Integrase strand transfer inhibitors (INSTIs) have emerged as clinically effective therapeutics that inhibit HIV-1 replication by blocking the strand transfer reaction catalyzed by HIV-1 integrase (IN). Of the three FDA-approved INSTIs, dolutegravir (DTG) is the least apt to select for resistance.
KEYWORDS: HIV-1, integrase strand transfer inhibitors, resistance, antiviral activity, susceptibility, infectivity
ABSTRACT
Integrase strand transfer inhibitors (INSTIs) have emerged as clinically effective therapeutics that inhibit HIV-1 replication by blocking the strand transfer reaction catalyzed by HIV-1 integrase (IN). Of the three FDA-approved INSTIs, dolutegravir (DTG) is the least apt to select for resistance. However, recent salvage therapy regimens had low response rates with therapies that included DTG, suggesting that DTG resistance can be selected in patients. Using a single-round infection assay, we evaluated a collection of our best inhibitors and DTG against a broad panel of INSTI-resistant mutants. Two of the new compounds, 4c and 4d, had antiviral profiles against the mutants we tested superior to that of DTG. The susceptibility profiles of 4c and 4d suggest that the compounds are candidates for development as INSTIs. Modeling the binding of 4d to HIV-1 IN reinforced the significance of mimicking the DNA substrate in developing compounds that are broadly effective in their abilities to inhibit HIV-1 INs with mutations in the active site.
INTRODUCTION
Drugs were developed that inhibited two of the three HIV-1 enzymes, reverse transcriptase and protease, long before there were drugs that inhibited integrase (IN). IN has two enzymatic activities: (i) 3′ processing (3′P), in which a GT dinucleotide is removed from each of the 3′ ends of the unintegrated linear viral DNA, and (ii) the strand transfer (ST) reaction, which catalyzes the integration of the viral DNA into the genome of the host cell (1, 2). Currently, all FDA-approved IN inhibitors target the ST reaction, and these compounds are, for that reason, called IN ST inhibitors (INSTIs) (3, 4). The three FDA-approved INSTIs are raltegravir (RAL), elvitegravir (EVG), and dolutegravir (DTG).
All three FDA-approved INSTIs bind to the IN active site, potently inhibit the replication of wild-type (WT) HIV-1, and are minimally toxic. However, INSTI-resistant mutants can emerge during treatment, many of which cause resistance to both of the approved first-generation INSTIs (5–9). Resistance mutations occur in and proximal to the IN active site, primarily in the β4α2 loop of the IN catalytic core domain (CCD) (10). Although resistance mutations reduce the activity of IN, integration of the viral DNA and viral replication require IN to catalyze only a total of four reactions (3′P of the two viral DNA ends and their subsequent insertion into the host DNA). Because only limited IN enzymatic activity is required, the virus is able to tolerate INSTI-resistant mutations that cause a reduction in the enzymatic activity of IN.
All INSTIs share two common characteristics: a central pharmacophore, which contains a chelating motif that interacts with the two Mg2+ ions at the IN active site, and a halogenated benzyl moiety that stacks against the nucleobase of the penultimate cytosine near the 3′ ends of the viral DNA (4, 11). Certain features of the first-generation INSTIs may help to explain some of their limitations. (i) The chelating motif comprises elements that reside partly on and partly off the central pharmacophore, whether the central pharmacophore is a pyrimidine (RAL) or a quinoline (EVG). (ii) The pharmacophore has appended to it modifications that are involved in contacts between the drug and the active site of WT HIV-1 IN in places where amino acid substitutions are readily tolerated (for example, RAL and Y143). (iii) The linker connecting the pharmacophore and the halogenated benzyl moiety is relatively short, which limits the ability of the compound to adjust its binding in response to changes in the IN active site. These limitations can compromise the ability of first-generation INSTIs to bind tightly to mutant forms of IN (12–14).
However, the structure of DTG addresses all three of the above-mentioned issues (see Fig. S1 in the supplemental material), helping it to retain tight binding despite changes in the IN active site (15). First, the entire chelating motif of DTG resides on a tricyclic scaffold. Second, the third ring of the central pharmacophore (on the “left” side of the molecule, away from the end of the viral DNA) is an oxazinane ring. The importance of the oxazinane ring was clarified by superposing the structure of the prototype foamy virus (PFV) intasome with DTG bound onto the structure of the PFV intasome with a target DNA (tDNA) bound (16, 17). This comparison showed that the oxazinane ring of DTG mimics aspects of the bound host DNA substrate (18, 19). Because IN needs to be able to bind its DNA substrates, mutations that would interfere with binding of the oxazinane ring of DTG are also likely to interfere with binding of the host DNA substrate. Accordingly, such mutations could impair the ability of IN to insert viral DNA into the host genome. Third, DTG has a longer linker group connecting the central metal-chelating pharmacophore and the halogenated benzyl moiety. This would allow DTG to adjust its structure and the details of its binding in ways that would allow it to maintain the key interactions with the penultimate cytosine and the catalytic Mg2+ ions in spite of mutations that change the geometry of the active site (Fig. 1). These structural features help to explain why DTG retains potency against HIV-1 that carries most of the known INSTI-resistant mutations (16). This also helps to explain why it has been relatively difficult to select for DTG resistance in cell culture and in treatment-naive patients (20–27).
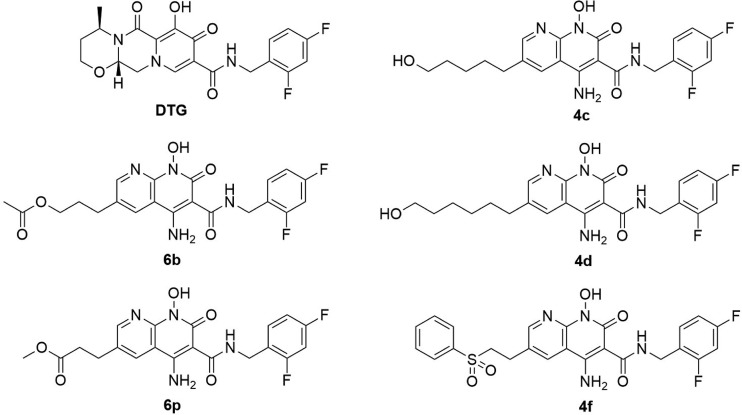
FIG 1.
Chemical structures of DTG and 4c, 4d, 4f, 6b, and 6p.
However, recent trials involving INSTI-experienced patients suggest that there are combinations of mutations in IN that can cause a dramatic decrease in susceptibility to DTG (28, 29). Many of the INSTI-experienced patients in these trials had been previously treated with RAL. RAL treatment is known to select for resistance mutations at Q148. In some cases, additional resistance mutations appeared at L74, E138, G140, or G163, which blunted the effectiveness of regimens that included DTG. These studies have shown that in patients infected with HIV with preexisting RAL resistance mutations who switch to a regimen that contains DTG, the virus can acquire additional resistance mutations in IN, which can reduce the susceptibility of the virus to DTG.
We recently developed five 4-amino-1-hydroxy-2-oxo-1,8-naphthyridine-containing compounds, 4c, 4d, 4f, 6b, and 6p, that featured different appendages at the 6′ position on the naphthyridine scaffold (Fig. 1). 4c and 4d include alcohol-derived attachments, 4f is a disulfonophenyl derivative (19), and 6b and 6p contain ester-derived substituents (18). All of the compounds potently inhibit the replication of WT HIV-1, and we showed that the best of these compounds retain their potency against a number of the well-known INSTI-resistant mutants (18, 19). That allowed us to choose the most promising of our new compounds for detailed analysis. Here, we describe experiments in which the potency of DTG and 4c, 4d, 4f, 6b, and 6p were compared against a more extensive panel of simple and complex INSTI-resistant mutants in single-round infection assays. Our data show that two of our synthetic inhibitors, 4c and 4d, were more broadly effective against the panel of IN mutants than DTG.
RESULTS
Antiviral activities of DTG and our compounds against a panel of INSTI-resistant single mutants.
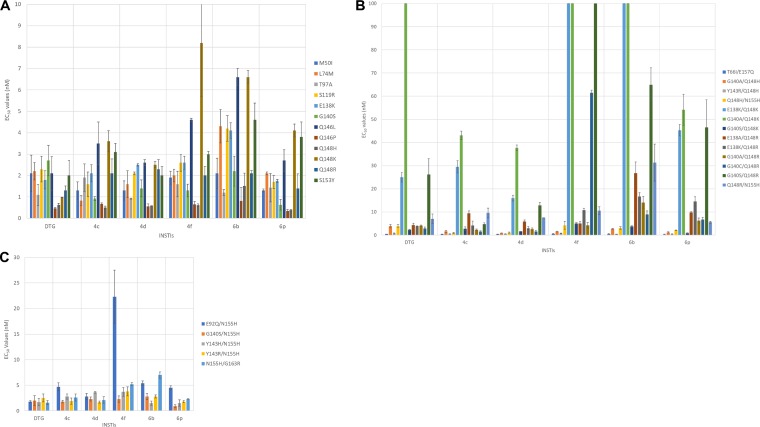
We previously measured the antiviral potencies of DTG and 4c, 4d, 4f, 6b, and 6p in single-round replication assays using WT HIV-1, well-established first-generation INSTI-resistant mutants, and selected putative DTG-resistant mutants (18, 19). However, we had not previously determined the 50% effective concentrations (EC50s) for DTG and our compounds against many of the emerging INSTI-resistant mutants that have recently been identified in cell culture selection studies and clinical trials (10, 30, 31). Therefore, we screened DTG and our compounds against a panel of INSTI-resistant single mutants that included M50I, L74M, T97A, S119R, E138K, G140S, Q146L, Q146P, Q148H, Q148K, Q148R, and S153Y mutants (Fig. 2A; see Table S1A in the supplemental material). We found that 4c, 4d, 4f, 6b, and 6p potently inhibited the INSTI-resistant M50I, L74M, T97A, S119R, E138K, G140S, Q146P, Q148H, Q148R, and S153Y single mutants, with antiviral potencies of <5.0 nM. (We have chosen to call the compounds reasonably effective if they retain an EC50 of <5.0 nM compounds against the mutant viruses [see Discussion]). We recently reported a comparison of the potencies of DTG and two compounds that are in late-stage clinical trials, cabotegravir and bictegravir, against the same broad panel of IN mutants we used to evaluate our compounds (32). The DTG data we used in the comparisons with our compounds are the same data we reported previously. The compounds effectively inhibited Q146L (<5.0 nM), with 6b (6.6 ± 0.4 nM) showing a minor loss of potency. Q148K was the only INSTI-resistant single mutant against which any of our compounds showed a loss of potency, and the reduction was minor. Compounds 4c, 4d, and 6p had antiviral activities of <5.0 nM, whereas 4f (8.2 ± 2.5 nM) and 6b (6.6 ± 0.3 nM) showed a small loss of potency. With the exception of Q146L and Q148K, DTG and our compounds had nearly equivalent antiviral profiles against this panel of INSTI-resistant single mutants. To compare the efficacies of the INSTIs and analyze the antiviral data more critically, Student's test was used to determine the statistical significance of the differences in the antiviral data for DTG and our compounds (see the supplemental material). Because the EC50s of DTG and our compounds were very similar against WT HIV-1, we could make direct comparisons of the antiviral data for the INSTI-resistant mutants. In this initial screen against 12 INSTI-resistant single mutants, DTG was significantly better than 4f and 6b against seven and five of the mutants, respectively (see Tables S2 and S1B in the supplemental material). Moreover, four of the seven times DTG was significantly better than 6b, the P values were <0.001. DTG had significantly better antiviral activities than 4c and 4d against three of the mutants in this group; however, 6p was significantly better than DTG against three other mutants in the group (once with a P value of <0.01). Although DTG and our compounds had potent antiviral activities against the single mutants, statistical analysis showed that DTG performed better against this panel of INSTI-resistant mutants.
FIG 2.
Antiviral activities of DTG, 4c, 6b, 4d, 6p, and 4f compounds against a panel of INSTI-resistant mutants. The EC50s were determined using a vector that carries the INSTI-resistant single mutants in a single-round infection assay. The DTG potency data used in the comparisons with the data for our compounds have been reported previously (32). The error bars represent the standard deviations of independent experiments; n = 4. (A) EC50s of DTG, 4c, 4d, 4f, 6b, and 6p against a panel of INSTI-resistant single mutants, with a maximum value of 10 nM. (B) EC50s of DTG, 4c, 4d, 4f, 6b, and 6p against a panel of INSTI-resistant double mutants, with a maximum value of 100 nM. (C) EC50s of DTG, 4c, 4d, 4f, 6b, and 6p against a panel of INSTI-resistant double mutants, with a maximum value of 30 nM.
Antiviral potencies of DTG and our compounds against a panel of INSTI-resistant double mutants based on a primary mutation at T66 or Q148.
Next, we tested DTG and our compounds against the T66I/E157Q double mutant and a panel of INSTI-resistant double mutants that had a mutation at position Q148 (H/K/R) plus an additional secondary mutation: E138A/K, G140A/S, Y143R, or N155H (Fig. 2B; see Table S3A in the supplemental material). DTG and all of our compounds showed strong antiviral activities (<1.0 nM) against the T66I/E157Q double mutant. Additionally, the INSTI-resistant double mutants that included a Q148H mutation, the G140A/Q148H, Y143R/Q148H, and Q148H/N155H mutants, were susceptible to DTG and our compounds (<5.0 nM). However, when DTG and our compounds were screened against the INSTI-resistant double mutants that included the Q148K primary mutation, there was a loss of potency for both DTG and our compounds. The INSTI-resistant E138K/Q148K double mutant caused a moderate reduction in susceptibility to 4d (16.0 ± 1.2 nM), DTG (25.0 ± 2.1 nM), and 4c (29.5 ± 2.7 nM); however, this mutant caused a substantial loss of susceptibility to 4f (127 ± 16.8 nM), 6b (134.4 ± 1.77 nM), and 6p (45.3 ± 2.5 nM). Similarly, the INSTI-resistant G140A/Q148K double mutant caused a considerable drop in susceptibility to 4c (43.1 ± 1.8 nM), 4d (37.7 ± 1.2 nM), and 6p (54.1 ± 6.7 nM) and very significant reductions in susceptibility to DTG (450.7 ± 58.8 nM), 6b (392.9 ± 13.9 nM), and 4f (789.0 ± 26.7 nM). Unlike the previous two INSTI-resistant double mutants that included Q148K, the G140S/Q148K double mutant was sensitive to DTG and our compounds (≤5.0 nM). Overall, DTG and our compounds were successful at inhibiting INSTI-resistant double mutants based on Q148R, which included E138A/Q148R, E138K/Q148R, G140A/Q148R, G140C/Q148R, G140S/Q148R, and Q148R/N155H. DTG and all of our compounds, with the exception of 6b (26.7 ± 4.9 nM), inhibited the INSTI-resistant E138A/Q148R double mutant with, at most, a modest loss of susceptibility (<10.0 nM). However, only DTG, 4c, and 4d potently inhibited the INSTI-resistant E138K/Q148R, G140A/Q148R, and G140C/Q148R double mutants (<5.0 nM). 6b and 6p inhibited the same mutants with a modest loss of potency (<15.0 nM). 4f retained a similar level of potency against two of these mutants; however, it exhibited a considerable reduction in efficacy against G140C/Q148R (61.5 ± 1.1 nM). 4c was the only INSTI that potently inhibited the INSTI-resistant G140S/Q148R double mutant, with an antiviral activity of <5.0 nM; 4d inhibited this mutant with a slightly higher EC50 (12.9 ± 1.2 nM). Conversely, the INSTI-resistant G140S/Q148R double mutant caused larger losses of susceptibility to DTG (26.2 ± 6.8 nM), 6b (64.9 ± 7.3 nM), 6p (46.5 ± 11.9 nM), and 4f (181.6 ± 12.0 nM). The INSTI-resistant Q148R/N155H double mutant was inhibited by all of the INSTIs with antiviral activities of <10.0 nM, except 6b, which inhibited the mutant with an EC50 of 31.3 ± 8.0 nM. DTG had significantly better antiviral potency against the double mutants in this panel than either 4f or 6b, seven and eight times better, respectively (see Tables S2 and S3B in the supplemental material). In five of the eight cases in which DTG was significantly better than 6b, the P values were <0.001. Conversely, 4c and 6p were both better than DTG against five of the mutants in the panel (both comparisons had P values of <0.001), and 4d was significantly better than DTG against eight of the mutants; thus, 4d was the most effective compound against this panel of double mutants.
Antiviral potencies of DTG and our compounds against a panel of INSTI-resistant double mutants that included a primary mutation at N155.
We measured the potencies of DTG and our compounds against a panel of INSTI-resistant double mutants that included an N155H primary mutation and an additional mutation at position E92, G140, Y143, or G163. The INSTI-resistant double mutants in this panel included E92Q/N155H, G140S/N155H, Y143H/N155H, Y143R/N155H, and N155H/G163R mutants (Fig. 2C; see Table S4A in the supplemental material). DTG, 4c, 4d, and 6p all potently inhibited these INSTI-resistant double mutants, with antiviral activities of <5.0 nM. 6b inhibited the entire panel with only a minor loss of potency (antiviral activities of <7.0 nM), whereas 4f retained most of its potency (<5.0 nM) against the INSTI-resistant G140S/N155H, Y143H/N155H, Y143R/N155H, and N155H/G163R double mutants but showed a moderate loss of potency against E92Q/N155H (22.3 ± 5.2 nM). There were no significant differences in efficacies between DTG and our compounds against the G140S/N155H and Y143R/N155H double mutants; however, DTG was significantly better than all of our compounds against at least two of the three remaining double mutants, especially versus 6b (see Tables S2 and S4B in the supplemental material) (this comparison, for two mutants, had P values of <0.001).
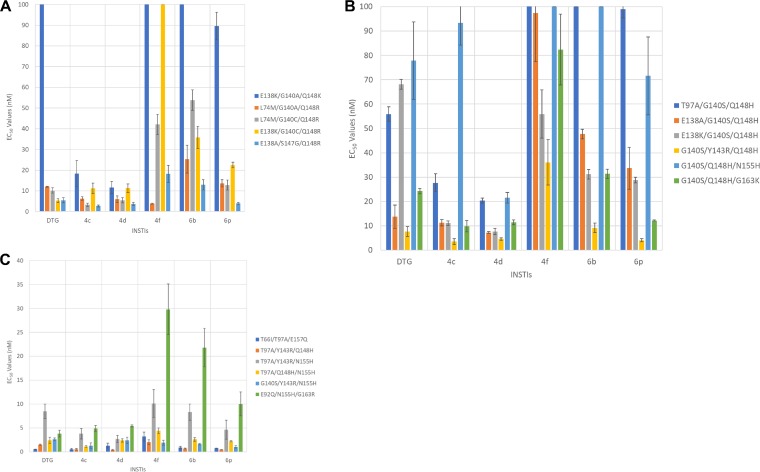
Antiviral potencies of DTG and our compounds against a panel of INSTI-resistant triple mutants that included a primary mutation at Q148.
We analyzed DTG and our compounds against a panel of INSTI-resistant triple mutants, the E138K/G140A/Q148K, L74M/G140A/Q148R, L74M/G140C/Q148R, E138K/G140C/Q148R, and E138A/S147G/Q148R mutants (Fig. 3A; see Table S5A in the supplemental material), that were identified in clinical trials in which DTG was used in a salvage regimen. The patients in this trial had a relatively low response rate to the salvage therapy. 4d and, to a certain extent, 4c were more effective than DTG and our other compounds in terms of their ability to inhibit the INSTI-resistant triple mutants in the panel. Both 4c and 4d retained most of their potency against the L74M/G140A/Q148R mutant, with antiviral activities of 6.3 ± 0.9 nM and 6.0 ± 1.6 nM, respectively, and the L74M/G140C/Q148R mutant, with antiviral activities of 3.3 ± 0.8 nM and 5.5 ± 1.3 nM, respectively, whereas DTG had antiviral activities of 12.0 ± 0.2 nM against the L74M/G140A/Q148R mutant and 10.2 ± 1.3 nM versus the L74M/G140C/Q148R mutant. 4f potently inhibited the L74M/G140A/Q148R mutant (3.8 ± 0.3 nM), but both 6b and 6p lost potency against this INSTI-resistant triple mutant, with efficacies at 25.3 ± 6.7 nM and 13.6 ± 1.8 nM, respectively. The INSTI-resistant L74M/G140C/Q148R triple mutant caused reductions in susceptibility to 6b (53.8 ± 4.94 nM), 6p (12.8 ± 2.4 nM), and 4f (42.1 ± 4.9 nM). DTG potently inhibited the INSTI-resistant E138K/G140C/Q148R triple mutant with an EC50 of 5.3 ± 1.0 nM, while minor reductions in potency were seen with 4c (11.2 ± 2.5 nM) and 4d (11.3 ± 2.1 nM); however, the mutant caused larger reductions in susceptibility to 4f (111.1 ± 22.5 nM), 6b (35.8 ± 5.4 nM), and 6p (22.5 ± 1.3 nM). DTG and all of our compounds inhibited the INSTI-resistant E138A/S147G/Q148R triple mutant with, in some cases, a minor loss of potency (<6.0 nM), except for 6b (12.9 ± 2.5 nM) and 4f (18.2 ± 4.2 nM). Finally, the INSTI-resistant E138K/G140A/Q148K triple mutant caused very significant drops in susceptibility to DTG (212.1 ± 46.0 nM), 6b (138.1 ± 23.5 nM), 6p (89.6 ± 6.6 nM), and 4f (500.1 ± 64.0 nM), whereas the mutant caused much smaller reductions in potency for 4c (18.3 ± 6.3 nM) and 4d (11.6 ± 3.0 nM). Thus, in terms of their abilities to inhibit these more complex INSTI-resistant mutants, 4c and 4d were shown to be more broadly effective than DTG. 4c and 4d were better able to inhibit four and three of the mutants, respectively, compared to DTG (see Tables S2 and S5B in the supplemental material). For two of the mutants, 4c was better than DTG, with P values of <0.001. Conversely, DTG was more effective than 6b and 4f against three and four of the mutants, respectively, with two of the 4f to DTG comparisons having P values of <0.001.
FIG 3.
Antiviral activities of DTG, 4c, 4d, 4f, 6b, and 6p versus panels of INSTI-resistant mutants. The EC50s were determined using a vector that carries the INSTI-resistant triple mutants in a single-round infection assay. The error bars represent the standard deviations of independent experiments; n = 4. (A) The EC50s shown in the graph have a maximum value of 100 nM; values higher than 100 nM are not shown. The EC50s of DTG against E138K/G140A/Q148K, 6b versus E138K/G140A/Q148K, and 4f against E138K/G140A/Q148K and E138K/G140C/Q148R INSTI-resistant triple mutants were >100 nM. (B) EC50s of the INSTIs against a panel of INSTI-resistant triple mutants that included the G140S and Q148H primary mutations. The graph has a maximum value of 100 nM. The EC50s of 6b versus T97A/G140S/Q148H and G140S/Q148H/N155H and 4f against T97A/G140S/Q148H and G140S/Q148H/N155H INSTI-resistant triple mutants were >100 nM. (C) EC50s were determined against a panel of INSTI-resistant triple mutants that included a mutation at position N155 and two additional mutations. The EC50s shown in the graph have a maximum value of 40 nM.
Antiviral potencies of DTG and our compounds against a panel of INSTI-resistant triple mutants that included the G140S and Q148H primary mutations.
Our compounds and DTG were tested against a panel of INSTI-resistant triple mutants that included the G140S/Q148H mutations plus an additional mutation. This panel of INSTI-resistant triple mutants included T97A/G140S/Q148H, E138A/G140S/Q148H, E138K/G140S/Q148H, G140S/Y143R/Q148H, G140S/Q148H/N155H, and G140S/Q148H/G163K mutants (Fig. 3B; see Tables S2 and S6A in the supplemental material). 4d was the most effective INSTI, in terms of its overall ability to retain potency against these INSTI-resistant mutants. 4d inhibited E138A/G140S/Q148H, E138K/G140S/Q148H, and G140S/Y143R/Q148H mutants with antiviral potencies of 7.3 ± 0.4 nM, 7.7 ± 1.3 nM, and 4.6 ± 0.5 nM, respectively. 4c was the second most effective compound, followed closely by DTG. Of the triple mutants in this panel, the G140S/Y143R/Q148H mutant proved to be the most susceptible to the compounds we tested. Conversely, the T97A/G140S/Q148H and G140S/Q148H/N155H mutants caused the largest drops in potencies, although 4c and 4d retained moderate efficacies (<30.0 nM) against T97A/G140S/Q148H and G140S/Q148H/N155H mutants; 4d lost a moderate amount of potency (21.6 ± 2.2 nM) against the G140S/Q148H/N155H mutant. 4c, 4d, and 6p inhibited the INSTI-resistant G140S/Q148H/G163K triple mutant with efficacies of 9.8 ± 2.3 nM, 11.4 ± 1.0 nM, and 12.2 ± 0.2 nM, respectively, whereas both DTG and 6b showed larger reductions in potencies (24.3 ± 1.1 nM and 31.4 ± 1.9 nM, respectively). 4c and DTG retained considerable potency against the E138A/G140S/Q148H and G140S/Y143R/Q148H mutants but showed considerable reductions in potencies against the other INSTI-resistant triple mutants. 4f was ineffective at inhibiting this panel of INSTI-resistant triple mutants, whereas both 6b and 6p showed substantial reductions in potencies. Both 4c and 4d were considerably better than DTG against the mutants in this panel, four and five times better, respectively, and, for each of our compounds, three of the comparisons had P values of <0.001 (see Tables S2 and S6B in the supplemental material). Conversely, DTG was better than 6b and 4f against three and five of the triple mutants in the panel, respectively.
Antiviral potencies of DTG and our compounds against a panel of INSTI-resistant triple mutants that included a mutation at position N155 and two additional mutations.
We analyzed DTG and our compounds using a panel of INSTI-resistant triple mutants that included the T66I/T97A/E157Q triple mutant and triple mutants having a mutation at position N155 plus 2 additional mutations at amino acid E92, T97, Y143, Q148, or G163. The panel of INSTI-resistant triple mutants consisted of T66I/T97A/E157Q, T97A/Y143R/Q148H, T97A/Y143R/N155H, T97A/Q148H/N155H, G140S/Y143R/N155H, and E92Q/N155H/G163R mutants (Fig. 3C; see Table S7A in the supplemental material). The INSTI-resistant T66I/T97A/E157Q, T97A/Y143R/Q148H, T97A/Q148H/N155H, and G140S/Y143R/N155H triple mutants were all susceptible to DTG and our compounds (EC50s of <5.0 nM). The INSTI-resistant T97A/Y143R/N155H triple mutant showed minor drops in susceptibility to DTG (8.5 ± 1.5 nM), 6b (8.3 ± 1.7 nM), and 4f (10.1 ± 2.9 nM), while it maintained susceptibility to 4c (3.8 ± 1.1 nM), 4d (2.7 ± 0.7 nM), and 6p (4.6 ± 2.0 nM). The INSTI-resistant E92Q/N155H/G163R triple mutant was sensitive to DTG (3.8 ± 0.7 nM), 4c (4.9 ± 0.6 nM), and 4d (5.4 ± 0.2 nM), although it exhibited a moderate drop in susceptibility to 4f (29.8 ± 5.3 nM), 6b (21.8 ± 4.0 nM), and 6p (10.0 ± 2.5 nM). Both 4d and 6b were better than DTG against two of the triple mutants in this panel, whereas both 4c and 6p were better than DTG against three of the mutants. For two of the 6p-DTG comparisons, the P values were <0.001 (see Tables S2 and S7b in the supplemental material). DTG was better than 4f against three of the triple mutants in the panel.
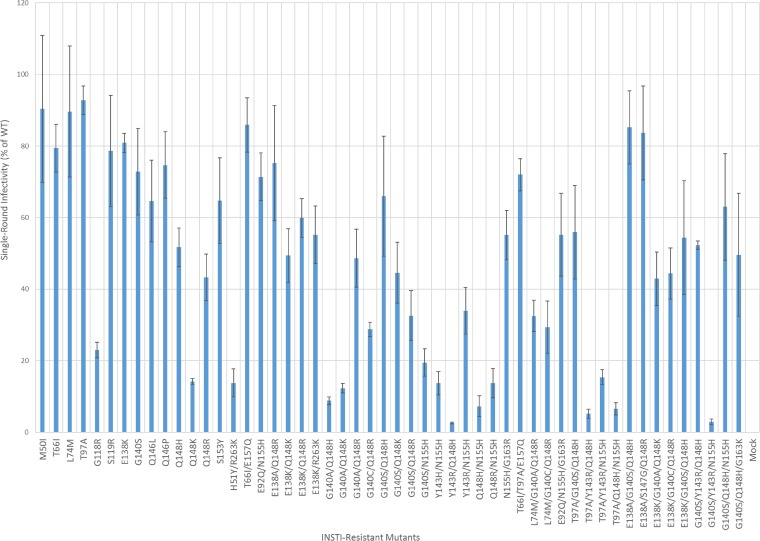
Replication capacities of the INSTI-resistant mutants using a single-round infection assay.
Because drug-resistant variants of HIV-1 are reduced in their infectivity, we compared the infectivities of the INSTI-resistant mutants used in this study to the infectivity of WT HIV-1 in single-round infectivity assays (Fig. 4; see Table S8 in the supplemental material). Of the 52 INSTI-resistant mutants we examined, 11 were found to have infectivities that were <20.0% of that of WT HIV-1. Most of the INSTI-resistant single mutants had >60.0% of the WT HIV-1 infectivity; the exceptions had mutations at Q148 (Q148H [51.7% ± 5.4%], Q148R [43.3% ± 6.5%], and Q148K [14.2% ± 0.8%]). Q148K was the only single mutant that had single-round infectivity that was less than 20.0% that of WT. The putative DTG-resistant mutant G118R (EC50 = 13.0 ± 5.0 nM), which caused a minor drop in susceptibility to our compounds, also showed relatively weak infectivity (23.0% ± 2.2%). Most of the INSTI-resistant double mutants displayed significant drops in infectivity (<60% of WT HIV-1 activity). However, when the E138A/K mutation was added to the Q148K/R single mutant or the G140S mutation was added to the Q148H/K single mutant, infectivity was enhanced. Adding the G140A/C mutations to the Q148H/K/R mutations did not restore infectivities to WT levels. All of the INSTI-resistant double mutants with mutations at the primary position Y143, Q148, or N155 had relatively low replication capacities (<33%), as was found with the putative DTG-resistant H51Y/R263K mutant (13.8% ± 3.9%). The E138K/Q148K and G140A/Q148K mutants, which caused significant reductions in susceptibility to DTG and our compounds, had infectivities of 49.4% ± 7.5% and 12.3% ± 1.3%, respectively. In some cases, the addition of a third mutation increased the infectivities of certain double mutants. Notably, adding a G140S mutation to the double mutants that had combinations of mutations at the primary position Y143, Q148, or N155 usually restored a portion of the replication capacity and had a more favorable effect on infectivity than a T97A mutation. The addition of a third mutation, E138K, to G140A/Q148K (E138K/G140A/Q148K; 42.9% ± 7.5%) and G140C/Q148R (E138K/G140C/Q148R; 44.4% ± 7.1%) increased the infectivities considerably relative to the parental double mutant, while the addition of E138A to G140S/Q148H (E138A/G140S/Q148H; 85.2% ± 10.2%) also enhanced infectivity, as measured in a single-round replication assay, relative to the parental INSTI-resistant double mutants. The INSTI-resistant triple mutant, which included the G140S and Q148H mutations plus a third mutation at either position T97A, E138K, or N155H, all of which caused major reductions in susceptibility to DTG and our compounds, had replication capacities of 55.9% ± 13.1%, 54.4% ± 15.9%, and 63.0% ± 14.9%, respectively, demonstrating that there are mutants that reduce the potency of the best available INSTIs, including DTG, and that still replicate reasonably well.
FIG 4.
Replication capacities of the INSTI-resistant mutants, using a single-round infection assay. The replication capacities of the INSTI-resistant mutants used in this study were measured using INSTI-resistant mutant vectors in a single-round infection assay. The luciferase activity of the WT virions was set to 100, and the infectivities of the mutant vectors (adjusted for the amount of p24/Gag used in the assay) were measured relative to WT infectivity. The error bars represent the standard deviations of independent experiments; n = 4.
DISCUSSION
Although INSTIs have emerged as important anti-HIV drugs that are widely used in combination antiretroviral therapy (cART), there are both clinical and cell culture data that show that, like all other anti-HIV drugs, INSTIs are susceptible to the development of resistance. Additionally, there have been reports of neural tube defects in babies of women who were undergoing DTG treatment during conception and early pregnancy (33). Therefore, not only do new INSTIs need to be developed and tested, but these new INSTIs should not have serious negative side effects and should be able to inhibit the resistant virus strains that are emerging in response to the current therapies. Here, we show that the best of our compounds, 4c and 4d, have a superior antiviral profile against a broad panel of INSTI-resistant mutants compared to the most broadly effective of the FDA-approved INSTIs, DTG. Overall, out of the 46 INSTI-resistant mutants tested, 4d had higher potency than DTG against 19 mutants (41%) (see Table S9 in the supplemental material), with 6 of the potency comparisons having P values of <0.001, whereas 4c was more potent than DTG against 18 of the 46 mutants (39%), with 8 of the potencies having P values of <0.001. Conversely, DTG was more broadly effective than our other compounds tested (6b, 6p, and 4f; 24, 16, and 27 times, respectively). Importantly, both 4c and 4d were much more potent than DTG against several INSTI-resistant triple mutants, with better efficacies against 11 and 10, respectively, out of the 17 INSTI-resistant triple mutants we tested. It is important to point out that a mutation, or group of mutations, that causes a minimal change in the susceptibility of the virus to a drug, leaving the EC50 below 5 nM, is likely to be much less problematic in a clinical setting than are mutations that cause a much greater loss of drug susceptibility. The degree to which a mutation, or group of mutations, is likely to be important clinically is also related to the impact of the mutation(s) on the ability of the virus to replicate (see below).
Some of the INSTI-resistant triple mutants that caused substantial reductions in susceptibility to DTG and/or our compounds have reduced replication capacities compared to WT HIV-1. However, the E138A/G140S/Q148H mutant has a relatively high replication capacity (85.2% ± 10.2%), which, based on the replication capacity of INSTI-resistant mutants isolated from patients, is sufficient to support active HIV-1 infections. There are several other INSTI-resistant triple mutants, including the E138K/G140A/Q148K, T97A/G140S/Q148H, E138K/G140S/Q148H, and G140S/Q148H/N155H mutants, that have replication capacities approximately 50% that of WT HIV-1. Even if these mutants are not able to replicate efficiently in patients, it is possible that they could acquire additional compensatory mutations during an active infection in the presence of the drug. There is evidence that INSTI-resistant quadruple and quintuple mutants that have significantly reduced susceptibilities to DTG can arise in patients in whom they are able to replicate well enough to sustain an active, ongoing infection (28, 29).
The recent data on INSTI resistance, taken together with better structural data, have improved our understanding of the interactions between the INSTIs and WT and mutant forms of HIV IN. This, in turn, has facilitated a better understanding of what makes an INSTI effective against a broad range of resistant mutants. It appears that it is important to have a relatively compact pharmacophore that is moderately flexible. The most successful compounds have, at a minimum, a bicyclic ring system that that binds the Mg2+ ions at the IN active site. In all cases, there is a halogenated benzyl ring, which interacts with the nucleobase of the penultimate nucleotide (cytosine) at the 3′ end of the viral DNA. It appears that having a longer linker joining the halogenated benzyl ring with the bi- or tricyclic pharmacophore improves the ability of compounds to effectively inhibit many of the simpler drug-resistant mutants. This is apparent when the abilities of the first-generation INSTIs, RAL and EVG, which have relatively short linkers, are compared to those of the second-generation INSTI, DTG, which has a longer linker. It is likely that compounds with longer linkers are better able to adapt to changes in the active-site geometry caused by resistance mutations. However, the most broadly effective of the approved INSTIs, DTG, loses significant potency against some of the INSTI-resistant mutants that we tested. Generally speaking, the most problematic mutants are complex and have mutations at both the G140 and Q148 positions of the IN β4α2 loop.
We did additional tests with four of the compounds we developed and show here that two, 4c and 4d, are more effective than DTG against the INSTI-resistant mutants we analyzed. We have data for 4c bound to the PFV intasome (19). These data helped us prepare models of 4d and 6p bound to the HIV-1 intasome (Fig. 5). The favorable antiviral profiles of these two compounds, taken together with the modeling data, provide information that can be used in the design of future INSTIs that have additional modifications of the pharmacophore. Comparing the binding of our compounds with DTG in the model is also helpful (Fig. 5). In particular, we are interested in generating and testing additional modifications at the 6′ position of the tricyclic core that would lead to additional interactions with IN. As we previously described (18, 19), modifications of the 6′ position can mimic the interactions that the host and/or viral DNA substrates have with IN (Fig. 5). Fortunately, for IN to be functional, the mutated active site must be able to interact appropriately with both host and viral DNAs. For this reason, we plan to optimize the interactions of moieties appended to the 6′ position with elements of IN that interact with the DNA substrates. These interactions should involve amino acid side chains where mutations would be deleterious to the ability of IN to bind its DNA substrates and catalyze the steps required for the integration reaction. Thus, modifications to the compounds that enhance their interactions with key residues of IN are likely to improve the abilities of the compounds to inhibit a broad range of resistant mutants. Based on this logic, we suggest that making similar modifications to other INSTIs that have different central pharmacophores could similarly broaden their efficacies against the known resistant mutants.
FIG 5.
Modeling 4d into the HIV-1 intasome using the available structural data. Using the structure of 6p bound to the PFV intasome (PDB ID, 5MMB) and the available HIV-1 IN structure (PDB ID, 5U1C), an HIV-1 IN model was constructed with 6p bound in the active site. 4d (green) was docked onto this 6p (orange) template to predict its binding and further superposed with the available DTG (magenta) structure (PDB ID, 3S3M). The Mg2+ ions (shown in maroon and labeled) interact with the chelating motifs of 6p, 4d, and DTG. The benzyl moiety of both 6p and 4d hydrophobically stacks with the penultimate cytosine (labeled dC and shaded in dark orange with line configurations) of the 3′ end of the viral DNA and align with DTG.
MATERIALS AND METHODS
INSTI synthesis.
DTG was obtained as previously described (16). The synthesis of 4c, 4d, 4f, 6b, and 6p has been reported (18, 19).
Cell-based assays.
WT and mutant HIV-based viral vectors were used in single-round infectivity assays to determine the antiviral activities (EC50s) of the compounds and the effects of the mutants on the EC50s, as previously described (34).
A modified version of the single-round infectivity assay was used to determine the replication capacities of the INSTI-resistant mutants. Briefly, 200 ng of a WT or INSTI-resistant mutant HIV-1-based vector was added to 96-well plates and incubated for 48 h, and luciferase activity was measured as was done previously (34). The luciferase activity of the WT virions was set to 100%, from which the infectivity of the mutant virions was measured as a percentage of the WT activity.
Vector constructs.
The vector pNLNgoMIVR-ΔENV.LUC has been described previously (18). To produce the new IN mutants used in this study, the IN open reading frame was removed from pNLNgoMIVR-ΔENV.LUC by digestion with KpnI and SalI, and the resulting fragment was inserted between the KpnI and SalI sites of pBluescript KS(+). Using that construct as the wild-type template, we prepared the following HIV-1 IN mutants using the QuikChange II XL site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) protocol: M50I, L74M, T97A, S119R, E138K, G140S, Q146L, Q146P, Q148H, Q148K, Q148R, S153Y, T66I/E157Q, E92Q/N155H, E138A/Q148R, E138K/Q148K, E138K/Q148R, E138K/R263K, G140A/Q148H, G140A/Q148K, G140A/Q148R, G140C/Q148R, G140S/Q148K, G140S/Q148R, G140S/N155H, Y143H/N155H, Y143R/Q148H, Y143R/N155H, Q148H/N155H, Q148R/N155H, N155H/G163R, T66I/T97A/E157Q, L74M/G140A/Q148R, L74M/G140C/Q148R, E92Q/N155H/G163R, T97A/G140S/Q148H, T97A/Y143R/Q148H, T97A/Y143R/N155H, T97A/Q148H/N155H, E138A/G140S/Q148H, E138A/S147G/Q148R, E138K/G140A/Q148K, E138K/G140C/Q148R, E138K/G140S/Q148H, G140S/Y143R/Q148H, G140S/Y143R/N155H, G140S/Q148H/N155H, and G140S/Q148H/G163K mutants. The following sense oligonucleotides were used with matching cognate antisense oligonucleotides (not shown) (Integrated DNA Technologies, Coralville, IA) in the mutagenesis: M50I, 5′-CAGCTAAAAGGGGAAGCCATTCATGGACAAGTAGACTGT-3′; T66I, 5′-ATATGGCAGCTAGATTGTATTCATTTAGAAGGAAAAGTT-3′; L74M, 5′-TTAGAAGGAAAAGTTATCATGGTAGCAGTTCATGTAGCC-3′; E92Q, 5′-GCAGAAGTAATTCCAGCACAAACAGGGCAAGAAACAGCA-3′; T97A, 5′-GCAGAGACAGGGCAAGAAGCTGCATACTTCCTCTTAAAA-3′; S119R, 5′-GTACATACAGACAATGGCCGTAATTTCACCAGTACTACA-3′; E138A, 5′-TGGGCGGGGATCAAGCAGGCTTTTGGCATTCCCTACAAT-3′; E138K, 5′-TGGGCGGGGATCAAGCAGAAATTTGGCATTCCCTACAAT-3′; G140A, 5′-GGGATCAAGCAGGAATTTGCTATTCCCTACAATCCCCAA-3′; G140C, 5′-GGGATCAAGCAGGAATTTTGTATTCCCTACAATCCCCAA-3′; G140S, 5′-GGGATCAAGCAGGAATTTTCCATTCCCTACAATCCCCAA-3′; Y143H, 5′-CAGGAATTTGGCATTCCCCATAATCCCCAAAGTCAAGGA-3′; Y143R, 5′-CAGGAATTTGGCATTCCCAGAAATCCCCAAAGTCAAGGA-3′; Q146L, 5′-GGCATTCCCTACAATCCCTTAAGTCAAGGAGTAATAGAA-3′; Q148H, 5′-TACAATCCCCAAAGTCACGGAGTAATAGAATCT-3′; Q148K, 5′-CCCTACAATCCCCAAAGTAAAGGAGTAATAGAATCTATG-3′; Q148R, 5′-CCCTACAATCCCCAAAGTCGTGGAGTAATAGAATCTATG-3′; S153Y, 5′-AGTCAAGGAGTAATAGAATATATGAATAAAGAATTAAAG-3′; N155H, 5′-GGAGTAATAGAATCTATGCATAAAGAATTAAAGAAAATT-3′; E157Q, 5′-ATAGAATCTATGAATAAACAATTAAAGAAAATTATAGGA-3′; G163K, 5′-GAATTAAAGAAAATTATAAAACAGGTAAGAGATCAGGCT-3′; G163R, 5′-GAATTAAAGAAAATTATACGTCAGGTAAGAGATCAGGCT-3′; E138K for G140S/Q148H, 5′-TGGTGGGCGGGGATCAAGCAGAAATTTTCCATTCCCTACAATCCC-3′; S147G for E138A/Q148R, 5′-ATTCCCTACAATCCCCAAGGTCGTGGAGTAATAGAATCT-3′; E138K for G140C/Q148R, 5′-TGGGCGGGGATCAAGCAGAAATTTTGTATTCCCTACAAT-3′; E138K for G140A/Q148K, 5′-TGGGCGGGGATCAAGCAGAAATTTGCTATTCCCTACAAT-3′; E138A for G140S/Q148H, 5′-TGGGCGGGGATCAAGCAGGCATTTTCCATTCCCTACAAT-3′; Y143R for Y143R/Q148H, 5′-CAGGAATTTGGCATTCCCAGAAATCCCCAAAGTCACGGA-3′; Y143R for G140S/Q148H, 5′-CAGGAATTTTCCATTCCCAGAAATCCCCAAAGTCACGGA-3′; Y143R for G140S/N155H, 5′-CAGGAATTTTCCATTCCCAGAAATCCCCAAAGTCAAGGA-3′.
The IN mutants shown in Fig. 2A (see Tables S1A and B in the supplemental material), which include M50I, L74M, T97A, S119R, E138K, G140S, Q146L, Q146P, Q148H, Q148K, Q148R, and S153Y mutants, were constructed as described above using the appropriate listed oligonucleotides.
The IN mutants shown in Fig. 2B (see Tables S3A and B in the supplemental material), were made as follows. The E138A/Q148R and E138K/Q148R double mutants were made using the previously generated Q148R mutant and the E138A and E138K oligonucleotides, respectively, to add the second mutation. The E138K/Q148K double mutant was constructed using the previously made E138K mutant and the appropriate Q148K oligonucleotides, which were used to add the second mutation. The G140A/Q148H and G140A/Q148K double mutants were made with the previously constructed G140A mutant and the appropriate oligonucleotides for the second mutation, either Q148H or Q148K, respectively. The G140A/Q148R and G140C/Q148R double mutants were made with the previously generated Q148R mutant and the oligonucleotides for the second mutation, either G140A or G140C, respectively. The G140S/Q148K and G140S/Q148R double mutants were made using the previously generated G140S mutant and appropriate oligonucleotides for the second mutation, either Q148K or Q148R, respectively. The Q148H/N155H and Q148R/N155H double mutants were made using the previously generated N155H mutant and appropriate oligonucleotides for the second mutation, either Q148H or Q148R, respectively. The Y143R/Q148H double mutant was made using the previously generated Q148H mutant and appropriate oligonucleotides to introduce the second mutation, Y143R. The T66I/E157Q double mutant was generated after the T66I mutant was made with the appropriate T66I oligonucleotides, which was used as the template to make the second mutation, E157Q, using the appropriate E157Q oligonucleotides.
The IN mutants shown in Fig. 2C (see Tables S4A and B in the supplemental material) were made as follows. The E92Q/N155H, G140S/N155H, Y143H/N155H, Y143R/N155H, and N155H/G163R double mutants were made using the previously generated N155H mutant and appropriate oligonucleotides for the second mutation, either E92Q, G140S, Y143H, Y143R, or G163R, respectively.
The IN mutants shown in Fig. 3A (see Tables S5A and B in the supplemental material) were constructed as follows. The L74M/G140A/Q148R triple mutant was made using the previously generated G140A/Q148R double mutant and the oligonucleotides for the third mutation, L74M. The L74M/G140C/Q148R triple mutant was made with the previously generated G140C/Q148R double mutant and the oligonucleotides for the third mutation, L74M. The E138K/G140C/Q148R triple mutant was made using the previously generated G140C/Q148R double mutant and the appropriate oligonucleotides to create the third mutation, E138K. The E138A/S147G/Q148R triple mutant was made with the previously generated E138A/Q148R double mutant and oligonucleotides to make the third mutation, S147G. The E138K/G140A/Q148K triple mutant was made using the previously constructed G140A/Q148K double mutant and the appropriate oligonucleotides to make the third mutation, E138K.
The IN mutants shown in Fig. 3B (see Tables S6A and B in the supplemental material), were constructed as follows. The T97A/G140S/Q148H, G140S/Q148H/N155H, and G140S/Q148H/G163K triple mutants were each made with the previously generated G140S/Q148H double mutant and the appropriate oligonucleotides for the third mutation, either T97A, N155H, or G163K, respectively. The E138A/G140S/Q148H triple mutant was made using the previously constructed G140S/Q148H double mutant and oligonucleotides to make the third mutation, E138A. The E138K/G140S/Q148H triple mutant was made using the previously generated G140S/Q148H double mutant and the correct oligonucleotides to make the third mutation, E138K. The G140S/Y143R/Q148H triple mutant was made using the previously constructed G140S/Q148H double mutant and the appropriate oligonucleotides to make the third mutation, Y143R.
The IN mutants shown in Fig. 3C (see Tables S7A and B in the supplemental material) were made as follows. The T66I/T97A/E157Q triple mutant was made using the previously generated T66I/E157Q double mutant and the oligonucleotides for the third mutation, T97A. The E92Q/N155H/G163R triple mutant was made using the previously generated E92Q/N155H double mutant and the oligonucleotides for the third mutation, G163R. The G140S/Y143R/N155H triple mutant was made using the previously constructed G140S/N155H double mutant and the correct oligonucleotides to create the third mutation, Y143R. The T97A/Y143R/N155H triple mutant was made with the previously generated Y143R/N155H double mutant and the appropriate oligonucleotides for the third mutation, T97A. The T97A/Y143R/Q148H triple mutant was constructed using the previously generated Y143R/Q148H double mutant and the appropriate oligonucleotides for the third mutation, T97A. The T97A/Q148H/N155H triple mutant was made using the previously constructed Q148H/N155H double mutant and the appropriate oligonucleotides for the third mutation, T97A.
The DNA sequence of each construct was verified independently by DNA sequence determination. The mutated IN coding sequences from pBluescript KS(+) were then subcloned into pNLNgoMIVR-ΔEnv.LUC (between the KpnI and SalI sites) to produce mutant HIV-1 constructs, which were also checked by DNA sequencing.
Computer modeling.
All modeling was conducted using MOE 2016.0802 (Chemical Computing Group, Montreal, Quebec, Canada). The sequences and structures of 6p bound in the PFV intasome (PDB ID, 5MMB) and HIV-1 IN (PDB ID, 5U1C) served as the structural templates to construct an HIV-1 IN model with 6p bound in the active site (18, 35). First, portions of the N-terminal domain (NTD), CCD, and C-terminal domain (CTD) of PFV and HIV-1 INs were constrained so that the domains were aligned properly. Next the sequences and structures of HIV-1 and PFV INs were aligned, and the HIV IN sequence was then added and matched to superpose the HIV-1 and PFV IN alignments. The coordinates of the HIV-1 IN structure (PDB ID, 5U1C) from the above-mentioned alignment were used as the IN template to construct the HIV-1 IN model. This structure was modified to fit the structural coordinates of 6p, Mg2+ cofactors, and the viral DNA, which were taken from the PFV intasome (PDB ID, 5MMB). The model of the HIV-1 intasome with 6p bound was energy minimized using a PFROSST force field with relative field solvation as recommended by the manufacturer (Chemical Computing Group). The HIV-1 IN model was aligned with the HIV-1 IN structure (PDB ID, 5U1C) from the above-mentioned alignment with the PFV structure (PDB ID, 5MMB) for root mean square deviation (RMSD) identification (0.83 Å), and the surface interaction (Van der Waals) of 6p was determined to locate possible steric clashes among the active-site residues in the model. To identify the potential contacts with 4d, docking was performed, using 6p as the template. 4d was placed using the triangle matcher method and scored with London dG (scoring function) with approximately 30 poses, and then the putative ligand poses were further refined using the rigid-receptor method in MOE and scored with the GBVI/WSA dG function. If the expected ligand poses were not created, a pharmacophore editor tool in the docking function was used to add certain features that made the appropriate docking of 4d to 6p easier, and the resulting structures were refined in the manner described above. The poses with the best docking scores were selected based on how well the bound compounds overlaid the 6p scaffold, bound to Mg2+, and how well their halogenated benzyl moieties interacted hydrophobically through π-π stacking with the penultimate cytosine on the 3′ end of the bound viral DNA.
Supplementary Material
ACKNOWLEDGMENTS
We thank Teresa Burdette for help in preparing the manuscript, Alan Kane for help with the figures, and Brian Luke for help with the statistical analysis.
This research was supported by the Intramural Research Programs of the National Cancer Institute and the Intramural AIDS Targeted Antiviral Program.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01035-18.
REFERENCES
- 1.Bushman FD, Craigie R. 1991. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc Natl Acad Sci U S A 88:1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engelman A, Mizuuchi K, Craigie R. 1991. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell 67:1211–1221. doi: 10.1016/0092-8674(91)90297-C. [DOI] [PubMed] [Google Scholar]
- 3.Hazuda DJ, Felock P, Witmer M, Wolfe A, Stillmock K, Grobler JA, Espeseth A, Gabryelski L, Schleif W, Blau C, Miller MD. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 4.Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. 2010. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464:232–236. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malet I, Delelis O, Valantin MA, Montes B, Soulie C, Wirden M, Tchertanov L, Peytavin G, Reynes J, Mouscadet JF, Katlama C, Calvez V, Marcelin AG. 2008. Mutations associated with failure of raltegravir treatment affect integrase sensitivity to the inhibitor in vitro. Antimicrob Agents Chemother 52:1351–1358. doi: 10.1128/AAC.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fransen S, Gupta S, Danovich R, Hazuda D, Miller M, Witmer M, Petropoulos CJ, Huang W. 2009. Loss of raltegravir susceptibility by human immunodeficiency virus type 1 is conferred via multiple nonoverlapping genetic pathways. J Virol 83:11440–11446. doi: 10.1128/JVI.01168-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goethals O, Clayton R, Van Ginderen M, Vereycken I, Wagemans E, Geluykens P, Dockx K, Strijbos R, Smits V, Vos A, Meersseman G, Jochmans D, Vermeire K, Schols D, Hallenberger S, Hertogs K. 2008. Resistance mutations in human immunodeficiency virus type 1 integrase selected with elvitegravir confer reduced susceptibility to a wide range of integrase inhibitors. J Virol 82:10366–10374. doi: 10.1128/JVI.00470-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimura K, Kodama E, Sakagami Y, Matsuzaki Y, Watanabe W, Yamataka K, Watanabe Y, Ohata Y, Doi S, Sato M, Kano M, Ikeda S, Matsuoka M. 2008. Broad antiretroviral activity and resistance profile of the novel human immunodeficiency virus integrase inhibitor elvitegravir (JTK-303/GS-9137). J Virol 82:764–774. doi: 10.1128/JVI.01534-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Margot NA, Hluhanich RM, Jones GS, Andreatta KN, Tsiang M, McColl DJ, White KL, Miller MD. 2012. In vitro resistance selections using elvitegravir, raltegravir, and two metabolites of elvitegravir M1 and M4. Antiviral Res 93:288–296. doi: 10.1016/j.antiviral.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 10.Metifiot M, Marchand C, Maddali K, Pommier Y. 2010. Resistance to integrase inhibitors. Viruses 2:1347–1366. doi: 10.3390/v2071347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lesbats P, Engelman AN, Cherepanov P. 2016. Retroviral DNA integration. Chem Rev 116:12730–12757. doi: 10.1021/acs.chemrev.6b00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mesplede T, Wainberg MA. 2013. Integrase strand transfer inhibitors in HIV therapy. Infect Dis Ther 2:83–93. doi: 10.1007/s40121-013-0020-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quashie PK, Mesplede T, Wainberg MA. 2013. HIV drug resistance and the advent of integrase inhibitors. Curr Infect Dis Rep 15:85–100. doi: 10.1007/s11908-012-0305-1. [DOI] [PubMed] [Google Scholar]
- 14.Hare S, Vos AM, Clayton RF, Thuring JW, Cummings MD, Cherepanov P. 2010. Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance. Proc Natl Acad Sci U S A 107:20057–20062. doi: 10.1073/pnas.1010246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johns BA, Kawasuji T, Weatherhead JG, Taishi T, Temelkoff DP, Yoshida H, Akiyama T, Taoda Y, Murai H, Kiyama R, Fuji M, Tanimoto N, Jeffrey J, Foster SA, Yoshinaga T, Seki T, Kobayashi M, Sato A, Johnson MN, Garvey EP, Fujiwara T. 2013. Carbamoyl pyridone HIV-1 integrase inhibitors 3. A diastereomeric approach to chiral nonracemic tricyclic ring systems and the discovery of dolutegravir (S/GSK1349572) and (S/GSK1265744). J Med Chem 56:5901–5916. doi: 10.1021/jm400645w. [DOI] [PubMed] [Google Scholar]
- 16.Hare S, Smith SJ, Metifiot M, Jaxa-Chamiec A, Pommier Y, Hughes SH, Cherepanov P. 2011. Structural and functional analyses of the second-generation integrase strand transfer inhibitor dolutegravir (S/GSK1349572). Mol Pharmacol 80:565–572. doi: 10.1124/mol.111.073189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maertens GN, Hare S, Cherepanov P. 2010. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature 468:326–329. doi: 10.1038/nature09517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao XZ, Smith SJ, Maskell DP, Metifiot M, Pye VE, Fesen K, Marchand C, Pommier Y, Cherepanov P, Hughes SH, Burke TR Jr. 2017. Structure-guided optimization of HIV integrase strand transfer inhibitors. J Med Chem 60:7315–7332. doi: 10.1021/acs.jmedchem.7b00596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao XZ, Smith SJ, Maskell DP, Metifiot M, Pye VE, Fesen K, Marchand C, Pommier Y, Cherepanov P, Hughes SH, Burke TR Jr. 2016. HIV-1 integrase strand transfer inhibitors with reduced susceptibility to drug resistant mutant integrases. ACS Chem Biol 11:1074–1081. doi: 10.1021/acschembio.5b00948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cahn P, Pozniak AL, Mingrone H, Shuldyakov A, Brites C, Andrade-Villanueva JF, Richmond G, Buendia CB, Fourie J, Ramgopal M, Hagins D, Felizarta F, Madruga J, Reuter T, Newman T, Small CB, Lombaard J, Grinsztejn B, Dorey D, Underwood M, Griffith S, Min S, extended SAILING Study Team. 2013. Dolutegravir versus raltegravir in antiretroviral-experienced, integrase-inhibitor-naive adults with HIV: week 48 results from the randomised, double-blind, non-inferiority SAILING study. Lancet 382:700–708. doi: 10.1016/S0140-6736(13)61221-0. [DOI] [PubMed] [Google Scholar]
- 21.Min S, Sloan L, DeJesus E, Hawkins T, McCurdy L, Song I, Stroder R, Chen S, Underwood M, Fujiwara T, Piscitelli S, Lalezari J. 2011. Antiviral activity, safety, and pharmacokinetics/pharmacodynamics of dolutegravir as 10-day monotherapy in HIV-1-infected adults. AIDS 25:1737–1745. doi: 10.1097/QAD.0b013e32834a1dd9. [DOI] [PubMed] [Google Scholar]
- 22.Raffi F, Jaeger H, Quiros-Roldan E, Albrecht H, Belonosova E, Gatell JM, Baril JG, Domingo P, Brennan C, Almond S, Min S, extended SAILING Study Team. 2013. Once-daily dolutegravir versus twice-daily raltegravir in antiretroviral-naive adults with HIV-1 infection (SPRING-2 study): 96 week results from a randomised, double-blind, non-inferiority trial. Lancet Infect Dis 13:927–935. doi: 10.1016/S1473-3099(13)70257-3. [DOI] [PubMed] [Google Scholar]
- 23.Raffi F, Rachlis A, Stellbrink HJ, Hardy WD, Torti C, Orkin C, Bloch M, Podzamczer D, Pokrovsky V, Pulido F, Almond S, Margolis D, Brennan C, Min S, SPRING-2 Study Group. 2013. Once-daily dolutegravir versus raltegravir in antiretroviral-naive adults with HIV-1 infection: 48 week results from the randomised, double-blind, non-inferiority SPRING-2 study. Lancet 381:735–743. doi: 10.1016/S0140-6736(12)61853-4. [DOI] [PubMed] [Google Scholar]
- 24.van Lunzen J, Maggiolo F, Arribas JR, Rakhmanova A, Yeni P, Young B, Rockstroh JK, Almond S, Song I, Brothers C, Min S. 2012. Once daily dolutegravir (S/GSK1349572) in combination therapy in antiretroviral-naive adults with HIV: planned interim 48 week results from SPRING-1, a dose-ranging, randomised, phase 2b trial. Lancet Infect Dis 12:111–118. doi: 10.1016/S1473-3099(11)70290-0. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi M, Yoshinaga T, Seki T, Wakasa-Morimoto C, Brown KW, Ferris R, Foster SA, Hazen RJ, Miki S, Suyama-Kagitani A, Kawauchi-Miki S, Taishi T, Kawasuji T, Johns BA, Underwood MR, Garvey EP, Sato A, Fujiwara T. 2011. In vitro antiretroviral properties of S/GSK1349572, a next-generation HIV integrase inhibitor. Antimicrob Agents Chemother 55:813–821. doi: 10.1128/AAC.01209-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quashie PK, Mesplede T, Han YS, Oliveira M, Singhroy DN, Fujiwara T, Underwood MR, Wainberg MA. 2012. Characterization of the R263K mutation in HIV-1 integrase that confers low-level resistance to the second-generation integrase strand transfer inhibitor dolutegravir. J Virol 86:2696–2705. doi: 10.1128/JVI.06591-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quashie PK, Mesplede T, Han YS, Veres T, Osman N, Hassounah S, Sloan RD, Xu HT, Wainberg MA. 2013. Biochemical analysis of the role of G118R-linked dolutegravir drug resistance substitutions in HIV-1 integrase. Antimicrob Agents Chemother 57:6223–6235. doi: 10.1128/AAC.01835-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castagna A, Maggiolo F, Penco G, Wright D, Mills A, Grossberg R, Molina JM, Chas J, Durant J, Moreno S, Doroana M, Ait-Khaled M, Huang J, Min S, Song I, Vavro C, Nichols G, Yeo JM, VIKING-3 Study Group. 2014. Dolutegravir in antiretroviral-experienced patients with raltegravir- and/or elvitegravir-resistant HIV-1: 24-week results of the phase III VIKING-3 study. J Infect Dis 210:354–362. doi: 10.1093/infdis/jiu051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eron JJ, Clotet B, Durant J, Katlama C, Kumar P, Lazzarin A, Poizot-Martin I, Richmond G, Soriano V, Ait-Khaled M, Fujiwara T, Huang J, Min S, Vavro C, Yeo J, VIKING-3 Study Group. 2013. Safety and efficacy of dolutegravir in treatment-experienced subjects with raltegravir-resistant HIV type 1 infection: 24-week results of the VIKING Study. J Infect Dis 207:740–748. doi: 10.1093/infdis/jis750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Markowitz M, Morales-Ramirez JO, Nguyen BY, Kovacs CM, Steigbigel RT, Cooper DA, Liporace R, Schwartz R, Isaacs R, Gilde LR, Wenning L, Zhao J, Teppler H. 2006. Antiretroviral activity, pharmacokinetics, and tolerability of MK-0518, a novel inhibitor of HIV-1 integrase, dosed as monotherapy for 10 days in treatment-naive HIV-1-infected individuals. J Acquir Immune Defic Syndr 43:509–515. doi: 10.1097/QAI.0b013e31802b4956. [DOI] [PubMed] [Google Scholar]
- 31.Johnson VA, Calvez V, Gunthard HF, Paredes R, Pillay D, Shafer RW, Wensing AM, Richman DD. 2013. Update of the drug resistance mutations in HIV-1: March 2013. Top Antivir Med 21:6–14. [PMC free article] [PubMed] [Google Scholar]
- 32.Smith SJ, Zhao XZ, Burke TR Jr, Hughes SH. 2018. Efficacies of Cabotegravir and Bictegravir against drug-resistant HIV-1 integrase mutants. Retrovirology 15:37. doi: 10.1186/s12977-018-0420-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zash R, Jacobson D, Mayondi G, Diseko M, Makhema J, Mmalane M, Gaolathe T, Petlo C, Holmes L, Essex M, Lockman S, Shapiro R. 2017. Dolutegravir/tenofovir/emtricitabine (DTG/TDF/FTC) started in pregnancy is as safe as efavirenz/tenofovir/emtricitabine (EFV/TDF/FTC) in nationwide birth outcomes surveillance in Botswana, abstr MOAX0202LB. 9th IAS Conf HIV Sci, 23 to 26 July 2017, Paris, France https://onlinelibrary.wiley.com/doi/full/10.7448/IAS.20.6.22253. [Google Scholar]
- 34.Smith SJ, Hughes SH. 2014. Rapid screening of HIV reverse transcriptase and integrase inhibitors. J Vis Exp doi: 10.3791/51400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Passos DO, Li M, Yang R, Rebensburg SV, Ghirlando R, Jeon Y, Shkriabai N, Kvaratskhelia M, Craigie R, Lyumkis D. 2017. Cryo-EM structures and atomic model of the HIV-1 strand transfer complex intasome. Science 355:89–92. doi: 10.1126/science.aah5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.