Abstract
Photolyases and cryptochromes form an almost ubiquitous family of blue light photoreceptors involved in the repair and maintenance of DNA integrity or regulatory control. We found that one cryptochrome from the green alga Chlamydomonas reinhardtii (CraCRY) is capable of both, control of transcript levels and the sexual cycle of the alga in a positive (germination) and negative manner (mating ability), as well as catalyzing the repair of UV-DNA lesions. Its 1.6 Å crystal structure shows besides the FAD chromophore an aromatic tetrad that is indispensable in animal-like type I cryptochromes for light-driven change of their signaling-active redox state and formation of a stable radical pair. Given CraCRY’s catalytic activity as (6-4) photolyase in vivo and in vitro, we present the first co-crystal structure of a cryptochrome with duplex DNA comprising a (6-4) pyrimidine–pyrimidone lesion. This 2.9 Å structure reveals a distinct conformation for the catalytic histidine His1, H357, that challenges previous models of a single-photon driven (6-4) photolyase mechanism.
INTRODUCTION
The photolyase/cryptochrome superfamily (PCSf) is a large family of photoactive enzymes, which occurs in all three domains of life. They are roughly classified by their function either as photolyases (PHL) or as cryptochromes (CRY). Photolyases repair UV-induced DNA lesions, namely either cyclobutane-pyrimidine dimers (CPD) or pyrimidine-(6-4)-pyrimidone photoadducts, (6-4)PP. Their close relatives, the cryptochromes, have mostly lost DNA repair functionality and act as signaling proteins for coupling various biological responses to light input, e.g. plant growth and development, initiation of flowering, or as triggers for the entrainment of circadian rhythms in plants and animals (1). CRYs are furthermore separated into the animal types I and II, plant CRYs and CRY-DASH photoreceptors (DASH: Drosophila, Arabidopsis, Synechocystis, Homo). During the last decade, several animal-like CRYs (aCRY) have been discovered and characterized outside the animal kingdom, especially in photosynthetic algae (2,3), where they may act not only in signaling, but also as (6-4) photolyases.
The green alga Chlamydomonas reinhardtii harbors one aCRY ortholog (CraCRY) and a plant CRY besides two DASH-type cryptochromes (4,5). Interestingly, although C. reinhardtii has a CPD class II photolyase, it lacks a further (6-4) photolyase as well as red light-dependent phytochromes (4–6). The photolyase-homology region (PHR) of CraCRY shows high sequence identity (40–52%) with the aCRY orthologs OtCPF1 from the green alga Ostreococcus tauri, PtCPF1 from the diatom Phaeodactylum tricornutum and an assigned (6-4) photolyase from the green algae Dunaliella salina (SI-S1). At least the diatom cryptochrome PtCPF1 is bifunctional as shown by its in vitro (6-4) photolyase activity and in vivo function as transcriptional regulator (7). Surprisingly, CraCRY regulates the transcript levels of various genes not only upon blue light due to absorption by its oxidized FAD chromophore but also upon yellow and red light as revealed by in vivo studies with a knock-down mutant (5). The latter suggests a light sensor function of the chromophore's semiquinoid FADH° state (8). However, till now, there has not yet been identified any direct interaction partner of CraCRY for downstream signaling. Unlike conventional photolyases, CraCRY harbors like other cryptochromes an elongated C-terminal extension (CTE, A457-E595) as an additional feature besides its light-receptive PHR. These CTE are generally highly variable in length and sequence in cryptochromes and predicted to be mostly disordered in the absence of an interaction partner. CTE are hence generally hypothesized to be key elements of cryptochrome-dependent signal transduction (9). For example, in plant cryptochromes the CTE are targets for extensive blue-light dependent phosphorylation, which is triggered by dimerization of the PHR domain in the lit, i.e. semiquinoid, state (10). Interestingly, the CTE of CraCRY was supposed to fulfill a similar function in homo-dimerization (11), although no direct evidence of its conformational change upon blue or red light illumination has been shown till now.
Almost all members of the PCSf undergo a blue light mediated reaction called photoactivation (1), in which the catalytically inactive fully-oxidized (FADox) or semi-reduced (FADH°) states abstract an electron from a nearby tryptophan upon photoexcitation. Exceptions may be represented by type-II cryptochromes, which act as light-unresponsive transcription-repressing enzymes in mouse, human and other vertebrates (1,12). Here, photoreduction can be observed under artificial in vitro conditions, but has not been proven to be physiologically relevant. In the reduced state the flavin chromophore acts either in DNA repair (FADH―) after further light absorption or in triggering signal transduction (FADH°). In CraCRY the light-dependent photoreduction process can be easily observed in vitro, starting from the fully oxidized (FADox) through the neutral radical (FADH°) up to the fully reduced form (FADH−). A common feature of PCSf orthologs is the presence of an aromatic triad for the fast replenishment of an electron from the protein surface. This intramolecular electron transfer (ET) pathway is elongated in CraCRY and related type I animal cryptochromes by a fourth aromatic residue (11,13), which can be either a tyrosine (CraCRY: Y373) or a tryptophan, W394 in the cryptochrome of Drosophila melanogaster. This elongated ET pathway is crucial not only for the photoreduction reactions of aCRY orthologs but also for the function of (6-4) photolyases (14). At least in CraCRY, the life time of the terminal Y373-O° radical is unusually long with 26 ms after photoreduction to the FADH° state (13) and even 2.6 s upon light-driven formation of the FADH― state (11). As stable tyrosyl radicals such as that of photosystem II or class I ribonucleotide reductases are known to reside in well-shielded protein environments the structural base of the relative longevity of the Y373-O° radical is unclear given its predicted proximity to the protein surface.
All photolyases and cryptochromes share a conserved bilobal architecture, with an FAD chromophore bound to their C-terminal, all α-helical domain in an unusual U-shaped conformation (15). The C-terminal domain is hence responsible for either providing DNA-interaction sites in photolyases or light-dependent/independent interaction sites for CTE or other down-stream signaling partners in cryptochromes. The function of the N-terminal domain appears often to bind a second chromophore as light-harvesting antenna for broadening absorption in the visible spectral region (1). However, this domain has been recently found to provide specific interaction sites for signaling partners in type II cryptochromes (16). Antenna chromophores identified so far include 5,10-methenyltetrahydrofolate (MTHF) (17,18), 8-hydroxydeazaflavin (8-HDF) (19), flavin mononucleotide (FMN) (20), 6,7-dimethyl-8-ribityllumazine (DMLR) (21) and FAD in its fully oxidized state (22). The nature of the antenna chromophore in non-animal aCRYs is still unknown, e.g. CraCRY lacks an additional chromophore besides FAD when overproduced in Escherichia coli (5,11,13).
In this study, we show that CraCRY indeed represents a bifunctional cryptochrome due to its additional in vivo and in vitro function as a (6-4) photolyase. Like for the class II CPD photolyase from C. reinhardtii the blue light sensitivity of CraCRY is boostered by its capability to incorporate 8-HDF as antenna chromophore. The co-crystal structure of CraCRY with duplex DNA comprising a (6-4)PP shows not only a higher degree of variable bending of duplex DNA when bound to this type of photolyase, but also a distinct conformation for a catalytically critical histidine within the active site. The latter is clearly inconsistent with several recent theoretical models of the (6-4) photolyase mechanism, but suggests instead a repair reaction via an oxetane intermediate. Furthermore, the structure of CraCRY shows an environment for shielding the terminus of the aromatic tetrad, which rationalizes the unusually long lifetime of the tyrosine radical state formed upon photoactivation.
MATERIALS AND METHODS
Multiple sequence alignment
1186 orthologs of the animal-like cryptochromes/(6-4) photolyases were derived from a sequence-similarity network generated on the whole photolyase-cryptochrome superfamily (23); their pairwise sequence identity for non-redundancy was restricted to <90% leaving 541 sequences for multiple-sequence alignment by Clustal Omega. The resulting alignment was used in WebLogo 3 (24) for visualization with the CraCRY sequence as reference.
Co-expression of CraCRYΔCTE·8-HDF and CraCRY·8-HDF
Escherichia coli BL21(DE3) Gold cells (Stratagene) were transformed with either pET28a-CraCRY (5) or pET28a-CraCRYΔCTE (11) and the cofactor plasmid pCDF-His6FbiC, encoding the FO synthase (7,8-didemethyl-8-hydroxy-5-deazariboflavin synthase) of Streptomyces coelicolor, following published procedures (25).
Expression and purification
Expression and purification of all CraCRY variants were carried out following a published protocol (5). An additional heparin affinity chromatography was performed, which was washed with four column volumes of buffer containing 50 mM sodium phosphate, pH 7.8 and 20% (v/v) glycerol before applying the protein solution. It was eluted with a salt gradient reaching a concentration of 2 M sodium chloride. For crystallization, the protein was further purified by size exclusion chromatography (Superdex 200, 120 ml CV) in 20 mM Tris, 200 mM NaCl, pH 8.5.
Crystallization and structure determination of CraCRYΔCTE
For crystallization CraCRYΔCTE was incubated overnight in the dark and pipetted under safe light using a crystallization robot (Digilab Honey Bee 963™) for screening of different conditions. Initial crystals were grown in 0.1 M 2-(N-morpholino)ethanesulfonic acid (MES), pH 5.6 and 35% PEG 4000 at 4°C. Crystals were then further improved by micro-seeding, and grown in 0.1 MES pH 5.6, 35% PEG3350. Crystals were soaked with 30% glycerol as a cryoprotectant and flash-frozen in liquid nitrogen. Diffraction data collection from a single crystal at 100 K was taking place at the BESSY II synchrotron light source in Berlin, or, alternatively, at the Taiwan Photon Source 05A microcrystallography beamline, National Synchrotron Radiation Research Center in Hsinchu. Diffraction images were indexed by XDS (26) with datasets derived form different crystals merged via a slightly modified version of the KAMO protocol (27). Briefly, datasets were first manually processed with XDS. Then, datasets were hierarchically clustered by their unit-cell parameters via the BLEND software within the CCP4 software suite. Next, clusters were merged via XSCALE (26). Finally, cluster data quality was assessed hierarchically by their 50% correlation coefficients (28). The structure was solved via molecular replacement (Phaser MR (29)) using a homology model that derived from the (6-4) photolyase of Arabidopsis thaliana (PDB: 3FY4). Refinement was performed by using a combination of REFMAC5 (CCP4 package (30,31)), phenix.refine (PHENIX) and COOT (32). Data processing and refinement statistics are presented in Table 1.
Table 1.
Data collection and refinement statistics
| CraCRYΔ496* 5ZM0 | CraCRYΔ496 6FN3 | CraCRYΔ496•8-HDF 6FN2 | CraCRYΔ496•(6-4) DNA 6FN0 | |
|---|---|---|---|---|
| Data collection and processing | ||||
| X-ray source, beamline | BESSY II Berlin, 14.3 | BESSY II Berlin, 14.3 | ESRF, Grenoble, | ESRF, Grenoble |
| NSRRC, Hsinchu, 05A | ID23-2 | ID23-1 | ||
| Detector | MX-225 and MX-300 | MX-225 | Pilatus6m | Pilatus6m |
| Wavelength (Å) | 0.894 and 0.998 | 0.894 | 0.992 | 1.033 |
| Space group | P212121 | P212121 | P212121 | P42212 |
| Cell dimensions (a, b, c) | 50.21, 65.12, 151.98 | 151.80, 50.13, 65.07 | 150.25, 50.53, 64.97 | 146.24, 146.24, 67.44 |
| Resolution (Å) | 39.8–1.60 (1.66–1.60) | 75.9–1.9 (2.0–1.9) | 29.8–2.3 (2.4–2.3) | 103.4–2.9 (3.0–2.9) |
| Total reflections‡ | 886 219 (66 592) | 74 526 (7462) | 43 054 (4233) | 33 333 (3269) |
| Multiplicity‡ | 13.3 (10.2) | 2.0 (2.0) | 2.0 (2.0) | 2.0 (2.0) |
| Unique reflections‡ | 66 645 (6554) | 38 187 (3803) | 22 003 (2159) | 16 737 (1639) |
| Completeness (%)‡ | 100 (100) | 95.5 (96.6) | 96.6 (97.2) | 99.8 (99.9) |
| R merge *,‡ | 0.275 (1.68) | 0.058 (0.211) | 0.070 (0.205) | 0.050 (0.307) |
| CC1/2 *,‡ | 0.992 (0.506) | |||
| I/σ(I) ‡ | 8.5 (1.3) | 13.7 (4.4) | 7.2 (3.5) | 12.5 (2.7) |
| Mosaicity (°) | 0.150 | 0.15 | 1.53 | 0.134 |
| Wilson B-factor (Å2) | 10.01 | 11.74 | 19.75 | 61.95 |
| Refinement | ||||
| Resolution (Å) | 39.8–1.6 | 75.9–1.9 | 29.8–2.3 | 61.2–2.9 |
| R factor, Rfree | 0.159, 0.188 | 0.163, 0.206 | 0.170, 0.228 | 0.155, 0.215 |
| Reflections (working, test set) | 66 645, 3302 | 37 003, 1184 | 21 265, 682 | 16 201, 536 |
| Completeness for range (%) | 100 | 95.6 | 96.5 | 99.8 |
| r.m.s.d. from ideal: | ||||
| Bond lengths (Å) | 0.008 | 0.009 | 0.009 | 0.010 |
| Bond angles (°) | 1.280 | 1.416 | 1.436 | 1.507 |
| Total number of atoms | 4907 | 4527 | 4306 | 4754 |
| Mean B-value (Å2) | 15.0 | 13.6 | 21.5 | 63.8 |
*5ZM0 corresponds to a composite dataset based on a total of four crystals collected at BESSY II and NSRRC achieving a multiplicity of 13.29. As described before (28), high multiplicity datasets suffer profoundly from Rmerge-derived resolution cutoffs. Instead, the 50% correlation coefficient (CC1/2) should be used (28). In this case, the maximum resolution cutoff was determined by a minimal CC1/2 value of 0.5 at the highest resolution shell.
‡Values in parentheses correspond to highest resolution shell.
Crystallization of CraCRYΔCTE·8-HDF
Isomorphous crystals of the CraCRYΔCTE·8-HDF complex were grown in the dark at 18°C in 0.1 M MES, pH 6.0 with 15% PEG 5000. The crystals were mounted with 30% glycerol as cryoprotectant and flash-frozen in liquid nitrogen. Diffraction data were collected at 100 K from a single crystal at beamline ID23-2 in the European Synchrotron Radiation Facility (ESRF). Diffraction images were processed as described before.
Crystallization of CraCRYΔCTE in complex with DNA
For crystallizing the complex with (6-4)PP damaged DNA a synthetic DNA strand 5′-CAG CGG T(6-4)TG CCG TG-3′ was hybridized with its counter strand 3′-GTC GCC AAC GGC AC-5′ (96°C, shaking 10 min at 300 rpm) resulting in a blunt-ended double strand. Purified CraCRY was concentrated up to 10 mg/ml and incubated for at least 2 h in the dark, before the ds(6-4)DNA was added in 1.25 folded excess under safe light. Initial crystals grew under conditions with 0.1 M MES, PEG 6000 and different pH as well as precipitant concentrations. These conditions were used for further optimization screens, which resulted in crystals grown in dark at 4°C using 0.1 M MES, pH 5.5 and 10% (w/v) PEG 6000. Crystals were mounted with 30% glycerol as cryoprotectant and flash-frozen in liquid nitrogen. Diffraction data were collected at beamline ID23-1 at the ESRF resulting in 720 processed diffraction images.
Steady-state UV/Vis spectroscopy and photoreduction assay
Absorption spectra of CraCRY variants were recorded using a V-660 spectrometer (JASCO). The protein solutions were measured in buffer containing 50 mM NaH2PO4 pH 7.8, 100 mM NaCl and 20% (v/v) glycerol. The samples were incubated for 5 min in the dark before first spectra were recorded. Spectra were measured after different illumination times using a high power LED 450 nm (9.7 mW cm−2 at a distance of 10 cm, Roithner Lasertechnik) at 10°C.
Generation of (6-4)PP and in vitro photorepair assay
The in vitro repair assay of wild type CraCRY was done following published procedures (33). All steps were carried out under safe light (red) conditions. To generate fully reduced CraCRY capable of photorepair, samples of 120 μM protein solution were illuminated at a distance of 5.5 cm for 60 min by an LED (λmax = 450 nm, 9.7 mW cm−2; Roithner Lasertechnik) in the presence of 25 mM DTT. The (6-4)PP repair assay was performed with 39.5 μM of the irradiated oligo (dT)18, 10 μM of fully reduced CraCRY and 25 mM DTT. The mixture was illuminated at a λmax of 385 nm (LED, Roithner Lasertechnik; 3.5 mW cm−2 at 5.5 cm) and 15°C for 60 min. For control experiments, assays were performed without illumination and without CraCRY, respectively. A Maya 2000 Pro spectrometer (Ocean Optics) was used for recording of UV/Vis absorption spectra.
In vivo photorepair assay
The following Chlamydomonas reinhardtii strains were used: wild type SAG73.72 (mt+), a knockdown acry mutant SAG73.72:acry1A (acrymut (5)) and a complemented strain (acrycompl) that has been transformed with vector pKP39 (5). The strain acrycompl, was analyzed by immunoblots using anti-aCRY antibodies and showed a rescue of CraCRY protein levels of ∼140% as compared to wild-type (100%; SI-S2b). UV-B survival assays were modified from Petersen and Ronan, 2010. Cells were grown under a 12-h light/12-h dark cycle (LD12:12) at a light intensity of 75 μmol photons m−2 s−1 and 23°C in Tris-acetate phosphate (TAP) medium (Harris, 1989) until they reached a cell density of 4–6 × 106 cells/ml. Cells were normalized to 2.6 × 106 cells/ml, three tenfold dilutions were made and 10 μl of each dilution were spotted onto TAP plates. The plates were either non UV-B treated or illuminated for 0.5, 2 and 3 min, respectively, with an UV-B lamp (XX-15M model from UVP) at an intensity of 2.2 mW/cm2 with a peak at 312 nm as experimentally determined. The UV-B lamp was pre-run for 20 min to ensure stability in the UV spectrum and the energy output, which was measured with an UV-X radiometer (UVP). Afterwards the plates were immediately placed under the above described LD conditions with cool-white fluorescent light at an intensity of 75 μmol photons m−2 s−1 for recovery. After 7 days of cell growth under these conditions, pictures of the plates were taken.
RESULTS
Overall architecture of CraCRY and its chromophore binding sites
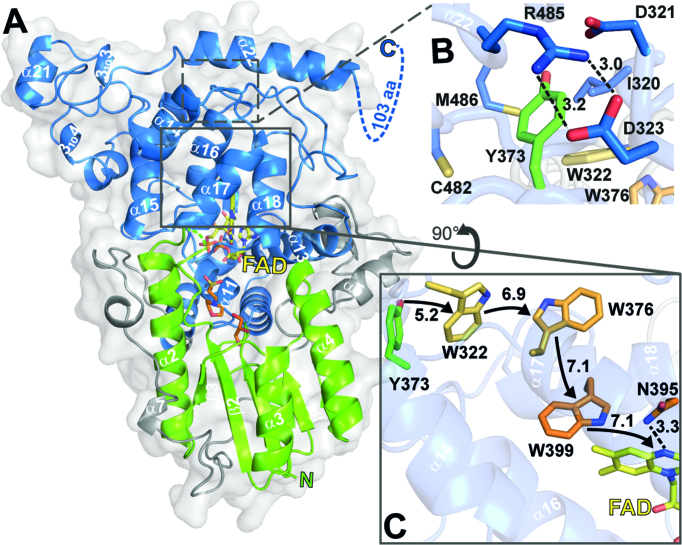
For structure determination of CraCRY, its wild type (WT) and the CTE deletion mutant (CraCRYΔCTE) were screened and optimized for crystallization. Despite its homogenous characteristics full-length CraCRY failed to crystallize, most likely due to a disordered CTE, whereas we obtained well-ordered orthorhombic crystals of CraCRYΔCTE diffracting up to 1.6 Å resolution (Table 1). The CraCRYΔCTE structure (Figure 1) displays the typical two-domain architecture, which is common for all members of the PCSf. In this bilobal structure, a typical Rossmann fold is defined by the N-terminal domain (K5-V129). This domain lacks an antenna chromophore but harbors in its putative binding site two additive molecules, 2-(N-morpholino)-ethanesulfonic acid (MES) and glycerol (GOL), from the crystallization condition (Figure 1A). The catalytic C-terminal domain harbors the flavin cofactor FAD in a U-shaped conformation like other PCSf members (18,34–36). The N5 nitrogen of the oxidized isoalloxazine moiety forms a rather weak hydrogen bond with N395 as judged by the N5-ND2 distance (3.3 Å). This asparagine or a structurally corresponding residue is known to control the photochemistry of flavins in all photolyases and cryptochromes and to be well conserved within all members of the PCSf capable of DNA repair apart class II and bacterial (6-4) photolyases (17,18,37). Despite this role, N395 is unsuitable to act as a proton donor for the FAD°― intermediate formed after photoreduction of the FAD chromophore because of its high pKa value (∼25,5 (38)). As no other deprotonatable residues are close to the N5 nitrogen, the proton is probably derived from the solvent. Here, N395 may be a gate keeper to the solvent because it forms a second H-bond with a surface-bound water that is coordinated to the surface-exposed residues E384 and Q390. This water is conserved in the structure of the D. melanogaster (6-4) photolyase as well, but replaced in cryptochromes failing to achieve the semiquinoid FADH° state like DmCRY by a hydrophobic leucine moiety.
Figure 1.
Cartoon representation of CraCRY structure with highlighted regions of interest. (A) Overall structure of CraCRYΔCTE (6FN3) with FAD (yellow) bound to the C-terminal domain (blue). The C- and N-terminal domain (green) are connected through a flexible loop region (grey). The antenna binding site is occupied by glycerol (GOL, orange) and 2-(N-morpholino) ethanesulfonic acid (MES, orange). The C-terminal extension (494–595) is displayed as dotted lines. (B) Detail environment of the fourth aromatic residue, Y373. (C) The electron transfer pathway of CraCRY is marked with arrows, centroid-centroid distances are shown in Å. The Trp triad is shown in orange and the fourth aromatic residue, a terminal tyrosine (Y373), is shown in green. Also the potential proton donorof the FAD, N395, is presented as sticks.
The tryptophan triad that is commonly found in the PCSf was identified as W399, W376 and W322 by structural comparison and multiple sequence alignment. These three aromatic residues catalyze ET from the surface of the protein to the excited flavin chromophore (39,40). In CraCRY this ET pathway is elongated by Y373 to an aromatic tetrad (Figure 1C) (11,13). The structure shows that Y373, which is only 5.5 Å distant from the third tryptophan, W322, resides close to the protein surface. Its side chain is surrounded by a network of charged residues, most prominently by a salt bridge formed between D323 and R485 (Figure 1B), which appears to stabilize the formation of the long-lived radical by shielding the deprotonated Tyr-O° from solvent access. Rapid deprotonation of the terminal tyrosyl radical may be supported by D321 as proton acceptor due to its H-bond formation with the Y373 hydroxyl group (2.9 Å). Like R485, two other residues contacting the phenolic moiety of Y373, C482 and M486 are part of the long C-terminal helix α22 (H475-K494, Figure 1A, B). Mutagenesis of C482 was found to affect red-light dependent, FADH°→ FADH― driven dimerization of CraCRY under in vitro conditions, maybe due to the ablation of disulfide bond formation (11).
CraCRY shows DNA repair activity in vitro and in vivo
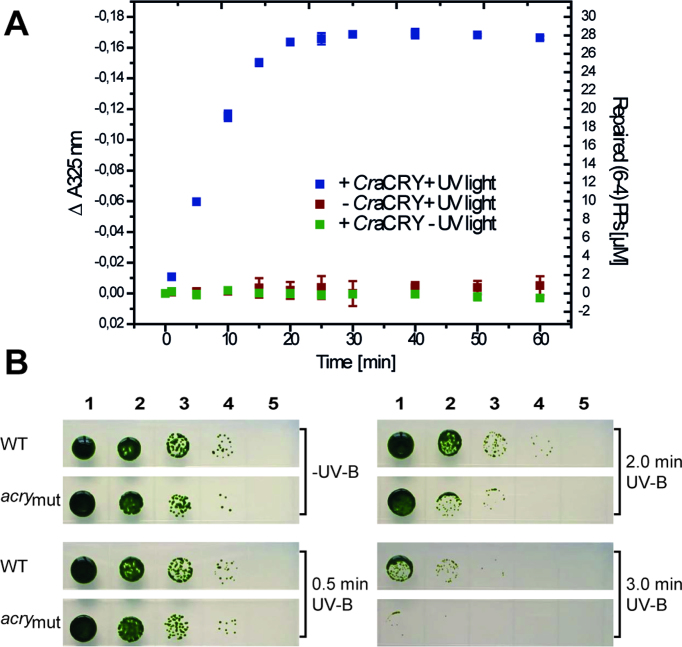
To address, whether CraCRY can act as (6-4) photolyase, we performed in vitro repair activity assays as established before (33,41). Incubation of the catalytically competent CraCRY in its FADH― state with a (6-4)PP comprising oligo(dT)18 shows, that illumination at 385 nm causes decreasing absorption at 325 nm, which corresponds to repair of the (6-4)PP (Figure 2A). From the specific decrease, we calculated that after 60 min of illumination almost 70% of the single-stranded DNA with (6-4)PP has been repaired. These data lead to the conclusion that CraCRY may act as efficient (6-4) photolyase in Chlamydomonas. To analyze the in vivo activity of CraCRY by an UV-B survival assay, we used the following C. reinhardtii strains: wild type SAG73.72 (mt+), the knockdown acry mutant SAG73.72:acry1A (acrymut (5)) and a complemented strain of acrymut that has been transformed with vector pKP39 (acrycompl (5)). The different algae strains were exposed to UV-B light for certain times and grown under white light within a light-dark cycle for 7 days. As predicted, survival of the knockdown mutant (CraCRY expression level: ∼20% of WT) decreases with longer UV-B illumination times (0.5, 2.0 and 3.0 min) when compared to WT (Figure 2B). In contrast, the complemented strain acrycompl (CraCRY expression level: ∼140% of WT) shows WT-like resilience against UV-B light even when illuminated for 3.0 min (SI-S2A). These results correlate with the in vitro data and confirm the important role of the animal-like cryptochrome as a repair enzyme for UV-B induced DNA lesions in C. reinhardtii.
Figure 2.
In vitro and in vivo repair of (6-4)PP by CraCRY. (A) In vitro DNA repair activity assay with 30 μM of the irradiated oligo-(dT)18, abbreviated as (6-4)PPs, 10 μM of CraCRY in the FADH− state and 25 mM DTT. Controls are shown without CraCRY or illumination. (B) UV-B survival of C. reinhardtii is reduced in the acry mutant compared to WT. Equal numbers of cells starting from 2.6 × 106 cells/ml (lane 1) and three subsequent tenfold dilutions (lanes 2–4) or no cells (lane 5) were spotted onto TAP plates and illuminated for 0.5, 2 and 3 min, respectively, with 2.2 mW/cm2 UV-B light. Cells from wild type (WT) and an acry knockdown mutant (acrymut) with a reduced rate down to ∼20% compared to WT were used for the assay. A plate with no UV-B treatment was used as growth control (-UV-B). Pictures were taken after 7 days of growth.
Structural basis for (6-4)PP repair by CraCRY
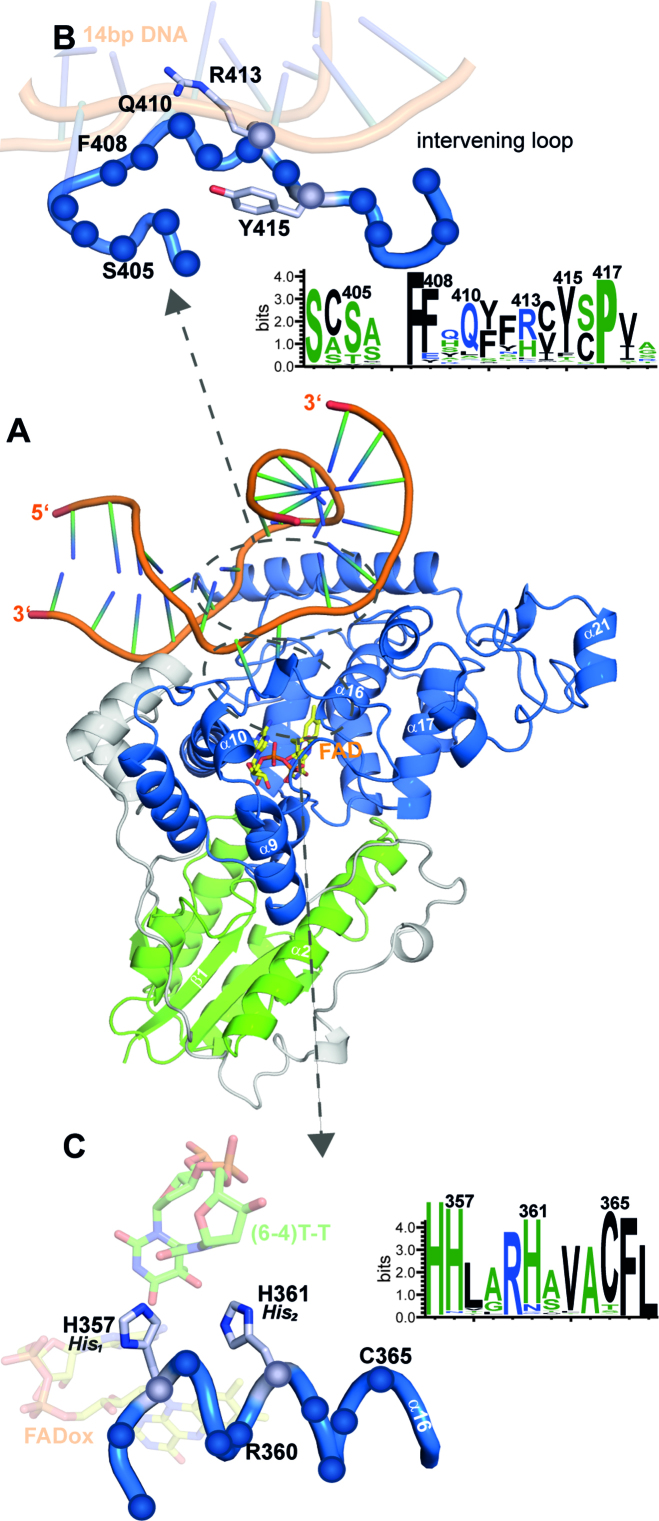
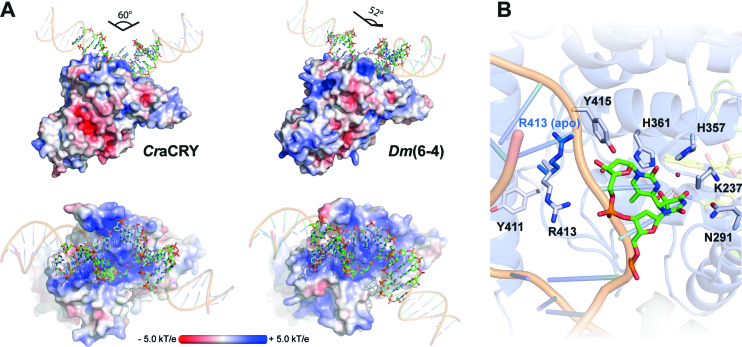
Co-crystals of CraCRYΔCTE in complex with blunt end 14mer duplex DNA diffracted to a resolution of 2.9 Å. The CraCRYΔCTE/(6-4)PP-DNA complex shows a highly kinked duplex DNA bound to the C-terminal domain, with the (6-4)PP flipped into the active site (Figure 3A) thus giving rise to a large unpaired bubble within the duplex DNA. With a kinking angle of 60°CraCRY causes stronger bending of (6-4)PP-comprising duplex DNA than the previously characterized structures of D. melanogaster (6-4) photolyase/DNA complexes (52°, 3CVU). When comparing CraCRY and the Dm(6-4) the conformation of the lesion-comprising DNA strand is almost the same for the bound (6-4)PP and the backbone between the −1 and +3 phosphates (Figure 4A). However, conformational differences between DNA bound to CraCRY and Dm(6-4) are not only observed for the flanking ends of the duplexes, but also for the counter bases of the thymines corresponding to the (6-4)PP (SI-S3B).
Figure 3.
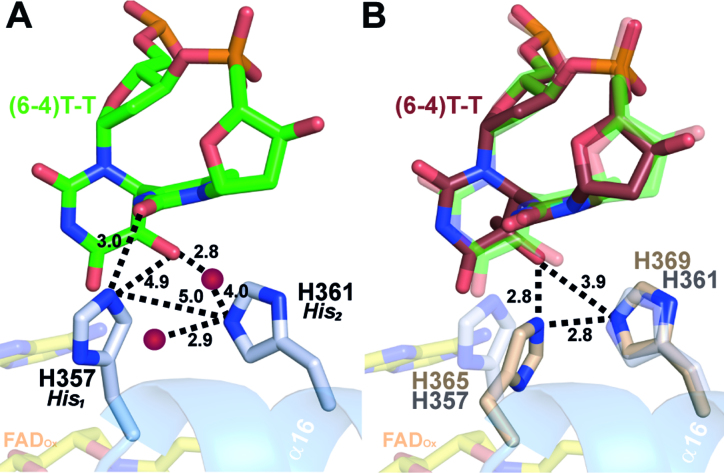
Co-crystal structure of CraCRY in complex with (6-4)PP comprising duplex DNA. (A) Overall structure of CraCRYΔCTE with 14 bp duplex DNA (6FN0) illustrated as cartoon model. (B) Detailed view of the bubble-intruding region of CraCRY highlighting arginine residue R413. In contrast to Dm(6-4) photolyase this residue appears to form only a distant salt bridge (3.9 Å versus 3.1 Å) with the intra-lesion phosphodiester group of the (6-4)PP. Conservation of the BIR within the animal-like cryptochromes/(6-4) photolyases was obtained from 541 non-redundant sequences with less than 90% pairwise sequence identity. The size of the letter correlates with the degree of conservation, for details refer to Supplementary Figure S4. (C) Detailed view of helix α16, which harbors the catalytic histidine residues H357 (His1) and H361 (His2), nomenclature in parentheses according to Yamamoto et al. (43). The HHLARH motif commonly found in (6-4) photolyases is also part of the CraCRYΔCTE active site.
Figure 4.
(A) Comparison of the (6-4)PP damaged duplex DNA binding to D. melanogaster (6-4) photolyase (3CVU) and CraCRY (6FN0). Electrostatic potentials were calculated with APBS (c = 0.1 M, solvent radius = 1.4 Å). The DNA ends were prolonged by regular B-type DNA using PyMOL and the overall binding angles of the DNA therefrom derived. (B) Detailed view of the (6-4)PP binding depicting the flipping of R413 upon DNA binding. The side chain of R413 in the unbound CraCRY structure is shown in blue.
The wider opening of the double strand along the lesion site in the CraCRYΔCTE/(6-4)PP-DNA complex and the flipping of the damaged DNA is supported by a bubble-intruding region (BIR, S409-R413) that is part of the loop linking the α18 and α19 helices. The BIR intervenes between the double strand by forming hydrophobic interactions between the aromatic side chain of F412 and a cytosine preceding the adenine counter bases (π–π interaction) as well as between R413 and the guanine base (CH–π) that is 3′ of the thymine residues of the (6-4)PP. The latter residue, whose side chain conformation depends on DNA binding (Figure 4B), is highly conserved in most (6-4) photolyases and forms Coulombic interactions with the intra-lesion and the 3′ phosphate groups. QM/MM calculations and mutagenesis of Xenopus laevis (6-4) photolyase, Xl(6-4), as shown in an accompanying paper (42) corroborate the notion that the Coulombic R410-phosphate (R410 in Xl(6-4) corresponding to R413 in CraCRY) interactions serve as anchor during early formation of the CraCRY/DNA complex, whereas the CH–π interaction with the base following the (6-4)PP are required to form the maturely complex. Due to the local opening of the DNA duplex the (6-4)PP enters the active pocket for interacting directly with the FAD. There, the photoproduct forms a hydrogen bond (2.8 Å) between the C4 carbonyl group of the 5′-thymine moiety with the N6-amino group of the FAD’s adenine. A glutamine (Q291) provides specific interactions with the 5′-thymine via hydrogen bonds. The 3′-thymine is stabilized by a water network (Figure 5A) also involving two highly conserved histidine residues (Figure 3C).
Figure 5.
Conformation of the active site histidines in (6-4) photolyases. (A) Enhanced view of the (6-4)PP interactions with the two prominent residues H357 (His1) and H361 (His2) of CraCRY (gray). Distances (in Å) were measured between the histidines, the DNA lesion and two water atoms (red). (B) Comparison of the active site of CraCRY (6FN0, gray) with Dm(6-4) (3CVU, wheat) (r.m.s.d. 0.658 Å). The (6-4)PP bound to Dm(6-4) (red) is slightly tilted compared to the (6-4)PP bound to CraCRY (green).
Both catalytically crucial histidines, H357 (His1) and H361 (His2, for nomenclature see Yamamoto et al. (43)), and the (6-4)PP are clearly defined by OMIT electron density maps. His2 of CraCRY shows the exact position as H369 in Dm(6-4) and forms with its imidazole side chain two hydrogen bonds (Figure 5). One is formed via its Nϵ2 atom to a nearby tyrosine, Y415 (2.7 Å; Dm(6-4): Y423), the other via Nδ1 to water molecule WAT2, which is centrally located within the active site by bridging both to the O4 carbonyl and 5-hydroxyl groups of the (6-4)PP as well as to the N6 amino group. The important difference is presented by the conformation of the His1 side chain. In the CraCRYΔCTE/(6-4)PP-DNA complex His1 adopts a gauche― rotamer for its χ1 torsion (77.3°), whereas the corresponding residue in the Dm(6-4)/(6-4)PP complex, H365, shows a trans conformer (177.2°). Consequently, His1/H357 points in CraCRY towards the preceding histidine, H356, and the 3′ base of the (6-4)PP, but away from the adenine moiety of the FAD cofactor. Accordingly, H357 cannot form a hydrogen bond with the O5-hydroxyl of the 5′-base anymore as observed before for His1 of Dm(6-4). Instead it swivels away up to 4 Å from the 5′-base and His2 into a highly conserved subpocket that is walled by the side chains of K237, T240, H356, L358 and the 3′-base, but unoccupied in Dm(6-4). The freed space is instead filled in CraCRY by water molecule WAT1 that forms an H-bond with His2. Most importantly, His1 is now in H-bonding distance to the carbonyl O2 of the 3′-base (NE2-O2: 3.0 Å; Figure 5A). For Dm(6-4) it has been shown that the complete triad of His1-His2-Tyr (Y423, CraCRY: Y415) is important for catalysis (36). Especially, His1 is found to be crucial, because mutagenesis to alanine in the Xl(6-4) reduces the quantum yield of repair by ∼500% (43). This triad is also present in CraCRY, but as the conformation for His1 is clearly different, our crystal form may have trapped another catalytically competent conformation of the active site.
The antenna chromophore in CraCRY
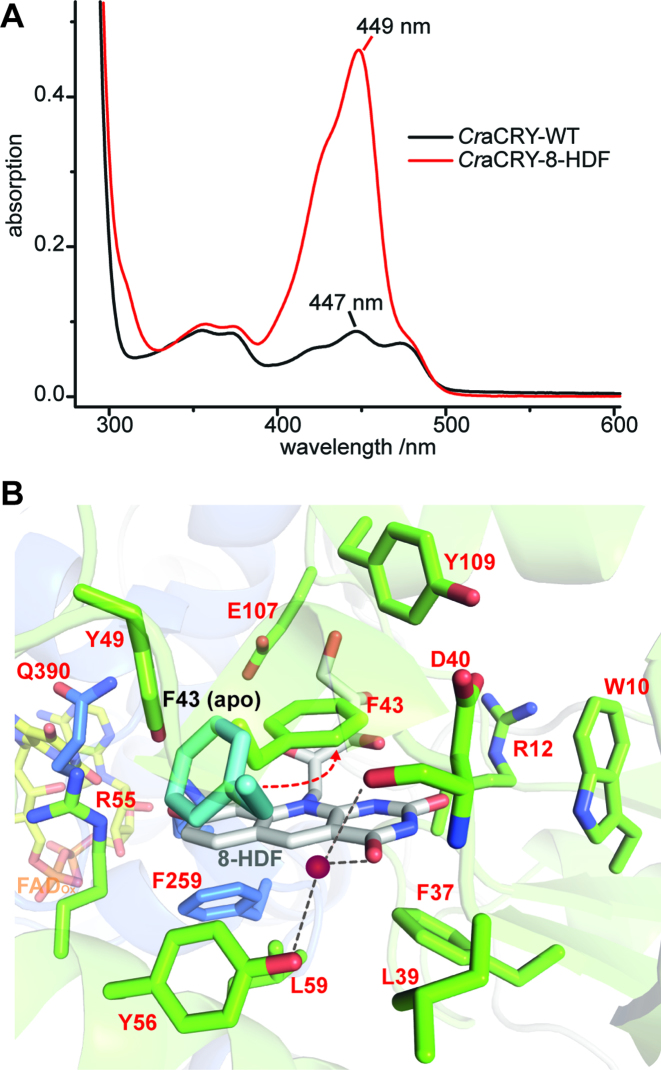
As recombinant CraCRY lacks any antenna pigment, the identity of its light harvesting chromophore was elusive before. Interestingly, Petersen et al. showed that not only the phr2 gene coding for a class II CPD-photolyase, but also phr1 from Chlamydomonas is essential for photoreactivation, i.e. light-dependent repair of UV-lesions in DNA (44,45). However, this gene does not code for a photolyase but a bifunctional FO synthase, which catalyzes the synthesis of 8-HDF, a chromophore that serves in many members of the PCSf as antenna and is particularly important for photolyases because of the low blue-light extinction coefficient of the catalytically competent FADH― cofactor. We performed in vivo reconstitution using a helper plasmid coding for the bifunctional FO synthase from Streptomyces coelicolor (19). UV/Vis spectroscopy reveals a significant peak at 449 nm for a CraCRY•8-HDF complex (Figure 6A), where the incorporated 8-HDF cofactor is bathochromically shifted by 29 nm relative to free 8-HDF (λmax = 420 nm) (19). Similar shifts of the antenna chromophore absorption have been described for other 8-HDF binding members of the PCSf (25,46–49).
Figure 6.
8-HDF as light harvesting antenna chromophore of CraCRY. (A) Comparison of the absorption spectra of WT CraCRY and CraCRY•8-HDF in solution. (B) Binding pocket of 8-hydroxydeazaflavin (8-HDF, gray) and comparison of the conformation of F43 in the apo (6FN3) and holo (6FN2) structure. F43 flips to engage a π–π-stacking with the aromatic rings of 8-HDF. A water molecule (red) is present in the pocket.
To characterize CraCRY/8-HDF interactions we generated and analyzed the co-crystal structure of the CraCRYΔCTE/8-HDF complex (Figure 6B). The binding motif within the N-terminal domain that surrounds the ribityl moiety of the 8-HDF is composed of several highly conserved amino acids, namely D105, E107, R113 and D114, which form H-bonds with the ribityl hydroxy-groups. The C2-carbonyl group of the deazaflavin moiety is coordinated by two water molecules, which form H-bonds with W10, the backbone of F11 and D40. The second carbonyl group (C4) forms a hydrogen bond to a water molecule, which is coordinated by Y56 and the carbonyl-group of D40. The deprotonated hydroxy-group of the benzo ring system makes salt bridges to two basic amino acids, R55 and K258, and is further stabilized by Y49. The two basic amino acids are highly conserved in this class of aCRY/(6-4) photolyases as shown in multiple sequence alignments (SI-S4), whereas in class II photolyases the lysine is always replaced by a histidine (19). Unlike the apo-structure, the antenna loop is completely defined by electron density as F43 performs a flip, to engage π-π-stacking interactions with the deazaflavin moiety of 8-HDF. The bottom of the binding pocket is flanked by some small, unpolar amino acids (L59, L39), which are similarly found in other photolyases (25,46).
DISCUSSION
Our study shows that the full length animal-like cryptochrome of C. reinhardtii exerts DNA repair activity for (6-4)PP in vitro and in vivo. Before, CraCRY has been annotated due to its biological function as a cryptochrome. However, our sequence similarity network analysis (SI-S1) clearly shows that a distinction between (6-4) photolyase and cryptochrome in this common subgroup of the PCSf (1186 sequences) is non-trivial. Only members of the subcluster of insect cryptochromes (89 seq.), including DmCRY, function unambiguously only as signaling proteins due to a cysteine that replaces the conserved asparagine interacting with the N5 nitrogen of FAD (CraCRY: N395). This replacement is known to arrest insect cryptochromes in the non-protonated FAD°― state after the first photoreduction event, so that these members of the PCSf cannot form the fully reduced and catalytically active FADH― state required for (6-4) photolyase activity (50). In contrast, CraCRY can be found like other algal cryptochromes/(6-4)photolyases between a large subcluster of cryptochromes (480 seq.), mainly of animal origin including vertebrates and mammals, and a subcluster of assigned (6-4) photolyases from plants (77 seq.). In the fungal subcluster (213 seq.) annotations as (6-4) photolyases and cryptochromes are almost evenly split (SI-S1). A prevalence of bifunctional cryptochromes among fungi may be hence similarly given as for algal cryptochromes like CraCRY, e.g. the ortholog from Trichoderma atroviride that is designated as cry1 was found to mediate both (6-4) photolyase-dependent photoreactivation and control of gene expression (51). Interestingly, in the plantal subcluster of (6-4) photolyases the forth redox-active residue of the aromatic tetrad is replaced by a phenylalanine. Accordingly, one may suggest that cryptochrome function or bifunctionality depend on the presence of an elongated intramolecular electron transfer chain and thereby increased stabilities of radical pair states, whereas (6-4) photolyase activity may be also driven by a ‘conventional’ aromatic triad as found in other photolyase families of the PCSf.
The specific repair function for (6-4)PP damaged DNA was observed before in algae for the CPF1 orthologs from P. tricornutum and O. tauri (7,52). Both CPF1s act as transcriptional regulators and have been therefore assigned as bifunctional photoreceptors (52). CraCRY also shows this bifunctional behavior, as it exerts not only repair of (6-4)PP but also regulates the expression of various genes (5). Recent studies showed that CraCRY controls the sexual life cycle of C. reinhardtii together with pCRY (53) at steps affecting mating ability in a negative manner in contrast to PHOT (54). Together with phototropin and pCRY, CraCRY also controls positively zygote germination (55). CraCRY was shown to be enriched in the nucleus during daytime, whereas it is delocalized over the whole cell body at night (55). The latter is consistent with the repair of nuclear (6-4)PP as these are only formed upon exposure to ultraviolet light as provided by sunlight during the day period. Given its red/yellow-light activity (5) that is clearly distinct from other characterized cryptochromes, the signaling function of CraCRY may highly rely on the light-dependent formation of the FADH― state rather than that of the FADH° state. Accordingly, bifunctionality as realized by CraCRY may be a highly economic way by exploiting the photoactivation reaction, FADox→FADH°→FADH―, 2-fold: First, for monitoring the illumination status of the cell and, secondly, for keeping the (6-4) photolyase in its active fully reduced state. Clearly, such a scheme is expected to give rise to rather complex regulation with down-stream interaction partners under blue/near-UV light as CraCRY binds like other bifunctional cryptochromes/photolyases to UV-damaged DNA also in its oxidized states. Signaling as provided by CraCRY may hence not only simply reflect changes of light exposure, but also the UV-damaging status of the genomic DNA.
Our first crystal structure of a cryptochrome from a green algae, CraCRY, shows besides the essential catalytic FAD cofactor, 8-hydroxydeazaflavin as antenna chromophore. For its biosynthesis the bifunctional synthase, Phr1, is required in C. reinhardtii and other green algae. Interestingly, deletion of the PHR1 gene causes loss of photoreactivation in C. reinhardtii, which cannot be rescued by high-level overexpression of the PHR2 gene coding for an endogenous class II CPD photolyase, which might harbor 8-HDF (25,44). Given the capability of CraCRY to incorporate 8-HDF in its N-terminal antenna binding domain the loss of photoreactivation by PHR1 deletion may not only be caused by insufficient in vivo activity of the class II CPD photolyase Phr2, but also by a loss of (6-4) photolyase activity as provided by CraCRY. Indeed, a multiple sequence alignment covering 541 non-redundant animal-like CRYs and (6-4) photolyases (SI-S4) delineates well conserved residues within the antenna binding site and hence a wide-spread usage of 8-HDF, including several (6-4) PHL from animals (46). Interestingly, the biosynthesis pathway for 8-HDF is preserved in the green lineage also in mosses, but not any more in higher, vascular plants (23). This distribution can be explained by the biological role of the second chromophore, which is to broaden the absorption range and efficiency of the photolyases especially in their catalytically competent FADH― state, e.g. 8-HDF has an almost tenfold higher extinction coefficient at 400 nm than FADH― (25 mM−1 cm−1 versus 2.8 mM−1 cm−1). Apparently, algae face a greater challenge than higher plants to collect light at short wavelengths for non-photosynthetic, but essential reactions such as DNA repair, as they have to thrive in aqueous environments.
The photoreduction of the catalytic flavin cofactor CraCRY from the fully oxidized (FADox) over the neutral radical (FADH°) to the fully reduced (FADH―) state, is not catalyzed by the canonical tryptophan triad known from other PCSf subfamilies, but by an extended tetrad that depends on a fourth aromatic residue (11,13), in the case of CraCRY ending on a terminal tyrosine, Y373. This extended tetrad is uniquely conserved in the family of animal-like cryptochromes/(6-4) photolyases and was first described for members harboring a terminal tryptophan (56). Upon excitation of the FAD in CraCRY an electron gets abstracted from the proximal tryptophan (W399) resulting in an electron hole at this residue. Then the hole skips towards the surface (W399←W376←W322), where tyrosine (Y373) functions as terminal electron donor. We could show before, that Y373 forms a long-lived deprotonated tyrosyl radical (2.6 s) during formation of the FADH― state (11). In vivo, the unusually long lifetime could be physiologically advantageous, because it allows formation of the catalytically competent FADH― cofactor even when extrinsic reductants for reduction of the tyrosyl radical are in scarce supply. Subsequently, CraCRY with accumulated FADH― state can act as photolyase for repairing (6-4)PP lesions. Sequence analysis of 541 non-redundant members of the animal-like cryptochrome/(6-4) photolyase family shows that about 1/3 (162/541) harbor tyrosine as terminal aromatic residue, almost all of them belonging to fungal orthologs and some algae and a few plants including C. reinhardtii and Zea mays. Those ending their aromatic tetrad with a tryptophan (333/541) mostly belong to animals and a few algae like CPF1 from P. tricornutum (11). Interestingly, in the green lineage including most higher plants and also some algae like O. tauri, the terminal aromatic residue is replaced by a non-redox active phenylalanine (44/542) indicating that the presence of a catalytic tetrad may have gone lost during further evolution and is dispensible at least for plantal (6-4) photolyase activity. Recent studies on the Xl(6-4) photolyase showed that the loss of the fourth redox-active aromatic residue slows indeed photoreduction by up to three orders of magnitude under steady-state conditions and hence impedes recovery of Xl(6-4) photolyase from accidental electron loss of its FAD cofactor (14).
Animal-like cryptochromes have apparently evolved a plethora of different docking sites for down-stream signaling partners. In DmCRY the active site itself interacts in a light-dependent manner with the CTE, whereas murine CRY2 uses this site and the emptied FAD-binding pocket for binding to the ubiquitin-ligase FBXL3 (57,58). Another interaction site that has been assigned in type I and type II animal cryptochromes is the 8-HDF binding site of CraCRY, which lost the ability to bind this or another antenna chromophore during their evolution (16). Interestingly, the C-terminal helix α22 acts in animal cryptochromes like murine CRY1 as major docking site for several circadian rhythm factors like TIM, PER2, BMAL1 and FBXL3 (57,59–62). Given that no CraCRY interaction partners have been identified till date, one may suggest that the rather slow light-driven change of the redox and protonation state of Y373 may distort the packing of helix α22 to the photolyase domain, e.g. by proton transfer and subsequent breakage of the hydrogen bond to D321 (3.1 Å). Such a mechanism for an altered interaction with down-stream partners is of some reminiscence to that of phototropin-like LOV domains, where the C-terminal Jα-helix dissociates from the photoreceptor domain upon lit state formation (63).
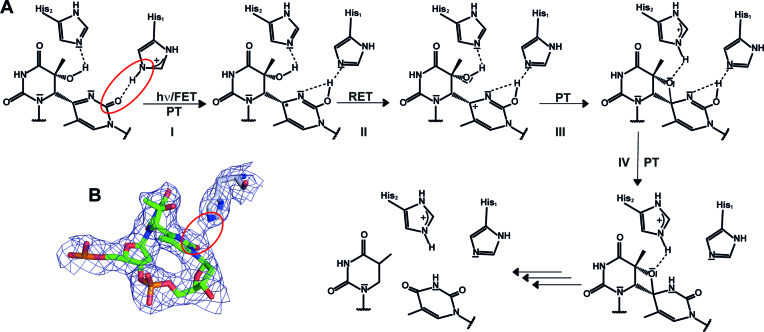
Although the binding mode of the (6-4)PP in CraCRY resembles that of the (6-4) photolyase from D. melanogaster, there are some striking structural differences: First, the overall kinking angle of the DNA duplex differs by ∼8° and coincides with a wider opening of the duplex at the lesion site and formation of specific interactions between the counter bases of the (6-4)PP and the C-terminal helix. The (6-4)PP is likewise positioned in the active site, but an unexpected difference is found for the conformation of His1 in CraCRY (H357). Although adopting only a different rotamer for its side chain than the corresponding His1 residue of Dm(6-4), H365, this structural detail challenges mechanisms of (6-4) photolyases, which were based on previous structures of Dm(6-4) with (6-4) lesion-comprising DNA. In CraCRY, the Nϵ2 atom of the imidazole moiety of His1 is in H-bonding distance to the C2 carbonyl group of the 3′-base (Figure 7B). Therefore, we propose that His1 is protonated and acts as general acid for the C2 carbonyl oxygen after light-driven formation of the radical anion by forward electron transfer (FET). A protonated state of His1 in the substrate complex is corroborated by earlier studies on Xl(6-4) (64,65) and theoretical calculations done for Dm(6-4) (66). As a result of FET, which may involve additional intramolecular electron transfer from the electron-rich 5′ ring, the radical anion is formed at 3′ base. Fast proton transfer onto its C2 carbonyl oxygen (PT, step I in Figure 7A) results in formation of the enol form of the 3′-thymine moiety of the (6-4)PP. Through reverse electron transfer (RET, step II) back to FADH−, C4 gets highly nucleophilic which may lead to concomitant formation of the oxetane intermediate (step III) under proton transfer (PT) to His2. Light-driven formation of the oxetane intermediate is accomplished after intramolecular proton transfer to form the final keto-form (step IV). In this model, the first photoactivation and electron transfer step from the FADH− to the (6-4)PP and back is merely required to overcome the high activation barrier for thermal formation of the oxetane intermediate (65). According to this model, DNA repair by (6-4) photolyases requires a second photon to proceed from the bound oxetane intermediate to breaking the C6-C4 and C5-O4 bonds. This second step would be analogous to the single-photon driven repair of CPD lesions by CPD photolyases, where the light-generated CPD radical-anion undergoes almost barrier-free breakage of the C5-C5 and C6-C6 bonds (36,43,67,68).
Figure 7.
Proposed two-photon mechanism of (6-4)PP repair by CraCRY. (A) This mechanism resembles the two photon mechanism suggested by Sadeghian et al.(66) without the direct involvement of a water molecule. Both histidine residues are involved in the proton transfer (PT) and the stabilization of the oxetane intermediate. The second forward electron transfer (FET) as well as the return electron transfer (RET) are not shown in detail. (B) Given the difference electron density map (Fobs - Fcalc, contoured at 1.25σ) a hydrogen bond between the carbonyl oxygen of the 3′ base and the nitrogen N2 of His1 can be predicted (3.0 Å distance).
The shown mechanism is in major parts based on the two-photon mechanism as proposed by Sadeghian et al. (66) and Yamamoto et al. (43,69) and experimentally supported by recent time-resolved spectroscopy experiments (41). However, as a consequence of the observed His1 conformation and hydrogen bonding pattern, a major difference to the model of Schütz and coworkers (66) is the transient proton transfer from His1 to the C2-carbonyl group of the 3′-thymine instead to the N3 nitrogen. The nucleophilicity of the C2-carbonyl oxygen is apparently higher than that of the nitrogen, as 15N-labeling of the (6-4)PP and NMR-monitored titration showed that the N3 nitrogen lacks any proton accepting properties in the ground state due to a pKa < 1 (70). Given the limited lifetime of an enzyme-bound oxetane-like intermediate, the high propensity to form the fully reduced and catalytically active FADH− cofactor by an aromatic tetrade, even when external reductants are limiting, may foster the efficiency of DNA repair under physiological conditions. The contradiction to most previous computational studies on the (6-4)PP repair mechanism, which claim a single-photon based mechanism for (6-4) photolyases, can be now understood by their high reliance on the available structural information for Dm(6-4) (36,71). As the same can be assumed for our structure-based two-photon model and to overcome the one- vs. two-photon controversy we require either time-resolved X-ray crystallographic data of the (6-4) lesion repair process or bioanalytical identification of the reaction product as formed after first photon absorption by (6-4) photolyase/(6-4)PP complexes.
DATA AVAILABILITY
The multiple sequence alignment was analyzed with SeaView v3.2 (http://doua.prabi.fr/software/seaview). WebLogo 3.0 was used for the visualization of conserved residues (http://weblogo.threeplusone.com/create.cgi). For structure solution and refinement tools of the CCP4i package were used (www.ccp4.ac.uk). Electrostatic potentials were calculated with APBS (http://www.poissonboltzmann.org). Data sets were merged following the KAMO protocol (https://github.com/keitaroyam/yamtbx/blob/master/doc/kamo-en.md). Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data bank under accession number 5ZM0, 6FN3, 6FN0 and 6FN2.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Benedikt Beel for plasmids, Tilman Kottke for support of the in vivo measurements, Yann Geisselbrecht for initial crystallization experiments and Laura Sinatra, Filipp Bezold and Ralf Pöschke for technical assistance. We thank the beamline staff of Berlin Electron Storage Ring Society for Synchrotron Radiation (BESSY-II, Berlin, Germany), European Synchrotron Radiation Facility (ESRF, Grenoble, France) and the National Synchrotron Radiation Research Center (NSRRC, Hsinchu, Taiwan). We thank Silke von Horsten and Vitali Kalugin for data collection and the group of Alfred Batschauer for help with the in vitro assays.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
International Max Planck research school for Environmental, Cellular and Molecular Biology [SF]; Deutsche Forschungsgemeinschaft [LOE: FOR 1261-2, ES 152/12-1, MM: Mi373/12-2]; Air Force Office of Scientific Research (AFOSR) [FA9550-14-1-0409]; Taiwanese Ministry for Science and Technology [MOST107-0210-01-19-02]. Funding for open access charge: DFG FOR1261.
Conflict of interest statement. None declared.
REFERENCES
- 1. Chaves I., Pokorny R., Byrdin M., Hoang N., Ritz T., Brettel K., Essen L.-O., van der Horst G.T.J., Batschauer A., Ahmad M.. The cryptochromes: blue light photoreceptors in plants and animals. Annu. Rev. Plant Biol. 2011; 62:335–364. [DOI] [PubMed] [Google Scholar]
- 2. Kottke T., Oldemeyer S., Wenzel S., Zou Y., Mittag M.. Cryptochrome photoreceptors in green algae: unexpected versatility of mechanisms and functions. J. Plant Physiol. 2017; 217:4–14. [DOI] [PubMed] [Google Scholar]
- 3. Fortunato A.E., Annunziata R., Jaubert M., Bouly J., Falciatore A.. Dealing with light: the widespread and multitasking cryptochrome/photolyase family in photosynthetic organisms. J. Plant Physiol. 2015; 172:42–54. [DOI] [PubMed] [Google Scholar]
- 4. Merchant S.S., Prochnik S.E., Vallon O., Harris E.H., Karpowicz S.J., Witman G.B., Terry A., Salamov A., Fritz-Laylin L.K., Maréchal-Drouard L. et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007; 318:245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beel B., Prager K., Spexard M., Sasso S., Weiss D., Müller N., Heinnickel M., Dewez D., Ikoma D., Grossman A.R. et al. A flavin binding cryptochrome photoreceptor responds to both blue and red light in Chlamydomonas reinhardtii. Plant Cell. 2012; 24:2992–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mittag M., Kiaulehn S., Johnson C.H.. The circadian clock in Chlamydomonas reinhardtii. What is it for? What is it similar to. Plant Physiol. 2005; 137:399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coesel S., Mangogna M., Ishikawa T., Heijde M., Rogato A., Finazzi G., Todo T., Bowler C., Falciatore A.. Diatom PtCPF1 is a new cryptochrome/photolyase family member with DNA repair and transcription regulation activity. EMBO Rep. 2009; 10:655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Spexard M., Thöing C., Beel B., Mittag M., Kottke T.. Response of the sensory animal-like cryptochrome aCRY to blue and red light as revealed by infrared difference spectroscopy. Biochemistry. 2014; 53:1041–1050. [DOI] [PubMed] [Google Scholar]
- 9. Liu B., Zuo Z., Liu H., Liu X., Lin C.. Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 2011; 25:1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sang Y., Li Q.-H., Rubio V., Zhang Y.-C., Mao J., Deng X., Yang H.-Q.. N-Terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsis CRYPTOCHROME 1. Plant Cell. 2005; 17:1569–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oldemeyer S., Franz S., Wenzel S., Essen L.-O., Mittag M., Kottke T.. Essential role of an unusually long-lived tyrosyl radical in the response to red light of the animal-like cryptochrome aCRY. J. Biol. Chem. 2016; 291:14062–14071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ozturk N. Phylogenetic and functional classification of the photolyase/cryptochrome family. Photochem. Photobiol. 2017; 93:104–111. [DOI] [PubMed] [Google Scholar]
- 13. Nohr D., Franz S., Rodriguez R., Paulus B., Essen L.-O., Weber S., Schleicher E.. Extended electron-transfer in animal cryptochromes mediated by a tetrad of aromatic amino acids. Biophys. J. 2016; 111:301–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yamamoto J., Shimizu K., Kanda T., Hosokawa Y., Iwai S., Plaza P., Müller P.. Loss of fourth electron-transferring tryptophan in animal (6-4) photolyase impairs DNA repair activity in bacterial cells. Biochemistry. 2017; 56:5356–5364. [DOI] [PubMed] [Google Scholar]
- 15. Mees A., Klar T., Gnau P., Hennecke U., Eker A.P.M., Carell T., Essen L.-O.. Crystal structure of a photolyase bound to a CPD-like DNA lesion after in situ repair. Science. 2004; 306:1789–1793. [DOI] [PubMed] [Google Scholar]
- 16. Rosensweig C., Reynolds K.A., Gao P., Laothamatas I., Shan Y., Ranganathan R., Takahashi J.S., Green C.B.. An evolutionary hotspot defines functional differences between CRYPTOCHROMES. Nat. Commun. 2018; 9:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Klar T., Pokorny R., Moldt J., Batschauer A., Essen L.-O.. Cryptochrome 3 from Arabidopsis thaliana: structural and functional analysis of its complex with a folate light antenna. J. Mol. Biol. 2007; 366:954–964. [DOI] [PubMed] [Google Scholar]
- 18. Park H.W., Kim S.T., Sancar A., Deisenhofer J.. Crystal structure of DNA photolyase from Escherichia coli. Science. 1995; 268:1866–1872. [DOI] [PubMed] [Google Scholar]
- 19. Kiontke S., Gnau P., Haselsberger R., Batschauer A., Essen L.-O.O.. Structural and evolutionary aspects of antenna chromophore usage by class II photolyases. J. Biol. Chem. 2014; 289:19659–19669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Klar T., Kaiser G., Hennecke U., Carell T., Batschauer A., Essen L.-O.. Natural and non-natural antenna chromophores in the DNA photolyase from Thermus thermophilus. ChemBioChem. 2006; 7:1798–1806. [DOI] [PubMed] [Google Scholar]
- 21. Zhang F., Scheerer P., Oberpichler I., Lamparter T., Krauss N., Krauß N., Krauss N.. Crystal structure of a prokaryotic (6-4) photolyase with an Fe-S cluster and a 6,7-dimethyl-8-ribityllumazine antenna chromophore. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:7217–7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fujihashi M., Numoto N., Kobayashi Y., Mizushima A., Tsujimura M., Nakamura A., Kawarabayasi Y., Miki K.. Crystal structure of archaeal photolyase from Sulfolobus tokodaii with two FAD molecules: implication of a novel light-harvesting cofactor. J. Mol. Biol. 2007; 365:903–910. [DOI] [PubMed] [Google Scholar]
- 23. Essen L.-O., Franz S., Banerjee A.. Structural and evolutionary aspects of algal blue light receptors of the cryptochrome and aureochrome type. J. Plant Physiol. 2017; 217:27–37. [DOI] [PubMed] [Google Scholar]
- 24. Crooks G., Hon G., Chandonia J., Brenner S.. WebLogo: a sequence logo generator. Genome Res. 2004; 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kiontke S., Gnau P., Haselsberger R., Batschauer A., Essen L.-O.. Structural and evolutionary aspects of antenna chromophore usage by class II photolyases. J. Biol. Chem. 2014; 289:19659–19669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry land cell constants. J. Appl. Crystallogr. 1993; 26:795–800. [Google Scholar]
- 27. Yamashita K., Hirata K., Yamamoto M.. KAMO: towards automated data processing for microcrystals. Acta Crystallogr. Sect. D Struct. Biol. 2018; 74:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Karplus P.A., Diederichs K.. Linking crystallographic model and data quality. Science. 2012; 336:1030–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Project C.C. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994; 50:760–763. [DOI] [PubMed] [Google Scholar]
- 31. Murshudov G.N., Vagin A.A., Dodson E.J.. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997; 53:240–255. [DOI] [PubMed] [Google Scholar]
- 32. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brych A., Mascarenhas J., Jaeger E., Charkiewicz E., Pokorny R., Bölker M., Doehlemann G., Batschauer A.. White collar 1-induced photolyase expression contributes to UV-tolerance of Ustilago maydis. Microbiologyopen. 2016; 5:224–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zoltowski B.D., Vaidya A.T., Top D., Widom J., Young M.W., Crane B.R.. Structure of full-length Drosophila cryptochrome. Nature. 2011; 480:396–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brautigam C.A., Smith B.S., Ma Z., Palnitkar M., Tomchick D.R., Machius M., Deisenhofer J.. Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:12142–12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maul M.J., Barends T.R.M., Glas A.F., Cryle M.J., Domratcheva T., Schneider S., Schlichting I., Carell T.. Crystal structure and mechanism of a DNA (6-4) photolyase. Angew. Chem. Int. Ed. 2008; 47:10076–10080. [DOI] [PubMed] [Google Scholar]
- 37. Kiontke S., Geisselbrecht Y., Pokorny R., Carell T., Batschauer A., Essen L.-O.. Crystal structures of an archaeal class II DNA photolyase and its complex with UV-damaged duplex DNA. EMBO J. 2011; 30:4437–4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bordwell F.G., Bartmess J.E., Hautala J.A.. Alkyl effects on equilibrium acidities of carbon acids in protic and dipolar aprotic media and the gas phase. J. Org. Chem. 1978; 43:3095–3101. [Google Scholar]
- 39. Li Y.F., Heelis P.F., Sancar A.. Active site of DNA photolyase: Tryptophan-306 is the intrinsic hydrogen atom donor essential for flavin radical photoreduction and DNA repair in vitro. Biochemistry. 1991; 30:6322–6329. [DOI] [PubMed] [Google Scholar]
- 40. Kleine T., Lockhart P., Batschauer A.. An Arabidopsis protein closely related to Synechocystis cryptochrome is targeted to organelles. Plant J. 2003; 35:93–103. [DOI] [PubMed] [Google Scholar]
- 41. Yamamoto J., Martin R., Iwai S., Plaza P., Brettel K.. Repair of the (6-4) photoproduct by DNA photolyase requires two photons. Angew. Chem. Int. Ed. 2013; 52:7432–7436. [DOI] [PubMed] [Google Scholar]
- 42. Terai Y., Sato R., Yumiba T., Harada R., Shimizu K., Toga T., Ishikawa-Fujiwara T., Todo T., Iwai S., Shigeta Y. et al. Coulomb and CH – π interactions in (6 – 4) photolyase – DNA complex dominate DNA binding and repair abilities. Nucleic Acids Res. 2018; doi:10.1093/nar/gky364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yamamoto J., Plaza P., Brettel K.. Repair of (6-4) lesions in DNA by (6-4) photolyase: 20 years of quest for the photoreaction mechanism. Photochem. Photobiol. 2016; 38:51–66. [DOI] [PubMed] [Google Scholar]
- 44. Petersen J.L., Small G.D.. A gene required for the novel activation of a class II DNA photolyase in Chlamydomonas. Nucleic Acids Res. 2001; 29:4472–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Petersen J.L., Ronan P.J.. Critical role of 7,8-Didemethyl-8-hydroxy-5-deazariboflavin for photoreactivation in Chlamydomonas reinhardtii. J. Biol. Chem. 2010; 285:32467–32475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Glas A.F., Maul M.J., Cryle M., Barends T.R.M., Schneider S., Kaya E., Schlichting I., Carell T.. The archaeal cofactor F 0 is a light-harvesting antenna chromophore in eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:11540–11545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Malhotra K., Kim S.T., Walsh C., Sancar A.. Roles of FAD and 8-hydroxy-5-deazaflavin chromophores in photoreactivation by Anacystis nidulans DNA photolyase. J. Biol. Chem. 1992; 267:15406–15411. [PubMed] [Google Scholar]
- 48. Kort R., Komori H., Adachi S.I., Miki K., Eker A.. DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction. Acta Crystallogr. D. Biol. Crystallogr. 2004; 60:1205–1213. [DOI] [PubMed] [Google Scholar]
- 49. Eker A.P.M., Kooiman P., Hessels J.K.C., Yasui A.. DNA photoreactivating enzyme from the cyanobacterium Anacystis nidulans. J. Biol. Chem. 1990; 265:8009–8015. [PubMed] [Google Scholar]
- 50. Oztürk N., Song S.-H., Selby C.P., Sancar A.. Animal type 1 cryptochromes. Analysis of the redox state of the flavin cofactor by site-directed mutagenesis. J. Biol. Chem. 2008; 283:3256–3263. [DOI] [PubMed] [Google Scholar]
- 51. García-Esquivel M., Esquivel-Naranjo E.U., Hernández-Oñate M.A., Ibarra-Laclette E., Herrera-Estrella A.. The Trichoderma atroviride cryptochrome/photolyase genes regulate the expression of blr1 -independent genes both in red and blue light. Fungal Biol. 2016; 120:500–512. [DOI] [PubMed] [Google Scholar]
- 52. Heijde M., Zabulon G., Corellou F., Ishikawa T., Brazard J., Usman A., Sanchez F., Plaza P., Martin M., Falciatore A. et al. Characterization of two members of the cryptochrome/photolyase family from Ostreococcus tauri provides insights into the origin and evolution of cryptochromes. Plant, Cell Environ. 2010; 33:1614–1626. [DOI] [PubMed] [Google Scholar]
- 53. Müller N., Wenzel S., Zou Y., Künzel S., Sasso S., Weiß D., Prager K., Grossman A., Kottke T., Mittag M.. A plant cryptochrome controls key features of the Chlamydomonas circadian clock and its life cycle. Plant Physiol. 2017; 174:185–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Huang K., Beck C.F.. Phototropin is the blue-light receptor that controls multiple steps in the sexual life cycle of the green alga Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:6269–6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zou Y., Wenzel S., Müller N., Prager K., Jung E.-M., Kothe E., Kottke T., Mittag M.. An Animal-Like cryptochrome controls the Chlamydomonas sexual cycle. Plant Physiol. 2017; 174:1334–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Müller P., Yamamoto J., Martin R., Iwai S., Brettel K.. Discovery and functional analysis of a 4th electron-transferring tryptophan conserved exclusively in animal cryptochromes and (6-4) photolyases. Chem. Commun. 2015; 51:15502–15505. [DOI] [PubMed] [Google Scholar]
- 57. Xing W., Busino L., Hinds T.R., Marionni S.T., Saifee N.H., Bush M.F., Pagano M., Zheng N.. SCF(FBXL3) ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature. 2013; 496:64–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Levy C., Zoltowski B.D., Jones A.R., Vaidya A.T., Top D., Widom J., Young M.W., Scrutton N.S., Crane B.R., Leys D.. Updated structure of Drosophila cryptochrome. Nature. 2013; 495:E3–E4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Engelen E., Janssens R.C., Yagita K., Smits V.A.J., van der Horst G.T.J., Tamanini F.. Mammalian TIMELESS is involved in period determination and DNA damage-dependent phase advancing of the circadian clock. PLoS One. 2013; 8:e56623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Busza A., Emery-Le M., Rosbash M., Emery P.. Roles of the two Drosophila CRYPTOCHROME structural domains in circadian photoreception. Science. 2004; 304:1503–1506. [DOI] [PubMed] [Google Scholar]
- 61. Schmalen I., Reischl S., Wallach T., Klemz R., Grudziecki A., Prabu J.R., Benda C., Kramer A., Wolf E.. Interaction of circadian clock proteins CRY1 and PER2 is modulated by zinc binding and disulfide bond formation. Cell. 2014; 157:1203–1215. [DOI] [PubMed] [Google Scholar]
- 62. Xu H., Gustafson C.L., Sammons P.J., Khan S.K., Parsley N.C., Ramanathan C., Lee H.W., Liu A.C., Partch C.L.. Cryptochrome 1 regulates the circadian clock through dynamic interactions with the BMAL1 C terminus. Nat. Struct. Mol. Biol. 2015; 22:476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Harper S.M., Christie J.M., Gardner K.H.. Disruption of the LOV-Jalpha helix interaction activates phototropin kinase activity. Biochemistry. 2004; 43:16184–16192. [DOI] [PubMed] [Google Scholar]
- 64. Schleicher E., Hitomi K., Kay C.W.M., Getzoff E.D., Todo T., Weber S.. Electron nuclear double resonance differentiates complementary roles for active site histidines in (6-4) photolyase. J. Biol. Chem. 2007; 282:4738–4747. [DOI] [PubMed] [Google Scholar]
- 65. Hitomi K., Nakamura H., Kim S.T., Mizukoshi T., Ishikawa T., Iwai S., Todo T.. Role of two histidines in the (6-4) photolyase reaction. J. Biol. Chem. 2001; 276:10103–10109. [DOI] [PubMed] [Google Scholar]
- 66. Sadeghian K., Bocola M., Merz T., Schuütz M.. Theoretical study on the repair mechanism of the (6−4) photolesion by the (6−4) photolyase. J. Am. Chem. Soc. 2010; 132:16285–16295. [DOI] [PubMed] [Google Scholar]
- 67. Glas A.F., Kaya E., Schneider S., Heil K., Fazio D., Maul M.J., Carell T.. DNA (6-4) photolyases reduce dewar isomers for isomerization into (6-4) lesions. J. Am. Chem. Soc. 2010; 132:3254–3255. [DOI] [PubMed] [Google Scholar]
- 68. Li J., Liu Z., Tan C., Guo X., Wang L., Sancar A., Zhong D.. Dynamics and mechanism of repair of ultraviolet-induced (6-4) photoproduct by photolyase. Nature. 2010; 466:887–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yamamoto J., Hitomi K., Hayashi R., Getzoff E.D., Iwai S.. Role of the carbonyl group of the (6-4) photoproduct in the (6-4) photolyase reaction. Biochemistry. 2009; 48:9306–9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yamamoto J., Tanaka Y., Iwai S.. Spectroscopic analysis of the pyrimidine(6-4)pyrimidone photoproduct: insights into the (6-4) photolyase reaction. Org. Biomol. Chem. 2009; 7:161–166. [DOI] [PubMed] [Google Scholar]
- 71. Dokainish H.M., Kitao A.. Computational assignment of the histidine protonation state in (6-4) photolyase enzyme and its effect on the protonation step. ACS Catal. 2016; 6:5500–5507. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The multiple sequence alignment was analyzed with SeaView v3.2 (http://doua.prabi.fr/software/seaview). WebLogo 3.0 was used for the visualization of conserved residues (http://weblogo.threeplusone.com/create.cgi). For structure solution and refinement tools of the CCP4i package were used (www.ccp4.ac.uk). Electrostatic potentials were calculated with APBS (http://www.poissonboltzmann.org). Data sets were merged following the KAMO protocol (https://github.com/keitaroyam/yamtbx/blob/master/doc/kamo-en.md). Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data bank under accession number 5ZM0, 6FN3, 6FN0 and 6FN2.