Abstract
Familial dysautonomia (FD) is a severe genetic disorder causing sensory and autonomic dysfunction. It is predominantly caused by a c.2204+6T>C mutation in the IKBKAP gene. This mutation decreases the 5′ splice site strength of IKBKAP exon 20 leading to exon 20 skipping and decreased amounts of full-length IKAP protein. We identified a binding site for the splicing regulatory protein hnRNP A1 downstream of the IKBKAP exon 20 5′-splice site. We show that hnRNP A1 binds to this splicing regulatory element (SRE) and that two previously described inhibitory SREs inside IKBKAP exon 20 are also bound by hnRNP A1. Knockdown of hnRNP A1 in FD patient fibroblasts increases IKBKAP exon 20 inclusion demonstrating that hnRNP A1 is a negative regulator of IKBKAP exon 20 splicing. Furthermore, by mutating the SREs in an IKBKAP minigene we show that all three SREs cause hnRNP A1-mediated exon repression. We designed splice switching oligonucleotides (SSO) that blocks the intronic hnRNP A1 binding site, and demonstrate that this completely rescues splicing of IKBKAP exon 20 in FD patient fibroblasts and increases the amounts of IKAP protein. We propose that this may be developed into a potential new specific treatment of FD.
INTRODUCTION
Familial dysautonomia (FD) (OMIM no. 223900) is a rare fatal recessive genetic disorder nearly completely restricted to Ashkenazi Jews. FD causes sensory and autonomic dysfunction due to abnormal development and progressive degeneration of the sensory and autonomic nervous system. The disease is most frequently caused by an intronic T>C mutation at position 6 in intron 20 (c.2204+6T>C) of the IKBKAP gene, which has a carrier frequency of ∼1/32 in Ashkenazi Jews (1,2). The IKBKAP c.2204+6T>C mutation causes exon 20 skipping in a tissue-specific manner with high levels of exon skipping in neuronal tissue, while e.g. lymphoblasts show less exon 20 skipping (3). Skipping of IKBKAP exon 20 leads to a frame-shift in the reading frame and introduction of a premature stop codon (2,4), and hence the IKBKAP transcript is a candidate for degradation by the nonsense-mediated decay (NMD) pathway.
mRNA splicing is a crucial step in gene expression and errors in mRNA splicing may lead to altered gene expression and disease. mRNA splicing depends on recognition of short well-conserved splice site sequences at the exon–intron boundaries. However, because of the degeneracy of the splice site sequences, accurate and efficient mRNA splicing is also dependent on additional splicing regulatory elements (SREs) (5,6). Splicing enhancer (SE) elements attract proteins which enhance exon inclusion, while splicing silencer (SS) elements attract proteins which inhibit exon inclusion (7). The exact regulation of IKBKAP exon 20 splicing is not known, though some SREs with SE or SS function have been reported.
A large number of small-molecule drugs have been shown to increase the inclusion of IKBKAP exon 20 (8–10). Among the most investigated drugs is the plant cytokinin kinetin, which has been shown to be efficient in a FD patient lymphoblast cell line (11), in FD-iPSC-derived neural crest cells (12), transgenic mice (13) and FD patients (14). Unfortunately, the small-molecule drugs also cause more broad effects on mRNA splicing and transcription rather than specific correction of only IKBKAP exon 20 splicing. For instance kinetin and digoxin improves exon inclusion of several other exons (15–17). Recently, another drug, RECTAS, was shown to improve splicing of IKBKAP (18) and also EGCG has a positive effect on IKBKAP exon 20 splicing (9). Phosphatidylserine treatment affects the MAPK/ERK signaling pathway resulting in increased transcription of IKBKAP (19). Overall, the effect of these treatments is not specific to IKBKAP, and a treatment of FD, which specifically targets IKBKAP exon 20 splicing is thus highly desirable to avoid potential off-target effects.
Splice switching oligonucleotides (SSOs) can be designed to enhance exon inclusion by blocking SSs (20,21). Likewise, SSOs can be designed to decrease exon inclusion by blocking splice site sequences or by blocking SEs (22,23). SSOs may thus serve as a highly specific way of reversing aberrant splicing by targeting unique SREs. Several examples of promising SSO-based therapies have been reported (24). The most prominent example so far is the FDA-approved treatment of spinal muscular atrophy (SMA). Patients suffering from SMA lack a functional SMN1 gene; instead they depend on the highly similar SMN2 gene, which, however, predominantly skips exon 7 and therefore mainly produces a truncated protein. SSO-based blocking of an hnRNP A1-binding SS, ISS-N1, located immediately downstream of the 5′ splice site of SMN2 successfully corrects exon 7 splicing and when administered in patients it slows down the development of disease (25,26).
hnRNP A1 is one of the major splicing regulatory proteins, which traditionally has been considered a splicing repressor (27), though it may also work as a splicing activator (20,28). hnRNP A1 may repress splicing by several different mechanisms including antagonizing positive splicing regulatory proteins and sterically blocking splice site recognition (27). Recently, we mapped in vivo hnRNP A1 binding sites in HeLa cells by performing individual nucleotide resolution crosslinking and immunoprecipitation (iCLIP) and identified thousands of binding sites in the human genome (20). We observed that binding of hnRNP A1 downstream of 5′ splice sites is especially important for hnRNP A1-mediated exon repression, and that intronic hnRNP A1 binding sites close to the 5′ splice sites may generally be suitable for SSO-based improvement of exon inclusion (20).
Here, we identify a fundamental hnRNPA1-binding SS located downstream of IKBKAP exon 20. Furthermore, we demonstrate that hnRNP A1 also binds to two previously reported exonic SSs, ESS1 and ESS2, which repress IKBKAP exon 20 inclusion. Knockdown of hnRNP A1 in FD patient fibroblasts improves splicing of IKBKAP exon 20 supporting that hnRNP A1 represses its splicing. Site-specific mutagenesis of an IKBKAP minigene confirms that both the exonic and the intronic hnRNP A1 binding sites cause hnRNP A1-mediated exon repression. This suggests that hnRNP A1 binds in a cooperative way to all three silencer elements to repress IKBKAP exon 20 inclusion in the FD IKBKAP gene. Finally, we show that SSO-mediated blocking of the intronic hnRNP A1 binding SS completely restores normal splicing of IKBKAP exon 20 even at low doses, and that this is accompanied by an increased IKAP protein level. We propose that this may be developed into a potent and specific future therapeutic strategy to restore IKBKAP exon 20 splicing in patients suffering from FD.
MATERIALS AND METHODS
RNA affinity purification
High-performance liquid chromatography-purified RNA oligonucleotides with 3′-biotin were purchased from LGC Biosearch Technologies. The association of hnRNP A1 and hnRNP A2/B1 with IKBKAP were analyzed using the following RNA oligonucleotides: intronic splicing silencer (ISS) wild-type (WT) corresponding to positions IKBKAP intron 20 +11−36: UUGUACUGUUUGCGACUAGUUAGCUU-Biotin, and ISS MUT where the putative hnRNP A1 binding sites were mutated: UUGUACUGUUUGCGACUCGUUCGCUU-Biotin. IKBKAP exon 20 ESS1 WT: GGAAACUUAGAAGUUGUUCA-Biotin containing the first putative hnRNP A1 binding site in IKBKAP exon 20 (ESS1), and IKBKAP exon 20 ESS1 MUT: GGAAACUUCGAAGUUGUUCA-Biotin, where the binding site is disrupted. IKBKAP exon 20 ESS2 WT: UGGUUUUAGCUCAGAUUCGG-Biotin contains the second putative hnRNP A1 binding site in IKBKAP exon 20, which is mutated in the IKBKAP exon 20 ESS2 MUT: UGGUUUUCGCUCAGAUUCGG-Biotin.
For each purification, 100 pmol of RNA oligonucleotide was coupled to 50 μl of streptavidin-coupled magnetic beads (PureProteome™ Streptavidin Magnetic Bead System, Millipore) for 15 min in 1 × binding buffer (20 mM Hepes/KOH [pH 7.9], 72 mM KCl, 1.5 mM MgCl, 1.56 mM MgAc, 0.5 mM Dithiothreitol (DTT), 4 mM glycerol, 0.75 mM adenosine triphosphate and 0.2 μg/μl bulk tRNA). The suspension was then placed in a magnet, and the supernatant was removed. The oligonucleotide–bead complexes were then resuspended in 500 μl of binding buffer containing either 100 μl of HeLa cell nuclear extract (Cilbiotech) or 5 or 10 pmol pf recombinant hnRNP A1 (Origene) and incubated for 25 min at room temperature. Then, the supernatant was removed, and the beads were washed three-times in binding buffer containing 600 mM KCl or 1200 mM KCl. At last, the proteins bound to the RNA were eluted by the addition of 50 μl protein XT sample buffer (BioRad) and heated for 4 min at 90°C. The biotinylated RNA oligonucleotides were immobilized in a magnet, and the supernatant proteins were separated on a 4–12% sodium dodecyl sulphate (SDS)-polyacrylamide gel and were electroblotted onto a nitrocellulose membrane. The membranes were then probed with an anti-hnRNP A1 antibody (R9778, Sigma-Aldrich) or anti-hnRNP A2/B1 (sc-53531, Santa Cruz Biotechnology) or anti-RBM24 (ab94567, Abcam) and the control anti-hnRNP H (N16) (sc-10042, Santa Cruz Biotechnology). All RNA affinity purifications were performed at least four times employing two different batches of HeLa cell nuclear extract (Cilbiotech).
Surface plasmon resonance imaging (SPRi)
Biotinylated oligonucleotides were immobilized on a Senseye G strep (SSENS) sensorchip in a 2 × 4 × 12 array by continuous flow in a CFM 2.0 printer (Wasatch microfluidics). The oligonucleotides were diluted in 1 × Tris Buffered Saline (TBS) to a concentration of 1 μM and spotted for 20 min followed by 5 minutes washing with TBS + 0.05% Tween-20. The sensorchip was transferred to the MX96 (IBIS technologies), and the system was primed with Surface Plasmon Resonance imaging (SPRi) buffer (10 mM Hepes/KOH pH 7.9, 150 mM KCl, 10 mM MgCl2 and 0.075% Tween-80). SPRi by IBIS MX-96 was used to measure the kinetics of recombinant hnRNP A1 (ab224866, Abcam) binding to the immobilized RNA oligonucleotides. Binding was measured in real time by following changes of the SPR angles at all printed positions of the array during 10 min injections of hnRNP A1 protein over the entire surface. Six injections of a 2-fold titration series from 6.25 to 200 nM hnRNP A1 were injected in sequence from the lowest concentration to the highest. Before adding protein to the chip, residual background binding was blocked by injecting 1 mg/ml bovine serum albumin in SPR buffer onto the chip for 10 min. A continuous flow of SPR buffer flowed over the surface before, between and after the hnRNP A1 injections, to measure baseline and dissociation kinetics. Dissociation was measured for 5 min, by injecting SPR buffer over the chip at a rate of 8 μl/s. Responses for a calibration curve were created after the concentration series by measuring SPR responses from defined dilutions of glycerol in running buffer (ranging from 5 to 0% glycerol) and of pure water as defined by the automated calibration routine of IBIS MX-96.
For data analysis the SPRi data were imported into SPRINTX software (v. 2.1.1.0, IBIS technologies), calibrated, reference subtracted and the baseline of the responses before all hnRNP A1 injections were zeroed. The time starting point was aligned at the beginning of each new injection. Then the data were exported to Scrubber 2 (Biologics Inc.). Binding curves for all positions where binding was observed were fitted globally to the integrated rate equation that describes simple first order 1:1 binding kinetics in order to obtain kinetic association rate (ka), dissociation rate (kd) and equilibrium dissociation (KD = kd/ka) constants. To obtain a bimodal binding for 1:2 kinetics the simulations were calculated using the pbm package for R, and using ggplot2 for plotting the resulting binding curves.
Knockdown of hnRNP A1 and hnRNP A2/B1 in patient fibroblasts
FD patient fibroblasts (GMO4589, GMO4899, GMO2343 and GMO4663) (Coriell Cell Repositories) and control fibroblasts (GMO2912 (Coriell Cell Repositories)) were grown to approximately 70% confluency and transfected with siRNA smartpools targeting hnRNP A1 (L-008221-00, Dharmacon), hnRNP A2/B1 (L-011690-01, Dharmacon), the hnRNP A1 and hnRNP A2/B1 siRNAs combined or control non-targeting siRNA (D-001810-10-20, Dharmacon) using Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher Scientific). The cells were transfected twice and harvested 72 h after the first transfection. RNA was extracted as described below. Protein for western blot analysis of fibroblasts was harvested using M-PER Mammalian Protein Extraction Reagent (ThermoFisher Scientific) supplemented with Complete Protease Inhibitor Cocktail (11836145001, Roche) and PMSF (93482, Sigma-Aldrich). Knockdown was validated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and/or quantitative polymerase chain reaction (qPCR) analysis. Proteins were separated on a 4–12% SDS-polyacrylamide gel and were electroblotted onto a nitrocellulose membrane. The membranes were then probed with an anti-hnRNP A1 antibody (R9778, Sigma-Aldrich) or anti-hnRNP A2/B1 (sc-53531, Santa Cruz Biotechnology) and the control anti-hnRNP H (N16) (sc-10042, Santa Cruz Biotechnology) or an anti-HPRT antibody (HPA006360, Sigma-Aldrich). Cycloheximide-treatment was carried out supplementing the cell culturing media with 40 μg/ml cycloheximide for 6 h prior to cell harvest.
Transfection of splice switching oligonucleotides
FD patient fibroblasts (GMO4663, GMO4589, GMO4899, GMO2343 (Coriell Cell Repositories)) or control fibroblasts (GMO2912 (Coriell Cell Repositories) or an internal fibroblast control cell line FB-01) were grown to approximately 70% confluency and transfected with SSOs using Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions either performing reverse transfection using 6, 12 or 24 nM of SSO or forward transfection of an SSO concentration gradient (0.046, 0.188, 0.75, 3 or 12 nM). SSOs were phosphorothioate oligonucleotides with 2′-O-methyl modification on each sugar moiety (LGC Biosearch Technologies). The IKBKAP SSOs targets IKBKAP intron 20; SSO1 (pos.+11−35): 5′-AGCUAACUAGUCGCAAACAGUACAA-3′; SSO2 (pos.+19−43): 5′-AAAUCACAAGCUAACUAGUCGCAAA-3′; SSO3 (pos.+27−51): 5′-UCACACAUAAAUCACAAGCUAACUA-3′; SSO4 (pos.+44−68): 5′-UAAAAUACUUAUUGUCUUCACACAU-3′; SSO5 (pos.+53−72): 5′-AAAUUGUAAUAAAAUACUUAUUGUC-3′. The control SSO does not target any human genes: 5′-GCUCAAUAUGCUACUGCCAUGCUUG-3′. After transfection, cells were incubated for 48 h before RNA extraction. Western blot analysis of SSO treated fibroblasts: 72 h post-transfection proteins for western blot analysis of fibroblasts was harvested using M-PER Mammalian Protein Extraction Reagent (ThermoFisher Scientific) supplemented with Complete Protease Inhibitor Cocktail (11836145001, Roche) and PMSF (93482, Sigma-Aldrich). Proteins were separated on a 4–12% SDS-polyacrylamide gel and electroblotted onto a nitrocellulose membrane. The membranes were then probed with an anti-IKAP antibody (sc-136412, Santa Cruz Biotechnology) or anti-hnRNP A2/B1 (sc-53531, Santa Cruz Biotechnology) and the control anti-hnRNP H (N16) (sc-10042, Santa Cruz Biotechnology).
Transfection of IKBKAP and SMN1/2 minigenes
IKBKAP minigene covered the unaltered sequence of exon 19–21 of IKBKAP (genomic location 108 900 438–108 898 593). The minigene was cloned into pJet1.2 and subcloned into pcDNA3.1 using heterozygous (IKBKAP c.2204+6T>C) genomic DNA as template (NA05044, NA05045 (Coriell). Mutagenesis of the IKBKAP minigene was performed by Genscript.
The IKBKAP ISS (ctagttagctt) was introduced into the pCI SMN1 and pCI SMN2 minigenes by in vitro mutagenesis (GeneScript) (Figure 5), replacing nucleotides +10 to +19 of intron 20 from the ISS-N1 sequence in SMN1 and SMN2 (ccagcattatg) (pCI SMN1 and pCI SMN2 were kindly provided by Prof. Adrian Krainer, Cold Spring Harbor Labory, NY, USA). The pCI SMN mutant minigenes with the IKBKAP ISS were named pCI SMN1 IKBKAP ISS and pCI SMN2 IKBKAP ISS. Minigenes were transfected into HeLa cells using XtremeGene9 (Roche). Forty-eight hours after transfection cells were harvested for RNA analysis.
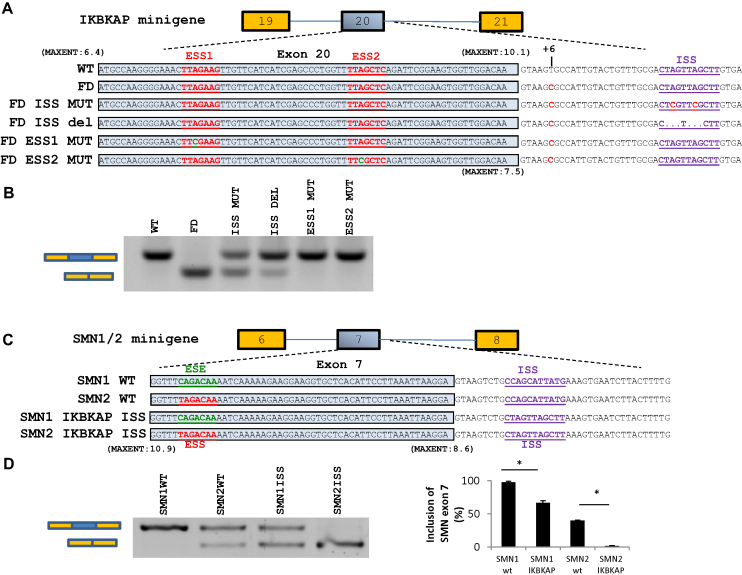
Figure 5.
Mutation of ESS1 or ESS2 or ISS corrects splicing of a FD-IKBKAP minigene and insertion of the IKBKAP ISS in SMN1/2 minigenes confirms that it is a strong SS. (A) Schematic of the IKBKAP minigenes. (B) Splicing of IKBKAP exon 20 after transfection of IKBKAP minigenes into HeLa cells. Disruption of the hnRNP A1 motifs restores IKBKAP exon 20 splicing. The transfections were done twice in triplicates. (C) Schematic of the SMN1/2 minigenes where the ISS-N1 SS was disrupted by insertion of the IKBKAP ISS sequence. The IKBKAP ISS sequence inhibits inclusion of SMN1/2 exon 7 stronger than the endogenous hnRNP A1 binding ISS-N1. (D) Inclusion of SMN1/2 exon 7 was quantified using the fragment analyzer. Error bars indicates SEM, n ≥ 3 ***P-value < 0.001, **P-value < 0.01, *P-value < 0.05.
RNA extraction and analysis
RNA was harvested using Isol (5Prime) or Qiazol (Qiagen) and phenol/chloroform extraction. RNA was used as template for cDNA synthesis using High Capacity cDNA Reverse Transcription Kit (Thermo Scientific). Splicing of IKBKAP exon 20 was examined by PCR using Tempase Hot Start DNA Polymerase (Ampliqon) and primers for analysis of endogenous IKBKAP splicing: IKBKAP exon 19 F: GGGGTTCACGGATTGTCACT and IKBKAP exon 21 R: TCTCAGCTTTCTCATGCATTCAA. For analysis of minigene splicing the IKBKAP exon19 F primer was used together with BGHREV pcDNA3.1 modified: AACTGAAAGGCACAGTCGAGGCTG, which binds the pcDNA3.1 vector backbone. Splicing of exon 7 from pCI SMN1 and SMN2 mini genes were analyzed using vector specific pCIFwdB 5′-GACTCACTATAGGCTAGCCTCG-3′ and SMNtestex8as 5′-GTGGTGTCATTTAGTGCTGCTC-3′. Control PCR was performed using primers amplifying HPRT: HPRT F: TGACACTGGCAAAACAATGCA and HPRT R: GGTCCTTTTCACCAGCAAGCT. PCR products were analyzed either by agarose gel electrophoresis or by capillary gel electrophoresis using the fragment analyzer (Advanced Analytical). The total expression of IKBKAP was analyzed by quantitative PCR using primers IKBKAP F: ACGAGCTTCAAGGAAATGCTC and IKBKAP R: TCAAAGTCATAGGTCCAAGAGA and normalized to the expression of TBP using primers TBP F123 (exon 1/2): GTGACCCAGCATCACTGTTTC and TBP R260 (exon 2): GCAAACCAGAAACCCTTGCG. The expression of hnRNPA1, hnRNPA2/B2 and NRG1 was analyzed by quantitative PCR using the following primers: hnRNPA1hum1s: CACCCTGCCGTCATGTCTAAGTCACA and hnRNPA1hum1as: AAAGCCCCTGGAGCGCTTGG; hnRNPA2hum1s: CCATGGCTGCAAGACCTCAT and hnRNPA2hum1as: TCCCTCATTACCTCTCTTGCT; NRG1-1s: CACCACTGGGACAAGCCATC and NRG1-1as 5-GTAGTTTTGGCAGCGATCACC and normalized to the expression of TBP. The qPCR reactions were carried out on the LightCycler 480 Instrument II (Roche) using essential DNA Green Master mix (Roche).
RESULTS
HnRNP A1 binds sequences in IKBKAP exon 20 and sequences flanking the 5′ splice site
IKBKAP exon 20 is partially skipped in the c.2204+6T>C FD patient fibroblasts (Figure 1) (2). Analysis of the IKBKAP exon 20 splice sites using MaxEntScan (29) shows that IKBKAP exon 20 contains a weak 3′ splice site with a maximum entropy score of 6.36, but a relatively high 5′ splice site score of 10.08, which is only decreased to 7.47 by the c.2204+6T>C mutation. Because this relatively slight decrease in 5′ splice site strength causes a dramatic decrease in splicing efficiency, we hypothesized that exon 20 definition is challenged by SSs. It is well known that exons with weak splice sites are particularly sensitive to a shifted balance between positive and negative SREs (21,30). Therefore, we examined our previous data generated by iCLIP using HeLa cells stably expressing T7-tagged hnRNP A1 (20), and we identified hnRNP A1 iCLIP reads downstream of the 5′ splice site of IKBKAP exon 20 (Figure 1), which overlapped two TAG motives characteristic of hnRNP A1 binding (20,31). Since iCLIP reads pinpoint protein binding sites, this suggests that an hnRNP A1 binding site exists immediately downstream of the 5′ splice site. Furthermore, we analyzed data from an hnRNP A1 HITS-CLIP study performed in HEK293T cells, and identified hnRNP A1 reads within IKBKAP exon 20 (28) (Supplementary Figure S1). These reads cover a previously described exonic splicing silencer (ESS2) (32). In the same study, an additional exonic splicing silencer (ESS1) was also identified. However, deletion and mutagenesis studies indicate that ESS1 has lower silencing capability than ESS2, possibly due to the existence of an overlapping SE element (17,32). We hypothesized that ESS1 and ESS2 may function by binding hnRNP A1, and that the intronic hnRNP A1 binding site could represent an ISS. Thus, binding of hnRNP A1 to these three silencers would have a repressive effect on IKBKAP exon 20 inclusion, which could result in exon 20 skipping when the IKBKAP exon 20 5′ splice site is compromised by the c.2204+6T>C mutation. To confirm binding of hnRNP A1 to these sites, we first performed RNA affinity purification with biotinylated RNA oligonucleotides either harboring the sequence around the ESS1, the ESS2 or the intronic hnRNP A1 binding site (ISS) (Figure 2). The sequence-specific binding of hnRNP A1 was evaluated by including mutant oligonucleotides (Mut), where the important UAG of the hnRNP A1-binding motif was mutated to UCG. We have previously documented that this type of A>C mutation dramatically reduces binding of hnRNP A1 (20,30,33,34). We initially used recombinant hnRNP A1 and analyzed the bound protein by SDS-PAGE and western blotting (Supplementary Figure S2). This showed that recombinant hnRNP A1 binds these sequences, thus hnRNP A1 is able to recognize and bind these sequences independently of other RNA binding proteins. Furthermore, the binding of hnRNP A1 was markedly reduced when using mutant oligonucleotides suggesting that hnRNP A1 interacts with these elements in a sequence-dependent way. Next, we performed the same experiment using HeLa cell nuclear extract for the binding reaction, thus better mimicking the environment of splicing in the cell nucleus, where protein binding is competitive and cooperative among several splicing regulatory proteins (35) (Figure 2 and Supplementary Figure S2). hnRNP A1 also bound to the IKBKAP oligonucleotides in a sequence-specific way in this context, suggesting that hnRNP A1 both on its own and in competition with other RNA binding proteins specifically binds these three regulatory elements.
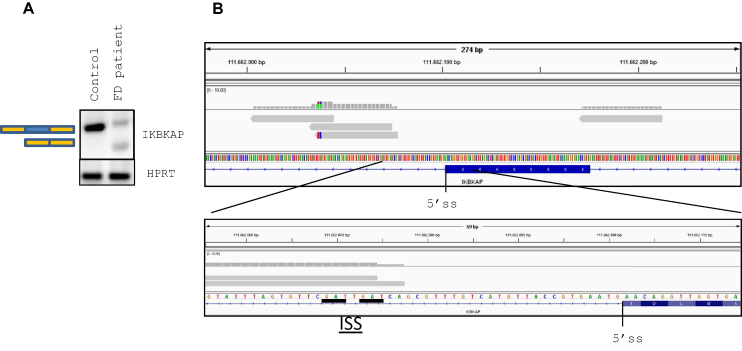
Figure 1.
hnRNP A1 iCLIP reads at IKBKAP exon 20. (A) Analysis of the splicing of IKBKAP exon 20 in FD-derived fibroblasts (GMO4589) and control fibroblasts (GMO2912). FD-derived fibroblasts contain partially skipped IKBKAP exon 20 and reduced expression levels compared with control fibroblasts. (B) Screenshot from the IGV genome browser. Upper: hnRNP A1 iCLIP reads (gray) surrounding IKBKAP exon 20. Lower: enlargement of the 5′ splice site region. The iCLIP technique results in reads that begin at or close to the cross-linked nucleotide. As such, the beginning of the reads indicates the most likely location of hnRNP A1 binding. Two hnRNP A1 binding motives (underscored) are present downstream of the 5′ splice site overlapping with the iCLIP reads. IKBKAP is located on the reverse strand.
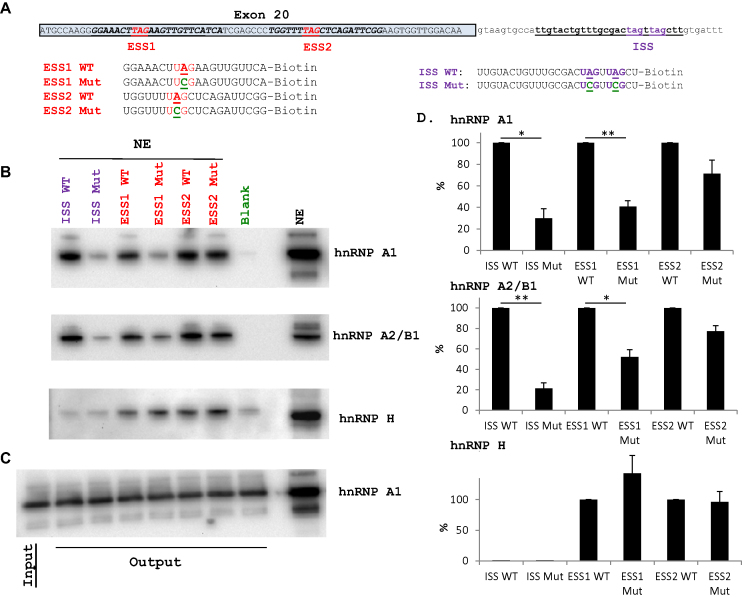
Figure 2.
hnRNP A1 and hnRNP A2/B1 interacts with IKBKAP inside exon 20 and downstream of the 5′ splice site. (A) The position of the ESS1 and ESS2 in IKBKAP exon 20 and the position of the ISS downstream of the exon 20 5′ splice site. hnRNP A1 motives (TAG) are indicated. The sequences analyzed by RNA affinity purification are shown. For evaluation of the sequence-specific binding, the important hnRNP A1-binding UAG triplet is mutated to UCG. (B) RNA affinity purification of HeLa cell nuclear extract using the IKBKAP oligonucleotides Purified proteins were analyzed by western blotting using anti-hnRNP A1 or anti-hnRNP A2/B1 antibodies. hnRNP H was used as loading control. Results are representative for four experiments. (C) Western blot (anti-hnRNP A1 antibody) a gel with a lane for input (binding solution with NE before addition of beads with oligonucleotides) and lanes with output from each of the pull downs, demonstrating equal content of hnRNP A1 in all binding assays. (D) Quantification of bands using IMAGE J from three representative blots. WT was set to 100% and the corresponding mutant expressed in % of WT. Error bars indicate SEM, n = 3, **P-value < 0.01, *P-value > 0.05 (one-sample t-test).
Since hnRNP A1 and hnRNP A2/B1 have been reported to have overlapping binding motifs (28), we also characterized the binding of hnRNP A2/B1 to these elements. As expected, we observe that hnRNP A2/B1 binds in a manner similar to hnRNP A1 (Figure 2), suggesting that hnRNP A1 and hnRNP A2/B1 both regulate IKBKAP exon 20 splicing. Recently, it has been proposed that RBM24 can stimulate IKBKAP exon 20 inclusion from the c.2204+6T>C mutant splice site through binding to an intronic SE located in the region +13 to +29 (36). Consequently, we also checked for binding of RBM24 to all six different biotinylated RNA oligonucleotides, but we were unable to observe RBM24 binding above background (results not shown), possibly because RBM24 expression is low in HeLa cells (https://www.proteinatlas.org/ENSG00000112183-RBM24/cell).
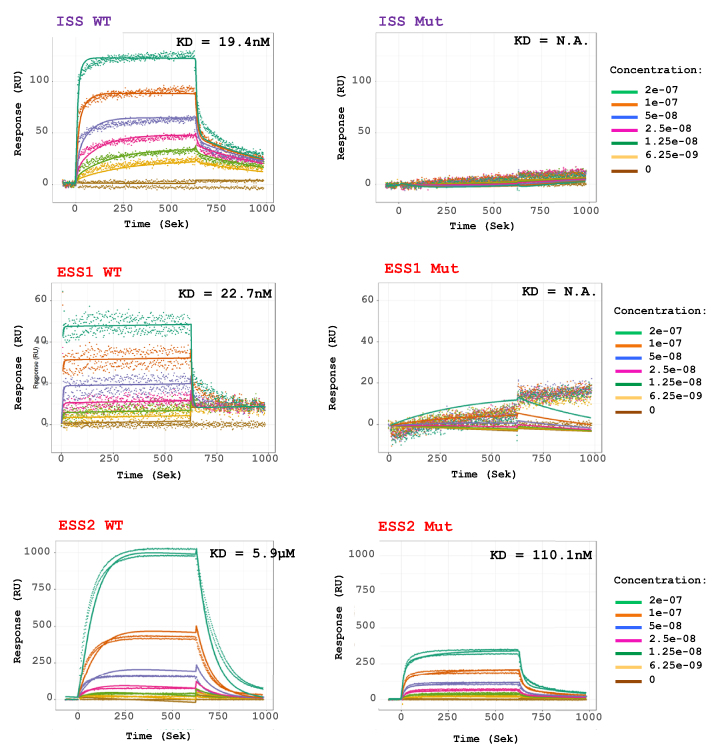
In order to further investigate the binding of hnRNP A1 to the ISS and ESS’s in IKBKAP exon 20 and intron 20, the biotinylated oligonucleotides used for RNA affinity analysis were analyzed by SPRi, using recombinant hnRNP A1 protein to analyze the kinetics that occur during RNA–protein interaction. For all three set of oligonucleotides, we observe specific and concentration dependent increase in hnRNPA1 binding (Figure 3 and Supplementary Figure S3). For the ISS containing oligonucleotides binding of hnRNP A1 was completely disrupted in the mutant oligonucleotide, and when fitting the association curves to a bimodal binding model, we observe two different association and dissociations (Figure 3 and Supplementary Figure S3) which is consistent with the two hnRNP A1 binding motifs located closely to each other in the IKBKAP ISS (37). When we abolish both sites in the mutant ISS oligonucleotide all hnRNP binding is lost, and only a very poor fit can be made (Figure 3 and Supplementary Figure S3). For the two ESSs in IKBKAP exon 20, we observe very different binding reactions. The ESS1 containing oligonucleotide shows some binding of hnRNP A1, which is completely lost in the mutant oligonucleotide. The ESS2 containing oligonucleotide shows a very strong bimodal binding of hnRNP A1 that is not completely disrupted in the mutant oligonucleotide. While binding is severely decreased the mutant ESS2 oligonucleotide can still bind more hnRNP A1 than the WT ISS oligonucleotide, consistent with previous reports of ESS2 being a strong SS (32) (Figure 3 and Supplementary Figure S3). We were a bit puzzled by the fact that the mutant ESS2 oligonucleotide still had significant affinity toward hnRNP A1, but a closer examination shows that a potential second low score hnRNP A1 binding site (UCAGAUU) is present immediately downstream. It is thus likely that the strong binding to the ESS2 oligonucleotide is caused by the simultaneous presence of two hnRNPA1 binding sites, perhaps working synergistically.
Figure 3.
SPRi analysis confirms hnRNP A1 binding to the IKBKAP SSs. SPRi of recombinant hnRNP A1 protein binding to biotinylated RNA oligonucleotides harboring WT and mutant ESS1, ESS2 and ISS. Protein was injected in increasing 2-fold concentrations from 6.25 to 200 nM. ISS and ESS2 oligonucleotide measurements were fitted to a bimodal 1:2 binding model, to account for multiple hnRNP binding sites on the oligonucleotide. ESS1 oligonucleotide measurements could only be fitted to a monomodal 1:1 binding model. KD is based on kd/ka. It is the best indicator of how much protein the oligonucleotides can bind at equilibrium. It was not possible to calculate KD for ISS MUT and ESS1 MUT due to low binding.
hnRNP A1 regulates the splicing of IKBKAP exon 20
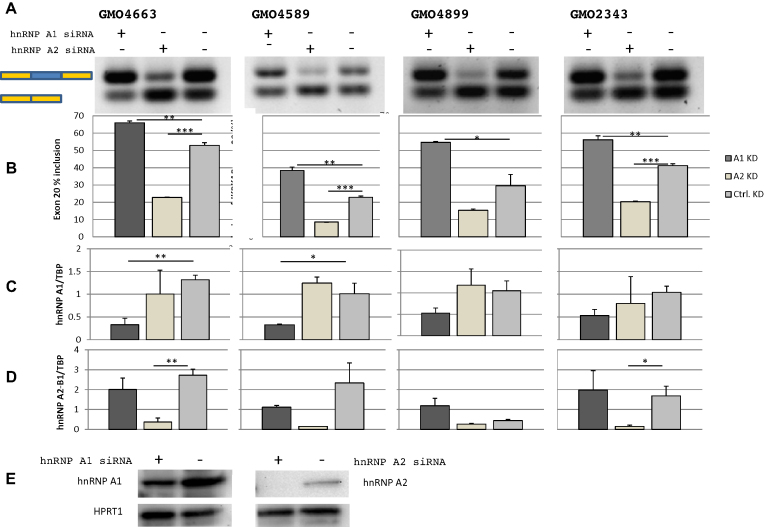
To evaluate whether the binding of hnRNP A1 and hnRNP A2/B1 regulate IKBKAP exon 20 splicing, we performed siRNA-mediated knockdown of hnRNP A1, hnRNP A2/B1 or both proteins combined in FD patient fibroblast (Figure 4; Supplementary Figures S4 and 5). Knockdown of hnRNP A1 increases the inclusion of IKBKAP exon 20 in all four tested patient cell lines, supporting that hnRNP A1 has a negative effect on IKBKAP exon 20 splicing. This reflects the established role of hnRNP A1 as a repressive splicing regulatory protein. The fact that knockdown of hnRNP A1 does not fully restore splicing suggests that also other splicing repressors can inhibit exon 20 inclusion, but it can also be explained by the fact that we were unable to achieve complete hnRNP A1 knockdown.
Figure 4.
hnRNP A1 and hnRNP A2/B1 regulate inclusion of IKBKAP exon 20. Knockdowns of hnRNP A1 or hnRNP A2/B1 in four different FD patient fibroblasts GMO4663, GMO4589, GMO4899 and GMO2343. (A) hnRNP A1 knockdown increases IKBKAP exon 20 inclusion, while hnRNP A2/B1 decreases IKBKAP exon 20 inclusion. (B) IKBKAP exon 20 inclusion was quantified using the fragment analyzer. Error bars indicates SEM, n ≥ 2 ***P-value < 0.001, **P-value < 0.01, *P-value < 0.05 knockdown was validated by qPCR where expression of hnRNP A1 (C) and hnRNP A2/B1 (D) was normalized to the expression of TBP. Error bars indicates SEM, n ≥ 3 ***P-value < 0.001, **P-value < 0.01, *P-value < 0.05. (E) Knockdown of hnRNP A1 and hnRNP A2/B1 was confirmed by western blotting (GMO4663) and HPRT was used as a loading control.
Interestingly, knockdown of hnRNP A2/B1 decreases IKBKAP exon 20 inclusion, indicating that hnRNP A2/B1 enhances splicing of exon 20. Consistent with the opposite effects of hnRNP A1 and hnRNP A2/B1 knockdown, the combined knockdown of both proteins has little or no net effect on IKBKAP exon 20 splicing (Supplementary Figures S4 and 5). Since the IKBKAP transcript without exon 20 is an NMD-candidate, a proportion of these transcripts will be quickly degraded and analysis of the relative inclusion level will be skewed. Cycloheximide-treatment of cells 6 hours prior to harvesting blocks translation and thus blocks the NMD pathway. In line with this, we detect an increase of the transcript with exon 20 skipped in cycloheximide-treated cells compared to untreated cells (Supplementary Figures S4 and 5).
Disruption of hnRNP A1 binding sites restores splicing of IKBKAP exon 20 in a FD-IKBKAP minigene
To further characterize the regulation of IKBKAP exon 20 splicing, we constructed an IKBKAP minigene harboring the region from exon 19 to exon 21 cloned into pcDNA3.1. Splicing of the FD-IKBKAP minigene containing the c.2204+6T>C mutation mimicked the endogenous FD-IKBKAP gene when transfected into HeLa cells (Figure 5). Therefore, we constructed FD-IKBKAP minigenes with mutations in the hnRNP A1 binding sites disrupting the hnRNP A1 binding silencer motifs (Figure 5). Disruption of the exonic splicing silencers ESS1 and ESS2 completely restored splicing, and deletion of the ISS hnRNP A1 binding motif leads to a nearly complete restoration, whereas mutation of the ISS (ISS MUT) causes a high, but less efficient correction (Figure 5). It has been proposed that hnRNP A1 binds high-affinity binding sites and through protein-protein interactions spread across an exon to inhibit exon recognition (38). We therefore hypothesized that hnRNP A1 binds cooperatively to all three silencers and spreads across the exon to inhibit exon recognition. Thus disruption of any of these silencers may reduce the association of hnRNP A1 with IKBKAP exon 20 allowing the U1 snRNP to bind to the weakened c.2204+6T>C 5′ splice site of FD-IKBKAP. Disrupting the ISS silencer by UAG>UCG mutations is less efficient in restoring splicing of IKBKAP exon 20 than deleting the hnRNP A1 ISS motif, suggesting that additional overlapping inhibitory sequences were disrupted in the deleted sequence or that introduction of the UAG>UCG mutations in the ISS does not abolish binding of positive factors, like for instance RBM24 to the intronic SE (36).
In order to further characterize the IKBKAP ISS, we tested its effect by using it to substitute one of the best-known ISSs, ISS-N1, in SMN1/2 genes. ISS-N1 also functions by recruiting hnRNP A1 in order to repress recognition of a weak 5′ splice site (39). We replaced nucleotides +10 to +20 of the ISS-N1 in the SMN1 and SMN2 minigenes with the IKBKAP ISS sequence (nucleotides +26 to +36 of the IKBKAP intron 20). This showed that the IKBKAP ISS repressed splicing of SMN1 and SMN2 even stronger than the known hnRNPA1 binding ISS-N1 silencer (Figure 5) confirming that the IKBKAP ISS is a strong ISS.
Blocking of the ISS completely corrects IKBKAP exon 20 splicing
Since the hnRNP A1 binding silencers are important for the skipping of IKBKAP exon 20 in FD patient fibroblasts, we hypothesized that blocking any one of these binding sites with SSOs could improve splicing. Others and we have previously shown that this can be an effective way of correcting deregulated splicing (20,21,39). SSOs targeting exonic splicing silencers may unintentionally interfere with recognition of other important regulatory elements such as exonic splicing enhancers (ESEs), moreover blocking of the hnRNP A1 binding ISS-N1 silencer (32) in SMN2 using an SSO corrects splicing of exon 7 and is currently in clinical use.
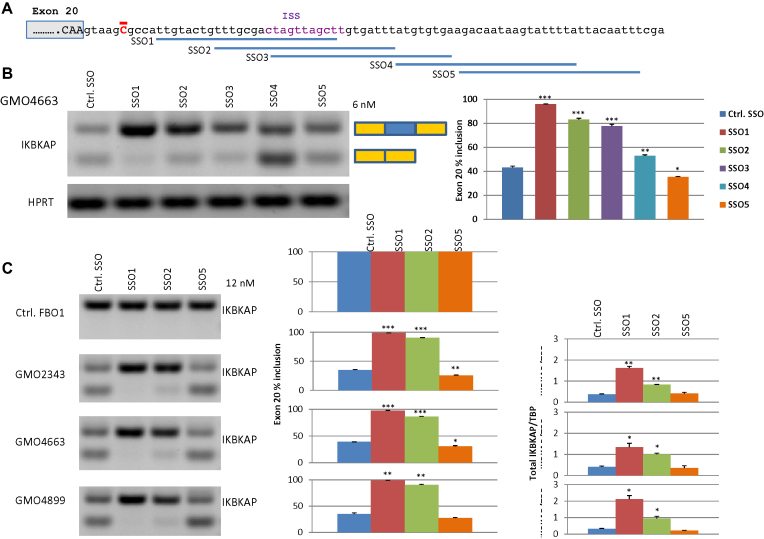
Therefore we designed SSOs located downstream of IKBKAP exon 20 (Figure 6). The overlapping SSO1, SSO2 and SSO3 all cover the ISS hnRNP A1 binding site, whereas SSO4 and SSO5 are located further downstream (Figure 6). We transfected FD patient fibroblast cell lines with the IKBKAP SSOs or a control SSO (Figure 6). All three SSOs that cover the hnRNP A1 binding ISS resulted in a dramatic correction of the impaired splicing of IKBKAP exon 20 in FD patient cells, and also increased total IKBKAP mRNA amounts (Figure 6) as the transcript without exon 20 is subject to degradation by the NMD pathway (40).
Figure 6.
SSOs that block the IKBKAP ISS restores splicing of IKBKAP exon 20 in FD patient cell lines. (A) Schematic of IKBKAP intron 20 and the binding sites for the SSOs. (B) Transfection of 12 nM IKBKAP SSOs or a control SSO into FD patient fibroblasts (GMO4663) was analyzed by gel electrophoresis of PCR products and inclusion of exon 20 was measured on the fragment analyzer. (C) Transfection of 12 nM IKBKAP SSO1, SSO2 or SSO5 or a control SSO into three different FD patient fibroblasts (GMO2343, GMO4663 and GMO4899) was analyzed by gel electrophoresis of PCR products and inclusion of exon 20 was measured on the fragment analyzer. The total IKBKAP expression was quantified by qPCR. Error bars indicates SEM, n ≥ 2 ***P-value < 0.001, **P-value < 0.01, *P-value < 0.05. Compared to control SSO.
In contrast, the two SSOs that did not block access, had no (SSO5) or even a slightly negative effect (SSO4). SSO1 performed slightly better than SSO2 and SSO3. This could indicate that the effect of the SSOs is also influenced by blocking other regulatory sites and/or that they bind with different efficiencies. Examination of the SSO designs by the IDT software (https://eu.idtdna.com/calc/analyzer) did not reveal any obvious differences, except that SSO1 has the highest melting temperature (56.2°C), perhaps indicating a more efficient binding.
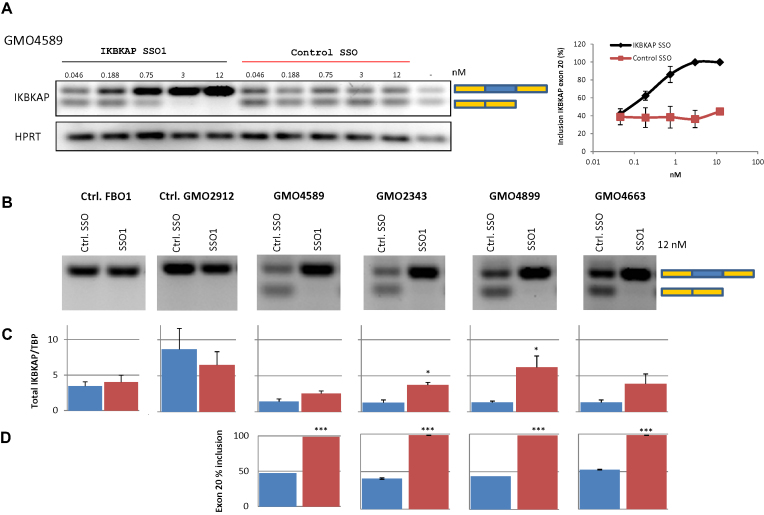
To further characterize the potency of the IKBKAP SSO1, we performed a gradient transfection using decreasing concentrations of SSOs (Figure 7). This generated a dose-response curve, which reached 100% inclusion already at ∼3 nM SSO1. Compared with other SSOs used in our laboratory this is a very potent SSO (20). SSO1 treatment at 12 nM caused complete correction of splicing in all four patient cell lines and IKBKAP mRNA levels were increased between 1.9- and 4.7-fold.
Figure 7.
SSO1 treatment of FD patient cell lines restores splicing of IKBKAP exon 20. (A) FD-patient cells GMO4589 were transfected with increasing amounts of IKBKAP SSO1 or control SSO. The splicing of IKBKAP exon 20 was then analyzed by PCR. The inclusion of exon 20 was quantified using the fragment analyzer. The range of inclusion for two biological replicates in duplicates is shown. (B) Transfection of 12 nM IKBKAP SSO1 or a control SSO into four different FD patient fibroblasts (GMO2343, GMO4663, GMO4589 and GMO4899) or two control fibroblasts (FB01 and GMO2912) was analyzed by gel electrophoresis of PCR products. (C) The total IKBKAP expression was quantified by qPCR. Error bars indicates SEM, n ≥ 3 ***P-value < 0.001, **P-value < 0.01, *P-value < 0.05. (D) Inclusion of exon 20 was measured on the fragment analyzer. Error bars indicates SEM, n ≥ 2 ***P-value < 0.001, **P-value < 0.01, *P-value < 0.05.
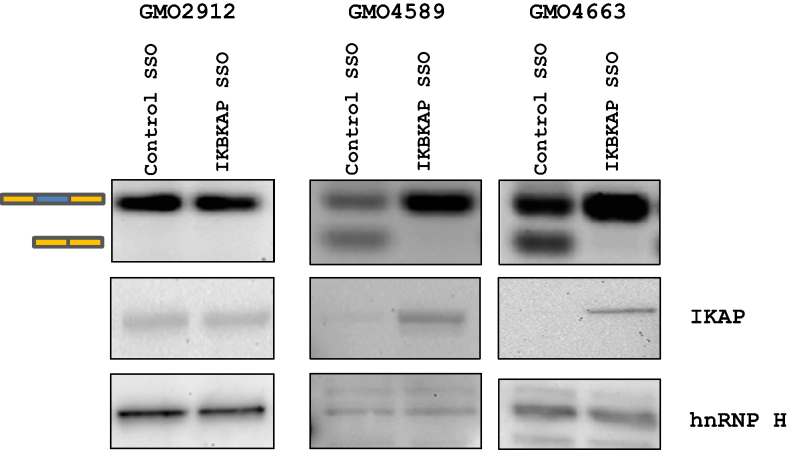
Next, we transfected the IKBKAP SSO1 into two FD patient cell lines and a control fibroblast cell line to evaluate the effect of IKBKAP SSO1 treatment on the IKAP protein expression (Figure 8). As previously observed, the IKBKAP SSO1 completely restores IKBKAP exon 20 splicing in patient cells and does not disturb splicing in non-FD fibroblasts. Importantly, we also observe that the positive effect of SSO1 treatment is reflected in a dramatic increase in IKAP protein levels in FD patient cells, indicating that this SSO could potentially be used to increase the level of functional IKAP protein in patients.
Figure 8.
IKBKAP SSO treatment increases IKAP protein expression. Transfections of IKBKAP SSO (12 nM) into control (GMO2912) or FD fibroblasts (GMO4589, GMO4663) restore splicing of IKBKAP exon 20 (top). Western blotting shows an increase in IKAP protein expression after transfection with IKBKAP SSO into FD fibroblasts but not control fibroblasts. hnRNP H was used as a loading control. Transfections were performed in triplicates.
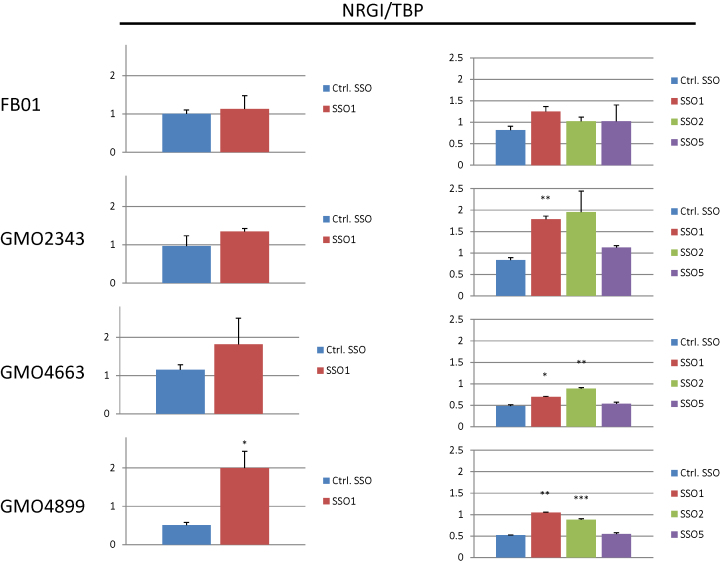
At last, we wanted to evaluate downstream effects of SSO based IKBKAP splicing correction in patient cells. One of the hallmarks of FD, which is also observed in a mouse model (41), is a reduction of pain and temperature-sensing TrkA+ neurons in the dorsal root ganglia (DRG), which is a primary site of IKBKAP expression. Interestingly, Neuregulin 1 (NRG1) knock-out mice also show a reduction of pain and temperature-sensing TrkA+ neurons in the DRG (42), and NRG1 has been reported to be downregulated in FD patient nasal mucosa cells (43). Both SSO1 and SSO2, but not SSO5 or a control SSO increased NRG1 expression in FD patient fibroblasts (Figure 9), consistent with the observed effects on IKBKAP expression (Figure 6). The fact that similar results are observed in different patient cell lines, and by treatment with different IKBKAP SSOs, reflecting their efficiency in restoring IKBKAP splicing, suggests that they are in fact a result of IKBKAP SSO based restoration of IKAP function.
Figure 9.
SSO treatment of FD patient cell lines restores NRG1 expression. Transfection of 12 nM IKBKAP SSOs or a control SSO into three different FD patient fibroblasts (GMO2343, GMO4663 and GMO4899) or a control fibroblast (FB01) was analyzed for NRG1 expression by qPCR and normalized to TBP. Left and right panel represent different experiments. Left: error bars indicates SEM, n ≥ 3 ***P-value < 0.001, **P-value < 0.01, *P-value < 0.05 compared to control SSO. Right: error bars indicates SEM, n ≥ 2 ***P-value < 0.001, **P-value < 0.01, *P-value < 0.05 compared to control SSO.
DISCUSSION
An estimated 25% of all disease-causing mutations may interfere with mRNA splicing (44,45). Therefore, the ability to control mRNA splicing has great potential in disease therapy. In recent years, a growing number of cases proves that manipulation of mRNA splicing using SSOs is a feasible approach for correcting disease-associated deregulated splicing (24). However, for specific and efficient design of SSOs insight into the mechanism of splicing regulation is necessary.
Here, we show that hnRNP A1 binds two silencer elements ESS1 and ESS2 inside IKBKAP exon 20 and to an ISS immediately downstream of the 5′ splice site. Knockdown of hnRNP A1 increases the inclusion of IKBKAP exon 20 in FD patient fibroblasts, and disruption of any of the hnRNP A1 binding sites in an FD-IKBKAP minigene restores exon 20 splicing, showing that hnRNP A1 plays an important role in repressing exon 20 in FD fibroblasts. The two exonic splicing silencers have previously been described, and the functionality of these were analyzed by minigene mutagenesis studies and in vitro splicing (32). Both silencers were shown to inhibit exon 20 inclusion, though ESS2 appeared to be a stronger inhibitor than ESS1, which is consistent with the fact that our SPRi analysis demonstrated more hnRNP A1 binding to the ESS2 than to ESS1. Furthermore, in another study investigating splicing of deletion mutants of an FD-IKBKAP minigene, only a deletion that disrupted ESS2, but not a deletion that disrupted the ESS1 silencer, restored splicing of exon 20 (17). We show that specifically mutating the hnRNP A1 binding sequence in either of the two exonic silencers, ESS1 and ESS2, completely restores splicing. The divergence regarding ESS1 may be caused by different experimental conditions. Our UUAG>UUCG ESS-disrupting mutations introduced into the FD-IKBKAP minigene may be a much more efficient disruption of the silencer element than the previously investigated UUAG>AUAG mutation in ESS1 (32). Moreover, deletion of the ESS1 silencer may not increase exon inclusion, because this at the same time disrupts the ESE1 SE (32), which overlaps the ESS1 silencer. Our study suggests that the ESS1, ESS2, as well as the ISS downstream of the 5′ splice site all inhibit inclusion of FD-IKBKAP exon 20. We propose that hnRNP A1 cooperatively binds to the three elements and spreads across the exon to prevent recognition of the weakened FD c.2204+6T>C 5′ splice site of IKBKAP exon 20 and replace positive splicing factors (Figure 10) (38). Disruption of any of the hnRNP A1-binding silencers may shift a finely tuned balance between positive and negative splicing elements and prevent the cooperative spreading of hnRNP A1. The loss of hnRNP A1 binding may then enable binding of positive splicing factors, like SR proteins or RBM24 (36), to SEs, which would help attract U1 snRNP to the weak 5′ splice site of c.2204+6T>C IKBKAP exon 20 and thereby restore splicing. On the other hand, the WT 5′ splice site is sufficiently strong to overcome hnRNP A1 binding allowing U1 snRNP to bind and it may therefore also prevent the cooperative spreading of hnRNP A1 across the exon. We have previously shown that splicing is a delicate process and single nucleotide variants outside the canonical splice sites can have strong effects on splicing. In the ACADM gene, a pathogenic mutation disrupting an ESE element causes exon skipping, but this can be counteracted by a nearby Single Nucleotide Polymorphism (SNP) that disrupts an hnRNPA1 binding ESS and thus restores the balance between positive and negative splicing elements (30).
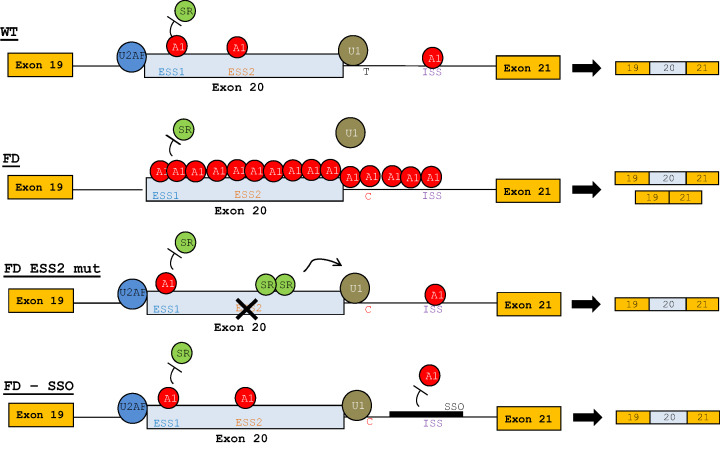
Figure 10.
Model of the hnRNP A1-dependent regulation of IKBKAP exon 20. WT: hnRNP A1 binds ESS1, ESS2 and ISS, however, the WT 5′ splice site is strong enough to attract the U1 snRNP, which prevents hnRNP A1 multimerization across the exon and ensures complete inclusion of IKBKAP exon 20. FD: the reduced splice site strength of the FD 5′ splice site makes U1 snRNP less capable of competing with hnRNP A1 for binding, resulting in multimerization of hnRNP A1 across the exon, inhibiting U1 snRNP and SR protein binding, which results in only partial exon inclusion. FD ESS2 mut: disruption of the ESS2 allows SR proteins to bind and attract U1 snRNP to the FD 5′ splice site. FD–SSO: treatment with the IKBKAP SSO inhibits hnRNP A1 binding to the ISS which enables U1 snRNP to bind to the 5′ splice site and therefore induce complete inclusion of IKBKAP exon 20.
To further analyze the WT and the FD 5′ splice site, we extracted 284213 donor splice sites from the Ensembl v79 annotation of the hg38 genome and found that of these, 1319 matched the CAAgtaagt sequence of the WT IKBKAP exon 20 5′ splice site, while only 254 matched the CAAgtaagc site of the FD 5′ splice site indicating that the WT splice site is indeed a more frequently used splice site. We counted the number of UAG motifs in the immediate downstream intronic region (+10 to +40) where the IKBKAP ISS is located and found that 413 of the 1319 (31.3%) WT splice sites harbored at least one UAG motif, whereas only 58 (22.8%) of the 254 exons with the weaker FD 5′ splice site harbored one UAG motif, suggesting that the stronger WT splice site may generally tolerate a higher number of silencer elements downstream of the splice site. This further illustrates that exon inclusion depends on a finely tuned balance of SREs in addition to splice site strength, and as we show here, that splicing can be affected by changing this balance of positive and negative SREs.
In our previous hnRNP A1 iCLIP study, we showed that hnRNP A1 binding immediately downstream of the 5′ splice site is generally important for exon repression (20), and that SSO-mediated blocking of hnRNP A1 binding sites in this region is effective in improving exon inclusion. In addition, SSOs targeting intronic regions have lower risk of interfering with important regulatory elements than SSOs targeting exons. An SSO targeting downstream of exon 16 of the CFTR gene corrects splicing in a c.2657+5G>A mutant minigene (46), and close inspection revealed a UAG motif in the sequence blocked by the SSO. The FDA-approved SSO correcting the SMN2 exon 7 exclusion also targets the hnRNP A1 binding ISS-N1 element immediately downstream the 5′ splice site (47). We have previously shown that inclusion of the cassette exon 3 in the SKA2 gene and a pseudoexon in MTRR can be improved by blocking an iCLIP-identified hnRNP A1 binding site immediately downstream of the 5′ splice site (20). Here, we employed three different overlapping SSOs targeting the hnRNP A1-binding ISS, which is also located immediately downstream of IKBKAP exon 20. Transfection of these IKBKAP SSOs into FD fibroblasts restored exon 20 splicing even at low doses. For the most efficient SSO (SSO1) we performed a dose response curve and showed a dose-dependent inclusion of exon 20. This underscores the importance of the IKBKAP ISS in repressing IKBKAP exon 20 splicing. We further confirmed the importance and potency of the IKBKAP ISS by demonstrating that it also functions very efficiently to suppress inclusion of exon 7 from the SMN1/2 genes when it substitutes the ISS-N1 in the SMN1/2 genes. This further strengthens the notion that blocking of hnRNP A1 binding sites downstream of weak 5′ splice sites is an efficient way of increasing exon inclusion, and it demonstrates how our previously generated iCLIP hnRNP A1 binding map can be used to pinpoint efficient target sites for SSO-based modulation of splicing. The fact that we were unable to completely restore IKBKAP exon 20 splicing by hnRNP A1 knockdown and by mutating the IKBKAP ISS in a minigene, may suggest that also other splicing inhibitory proteins can bind to the ISS or the flanking sequences in the region blocked by the SSOs.
Compared to current treatment strategies against FD, the IKBKAP SSO approach is very specific. Many of the small-molecule drugs that have been shown to improve IKBKAP exon 20 splicing, have broad non-specific effects on mRNA splicing or transcription (15–17,19). These drugs therefore pose the risk of off-target effects. Moreover, the successful therapy against SMA indicates that SSO-based therapy is well-tolerated in vivo (25), and that delivery of SSOs to CNS, which is also the primary target site for the IKBKAP SSO, is possible in vivo.
EGCG has been shown to rescue IKBKAP exon 20 splicing. Since EGCG treatment decreases hnRNP A2/B1 expression, it was suggested that hnRNP A2/B1 is a negative regulator of IKBKAP exon 20 splicing (9). Interestingly, we observed that knockdown of hnRNP A2/B1 has a negative effect on IKBKAP exon 20 inclusion, showing that hnRNP A2/B1 is a positive regulator of IKBKAP exon 20 splicing. This suggests that the positive effect of EGCG is most likely mediated through a different mechanism than hnRNP A2/B1 downregulation. HnRNP A1 and hnRNP A2/B1 have previously been shown to have opposite effects on the same splicing events (28,48). When hnRNP A2/B1 and hnRNP A1 bind the same splicing elements, hnRNP A2/B1 may have a positive effect on splicing by competing with and inhibiting the binding of hnRNP A1 to the SSs. This correlates with the improvement of IKBKAP exon 20 splicing in the FD-IKBKAP minigene when disrupting the ESS1, ESS2 or the ISS, suggesting that predominantly negative splicing factors bind these splicing elements. In particular the positive effect of blocking the ISS with SSOs indicates that the ISS mainly binds negative splicing factors.
At last, we demonstrate that SSO-based blocking of the ISS increases IKAP protein levels in FD patient cell lines, and interestingly, that SSO treatment increases the suppressed neuregulin 1 (NRG1) expression in patient cell lines investigated. Decreased expression of NRG1 in FD patient nasal mucosa cells cells has previously been reported (43). Neuregulin 1 is a key regulator of the peripheral nervous system and has been shown to be essential for survival of TrkA+ sensory neurons. Because increased death of TrkA+ sensory neurons is a hallmark of FD, it could be speculated that some of the clinical symptoms in FD may be due to IKBKAP deficiency-mediated NRG1 downregulation.
Altogether, we demonstrate that hnRNP A1 is an important inhibitor of inclusion of FD-IKBKAP exon 20 carrying the weak c.2204+6T>C 5′ splice site, which is the most frequent cause of FD. We take advantage of this by SSO-mediated blocking of the hnRNP A1-binding ISS, which completely restores splicing of IKBKAP exon 20 in FD patient cell lines.
However, before this approach can be developed into an efficient therapy for FD patients it is clear that further preclinical testing for instance in the available FD mouse model (49) is required. Although a BLAST search did not reveal any other potential targets in the human genome, testing for potential side effects resulting from unspecific binding of SSO1 to other targets should also be evaluated for instance by RNA-seq analysis of human cells.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Tanja Bruun, Kristian Traantoft Rasmussen, Margrethe Thusholdt, Krystyna Giemza and Aleksandra Kulus for technical assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natur og Univers, Det Frie Forskningsråd [4181-00515 to B.S.A.]; Novo Nordisk Fonden (DK) [61310-0128, NNF17OC0029240 to B.S.A.]. Funding for open access charge: University of Southern Denmark.
Conflict of interest statement. None declared.
REFERENCES
- 1. Dong J., Edelmann L., Bajwa A.M., Kornreich R., Desnick R.J.. Familial dysautonomia: detection of the IKBKAP IVS20(+6T → C) and R696P mutations and frequencies among Ashkenazi Jews. Am. J. Med. Genet. 2002; 110:253–257. [DOI] [PubMed] [Google Scholar]
- 2. Slaugenhaupt S.A., Blumenfeld A., Gill S.P., Leyne M., Mull J., Cuajungco M.P., Liebert C.B., Chadwick B., Idelson M., Reznik L. et al. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am. J. Hum. Genet. 2001; 68:598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cuajungco M.P., Leyne M., Mull J., Gill S.P., Lu W., Zagzag D., Axelrod F.B., Maayan C., Gusella J.F., Slaugenhaupt S.A.. Tissue-specific reduction in splicing efficiency of IKBKAP due to the major mutation associated with familial dysautonomia. Am. J. Hum. Genet. 2003; 72:749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anderson S.L., Coli R., Daly I.W., Kichula E.A., Rork M.J., Volpi S.A., Ekstein J., Rubin B.Y.. Familial dysautonomia is caused by mutations of the IKAP gene. Am. J. Hum. Genet. 2001; 68:753–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shapiro M.B., Senapathy P.. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987; 15:7155–7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Roca X., Olson A.J., Rao A.R., Enerly E., Kristensen V.N., Borresen-Dale A.L., Andresen B.S., Krainer A.R., Sachidanandam R.. Features of 5′-splice-site efficiency derived from disease-causing mutations and comparative genomics. Genome Res. 2008; 18:77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang Z., Burge C.B.. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008; 14:802–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee G., Ramirez C.N., Kim H., Zeltner N., Liu B., Radu C., Bhinder B., Kim Y.J., Choi I.Y., Mukherjee-Clavin B. et al. Large-scale screening using familial dysautonomia induced pluripotent stem cells identifies compounds that rescue IKBKAP expression. Nat. Biotechnol. 2012; 30:1244–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Anderson S.L., Qiu J., Rubin B.Y.. EGCG corrects aberrant splicing of IKAP mRNA in cells from patients with familial dysautonomia. Biochem. Biophys. Res. Commun. 2003; 310:627–633. [DOI] [PubMed] [Google Scholar]
- 10. Anderson S.L., Liu B., Qiu J., Sturm A.J., Schwartz J.A., Peters A.J., Sullivan K.A., Rubin B.Y.. Nutraceutical-mediated restoration of wild-type levels of IKBKAP-encoded IKAP protein in familial dysautonomia-derived cells. Mol. Nutr. Food Res. 2012; 56:570–579. [DOI] [PubMed] [Google Scholar]
- 11. Slaugenhaupt S.A., Mull J., Leyne M., Cuajungco M.P., Gill S.P., Hims M.M., Quintero F., Axelrod F.B., Gusella J.F.. Rescue of a human mRNA splicing defect by the plant cytokinin kinetin. Hum. Mol. Genet. 2004; 13:429–436. [DOI] [PubMed] [Google Scholar]
- 12. Lee G., Papapetrou E.P., Kim H., Chambers S.M., Tomishima M.J., Fasano C.A., Ganat Y.M., Menon J., Shimizu F., Viale A. et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009; 461:402–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shetty R.S., Gallagher C.S., Chen Y.T., Hims M.M., Mull J., Leyne M., Pickel J., Kwok D., Slaugenhaupt S.A.. Specific correction of a splice defect in brain by nutritional supplementation. Hum. Mol. Genet. 2011; 20:4093–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Axelrod F.B., Liebes L., Gold-Von Simson G., Mendoza S., Mull J., Leyne M., Norcliffe-Kaufmann L., Kaufmann H., Slaugenhaupt S.A.. Kinetin improves IKBKAP mRNA splicing in patients with familial dysautonomia. Pediatr. Res. 2011; 70:480–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hims M.M., Ibrahim E.C., Leyne M., Mull J., Liu L., Lazaro C., Shetty R.S., Gill S., Gusella J.F., Reed R. et al. Therapeutic potential and mechanism of kinetin as a treatment for the human splicing disease familial dysautonomia. J. Mol. Med. (Berl). 2007; 85:149–161. [DOI] [PubMed] [Google Scholar]
- 16. Pros E., Fernandez-Rodriguez J., Benito L., Ravella A., Capella G., Blanco I., Serra E., Lazaro C.. Modulation of aberrant NF1 pre-mRNA splicing by kinetin treatment. Eur. J. Hum. Genet. 2010; 18:614–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu B., Anderson S.L., Qiu J., Rubin B.Y.. Cardiac glycosides correct aberrant splicing of IKBKAP-encoded mRNA in familial dysautonomia derived cells by suppressing expression of SRSF3. FEBS J. 2013; 280:3632–3646. [DOI] [PubMed] [Google Scholar]
- 18. Yoshida M., Kataoka N., Miyauchi K., Ohe K., Iida K., Yoshida S., Nojima T., Okuno Y., Onogi H., Usui T. et al. Rectifier of aberrant mRNA splicing recovers tRNA modification in familial dysautonomia. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:2764–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Donyo M., Hollander D., Abramovitch Z., Naftelberg S., Ast G.. Phosphatidylserine enhances IKBKAP transcription by activating the MAPK/ERK signaling pathway. Hum. Mol. Genet. 2016; 25:1307–1317. [DOI] [PubMed] [Google Scholar]
- 20. Bruun G.H., Doktor T.K., Borch-Jensen J., Masuda A., Krainer A.R., Ohno K., Andresen B.S.. Global identification of hnRNP A1 binding sites for SSO-based splicing modulation. BMC Biol. 2016; 14:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Palhais B., Praestegaard V.S., Sabaratnam R., Doktor T.K., Lutz S., Burda P., Suormala T., Baumgartner M., Fowler B., Bruun G.H. et al. Splice-shifting oligonucleotide (SSO) mediated blocking of an exonic splicing enhancer (ESE) created by the prevalent c.903+469T>C MTRR mutation corrects splicing and restores enzyme activity in patient cells. Nucleic Acids Res. 2015; 43:4627–4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hartung A.M., Swensen J., Uriz I.E., Lapin M., Kristjansdottir K., Petersen U.S., Bang J.M., Guerra B., Andersen H.S., Dobrowolski S.F. et al. The splicing efficiency of activating HRAS mutations can determine costello syndrome phenotype and frequency in cancer. PLoS Genet. 2016; 12:e1006039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Palhais B., Dembic M., Sabaratnam R., Nielsen K.S., Doktor T.K., Bruun G.H., Andresen B.S.. The prevalent deep intronic c. 639+919 G>A GLA mutation causes pseudoexon activation and Fabry disease by abolishing the binding of hnRNPA1 and hnRNP A2/B1 to a splicing silencer. Mol. Genet. Metab. 2016; 119:258–269. [DOI] [PubMed] [Google Scholar]
- 24. Havens M.A., Hastings M.L.. Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res. 2016; 44:6549–6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Finkel R.S., Chiriboga C.A., Vajsar J., Day J.W., Montes J., De Vivo D.C., Yamashita M., Rigo F., Hung G., Schneider E. et al. Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet. 2016; 388:3017–3026. [DOI] [PubMed] [Google Scholar]
- 26. Corey D.R. Nusinersen, an antisense oligonucleotide drug for spinal muscular atrophy. Nat. Neurosci. 2017; 20:497–499. [DOI] [PubMed] [Google Scholar]
- 27. Jean-Philippe J., Paz S., Caputi M.. hnRNP A1: the Swiss army knife of gene expression. Int. J. Mol. Sci. 2013; 14:18999–19024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huelga S.C., Vu A.Q., Arnold J.D., Liang T.Y., Liu P.P., Yan B.Y., Donohue J.P., Shiue L., Hoon S., Brenner S. et al. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep. 2012; 1:167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yeo G., Burge C.B.. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004; 11:377–394. [DOI] [PubMed] [Google Scholar]
- 30. Nielsen K.B., Sorensen S., Cartegni L., Corydon T.J., Doktor T.K., Schroeder L.D., Reinert L.S., Elpeleg O., Krainer A.R., Gregersen N. et al. Seemingly neutral polymorphic variants may confer immunity to splicing-inactivating mutations: a synonymous SNP in exon 5 of MCAD protects from deleterious mutations in a flanking exonic splicing enhancer. Am. J. Hum. Genet. 2007; 80:416–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Burd C.G., Dreyfuss G.. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994; 13:1197–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ibrahim E.C., Hims M.M., Shomron N., Burge C.B., Slaugenhaupt S.A., Reed R.. Weak definition of IKBKAP exon 20 leads to aberrant splicing in familial dysautonomia. Hum. Mutat. 2007; 28:41–53. [DOI] [PubMed] [Google Scholar]
- 33. Olsen R.K., Broner S., Sabaratnam R., Doktor T.K., Andersen H.S., Bruun G.H., Gahrn B., Stenbroen V., Olpin S.E., Dobbie A. et al. The ETFDH c.158A>G variation disrupts the balanced interplay of ESE- and ESS-binding proteins thereby causing missplicing and multiple Acyl-CoA dehydrogenation deficiency. Hum. Mutat. 2014; 35:86–95. [DOI] [PubMed] [Google Scholar]
- 34. Doktor T.K., Schroeder L.D., Vested A., Palmfeldt J., Andersen H.S., Gregersen N., Andresen B.S.. SMN2 exon 7 splicing is inhibited by binding of hnRNP A1 to a common ESS motif that spans the 3′ splice site. Hum. Mutat. 2011; 32:220–230. [DOI] [PubMed] [Google Scholar]
- 35. Fu X.D., Ares M. Jr.. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 2014; 15:689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ohe K., Yoshida M., Nakano-Kobayashi A., Hosokawa M., Sako Y., Sakuma M., Okuno Y., Usui T., Ninomiya K., Nojima T. et al. RBM24 promotes U1 snRNP recognition of the mutated 5′ splice site in the IKBKAP gene of familial dysautonomia. RNA. 2017; 23:1393–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu B., Su S., Patil D.P., Liu H., Gan J., Jaffrey S.R., Ma J.. Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat. Commun. 2018; 9:420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Okunola H.L., Krainer A.R.. Cooperative-binding and splicing-repressive properties of hnRNP A1. Mol. Cell. Biol. 2009; 29:5620–5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hua Y., Vickers T.A., Okunola H.L., Bennett C.F., Krainer A.R.. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet. 2008; 82:834–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Boone N., Loriod B., Bergon A., Sbai O., Formisano-Treziny C., Gabert J., Khrestchatisky M., Nguyen C., Feron F., Axelrod F.B. et al. Olfactory stem cells, a new cellular model for studying molecular mechanisms underlying familial dysautonomia. PLoS One. 2010; 5:e15590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. George L., Chaverra M., Wolfe L., Thorne J., Close-Davis M., Eibs A., Riojas V., Grindeland A., Orr M., Carlson G.A., Lefcort F.. Familial dysautonomia model reveals Ikbkap deletion causes apoptosis of Pax3+ progenitors and peripheral neurons. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:18698–18703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hancock M.L., Nowakowski D.W., Role L.W., Talmage D.A., Flanagan J.G.. Type III neuregulin 1 regulates pathfinding of sensory axons in the developing spinal cord and periphery. Development. 2011; 138:4887–4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Boone N., Bergon A., Loriod B., Devèze A., Nguyen C., Axelrod F.B., Ibrahim E.C.. Genome-wide analysis of familial dysautonomia and kinetin target genes with patient olfactory ecto-mesenchymal stem cells. Hum. Mutat. 2012; 33:530–540. [DOI] [PubMed] [Google Scholar]
- 44. Lim K.H., Ferraris L., Filloux M.E., Raphael B.J., Fairbrother W.G.. Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:11093–11098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sterne-Weiler T., Howard J., Mort M., Cooper D.N., Sanford J.R.. Loss of exon identity is a common mechanism of human inherited disease. Genome Res. 2011; 21:1563–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Igreja S., Clarke L.A., Botelho H.M., Marques L., Amaral M.D.. Correction of a cystic fibrosis splicing mutation by antisense oligonucleotides. Hum. Mutat. 2016; 37:209–215. [DOI] [PubMed] [Google Scholar]
- 47. Singh N.N., Howell M.D., Androphy E.J., Singh R.N.. How the discovery of ISS-N1 led to the first medical therapy for spinal muscular atrophy. Gene Ther. 2017; 24:520–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bonomi S., di Matteo A., Buratti E., Cabianca D.S., Baralle F.E., Ghigna C., Biamonti G.. HnRNP A1 controls a splicing regulatory circuit promoting mesenchymal-to-epithelial transition. Nucleic Acids Res. 2013; 41:8665–8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hims M.M., Shetty R.S., Pickel J., Mull J., Leyne M., Liu L., Gusella J.F., Slaugenhaupt S.A.. A humanized IKBKAP transgenic mouse models a tissue-specific human splicing defect. Genomics. 2007; 90:389–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.