Zika virus (ZIKV) infection is associated with serious, long-term neurological manifestations. There are currently no approved therapies for the treatment or prevention of ZIKV infection.
KEYWORDS: mathematical modeling, Zika virus, antiviral therapy, favipiravir, hollow-fiber infection model, pharmacodynamics, pharmacokinetics
ABSTRACT
Zika virus (ZIKV) infection is associated with serious, long-term neurological manifestations. There are currently no approved therapies for the treatment or prevention of ZIKV infection. Favipiravir (FAV) is a viral polymerase inhibitor with broad-spectrum activity. Our prior studies used static FAV concentrations and demonstrated promising activity. However, the anti-ZIKV activity of dynamic FAV concentrations has never been evaluated in a human cell line. Here we employed the hollow-fiber infection model (HFIM) to simulate the human pharmacokinetic (PK) profiles associated with the clinically utilized FAV dosage regimens against influenza and Ebola viruses and assessed the viral burden profiles. Clinically achievable FAV concentrations inhibited ZIKV replication in HUH-7 cells in a dose-dependent fashion (50% effective concentration = 236.5 μM). The viral burden profiles under dynamic FAV concentrations were predicted by use of a mechanism-based mathematical model (MBM) and subsequently successfully validated in the HFIM. This validated, translational MBM can now be used to predict the anti-ZIKV activity of other FAV dosage regimens in the presence of between-patient variability in pharmacokinetics. This approach can be extended to rationally optimize FAV combination dosage regimens which hold promise to treat ZIKV infections in nonpregnant patients.
INTRODUCTION
Zika virus (ZIKV), a single-stranded RNA virus belonging to the genus Flavivirus (Flaviviridae family), has become a significant threat to public health due to the widespread 2015 epidemic throughout the Western Hemisphere. Viral transmission most commonly occurs through the bite of an infected mosquito, but spread via sexual contact as well as vertical (i.e., mother-to-fetus) transmission has also been widely reported (1–3). ZIKV disease is usually mild and self-limiting (4); however, severe neurological consequences, such as congenital disabilities and Guillain-Barré syndrome, are associated with infection (5–7). Currently, neither vaccines nor antiviral drugs are available to help treat or prevent ZIKV infections, and the development of new medical countermeasures will take years until approval for human use. Due to the severity and chronic nature of neurological ZIKV disease, there is an urgent medical need to identify effective antiviral therapies against viral infection.
Drug repurposing using agents that are already approved by the FDA or other regulatory agencies for different indications is one strategy to expedite drug development timelines and deliver a much needed therapy to patients faster. Favipiravir (FAV) is a nucleoside precursor RNA polymerase inhibitor that exhibits antiviral activity against a wide variety of RNA viruses (8–10). FAV is approved for the treatment of human influenza virus infections in Japan and is currently in phase III clinical trials for the treatment of uncomplicated influenza in the United States. Moreover, it was used with some success to treat Ebola virus-infected patients during the 2014-2015 Ebola outbreak in West Africa (11). This broad-spectrum activity makes FAV an attractive candidate for repurposing for the treatment of ZIKV disease.
Our previous work showed that FAV extensively inhibits the production of infectious ZIKV (12). We developed a translational mechanism-based mathematical model (MBM) which predicted that drug concentration-time profiles associated with clinically relevant FAV regimens are effective at suppressing ZIKV replication (12). This work employed Vero cells (i.e., African green monkey kidney cells) as the host cell line. While Vero cells are suitable for initial drug evaluations, they may differ from human cell lines due to potential differences in FAV cellular uptake and metabolism.
In this study, we aimed to characterize the antiviral activity of FAV on host cells of human origin using traditional tissue culture assays and the pharmacodynamic hollow-fiber infection model (HFIM) system, which can simulate clinically relevant drug and viral concentration profiles. As a second objective, we refined our translational MBM to characterize the time course of the ZIKV burden in the face of FAV therapy in both experimental assay formats. The refined MBM was then used to predict the virologic outcome when the plasma concentration-time profiles associated with clinically utilized FAV regimens were simulated in vitro. Our findings demonstrate the clinical potential of FAV as a therapeutic strategy for the treatment of patients infected with ZIKV.
RESULTS
Antiviral activity of static FAV concentrations against ZIKV.
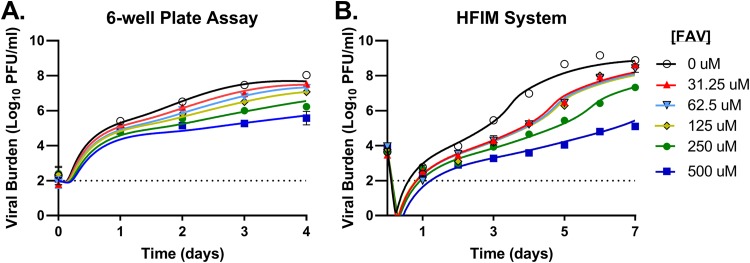
The antiviral activity of FAV was evaluated on HUH-7 cells (HUH-7 is a human hepatoma cell line) against ZIKV using a traditional tissue culture drug assay (conducted in 6-well plates). ZIKV production was robust in HUH-7 cells, reaching titers of 8 log10 PFU/ml on day 4 postinfection in the control arm (Fig. 1A). FAV inhibited ZIKV replication in a dose-dependent manner, with concentrations of 250 μM and 500 μM suppressing the viral burden by 1.8 log10 PFU/ml and 2.5 log10 PFU/ml, respectively (Fig. 1A). Interestingly, concentrations ranging from 31.25 μM to 125 μM provided similar levels of inhibition and decreased the viral titers by approximately 0.7 log10 PFU/ml. The concentration yielding a half-maximal effect (the 50% effective concentration [EC50]) in this system over 4 days was 251.3 μM, as determined by a Hill-type effect model. Cytotoxicity was not observed at any FAV concentration evaluated in these studies (data not shown).
FIG 1.
Antiviral activity of favipiravir (FAV) against Zika virus (ZIKV) on HUH-7 cells in a 6-well plate assay and the hollow-fiber infection model (HFIM) system. HUH-7 cells were infected with ZIKV and exposed to increasing concentrations of FAV for 4 days in a 6-well plate assay (A) or 7 days in the HFIM system (B). Cell culture supernatant was collected daily, and the infectious virus burden, reported as the log10 number of PFU per milliliter, was quantified by plaque assay on Vero cells. The symbols correspond to the mean for three independent samples for the 6-well plate assay and two samples for the HFIM system. Lines through the data points signify the predicted viral burden determined by the mathematical model.
The effectiveness of FAV at static drug concentrations against ZIKV was also assessed in HUH-7 cells in the hollow-fiber infection model (HFIM) system. For these studies, 108 HUH-7 cells were mixed with 105 PFU of ZIKV before inoculation into hollow-fiber cartridges; FAV was administered via continuous infusion, which yielded a constant drug concentration. ZIKV replicated efficiently in the HFIM, as peak viral titers of 9.2 log10 PFU/ml were achieved on day 6 postinoculation in the control arm (Fig. 1B). FAV concentrations ranging from 31.25 μM to 125 μM yielded a nearly identical extent of viral suppression (∼1.2 log10 PFU/ml on day 6). FAV at 250 μM and 500 μM provided greater viral inhibition in a dose-dependent manner, resulting in a viral burden reduction of 2.8 log10 PFU/ml and 4.4 log10 PFU/ml, respectively (Fig. 1B). The EC50 of FAV against ZIKV over 7 days of therapy in the HFIM was 236.5 μM, in good agreement with the results of the well plate assay.
Mechanism-based PD modeling.
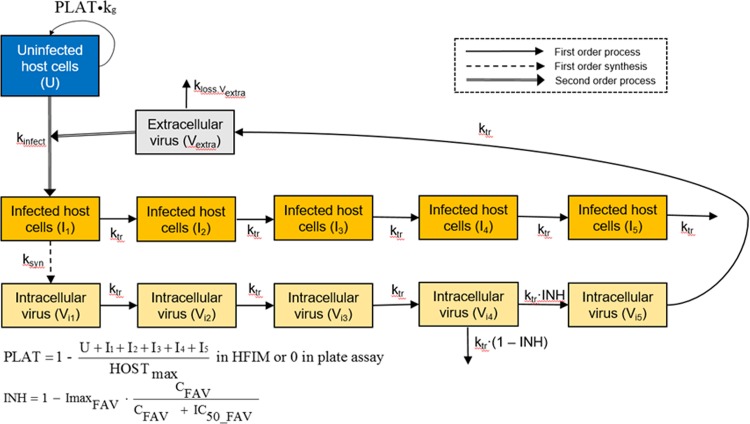
Previously, we developed a pharmacodynamic (PD) mechanism-based mathematical model (MBM) to describe the inhibitory effect of FAV on the production of infectious ZIKV in a 6-well-plate drug assay using Vero cells (12). We have now adapted this model to HUH-7 cells, which were studied under static FAV concentrations in a plate assay and in the HFIM. We extended our previous MBM to reflect the experimental infection and inoculation processes in the HFIM. First, we accounted for the loading of extracellular virus in the HFIM and thereby allowed the model to simultaneously describe both experimental systems (Fig. 2). For the plate assay, virus was allowed to first attach to uninfected host cells for 1 h before all unbound virus was removed. Therefore, at time zero, only limited cell-free virus (∼2 log10 PFU/ml) remained in the medium. In contrast, virus and uninfected host cells were inoculated into the HFIM at time zero; consequently, the viral burden was detectable in the medium and infected host cells did not yet exist at this time point. The initial uninfected host cell density was 106.82 cells/ml in the HFIM and 105.82 cells/ml in the plate assay.
FIG 2.
Mechanism-based model for the activity of FAV against ZIKV-infected cells. The model describes the inhibitory effect of FAV (INH) on viral maturation in the host cells. See Table 1 for parameter explanations.
As a second modification of our previous MBM (12), we introduced a replication term for uninfected host cells in the HFIM. This dynamic in vitro model provides more nutrients and thus can sustain higher host cell densities than a static plate assay. In our refined MBM, we included five stages of infected host cells in parallel to the stages of intracellular viral maturation; the last (i.e., 5th) stage represents infected host cells which die due to release of the virus. Both infected and uninfected host cells were assumed to require nutrients and to thus contribute to the maximum sustainable host cell population. We estimated different maximum cell densities for the HFIM and plate assays.
Our refined MBM successfully described the viral dynamics data from the plate and HFIM assays using a consistent model structure. For both assays, the maximum extent of inhibition for ZIKV production was estimated to be 0.9998 for FAV, indicating nearly complete viral suppression (Table 1). The FAV concentration yielding half-maximal inhibition (50% inhibitory concentration [IC50]) of virus release from the last intracellular compartment was estimated to be 61.6 μM for both models. This estimate reflects the intracellular inhibition of ZIKV replication and was therefore smaller than the macroscopic inhibition of viral titers over 7 days (EC50). Most model parameter estimates were shared (i.e., they had identical estimates) or had similar estimates between the two experimental systems (Table 1); the synthesis rate constant for viral production was 39.9 h−1 in the plate assay and 39.7 h−1 in the HFIM. Larger differences between the two systems were estimated for the mean survival time of host cells (94.0 h for HFIM versus 32.4 h for the plate assay) and the mean survival time of extracellular virus (10.6 h for HFIM versus 15.7 h for the plate assay). Both of the latter estimates agree with previous literature reports on ZIKV physiology (13).
TABLE 1.
Parameter estimates for the PD MBM of FAV against ZIKV in plaque and HFIM assays
| PD parameter | Symbol (units) | Population mean (% SE)a |
|
|---|---|---|---|
| Plate assay | HFIM system | ||
| Log10 of 2nd-order infection rate constant | kinfect (log10) | −7.36 (0.985) | −7.03 (1.92) |
| Synthesis rate constant of virus | ksyn (1/h) | 39.9 (2.89) | 39.7 (31.4) |
| Mean delay time until release of virus in the absence of drug, equivalent to the mean survival time of infected cells | Tdelay = 5/ktr (h) | 32.4 (5.03) | 94.0 (18.8) |
| Mean survival time for extracellular virus | MSTvirus = 1/kloss,virus (h) | 15.7 (3.57) | 10.6 (27.9) |
| Log10 of initial no. of uninfected cells | log_U | 5.82 (fixed) | 6.82 (fixed) |
| Log10 of initial no. of infected cells | log_I | 3.61 (1.67) | 0c (fixed) |
| Log10 of plateau of host cells | log_max | NAd | 7.39 (0.849) |
| Mean doubling time for host cell replication | Trepl (h) | 24 (fixed) | 24 (fixed) |
| Initial virus loading | Virus_Load | 0 (fixed) | 6670 (fixed) |
| Maximum extent of inhibition by FAV | ImaxFAV (normal scale) | 0.9998 (0.983 to 1.00)b | 0.9998 (0.983 to 1.00)b |
| FAV concn causing 50% of ImaxFAV | IC50_FAV (μM) | 61.6 (18.1) | 61.6 (18.1) |
| Additive error for viral load on log10 scale | SDin | 0.286 (5.09) | 0.286 (5.09) |
The between-curve variability for in vitro assays was assumed to be small, and therefore, the variance was eventually fixed at 0.01.
Imax was assumed to be normally distributed on the logistically transformed scale (Imaxtransformed). The population mean of Imax was reported on a normal scale (i.e., from 0 to 1).
No infected cells, i.e., 0 cells on a linear scale.
NA, not applicable.
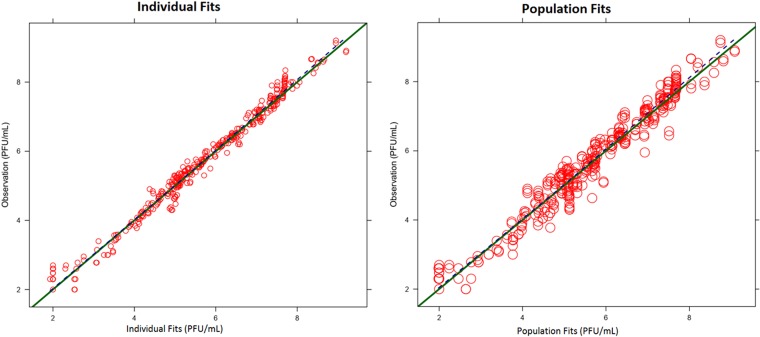
The proposed model simultaneously described virus burden-time profiles in the plate assay and HFIM system (Fig. 1). Curve fits were unbiased and reasonably precise, with linear regression analysis of fitted-versus-observed plots for viral burden yielding correlation coefficients (r) of 0.981 for the individual counts and 0.970 for population fitted-versus-observed counts (Fig. 3). The model's predictive performance was assessed by the use of normalized prediction distribution errors (14) and showed adequate predictive performance (results not shown).
FIG 3.
Predicted-versus-observed plots for ZIKV burden. The individual (Bayesian; left) and population (pre-Bayesian; right) fitted viral burdens for FAV against ZIKV are shown.
Model simulations and prospective validation of FAV activity against ZIKV in the HFIM.
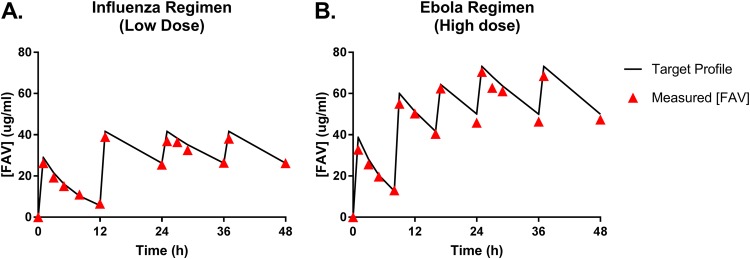
We then performed deterministic simulations (i.e., simulations of the mean profiles) based on the refined MBM to predict the viral burden profiles following clinically relevant FAV dosage regimens. Free-drug concentration-time profiles (Fig. 4) were simulated for the standard FAV influenza regimen (low dose, 1,800 mg at 0 h and 12 h on day 1 of dosing, followed by 800 mg every 12 h starting at day 2) and the Ebola regimen (high dose, 2,400 mg at 0 h and 8 h and 1,800 mg at 16 h on day 1 of dosing, followed by 1,200 mg every 12 h starting on day 2). The viral burden profiles associated with each FAV regimen are depicted by the lines in Fig. 5. We then experimentally validated these MBM predictions in the HFIM and quantified the virus burden and drug concentrations in the medium over 7 days. The observed and simulated FAV concentration pharmacokinetic (PK) profiles in the HFIM system matched reasonably (Fig. 4).
FIG 4.
Favipiravir (FAV) concentrations in the hollow-fiber infection model (HFIM) system for the low-dose influenza regimen (A) and the high-dose Ebola regimen (B). The lines correspond to the target pharmacokinetic profiles, and the triangles represent the measured FAV concentrations from medium samples harvested from the HFIM system. FAV was measured using LC-MS/MS.
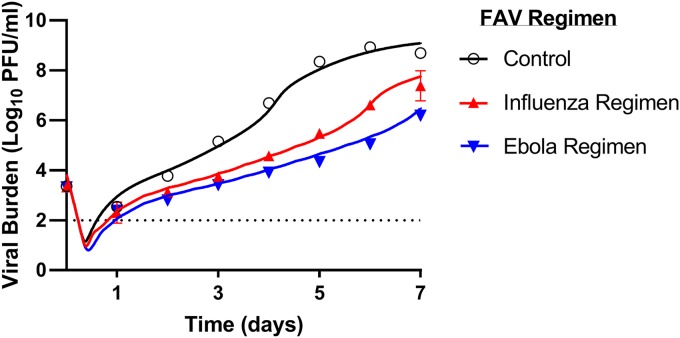
FIG 5.
Prospective validation showing predicted and measured ZIKV burden profiles during FAV treatment with clinically relevant regimens in the hollow-fiber infection model (HFIM) system. HUH-7 cells were mixed with ZIKV at a multiplicity of infection of 10−3 PFU/cell and inoculated into the HFIM system. FAV was administered via syringe pumps to simulate the free-drug concentration-time profiles associated with the low-dose influenza regimen or the high-dose Ebola regimen for 7 days. Viral supernatants were collected daily, and the infectious ZIKV burden, reported as the log10 number of PFU per milliliter, was quantified by plaque assay on Vero cells. Symbols correspond to measured mean viral titers from two samples in the HFIM system. Error bars represent 1 standard deviation. Error bars were generally smaller than the symbol size and are thus not visible. The lines signify the viral burden predicted by the mathematical model.
Both FAV regimens inhibited the viral burden over 7 days. The degree of inhibition was similar between the two regimens for the first 3 days (Fig. 5). From days 4 to 7, viral replication was faster in the low-dose regimen than in the high-dose regimen. The maximum extent of inhibition achieved in both regimens was 2.9 log10 PFU/ml for the low-dose regimen (day 5) and 4.0 log10 PFU/ml for the high-dose regimen (day 5). The MBM-simulated viral burden profiles excellently matched the observed profiles (Fig. 5) for the growth control, the low-dose (influenza), and the high-dose (Ebola) FAV regimens; the r value for the observed-versus-predicted viral burden was 0.992. Overall, this study prospectively validated the MBM predictions for viral burden in the HFIM under dynamic, human-like drug concentrations.
DISCUSSION
There is an urgent need for the development of antiviral agents against ZIKV infections that are effective at clinically achievable concentrations. A drug repurposing approach offers significant advantages for the identification of new antiviral strategies over the development of new drugs, since the safety and pharmacology in humans are already well-known for repurposed agents. Therefore, if a current clinically utilized drug demonstrates considerable activity against ZIKV, there is great potential to expedite the approval process for this agent to be used for the treatment of ZIKV infections, thus bringing a much needed treatment to patients faster. In our previous work, we identified FAV to be an agent that has promising antiviral activity against ZIKV alone and in combination with alpha interferon when Vero cells were employed as the host cell line (12). Since the kinetics of FAV metabolic activation may differ between nonhuman primate and human cell lines, we evaluated the antiviral activity of FAV in a human cell line in the present study. Furthermore, we assessed the potential of clinical FAV regimens to inhibit ZIKV replication using our MBM and the HFIM system.
Clinical trials with FAV during the Ebola outbreak have shown that average concentrations of 390 μM are achievable in humans (15). This drug exposure is nearly twice the exposure associated with the standard influenza regimen, which has an average concentration of 206 μM (16). The safety of this high-dose Ebola regimen is unknown, as it was impossible to discern between drug-related toxicities and Ebola disease. Nevertheless, our initial drug susceptibility assays using two different experimental systems with FAV on the human HUH-7 cell line revealed EC50s that were considerably lower than the maximum examined concentration of 390 μM (plate assay EC50 = 251.3 μM, HFIM assay EC50 = 236.5 μM). The refined MBM estimated an IC50 of 63.6 μM, referring to the inhibition of the release of infectious cell-free ZIKV from the last intracellular virus compartment (Fig. 2). Taken together, FAV markedly inhibited the production of infectious ZIKV in a human cell line at physiologically achievable concentrations, thus highlighting the clinical promise of FAV for the treatment of ZIKV infections.
The drug effect assays, regardless of assay format, were conducted under conditions in which FAV concentrations were kept constant. This experimental situation does not accurately reflect the drug concentration-time (i.e., pharmacokinetic) profiles in humans after administration. In humans, drug concentrations constantly fluctuate due to dosing, absorption, distribution, metabolism, and other elimination routes. Human pharmacokinetic profiles for FAV associated with the standard influenza regimen and the higher-dose Ebola regimen have been published (15–17). We simulated the human pharmacokinetic profiles associated with influenza and Ebola FAV regimens to predict the virologic outcome against ZIKV in humans over time (Fig. 4). These simulations revealed that both FAV regimens markedly inhibited the production of infectious ZIKV over the 7-day treatment. The higher-dose Ebola regimen provided approximately 1.5-log10 more suppression than the standard influenza regimen at the later time points.
These model predictions were experimentally validated using the HFIM system. Our findings suggest that clinically relevant FAV regimens may maximally reduce the ZIKV burden by 99.9% for the influenza regimen (day 5) and 99.99% for the Ebola regimen (day 5). Given that this efficacy was predicted and observed in the absence of an immune response, the overall antiviral efficacy in humans may be even better due to a contribution from the immune system. Thus, the degree of antiviral activity provided by the standard influenza regimen may be sufficient to allow the immune system to overcome and clear any residual viral burden not inhibited by FAV treatment.
There are several weaknesses associated with FAV treatment, despite its promising antiviral activity against ZIKV, as we described previously (12). First, FAV is contraindicated during pregnancy due to the potential for teratogenic and embryotoxic effects (17). Although FAV cannot be used to prevent the congenital disabilities associated with ZIKV infection, it may be effective in controlling sexual transmission, since FAV can be detected in semen (17). Although ZIKV viremia has been reported to clear by 10 days postinfection, ZIKV has been shown to persist in the central nervous system (CNS) and lymphoid tissues for weeks (18). To our knowledge, no published studies have assessed the CNS concentrations of FAV in humans; thus, predicting the efficacy of FAV against ZIKV at these peripheral infection sites carries considerable uncertainty at this time.
The present study has several potential limitations. First, data on the between-patient variability for FAV pharmacokinetic profiles are not available. Additionally, human pharmacokinetic profiles for FAV were obtained from healthy volunteers and not ZIKV-infected patients. There is a potential that FAV concentration-time profiles may differ in individuals experiencing ZIKV disease. As more information regarding FAV pharmacokinetics becomes available, we will be able to update our simulations. Second, our experimental analyses utilized only FAV concentrations and did not consider the concentration of the intracellular triphosphorylated FAV metabolite, which is the active form of the drug. Finally, we chose to use HUH-7 cells as a human cellular model. This cell line may not be the most clinically relevant since the effects of ZIKV infection on the human liver are not well characterized. HUH-7 cells were chosen because they are of human origin and support robust ZIKV replication. We have examined other human cell lines derived from various tissues, but these cells were not permissive to infection or failed to yield suitable viral titers necessary for antiviral evaluations.
In conclusion, we have demonstrated that clinically utilized FAV regimens hold promise as a therapeutic strategy to treat nonpregnant patients infected with ZIKV. Moreover, to our knowledge, the developed MBM is the first to successfully estimate the drug effect consistently between the plate and HFIM assays. We identified differences between virus infection, viral production, and host cell dynamics in the two systems. This combination model strategy renders the substantial benefit of improving the confidence in the estimated parameters. Ultimately, our MBM is capable of simulating viral burden profiles which excellently matched the profiles observed for the growth control and two clinically relevant dosage regimens. The MBM is also excellently suited to investigate the impact of between-patient variability, including different FAV half-lives in humans. The latter may arrive from polymorphism in the drug-metabolizing enzymes (19). Overall, our encouraging results suggest that further preclinical (in vitro and in vivo) and clinical studies of the activity of FAV against ZIKV are warranted.
MATERIALS AND METHODS
Cells and compound.
HUH-7 cells were cultured at 37°C in 5% CO2 in Dulbecco's modified Eagle's medium, high glucose (DMEM; HyClone, Logan City, UT), supplemented with 5% fetal bovine serum (FBS; HyClone, Logan City, UT) and 1% penicillin-streptomycin solution (HyClone, Logan City, UT). Vero cells were maintained at 37°C in 5% CO2 in Eagle's minimum essential medium (MEM; Corning Cellgro; Mediatech, Manassas, VA) in the presence of 5% fetal bovine serum (FBS; HyClone, Logan City, UT) and 1% penicillin-streptomycin solution (HyClone, Logan City, UT).
FAV was purchased from MedKoo Biosciences, Inc. (Research Triangle Park, NC), and was stored according to the manufacturer's recommendation. FAV drug stocks were freshly prepared before each assay by reconstituting the powder in 100% dimethyl sulfoxide (DMSO) and sonicating for 15 min. A 1% DMSO concentration was maintained in all assay media, including the control, to ensure drug solubility.
ZIKV and plaque assay.
The PRVABC59 ZIKV strain, isolated from a patient in Puerto Rico in 2015, was obtained from BEI Resources (Manassas, VA). ZIKV stocks were propagated on Vero cells as previously described (12).
The infectious ZIKV burden in viral supernatant samples was quantified by plaque assay on Vero cells as previously described (12).
Antiviral evaluations of FAV in cell culture 6-well plates.
Antiviral evaluations of FAV using traditional tissue culture methods were conducted as previously described (12), with the exception that ZIKV was inoculated onto confluent HUH-7 cell monolayers at a multiplicity of infection (MOI) equivalent to 0.1 PFU/cell. Serial 2-fold dilutions of FAV were evaluated at concentrations ranging from 31.25 μM to 500 μM. A no-treatment control was also included for a total of six concentrations per assay. DMSO at a final concentration of 1% was maintained in the assay medium to ensure FAV solubility. Viral supernatant samples were collected daily for 4 days. Uninfected HUH-7 cells were exposed to the above-mentioned FAV concentrations, and cell monolayers were examined daily for 4 days to ensure that treatment did not result in cytotoxicity. At the end of the experiment, all samples were quantified for infectious viral burden by the plaque assay on Vero cells. GraphPad Prism software (La Jolla, CA) was used to estimate the EC50 over the entire time course of the study as previously described (12). Three antiviral studies were performed in three independent replicates.
Dose-ranging studies in the HFIM system.
The HFIM system has been described in detail elsewhere (20–23). For the present study, 108 HUH-7 cells were mixed with 105 PFU/ml of ZIKV (MOI, 0.001 PFU/cell) and inoculated into the extracapillary space (ECS) of a cellulosic hollow-fiber (HF) cartridge (FiberCell Systems, Frederick, MD). A total of six HF cartridges were employed for this study. FAV was administered as a continuous infusion into five cartridges at concentrations ranging from 31.25 μM to 500 μM. One cartridge served as a no-treatment control (without FAV treatment). A final concentration of 1% DMSO was maintained in the tissue culture medium, including that of the control, to ensure the solubility of FAV. The ECS of each HF cartridge was sampled daily for 7 days. Samples were clarified by high-speed centrifugation and frozen at −80°C until the end of the study. The viral burden in all samples was quantified by plaque assay on Vero cells. Medium was sampled from the central reservoir of the HFIM system over the course of the 7-day experiment, and the samples were frozen at −80°C. The FAV concentrations in these samples were measured by liquid chromatography-tandem mass spectrometry (LC-MS/MS). All measured FAV concentrations were within 10% of the targeted value, demonstrating that the desired profiles were achieved in the HFIM system. Two independent studies were performed.
Mechanism-based mathematical modeling.
We refined a previously developed PD MBM to simultaneously describe and predict the effect of FAV against ZIKV in the plate assay and the HFIM. This MBM integrated the host cell dynamics, viral replication, and drug effect. The drug effect parameters were shared between the plate assay and the HFIM. To comodel the plate assay and the dynamic HFIM systems, host cell dynamics and viral replication were allowed to differ.
Host cell dynamics.
Uninfected host cells (U) were infected by extracellular virus (Vextra) via a second-order process with the infection rate constant (kinfect). Replication of uninfected host cells was accounted for in the HFIM via a logistic growth model (24, 25). As the HFIM provides a continuous flow of nutrients via fresh medium, the maximum number of host cells (HOSTmax) was allowed to differ between the HFIM and the plate assay. The differential equation for U was
| (1) |
where t is time and I1 to I5 refer to the five stages in infected host cells. PLAT refers to the plateau function which limits cell growth to a maximum cell density. The term k21 corresponds to the cell replication rate and was fixed to 1/24 h−1. The initial conditions (IC) were set to 105.82 cells/ml in the plate assay and 106.82 cells/ml in the HFIM.
Viral replication.
After host cells were infected, intracellular virus (Vi1) was generated via a first-order synthesis rate constant (ksyn). Consistent with our previous MBM, a series of five transit compartments was used to represent intracellular virus (Vi1, Vi2, Vi3, Vi4, and Vi5, linked by the transit rate constant [ktr]) and describes viral maturation and replication. Infected host cells were modeled to die upon virus release. The differential equations for infected host cells were (initial conditions of I2 to I5):
| (2) |
The initial condition for the first intracellular host cell compartment (I1) was log10 InocI (the initial estimated amount of infected host cells) for the plate assay and 0 for the HFIM. Initial conditions for I2 to I5 were 0.
The differential equations for the intracellular virus were (initial conditions for Vi1 to Vi5, all of which were equal to 0)
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
FAV inhibits the replication of virus RNA in infected host cells. This inhibitory drug effect (INHFAV) was described by a Hill function and implemented between the 4th and the 5th intracellular virus compartments. The maximum extent of inhibition (ImaxFAV), FAV concentrations causing 50% of ImaxFAV (IC50_FAV), and the respective parameters were estimated as follows:
| (8) |
where CFAV is the FAV concentration.
In the absence of drug, INHFAV was 1, and therefore, all virus leaving compartment Vi4 reached compartment Vi5. At high FAV concentrations, INHFAV approached 0, and therefore, intracellular virus was lost from compartment Vi4 and did not reach compartment Vi5.
Initial extracellular virus (Vextra) at time zero originated either from the virus inoculation in the HFIM or from the limited remaining cell-free virus in the plate assay. After host cell infection, extracellular virus arose from the egress of intracellular virus from the last transit compartment (Vi5) and was subject to a first-order loss rate constant (kloss,Vextra) and kinfect (as defined above). The differential equation for extracellular virus was (for an initial condition of Vextra of 3.82 log10 PFU/ml in the hollow-fiber assay or 2 log10 PFU/ml in the plate assay)
| (9) |
System outputs and residual error model.
The logarithm of virus burden units per milliliter (log10 number of PFU per milliliter) served as the dependent variable for modeling. We used an additive residual error on log-transformed scale. To account for observations below the lower limit of quantification (e.g., at time zero for the plate assay), we employed the Beal M3 method (26).
Parameter variability model and model qualification.
A standard exponential parameter variability model was used for most parameters to describe the small between-curve variability. A logistic transformation was utilized to constrain Imax between 0 and 1 (27). Parameters estimated on the log10 scale were modeled via a normal distribution. Models were compared via standard diagnostic plots, the objective function (negative log-likelihood in S-ADAPT software) (28), and the plausibility of parameter estimates (21, 29).
Software and algorithm.
To estimate the PD model parameters and variability, we used importance sampling in parallelized S-ADAPT (version 1.57) software. The SADAPT-TRAN facilitator tool was also employed (27, 30), and deterministic simulations were performed in Berkeley Madonna (version 8.23.3.0) software.
Prospective experimental validation of MBM predictions in the HFIM system.
The ECS of three cellulosic HF cartridges were each inoculated with a mixture of 108 HUH-7 cells and 105 PFU/cell of ZIKV (MOI, 0.001 PFU/cell), as described above. FAV was administered into two cartridges via a 1-h infusion to simulate the free-drug pharmacokinetic profiles associated with two clinically relevant regimens. The concentration-time profiles simulated in these studies are depicted in Fig. 4. The first regimen is the standard regimen used to treat uncomplicated human influenza virus infections: 1,800 mg at 0 h and 12 h on day 1 of treatment, followed by 800 mg every 12 h starting on day 2 of treatment (termed the low-dose regimen). The second regimen corresponds to the dosage regimen used to treat Ebola virus-infected patients: 2,400 mg at 0 h, 2,400 mg at 8 h, and 1,800 mg at 16 h on the first day of treatment, followed by 1,200 mg every 12 h on day 2 of treatment (termed the high-dose regimen). The third cartridge did not receive drug administration and served as the growth control. A final concentration of 1% DMSO was maintained in the assay medium in all three treatment or control arms. Viral supernatants were sampled from the ECS daily for 7 days and processed as described above for the dose-ranging studies. Viral load was quantified by the plaque assay on Vero cells. Intensive medium sampling from the central reservoir of the HFIM system was conducted over the first 48 h of the study, and samples were frozen at −80°C. The FAV concentrations in these samples were measured by liquid chromatography-tandem mass spectrometry (LC-MS/MS).
Pharmacokinetic analyses of FAV.
FAV concentrations in the HFIM system were measured by LC-MS/MS with an assay system consisting of a Prominence high-performance liquid chromatograph (HPLC; Shimadzu) and a TSQ Vantage triple-quadrupole mass spectrometer (Thermo Scientific). Separation was achieved using a Thermo HyperGold HPLC column (100 by 4.6 mm; particle size, 5 μm) at room temperature with a run time of 4.5 min. Mobile phases consisted of water (mobile phase A) and acetonitrile (mobile phase B) at a flow rate of 0.750 ml/min in gradient mode.
Samples in DMEM were stored at −80°C until analysis. After thawing at room temperature, 0.020 ml of each sample and 0.010 ml of internal standard (5-fluorocytosine, 50.0 μg/ml in water) were added to a 1.5-ml microtube, followed by addition of 0.500 ml of acetonitrile. Samples were vortexed well and centrifuged for 10 min at 16,000 × g at 4°C. One hundred microliters of the resulting sample supernatant was transferred into a 96-well plate, and samples were analyzed by LC-MS/MS using a 1-μl injection volume.
The mass spectrometer was operated in negative ion mode (FAV) and positive ion mode (5-fluorocytosine) using the heated electrospray ionization (HESI) probe interface. Selected reaction monitoring (SRM) m/z 156.0/113 (quantifier) and SRM m/z 156.0/66 (qualifier) were used for FAV; and SRM m/z 129.9/58 was used for the internal standard, 5-fluorocytosine. TSQ Vantage triple-quadrupole mass spectrometer parameters were as follows: spray voltage, 3,500 V; temperature, 350°C; sheath gas, 50 arbitrary units (AU); auxiliary gas, 10 AU; capillary temperature, 300°C; for SRM m/z 156.0/113, width 0.01 full width at half maximum (FWHM), scan 0.100 s, collision energy (CE) −17 V, and S-lens 65 V; for SRM m/z 156.0/66, width 0.01 FWHM, scan 0.100 s, CE −27 V, and S-lens 65 V; for SRM m/z 129.9/58, width 0.01 FWHM, scan 0.100 s, CE 37 V, and S-lens 42 V. Concentrations were calculated using the Xcalibur Quan Browser (version 2.2; Thermo Scientific).
ACKNOWLEDGMENTS
This work was supported by grant 7ZK30 from the Florida Department of Health Biomedical Research Program.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We declare that no conflict of interest exists.
REFERENCES
- 1.Ferreira-de-Brito A, Ribeiro IP, Miranda RM, Fernandes RS, Campos SS, Silva KA, Castro MG, Bonaldo MC, Brasil P, Lourenco-de-Oliveira R. 2016. First detection of natural infection of Aedes aegypti with Zika virus in Brazil and throughout South America. Mem Inst Oswaldo Cruz 111:655–658. doi: 10.1590/0074-02760160332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foy BD, Kobylinski KC, Chilson Foy JL, Blitvich BJ, Travassos da RA, Haddow AD, Lanciotti RS, Tesh RB. 2011. Probable non-vector-borne transmission of Zika virus, Colorado, USA. Emerg Infect Dis 17:880–882. doi: 10.3201/eid1705.101939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Musso D, Roche C, Robin E, Nhan T, Teissier A, Cao-Lormeau VM. 2015. Potential sexual transmission of Zika virus. Emerg Infect Dis 21:359–361. doi: 10.3201/eid2102.141363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmad SS, Amin TN, Ustianowski A. 2016. Zika virus: management of infection and risk. BMJ 352:i1062. doi: 10.1136/bmj.i1062. [DOI] [PubMed] [Google Scholar]
- 5.Cao-Lormeau VM, Blake A, Mons S, Lastere S, Roche C, Vanhomwegen J, Dub T, Baudouin L, Teissier A, Larre P, Vial AL, Decam C, Choumet V, Halstead SK, Willison HJ, Musset L, Manuguerra JC, Despres P, Fournier E, Mallet HP, Musso D, Fontanet A, Neil J, Ghawche F. 2016. Guillain-Barre syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet 387:1531–1539. doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cauchemez S, Besnard M, Bompard P, Dub T, Guillemette-Artur P, Eyrolle-Guignot D, Salje H, Van Kerkhove MD, Abadie V, Garel C, Fontanet A, Mallet HP. 2016. Association between Zika virus and microcephaly in French Polynesia, 2013–15: a retrospective study. Lancet 387:2125–2132. doi: 10.1016/S0140-6736(16)00651-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kleber de Oliveira W, Cortez-Escalante J, De Oliveira WT, do Carmo GM, Henriques CM, Coelho GE, Araujo de Franca GV. 2016. Increase in reported prevalence of microcephaly in infants born to women living in areas with confirmed Zika virus transmission during the first trimester of pregnancy—Brazil, 2015. MMWR Morb Mortal Wkly Rep 65:242–247. doi: 10.15585/mmwr.mm6509e2. [DOI] [PubMed] [Google Scholar]
- 8.Furuta Y, Takahashi K, Fukuda Y, Kuno M, Kamiyama T, Kozaki K, Nomura N, Egawa H, Minami S, Watanabe Y, Narita H, Shiraki K. 2002. In vitro and in vivo activities of anti-influenza virus compound T-705. Antimicrob Agents Chemother 46:977–981. doi: 10.1128/AAC.46.4.977-981.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gowen BB, Wong MH, Jung KH, Sanders AB, Mendenhall M, Bailey KW, Furuta Y, Sidwell RW. 2007. In vitro and in vivo activities of T-705 against arenavirus and bunyavirus infections. Antimicrob Agents Chemother 51:3168–3176. doi: 10.1128/AAC.00356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendenhall M, Russell A, Smee DF, Hall JO, Skirpstunas R, Furuta Y, Gowen BB. 2011. Effective oral favipiravir (T-705) therapy initiated after the onset of clinical disease in a model of arenavirus hemorrhagic fever. PLoS Negl Trop Dis 5:e1342. doi: 10.1371/journal.pntd.0001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sissoko D, Laouenan C, Folkesson E, M'Lebing AB, Beavogui AH, Baize S, Camara AM, Maes P, Shepherd S, Danel C, Carazo S, Conde MN, Gala JL, Colin G, Savini H, Bore JA, Le Marcis F, Koundouno FR, Petitjean F, Lamah MC, Diederich S, Tounkara A, Poelart G, Berbain E, Dindart JM, Duraffour S, Lefevre A, Leno T, Peyrouset O, Irenge L, Bangoura N, Palich R, Hinzmann J, Kraus A, Barry TS, Berette S, Bongono A, Camara MS, Chanfreau MV, Doumbouya L, Souley H, Kighoma PM, Koundouno FR, Rene L, Loua CM, Massala V, Moumouni K, Provost C, Samake N, Sekou C, Soumah A, Arnould I, Komano MS, Gustin L, Berutto C, Camara D, Camara FS, Colpaert J, et al. . 2016. Experimental treatment with favipiravir for Ebola virus disease (the JIKI trial): a historically controlled, single-arm proof-of-concept trial in Guinea. PLoS Med 13:e1001967. doi: 10.1371/journal.pmed.1001967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pires de Mello CP, Tao X, Kim TH, Bulitta JB, Rodriquez JL, Pomeroy JJ, Brown AN. 2018. Zika virus replication is substantially inhibited by novel favipiravir and interferon alpha combination regimens. Antimicrob Agents Chemother 62:e01983-17. doi: 10.1128/AAC.01983-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goo L, Dowd KA, Smith AR, Pelc RS, DeMaso CR, Pierson TC. 2016. Zika virus is not uniquely stable at physiological temperatures compared to other flaviviruses. mBio 7:e01396-16. doi: 10.1128/mBio.01396-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brendel K, Comets E, Laffont C, Laveille C, Mentre F. 2006. Metrics for external model evaluation with an application to the population pharmacokinetics of gliclazide. Pharm Res 23:2036–2049. doi: 10.1007/s11095-006-9067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen TH, Guedj J, Anglaret X, Laouenan C, Madelain V, Taburet AM, Baize S, Sissoko D, Pastorino B, Rodallec A, Piorkowski G, Carazo S, Conde MN, Gala JL, Bore JA, Carbonnelle C, Jacquot F, Raoul H, Malvy D, de LX, Mentre F. 2017. Favipiravir pharmacokinetics in Ebola-infected patients of the JIKI trial reveals concentrations lower than targeted. PLoS Negl Trop Dis 11:e0005389. doi: 10.1371/journal.pntd.0005389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pharmaceuticals and Medical Devices Agency. 2014. Review report. Report on the deliberation results of favipiravir (English version). Pharmaceuticals and Medical Devices Agency, Tokyo, Japan. [Google Scholar]
- 17.Nagata T, Lefor AK, Hasegawa M, Ishii M. 2015. Favipiravir: a new medication for the Ebola virus disease pandemic. Disaster Med Public Health Prep 9:79–81. doi: 10.1017/dmp.2014.151. [DOI] [PubMed] [Google Scholar]
- 18.Aid M, Abbink P, Larocca RA, Boyd M, Nityanandam R, Nanayakkara O, Martinot AJ, Moseley ET, Blass E, Borducchi EN, Chandrashekar A, Brinkman AL, Molloy K, Jetton D, Tartaglia LJ, Liu J, Best K, Perelson AS, De La Barrera RA, Lewis MG, Barouch DH. 2017. Zika virus persistence in the central nervous system and lymph nodes of rhesus monkeys. Cell 169:610–620. doi: 10.1016/j.cell.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka E. 1999. Update: genetic polymorphism of drug metabolizing enzymes in humans. J Clin Pharm Ther 24:323–329. doi: 10.1046/j.1365-2710.1999.00236.x. [DOI] [PubMed] [Google Scholar]
- 20.Brown AN, McSharry JJ, Weng Q, Driebe EM, Engelthaler DM, Sheff K, Keim PS, Nguyen J, Drusano GL. 2010. In vitro system for modeling influenza A virus resistance under drug pressure. Antimicrob Agents Chemother 54:3442–3450. doi: 10.1128/AAC.01385-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown AN, Bulitta JB, McSharry JJ, Weng Q, Adams JR, Kulawy R, Drusano GL. 2011. Effect of half-life on the pharmacodynamic index of zanamivir against influenza virus delineated by a mathematical model. Antimicrob Agents Chemother 55:1747–1753. doi: 10.1128/AAC.01629-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown AN, McSharry JJ, Weng Q, Adams JR, Kulawy R, Drusano GL. 2011. Zanamivir, at 600 milligrams twice daily, inhibits oseltamivir-resistant 2009 pandemic H1N1 influenza virus in an in vitro hollow-fiber infection model system. Antimicrob Agents Chemother 55:1740–1746. doi: 10.1128/AAC.01628-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown AN, Adams JR, Baluya DL, Drusano GL. 2015. Pharmacokinetic determinants of virological response to raltegravir in the in vitro pharmacodynamic hollow-fiber infection model system. Antimicrob Agents Chemother 59:3771–3777. doi: 10.1128/AAC.00469-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luma HN, Eloumou SA, Ekaney DS, Lekpa FK, Donfack-Sontsa O, Ngahane BH, Mapoure YN. 2016. Sero-prevalence and correlates of hepatitis B and C co-infection among HIV-infected individuals in two regional hospitals in Cameroon. Open AIDS J 10:199–208. doi: 10.2174/1874613601610010199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yano Y, Oguma T, Nagata H, Sasaki S. 1998. Application of logistic growth model to pharmacodynamic analysis of in vitro bactericidal kinetics. J Pharm Sci 87:1177–1183. doi: 10.1021/js9801337. [DOI] [PubMed] [Google Scholar]
- 26.Beal SL. 2001. Ways to fit a PK model with some data below the quantification limit. J Pharmacokinet Pharmacodyn 28:481–504. doi: 10.1023/A:1012299115260. [DOI] [PubMed] [Google Scholar]
- 27.Bulitta JB, Bingolbali A, Shin BS, Landersdorfer CB. 2011. Development of a new pre- and post-processing tool (SADAPT-TRAN) for nonlinear mixed-effects modeling in S-ADAPT. AAPS J 13:201–211. doi: 10.1208/s12248-011-9257-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bauer RJ, Guzy S, Ng C. 2007. A survey of population analysis methods and software for complex pharmacokinetic and pharmacodynamic models with examples. AAPS J 9:E60–E83. doi: 10.1208/aapsj0901007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bulitta JB, Okusanya OO, Forrest A, Bhavnani SM, Clark K, Still JG, Fernandes P, Ambrose PG. 2013. Population pharmacokinetics of fusidic acid: rationale for front-loaded dosing regimens due to autoinhibition of clearance. Antimicrob Agents Chemother 57:498–507. doi: 10.1128/AAC.01354-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bulitta JB, Landersdorfer CB. 2011. Performance and robustness of the Monte Carlo importance sampling algorithm using parallelized S-ADAPT for basic and complex mechanistic models. AAPS J 13:212–226. doi: 10.1208/s12248-011-9258-9. [DOI] [PMC free article] [PubMed] [Google Scholar]