The highly variable pharmacokinetics of β-lactam antibiotics and β-lactamase inhibitors poses a significant challenge to clinicians in ensuring appropriate antibiotic doses in critically ill patients. Therefore, routine monitoring of plasma concentrations is important for individualization of antimicrobial therapy.
KEYWORDS: LC-MS/MS, assay development, β-lactams, human plasma, validation
ABSTRACT
The highly variable pharmacokinetics of β-lactam antibiotics and β-lactamase inhibitors poses a significant challenge to clinicians in ensuring appropriate antibiotic doses in critically ill patients. Therefore, routine monitoring of plasma concentrations is important for individualization of antimicrobial therapy. Accordingly, a simple and robust analytical method for the simultaneous measurement of multiple β-lactam antibiotics and β-lactamase inhibitors is highly desirable to ensure quick decisions on dose adjustments. In this study, a sensitive, simple, and robust method for the simultaneous quantification of cefepime, meropenem, piperacillin, and tazobactam in human plasma was developed and rigorously validated according to FDA guidance. Sample extraction was accomplished by simple protein precipitation. Chromatographic separation of analytes was achieved using stepwise gradient elution. Analytes were monitored using tandem mass spectrometry (MS/MS) with a turbo ion spray source in positive multiple-reaction-monitoring mode. The calibration curve ranged from 0.5 to 150 μg/ml for cefepime, 0.1 to 150 μg/ml for meropenem and piperacillin, and 0.25 to 150 μg/ml for tazobactam. Inter- and intraday precision and accuracy, sensitivity, selectivity, dilution integrity, matrix effect, extraction recovery, and hemolysis effect were investigated for all four analytes, and the results met the acceptance criteria. Compared to other reported methods, our method is more robust because of the combination of the following features: (i) a simple sample extraction procedure, (ii) a short sample run time, (iii) a wide dynamic range, and (iv) the small plasma sample volume needed. Since our method already covers β-lactams and a β-lactamase inhibitor with highly heterogeneous physicochemical properties, further antibiotic candidates may easily be incorporated into this multianalyte method.
INTRODUCTION
Severe infection and associated sepsis represent the most common cause of mortality in critically ill patients (1). Among the antibiotics available on the market, β-lactam antibiotics and β-lactamase inhibitors are the most frequently prescribed antimicrobial agents for the treatment of bacterial infections. Appropriate and adequate antimicrobial therapy, including early administration of effective antibiotics at appropriate doses, is of utmost importance for reducing mortality in critically ill patients. However, it has been reported that the plasma concentrations of β-lactam antibiotics and β-lactamase inhibitors are very variable and hard to predict in critically ill patients, which makes the development of an optimal antibiotic dosing regimen a big challenge (2, 3). Patients with sepsis frequently have increased interstitial fluid volumes due to capillary leak syndrome, which can lead to an increased volume of distribution of most β-lactam antibiotics (4). In addition, patients with sepsis are known to become hyperdynamic and therefore have increased renal blood flow and glomerular filtration, leading to increased clearance of many β-lactam antibiotics (4, 5). Considering the highly variable pharmacokinetics of β-lactam antibiotics and β-lactamase inhibitors, together with the strong association between their exposure in infected critically ill patients and mortality (6, 7), therapeutic drug monitoring (TDM) represents a valuable tool for enabling personalized therapy, optimizing β-lactam antibiotic concentrations in each patient, and improving clinical outcomes (8).
Accompanying the increased interest in β-lactam TDM, there has been an increasing effort to develop a rapid and robust method to measure plasma concentrations of β-lactam antibiotics and β-lactamase inhibitors. Many methods for high-performance liquid chromatography with UV detection (HPLC-UV) have been reported for quantification of β-lactam antibiotics in human plasma (9–12). However, because of their inherently low sensitivity and poor selectivity, HPLC-UV methods have many disadvantages, such as a large sample volume, long sample run times, and rather unspecific UV detection (9, 11, 12). The unspecific UV detection can be a particular problem for samples collected from critically ill patients because of the presence of various comedications in such patients. Several liquid chromatography-tandem mass spectrometry (LC-MS/MS) methods have been reported as well (13, 14). However, only a few LC-MS/MS methods are available for the simultaneous quantification of structurally unrelated β-lactams, such as penicillins, carbapenems, and cephalosporins (13, 15). In addition, most of these methods have a fairly narrow linearity range of the calibration standards and require a relatively large sample volume (14, 15). Furthermore, none of the reported methods have been evaluated thoroughly to determine the stability of β-lactams (13, 16). As many β-lactams have stability issues, it is important to evaluate stability-related assay validation parameters, such as stock solution stability, whole-blood stability, short-term stability, long-term stability, processed sample stability, and freeze-thaw stability. Unfortunately, most of these stability data are missing for previously reported methods. The aim of the present study was to develop and validate a rapid, sensitive, reproducible, and robust LC-MS/MS method for the simultaneous analysis of cefepime, meropenem, piperacillin, and tazobactam in human plasma. These four antibiotics are the most commonly prescribed of the β-lactams and β-lactamase inhibitors in infected critically ill patients, such as those in the intensive care unit (ICU) or patients with cystic fibrosis (CF) who have severe infection. In our study, much effort was devoted toward the establishment of a robust LC-MS/MS method. During the assay development process, we tested several LC columns, evaluated various gradient elution profiles, tested different sample preparation procedures, and finally developed a highly sensitive and robust LC-MS/MS method. After the method was established, we subsequently conducted a comprehensive assay validation by applying an extensive validation protocol based on FDA guidance, with which 17 validation parameters were evaluated. The assay validation was performed in a good laboratory practice (GLP) environment. The assay development and a majority of the assay validation data are reported in the current article, which represents part 1 of our work. Due to the importance of the stability information, all stability-related assay validation parameters are highlighted in a separate article (part 2 of our work).
RESULTS
Assay development. (i) MS/MS analysis.
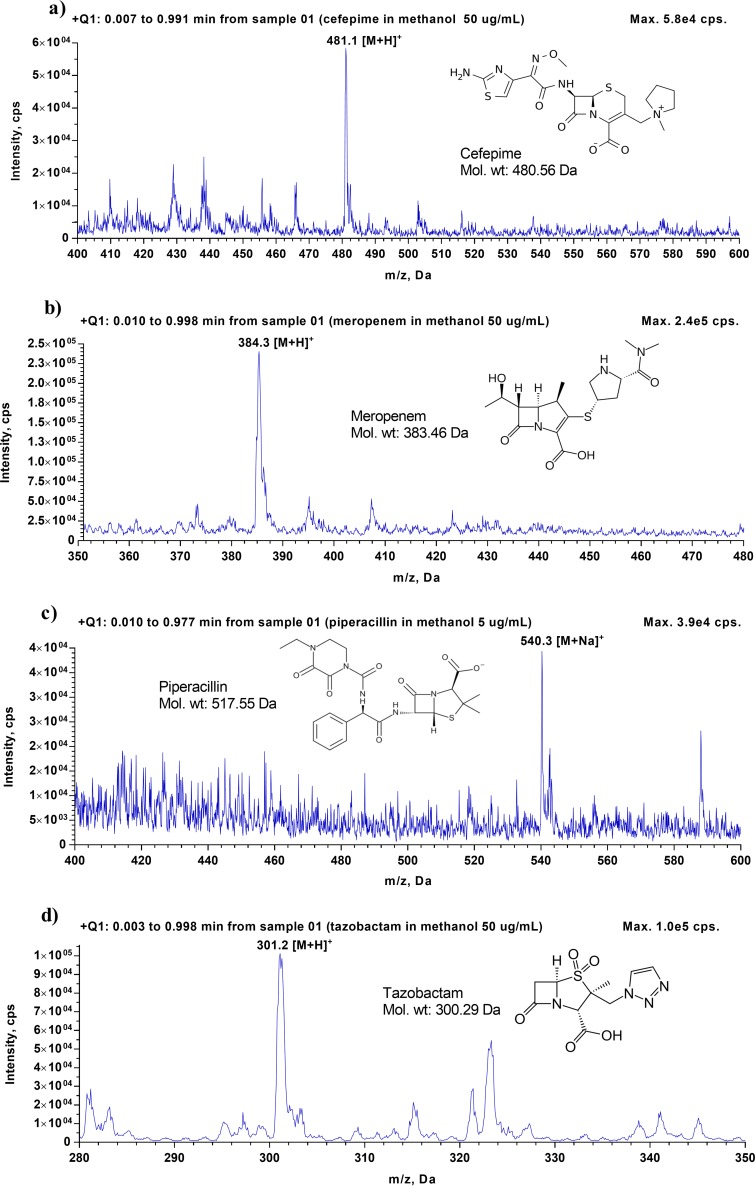
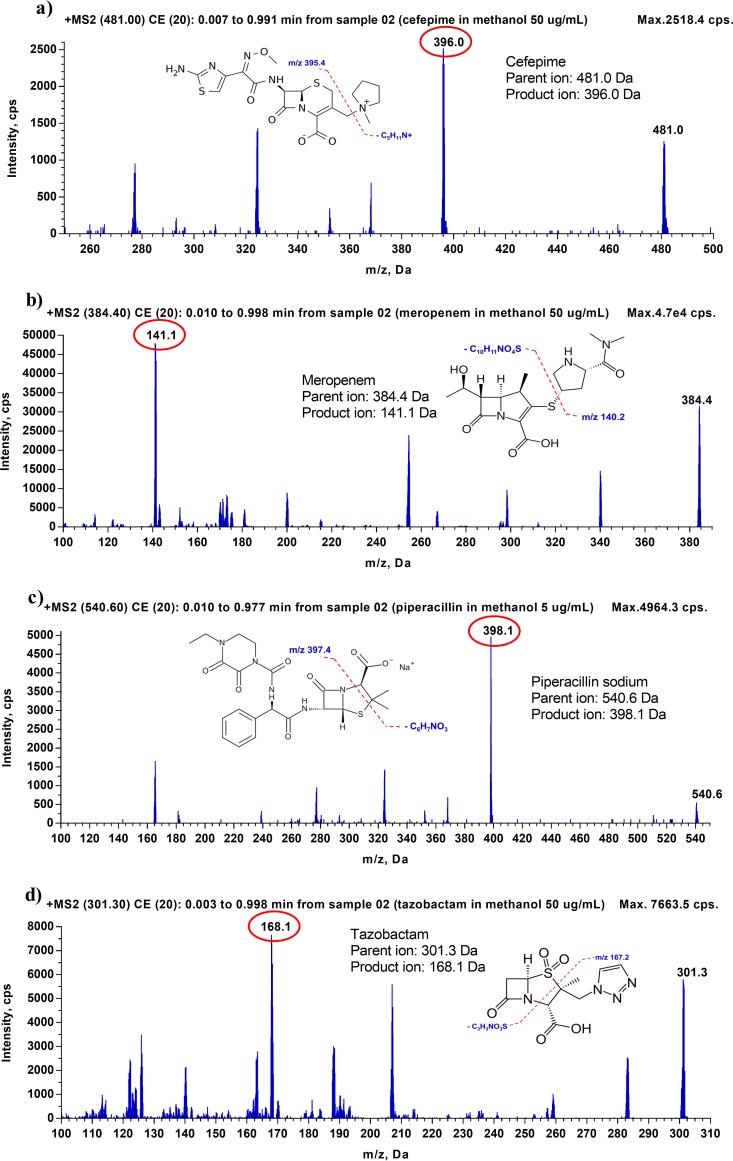
To identify the mass spectra of cefepime, meropenem, piperacillin, and tazobactam, as well as their isotope-labeled internal standards (ISs), 50 μg/ml of each analyte, except for piperacillin (for which 5 μg/ml was used), was prepared individually in a mixture of methanol-water (50:50 [vol/vol]) and injected into the LC-MS/MS. A Q1 scan and a Q3 scan were performed in positive-ion mode to identify the precursor ions and product ions, respectively. The Q1 scan showed that precursor ions of cefepime, meropenem, and tazobactam have m/z values of 481.400, 384.300, and 301.400, respectively, which correspond to their protonated molecules ([M + H]+). For piperacillin, despite its molecular weight of 517.555, the monitored precursor ion had an m/z of 540.300 because this compound formed a sodium adduct ([M + Na]+) (Table 1). The Q3 scan showed that after collision-induced dissociation, the most abundant fragments observed for cefepime and meropenem were at m/z ratios of 396.100 and 141.100, respectively, generated by the loss of the N-methylpyrrolidine ring in cefepime and the cleavage of the C-S bond in meropenem. The most abundant fragment observed for piperacillin was at m/z 398.100, due to the loss of CO2 and C5H7NO, and the dominating fragment of tazobactam was at m/z 168.100, due to the loss of C2H4NO3S. The mass spectra of cefepime, meropenem, piperacillin, and tazobactam and their corresponding fragments are illustrated in Fig. 1 and 2. For each analyte, after the precursor ion and the most abundant product ion were identified, all MS parameters, including both source- and compound-dependent parameters, were optimized using the FIA method, a built-in, automatically optimizing function in the Analyst software. The compound-dependent MS parameters of the tested compounds are provided in Table 1. The values of source-dependent parameters, which are the same for all tested compounds, are mentioned in Materials and Methods.
TABLE 1.
Compound-specific parametersa
| Antibiotic | Corresponding IS | Mol wt (g/mol) | Form of ion | Retention time (min) | Precursor ion m/z (Q1) | Product ion m/z (Q3) | Collision energy (V) |
|---|---|---|---|---|---|---|---|
| Cefepime | 2H6-meropenem | 480.560 | [M + H]+ | 2.76 | 481.400 | 396.100 | 15.00 |
| Meropenem | 2H6-meropenem | 383.464 | [M + H]+ | 2.87 | 384.300 | 141.100 | 20.00 |
| Piperacillin | 2H5-piperacillin | 517.555 | [M + Na]+ | 3.45 | 540.300 | 398.100 | 20.00 |
| Tazobactam | 2H5-piperacillin | 300.289 | [M + H]+ | 2.75 | 301.400 | 168.100 | 20.00 |
| 2H6-meropenem | 389.500 | [M + H]+ | 2.85 | 390.300 | 147.100 | 20.00 | |
| 2H5-piperacillin | 521.567 | [M + Na]+ | 3.42 | 545.300 | 398.100 | 20.00 |
For all antibiotics, the declustering potential was 40.00 V, the collision cell exit potential was 12.00 V, and the dwell time was 200 ms.
FIG 1.
Full-scan MS1 spectra for cefepime (a), meropenem (b), piperacillin (c), and tazobactam (d).
FIG 2.
Fragmentation spectra (MS2) for cefepime (a), meropenem (b), piperacillin (c), and tazobactam (d), along with the fragmentation pattern for each analyte.
(ii) Chromatographic separation.
To ensure good peak shapes for all analytes, maintain their retention time, and minimize the total run time, great effort was spent on column selection, gradient profile development, and organic solvent selection. We tested several LC columns, including the Phenomenex Synergy C18 column (4.0 μm × 150 mm × 2 mm), the Waters xBridge BEH C18 column (2.5 μm × 100 mm × 2.1 mm), the Phenomenex Kinetex C18 column (2.6 μm × 50 mm × 2.1 mm), and the Phenomenex Kinetex F5 column (5.0 μm × 100 mm × 2.1 mm). Among the columns evaluated, only the Phenomenex Kinetex C18 column provided excellent peak shapes for all analytes, while other columns led to poor peak shapes for some analytes, especially cefepime. Therefore, Phenomenex Kinetex 2.6 μm C18 columns were used for our formal assay validation. Regarding organic solvent selection (mobile phase B), both methanol and acetonitrile were evaluated. Acetonitrile was chosen as mobile phase B in the final method due to very good peak shapes for all analytes as well as a significant reduction in the pump pressure in comparison to that with methanol.
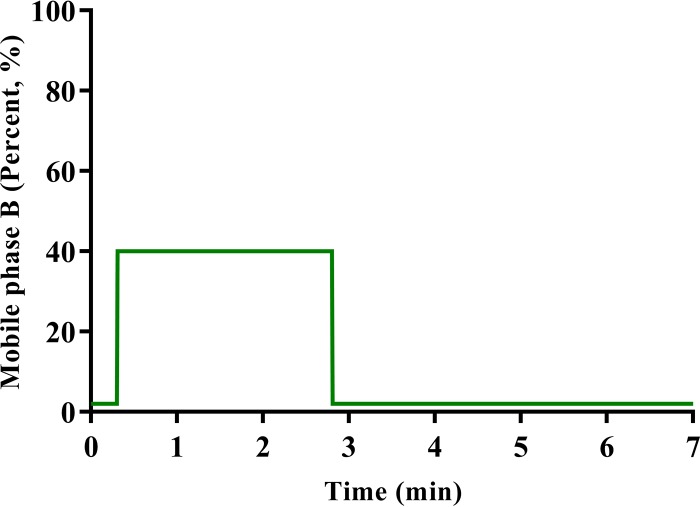
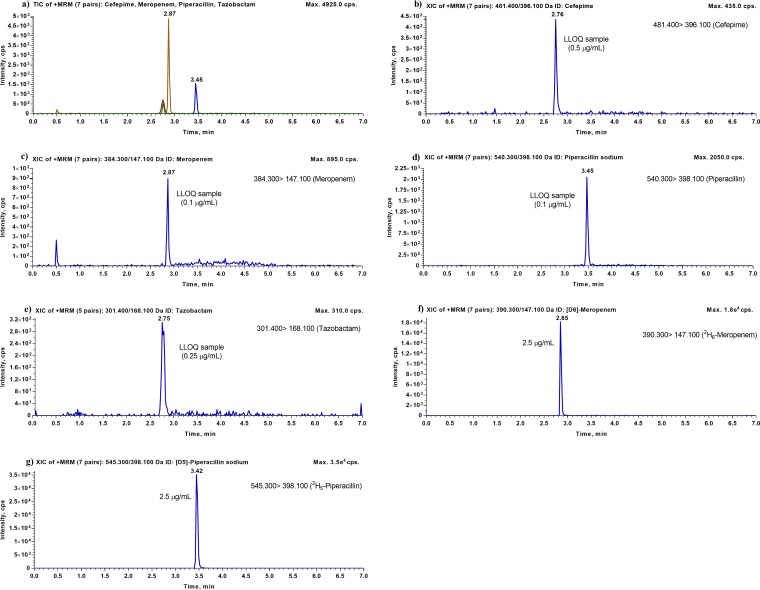
In terms of the final elution conditions, both isocratic and gradient elution conditions were evaluated. Because the analytes have very different physicochemical properties, we were unable to detect all four analytes simultaneously in a relatively short run time by using isocratic elution conditions. Consequently, we switched to gradient elution conditions for chromatographic separation. Among various gradient profiles evaluated, a novel stepwise gradient profile was developed, which provided very narrow and excellent peaks for all analytes and maintained the retention times of all analytes. Using these conditions for gradient elution (Fig. 3), the total run time is only 7.0 min. Representative chromatograms showing the typical peak shapes for the analytes are presented in Fig. 4. All of the compounds, including both analytes and their ISs, were eluted after 2.7 min, within a 1-min interval. Further chromatographic separation was not needed, since the m/z ratios were distinct, with no cross talk, which allowed for monitoring of unique transitions for each compound in multiple-reaction-monitoring (MRM) mode. In addition, a number of other chromatographic parameters, such as column temperature and flow rate, were also studied, and the best results were obtained when the column temperature was set at 35°C and the flow rate at 0.25 ml/min.
FIG 3.
Gradient elution profile between mobile phase B (acetonitrile plus 0.1% formic acid) and mobile phase A (water plus 0.1% formic acid) for simultaneous analysis of cefepime, meropenem, piperacillin, and tazobactam in human plasma.
FIG 4.
Representative LC-MS/MS chromatograms for cefepime, meropenem, piperacillin, and tazobactam. (a) Total ion chromatogram (TIC) for human plasma with lithium heparin in a single sample spiked with the lowest calibration standards (LLOQ) for cefepime, meropenem, piperacillin, and tazobactam. (b) Typical chromatogram for human plasma with lithium heparin spiked with cefepime at the LLOQ (0.5 μg/ml). (c) Typical chromatogram for human plasma with lithium heparin spiked with meropenem at the LLOQ (0.1 μg/ml). (d) Typical chromatogram for human plasma with lithium heparin spiked with piperacillin at the LLOQ (0.1 μg/ml). (e) Typical chromatogram for human plasma with lithium heparin spiked with tazobactam at the LLOQ (0.25 μg/ml). (f) Typical chromatogram for human plasma with lithium heparin spiked with 2H6-meropenem (2.5 μg/ml). (g) Typical chromatogram for human plasma with lithium heparin spiked with 2H5-piperacillin (2.5 μg/ml).
(iii) Optimization of the extraction procedure.
A simple and efficient extraction procedure is important for a multiclass antibiotic method in order to minimize sample turnover time and maintain extraction sufficiency. Acetonitrile has been recommended as the precipitant of choice among organic solvents due to its very low ionization suppression effect (17). In the present method, acetonitrile was added to plasma at a 4:1 ratio for protein precipitation; this facilitated a simple and efficient sample extraction process with excellent analyte recovery rates. To ensure good peak shapes for all analytes and to reduce the matrix effect, the supernatant was diluted with water before the LC injection. For this purpose, several dilution factors were evaluated, including 1:1, 1:2, 1:3, 1:4, 1:6, and 1:9, and the best results, in terms of peak shape, calibration range, and matrix effect, were obtained when a dilution factor of 1:9 (supernatant-water [vol/vol]) was employed. Therefore, a dilution factor of 1:9 was used in our formal assay validation.
Assay validation. (i) Linearity and sensitivity.
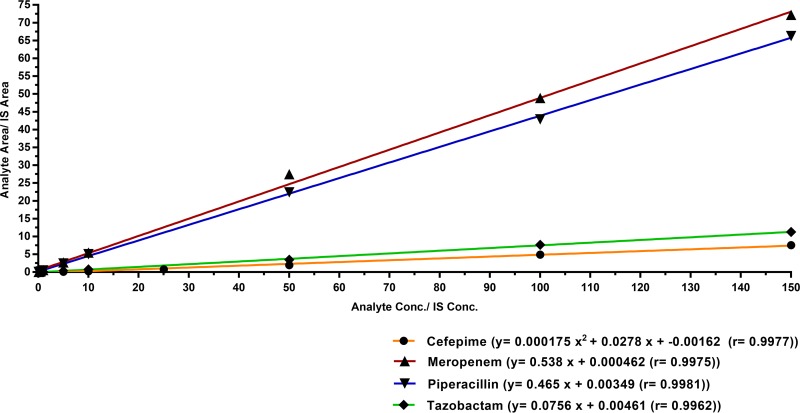
Plasma calibration curves for meropenem over the concentration range of 0.1 to 150 μg/ml, piperacillin over the concentration range of 0.1 to 150 μg/ml, and tazobactam over the concentration range of 0.25 to 150 μg/ml were established by weighted (1/x2) linear regression analysis, and R2 values were >0.990. The plasma calibration curve for cefepime was constructed over a concentration range of 0.5 to 150 μg/ml, using weighted (1/x2) quadratic regression. During the development of this assay, the use of quadratic regression with 1/x2 showed the most consistent and best fit for the analytical measurement of cefepime in human plasma. Typical calibration equations and regression parameters for all four antibiotics are presented in Table 2. Representative calibration curves for cefepime, meropenem, piperacillin, and tazobactam are shown in Fig. 5. The lower limits of quantification (LLOQ) for cefepime, meropenem, piperacillin, and tazobactam in human plasma were confirmed to be 0.50 μg/ml, 0.1 μg/ml, 0.1 μg/ml, and 0.25 μg/ml, respectively, at which accuracy and precision were within ±20% for each antibiotic. The results of the sensitivity validation are presented in Table 3, and typical chromatograms obtained for human plasma samples spiked with cefepime, meropenem, piperacillin, and tazobactam at their LLOQ, along with their respective ISs, are shown in Fig. 4.
TABLE 2.
Linearitya
| Antibiotic | Linear equation (y = mx + b) |
Quadratic equation (y = ax2 + bx + c) |
R2 (mean ± SD) | Range (μg/ml) | |||
|---|---|---|---|---|---|---|---|
| Slope (mean ± SD) | Intercept (mean ± SD) | a (mean ± SD) | b (mean ± SD) | c (mean ± SD) | |||
| Meropenem | 0.511 ± 0.088 | 0.008 ± 0.014 | 0.997 ± 0.001 | 0.100–150 | |||
| Piperacillin | 0.398 ± 0.194 | 0.002 ± 0.006 | 0.997 ± 0.001 | 0.100–150 | |||
| Tazobactam | 0.117 ± 0.074 | 0.003 ± 0.006 | 0.997 ± 0.002 | 0.250–150 | |||
| Cefepime | 0.00019 ± 0.0005 | 0.037 ± 0.017 | −0.003 ± 0.008 | 0.996 ± 0002 | 0.500–150 | ||
For the linear equation, y represents the ratio of the peak area for the analyte to that for the IS, x represents the plasma concentration, m is the slope, and b is the intercept. For the quadratic equation, y represents the ratio of the peak area for the analyte to that for the IS, x represents the plasma concentration, a is the quadratic coefficient, b is the linear coefficient, and c is the intercept.
FIG 5.
Representative calibration curves for cefepime, meropenem, piperacillin, and tazobactam analysis in human plasma. For cefepime, the curve was fit to the calibrators by quadratic regression using 1/x2 weighting. For meropenem, piperacillin, and tazobactam, the curve was fit to the calibrators by linear regression using 1/x2 weighting.
TABLE 3.
Accuracy and precision for intraday and interday runsa
| Antibiotic | Concn (μg/ml) (QC level) | n | Intraday runs |
Interday runs |
||
|---|---|---|---|---|---|---|
| Bias (%) | CV (%) | Bias (%) | CV (%) | |||
| Cefepime | 0.500 (QC LLOQ) | 18 | −6.1 to 19.4 | 6.0 to 7.8 | 10.9 | 12.8 |
| 1.50 (QC low) | 18 | −9.0 to −2.9 | 6.6 to 8.1 | −6.6 | 7.6 | |
| 75.0 (QC medium) | 18 | −3.6 to 12.6 | 2.0 to 3.2 | 4.0 | 7.0 | |
| 120 (QC high) | 18 | −13.6 to −2.5 | 4.2 to 5.1 | −6.5 | 7.0 | |
| Meropenem | 0.100 (QC LLOQ) | 18 | −12.4 to 18.3 | 8.3 to 17.5 | 5.6 | 18.9 |
| 0.300 (QC low) | 18 | 3.3 to 11.3 | 5.3 to 10.1 | 8.4 | 8.5 | |
| 75.0 (QC medium) | 18 | −9.8 to −5.5 | 2.9 to 4.4 | −8.2 | 4.0 | |
| 120 (QC high) | 18 | −14.2 to −4.6 | 6.4 to 8.5 | −8.0 | 7.8 | |
| Piperacillin | 0.100 (QC LLOQ) | 18 | −2.5 to 15.3 | 6.1 to 11.0 | 4.5 | 10.8 |
| 0.300 (QC low) | 16b | 8.8 to 10.8 | 7.8 to 14.6 | 10.0 | 11.7 | |
| 75.0 (QC medium) | 18 | −10.2 to −5.0 | 4.9 to 7.9 | −7.1 | 7.0 | |
| 120 (QC high) | 18 | −10.6 to −3.6 | 3.8 to 7.2 | −6.1 | 6.2 | |
| Tazobactam | 0.250 (QC LLOQ) | 18 | −4.9 to 10.1 | 4.2 to 12.0 | 1.3 | 9.6 |
| 0.750 (QC low) | 18 | 2.7 to 14.3 | 6.3 to 8.1 | 9.9 | 8.4 | |
| 75.0 (QC medium) | 18 | −2.0 to 10.1 | 4.9 to 9.4 | 5.8 | 8.3 | |
| 120 (QC high) | 18 | −0.7 to 9.7 | 6.5 to 11.4 | 5.8 | 9.8 | |
Intraday accuracy and precision were evaluated on three different days, with six samples for each QC level on each day. Interday accuracy and precision were assessed by using the results from these three intraday runs.
Outliers were excluded.
(ii) Selectivity.
Under the current conditions, the present method was found to be selective for cefepime, meropenem, piperacillin, and tazobactam. The response of any endogenous compounds that might have coeluted with the four antibiotics was less than 10% of the LLOQ standard for all four analytes and less than 0.5% for the ISs. These values are well within the acceptance criteria for both analytes and ISs and demonstrated that there was no significant interference from endogenous compounds in blank plasma samples at the retention times of the four antibiotics or those of their respective internal standards. This confirms the high selectivity of the assay in the presence of matrix components.
(iii) Interday and intraday accuracy and precision.
Intraday and interday precision and accuracy were assessed at the LLOQ and at the three quality control (QC) levels, QC low, QC medium, and QC high, for cefepime, meropenem, piperacillin, and tazobactam. Accuracy and precision data for human plasma samples for the four antibiotics are summarized in Table 3. The results are all within the acceptable limits of variation and deviation recommended by FDA guidance. This clearly shows that this assay possesses satisfactory accuracy and precision.
(iv) Injection carryover.
Carryover was investigated during validation by running a postinjection double-blank following the highest calibrator (upper limit of quantification [ULOQ]). Carryover rates for cefepime, meropenem, piperacillin, and tazobactam were found to be 1.8%, 23.4%, 24.4%, and 6.5%, respectively, compared to the analyte peak area for the LLOQ. Carryover rates for the internal standards, 2H6-meropenem and 2H5-piperacillin, were found to be 0.2% and 0.1%, respectively. From these results, injection carryover did not meet the acceptance criteria for meropenem and piperacillin. Based on our experience during assay validation, injecting the double-blank thrice was sufficient to reduce carryover of meropenem and piperacillin to within acceptable limits. Hence, when this assay is used to analyze clinical samples in the future, a double-blank sample must be injected at least thrice, followed by the reinjection of clinical samples that have concentrations at or near the LLOQ.
(v) Dilution integrity.
Plasma samples that were spiked with 4 times the QC high concentration for each antibiotic were used for dilution integrity tests, and the results showed that the accuracy and precision for these samples were within the acceptance criteria of ±15.0% for all four compounds. These data are summarized in Table 4. This confirms that if the study sample concentrations are higher than the range of linearity, the samples can be diluted a maximum of 4-fold and quantified accurately.
TABLE 4.
Dilution integritya
| Antibiotic | Dilution integrity |
|
|---|---|---|
| Bias (%) | CV (%) | |
| Cefepime | −0.1 | 1.0 |
| Meropenem | −8.5 | 4.3 |
| Piperacillin | 4.4 | 5.9 |
| Tazobactam | 10.3 | 2.9 |
The dilution integrity of each antibiotic was determined by using a dilution factor of 4, an antibiotic concentration of 480 μg/ml, and six replicates for each antibiotic.
(vi) Matrix effect and matrix factor.
As shown in Table 5, matrix effects in samples prepared at low and high QC concentrations for cefepime, meropenem, piperacillin, and tazobactam were within the acceptable limits of a ±15.0% difference from the theoretical nominal concentration and a matrix factor coefficient of variation percentage (CV %) of <10%. These data suggest that ion suppression or enhancement from human plasma is negligible for all four analytes with the current method.
TABLE 5.
Matrix effect, extraction recovery, and hemolysis effecta
| Antibiotic | Concn (μg/ml) (QC level) | Matrix factor |
Extraction recovery |
Hemolysis effect |
|||
|---|---|---|---|---|---|---|---|
| Mean | CV (%) | Recovery (%) | CV (%) | Bias (%) | CV (%) | ||
| Cefepime | 1.50 (QC low) | 1.0 | 5.3 | 84.2 | 5.3 | 3.6 | 7.5 |
| 120.0 (QC high) | 1.0 | 0.8 | 108 | 0.8 | 4.6 | 3.0 | |
| Meropenem | 0.300 (QC low) | 0.8 | 6.3 | 115 | 6.3 | −1.0 | 7.8 |
| 120.0 (QC high) | 1.1 | 8.2 | 135 | 8.2 | −13.7 | 4.3 | |
| Piperacillin | 0.300 (QC low) | 1.0 | 7.2 | 102 | 7.2 | 4.4 | 11.9 |
| 120.0 (QC high) | 1.0 | 5.7 | 116 | 5.7 | −12.5 | 10.4 | |
| Tazobactam | 0.750 (QC low) | 1.0 | 8.3 | 83.3 | 8.3 | 11.6 | 7.8 |
| 120.0 (QC high) | 0.8 | 2.7 | 139 | 2.7 | −1.9 | 7.9 | |
n = 6 for each concentration of each antibiotic.
(vii) Extraction recovery.
The mean extraction recovery rates for cefepime, meropenem, piperacillin, and tazobactam in human plasma were calculated to be 96.1%, 125%, 109%, and 111%, respectively, with a precision of <10%. Recovery values were within acceptance limits, and hence the extraction procedure for cefepime, meropenem, piperacillin, and tazobactam was consistent and reproducible. These results are presented in Table 5.
(viii) Hemolysis effect.
Hemolyzed plasma samples that were spiked with the QC low and QC high concentrations for each antibiotic separately had accuracy and precision values within the acceptance limits of ±15.0% for all four compounds. The results are presented in Table 5 and indicate that study samples with 2% or less hemolysis can be quantified accurately.
(ix) Interference of analytes with ISs.
Samples that were prepared for testing of interference of ISs by the analytes had an observed mean peak area that was <0.2% for all four antibiotics, which is well below the acceptance limit of 5.0%. Hence, there was no interference detected for cefepime, meropenem, piperacillin, and tazobactam and their respective ISs.
DISCUSSION
We developed and validated a fast, sensitive, and robust LC-MS/MS method for the simultaneous measurement of three β-lactam antibiotics and a β-lactamase inhibitor that are very commonly used in critically ill patients, such as those in the ICU or with CF. The ability to measure several antibiotics from different β-lactam classes simultaneously in a single analytical run is a great advantage because it enables samples from different patients receiving different antibiotics to be analyzed together at the same time, which greatly increases the practicality of future, routine use of the method. In addition, simultaneous measurement of multiple antibiotics in one method can avoid the time-consuming and error-prone switching of sample extraction procedures, solvents, and columns which is required if several distinct monoanalyte methods are utilized in a specific bioanalytical laboratory. Furthermore, the antibiotics included in our method are from three different β-lactam classes (penicillins, cephalosporins, and carbapenems) and the β-lactamase inhibitors, among which antibiotics have very different structures and physicochemical properties. Since our method already covers β-lactams, with their highly heterogeneous physicochemical properties, further antibiotic candidates may easily be incorporated into this multianalyte method.
Many β-lactam antibiotics are hydrophilic and have very limited retention on conventional C18 columns. The antibiotics included in our study (cefepime, meropenem, piperacillin, and tazobactam) are hydrophilic and have highly variable physicochemical characteristics, which posed difficulty in terms of the choice of the column, mobile phase, and ideal gradient elution profile. Much effort was put into assay development, especially column selection and gradient condition optimization, resulting in a highly robust method.
Compared to other HPLC or LC-MS/MS methods available in the literature for simultaneous measurement of multiple antibiotics, our method has several advantages, as follows.
Simple sample preparation procedure and short sample run time.
In our method, a simple protein precipitation method using acetonitrile with a dilution factor of 1:9 (supernatant-water [vol/vol]) was employed. With this sample preparation procedure, excellent extraction recovery and minimal matrix effects were obtained for all four compounds. Our sample preparation procedure is simple and fast and is much less labor-intensive than the solid-phase extraction procedure used in many other methods (14, 18). In addition, using a stepwise gradient elution profile, the total LC run time for each sample is only 7 min. The combination of rapid sample preparation and a short sample run time ensures high-throughput sample quantification. For example, using our method, 30 to 40 samples can be extracted within 2 h, and the extracts can be analyzed in 4 to 5 h. Because of its high-throughput potential, our method is suitable for routine drug analysis, such as TDM, where an efficient and rapid assay is needed for a short turnover time.
Wide dynamic range.
Compared to other reported HPLC or LC-MS/MS methods for simultaneous measurement of multiple antibiotics, our method has the widest dynamic range. For our method, the ULOQ is 150 μg/ml for all four drugs, and the LLOQ is 0.1 μg/ml for meropenem and piperacillin, 0.25 μg/ml for tazobactam, and 0.5 μg/ml for cefepime. Most reported LC-MS/MS methods have similar LLOQ but lower ULOQ. For example, Sime et al. reported a method for measurement of multiple β-lactam antibiotics, including piperacillin and meropenem (15). The linearity range for meropenem was 0.1 to 50 μg/ml, and the linearity range for piperacillin was 0.1 to 25 μg/ml (15). With that method, sample dilution and subsequent sample repreparation and reanalysis are frequently required when samples with high concentrations of meropenem and piperacillin are analyzed. On the other hand, the reported HPLC methods have good coverage of ULOQ but much higher LLOQ. For example, Verdier et al. reported an HPLC-UV method for measurement of multiple β-lactam antibiotics, including cefepime, meropenem, and piperacillin (9). The ULOQ of that method was 250 μg/ml for those three compounds, and the LLOQ was 2 μg/ml for cefepime and meropenem and 5 μg/ml for piperacillin (9). Due to its poor sensitivity (i.e., high LLOQ values), that method is not suitable for samples with relatively low drug concentrations.
Requirement of small sample volume.
In our method, 90 μl plasma samples are used for sample extraction, and only 5 μl of final extracted sample is used for sample injection. The plasma sample volume needed in our method is smaller than those for a number of other reported HPLC and LC-MS/MS methods for simultaneous measurement of multiple antibiotics (15, 19). In addition, as we use a simple protein precipitation method (1:4 [vol/vol] plasma sample-acetonitrile) for sample extraction, our method still works even when the plasma volume is as small as 20 μl. Because of the ability to measure drug concentrations in a small plasma volume, our method can be applied to measurements of antibiotics in samples collected from very sick patients and pediatric patients, especially from premature infants, from whom extremely limited sample volumes can be collected.
Use of isotope-labeled internal standards.
As no isotope-labeled tazobactam was available and no reliable isotope-labeled cefepime was found, in our method 2H5-piperacillin was used as the IS for both piperacillin and tazobactam, while 2H6-meropenem was used as the IS for both cefepime and meropenem. The use of isotope-labeled internal standards presents a clear advantage, as they are known to provide the highest attainable level of reliability (20). The only drawback is that these isotope-labeled compounds are expensive. For most reported HPLC and LC-MS/MS methods for simultaneous measurement of multiple antibiotics, structurally different compounds were selected as the ISs. It is worth pointing out that the selection of ISs should be done carefully if isotope-labeled compounds are not used. Since the method is targeted for measurement of antibiotics in critically ill patients, who usually receive various other medications, a compound that is rarely used in those patient populations should be selected as the IS. The compounds that have been used as ISs in previously published HPLC and LC-MS/MS methods include fluconazole (15), dicloxacillin (16), and ethylparaben (14), most of which are not ideal because of the high possibility of prescription of these drugs in critically ill patients.
After the LC-MS/MS method was successfully developed, we performed a comprehensive assay validation, in which 17 parameters were evaluated. In the current article, we report the results for 10 parameters. Except for carryover effect, the other 9 parameters passed the predefined acceptance criteria. Meropenem and piperacillin were found to have small carryover effects with our method. When double-blank samples were injected after the ULOQ sample, meropenem and piperacillin were detected in the double-blank samples, with peak areas that were <30% of the LLOQ. This small carryover effect was significantly reduced when a double-blank sample was injected three times. Based on the meropenem and/or piperacillin doses used in the clinic, we do not anticipate many clinical samples that have meropenem and/or piperacillin plasma concentrations close to the LLOQ (i.e., 0.1 μg/ml).
The cefepime dosage in ICU patients is either 2 g every 12 h or 2 g every 24 or 36 h, for patients with creatinine clearance (CLCR) of ≥50 ml/min or CLCR of <50 ml/min, respectively (21). The average steady-state median peak and trough concentrations in ICU patients treated with a 2 g dose of cefepime every 12 h were reported to be 97 ± 8 μg/ml and 2.68 ± 3.06 μg/ml, respectively (21). The standard meropenem dosing regimen is 1,000 mg every 8 h administered via intravenous infusion, which is adjusted based on the patient's renal function (22). In a study in which ICU patients were administered various doses of meropenem—500 mg every 8 h, 1,000 mg every 8 h, 500 mg every 12 h, and 1,000 mg every 12 h—the mean meropenem peak concentration was found to be 35.2 (9.6 to 70.5) μg/ml, and the mean trough concentration was found to be 5.2 (0.02 to 14.2) μg/ml (22). For piperacillin-tazobactam, ICU patients receive 4.5 g of piperacillin-tazobactam by intermittent infusion two or three times daily, depending on their renal function, according to clinical guidelines (23). The median peak plasma concentrations of tazobactam and piperacillin in ICU patients receiving 4 g/0.5 g piperacillin-tazobactam thrice daily were reported to be 29.4 μg/ml (27.9 to 32.0) and 210.5 (161.5 to 229.0) μg/ml, respectively, while median trough concentrations were reported to be 12.3 (7.7 to 13.7) μg/ml and 64.3 (49.0 to 68.9) μg/ml, respectively (24). The developed method is able to accurately measure levels of cefepime, meropenem, piperacillin, and tazobactam within their clinical ranges.
Although the stability of β-lactam antibiotics is a known issue, extremely limited information is available regarding their short-term and long-term stabilities, stock solution stability, and whole-blood stability. As most of these stability parameters are method independent, this information, once obtained, can be used as a reference by other research groups, even when different HPLC or LC-MS/MS methods are used. Because the stability information on β-lactam antibiotics is highly valuable and very important, we performed a thorough assay validation of various stability parameters and present our results in a separate article (part 2 of our study).
Conclusions.
A sensitive, simple, reproducible, and robust method for the simultaneous quantification of cefepime, meropenem, piperacillin, and tazobactam in lithium-heparinized human plasma was developed and rigorously validated according to FDA guidance. This validated method permits high-throughput analysis of samples and can be applied successfully to analysis of patient samples in the clinical setting or in large clinical studies.
MATERIALS AND METHODS
Chemicals and reagents.
Cefepime, meropenem, piperacillin sodium salt, and tazobactam were purchased from Sigma-Aldrich Co. (St. Louis, MO, USA). The deuterated internal standards, 2H5-piperacillin sodium salt and 2H6-meropenem, were purchased from Alsachim Lab (Strasbourg, France). Formic acid (>99.5%), water, acetonitrile, and methanol (each LC-MS grade) were obtained from Fisher Scientific (Fairlawn, NJ, USA). Human plasma with lithium heparin and 2% hemolyzed human plasma with lithium heparin were purchased from BioreclamationIVT (Westbury, NY, USA). All chemicals were of the highest purity available from commercial providers.
Instrumentation and analytical conditions. (i) HPLC conditions.
High-performance liquid chromatography (HPLC) was performed with a Shimadzu UFLC-20AD system (Shimadzu, Japan), which includes an LC-20AD binary pump, a SIL-20AC UFLC autosampler, a CTO-20A column oven, and a DGU-20A3 degasser. Analytical separation of cefepime, meropenem, piperacillin, tazobactam, 2H6-meropenem, and 2H5-piperacillin was achieved using a Phenomenex Kinetex C18 100-Å LC column (50 mm × 2.1 mm × 2.6 μm) coupled with a Phenomenex SecurityGuard Ultra UPLC Evo C18 cartridge (both from Phenomenex, Torrance, CA, USA) under gradient elution conditions. The mobile phase used for analysis was a mixture of water with 0.1% formic acid (solvent A) and acetonitrile with 0.1% formic acid (solvent B). Solvents A and B were combined in a gradient as follows: 2% solvent B (0 to 0.3 min), 40% solvent B (0.3 to 2.8 min), and held at 2% solvent B until the end of the run. The mobile phase was delivered at a total flow rate of 0.25 ml/min. The gradient elution profile is illustrated in Fig. 3.
The column was maintained at 35°C and the autosampler at 4°C, and the total injection volume was 5 μl for all compounds except meropenem, for which a 2 μl injection volume was used. The total run time was 7.0 min. Under these conditions, cefepime and meropenem eluted with typical retention times of 2.76 and 2.87 min, respectively. Piperacillin and tazobactam had typical retention times of 3.45 and 2.75 min, respectively. 2H6-meropenem, the internal standard (IS) for cefepime and meropenem, and 2H5-piperacillin, the IS for piperacillin and tazobactam, eluted with typical retention times of 2.85 and 3.42 min, respectively.
(ii) MS/MS conditions.
All compounds were detected using a TurboIonSpray probe in positive-ion mode by use of an API4000 triple-quadrupole mass spectrometer (AB Sciex LLC, Redwood City, CA, USA). The samples were analyzed by multiple-reaction monitoring (MRM) at an ion spray voltage of 5,000 V. Fragments were induced with a collision energy (CE) of 20 eV for meropenem, piperacillin, tazobactam, 2H6-meropenem, and 2H5-piperacillin and 15 eV for cefepime. Additional MS conditions included a curtain gas supply of 10 lb/in2, a source temperature of 500°C, a declustering potential (DP) of 40 V, an entrance potential (EP) of 10 V, and a collision cell exit potential (CXP) of 12 V. Collision gas was set to 9 lb/in2, and ion source gases 1 and 2 were set to 50 lb/in2 and 40 lb/in2, respectively. The instrument control and data processing software Analyst 1.6.2 (AB Sciex LLC, Redwood City, CA, USA) was used for data acquisition and processing. The MRM transitions for the antibiotics and their respective ISs, with the corresponding compound-specific parameters, are shown in Table 1.
Preparation of calibration standards and quality control samples.
Stock solutions of antibiotics were prepared by dissolving compounds in methanol to a concentration of 2 mg/ml. Stock solutions of 2H6-meropenem and 2H5-piperacillin were prepared by dissolving compounds in methanol to a final concentration of 1 mg/ml. All stock solutions were stored at −20°C. In order to make calibration standards and quality control (QC) samples, working solutions of cefepime, meropenem, piperacillin, and tazobactam were prepared by serial dilution in methanol. Calibration standards and QC samples for all four antibiotics were prepared by using the appropriate working solution to spike blank plasma containing lithium heparin, and exact concentrations are provided in Table 6. IS working solutions were also prepared by serial dilution with methanol to achieve a concentration of 25 μg/ml. The appropriate IS was then used to spike calibration standards and QC samples, and the final concentration of IS in each sample was 2.5 μg/ml.
TABLE 6.
Final concentrations of calibration standards and quality control levels for the antibiotics
| Antibiotic | Final concn (μg/ml) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Calibration standards |
Quality controls |
|||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | QC LLOQ | QC low | QC medium | QC high | QC ALOQa | |
| Cefepime | 0.500 | 1.00 | 5.00 | 10.0 | 25.0 | 50.0 | 100 | 150 | 0.500 | 1.50 | 75.0 | 120 | 480 | |
| Meropenem | 0.100 | 0.250 | 0.500 | 1.00 | 5.00 | 10.0 | 50.0 | 100 | 150 | 0.100 | 0.300 | 75.0 | 120 | 480 |
| Piperacillin | 0.100 | 0.500 | 1.00 | 5.00 | 10.0 | 50.0 | 100 | 150 | 0.100 | 0.300 | 75.0 | 120 | 480 | |
| Tazobactam | 0.250 | 0.500 | 1.00 | 5.00 | 10.0 | 50.0 | 100 | 150 | 0.250 | 0.750 | 75.0 | 120 | 480 | |
ALOQ, above the limit of quantification.
Sample preparation.
Sample preparation involved spiking 90 μl of blank human plasma containing lithium heparin with 10 μl of the appropriate antibiotic working solution and 10 μl of the 25 μg/ml IS working solution. Double-blank samples were prepared by spiking 90 μl of blank human plasma with 20 μl of methanol (i.e., no analyte and no IS), and blank samples were prepared by spiking blank human plasma with 10 μl of methanol and 10 μl of the 25 μg/ml IS working solution (i.e., no analyte). Samples were vortexed, and 400 μl of acetonitrile was added to each sample for protein precipitation. After further vortexing for 30 s, the samples were centrifuged at 17,000 × g for 15 min at 4°C. Following this, 10 μl of the supernatant of each sample was diluted with 90 μl of mobile phase A (water with 0.1% formic acid). The contents were then transferred to LC-MS vials for analysis.
Method validation.
The method was validated for cefepime, meropenem, piperacillin, and tazobactam, using human plasma with lithium heparin as the matrix, according to the FDA guidance for bioanalytical method validation.
(i) Linearity and sensitivity.
The linearity of the method was evaluated within the standard concentration range for human plasma. Linear regression (weighted 1/x2) was used to produce the best fit for the concentration/peak area ratio relationship for meropenem, piperacillin, and tazobactam in human plasma. Quadratic fitting (weighted 1/x2) was used to produce the best fit for the concentration/peak area relationship for cefepime. Use of a linear calibration curve for cefepime was attempted but resulted in failure of more than half of the calibration standards. The calibration curve was accepted if ≥75% of calibration standards (including the LLOQ and ULOQ) had accuracy values that fell within the ±15% range (±20% for the LLOQ). The validation of method sensitivity in human plasma was conducted using target LLOQ of 0.5 μg/ml for cefepime, 0.1 μg/ml for meropenem, 0.1 μg/ml for piperacillin, and 0.25 μg/ml for tazobactam. Sensitivity was evaluated separately for each antibiotic by analyzing six replicates of human plasma at the QC LLOQ concentration in three individual batch runs. Acceptance criteria were defined as having accuracy and precision values within ±20%.
(ii) Selectivity.
Selectivity was investigated by evaluation of six different lots of human plasma to test for potential interference with endogenous substances. This was done to ensure that precision, selectivity, and sensitivity were not compromised due to matrix effects at the retention times of the four antibiotics. The acceptance criteria were defined as the peaks for the blanks present on the chromatogram that could interfere with the analyte being no more than 20% of the response (peak area) of the LLOQ samples and, for the IS channel, no more than 5% of the IS response.
(iii) Interday and intraday accuracy and precision.
Intraday accuracy and precision for each antibiotic were determined separately within 1 day by analyzing six replicates of QC samples prepared at four different concentrations (QC LLOQ, QC low, QC med, and QC high). These concentrations were specifically selected to cover the entire range of the calibration curve. Interday accuracy and precision were assessed by repeating the intraday assay on three different days. Acceptance criteria were set based on FDA bioanalytical method validation guidance and were as follows: for accuracy, a ±15% standard deviation from the nominal values for all samples; and for precision, a ±15% coefficient of variation (CV) for replicates at each QC level, except for the LLOQ, which was allowed a deviation of ±20% for accuracy and precision. At each QC level, 50% of the samples had to meet the acceptance criteria and 2/3 of all QC samples had to pass in order for an individual batch run to be accepted.
(iv) Injection carryover.
Carryover was assessed during the validation by injecting a double-blank sample after the ULOQ calibrator. The carryover of analyte observed with the double-blank was compared to the analyte peak area for the LLOQ (see Table 6 for the exact concentration). Carryover is deemed to be acceptable if the concentration of analyte observed in the double-blank is not more than 20% of that in the LLOQ sample and the peak area for the IS in the double-blank after a ULOQ injection is not more than 5% of the average IS peak area for all other samples in the same batch run.
(v) Dilution integrity.
Per FDA recommendations, studies have to be performed to ensure the dilution integrity of specimens that may either be sample limited or contain analyte levels above the ULOQ. The dilution integrity of plasma samples containing 480 μg/ml of the antibiotic (i.e., 4 times the QC high concentration, above the limit of quantification [ALOQ]) was tested by performing 4-fold dilution of these samples with blank plasma to obtain a final concentration of 120 μg/ml. Samples were prepared and analyzed in replicates of 6, and the calculated concentrations were compared to the nominal concentration. Accuracy and precision should be within ±15%.
(vi) Matrix effects and matrix factors.
Matrix effects can occur if endogenous matrix components or preservative agents affect the chromatographic behavior and ionization of target compounds, resulting in ion suppression or enhancement. To evaluate matrix effects, blank human plasma samples from six different lots were processed according to the sample preparation procedure described above and then spiked with the antibiotic at two final concentrations, QC low and QC high (n = 6 each), and with the IS after extraction. The matrix effect of the plasma was investigated using the matrix factor, which is expressed as the ratio of the IS-normalized mean peak area for analytes spiked postextraction to that for the undiluted standard solution with mobile phase, at the same two concentration levels, as follows: matrix factor = IS-normalized mean peak area ratio in presence of matrix/IS-normalized mean peak area ratio in absence of matrix. Acceptance criteria require the CV % of IS-normalized matrix factors to be less than 15%.
(vii) Extraction recovery.
Extraction recovery of each of the four antibiotics was evaluated using six replicates each at two QC levels, QC low and QC high, by contrasting the IS-normalized peak areas for analytes in plasma samples before and after deproteinization, as follows: recovery = IS-normalized mean peak area ratio for extracted spiked samples/IS-normalized mean peak area ratio for samples spiked postextraction. The CV % of extraction recoveries should be <15%.
(viii) Hemolysis effect.
The hemolysis effect is considered a special category of matrix effects and is caused by a rupture of blood cells that leads to the release of hemoglobin and other endogenous cellular components into plasma samples. This can cause additional matrix interferences. To assess the blood hemolysis effect on the analytes of interest (the four antibiotics), QC low and QC high samples were prepared in 2% hemolyzed plasma. Six replicates of the QC low and QC high samples in hemolyzed plasma were extracted and analyzed with standards and intrarun QC samples prepared in nonhemolyzed plasma. The acceptance criteria require accuracy to be within ±15% of the nominal concentration, and the CV % should be no more than 15%.
(ix) Interference of analytes with ISs.
To evaluate the impact of interference of analytes with the ISs, three replicates of fortified samples containing analyte were prepared at the ULOQ concentration of each antibiotic separately and then processed and analyzed without adding the IS. The observed mean IS peak area for the interference samples should be ≤5% of the mean IS peak area for all the accepted calibration standards and intrarun QC samples. The formula used for this calculation is as follows: % interference = [(IS mean peak area for standards and QC samples − IS mean peak area for ULOQ)/IS peak area for standards and QC samples] × 100.
ACKNOWLEDGMENT
This work was supported by the Division of Microbiology and Infectious Disease, National Institute of Allergy and Infectious Diseases, National Institutes of Health (grant HHSN272200800008C).
Footnotes
For a companion article on this topic, see https://doi.org/10.1128/AAC.00861-18.
REFERENCES
- 1.Vincent JL, Rello J, Marshall J, Silva E, Anzueto A, Martin CD, Moreno R, Lipman J, Gomersall C, Sakr Y, Reinhart K, EPIC II Group of Investigators. 2009. International study of the prevalence and outcomes of infection in intensive care units. JAMA 302:2323–2329. doi: 10.1001/jama.2009.1754. [DOI] [PubMed] [Google Scholar]
- 2.Theuretzbacher U. 2012. Pharmacokinetic and pharmacodynamic issues for antimicrobial therapy in patients with cancer. Clin Infect Dis 54:1785–1792. doi: 10.1093/cid/cis210. [DOI] [PubMed] [Google Scholar]
- 3.Roberts JA, Lipman J. 2006. Antibacterial dosing in intensive care: pharmacokinetics, degree of disease and pharmacodynamics of sepsis. Clin Pharmacokinet 45:755–773. doi: 10.2165/00003088-200645080-00001. [DOI] [PubMed] [Google Scholar]
- 4.Roberts JA, Lipman J. 2009. Pharmacokinetic issues for antibiotics in the critically ill patient. Crit Care Med 37:840–851. doi: 10.1097/CCM.0b013e3181961bff. [DOI] [PubMed] [Google Scholar]
- 5.Gomez CM, Cordingly JJ, Palazzo MG. 1999. Altered pharmacokinetics of ceftazidime in critically ill patients. Antimicrob Agents Chemother 43:1798–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joukhadar C, Frossard M, Mayer BX, Brunner M, Klein N, Siostrzonek P, Eichler HG, Muller M. 2001. Impaired target site penetration of beta-lactams may account for therapeutic failure in patients with septic shock. Crit Care Med 29:385–391. doi: 10.1097/00003246-200102000-00030. [DOI] [PubMed] [Google Scholar]
- 7.Taccone FS, Laterre PF, Dugernier T, Spapen H, Delattre I, Wittebole X, De Backer D, Layeux B, Wallemacq P, Vincent JL, Jacobs F. 2010. Insufficient beta-lactam concentrations in the early phase of severe sepsis and septic shock. Crit Care 14:R126. doi: 10.1186/cc9091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts JA, Ulldemolins M, Roberts MS, McWhinney B, Ungerer J, Paterson DL, Lipman J. 2010. Therapeutic drug monitoring of beta-lactams in critically ill patients: proof of concept. Int J Antimicrob Agents 36:332–339. doi: 10.1016/j.ijantimicag.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 9.Verdier MC, Tribut O, Tattevin P, Le Tulzo Y, Michelet C, Bentue-Ferrer D. 2011. Simultaneous determination of 12 beta-lactam antibiotics in human plasma by high-performance liquid chromatography with UV detection: application to therapeutic drug monitoring. Antimicrob Agents Chemother 55:4873–4879. doi: 10.1128/AAC.00533-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopez KJ, Bertoluci DF, Vicente KM, Dell'Aquilla AM, Santos SR. 2007. Simultaneous determination of cefepime, vancomycin and imipenem in human plasma of burn patients by high-performance liquid chromatography. J Chromatogr B Analyt Technol Biomed Life Sci 860:241–245. doi: 10.1016/j.jchromb.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 11.Denooz R, Charlier C. 2008. Simultaneous determination of five beta-lactam antibiotics (cefepim, ceftazidim, cefuroxim, meropenem and piperacillin) in human plasma by high-performance liquid chromatography with ultraviolet detection. J Chromatogr B Analyt Technol Biomed Life Sci 864:161–167. doi: 10.1016/j.jchromb.2008.01.037. [DOI] [PubMed] [Google Scholar]
- 12.McWhinney BC, Wallis SC, Hillister T, Roberts JA, Lipman J, Ungerer JP. 2010. Analysis of 12 beta-lactam antibiotics in human plasma by HPLC with ultraviolet detection. J Chromatogr B Analyt Technol Biomed Life Sci 878:2039–2043. doi: 10.1016/j.jchromb.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 13.Zander J, Maier B, Suhr A, Zoller M, Frey L, Teupser D, Vogeser M. 2015. Quantification of piperacillin, tazobactam, cefepime, meropenem, ciprofloxacin and linezolid in serum using an isotope dilution UHPLC-MS/MS method with semi-automated sample preparation. Clin Chem Lab Med 53:781–791. doi: 10.1515/cclm-2014-0746. [DOI] [PubMed] [Google Scholar]
- 14.Ohmori T, Suzuki A, Niwa T, Ushikoshi H, Shirai K, Yoshida S, Ogura S, Itoh Y. 2011. Simultaneous determination of eight beta-lactam antibiotics in human serum by liquid chromatography-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 879:1038–1042. doi: 10.1016/j.jchromb.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Sime FB, Roberts MS, Roberts JA, Robertson TA. 2014. Simultaneous determination of seven beta-lactam antibiotics in human plasma for therapeutic drug monitoring and pharmacokinetic studies. J Chromatogr B Analyt Technol Biomed Life Sci 960:134–144. doi: 10.1016/j.jchromb.2014.04.029. [DOI] [PubMed] [Google Scholar]
- 16.Cohen-Wolkowiez M, White NR, Bridges A, Benjamin DK Jr, Kashuba AD. 2011. Development of a liquid chromatography-tandem mass spectrometry assay of six antimicrobials in plasma for pharmacokinetic studies in premature infants. J Chromatogr B Analyt Technol Biomed Life Sci 879:3497–3506. doi: 10.1016/j.jchromb.2011.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polson C, Sarkar P, Incledon B, Raguvaran V, Grant R. 2003. Optimization of protein precipitation based upon effectiveness of protein removal and ionization effect in liquid chromatography-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 785:263–275. doi: 10.1016/S1570-0232(02)00914-5. [DOI] [PubMed] [Google Scholar]
- 18.Colin P, De Bock L, T'Jollyn H, Boussery K, Van Bocxlaer J. 2013. Development and validation of a fast and uniform approach to quantify beta-lactam antibiotics in human plasma by solid phase extraction-liquid chromatography-electrospray-tandem mass spectrometry. Talanta 103:285–293. doi: 10.1016/j.talanta.2012.10.046. [DOI] [PubMed] [Google Scholar]
- 19.Cazorla-Reyes R, Romero-Gonzalez R, Frenich AG, Rodriguez Maresca MA, Martinez Vidal JL. 2014. Simultaneous analysis of antibiotics in biological samples by ultra high performance liquid chromatography-tandem mass spectrometry. J Pharm Biomed Anal 89:203–212. doi: 10.1016/j.jpba.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Vogeser M, Seger C. 2010. Pitfalls associated with the use of liquid chromatography-tandem mass spectrometry in the clinical laboratory. Clin Chem 56:1234–1244. doi: 10.1373/clinchem.2009.138602. [DOI] [PubMed] [Google Scholar]
- 21.Chapuis TM, Giannoni E, Majcherczyk PA, Chiolero R, Schaller MD, Berger MM, Bolay S, Decosterd LA, Bugnon D, Moreillon P. 2010. Prospective monitoring of cefepime in intensive care unit adult patients. Crit Care 14:R51. doi: 10.1186/cc8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Binder L, Schworer H, Hoppe S, Streit F, Neumann S, Beckmann A, Wachter R, Oellerich M, Walson PD. 2013. Pharmacokinetics of meropenem in critically ill patients with severe infections. Ther Drug Monit 35:63–70. doi: 10.1097/FTD.0b013e31827d496c. [DOI] [PubMed] [Google Scholar]
- 23.Zander J, Döbbeler G, Nagel D, Maier B, Scharf C, Huseyn-Zada M, Jung J, Frey L, Vogeser M, Zoller M. 2016. Piperacillin concentration in relation to therapeutic range in critically ill patients—a prospective observational study. Crit Care 20:79. doi: 10.1186/s13054-016-1255-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varghese JM, Jarrett P, Boots RJ, Kirkpatrick CM, Lipman J, Roberts JA. 2014. Pharmacokinetics of piperacillin and tazobactam in plasma and subcutaneous interstitial fluid in critically ill patients receiving continuous venovenous haemodiafiltration. Int J Antimicrob Agents 43:343–348. doi: 10.1016/j.ijantimicag.2014.01.009. [DOI] [PubMed] [Google Scholar]