A pan-azole-resistant Aspergillus fumigatus strain with the cyp51A mutations Gly138Ser and Asn248Lys was isolated from a patient receiving long-term voriconazole treatment. PCR fragments containing cyp51A with the mutations were introduced along with the Cas9 protein and single guide RNA into the azole-resistant/susceptible strains.
KEYWORDS: azole drugs, antifungal resistance, Cyp51A, Cas9, CRISPR, genome editing
ABSTRACT
A pan-azole-resistant Aspergillus fumigatus strain with the cyp51A mutations Gly138Ser and Asn248Lys was isolated from a patient receiving long-term voriconazole treatment. PCR fragments containing cyp51A with the mutations were introduced along with the Cas9 protein and single guide RNA into the azole-resistant/susceptible strains. Recombinant strains showed increased susceptibility via the replacement of Ser138 by glycine. Genetic recombination, which has been hampered thus far in clinical isolates, can now be achieved using CRISPR/Cas9 genome editing.
TEXT
The filamentous fungus Aspergillus fumigatus is the most common opportunistic human fungal pathogen, with a wide range of clinical features, including invasive pulmonary aspergillosis, chronic progressive pulmonary aspergillosis (CPPA), and allergic bronchopulmonary aspergillosis (1). Triazole antifungal drugs are the most common treatment for A. fumigatus infection. Itraconazole (ITC) and voriconazole (VRC) are the only oral drug treatment options for aspergillosis, which may lead to long-term administration. Since the discovery of the first ITC-resistant isolate in 1997 (2), epidemiological reports of new triazole-resistant isolates have been increasing worldwide (3). Mechanisms of acquired azole resistance may be explained by extended periods of azole exposure in the host or by environmental exposure of A. fumigatus to agricultural fungicides.
The primary molecular mechanisms of triazole resistance in A. fumigatus isolates are mutations that alter the target protein Cyp51A and prevent its interaction with the drug (4). Mutations in cyp51A may be classified as single-nucleotide polymorphisms (SNPs) and/or tandem repeats in the promoter region (3). The major SNPs affecting Cyp51A are positioned at Gly54, Gly138, Met220, and Gly448. The clinical isolates with these SNPs demonstrate various azole susceptibility profiles; for example, isolates with SNPs at Gly54 show resistance to ITC and varied susceptibility to posaconazole (POS) and VRC, whereas isolates with SNPs at Gly138 show pan-azole resistance, including resistance to ITC, POS, and VRC. Another alteration is a tandem repeat in the promoter region that results in the overexpression of cyp51A with specific SNPs. Two major classes of such azole-resistant mutations are TR34-Leu98His and TR46-Tyr121Phe-Thr289Ala, which carry a 34-bp and a 46-bp sequence duplication, respectively, as well as amino acid substitutions. Although many SNPs in cyp51A that may be linked to low susceptibility in azole-resistant isolates have been previously reported, few studies have been conducted to conclusively demonstrate the contribution of SNPs to decreased azole susceptibility in clinical isolates. One obstacle affecting the molecular analysis of clinical A. fumigatus isolates is the production of genetically manipulated mutants, as the efficiency of homologous recombination is extremely low.
Clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 has been applied as a powerful genome editing tool in various organisms (5). By forming a ribonucleoprotein complex with an artificial single guide RNA (sgRNA) designed to target a cellular gene, the Cas9 nuclease efficiently introduces double-stranded breaks (DSBs) at the corresponding target locus (6). The sgRNA hybridizes to its cDNA sequence immediately upstream of the protospacer-adjacent motif (PAM), which consists of NGG for the Streptococcus pyogenes Cas9 variant (7). DSBs in the target genomic DNA can be repaired by either homology-directed repair or nonhomologous end joining (NHEJ) (5, 8, 9). DNA repair via homology-directed repair requires a homologous DNA template with sequence similarity to that of the adjacent region of the DSB locus, whereas NHEJ ligates the DSB, leading to indels in a template-independent manner. The CRISPR/Cas9 system has also been successfully applied to A. fumigatus (10–12).
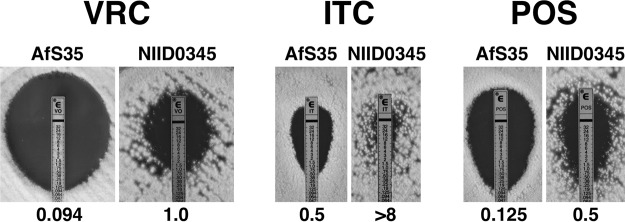
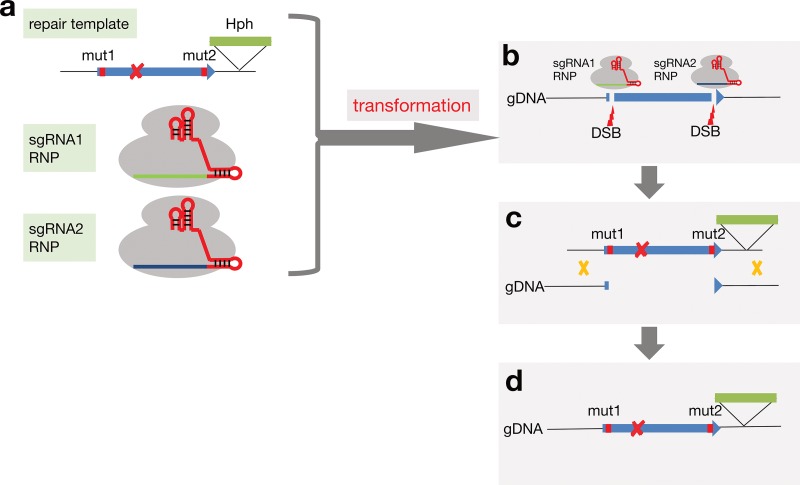
The A. fumigatus strains used in the present study are listed in Table 1. The clinical isolate NIID0345 was obtained in 2016 from the sputum sample of a 74-year-old male patient with CPPA who had received VRC treatment for 3 years. The isolate was not susceptible to VRC, ITC, or POS (Fig. 1) but was susceptible to amphotericin B, micafungin, and caspofungin (data not shown). A comparison of cyp51A from the azole-resistant isolate (NIID0345) with those from azole-susceptible strains (Af293 and AfS35) revealed that NIID0345 carried two amino acid substitutions, Gly138Ser (GGC→AGC) and Asn248Lys (AAT→AAA). To verify which SNP is involved in azole resistance, we substituted the nucleotide sequences corresponding to amino acid Ser138 and/or Lys248 in the clinical isolate NIID0345. We attempted to replace the genomic cyp51A gene locus by homologous recombination with a linear DNA fragment harboring mutations. For this purpose, Cas9/sgRNA ribonucleoproteins and the repair template were simultaneously transformed via the protoplast-polyethylene glycol method into the A. fumigatus azole-resistant clinical isolate (Fig. 2). For detailed methods for strain construction, see described in the text in the supplemental material.
TABLE 1.
Aspergillus fumigatus strains used in this study and their in vitro antifungal susceptibility profiles against three triazoles according to the Etest methoda
| Strain(s) | Parent | Genotypeb | MICs (μg/ml)c |
Source | ||
|---|---|---|---|---|---|---|
| VRC | ITC | POS | ||||
| NIID0345 | clinical isolate | 1.0 | >8 | 0.5 | Current study | |
| NIID0345-mut1-2 | NIID0345 | mut1 mut2 cyp51A hph | 1.5 | >8 | 0.75 | Current study |
| NIID0345-S138G | NIID0345 | mut1 S138G mut2 cyp51A hph | 0.5 | 1.0 | 0.25 | Current study |
| NIID0345-K248N | NIID0345 | mut1 K248N mut2 cyp51A hph | 2 | >8 | 0.5 | Current study |
| NIID0345-S138G-K248N | NIID0345 | mut1 mut2 cyp51A hph | 0.5 | 1.0 | 0.25 | Current study |
| AfS35 | D141 | akuAΔloxP | 0.094 | 0.5 | 0.125 | Fungal Genetics Stock Center |
| AfS35-mut1-2 | AfS35 | mut1 mut2 cyp51A hph | 0.125 | 0.5 | 0.125 | Current study |
| AfS35-G138S | AfS35 | mut1 G138S mut2 cyp51A hph | 0.38 | 1.5 | 0.19 | Current study |
FIG 1.
Antifungal susceptibility testing using Etest strips for voriconazole, itraconazole, and posaconazole in the azole-susceptible Aspergillus fumigatus strain AfS35 and clinical azole-resistant A. fumigatus strain NIID0345. The number below each photo represents the MIC (in μg/ml).
FIG 2.
Overview of the genetic modification via CRISPR/Cas9-promoted homology-directed repair. (a) Cas9 protein and in vitro-synthesized sgRNAs were mixed to form two RNPs. The repair template and two RNPs were transformed into Aspergillus fumigatus protoplasts. (b) The dual Cas9-sgRNA complex introduced two double-stranded breaks at the N and C termini of cyp51A (c and d). The cleaved cyp51A on the genomic DNA is replaced by the repair template, resulting in the introduction of the desired mutations and hph marker. The silent mutations mut1 and mut2 on the repair template and the replaced genomic DNA (gDNA) cannot be cleaved by the RNP nuclease.
Next, we examined azole susceptibility by using Etest strips on the constructed recombinant strains. The strains in which only nuclease-resistant silent mutations were introduced demonstrated an azole resistance profile similar to that of the parental strain, NIID0345 (Fig. 3A; Table 1), indicating that CRISPR/Cas9-mediated homologous recombination had no effect on azole susceptibility. Both of the recombinant strains with Ser138Gly and Ser138Gly/Lys248Asn amino acid substitutions showed increased susceptibility to all azoles tested, whereas the strain with only the Lys248Asn substitution showed an azole resistance profile similar to that of the parental clinical isolate. These results indicate that Lys248 is not associated with azole resistance and that Ser138 is responsible for azole resistance in this clinical isolate.
FIG 3.
Antifungal susceptibility testing using Etest strips for voriconazole, itraconazole, and posaconazole for the strains generated via CRISPR/Cas9-promoted gene replacement from the strains NIID0345 (A) and AfS35 (B). The number below each photo represents the MIC (in μg/ml).
To verify whether Gly138 in Cyp51A is responsible for azole resistance, an amino acid substitution of Gly138 to serine was introduced into the azole-susceptible strain AfS35. The method for producing the recombinant strain was the same as that described above; however, highly efficient homologous recombination was expected because strain AfS35 is deficient in the NHEJ repair system. As expected, almost all transformants exhibited ideal recombination. Azole susceptibility testing of the recombinant strains with Etest showed a slight decrease in azole susceptibility when the Ser138 mutation was introduced into the azole-susceptible strain AfS35 (Fig. 3B; Table 1). The strain with only nuclease-resistant silent mutations demonstrated an azole susceptibility profile similar to that of parental strain AfS35. From these results, we elucidated the direct involvement of Gly138 in Cyp51A in azole resistance, which had previously been supported only by indirect epidemiological evidence.
The CRISPR/Cas9 genome editing technique used in this study has enabled site-directed mutagenesis, altering Ser138 to glycine on the genomic Cyp51A locus in an azole-resistant clinical strain. This is, to our knowledge, the first example of site-directed mutagenesis performed in a clinical azole-resistant fungal isolate to elucidate whether azole susceptibility is altered by mutations in the genomic Cyp51A locus. Although genetically recombinant strains harboring mutations such as TR34-Leu98His (13), TR46-Tyr121Phe-Thr289Ala (14), Gly54Trp (13), and Thr301Ile (15) in the genomic Cyp51A locus have been reported, all these strains were constructed in the akuBKU80-deficient strain as a recipient. It is well known that wild-type strains, such as clinical isolates, tend to exhibit low efficiency in homologous recombination, largely because of high NHEJ activity. To overcome this limitation, the gene encoding either KU70 or KU80, which are the components of NHEJ machinery, was knocked out, leading to a significant increase in the frequency of homologous recombination (16, 17). In contrast, our CRISPR/Cas9 method can facilitate efficient homologous recombination without the inactivation of the NHEJ pathway, which is supported by previous studies, concluding that the frequency of homologous recombination can be increased by the CRISPR/Cas9 system in Candida glabrata (18).
To build the CRISPR/Cas9 system in A. fumigatus clinical isolates, we incorporated several additional methods to improve the efficiency and accuracy of cyp51A gene replacement events. Improved efficiency of cyp51A replacement was achieved by introducing two DSBs via the design of two target sequences for sgRNA at sites close to the N and C termini, repressing homologous recombination within the cyp51A coding region. Additionally, to avoid digestion of the repair template and redigestion of the edited target after the homologous recombination event, nuclease-resistant silent mutations were introduced in two loci of three codons immediately upstream from the PAM sites of the repair template, preventing it from being targeted by CRISPR/Cas9 (19). To minimize the off-target effects from continuous DNA-based Cas9 and sgRNA expression (which should be considered whenever the CRISPR/Cas9 system is used for genome editing), we introduced ribonucleoproteins consisting of commercially available recombinant Cas9 protein and in vitro-synthesized sgRNAs directly into protoplasts of clinical isolates. As one means of minimizing off-target effects, directly transfected Cas9 protein reduces the off-target cleavage rate when compared with Cas9 expression by a plasmid or mRNA transfection in mammalian cells (20). One recent study demonstrated that direct delivery of Cas9-guide RNA ribonucleoprotein can facilitate genome editing in A. fumigatus (10). Based on these improvements, we produced a simple, efficient, and accurate site-directed mutagenesis system for investigating structure-phenotype relationships of the azole target Cyp51A. Since this system can be applied to numerous genes other than cyp51A, this method will accelerate the progress of many pathogenic fungal studies.
In conclusion, we have developed a simple, efficient, and accurate gene replacement system using CRISPR/Cas9 genome editing techniques and applied these techniques to investigate the mechanisms of azole resistance via Cyp51A alteration. We confirmed at the molecular level that the Gly138Ser mutation is one reason for azole resistance in a clinical isolate. There are many cyp51A mutations that may result in potential but unconfirmed amino acid changes conferring azole resistance. Further investigation of Cyp51A using our CRISPR/Cas9 system is required to verify whether the diverse SNPs reported are in fact responsible for azole resistance.
Supplementary Material
ACKNOWLEDGMENTS
We thank Hiroko Tomuro for the technical assistance. We also thank Enago for the English language review.
This work was supported by MEXT KAKENHI grant number JP16K09954 and the Joint Usage/Research Program of the Medical Mycology Research Center, Chiba University (17–9).
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00894-18.
REFERENCES
- 1.Kohno S, Tamura K, Niki Y, Izumikawa K, Oka S, Ogawa K, Kadota J, Kamei K, Kanda Y, Kiuchi T, Shibuya K, Takakura S, Takata T, Takesue Y, Teruya K, Tokimatsu I, Fukuda T, Maesaki S, Makimura K, Mikamo H, Mitsutake K, Miyazaki Y, Mori M, Yasuoka A, Yano K, Yamanaka N, Yoshida M. 2016. Executive summary of Japanese domestic guidelines for management of deep-seated mycosis 2014. Med Mycol J 57:E117–E163. doi: 10.3314/mmj.16-00010. [DOI] [PubMed] [Google Scholar]
- 2.Denning DW, Venkateswarlu K, Oakley KL, Anderson MJ, Manning NJ, Stevens DA, Warnock DW, Kelly SL. 1997. Itraconazole resistance in Aspergillus fumigatus. Antimicrob Agents Chemother 41:1364–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Resendiz Sharpe A, Lagrou K, Meis JF, Chowdhary A, Lockhart SR, Verweij PE, ISHAM/ECMM Aspergillus Resistance Surveillance working group. 2018. Triazole resistance surveillance in Aspergillus fumigatus. Med Mycol 56:83–92. [DOI] [PubMed] [Google Scholar]
- 4.Diaz-Guerra TM, Mellado E, Cuenca-Estrella M, Rodriguez-Tudela JL. 2003. A point mutation in the 14alpha-sterol demethylase gene cyp51A contributes to itraconazole resistance in Aspergillus fumigatus. Antimicrob Agents Chemother 47:1120–1124. doi: 10.1128/AAC.47.3.1120-1124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doudna JA, Charpentier E. 2014. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096–1258096. [DOI] [PubMed] [Google Scholar]
- 6.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mojica FJM, Díez-Villaseñor C, García-Martínez J, Almendros C. 2009. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 8.Ran FA, Hsu PD, Lin C-Y, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. 2013. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al Abdallah Q, Ge W, Fortwendel JR. 2017. A simple and universal system for gene manipulation in Aspergillus fumigatus: in vitro-assembled Cas9-guide RNA ribonucleoproteins coupled with microhomology repair templates. mSphere 2:e00446-17. doi: 10.1128/mSphere.00446-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang C, Meng X, Wei X, Lu L. 2016. Highly efficient CRISPR mutagenesis by microhomology-mediated end joining in Aspergillus fumigatus. Fungal Genet Biol 86:47–57. doi: 10.1016/j.fgb.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Fuller KK, Chen S, Loros JJ, Dunlap JC. 2015. Development of the CRISPR/Cas9 system for targeted gene disruption in Aspergillus fumigatus. Eukaryot Cell 14:1073–1080. doi: 10.1128/EC.00107-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Snelders E, Karawajczyk A, Verhoeven RJA, Venselaar H, Schaftenaar G, Verweij PE, Melchers WJG. 2011. The structure-function relationship of the Aspergillus fumigatus cyp51A L98H conversion by site-directed mutagenesis: the mechanism of L98H azole resistance. Fungal Genet Biol 48:1062–1070. doi: 10.1016/j.fgb.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Snelders E, Camps SMT, Karawajczyk A, Rijs AJMM, Zoll J, Verweij PE, Melchers WJG. 2015. Genotype-phenotype complexity of the TR46/Y121F/T289A cyp51A azole resistance mechanism in Aspergillus fumigatus. Fungal Genet Biol 82:129–135. doi: 10.1016/j.fgb.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 15.Leonardelli F, Macedo D, Dudiuk C, Cabeza MS, Gamarra S, Garcia-Effron G. 2016. Aspergillus fumigatus intrinsic fluconazole resistance is due to the naturally occurring T301I substitution in Cyp51Ap. Antimicrob Agents Chemother 60:5420–5426. doi: 10.1128/AAC.00905-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krappmann S, Sasse C, Braus GH. 2006. Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end-joining-deficient genetic background. Eukaryot Cell 5:212–215. doi: 10.1128/EC.5.1.212-215.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.da Silva Ferreira ME, Kress MRVZ, Savoldi M, Goldman MHS, Härtl A, Heinekamp T, Brakhage AA, Goldman GH. 2006. The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot Cell 5:207–211. doi: 10.1128/EC.5.1.207-211.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enkler L, Richer D, Marchand AL, Ferrandon D, Jossinet F. 2016. Genome engineering in the yeast pathogen Candida glabrata using the CRISPR-Cas9 system. Sci Rep 6:35766. doi: 10.1038/srep35766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim H, Ishidate T, Ghanta KS, Seth M, Conte D, Shirayama M, Mello CC. 2014. A co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans. Genetics 197:1069–1080. doi: 10.1534/genetics.114.166389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang X, Potter J, Kumar S, Zou Y, Quintanilla R, Sridharan M, Carte J, Chen W, Roark N, Ranganathan S, Ravinder N, Chesnut JD. 2015. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J Biotechnol 208:44–53. doi: 10.1016/j.jbiotec.2015.04.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.