The aim of this study was to evaluate the antimicrobial susceptibilities of 866 Neisseria meningitidis invasive strains during 11 years of surveillance in Italy. Two and six strains were resistant to ciprofloxacin and rifampin, respectively.
KEYWORDS: antimicrobial resistance, Neisseria meningitidis, penA gene, Peni, penicillin-binding protein 2
ABSTRACT
The aim of this study was to evaluate the antimicrobial susceptibilities of 866 Neisseria meningitidis invasive strains during 11 years of surveillance in Italy. Two and six strains were resistant to ciprofloxacin and rifampin, respectively. Forty-five percent were penicillin intermediate, associated with hypervirulent serogroup C clonal complex 11. All of the strains were susceptible to cephalosporins.
TEXT
Invasive meningococcal disease (IMD) is a serious and rapidly progressive illness; third-generation cephalosporins or penicillin G are usually used for the treatment of patients with invasive diseases (1, 2). Ciprofloxacin or rifampin is recommended for chemoprophylaxis of close contacts of the case (2).
Although antimicrobial resistance in Neisseria meningitidis strains is rare (3), reduced susceptibility to third-generation cephalosporins has recently been reported (4). Moreover, meningococci with reduced susceptibility to penicillin G (penicillin intermediate [Peni]) have been described (3, 5–7). The Peni phenotype is mainly due to the presence of five amino acid substitutions (F504L, A510V, I515V, G541N, and I566V) in the transpeptidase region of the penicillin-binding protein 2 (PBP2), encoded by the penA gene (8–10).
This study was conducted to evaluate the antimicrobial susceptibilities of 866 meningococcal invasive strains isolated from 2006 to 2016 in Italy. Genotyping and determination of the penA gene of Peni strains from 2014 to 2016 were also performed.
Clinical data, strains, and/or clinical samples of each IMD case are collected throughout the country and sent to the National Reference Laboratory (NRL) at the Istituto Superiore di Sanità (ISS), within the activities of the National Surveillance System.
Strains were cultured on Thayer-Martin agar plates with IsoVitaleX 2% (Oxoid, Ltd.) in 5% CO2 atmosphere at 37°C. Serogroup by slide agglutination with commercial antisera (Remel Europe, Ltd., UK) or by multiplex PCR was determined (11).
Antimicrobial susceptibility testing for ceftriaxone, cefotaxime, ciprofloxacin, penicillin G, and rifampin was performed using Etest (bioMérieux, Sweden) and MIC test strip methods (Liofilchem Diagnostici, Italy) interpreted according to European Committee Antimicrobial Susceptibility Testing (EUCAST; v. 7.1, 2017-03-10) (12). In this study, MIC values ranging from 0.094 to 0.25 μg/ml define the Peni phenotype.
DNA was extracted using QIAamp DNA minikit (Qiagen, Hilden, Germany) for whole-genome sequencing (WGS) (13). Genomes were uploaded and analyzed on the Neisseria PubMLST database (http://pubmlst.org/neisseria/). Multilocus sequence typing (MLST), porin A (PorA) and ferric enterobactin transport protein A (FetA) typing, and the penA allele were identified as described in the database. The genotypic formula is identified as follows: capsular group: porA (P1); variable region 1 (VR1), VR2 : FetA VR: sequence type (ST) clonal complex (CC).
Statistical analysis was performed by the χ2 test. A P value of <0.05 was considered to be statistically significant.
From 1 January 2006 to 31 December 2016, a total of 1,188 samples from IMD cases were received at the NRL, of which 866 samples (866/1,188 [73%]) were culture positive. As shown in Table 1, all meningococci were susceptible to ceftriaxone (866/866 [100%]) and to cefotaxime (227/227 [100%]). Except for two samples, meningococci were susceptible to ciprofloxacin (864/866 [99.7%]). Those resistant (MICs, 0.064 μg/ml and 0.19 μg/ml, respectively) were from serogroups A and C, collected from unvaccinated adults with meningitis.
TABLE 1.
Antimicrobial and susceptibility categories identified in 866 Neisseria meningitidis strains by year, 2006 to 2016
| Susceptibility category (MIC) by antimicrobial (μg/ml)a | No. of strains by yr (n) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2006 (88) | 2007 (80) | 2008 (101) | 2009 (104) | 2010 (69) | 2011 (66) | 2012 (62) | 2013 (69) | 2014 (60) | 2015 (72) | 2016 (95) | Total (866) | |
| Ceftriaxone | ||||||||||||
| S ≤ 0.125 | 88 | 80 | 101 | 104 | 69 | 66 | 62 | 69 | 60 | 72 | 95 | 866 |
| Cefotaxime | ||||||||||||
| S ≤ 0.125 | NAb | NA | NA | NA | NA | NA | NA | NA | 60 | 72 | 95 | 227 |
| Ciprofloxacin | ||||||||||||
| S ≤ 0.03 | 88 | 80 | 101 | 103 | 68 | 66 | 62 | 69 | 60 | 72 | 95 | 864 |
| R > 0.03 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| Rifampin | ||||||||||||
| S ≤ 0.25 | 88 | 79 | 101 | 101 | 69 | 66 | 60 | 69 | 60 | 72 | 95 | 860 |
| R > 0.25 | 0 | 1 | 0 | 3 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 6 |
| Penicillin G | ||||||||||||
| S ≤ 0.06 | 49 | 54 | 69 | 69 | 38 | 39 | 25 | 34 | 22 | 32 | 41 | 472 |
| 0.094 < I < 0.25 | 34 | 26 | 32 | 34 | 31 | 27 | 37 | 35 | 38 | 40 | 54 | 388 |
| R > 0.25 | 5 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 |
S, susceptible; R, resistant; I, intermediate.
NA, not applicable; antimicrobial susceptibility testing for cefotaxime was evaluated starting from 2014.
Six strains were rifampin resistant (6/866 [0.7%]), with 3 strains from serogroups B, C, and NG, with MIC values ranging from 0.38 μg/ml to 2 μg/ml, and 3 strains of serogroup C with a high level of resistance (MIC, 32 μg/ml). Rifampin-resistant strains were isolated from unvaccinated patients (from 5 to 54 years of age), one of whom died.
A total of 472 strains (472/866 [55%]) were susceptible to penicillin G (Pens) (MIC, ≤0.06 μg/ml), and 388 strains (388/866 [45%]) were penicillin G intermediate (Peni), with an MIC range of 0.094 to 0.25 μg/ml (Table 1). Peni strains were collected from unvaccinated (184/388 [47%]) and vaccinated (23/388 [6%]) patients through all the age groups. Forty-two percent presented with meningitis, 32% presented with sepsis, 10% presented with meningitis plus sepsis, and the data for the remaining strains were unknown. Eleven percent (44/388) of the patients with Peni strains died, of which 68% (30/44) had sepsis.
A total of 6 penicillin G-resistant (Penr) strains, with 5 strains in 2006 and 1 strain in 2009, with an MIC range of 0.38 to 0.5 μg/ml (Table 1), were detected. Penr meningococci were isolated from unvaccinated patients (1 patient with meningitis, 4 patients with sepsis, and 1 patient with unknown clinical presentation), with an age range of 1 to 83 years. The 83-year-old patient, who had sepsis, died.
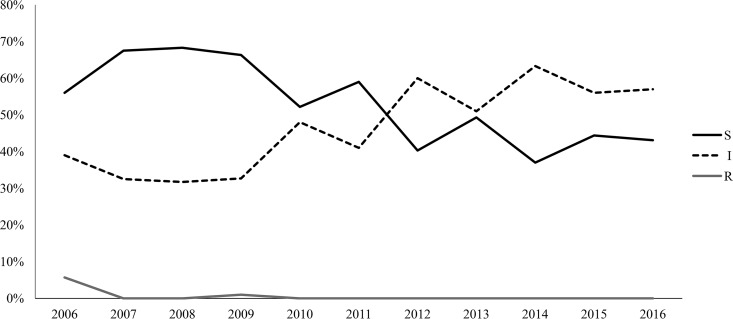
As shown in Fig. 1, the antimicrobial susceptibility trend of penicillin G changed over the time frame. In particular, starting from 2012, a statistically significant increase in Peni strains (P < 0.05) has been observed.
FIG 1.
Trend of antimicrobial susceptibility to penicillin G in 866 invasive meningococcal strains, Italy, from 2006 to 2016. S, susceptible; I, intermediate; R, resistant.
The sequence of a 402-bp DNA fragment of the 3′ part of penA was obtained for 132 Peni strains of more recent isolation (2014 to 2016). Twenty-three penA alleles were identified, of which penA248 was the most prevalent. Out of 23 penA alleles, 20 alleles coded for a peptide with 5 amino acid substitutions in the C-terminal region of PBP2 (Table 2). penA327 and penA648 harbored 4 substitutions (lacking I566V). The penA1 wild-type allele was found in 3 Peni strains (MIC, 0.094 to 0.25 μg/ml) (Table 2).
TABLE 2.
Characterization of 132 Peni meningococci isolated in Italy, from 2014 to 2016
| penA allele | No. of strains | No. of substitutions in PBP2 | No. of strainsa |
No. of strains by MIC (μg/ml) |
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MenC |
MenB |
MenY CC23 | MenW CC22 | |||||||||||||||||||||
| CC11 | CC175 | CC22 | CC334 | CC865 | Unknown | CC162 | CC167 | CC18 | CC213 | CC269 | CC32 | CC41/44 | CC461 | CC865 | Unknown | 0.094 | 0.125 | 0.19 | 0.25 | |||||
| 1 | 3 | 0 | 1 | 1 | 1 | 2 | 1 | |||||||||||||||||
| 7 | 19 | 5 | 19 | 5 | 8 | 6 | ||||||||||||||||||
| 9 | 9 | 5 | 1 | 1 | 2 | 4 | 1 | 1 | 4 | 3 | 1 | |||||||||||||
| 12 | 1 | 5 | 1 | 1 | ||||||||||||||||||||
| 13 | 2 | 5 | 1 | 1 | 1 | 1 | ||||||||||||||||||
| 14 | 13 | 5 | 1 | 9 | 1 | 1 | 1 | 3 | 7 | 3 | ||||||||||||||
| 15 | 4 | 5 | 1 | 3 | 3 | 1 | ||||||||||||||||||
| 19 | 1 | 5 | 1 | 1 | ||||||||||||||||||||
| 20 | 15 | 5 | 15 | 8 | 3 | 3 | 1 | |||||||||||||||||
| 25 | 1 | 5 | 1 | 1 | ||||||||||||||||||||
| 33 | 4 | 5 | 4 | 2 | 2 | |||||||||||||||||||
| 54 | 1 | 5 | 1 | 1 | ||||||||||||||||||||
| 100 | 1 | 5 | 1 | 1 | ||||||||||||||||||||
| 144 | 1 | 5 | 1 | 1 | ||||||||||||||||||||
| 248 | 36 | 5 | 35 | 1 | 9 | 11 | 12 | 4 | ||||||||||||||||
| 295 | 2 | 5 | 2 | 1 | 1 | |||||||||||||||||||
| 327 | 3 | 4 | 3 | 2 | 1 | |||||||||||||||||||
| 599 | 9 | 5 | 9 | 4 | 4 | 1 | ||||||||||||||||||
| 648 | 1 | 4 | 1 | 1 | ||||||||||||||||||||
| 685 | 1 | 5 | 1 | 1 | ||||||||||||||||||||
| 773b | 2 | 5 | 2 | 1 | 1 | |||||||||||||||||||
| 774b | 2 | 5 | 2 | 1 | 1 | |||||||||||||||||||
| 775b | 1 | 5 | 1 | 1 | ||||||||||||||||||||
| Total | 132 | 59 | 1 | 1 | 9 | 1 | 2 | 11 | 1 | 1 | 2 | 2 | 3 | 6 | 4 | 2 | 9 | 16 | 2 | 42 | 45 | 34 | 11 | |
MenC, meningococcal of serogroup C; MenB, meningococcal of serogroup B; MenY, meningococcal of serogroup Y; MenW, meningococcal of serogroup W.
New penA allele.
As shown in Table 2, 55% (73/132) of the Peni strains belonged to serogroup C, of which 81% (59/73) of the strains were associated with clonal complex 11 (CC11); serogroup B, comprising 31% (41/132) of the Peni strains, was mostly associated with CC162; serogroup Y was associated with CC23 (16/132 [12%]), and serogroup W was associated with CC22 (2/132 [2%]).
Here, in 11 years of IMD surveillance in Italy, invasive meningococcal strains showed a wide range susceptibility to the antimicrobials used for treatment and chemoprophylaxis. The exception was 6 rifampin-resistant strains, of which 3 strains were highly resistant and 2 strains were ciprofloxacin resistant. Of note, an increase in the proportion of Peni strains, starting from 2012, has been observed.
It is likely that the increase in Peni strains was due to the spread of the hypervirulent strain C-CC11 that is of a concern in our country (13). The penA248 allele was the predominant allele and was associated with the finetype C: P1. 5-1, 10-8:F3-6: ST-11 CC11) (data not shown), which is responsible for severe sporadic cases and outbreaks in Italy (13).
Interestingly, 3 Peni strains harboring the penA327 allele showed an increased MIC to cefotaxime even though they were within the susceptibility category. Two of these strains were isolated from men who have sex with men (MSM) with sepsis. This occurrence has been already reported by others (4), underlying that the similarity between penA327 of N. meningitidis and penA-XXXIV of N. gonorrhoeae might determine a genetic exchange between the two Neisseria spp. in the urethra (4, 9).
To conclude, resistant meningococci are rare in this country; however, an increase in Peni strains was observed mainly associated with the spread of C-CC11 meningococci. Because of the concern over the epidemic potential of this strain, it is crucial to link the molecular traits of invasive meningococcal strains with antimicrobial susceptibility, with a particular attention to the emergence of meningococci with reduced susceptibility to cephalosporins (4).
ACKNOWLEDGMENTS
The members of the following groups are: National Surveillance System of Invasive Meningococcal Diseases, Flavia Riccardo, Martina Del Manso, and Maria Grazia Caporali, Department of Infectious Diseases, Istituto Superiore di Sanità, Rome, Italy; Fortunato Paolo D'Ancona, Ministry of Health, Rome, Italy; Milena Arghittu, Laura Daprai, and Damiano Picicco, Microbiology Laboratory, Fondazione IRCCS Cà Granda Ospedale Maggiore Policlinico, Milan, Italy; Daniela Lombardi, SeREMI, ASL AL, Regional reference Centre for Infectious Disease Surveillance, Alessandria, Italy; Anna Maria Barbui, Microbiology and Laboratory, Molinette Hospital, Turin, Italy; Maria Paola Landini, Maria Carla Re, and Caterina Vocale, CRREM, Unit of Clinical Microbiology, St. Orsola Malpighi University Hospital, Bologna, Italy; Paola Bernaschi, Microbiology Unit, Children's Hospital Bambino Gesù, Rome, Italy; Teresa Spanu, Department of Infectious Diseases, Catholic University of Rome, Rome, Italy; Camilla Ajassa, Department of Public Health and Infectious Diseases, Sapienza University, Rome, Italy; Carla Fontana and Maria Cristina Bossa, Microbiology and Virology Laboratory, Tor Vergata University Hospital of Rome, Italy; Carmen Bonanno and Maria Carmela Cava, Microbiology and Virology Laboratory, Sandro Pertini Hospital, Rome, Italy; Iolanda Santino, Department of Clinical and Molecular Medicine, Sapienza University, Microbiology Unit, Sant'Andrea Hospital, Rome, Italy; Maria Grazia Paglia and Antonella Vulcano, National Institute for the Infectious Diseases Lazzaro Spallanzani, Microbiology Laboratory and Infectious Diseases Biorepository, Rome, Italy; Gian Maria Rossolini and Anna Maria Bartolesi, Clinical Microbiology and Virology Unit, Florence Careggi University Hospital, Florence, Italy; Chiara Azzari and Maria Moriondo, Department of Health Sciences, University of Florence, Anna Meyer Children's University Hospital, Florence, Italy; Patrizia Isola, UOC Medicina di Laboratorio Livorno, Azienda USL Toscana Nord Ovest, Livorno, Italy; Irene Galanti, San Donato Hospital, Arezzo, Italy; Roberto Degli Innocenti, Nuovo Ospedale Santo Stefano, Prato, Italy; Paolo Lanzafame, Department of Microbiology, Santa Chiara Regional Hospital, Trento, Italy; Patrizia Innocenti and Elisabetta Pagani, Laboratory of Microbiology and Virology, Comprensorio Sanitario di Bolzano, Italy; Paolo Castiglia, Department of Clinical and Sperimental Medicine-Hygiene and Preventive Medicine Unit, University-AOU of Sassari, Hospital of Sassari, Italy; Elena Poma, Laboratory of San Martino Hospital, Oristano, Italy; Luciana Contini, Department of Public Health, Sassari, Italy; Carlo Tascini and Novella Carannante, First Division of Infectious Diseases, Cotugno Hospital, Naples, Italy; Antonella Mencacci, Medical Microbiology Section, Department of Medicine, University of Perugia, Santa Maria della Misericordia Hospital, Perugia, Italy; Lucia Rossi, Microbiology and Virology Unit, University Hospital, Padua, Italy; Alessandro Camporese, Microbiology and Virology Department, Santa Maria degli Angeli Regional Hospital, Pordenone, Italy; Francesca Orecchioni, Valeria Travaglini, Clinical Analysis Laboratory, Section of Microbiology, Ospedali Riuniti di Ancona, Italy; Maria Chironna, Department of Biomedical Science and Human Oncology, Aldo Moro University of Bari, Italy; Anna Di Taranto, Department of Clinical Pathology, University Hospital OORR, Foggia, Italy; Anna Giammanco, Teresa Fasciana, and Chiara Mascarella, Department of Sciences for Health Promotion and Mother-Child Care G. D'Alessandro, Palermo, Italy; Paolo Fazii, S. Spirito Hospital, Pescara, Italy; Graziella Soldato, Department of Prevention, Epidemiology and Public Health, ASL Pescara, Italy; Maria Golato, Unit of Clinical Pathology, SS Annunziata University Hospital, ASL Lanciano-Vasto-Chieti, Chieti, Italy; Teresa Lopizzo and Letizia Camardese, Unit of Clinical Microbiology and Virology, San Carlo Hospital, Potenza, Italy; Cristina Giraldi, Unit of Microbiology and Virology, Annunziata Hospital, Cosenza, Italy; and Alessandro Bisbano, Epidemiology Unit ASP Crotone, Calabria, Italy.
This publication made use of the Neisseria MLST website (http://pubmlst.org/neisseria/), developed by Keith Jolley and sited at the University of Oxford, UK (14). The development of this site has been funded by the Wellcome Trust and European Union.
This work was partly funded by the Italian Ministry of Health-CCM project 6M22 “Sorveglianza delle malattie invasive da Neisseria meningitidis, Streptococcus pneumoniae ed Haemophilus influenzae.”
P.S. reports research grants from GSK and Pfizer for unrelated scientific projects.
REFERENCES
- 1.Nadel S. 2016. Treatment of meningococcal disease. J Adolesc Health 59:S21–S28. doi: 10.1016/j.jadohealth.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 2.Pickering LK, Baker CJ, Kimberlin DW, Long SS. 2012. Report of the Committee on Infectious Diseases, 28th ed American Academy of Pediatrics, Grove Village, IL. [Google Scholar]
- 3.Harcourt BH, Anderson RD, Wu HM, Cohn AC, MacNeil JR, Taylor TH, Wang X, Clark TA, Messonnier NE, Mayer LW. 2015. Population-based surveillance of Neisseria meningitidis antimicrobial resistance in the United States. Open Forum Infect Dis 2:ofv117. doi: 10.1093/ofid/ofv117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deghmane AE, Hong E, Taha MK. 2017. Emergence of meningococci with reduced susceptibility to third-generation cephalosporins. J Antimicrob Chemother 72:95–98. doi: 10.1093/jac/dkw400. [DOI] [PubMed] [Google Scholar]
- 5.Brown EM, Fisman DN, Drews SJ, Dolman S, Rawte P, Brown S, Jamieson F. 2010. Epidemiology of invasive meningococcal disease with decreased susceptibility to penicillin in Ontario, Canada, 2000 to 2006. Antimicrob Agents Chemother 54:1016–1021. doi: 10.1128/AAC.01077-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hedberg ST, Fredlund H, Nicolas P, Caugant DA, Olcén P, Unemo M. 2009. Antibiotic susceptibility and characteristics of Neisseria meningitidis isolates from the African meningitis belt, 2000 to 2006: phenotypic and genotypic perspectives. Antimicrob Agents Chemother 53:1561–1566. doi: 10.1128/AAC.00994-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertrand S, Carion F, Wintjens R, Mathys V, Vanhoof R. 2012. Evolutionary changes in antimicrobial resistance of invasive Neisseria meningitidis isolates in Belgium from 2000 to 2010: increasing prevalence of penicillin nonsusceptibility. Antimicrob Agents Chemother 56:2268–2272. doi: 10.1128/AAC.06310-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karch A, Vogel U, Claus H. 2015. Role of penA polymorphisms for penicillin susceptibility in Neisseria lactamica and Neisseria meningitidis. Int J Med Microbiol 305:729–735. doi: 10.1016/j.ijmm.2015.08.025. [DOI] [PubMed] [Google Scholar]
- 9.Zapun A, Morlot C, Taha MK. 2016. Resistance to β-lactams in Neisseria ssp. due to chromosomally encoded penicillin-binding proteins. Antibiotics (Basel) 5:E35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thulin S, Olcén P, Fredlund H, Unemo M. 2006. Total variation in the penA gene of Neisseria meningitidis: correlation between susceptibility to beta-lactam antibiotics and penA gene heterogeneity. Antimicrob Agents Chemother 50:3317–3324. doi: 10.1128/AAC.00353-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu H, Wang Q, Wen L, Xu J, Shao Z, Chen M, Reeves PR, Cao B, Wang L. 2012. Development of a multiplex PCR assay for detection and genogrouping of Neisseria meningitidis. J Clin Microbiol 50:46–51. doi: 10.1128/JCM.00918-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.EUCAST. 2017. Breakpoint tables for interpretation of MICs and zone diameters version 7.1. European Committee on Antimicrobial Susceptibility Testing, Växjö, Sweden: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_7.1_Breakpoint_Tables.pdf. [Google Scholar]
- 13.Stefanelli P, Fazio C, Neri A, Ciammaruconi A, Balocchini E, Anselmo A, Azzari C, Rossolini GM, Vacca P, Fortunato A, Palozzi A, Fillo S, Lista F, Moriondo M, Nieddu F, Rezza G. 2016. Genome-based study of a spatio-temporal cluster of invasive meningococcal disease due to Neisseria meningitidis serogroup C, clonal complex 11. J Infect 73:136–144. doi: 10.1016/j.jinf.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Jolley KA, Maiden MCJ. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]