoculum was diluted from a preculture at the mid-log phase (OD600, 0.3 to 0.7). The plates were sealed with parafilm, put in a container with moist tissue, and incubated for 6 days at 37°C. Each plate had a negative control (1% dimethyl sulfoxide [DMSO]) and a positive control (175 μM gentamicin). After incubation, the plates were monitored by fluorescence microscopy (EVOS Fl Auto Imaging System), and an image of each well was taken (10× magnification). Based on the image data, the mycobacterial growth was quantified by image analysis with ImageJ software. Images were converted to 8-bit format, and a threshold was applied. Particles have been analyzed, and the percentage of area was used as signal for quantification. The assay was performed in duplicate. A growth inhibition of >90% is considered activity.
MIC determination in vitro.
MICs were determined by the broth microdilution method. Flat-bottom 96-well Corning Costar plates were used. In the first well in each row, two times the desired highest concentration of each compound was added in 7H9 medium supplemented with 10% ADS and 0.05% Tween 80, respectively, in MHII, medium supplemented with 0.05% tyloxapol (MIC of MMV687807 was determined without addition of tyloxapol). Each compound was diluted 2-fold in a 10-point serial dilution (100-μl final concentration). The concentration of the starting inoculum was 5 × 105 cells/ml. The starting inoculum was diluted from a preculture at the mid-log phase (OD600, 0.3 to 0.7). The assay was performed in the same way as the in vitro single-point activity determination described above. The assay was performed in duplicate.
Data analysis.
Every assay plate contained DMSO (1%) as the negative control, which corresponds to 100% bacterial growth, and gentamicin (175 μM) as positive controls in which 100% inhibition of bacterial growth was reached. Controls were used to monitor assay quality through determination of the Z′ score and for normalizing the data on a per-plate basis. The Z′ factor was determined using the following formula:
where SD is the standard deviation and M is the mean.
The percentage of inhibition was calculated as
For MIC90 determination, killing curves were calculated with OriginPro 2017G (OriginLab Corporation). The curves were fitted using the formula
where y is growth, A1 is the initial value for y, A2 is the final value for y, x is the concentration of the test compound, x0 is the concentration of the test compound at the center of the curve, and P is power.
Supplementary Material
ACKNOWLEDGMENTS
Funding for this research was provided by Cystic Fibrosis Canada (grant 493021 to Y.A.-G).
We thank the Medicines for Malaria Venture and GlaxoSmithKline for providing us access to their libraries. We also thank Tirosh Shapira for providing insightful comments and Laurent Kremer (Montpellier, France) for sharing vectors and expertise.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00828-18.
REFERENCES
- 1.Petrini B. 2006. Mycobacterium abscessus: an emerging rapid-growing potential pathogen. 2006. APMIS 114:319–328. doi: 10.1111/j.1600-0463.2006.apm_390.x. [DOI] [PubMed] [Google Scholar]
- 2.Lee MR, Sheng WH, Hung FCC, Yu CJ, Lee LN, Hsueh PR. 2015. Mycobacterium abscessus complex infections in humans. Emerg Infect Dis 21:638–1646. doi: 10.3201/eid2104.141562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryant JM, Grogono DM, Rodriguez-Rincon D, Everall I, Brown KP, Moreno P, Verma D, Hill E, Drijkoningen J, Gilligan P, Esther CR, Noone PG, Giddings O, Bell SC, Thomson R, Wainwright CE, Coulter C, Pandey S, Wood ME, Stockwell RE, Ramsay KA, Sherrard LJ, Kidd TJ, Jabbour N, Johnson GR, Knibbs LD, Morawska L, Sly PD, Jones A, Bilton D, Laurenson I, Ruddy M, Bourke S, Bowler IC, Chapman SJ, Clayton A, Cullen M, Daniels T, Dempsey O, Denton M, Desai M, Drew RJ, Edenborough F, Evans J, Folb J, Humphrey H, Isalska B, Jensen-Fangel S, Jönsson B, Jones AM, Katzenstein TL, Lillebaek T, MacGregor G, Mayell S, Millar M, Modha D, Nash EF, O'Brien C, O'Brien D, Ohri C, Pao CS, Peckham D, Perrin F, Perry A, Pressler T, Prtak L, Qvist T, Robb A, Rodgers H, Schaffer K, Shafi N, van Ingen J, Walshaw M, Watson D, West N, Whitehouse J, Haworth CS, Harris SR, Ordway D, Parkhill J, Floto RA. 2016. Emergence and spread of a human-transmissible multidrug-resistant nontuberculous mycobacterium. Science 354:751–757. doi: 10.1126/science.aaf8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nessar R, Cambau E, Reyrat JM, Murray A, Gicquel B. 2012. Mycobacterium abscessus: a new antibiotic nightmare. J Antimicrob Chemother 67:810–818. doi: 10.1093/jac/dkr578. [DOI] [PubMed] [Google Scholar]
- 5.Park S, Kim S, Park EM, Kim H, Kwon OJ, Chang CL, Lew WJ, Park YK, Koh WJ. 2008. In vitro antimicrobial susceptibility of Mycobacterium abscessus in Korea. J Korean Med Sci 23:49–52. doi: 10.3346/jkms.2008.23.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cortes M, Singh AK, Reyrat JM, Gaillard JL, Nassif X, Herrmann JL. 2011. Conditional gene expression in Mycobacterium abscessus. PLoS One 6:e29306. doi: 10.1371/journal.pone.0029306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aksamit TR, Philley JV, Griffith DE. 2014. Nontuberculous mycobacterial (NTM) lung disease: the top ten essentials. Respir Med 108:417–425. doi: 10.1016/j.rmed.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 8.Medjahed H, Gaillard JL, Reyrat JM. 2010. Mycobacterium abscessus: a new player in the mycobacterial field. Trends Microbiol 18:117–123. doi: 10.1016/j.tim.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 9.Low JL, Wu ML, Aziz DB, Laleu B, Dick T. 2017. Screening of TB actives for activity against nontuberculous mycobacteria delivers high hit rates. Front Microbiol 8:1539. doi: 10.3389/fmicb.2017.01539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aziz DB, Low JL, Wu ML, Gengenbacher M, Teo JWP, Dartois V, Dick T. 2017. Rifabutin is active against Mycobacterium abscessus complex. Antimicrob Agents Chemother 61:e00155-17. doi: 10.1128/AAC.00155-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia de Carvalho NF, Sato DN, Pavan FR, Ferrazoli L, Chimara E. 2016. Resazurin microtiter assay for clarithromycin susceptibility testing of clinical isolates of Mycobacterium abscessus group. J Clin Lab Anal 30:751–755. doi: 10.1002/jcla.21933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lavollayabce M, Dubéeabc M, Heymef B, Herrmanneg JL, Gaillardef JL, Gutmannabce L, Arthurabc M, Mainardiabce JL. 2014. In vitro activity of cefoxitin and imipenem against Mycobacterium abscessus complex. Clin Microbiol Infect 20:439–440. doi: 10.1111/1469-0691.12666. [DOI] [PubMed] [Google Scholar]
- 13.Sorrentino F, Gonzalez del Rio R, Zheng X, Presa Matilla J, Torres Gomez P, Martinez Hoyos M, Perez Herran ME, Mendoza Losana A, Av-Gay Y. 2016. Development of an intracellular screen for new compounds able to inhibit Mycobacterium tuberculosis growth in human macrophages. Antimicrob Agents Chemother 60:640–645. doi: 10.1128/AAC.01920-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernut A, Le Moigne V, Lesne T, Lutfalla G, Herrmann JL, Kremer L. 2014. in vivo assessment of drug efficacy against Mycobacterium abscessus using the embryonic zebrafish test system. Antimicrob Agents Chemother 58:4054–4063. doi: 10.1128/AAC.00142-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cambier CJ, Takaki KK, Larson RP, Hernandez RE, Tobin DM, Urdahl KB, Cosma CL, Ramakrishnan L. 2014. Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature 505:218–222. doi: 10.1038/nature12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng X, Av-Gay Y. 2016. New era of TB drug discovery and its impact on disease management. Curr Treat Options Infect Dis 8:299–310. doi: 10.1007/s40506-016-0098-0. [DOI] [Google Scholar]
- 17.Jönsson BE, Gilljam M, Lindblad A, Ridell M, Wold AE, Welinder-Olsson C. 2007. Molecular epidemiology of Mycobacterium abscessus, with focus on cystic fibrosis. J Clin Microbiol 45:1497–1504. doi: 10.1128/JCM.02592-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ollinger J, Bailey MA, Moraski GC, Casey A, Florio S, Alling T, Miller MJ, Parish T. 2013. A dual read-out assay to evaluate the potency of compounds active against Mycobacterium tuberculosis. PLoS One 8:e60531. doi: 10.1371/journal.pone.0060531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang JH, Chung TD, Oldenburg KR. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards. 2003. Susceptibility testing of mycobacteria, Nocardiae and other aerobic actinomycetes; approved standard. Document M24-A. National Committee for Clinical Laboratory Standards, Wayne, PA. [PubMed] [Google Scholar]
- 21.Pryjma M, Burian J, Kuchinski K, Thompson CJ. 2017. Antagonism between front-line antibiotics clarithromycin and amikacin in the treatment of Mycobacterium abscessus infections is mediated by the whiB7 gene. Antimicrob Agents Chemother 61:e01353-17. doi: 10.1128/AAC.01353-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diacon AH, Pym A, Grobusch M, Patientia R, Rustomjee R, Page-Shipp L, Pistorius C, Krause R, Bogoshi M, Churchyard G, Venter A, Allen J, Palomino JC, De Marez T, van Heeswijk RP, Lounis N, Meyvisch P, Verbeeck J, Parys W, de Beule K, Andries K, McNeeley DF. 2009. The diarylquinoline TMC207 for multidrug-resistant tuberculosis. N Engl J Med 360:2397–2405. doi: 10.1056/NEJMoa0808427. [DOI] [PubMed] [Google Scholar]
- 23.Mingeot-Leclercq MP, Glupczynski Y, Tulkens PM. 1999. Aminoglycosides: activity and resistance. Antimicrob Agents Chemother 43:727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swaney SM, Aoki A, Clelia Ganoza M, Shinabarger D. 1998. The oxazolidinone linezolid inhibits initiation of protein synthesis in bacteria. Antimicrob Agents Chemother 42:3251–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dupont C, Viljoen A, Thomas S, Roquet-Banères F, Herrmann JL, Pethe K, Kremer L. 2017. Bedaquiline inhibits the ATP synthase in Mycobacterium abscessus and is effective in infected zebrafish. Antimicrob Agents Chemother 61:e01225-17. doi: 10.1128/AAC.01225-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ballell L, Bates RH, Young RJ, Alvarez-Gomez D, Alvarez-Ruiz E, Barroso V, Blanco D, Crespo B, Escribano J, González R, Lozano S, Huss S, Santos-Villarejo A, Martín-Plaza JJ, Mendoza A, Rebollo-Lopez MJ, Remuiñan-Blanco M, Lavandera JL, Pérez-Herran E, Gamo-Benito FJ, García-Bustos JF, Barros D, Castro JP, Cammack N. 2013. Fueling open-source drug discovery: 177 small-molecule leads against tuberculosis. Chem Med Chem 8:313–321. doi: 10.1002/cmdc.201200428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dupont C, Viljoen A, Dubar F, Blaise M, Bernut A, Pawlik A, Bouchier C, Brosch R, Guérardel Y, Lelièvre J, Ballell L, Herrmann JL, Biot C, Kremer L. 2016. A new piperidinol derivative targeting mycolic acid transport in Mycobacterium abscessus. Mol Microbiol 101:515–529. doi: 10.1111/mmi.13406. [DOI] [PubMed] [Google Scholar]
- 28.Kozikowski AP, Onajole OK, Stec J et al, Kozikowski AP, Onajole OK, Stec J, Dupont C, Viljoen A, Richard M, Chaira T, Lun S, Bishai W, Raj VS, Ordway D, Kremer L. 2017. Targeting mycolic acid transport by indole-2-carboxamides for the treatment of Mycobacterium abscessus infections. J Med Chem 60:5876–5888. doi: 10.1021/acs.jmedchem.7b00582. [DOI] [PubMed] [Google Scholar]
- 29.Felson DT, Anderson JJ, Meenan RF. 1990. The comparative efficacy and toxicity of second-line drugs in rheumatoid arthritis results of two metaanalyses. Arthritis Rheumatism 33:1449–1461. doi: 10.1002/art.1780331001. [DOI] [PubMed] [Google Scholar]
- 30.Harbut MB, Vilcheze C, Luo X, Hensler ME, Guo H, Yang B, Chatterjee AK, Nizet V, Jacobs WR, Schultz PG, Wang F. 2015. Auranofin exerts broad-spectrum bactericidal activities by targeting thiol-redox homeostasis. Proc Natl Acad Sci U S A 112:4453–4458. doi: 10.1073/pnas.1504022112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Onajole OK, Pieroni M, Tipparaju SK, Lun S, Stec J, Chen G, Gunosewoyo H, Guo H, Ammerman NC, Bishai WR, Kozikowski AP. 2013. Preliminary structure-activity relationships and biological evaluation of novel antitubercular indolecarboxamide derivatives against drug-susceptible and drug-resistant Mycobacterium tuberculosis strains. J Med Chem 56:4093–4103. doi: 10.1021/jm4003878. [DOI] [PubMed] [Google Scholar]
- 32.Stec J, Onajole OK, Lun S, Guo H, Merenbloom B, Vistoli G, Bishai WR, Kozikowski AP. 2016. Indole-2-carboxamide-based MmpL3 inhibitors show exceptional antitubercular activity in an animal model of tuberculosis infection. J Med Chem 59:6232–6247. doi: 10.1021/acs.jmedchem.6b00415. [DOI] [PubMed] [Google Scholar]
- 33.Poce G, Consalvi S, Biava M. 2016. MmpL3 inhibitors: diverse chemical scaffolds inhibit the same target. Mini Rev Med Chem 16:1274–1283. doi: 10.2174/1389557516666160118105319. [DOI] [PubMed] [Google Scholar]
- 34.Esposito M, Szadocka S, Degiacomi G, Orena BS, Mori G, Piano V, Boldrin F, Zemanová J, Huszár S, Barros D, Ekins S, Lelièvre J, Manganelli R, Mattevi A, Pasca MR, Riccardi G, Ballell L, Mikušová K, Chiarelli LR. 2017. A phenotypic based target screening approach delivers new antitubercular CTP synthetase inhibitors. ACS Infect Dis 3:428–437. doi: 10.1021/acsinfecdis.7b00006. [DOI] [PubMed] [Google Scholar]
- 35.Remuiñán MJ, Pérez-Herrán E, Rullás J, Alemparte C, Martínez-Hoyos M, Dow DJ, Afari J, Mehta N, Esquivias J, Jiménez E, Ortega-Muro F, Fraile-Gabaldón MT, Spivey VL, Loman NJ, Pallen MJ, Constantinidou C, Minick DJ, Cacho M, Rebollo-López MJ, González C, Sousa V, Angulo-Barturen I, Mendoza-Losana A, Barros D, Besra GS, Ballell L, Cammack N. 2013. Tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide and N-benzyl-6′,7′-dihydrospiro[piperidine-4,4′-thieno[3,2-c]pyran] analogues with bactericidal efficacy against Mycobacterium tuberculosis targeting MmpL3. PLoS One 8:e60933. doi: 10.1371/journal.pone.0060933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rebollo-Lopez MJ, Lelièvre J, Alvarez-Gomez D, Castro-Pichel J, Martínez-Jiménez F, Papadatos G, Kumar V, Colmenarejo G, Mugumbate G, Hurle M, Barroso V, Young RJ, Martinez-Hoyos M, González del Río R, Bates RH, Lopez-Roman EM, Mendoza-Losana A, Brown JR, Alvarez-Ruiz E, Marti-Renom MA, Overington JP, Cammack N, Ballell L, Barros-Aguire D. 2015. Release of 50 new, drug-like compounds and their computational target predictions for open source anti-tubercular drug discovery. PLoS One 10:e0142293. doi: 10.1371/journal.pone.0142293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li D, Chi B, Wang WW, Gao JM, Wan J. 2016. Exploring the possible binding mode of trisubstituted benzimidazoles analogues in silico for novel drug design targeting Mtb FtsZ. Med Chem Res 26:153–169. doi: 10.1007/s00044-016-1734-4. [DOI] [Google Scholar]
- 38.Kumar K, Awasthi D, Lee SY, Zanardi I, Ruzsicska B, Knudson S. 2011. Novel trisubstituted benzimidazoles, targeting Mtb FtsZ, as a new class of antitubercular agents. J Med Chem 54:374–381. doi: 10.1021/jm1012006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee IJ, Gruber TD, Samuels A, Yun M, Nam B, Kang M, Crowley K, Winterroth B, Boshoff HI, Barry CI. 2013. Structure-activity relationships of antitubercular salicylanilides consistent with disruption of the proton gradient via proton shuttling. Bioorg Med 21:114–126. doi: 10.1016/j.bmc.2012.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schaadt R, Sweeney D, Shinabarger D, Zurenko G. 2009. In vitro activity of TR-700, the active ingredient of the antibacterial prodrug TR-701, a novel oxazolidinone antibacterial agent. Antimicrob Agents Chemother 53:3236–3239. doi: 10.1128/AAC.00228-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar A, Guardia A, Colmenarejo G, Pérez E, Gonzalez RR, Torres P, Calvo D, Gómez RM, Ortega F, Jiménez E, Gabarro RC, Rullás J, Ballell L, Sherman DR. 2015. A Focused screen identifies antifolates with activity on Mycobacterium tuberculosis. ACS Infect Dis 1:604–614. doi: 10.1021/acsinfecdis.5b00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Childs GE, Lambros C. 1986. Analogues of N-benzyloxydihydrotriazines: in vitro antimalarial activity against Plasmodium falciparum. Ann Trop Med Parasitol 80:177–181. doi: 10.1080/00034983.1986.11812002. [DOI] [PubMed] [Google Scholar]
- 43.Swenson JM, Thornsberry C. 1978. Susceptibility tests for sulfamethoxazole-trimethoprim by a broth microdilution procedure. Curr Microbiol 1:189–193. doi: 10.1007/BF02601676. [DOI] [PubMed] [Google Scholar]
- 44.Thompson MG, Corey BW, Si Y, Craft DW, Zurawski DV. 2012. Antibacterial activities of iron chelators against common nosocomial pathogens. Antimicrob Agents Chemother 56:5419–5421. doi: 10.1128/AAC.01197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.