To guide the timely selection of antibiotic combinations against carbapenem-resistant Gram-negative bacteria (CR-GNB), an in vitro test with a short turnaround time is essential. We developed an in vitro ATP bioluminescence assay to determine effective antibiotic combinations against CR-GNB within 6 h.
KEYWORDS: in vitro combination testing, luciferin-luciferase reaction, multidrug resistance
ABSTRACT
To guide the timely selection of antibiotic combinations against carbapenem-resistant Gram-negative bacteria (CR-GNB), an in vitro test with a short turnaround time is essential. We developed an in vitro ATP bioluminescence assay to determine effective antibiotic combinations against CR-GNB within 6 h. We tested 42 clinical CR-GNB strains (14 Acinetobacter baumannii, 14 Pseudomonas aeruginosa, and 14 Klebsiella pneumoniae strains) against 74 single antibiotics and two-antibiotic combinations. Bacteria (approximately 5 log10 CFU/ml) were incubated with an antibiotic(s) at 35°C; ATP bioluminescence was measured at 6 h and 24 h; and the measurements were compared to viable counts at 24 h. Receiver operating characteristic (ROC) curves were used to determine the optimal luminescence thresholds (TRLU) for distinguishing between inhibitory and noninhibitory combinations. The areas under the 6-h and 24-h ROC curves were compared using the DeLong method. Prospective validation of the established thresholds was conducted using 18 additional CR-GNB. The predictive accuracy of TRLU for the 6-h ATP bioluminescence assay was 77.5% when all species were analyzed collectively. Predictive accuracies ranged from 73.7% to 82.7% when each species was analyzed individually. Upon comparison of the areas under the 6-h and 24-h ROC curves, the 6-h assay performed significantly better than the 24-h assay (P < 0.01). Predictive accuracy remained high upon prospective validation of the 6-h ATP assay (predictive accuracy, 79.8%; 95% confidence interval [CI], 77.6 to 81.9%), confirming the external validity of the assay. Our findings indicate that our 6-h ATP bioluminescence assay can provide guidance for prospective selection of antibiotic combinations against CR-GNB in a timely manner and may be useful in the management of CR-GNB infections.
INTRODUCTION
The treatment of infections caused by carbapenem-resistant Gram-negative bacteria (CR-GNB) has posed a major challenge to clinicians worldwide (1). While a number of novel antibiotics, such as ceftazidime-avibactam and ceftolozane-tazobactam, have recently been added to the pool of existing antibiotics against CR-GNB, most of these agents are effective only against a subset of CR-GNB; furthermore, limited clinical data are currently available for these agents (2). Hence, clinicians are still often forced to resort to toxic last-line agents, such as polymyxins, or to explore alternative options, such as combination antibiotic therapy (3, 4).
In routine clinical practice, a microbiological diagnosis of a pathogen, consisting of identification of the causative organism and phenotypic susceptibility testing, typically takes at least 48 to 72 h (5). In the case of CR-GNB infections, the conduct of in vitro combination testing to guide the selection of antibiotic combination therapy may be further required, resulting in an additional delay of approximately 3 days before targeted treatment against the causative organism can be identified (6, 7). Such a delay in the time to appropriate therapy not only is detrimental to patient outcomes but can also promote the development of further antibiotic resistance if unnecessarily broad spectrum or suboptimal empirical treatment options are employed (5, 8).
ATP bioluminescence has been widely employed to assess bacterial contamination in the food and health care settings and appears to be a rapid and sensitive alternative technique for antibiotic combination testing (9–11). This assay is based on the ability of firefly luciferase to catalyze the oxidation of d-luciferin in the presence of a magnesium salt and ATP, and the light intensity emitted has been shown to be proportional to the ATP content and hence to the viable bacterial biomass in the sample (12). A main advantage of using ATP bioluminescence to quantify bacteria is that unlike viable plating, ATP bioluminescence bypasses the additional incubation time needed for the bacteria to form visible CFU, thereby shortening the turnaround time by at least 24 h (6, 11). Furthermore, since the need for the tedious and labor-intensive process of viable plating is eliminated, the use of ATP bioluminescence can allow a large number of antibiotic combinations to be tested in a high-throughput manner, making it suitable for prospective combination testing to guide the selection of combinations against individual CR-GNB strains (6, 11).
We have previously developed a robust and reproducible ATP bioluminescence assay to determine effective antibiotic combinations against CR-GNB within 24 h (11). However, a number of published studies have proposed that with ATP bioluminescence, shorter incubations (2 to 8 h) of the bacteria with the tested antibiotic(s) may be sufficient (13, 14). In this study, we developed a combination testing assay using ATP bioluminescence to predict effective antibiotic combinations against CR-GNB within 6 h, and we compared this new assay to the 24-h ATP bioluminescence assay. We further validated the predictive accuracy of both the 6-h and the 24-h ATP bioluminescence assay prospectively, using additional CR-GNB strains.
RESULTS
Characteristics of the CR-GNB isolates.
The genotypic characteristics and MICs of the 42 CR-GNB isolates are shown in Tables 1 and 2, respectively. As shown, all CR-GNB isolates were nonsusceptible to cefepime, piperacillin-tazobactam, imipenem, meropenem, and doripenem. In addition, all Acinetobacter baumannii isolates were nonsusceptible to amikacin and levofloxacin. Most CR-GNB isolates remained susceptible to polymyxin B (73.8%); polymyxin B MICs ranged from 0.5 to ≥16 mg/liter. The most common carbapenemase genes detected among Klebsiella pneumoniae isolates were blaOXA-48-like (57.1%) and blaNDM (42.9%). Five (35.7%) K. pneumoniae isolates coproduced NDM and OXA-48-like carbapenemases.
TABLE 1.
Genotypic characteristics of the 42 CR-GNB used in establishing TRLU
| Organism (n) | Carbapenemase genes (n)a |
|---|---|
| A. baumannii (14) | blaOXA-23-like (14), blaOXA-51-like (14) |
| P. aeruginosa (14) | blaIMP (5), blaVIM (4), blaVEB (1) |
| K. pneumoniae (14) | blaOXA-48-like (8), blaNDM (6), blaKPC (2) |
All 14 A. baumannii strains harbored both the blaOXA-23-like and blaOXA-51-like carbapenemase genes. Five K. pneumoniae strains coharbored the blaOXA-48-like and blaNDM carbapenemase genes.
TABLE 2.
Phenotypic characteristics of the 42 CR-GNB used in establishing TRLU
| Antibiotic |
A. baumannii (n = 14) |
P. aeruginosa (n = 14) |
K. pneumoniae (n = 14) |
|||
|---|---|---|---|---|---|---|
| No. (%) nonsusceptible | MIC range (mg/liter) | No. (%) nonsusceptible | MIC range (mg/liter) | No. (%) nonsusceptible | MIC range (mg/liter) | |
| Amikacin | 14 (100.0) | ≥128 | 9 (64.3) | ≤1 to ≥128 | 9 (64.3) | 2 to ≥128 |
| Aztreonam | 1 to ≥128 | 12 (85.7) | 4 to ≥128 | 14 (100.0) | ≥128 | |
| Polymyxin B | 2 (14.3) | 1 to ≥16 | 3 (21.4) | 1 to 8 | 6 (42.9) | 0.5 to 16 |
| Cefepime | 14 (100.0) | ≥64 | 14 (100.0) | 32 to ≥64 | 14 (100.0) | ≥64 |
| Piperacillin-tazobactam | 14 (100.0) | ≥256 | 14 (100.0) | 32 to ≥256 | 14 (100.0) | ≥256 |
| Doripenem | 14 (100.0) | ≥16 | 14 (100.0) | ≥16 | 14 (100.0) | ≥16 |
| Imipenem | 14 (100.0) | ≥16 | 14 (100.0) | ≥16 | 14 (100.0) | 8 to ≥16 |
| Meropenem | 14 (100.0) | ≥16 | 14 (100.0) | ≥16 | 14 (100.0) | ≥16 |
| Levofloxacin | 14 (100.0) | 8 to ≥64 | 14 (100.0) | 32 to ≥64 | 12 (85.7) | 2 to ≥64 |
| Tigecycline | 10 (71.4) | 0.5 to ≥32 | NDa | 4 (28.6) | 0.5 to 4 | |
| Rifampin | 1 to ≥64 | ND | ≥64 | |||
ND, not done.
Changes in ATP bioluminescence and viable bacterial counts over 24 h.
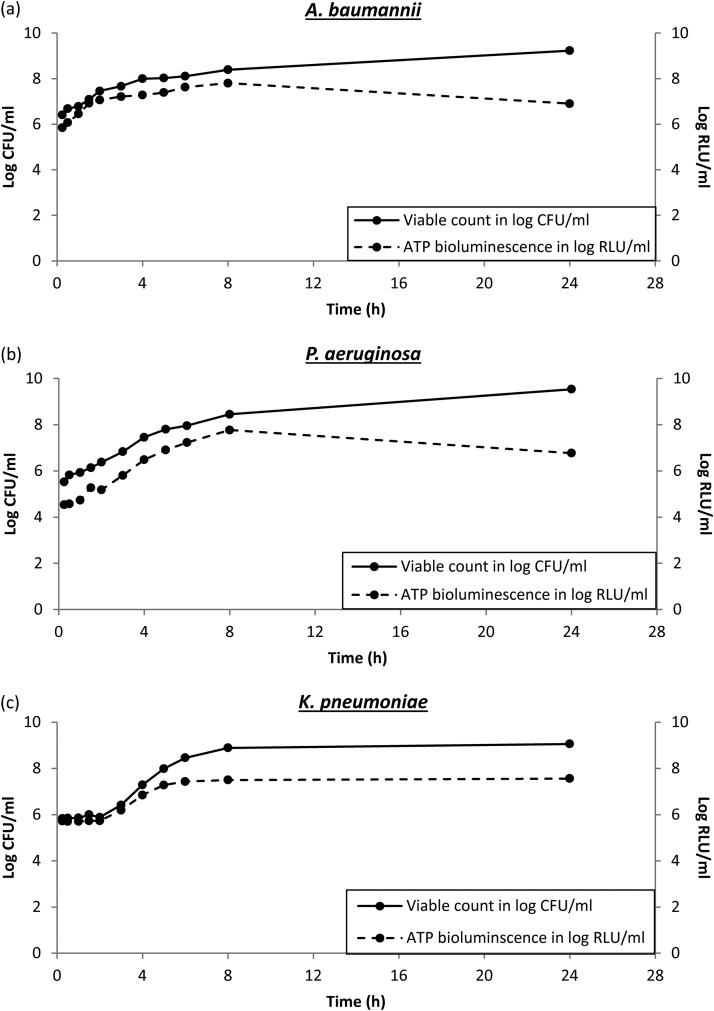
The changes in viable bacterial counts over 24 h appeared to be similar for all CR-GNB (Fig. 1). Viable bacterial counts increased exponentially during log-phase growth until 6 h, followed by corresponding increases in ATP bioluminescence for all CR-GNB. Interestingly, for A. baumannii and Pseudomonas aeruginosa, when the viable bacterial counts plateaued from 8 h to 24 h, we observed a ∼10-fold reduction in ATP bioluminescence (from 7.8 log10 relative light units [RLU]/ml at 8 h to 6.9 log10 RLU/ml at 24 h for A. baumannii and from 7.8 log10 RLU/ml at 8 h to 6.8 log10 RLU/ml at 24 h for P. aeruginosa). For K. pneumoniae, both viable bacterial counts and ATP bioluminescence plateaued and remained constant from 8 h to 24 h.
FIG 1.
Changes in ATP bioluminescence and viable bacterial counts for CR A. baumannii (a), CR P. aeruginosa (b), and CR K. pneumoniae (c) without exposure to antibiotics over 24 h.
Combination testing by viable plating.
A total of 74 different single antibiotics and two-antibiotic combinations were tested against each of the 42 CR-GNB isolates (for a total of 3,108 distinct antibiotic-isolate observations). Of these, 1,478 (47.6%) antibiotic combinations were found to be inhibitory against the CR-GNB isolates tested. The most effective antibiotic combinations against all CR-GNB isolates tested were polymyxin B plus amikacin (inhibitory against 34/42 [90.0%] CR-GNB isolates) and polymyxin B plus rifampin (inhibitory against 34/42 [90.0%] CR-GNB isolates) (see Table S1 in the supplemental material). Against CR A. baumannii, the most effective antibiotic combinations in vitro, based on the results of viable plating, were polymyxin B plus rifampin and polymyxin B plus cefepime (each combination was inhibitory against 13/14 [92.9%] CR A. baumannii isolates). Against CR P. aeruginosa, the most effective antibiotic combination in vitro was polymyxin B plus amikacin (inhibitory against 13/14 [92.9%] CR P. aeruginosa isolates). Polymyxin B plus tigecycline was the most effective antibiotic combination in vitro against K. pneumoniae (inhibitory against 14/14 [100%] CR K. pneumoniae isolates).
Determination of inhibitory-noninhibitory thresholds for the 6-h and 24-h ATP bioluminescence assays.
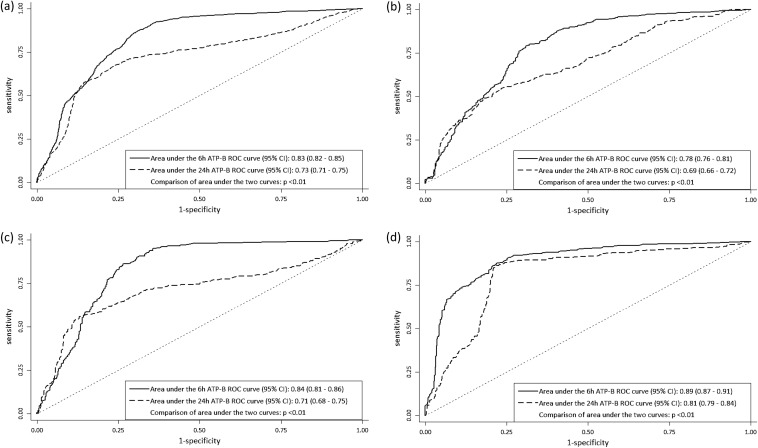
The receiver operating characteristic (ROC) curves for the 6-h and 24-h ATP bioluminescence assays for all CR-GNB and for each species are shown in Fig. 2. Upon comparison of the ROC curves for the two assays using the DeLong method (15), we found that the 6-h ATP bioluminescence assay performed significantly better than the 24-h ATP bioluminescence assay overall, as well as by bacterial species.
FIG 2.
Six-hour and 24-h ROC plots for all CR-GNB (a), A. baumannii (b), P. aeruginosa (c), and K. pneumoniae (d), for all single drugs and two-drug combinations. Upon comparison of the areas under the ROC curves, the 6-h ATP assay performed significantly better than the 24-h assay for all CRGNB and for each species.
We summarized the sensitivities, specificities, and unweighted accuracies of the optimal luminescence thresholds that distinguish between inhibitory and noninhibitory combinations (TRLU) for the 6-h and 24-h ATP bioluminescence assays in Table 3 and further described the sensitivities, specificities, and unweighted accuracies of the TRLU for selected antibiotic combinations in the 6-h and 24-h ATP bioluminescence assays in Tables S2 to S4 in the supplemental material. When all GNB organisms were collectively analyzed, the overall TRLU value that provided maximum unweighted accuracy (77.5%) for the 6-h ATP bioluminescence assay was −1.12, with a sensitivity of 82.7% and a specificity of 72.3%. For the 24-h ATP assay, the overall TRLU value that provided maximum unweighted accuracy was −0.26, with an unweighted accuracy of 71.5% (sensitivity, 69.3%; specificity, 73.7%). When individual TRLU values were established for each species, the unweighted accuracies of thresholds remained relatively high. The individual TRLU values appeared to differ between species and antibiotic combinations, as well as between the 6-h and 24-h assay. Notably, the unweighted accuracies of the 6-h assay TRLU were higher than those of the 24-h TRLU for all species.
TABLE 3.
Sensitivity, specificity, and unweighted accuracy of TRLU in distinguishing between inhibitory and noninhibitory antibiotic combinations at 6 h and 24 h
| Organism (no. of antibiotic-isolate observations) and time of ATP bioluminescence measurement | No. (%) of antibiotic combinations |
TRLU | Accuracy of the ATP bioluminescence assay (%) |
P value for comparison of 6-h and 24-h AUROC curvesb | |||
|---|---|---|---|---|---|---|---|
| Inhibitorya | Noninhibitorya | Sensitivity | Specificity | Unweighted accuracy | |||
| All organisms (3,108) | 1,478 (47.6) | 1,630 (52.5) | <0.01 | ||||
| 6 h | −1.12 | 82.7 | 72.3 | 77.5 | |||
| 24 h | −0.26 | 69.3 | 73.7 | 71.5 | |||
| A. baumannii (1,036) | 522 (50.4) | 514 (49.6) | <0.01 | ||||
| 6 h | −1.03 | 76.2 | 71.2 | 73.7 | |||
| 24 h | −0.20 | 51.2 | 76.3 | 63.8 | |||
| P. aeruginosa (1,036) | 477 (46.0) | 559 (54.0) | <0.01 | ||||
| 6 h | −1.26 | 86.2 | 73.5 | 79.9 | |||
| 24 h | −0.36 | 66.7 | 71.7 | 69.2 | |||
| K. pneumoniae (1,036) | 479 (46.2) | 557 (55.7) | <0.01 | ||||
| 6 h | −1.43 | 85.8 | 79.5 | 82.7 | |||
| 24 h | −0.72 | 86.0 | 78.6 | 82.3 | |||
Based on viable counts.
AUROC curves, areas under the ROC curves.
Prospective validation of established TRLU.
A total of 18 prospectively collected clinical CR-GNB isolates were employed for the validation of the established TRLU (1,332 distinct antibiotic-isolate observations). Like the 42 isolates employed for establishing the TRLU, all 18 CR-GNB isolates were not susceptible to cefepime, piperacillin-tazobactam, imipenem, meropenem, or doripenem. Eleven (61.1%) of the isolates were susceptible to polymyxin B; polymyxin B MICs ranged from 0.5 to ≥16 mg/liter. All A. baumannii isolates harbored blaOXA-23 and blaOXA-51-like genes; 4 (66.7%) P. aeruginosa isolates harbored genes encoding metallo-β-lactamases (MBLs). OXA-48-like carbapenemases (4/6 [66.7%]) were the most common carbapenemases detected among the K. pneumoniae isolates.
The results of the prospective validation are shown in Table 4. When all organisms were collectively analyzed, the predictive unweighted accuracies (95% confidence intervals [CI]) of the established TRLU for the 6-h and 24-h ATP bioluminescence assays were 79.8% (77.6% to 81.9%) and 81.0% (78.8% to 83.1%), respectively. The positive predictive value and negative predictive value of the 6-h ATP bioluminescence assay when all CR-GNB were collectively analyzed were 77.1% (95% CI, 74.6% to 79.4%) and 83.0% (95% CI, 80.3 to 85.4%), respectively. The accuracies of the individual TRLU values for each species appeared to be similar across all species for the 6-h assay, with the unweighted accuracy ranging from 79.8% to 80.4%. In contrast, the accuracies of the individual TRLU values for the 24-h assay upon external validation appeared to differ slightly for each species, which may be attributed to the differences in basal ATP content between different bacterial species upon entry into stationary phase. The unweighted accuracy of the 24-h TRLU value appeared to be highest for K. pneumoniae (91.4% [95% CI, 88.4% to 93.9%]) and lowest for P. aeruginosa (76.1 [95% CI, 71.9 to 80.0]).
TABLE 4.
Prospective validation of established TRLU
| Time of ATP bioluminescence measurement and organism(s) | Value (% [95% CI]) |
||||
|---|---|---|---|---|---|
| Sensitivity | Specificity | Unweighted accuracy | Positive predictive value | Negative predictive value | |
| 6 h | |||||
| All CR-GNB | 84.2 (81.2–86.9) | 75.5 (72.0–78.7) | 79.8 (77.6–81.9) | 77.1 (74.6–79.4) | 83.0 (80.3–85.4) |
| A. baumannii | 75.3 (69.1–80.9) | 86.2 (81.0–90.4) | 80.9 (76.9–84.4) | 84.2 (79.2–88.2) | 78.2 (73.9–82.0) |
| P. aeruginosa | 86.1 (81.3–90.0) | 70.8 (63.5–77.4) | 80.0 (75.9–83.6) | 81.5 (77.7–84.8) | 77.3 (71.3–82.3) |
| K. pneumoniae | 83.4 (77.1–88.6) | 78.4 (73.0–83.2) | 80.4 (76.4–84.0) | 71 6 (66.5–76.1) | 87.9 (83.8–91.1) |
| 24 h | |||||
| All CR-GNB | 82.0 (78.8–84.8) | 80.0 (76.8–83.0) | 81.0 (78.8–83.1) | 80.2 (77.6–82.5) | 81.9 (79.3–84.2) |
| A. baumannii | 72.2 (65.7–78.0) | 80.4 (74.7–85.4) | 76.4 (72.1–80.2) | 78.2 (73.1–82.6) | 74.8 (72.1–80.2) |
| P. aeruginosa | 79.3 (73.4–84.0) | 71.4 (64.1–77.9) | 76.1 (71.9–80.0) | 80.5 (76.5–84.0) | 69.8 (64.2–74.8) |
| K. pneumoniae | 97.7 (94.3–99.4) | 87.4 (82.8–91.1) | 91.4 (88.4–93.9) | 83.4 (78.6–87.3) | 98.3 (95.7–99.4) |
DISCUSSION
This study builds on previously published work that applied ATP bioluminescence to antimicrobial susceptibility and combination testing (9, 11, 13, 14). Our ATP bioluminescence assay for combination testing determined effective antibiotic combinations against CR-GNB within a rapid turnaround time of 6 h, with a reasonably high degree of accuracy. The assay was found to be accurate across a wide array of antibiotic combinations, as well as across different species of CR-GNB. Using prospective validation; we further ascertained the external validity of our assay. Given the reduced turnaround time of our method relative to those of conventional viable plating methods (which require at least 48 to 72 h), our 6-h ATP bioluminescence assay may be employed to prospectively guide the selection of antibiotic combinations against individual CR-GNB strains in a timely fashion.
The use of rapid susceptibility testing methods, including rapid antibiotic combination testing methods, can facilitate the early initiation of targeted treatment, which can potentially improve patient outcomes and contribute to reducing health care costs (5). To date, several different phenotypic approaches, including turbidimetry, conductance, and ATP bioluminescence, have been employed (16). ATP bioluminescence, in particular, has been frequently proposed as a rapid alternative to conventional microbiological testing methods, including susceptibility tests on Gram-positive and Gram-negative bacteria, Mycobacterium spp., and bacteria in biofilms, and for the assessment of postantibiotic effects (9, 17, 18). In most of these studies, ATP bioluminescence is rapid and relatively simple to perform and has demonstrated good concordance with traditional susceptibility testing methods.
A number of published studies have suggested that bacterial ATP bioluminescence, measured as early as 2 to 6 h, could be employed to predict the results of conventional 24-h susceptibility testing (13, 14). The growth curves generated in our study showed that bacteria were in log-phase growth up to 6 h; during log-phase growth, we observed that the ATP contents of the bacterial cells increased and demonstrated a linear relationship with viable bacterial counts. Interestingly, when the bacteria entered stationary phase at 8 to 24 h, we observed consequent reductions in bacterial ATP contents, particularly in A. baumannii and P. aeruginosa; this may be attributed to the fact that in stationary phase, ATP production is reduced as bacterial cell division slows. Our observations corroborated the findings by Vogel et al., who suggested that the bacterial growth phase affects ATP bioluminescence measurements and, consequently, the correlation with viable counts (19). Hence, we hypothesized that the 6-h bacterial ATP bioluminescence may potentially be associated with higher accuracy than the 24-h bacterial ATP bioluminescence in the prediction of inhibitory combinations on conventional viable plating.
In this study, we compared our ATP bioluminescence assay to viable plating results. This is because viable plating is the most common method employed for determining the activity of antibiotic combinations in in vitro combination testing against CR-GNB and is also the testing method currently employed in Singapore for patients infected with CR-GNB (7). We observed reasonable accuracy for the 6-h ATP bioluminescence assay upon internal and external validation. Most notably, upon comparison of the ROC curves of the 6-h and 24-h ATP bioluminescence assays, the 6-h ATP bioluminescence assay appeared to fare significantly better. Differences in accuracies and thresholds were observed between different CR-GNB species; such interspecies variation may be attributed to the difference in basal ATP content between different bacterial species (12). Inaccuracies in our 6-h ATP bioluminescence assay compared to viable plating could also be due to a number of other factors. First, the formation of bacterial spheroplasts with high ATP contents upon exposure to β-lactam antibiotics may cause inaccuracies when β-lactam-containing combinations are studied (20). Second, the potential presence of viable but nondividing dormant or persister cells, which produce basal amounts of ATP but cannot be detected by viable plating techniques, may contribute to the inaccuracies of our ATP bioluminescence assay (21). Third, and last, an incubation time of 6 h may be insufficient to detect bacterial regrowth, which can arise from selective amplification of a resistant subpopulation (22).
A notable strength of our study was that we accounted for potential differences in growth rates, and consequently in ATP contents, among different CR-GNB isolates by employing the antibiotic-minus-control difference (ΔΔRLU) to generate the TRLU differentiating between inhibitory and noninhibitory combinations. This design is similar to the study design published by Ivancic et al. and is critical, since differences in the growth rate between isolates can contribute to large variations in ATP bioluminescence measurements, with a resultant compromise of the accuracy of the ATP bioluminescence assay (13). Another advantage of our study was that we employed CR-GNB with a wide array of resistant mechanisms. This not only reflected the diversity in our local CR-GNB strains but also ensured that our established thresholds could be applied to CRGNB with a wide array of resistance mechanisms (23). Unfortunately, since we employed only a limited number of isolates to establish the TRLU, we were unable to generate individualized thresholds for each antibiotic combination, which may have inadvertently led to a reduction in the accuracy of our 6-h ATP bioluminescence assay.
Conclusion.
The emerging global spread of carbapenem resistance among Gram-negative organisms, combined with the lack of effective new agents, underscores the importance of identifying appropriate and effective targeted antibiotic therapy within the shortest period possible. Our 6-h ATP bioluminescence assay can provide guidance for prospective combination selection in a timelier manner than current, traditional combination testing methods. Further studies with larger numbers of specimens from multiple health care centers should be conducted to validate this approach and potentially to develop bioluminescence thresholds for individual antibiotic combinations.
MATERIALS AND METHODS
Microorganisms.
We employed 42 nonclonal clinical CR-GNB isolates (14 A. baumannii, 14 P. aeruginosa, and 14 K. pneumoniae isolates) collected from 5 public acute-care hospitals in Singapore from 2009 to 2012 as part of a national surveillance study. The MICs of multiple antibiotics were determined using custom-made broth microdilution panels (Trek Diagnostics, East Grinstead, UK), and susceptibility was defined in accordance with the breakpoints provided by the Clinical and Laboratory Standards Institute (24). All isolates were stored at −80°C and were subcultured twice on 5% blood agar plates (Thermo Scientific, Malaysia) for 24 h at 35°C before each experiment.
To describe the molecular mechanisms of carbapenem resistance, all A. baumannii isolates were screened for the blaOXA-23, blaOXA-24, blaOXA-51, and blaOXA-58 genes, while all P. aeruginosa isolates were screened for commonly acquired MBL genes (blaVIM, blaIMP, blaSIM, blaGIM, blaSPM, and blaNDM) using multiplex PCR assays (25). For K. pneumoniae, the presence of genes encoding extended-spectrum β-lactamases (ESBLs), plasmid-mediated AmpC β-lactamases, MBLs, and K. pneumoniae carbapenemases (KPCs) were determined using PCR (25).
Ethics.
This study was approved by the SingHealth institutional ethics review board prior to initiation (2012/110/D).
Antimicrobial agents and experimental reagents.
Eleven antibiotics were employed as single drugs and in two-antibiotic combinations for combination testing at clinically relevant concentrations (Table 5) (26–36). Stock solutions of all antimicrobial agents except rifampin were prepared in sterile water and aliquoted for storage at −80°C. Rifampin was dissolved in dimethyl sulfoxide (DMSO) and was then serially diluted in sterile water to the desired final drug concentration. The final DMSO concentration (<1%, vol/vol) had no effect on bacterial growth (37).
TABLE 5.
Antibiotic concentrations employed in combination testing and corresponding simulated dosing regimens
| Drug | Simulated dosing regimena | Concn (mg/liter) | Reference |
|---|---|---|---|
| Amikacin | 15–20 mg/kg of body wt every 24 h | 65 | 25 |
| Aztreonam | 8 g every 24 h (infused over 24 h) | 24 | 26 |
| Cefepime | 2 g every 8 h | 50 | 27 |
| Doripenem | 1 g every 8 h (infused over 4 h) | 13 | 28 |
| Imipenem | 1 g every 6 h (infused over 0.5 h) | 12.5 | 29 |
| Levofloxacin | 750 mg every 24 h | 8 | 30 |
| Meropenem | 2 g every 8 h (infused over 3 h) | 20 | 31 |
| Piperacillin-tazobactam | 4.5 g every 6 h (infused over 4 h) | 35 and 7 | 32 |
| Polymyxin B | 30,000 IU/kg/day or at least 1 MIU every 12 h | 2 | 33 |
| Rifampin | 600 mg every 12 h | 4 | 34 |
| Tigecycline | 100 mg every 12 h | 2 | 35 |
IU, international units; MIU, million international units.
To quantify ATP in bacterial samples, the BacTiter-Glo microbial cell viability assay (Promega, Madison, WI, USA) was prepared according to the manufacturer's instructions at the start of each experiment and was employed. Briefly, 100 ml of the BacTiter-Glo buffer was transferred to an amber bottle containing the substrate in order to reconstitute the lyophilized enzyme-substrate reagent after equilibration to room temperature, and the reagent was within 24 h.
Changes in ATP bioluminescence and viable bacterial counts over 24 h.
We described the changes in ATP bioluminescence (measured in relative light units [RLU] per milliliter) and counts obtained by viable plating (measured in CFU per milliliter) for 3 clinical CR-GNB strains (K. pneumoniae KP53879, P. aeruginosa PA14004, and A. baumannii AB8879) in the absence of antibiotics over 24 h. For each strain, an overnight bacterial culture was prepared using cation-adjusted Mueller-Hinton II broth (CA-MHB) and was incubated at 35°C until log-phase growth. The bacteria were further diluted to achieve a final inoculum concentration of approximately 5 log10 CFU/ml (1 × 105 CFU/ml to 5 × 105 CFU/ml), and 24 ml of the suspension was transferred to 50-ml sterile conical flasks. The flasks were then incubated in a shaker water bath at 35°C. Serial samples were obtained from each flask at 0 (baseline), 1, 2, 3, 4, 5, 6, and 24 h after incubation. At each time point, the total ATP content of the sample was determined by the addition of 100 μl BacTiter-Glo assay reagent, and the bioluminescence intensity was recorded using a GloMax integrated luminescence system (Promega, Madison, WI) with a 1-s integration time. Viable counts were also determined by quantitative cultures, performed by dropping serial 10-fold dilutions of the reconstituted samples onto Mueller-Hinton agar (MHA) plates (Thermo Scientific, Singapore) and enumerating bacteria visually after further incubation at 35°C for 18 to 24 h.
Combination testing by ATP bioluminescence and viable plating.
The procedures for combination testing by ATP bioluminescence and viable plating have been described in detail previously (11). All assays were repeated on the same and different days in order to ensure intraday and interday reproducibility of results. Briefly, for 6-h ATP bioluminescence, log-phase bacterial suspensions in CA-MHB were added to 96-well flat-bottom white microtiter plates (Greiner Bio-One, Frickenhausen, Germany) containing 50 μl of the test antibiotic(s) per well so as to obtain a final volume of 100 μl per well (final bacterial concentration, approximately 5 log10 CFU/ml [1 × 105 CFU/ml to 5 ×105 CFU/ml]) and were incubated with agitation at 35°C. At 6 h, the plates were removed; the total ATP content in each well was determined by the addition of the BacTiter-Glo assay reagent; and the bioluminescence intensity was recorded as described above. For 24-h ATP bioluminescence, the same procedure was repeated, but the 96-well plates were removed and measured after 24 h of incubation instead of 6 h.
For viable plating, 100 μl of the log-phase bacterial suspension in CA-MHB was added to each well of 96-well round-bottom clear microtiter plates (Greiner Bio-One, Frickenhausen, Germany) containing 100 μl of the test antibiotic(s) per well so as to obtain a final bacterial concentration of approximately 5 log10 CFU/ml (1 × 105 CFU/ml to 5 ×105 CFU/ml). The plates were incubated at 35°C with agitation for 24 h. At 24 h, samples were obtained from each well and were washed with sterile normal saline to minimize drug carryover. Viable counts were determined as described above. The lower limit of detection for the colony counts was 2.6 log10 CFU/ml. The pharmacodynamic endpoint for determining the efficacy of the antibiotic combination in vitro was the presence of inhibitory activity, defined as any decrease in the colony count on subculture of an organism in the presence of antibiotics from the colony count of the initial inoculum at 24 h.
Determination of inhibitory-noninhibitory thresholds for the 6-h and 24-h ATP bioluminescence assays.
The statistical methods employed to establish thresholds for distinguishing between inhibitory and noninhibitory combinations have been described previously (11). Briefly, for each specimen and antibiotic combination, background RLU values (obtained from blank CA-MHB) were first subtracted to obtain log10-corrected RLU values at time zero. To obtain the change in log10-corrected RLU values (ΔRLU) after exposure to antibiotics, the log10-corrected RLU at time zero was subtracted from the log10-corrected RLU obtained at 6 h (or at 24 h for 24-h ATP bioluminescence testing). To further account for possible differences in growth rates among the different bacterial isolates, the ΔRLU for the no-antibiotic control was subtracted from the ΔRLU for each antibiotic-bacterium combination to create an antibiotic-minus-control difference (ΔΔRLU). Receiver operating characteristic (ROC) curve analysis was carried out using the ΔΔRLU values to establish the optimal luminescence thresholds that distinguish between inhibitory and noninhibitory combinations (TRLU) as determined by viable plating (i.e., if ΔΔRLU values are less than or equal to TRLU, the combination will be classified as inhibitory). Individual ROC curves were also generated against each bacterial species in order to determine if the optimal threshold value differs across species. The presence of an inhibitory activity was defined as any decrease in colony count at 24 h upon subculture of an organism in viable plating, in the presence of antibiotics, when compared to initial inoculum. To compare the performances of the 6-h and 24-h ATP bioluminescence assays, we compared the 6-h and 24-h ROC curves using the DeLong method (15). All statistical analyses were performed using STATA 14 (StataCorp LP, College Station, TX, USA), and a final P value of ≤0.05 was considered to be significant.
Prospective validation of established TRLU.
Eighteen CR-GNB isolates (6 A. baumannii, 6 P. aeruginosa, and 6 K. pneumoniae isolates) were prospectively obtained from Singapore hospitals in 2015 to validate the 6-h and 24-h ATP bioluminescence assays. The isolates were described phenotypically and genotypically as described above. ATP bioluminescence combination testing was first performed at 6 h and 24 h, and each specimen-antibiotic combination was then classified as “inhibitory” or “noninhibitory” using the previously established TRLU. Combination testing via determination of viable counts was then performed as described above to determine the performances (sensitivity, specificity, overall unweighted accuracy, positive predictive value, and negative predictive value with 95% confidence intervals) of the assays in identifying inhibitory and noninhibitory antibiotic combinations.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by a National Medical Research Council Transition Award (TA16may012) and National Medical Research Council Center grants (NMRC/CG/M011/2017 and NMRC/CG/C005B/2017). This study was also supported by the SingHealth Allied Health Research Publication Fund. The funders had no role in study design, data collection, or the interpretation of the work for publication.
We thank the clinical microbiology staff of Singapore General Hospital, in particular Ong Lan Huay, for assistance in the collection of the bacterial isolates.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00183-18.
REFERENCES
- 1.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 2.van Duin D, Bonomo RA. 2016. Ceftazidime/avibactam and ceftolozane/tazobactam: second-generation beta-lactam/beta-lactamase inhibitor combinations. Clin Infect Dis 63:234–241. doi: 10.1093/cid/ciw243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zavascki AP, Bulitta JB, Landersdorfer CB. 2013. Combination therapy for carbapenem-resistant Gram-negative bacteria. Expert Rev Anti Infect Ther 11:1333–1353. doi: 10.1586/14787210.2013.845523. [DOI] [PubMed] [Google Scholar]
- 4.Cai Y, Lee W, Kwa AL. 2015. Polymyxin B versus colistin: an update. Expert Rev Anti Infect Ther 13:1481–1497. doi: 10.1586/14787210.2015.1093933. [DOI] [PubMed] [Google Scholar]
- 5.Pulido MR, Garcia-Quintanilla M, Martin-Pena R, Cisneros JM, McConnell MJ. 2013. Progress on the development of rapid methods for antimicrobial susceptibility testing. J Antimicrob Chemother 68:2710–2717. doi: 10.1093/jac/dkt253. [DOI] [PubMed] [Google Scholar]
- 6.Doern CD. 2014. When does 2 plus 2 equal 5? A review of antimicrobial synergy testing. J Clin Microbiol 52:4124–4128. doi: 10.1128/JCM.01121-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai Y, Chua NG, Lim TP, Teo JQ, Lee W, Kurup A, Koh TH, Tan TT, Kwa AL. 2016. From bench-top to bedside: a prospective in vitro antibiotic combination testing (iACT) service to guide the selection of rationally optimized antimicrobial combinations against extensively drug resistant (XDR) Gram negative bacteria (GNB). PLoS One 11:e0158740. doi: 10.1371/journal.pone.0158740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S, Suppes R, Feinstein D, Zanotti S, Taiberg L, Gurka D, Kumar A, Cheang M. 2006. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 34:1589–1596. doi: 10.1097/01.CCM.0000217961.75225.E9. [DOI] [PubMed] [Google Scholar]
- 9.Wheat PF, Hastings JG, Spencer RC. 1988. Rapid antibiotic susceptibility tests on Enterobacteriaceae by ATP bioluminescence. J Med Microbiol 25:95–99. doi: 10.1099/00222615-25-2-95. [DOI] [PubMed] [Google Scholar]
- 10.Shama G, Malik DJ. 2013. The uses and abuses of rapid bioluminescence-based ATP assays. Int J Hyg Environ Health 216:115–125. doi: 10.1016/j.ijheh.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Cai Y, Leck H, Lim TP, Teo J, Lee W, Hsu LY, Koh TH, Tan TT, Tan TY, Kwa AL. 2015. Using an adenosine triphosphate bioluminescent assay to determine effective antibiotic combinations against carbapenem-resistant Gram negative bacteria within 24 hours. PLoS One 10:e0140446. doi: 10.1371/journal.pone.0140446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chappelle EW, Levin GV. 1968. Use of the firefly bioluminescent reaction for rapid detection and counting of bacteria. Biochem Med 2:41–52. doi: 10.1016/0006-2944(68)90006-9. [DOI] [Google Scholar]
- 13.Ivancic V, Mastali M, Percy N, Gornbein J, Babbitt JT, Li Y, Landaw EM, Bruckner DA, Churchill BM, Haake DA. 2008. Rapid antimicrobial susceptibility determination of uropathogens in clinical urine specimens by use of ATP bioluminescence. J Clin Microbiol 46:1213–1219. doi: 10.1128/JCM.02036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lafond M, Vidal N, Letourneux Y, Brunel JM. 2010. A comparison of three rapid and accurate bioluminescent antibiotic susceptibility tests. J Pharmacol Toxicol Methods 61:16–19. doi: 10.1016/j.vascn.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 15.DeLong ER, DeLong DM, Clarke-Pearson DL. 1988. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44:837–845. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
- 16.Hattori N, Nakajima MO, O'Hara K, Sawai T. 1998. Novel antibiotic susceptibility tests by the ATP-bioluminescence method using filamentous cell treatment. Antimicrob Agents Chemother 42:1406–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kapoor R, Yadav JS. 2010. Development of a rapid ATP bioluminescence assay for biocidal susceptibility testing of rapidly growing mycobacteria. J Clin Microbiol 48:3725–3728. doi: 10.1128/JCM.01482-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isaksson B, Nilsson L, Maller R, Soren L. 1988. Postantibiotic effect of aminoglycosides on gram-negative bacteria evaluated by a new method. J Antimicrob Chemother 22:23–33. doi: 10.1093/jac/22.1.23. [DOI] [PubMed] [Google Scholar]
- 19.Vogel SJ, Tank M, Goodyear N. 2014. Variation in detection limits between bacterial growth phases and precision of an ATP bioluminescence system. Lett Appl Microbiol 58:370–375. doi: 10.1111/lam.12199. [DOI] [PubMed] [Google Scholar]
- 20.Hanberger H, Svensson E, Nilsson M, Nilsson LE, Hornsten EG, Maller R. 1993. Effects of imipenem on Escherichia coli studied using bioluminescence, viable counting and microscopy. J Antimicrob Chemother 31:245–260. doi: 10.1093/jac/31.2.245. [DOI] [PubMed] [Google Scholar]
- 21.Helaine S, Kugelberg E. 2014. Bacterial persisters: formation, eradication, and experimental systems. Trends Microbiol 22:417–424. doi: 10.1016/j.tim.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 22.Li J, Rayner CR, Nation RL, Owen RJ, Spelman D, Tan KE, Liolios L. 2006. Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 50:2946–2950. doi: 10.1128/AAC.00103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teo J, Cai Y, Lim TP, Tan TT, Kwa AL. 2016. Carbapenem resistance in Gram-negative bacteria: the not-so-little problem in the little red dot. Microorganisms 4(1):E13. doi: 10.3390/microorganisms4010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clinical and Laboratory Standards Institute. 2017. Performance standards for antimicrobial susceptibility testing; twenty-seventh informational supplement. CLSI document M100-27. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 25.Voets GM, Fluit AC, Scharringa J, Cohen Stuart J, Leverstein-van Hall MA. 2011. A set of multiplex PCRs for genotypic detection of extended-spectrum beta-lactamases, carbapenemases, plasmid-mediated AmpC beta-lactamases and OXA beta-lactamases. Int J Antimicrob Agents 37:356–359. doi: 10.1016/j.ijantimicag.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 26.Tod M, Lortholary O, Seytre D, Semaoun R, Uzzan B, Guillevin L, Casassus P, Petitjean O. 1998. Population pharmacokinetic study of amikacin administered once or twice daily to febrile, severely neutropenic adults. Antimicrob Agents Chemother 42:849–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LaPlante KL, Sakoulas G. 2009. Evaluating aztreonam and ceftazidime pharmacodynamics with Escherichia coli in combination with daptomycin, linezolid, or vancomycin in an in vitro pharmacodynamic model. Antimicrob Agents Chemother 53:4549–4555. doi: 10.1128/AAC.00180-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tam VH, McKinnon PS, Akins RL, Drusano GL, Rybak MJ. 2003. Pharmacokinetics and pharmacodynamics of cefepime in patients with various degrees of renal function. Antimicrob Agents Chemother 47:1853–1861. doi: 10.1128/AAC.47.6.1853-1861.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaruratanasirikul S, Wongpoowarak W, Kositpantawong N, Aeinlang N, Jullangkoon M. 2012. Pharmacodynamics of doripenem in critically ill patients with ventilator-associated Gram-negative bacilli pneumonia. Int J Antimicrob Agents 40:434–439. doi: 10.1016/j.ijantimicag.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 30.Sakka SG, Glauner AK, Bulitta JB, Kinzig-Schippers M, Pfister W, Drusano GL, Sorgel F. 2007. Population pharmacokinetics and pharmacodynamics of continuous versus short-term infusion of imipenem-cilastatin in critically ill patients in a randomized, controlled trial. Antimicrob Agents Chemother 51:3304–3310. doi: 10.1128/AAC.01318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sánchez Navarro A, Colino Gandarillas CI, Alvarez Lerma F, Menacho YA, Dominguez-Gil A. 2005. Pharmacokinetics and pharmacodynamics of levofloxacin in intensive care patients. Clin Pharmacokinet 44:627–635. doi: 10.2165/00003088-200544060-00004. [DOI] [PubMed] [Google Scholar]
- 32.Tam VH, Schilling AN, Nikolaou M. 2005. Modelling time-kill studies to discern the pharmacodynamics of meropenem. J Antimicrob Chemother 55:699–706. doi: 10.1093/jac/dki086. [DOI] [PubMed] [Google Scholar]
- 33.Shea KM, Cheatham SC, Wack MF, Smith DW, Sowinski KM, Kays MB. 2009. Steady-state pharmacokinetics and pharmacodynamics of piperacillin/tazobactam administered by prolonged infusion in hospitalised patients. Int J Antimicrob Agents 34:429–433. doi: 10.1016/j.ijantimicag.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 34.Sandri AM, Landersdorfer CB, Jacob J, Boniatti MM, Dalarosa MG, Falci DR, Behle TF, Bordinhao RC, Wang J, Forrest A, Nation RL, Li J, Zavascki AP. 2013. Population pharmacokinetics of intravenous polymyxin B in critically ill patients: implications for selection of dosage regimens. Clin Infect Dis 57:524–531. doi: 10.1093/cid/cit334. [DOI] [PubMed] [Google Scholar]
- 35.Gumbo T, Louie A, Deziel MR, Liu W, Parsons LM, Salfinger M, Drusano GL. 2007. Concentration-dependent Mycobacterium tuberculosis killing and prevention of resistance by rifampin. Antimicrob Agents Chemother 51:3781–3788. doi: 10.1128/AAC.01533-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodvold KA, Gotfried MH, Cwik M, Korth-Bradley JM, Dukart G, Ellis-Grosse EJ. 2006. Serum, tissue and body fluid concentrations of tigecycline after a single 100 mg dose. J Antimicrob Chemother 58:1221–1229. doi: 10.1093/jac/dkl403. [DOI] [PubMed] [Google Scholar]
- 37.Wadhwani T, Desai K, Patel D, Lawani D, Bahaley P, Joshi P, Kothari V.. October 2009. Effect of various solvents on bacterial growth in context of determining MIC of various antimicrobials. Internet J Microbiol 7(1):9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.