Abstract
The Repressor Element 1-silencing transcription factor (REST) represses a number of neuronal genes in non-neuronal cells or in undifferentiated neural progenitors. Here, we report that the DEAD box RNA helicase DDX17 controls important REST-related processes that are critical during the early phases of neuronal differentiation. First, DDX17 associates with REST, promotes its binding to the promoter of a subset of REST-targeted genes and co-regulates REST transcriptional repression activity. During neuronal differentiation, we observed a downregulation of DDX17 along with that of the REST complex that contributes to the activation of neuronal genes. Second, DDX17 and its paralog DDX5 regulate the expression of several proneural microRNAs that are known to target the REST complex during neurogenesis, including miR-26a/b that are also direct regulators of DDX17 expression. In this context, we propose a new mechanism by which RNA helicases can control the biogenesis of intronic miRNAs. We show that the processing of the miR-26a2 precursor is dependent on RNA helicases, owing to an intronic regulatory region that negatively impacts on both miRNA processing and splicing of its host intron. Our work places DDX17 in the heart of a pathway involving REST and miRNAs that allows neuronal gene repression.
INTRODUCTION
Cell fate decisions are regulated at both transcriptional and post-transcriptional levels, through complex gene circuits that often display feedforward and feedback regulatory loops involving transcription factors and other factors, in particular microRNAs (miRNAs) (1). Neuronal differentiation is a paradigm for biological processes that are finely controlled through multiple loops of regulation (2,3). A central factor in this process is the Repressor Element 1-silencing transcription factor (REST), which represses a large number of neuron-specific genes in non-neuronal cells or in undifferentiated neural progenitors (4–6). REST binds to a conserved 21–23 bp sequence and often recruits a repressive complex containing the REST corepressor 1 (RCOR1, also known as CoREST) and several chromatin modifying factors (7–11). Some of the genes targeted by the REST complex encode miRNAs that have critical functions for neural development (12,13). These miRNAs, like miR-9/9* and miR-124, have multiple targets during neurogenesis, in particular REST itself as well as REST cofactors (14–18). The decreased expression of REST, which relieves the repression of neuronal genes, coincides with the exit from cell cycle to license progenitors for their terminal neuronal differentiation process (10,17,19).
Another regulatory loop during neurogenesis involves the family of RNA polymerase II carboxyl-terminal domain (CTD) small phosphatase (CTDSP) proteins (also known as small CTD phosphatase or SCP1 to 3). It has been shown that CTDSP1 interacts with the REST complex and contributes to the transcriptional silencing of neuronal genes (20). Each of the three paralog genes CTDSP1, CTDSP2 and CTDSPL encodes an intronic miRNA precursor of the miR-26a/b family, respectively pri-mir-26b, 26a2 and 26a1. These miRNAs can target their host transcripts in a negative feedback loop of regulation (21). This feedback control is essential for neurogenesis in zebrafish, but it cannot take place in undifferentiated neural stem cells due to the incomplete maturation of the miR-26b precursor, the ortholog of mammalian miR-26a2 (21). The mechanism that blocked the processing of zebrafish miR-26b precursor remained uncharacterized.
Because of their multiple molecular activities, RNA helicases regulate virtually all gene expression steps (22–24). In particular, the highly related ATP-dependent DEAD box helicases 5 (DDX5, also known as p68) and DDX17 (p72) have multiple and partially redundant functions in the regulation of gene expression (25), and they emerge as key factors to regulate cell fate switches and biological transitions (26–28). One of their functions is to coregulate the activity of various transcription factors, including the Estrogen Receptor alpha, MyoD or RORγt (29,30). The interaction of DDX5 and DDX17 with MyoD is central during skeletal myoblast differentiation since it regulates the expression of master myogenic transcription factors and miRNAs (26,31). Other functions of DDX5 and DDX17 include the regulation of the nuclear maturation of some miRNA precursors through their interaction with the Microprocessor components Drosha and DGCR8 (32–36), and the regulation of pre-mRNA alternative splicing (26,37–43).
Here, we report a new function of DDX17 as a major regulator of the REST complex. DDX17 controls the binding of REST to its target promoters and corepresses neuronal gene expression. Moreover, DDX17 regulates the expression of miRNAs that are required for the coordinated loss of the REST complex during the early phases of neurogenesis. Our work also uncovers an intronic regulatory region that negatively impacts on both miR-26a2 processing and splicing of its host intron in absence of DDX5 and DDX17.
MATERIALS AND METHODS
Cell culture, differentiation and transfections
The human SH-SY5Y neuroblastoma cell line (ECACC) was cultured in DMEM/F12 medium complemented with 10% FBS, 1% glutamine and 1% penicilline/streptomycine. The CLBMA2 cell line (44) (see also Supplementary Materials and Methods for details) was cultured in RPMI medium complemented with 10% FBS, 1% glutamine and 1% penicilline/streptomycine. Cells were differentiated using 10 μM all-trans retinoic acid (ATRA, Sigma) for up to 2 weeks, as indicated in the figures. In standard transfections experiments on SH-SY5Y cells, 20 nM of siRNA were mixed with Lipofectamine RNAiMax (Thermo Fisher Scientific) following the manufacturer's instructions and cells were harvested 48 h after transfection, unless indicated. For double siRNA treatments, we used a total of 40 nM siRNA (20 nM siDDX5/DDX17 + 20 nM siREST). In this case the single depletion were also carried out with a total of 40 nM siRNA: 20 nM siDDX5/DDX17 or 20 nM siREST complemented with 20 nM of control siRNA. For experiments using 2′-O-methylated antisense RNA oligonucleotides (AON), 10 nM of siRNA were mixed with 100 nM of AONs. Finally, for overexpression of miRNAs, 10 nM of pre-miR miRNA precursors (#AM17100, Thermo Fisher Scientific) were transfected with Lipofectamine RNAiMax as for siRNAs.
Stable inducible MCF7 lines expressing DDX5 and DDX17 were cultured as previously described (26,43).
Luciferase assays
The 3′ half of the DDX17 3′ UTR, containing predicted binding sites for miR-26a/b and miR-9 (according to TargetScanHuman v6.2), was cloned in the psiCHEK-2 plasmid (Promega) downstream of the Renilla luciferase gene. This plasmids also contain the Firefly luciferase gene used for internal normalization of the data. A mutant derivative construct (made by Eurofins Genomics) was obtained by directed mutagenesis of the wild-type psiCHEK-DDX17 plasmid to delete four nucleotides within the predicted miR-26a/b binding site. Similarly, we cloned in the psiCHEK-2 plasmid the first 1000 bp of the REST 3′UTR, which contains the most conserved of the predicted binding sites for miR-26a/b, and we engineered a 4-bp deletion mutant from this wild-type construct.
SH-SY5Y cells were seeded (22 500 cells per well) in 96-well plates and transfected the day after with 7.5 ng of luciferase reporter along with 10 nM pre-miR precursor (control or pre-miR-26a or pre-miR-26b, ThermoFisher/Life Technologies), using JetPrime® (Polyplus transfection). Luciferase expression was measured 24 h after transfection directly in the plates using the Dual-Glo® Luciferase Assay System (Promega), as recommended by the manufacturer. Data were expressed as the ratio of Renilla (test) to Firefly (internal control) luminescence, and normalized to the ratio obtained with the control pre-miR.
Proximity ligation assay
In situ proximity ligation assay (PLA) were performed on SH-SY5Y cells using the Duolink in Situ Kit (Sigma-Aldrich), as recommended by the manufacturer. The following antibodies were used: DDX17 proteins (ab71958, Abcam), DDX5 (Pab204, Millipore) and REST (07-579, Millipore). Quantification of the PLA foci was based on a previously described macro (45) that was optimized to detect foci within nuclei of human cells. The macro was designed to automatically detect and process batches of microscope-coupled acquisitions, as detailed in Supplementary Materials and Methods.
Co-immunoprecipitation and western blotting
Total protein extraction was carried out as previously described (26). Primary antibodies used for Western-blotting: DDX5 (ab10261, Abcam), DDX17 (ab24601, Abcam), MYCN (sc-56729, SantaCruz), NCAM1 (ab18277, Abcam), REST (07-579 Millipore), CoREST (07-455 Millipore), Synapsin-1 (D12G5, Ozyme), Actin (sc-1616, SantaCruz) and GAPDH (sc-32233, SantaCruz).
For co-immunoprecipitation, SH-SY5Y cells (7 × 106 to 8 × 106 per assay) were harvested and lysed in a lysis buffer containing 20 mM Tris–HCl pH 7.5, 150 mM NaCl, 2 mM EDTA, 1% NP40, 10% glycerol, cOmplete protease inhibitor cocktail (Roche) (1 ml of lysis buffer was used for each 10 cm Petri dish). Lysis was carried out at 4°C for 30 min under rotation. The lysate was centrifuged for 15 min at 13 000 rpm to remove cell debris and then split in equal amounts for each assay. Each fraction received 5 μg of antibody and the incubation was left overnight at 4°C under rotation. The following antibodies were used for IP: rabbit anti-DDX17 (ProteinTech 19910-1-AP) or a control rabbit antibody (anti-SSU72, Cell Signalling D3I2D), and goat anti-DDX5 (Abcam ab10261) or control Rabbit IgG (Santa Cruz). The next day, the different lysate/antibody mixtures were incubated with 50 μl Dynabeads® Protein G (Thermo Fisher) blocked with salmon sperm DNA and bovine serum albumine, for 5 h at 4°C under rotation. Bead were then washed three times with lysis buffer. Elution was performed by boiling for 5 min in SDS-PAGE loading buffer prior to analysis by western-blotting.
Chromatin immunoprecipitation
The composition of all buffers is given in Supplementary Materials and Methods. SH-SY5Y cells were cross-linked with 1% formaldhehyde for 10 min. Cross-linking was stopped by addition of 0.125 M glycin and cells were harvested and centrifuged. Nuclei were isolated by sonication using a Covaris S220 (2′ peak power: 75; duty factor: 2; cycles/burst: 200), pelleted by centrifugation at 3000 rpm for 5 min at 4°C, washed once with FL buffer and resuspended in 1 ml Shearing buffer. Chromatin was sheared to fragments of about 200 bp using a Covaris S220 (20′ peak power: 140; duty factor: 5; cycles/burst: 200), and debris were eliminated by 10 minutes centrifugation at 13 200 rpm at 4°C. 30 μg of chromatin were diluted 10 times in Buffer D and pre-cleared for 30 min at 4°C on a rotating wheel with 30 μl Dynabeads® Protein A (Thermo Fisher) blocked for 1 h with 20 μg salmon sperm DNA. Beads were removed, 1% was collected as input fraction and the rest of the chromatin was immunoprecipitated overnight at 4°C with 5 μg of the following antibodies: rabbit anti-DDX17 (ProteinTech 19910-1-AP), rabbit anti REST (Millipore, 07-579), rabbit anti-EHMT2 (Cell Signaling Technologies, 3306S) or control rabbit IgG (Cell Signaling Technologies). Immunoprecipitates were bound to 30 μl Dynabeads® Protein A (Thermo Fisher) blocked as above, for 1.5 h at 4°C under rotation. Beads were washed under increasing stringency (see table below) and reverse cross-link was carried out by incubation in Elution buffer for 4 h at 65°C with 40 μg Proteinase K in a Thermomixer. DNA was purified by phenol–chloroform extraction and ethanol precipitation and resuspended in ultra-pure water for qPCR analysis.
RNA extraction and PCR analyses
Total RNA were isolated using TriPure Isolation Reagent (Roche). For reverse transcription, 1–2 μg of purifed RNAs were treated with Dnase I (ThermoFisher) and retrotranscribed using Maxima reverse transcriptase (ThermoFisher), as recommended by the supplier. Potential genomic DNA contamination was verified by performing negative RT controls in absence of enzyme, and by including controls with water instead of cDNA in PCR and qPCR assays. See also the Supplementary Materials and Methods for more details.
All PCR analyses were performed on 0.5 ng cDNA using 0.5 U GoTaq® DNA polymerase (Promega), as detailed (including primers sequences) in Supplementary Materials and Methods.
Quantitative PCR analyses
For quantitative analyses of global steady-state gene expression, primers were designed in regions of the transcript that are present in all mRNA isoforms and not submitted to alternative splicing changes, to be sure to quantify the overall expression of the gene. All primer sequences can be found in Supplementary Materials and Methods. The specificity and linear efficiency of all qPCR primers was first verified by establishing a standard expression curve with various amounts of human genomic DNA or cDNA. qPCR reactions were carried out on 0.625 ng cDNA using a LightCycler 480 System (Roche), with either the SYBR® Green I Master Mix (Roche) or the SYBR® Premix Ex Taq (Tli RNaseH Plus) (Takara), under thermocycling conditions that were recommenced by both manufacturers for this apparatus. Melting curves were systematically controlled to rule out the existence of non-specific products. Relative DNA levels were calculated using the ΔΔCt method (using the average Ct obtained from technical duplicates or triplicates) and were normalized to the expression of 18S RNA.
Quantification of mature miRNAs was carried out using the miRCURY LNA Universal cDNA synthesis and SYBR Green PCR kit (Exiqon), with specific LNA primer sets, in full compliance with the manufacturer's instructions. qRT-PCR values were normalized relative to U6 snRNA level.
RNA-Seq and bioinformatics
Stranded paired-end sequencing of total poly-A transcripts was performed on an Illumina HiSeq 2500 platform (Aros Applied Biotechnology, Aarhus, Denmark). Reads were mapped and analysed as detailed in Supplementary Materials and Methods. The differential gene analysis was carried out with DESeq2 package (46). Parameters for differential expression: adjusted P-value < 0.05 and [log2(FC)] ≥ 0.35. All enrichment analyses for biological functions and transcription factor binding sites were carried out using the Enrichr analysis tool (47).
Prediction of RNA secondary structures with Mfold
As the Mfold program (48) does not allow to predict the folding of long RNA sequences (maximum 800 bases), we used chimeric sequences of variable length that included the two regions regulated by DDX5 and DDX17, i.e. the exon 6 and the pri-mir-26a2. The default Mfold parameters were used. The picture presented in Figure 2 shows a part of a typical structure predicted from a 500 nt-long sequence composed of: (i) a 200-nt fragment that includes full exon 6 and the beginning of intron 6; (ii) a 300-nt fragment centered on the miRNA hairpin. Note that a competing base-pairing between the proximal intronic sequence and the lower part of the pri-mir-26a2 hairpin was predicted whatever the length of the tested sequence (up to 800 nt).
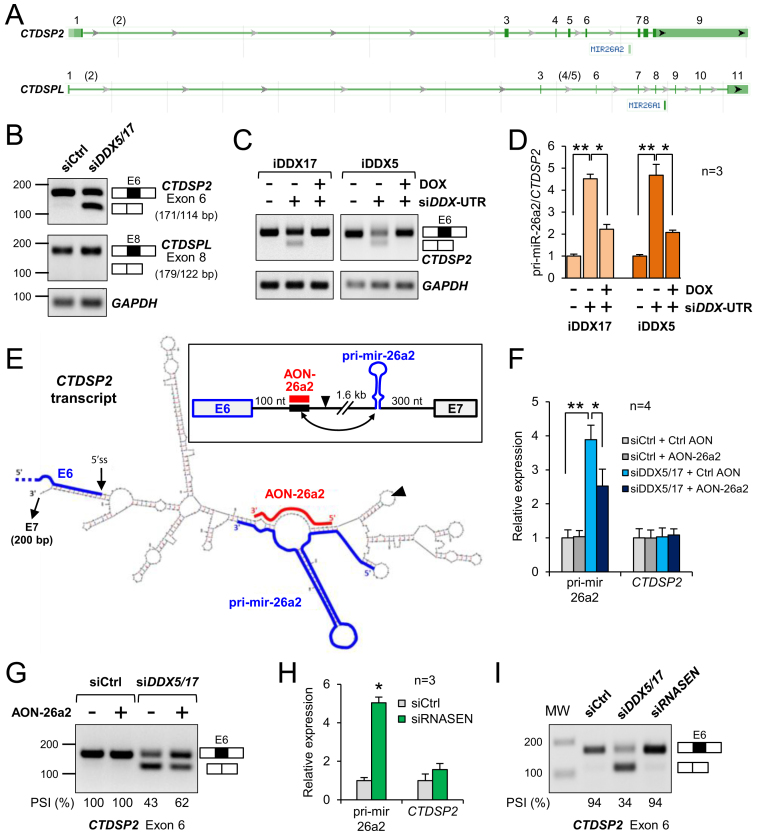
Figure 2.
DDX5 and DDX17 ensure the correct processing of CTDSP2/miR-26a2 transcripts. (A) Genomic organization of CTDSP2 and CTDSPL genes (NCBI Gene screenshots) showing the position of miR-26a2 and miR-26a1 within CTDSP2 intron 6 and CTDSPL intron 8, respectively. Exon numbering is based on the FasterDB database (78), which reports exons (in brackets) not annotated by the NCBI. (B) RT-PCR analysis of CTDSP2 and CTDSPL transcripts. The inclusion of the exon preceding the two miR-26a loci was monitored following treatment with control siRNA or siDDX5/DDX17. The size of the different PCR products (exon inclusion and exon skipping) is indicated. (C) RT-PCR analysis of CTDSP2 exon 6 inclusion in stable MCF7 cell lines. Cells were treated with siRNA to silence endogenous DDX5 and DDX17 genes, and treated or not with doxycyclin to induce the expression of recombinant DDX17 (iDDX17) or DDX5 (iDDX5). (D) RT-qPCR analysis of pri-miR-26a2 in stable MCF7 cell lines. Experimental details are as in C. The ratio of pri-mir-26a2 precursor to total CTDSP2 mRNA is represented as the mean value + S.E.M. of independent experiments (n = 3), normalized to the control condition (siCtrl without Doxycyclin). *P-value < 0.05; **P-value < 0.01 (Student's t-test). (E) Predicted folding of the RNA region encompassing CTDSP2 exon 6 and pri-mir-26a2. The framed inset on top of the figure represents a linear view of the region. The black arrowhead indicates the site of the junction between the 2 sequence fragments used for the Mfold prediction (see Materials and Methods for details). Exon 6 (E6) and the pri-mir-26a2 hairpin are highlighted by thick blue lines. The thick black element indicates the 5′-located intronic sequence predicted to base-pair (double-headed arrow in the inset) with the lower stem of the pri-miRNA. The antisense competitor RNA (AON-26a2, in red) was designed to perfectly hybridize with the repressor element, in order to restore the correct folding of the pri-miRNA hairpin. (F) Competing with the intronic inhibitory structure rescues the pri-mir-26a2 processing defect in absence of DDX5/DDX17. The AON-26a2 (or a control AON) was transfected in cells along with control siRNA (siCtrl) or siDDX5/DDX17. The pri-mir-26a2 precursor and the CTDSP2 mRNA were quantified by RT-qPCR. Data are represented as the mean values + S.E.M. of independent experiments (n = 4), normalized to the control condition (siCtrl + Ctrl AON), which was set to 1. *P-value < 0.05; **P-value < 0.01 (Student's t-test). (G) Competing with the intronic inhibitory structure rescues the inclusion defect of CTDSP2 exon 6 in absence of DDX5/DDX17. Experimental design is as in panel D. Inclusion of CTDSP2 exon 6 was analysed as in panel B. The percentage of spliced-in sequence (PSI) is indicated. (H) Inhibition of pri-mir-26a2 processing upon silencing of Drosha (RNASEN). Details are as in panel D. ( I) Silencing of Drosha (RNASEN) does not induce the skipping of CTDSP2 exon 6 (measured as in panel B), showing that splicing and pri-mir-26a2 processing are intrinsically uncoupled processes.
RESULTS
DDX5 and DDX17 control the expression of specific miRNAs during neuronal differentiation
Since DDX5 and DDX17 control the biogenesis of miRNAs during several differentiation processes (26), we asked whether those ubiquitously expressed proteins could play a similar role during neuronal differentiation. To this end, neuroblastoma SH-SY5Y cells were first treated for one day with a control siRNA or with an siRNA targeting both DDX5 and DDX17 genes (siDDX5/DDX17), and then with all-trans retinoic acid (ATRA) to induce their differentiation (Figure 1A and B). We monitored by RT-qPCR the expression of several neuron-enriched miRNAs, which were selected on the basis of their function in neuronal cells and/or their induction by ATRA in neuroblastoma cells (49,50). As expected, all tested miRNAs were induced by the ATRA treatment (Figure 1C, blue curves). However, in siDDX5/DDX17-treated cells the induction of eight out of the ten tested miRNAs was compromised (miR-26a/b, miR-9, miR-181a/b and miR-124) or strongly reduced (miR-212 and miR-132) after 3 days of ATRA treatment (Figure 1C, red curves). DDX5 and DDX17 silencing did not affect the expression of the other two miRNAs (miR-125b and miR-34a), denoting some selectivity (Figure 1C).
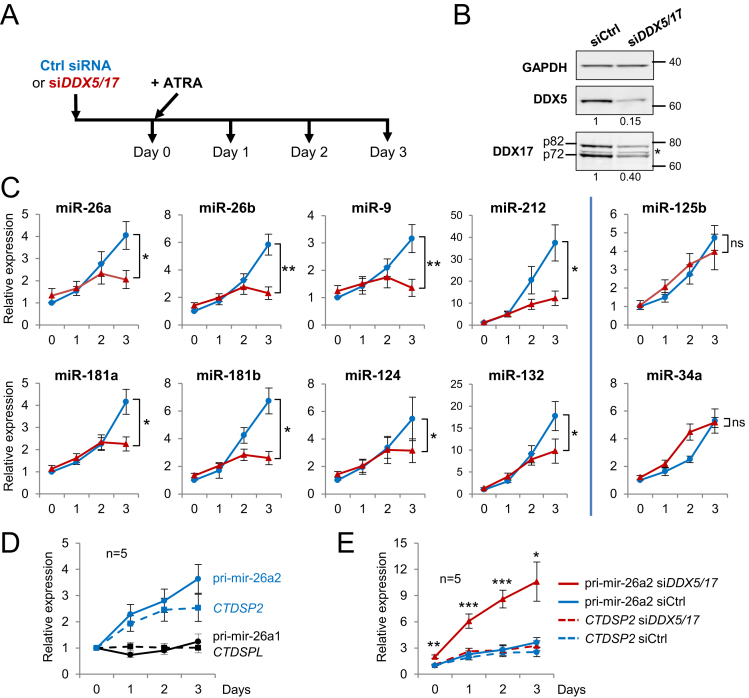
Figure 1.
DDX5 and DDX17 are required for the expression of specific miRNAs during neuronal differentiation. (A) Experimental outline. RNA analysis was carried out at day 0 (corresponding to undifferentiated SH-SY5Y cells pre-treated for 24 h with control siRNA or siDDX5/DDX17), and after 1, 2 and 3 days of ATRA treatment. Note that the combined silencing of DDX5 and DDX17 genes allows to study more easily the function of those proteins that have partially redundant functions and that cross-regulate each other (26,77). (B) Western-blot analysis showing the depletion of DDX5 and DDX17 at day 0 of the time-course. * non-specific band. (C) Quantitative RT-PCR (RT-qPCR) analysis showing the expression of mature miRNAs during time-course differentiation. Blue curves: Ctrl siRNA; red curves: siDDX5/DDX17. Data are represented as the mean values ± S.E.M. of independent experiments (n = 5) and normalized to the control sample (control siRNA, day 0), which was set to 1. *P-value < 0.05; **P-value < 0.01; ns: not significant (Student's t-test). (D) RT-qPCR analysis of the two miR-26a nuclear precursors (plain curves, black and dark blue respectively for pri-mir-26a1 and pri-mir-26a2) and their respective host genes (dashed curves, black and dark blue respectively for CTDSPL and CTDSP2 mRNAs) during differentiation. Data for each curve are represented as the mean values ± S.E.M. of independent experiments (n = 5) normalized to the control sample (day 0), which was set to 1. (E) RT-qPCR analysis showing the effect of DDX5/DDX17 silencing (red curves) on CTDSP2 mRNA (dashed curves) and pri-mir-26a2 (plain curves) expression during time-course differentiation. Details are as in C.
Among the DDX5/DDX17-dependent neuronal miRNAs, we focused on miR-26a since the processing of its zebrafish ortholog is a crucial rate-limiting step during neurogenesis (21). Thus, we sought to determine whether DDX5 and DDX17 control transcriptionally or post-transcriptionally the expression of miR-26a in differentiating SH-SY5Y cells. In mammals, miR-26a is expressed from two different loci (CTDSPL and CTDSP2) that generate the primary precursors pri-mir-26a1 and pri-mir-26a2, respectively. Neither the CTDSPL mRNA nor the corresponding pri-mir-26a1 was induced upon ATRA treatment (Figure 1D, black curves). In contrast, expression of both the CTDSP2 gene and the pri-mir-26a2 were steadily enhanced during differentiation, indicating that most of the induced miR-26a pool originates from the pri-mir-26a2 precursor in SH-SY5Y cells (Figure 1D, blue curves). We next analysed the ATRA-induced expression of CTDSP2/pri-mir-26a2 in absence of DDX5 and DDX17. Strikingly, we observed a strong accumulation of pri-mir-26a2 during differentiation upon DDX5 and DDX17 silencing (Figure 1E, compare red and blue plain curves), whereas CTDSP2 mRNA level was not altered (Figure 1E, dashed curves). This clearly indicated a defect in nuclear pri-mir-26a2 processing induced by DDX5 and DDX17 knockdown.
A DDX5/DDX17-regulated intronic region influences both pri-mir-26a2 processing and splicing of the miRNA host intron
Both miR-26a2 and miR-26a1 stem-loops are genomically located within an intron of their respective paralogous genes CTDSP2 and CTDSPL (Figure 2A). As described in the Introduction, a complex interplay exists between the CTDSP host genes and their products (miRNAs and proteins) during neurogenesis (21). Interestingly, we have previously shown that DDX5 and DDX17 regulate the splicing of CTDSP2 transcripts (26). As shown in Figure 2B, the exon upstream of the mir-26a2-containing intron (exon 6) is skipped in absence of both RNA helicases (Figure 2B). In contrast, the inclusion of the corresponding exon in the CTDSPL gene (exon 8) was not affected by siDDX5/DDX17 (Figure 2B). Since DDX5 and DDX17 also control the nuclear maturation of pri-mir-26a2 in differentiating cells (Figure 1), we explored in more details the mechanism that links the processing of the pri-mir-26a2 precursor to the splicing of its host CTDSP2 intron 6.
We first hypothesized that DDX5 and DDX17 may be required to ensure the correct folding of the miR-26a-encoding intronic region, to make it accessible to both the spliceosome and the Microprocessor complex. As a first step to test the activity of these RNA helicases on the processing of this region, we used previously described stable inducible MCF7 cell lines expressing DDX5 or DDX17 proteins (26,43). These cells were transfected with siRNAs targeting the 5′ or 3′ UTR of endogenous DDX5 and DDX17 mRNAs, respectively (siDDX-UTR). As in SH-SY5Y cells, this treatment resulted in CTDSP2 exon 6 skipping in both cell lines (Figure 2C) and in an increase of unprocessed pri-miR-26a2 (Figure 2D). Reexpression of DDX5 or DDX17 induced by doxycyclin rescued the CTDSP2 splicing pattern to a level comparable to the control level (full exon 6 inclusion), and also efficiently reduced the amount of pri-miR-26a2 (Figure 2C and D). This experiment supported a direct role of DDX17 and DDX5 in regulating the processing of CTDSP2/pri-miR-26a2 transcripts. Note that these results also underlined the functional redundancy between the two proteins, since the expression of either DDX5 or DDX17 could efficiently compensate the double DDX5/DDX17 knockdown.
Next, we used the Mfold web server (48) to predict whether RNA secondary structures may hinder the access to the 5′ splice site region of CTDSP2 exon 6 and/or to the region encompassing the pri-mir-26a2 stem-loop. Significantly, some of the most stable predicted structures involved an intronic sequence located shortly downstream of the 5′ splice site, and which was predicted to make stable base-pairing with nucleotides localized 1.6 kilobases further downstream, on either side of the basal part of the pri-mir-26a2 stem-loop precursor (Figure 2E and Supplementary Figure S1A). By preventing the expected folding of the miRNA precursor, such structure could likely impair the binding and/or activity of the Drosha/DGCR8 complex (51,52), and it may also be close enough to the exon 6 to sterically disturb the splicing process.
If RNA helicases are required to resolve this potential inhibitory structure, and thus to promote both exon 6 inclusion and pri-mir-26a2 cleavage, we reasoned that competing with the formation of the structure should contribute to restore normal splicing and pri-mir-26a2 processing when they are impaired upon knockdown of DDX5 and DDX17. To this aim, we designed an antisense 2′-O-methylated RNA oligonucleotide (AON-26a2) that is perfectly complementary to the region predicted to disrupt the lower stem of pri-mir-26a2, (Figure 2E and Supplementary Figure S1A, highlighted in red). We transfected SH-SY5Y cells with AON-26a2 or with a non-specific AON, with or without siDDX5/DDX17. As expected, depletion of DDX5/DDX17 resulted in a strong increase of pri-mir-26a2 (Figure 2F). The AON-26a2 did not have any effect in presence of DDX5/DDX17 (Figure 2F, grey bars), but it significantly reduced the pri-mir-26a2 maturation defect induced by DDX5 and DDX17 knockdown (Figure 2F, blue bars). In a remarkable manner, the AON-26a2 also partially restored the inclusion of CTDSP2 exon 6 in DDX5/DDX17-depleted cells (Figure 2G). This suggested that the same sequence can affect both miRNA biogenesis and splicing regulation. Importantly, the effect of AON-26a2 was specific of the CTDSP2 gene. Indeed, even though DDX5/DDX17 knockdown slightly increased the amount of unprocessed pri-mir-26a1, this defect was not restored by AON-26a2, and CTDSPL splicing remained unaffected in all conditions (Supplementary Figure S1B).
The restored inclusion of CTDSP2 exon 6 by AON-26a2 could indirectly result from the more efficient pri-mir-26a2 processing, as both splicing and maturation of intronic miRNAs are thought to be coupled (53). To test this hypothesis, we impaired pri-mir-26a2 processing using siRNAs against Drosha (RNASEN), the enzyme that cleaves the primary miRNA precursor (siRNASEN, Supplementary Figure S1C). As shown in Figure 2H, Drosha knockdown strongly impaired pri-mir-26a2 processing, but it had no effect on exon 6 splicing (Figure 2I). These results indicate that both exon 6 splicing and pri-mir-26a2 processing are mechanistically uncoupled, but they probably rely on the capacity of DDX5 and DDX17 helicases to resolve antagonistic secondary structures within CTDSP2 intron 6.
The expression of DDX17 and DDX5 is decreased during neuronal differentiation and DDX17 is a direct target of miR-26a/b
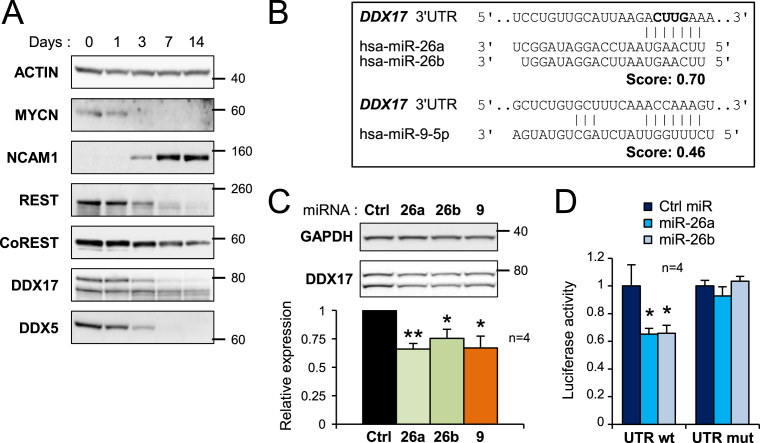
In the course of our work, we observed that the expression of both DDX17 and DDX5 was gradually repressed during ATRA-induced differentiation of two different neuroblastoma cell lines, as was REST expression that we used as a control (16) (Figure 3A and Supplementary Figure S2A and S2B). Investigating possible mechanisms that could explain this decrease, we found that potential sites for several neuron-enriched miRNAs are predicted within the 3′ untranslated regions (UTR) of both genes. Of particular interest, miR-26a/b and miR-9 were both predicted to target the DDX17 3′ UTR (Figure 3B). Overexpression of any of these 3 miRNAs in SH-SY5Y cells reproducibly decreased the endogenous DDX17 protein level compared to a control miRNA (Figure 3C). As positive controls, we confirmed that miR-26a (and also miR-26b) represses the expression of its host gene CTDSP2 (Supplementary Figure S2C), and that the expression of REST was efficiently repressed by miR-9 (Supplementary Figure S2D) (15,16,21). Interestingly, the 3′ UTR of REST also contains predicted miR-26a/b binding sites, and we observed that miR-26b significantly reduced endogenous REST protein level (Supplementary Figure S2D). A similar trend was observed with miR-26a, although it did not reach statistical significance. In contrast, neither miR-9, nor miR-26a/b had a significant effect on endogenous CoREST (Supplementary Figure S2D).
Figure 3.
Downregulation of DDX17 and DDX5 during neuronal differentiation. (A) Western-blot analysis showing the decreased expression of DDX17 and DDX5 during differentiation of SH-SY5Y cells. The pro-proliferative MYCN protein, the neural cell adhesion molecule (NCAM1) and the neuronal gene repressors REST and CoREST were used as controls of differentiation. A larger REST immunoblot is shown in Supplementary Figure S5. (B) Prediction of neuronal miRNA binding sites in the 3′ UTR of DDX17 transcripts (TargetScanHuman v6.2). The nucleotides deleted in the mutant DDX17-UTR construct are in bold. (C) Effect of overexpressed miR-26a/b and miR-9 on the expression of endogenous DDX17 in SH-SY5Y cells. The quantification of the experiment is shown at the bottom. The amount of DDX17 is represented as the mean intensity of the signal normalized to GAPDH level ± S.E.M. (n = 4 independent experiments) *P-value < 0.05; **P-value < 0.01 (Student's t test). The DDX17 signal corresponds to addition of both p72 and p82 bands. (D) Luciferase assays showing the effect of miR-26a and miR-26b on the expression of wild-type and mutant DDX17 3′ UTR reporters. Relative luciferase units data are represented as the mean values ± S.E.M. of four independent experiments, normalized to the control sample set to 1. *P-value < 0.05 (Student's t test).
We then carried out luciferase assays using reporters fused to the 3′ UTR of DDX17 to directly address the relationship between miR-26a/b and their potential target site in this transcript. Luciferase activity from the DDX17 reporter was significantly reduced upon expression of miR-26a or miR-26b, and fully restored when the predicted miR-26 binding site was mutated, demonstrating that DDX17 is a direct target for these two miRNAs (Figure 3D). We also carried out similar experiments using a reporter that contained a 1 kb-fragment of the REST 3′ UTR, but this gave more contrasted results. Both miR-26a and miR-26b reduced luciferase activity from the REST reporter to a similar extent (although statistical significance was reached only with miR-26b) (Supplementary Figure S2E). However, luciferase activity was not restored when we used a reporter carrying a mutation within the most conserved miR-26 binding site (data not shown), suggesting that the REST 3′UTR contains several miR-26 binding sites.
Beside their effect on DDX17 and REST transcripts, a large number of miR-26a/b direct targets correspond to genes involved in neurogenesis (54), so the overexpression of these miRNAs may stimulate some aspects of neuronal differentiation, as shown for other miRNAs (55–57). Supporting this hypothesis, overexpression of miR-26a/b tended to increase the expression of neuronal proteins NCAM1 and Synapsin-1, as did miR-9 (Supplementary Figure S2F), although more experiments would be necessary to confirm this point. Note that we tried to block the action of miR-26a/b by transfecting SH-SY5Y cells with specific antisense oligonucleotides, but this had no visible effect on DDX17 expression after 5 days of differentiation (data not shown).
As for DDX17, the expression of DDX5 was also markedly inhibited during neuronal differentiation (Figure 3A). Several neuron-enriched miRNAs (miR-124, the miR-181 family, miR-125a/b or miR-132/212) are predicted to target the DDX5 3′ UTR, and we have previously shown that miR-181b directly targets DDX5 3′ UTR in HMEC cells (26). However, none of those miRNAs had a significant effect on endogenous DDX5 expression when overexpressed in SH-SY5Y cells (data not shown), and we do not know presently the mechanism that causes DDX5 downregulation during differentiation. Altogether, results of Figures 1 to 3 indicate that during the early stages of neuronal diferentiation, DDX17 and DDX5 are required for the expression of several neuronal miRNAs, including miR-26a/b, and they suggest that these miRNAs may then contribute to the downregulation of DDX17.
DDX5, DDX17 and REST corepress the expression of neuronal genes
We next sought to understand the impact of DDX17 and DDX5 downregulation on gene expression, and we analysed the transcriptome of siDDX5/DDX17-treated SH-SY5Y cells by RNA-seq. In parallel, we analysed the transcriptome of cells treated with siRNA against REST. Neither REST protein nor REST RNA level was significantly affected by DDX5/DDX17 depletion, and REST silencing did not modify DDX5 or DDX17 expression levels (Figure 4A and Supplementary Figure S3A). Similarly, the expression of the REST cofactor CoREST was not modified upon silencing of any of those factors (Figure 4A). The silencing of REST significantly changed the expression of 130 genes, in an equivalent proportion of upregulated and downregulated genes (Supplementary Table S1). This included a number of previously known REST target genes, such as SCG2, SNAP25, SYP or VGF for example (Supplementary Table S1) (58).
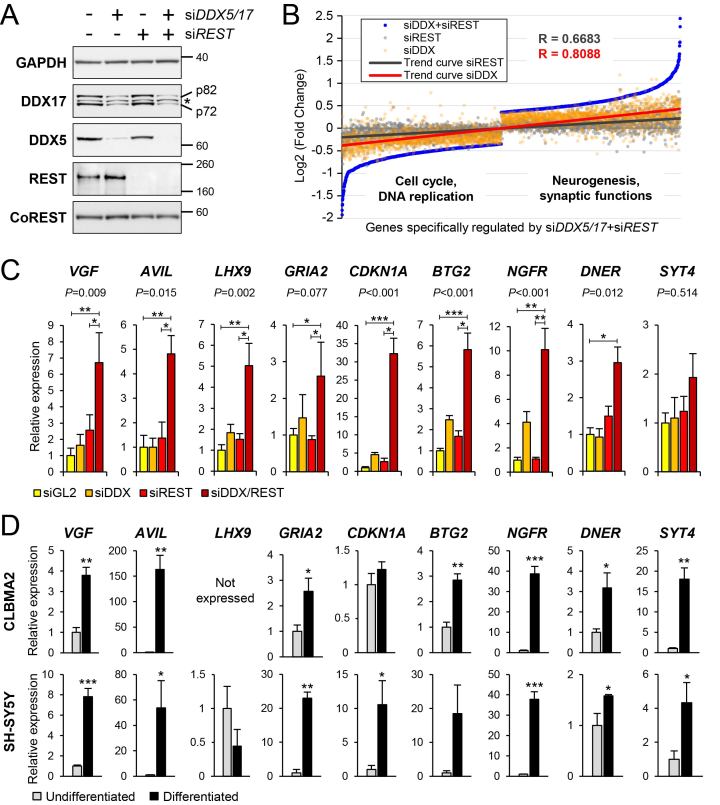
Figure 4.
Transcriptomic analysis of DDX5/DDX17 and REST-depleted cells. (A) Control western-blot for the knockdown of DDX5, DDX17 and REST upon treatment of SH-SY5Y cells with siRNA, as indicated. * non-specific band. Larger REST immunoblots are shown in Supplementary Figure S5. (B) Scatter plot representation of gene expression in absence of DDX5/DDX17 and/or REST. The 2507 genes of which steady-state expression level is significantly altered only in the siDDX5/DDX17+siREST condition were ranked and plotted according to their expression level (blue dots). The Log2(FC) value for the same genes in condition of single depletion of DDX5/DDX17 (siDDX, orange dots) or REST (siREST, gray dots) was then plotted on the same graph, showing a positive correlation between the 3 conditions. The respective trend curves are shown (red and black lines). Functions significantly associated to genes upregulated or downregulated in double-knockdown cells (siDDX5/DDX17+siREST) indicate a link with neurogenesis and cell cycle, respectively (see also Supplementary Tables S2 and S3). (C) Gene expression analysis of selected genes in condition of knockdown of REST and/or DDX5/DDX17. RT-qPCR data are represented as the mean values ± S.E.M. of independent experiments (n = 4), normalized to the expression of the control sample (siGL2) set to 1. Statistical significance was assessed by a one-way ANOVA (Kruskal-Wallis test), of which P-value is indicated above each graph. The siDDX5/DDX17+siREST condition was also compared to the control and siREST conditions (*P-value < 0.05; **P-value < 0.01; ***P-value < 0.001, Dunn's test). (D) Gene expression analysis of the same genes in undifferentiated and differentiated neuroblastoma cells (upper panel: CLBMA2 cells; lower panel: SH-SY5Y cells). RT-qPCR data are represented as the mean values ± S.E.M. of independent experiments (n = 3), normalized to the expression of the control sample (undifferentiated cells) set to 1. *P-value < 0.05; **P-value < 0.01; ***P-value < 0.001 (Student's t test). The LHX9 gene is not expressed in CLBMA2 cells.
To investigate a potential joint effect of REST and DDX5/DDX17 on gene expression, we analysed the expression of genes that were significantly regulated upon the combined silencing of both REST and DDX5/DDX17 (Supplementary Table S1). We found a positive correlation between their expression in both single siRNA conditions (siREST or siDDX5/DDX17) and their expression in the double siRNA treatment (Figure 4B). Furthermore, this analysis highlighted the more pronounced effect of the double siRNA treatment (siDDX5/DDX17+siREST) on gene expression compared to the absence of REST or DDX5/DDX17, for both activated and repressed genes (Figure 4B). Notably, many of the genes that are upregulated in absence of REST and DDX5/DDX17 are functionally associated with neurogenesis, with an enrichment in cellular compartments such as synapses or cellular junctions (Figure 4B and Supplementary Table S2). In contrast, functions associated with the downregulated genes in the same condition point at a regulation of cell cycle and DNA replication (Figure 4B and Supplementary Table S3).
To further validate the cooperation between REST and DDX5/DDX17, we tested by RT-qPCR the expression of several genes, including known REST-targeted genes (e.g. VGF, GRIA2, DNER, SYT4) (59). We observed that all these genes were more derepressed upon the codepletion of the 3 factors than upon the silencing of only REST or DDX5/DDX17 (Figure 4C). Finally, most of these genes are induced during neuronal differentiation of two different neuroblastoma cell lines (Figure 4D). These results strongly support the notion that a number of genes are derepressed during differentiation owing to the reduced expression of REST and DDX5/DDX17.
Interplay between DDX17 and REST recruitment at REST target promoters
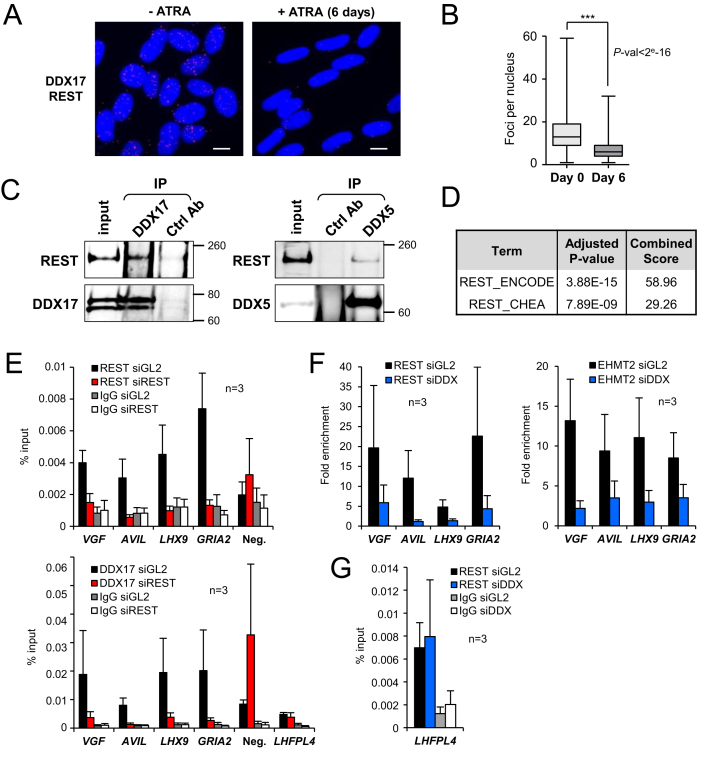
Since regulation of a number of REST-targeted genes appears to require the coordinated action of REST and DDX17/DDX5, we sought to determine whether these RNA helicases could be considered as transcriptional coregulators of REST. We first investigated a possible interaction between REST and DDX17 and DDX5 in SH-SY5Y cells. Using proximity ligation assays (PLA), we showed that DDX17 is in close proximity of REST in multiple sites within the nuclei of undifferentiated SH-SY5Y cells (Figure 5A and Supplementary Figure S4A). After 6 days of differentiation, the number of interaction loci was strongly reduced (Figure 5A and B), in line with the decreased expression of DDX17 and REST upon ATRA exposure (see Figure 3). Similar results were obtained when looking at proximity foci between DDX5 and REST (Supplementary Figure S4B). The interactions between endogenous REST and both DDX5 and DDX17 were confirmed by co-immunoprecipitation from SH-SY5Y cell extracts (Figure 5C), which further argued that at least a fraction of these factors are part of the same protein complex.
Figure 5.
DDX17 is a direct cofactor of REST. (A) Proximity-ligation assays (PLA) between REST and DDX17 proteins revealed numerous interaction sites, essentially restricted to DAPI-stained nuclei of undifferentiated SH-SY5Y cells (-ATRA, left). Upon 6 days of ATRA treatment, the signal was strongly diminished. Scale bar: 10 μm. (B) Quantification of PLA experiments. Statistical significance was calculated from a generalized linear model with Poisson regression. (C) Co-immunoprecipitation of endogenous REST with DDX17 and DDX5 in SH-SY5Y cells. Larger REST immunoblots are all shown in Supplementary Figure S5. (D) Enrichment of REST binding sites around genes that are upregulated in siDDX5/DDX17+siREST-treated cells (REST ChIP-seq datasets from ENCODE and CHEA, see also Supplementary Table S2). (E) Chromatin immunoprecipitation (ChIP)-qPCR experiments showing the binding of DDX17 and REST at the promoter of their target genes and at a control intergenic region (Neg.), in presence (black bars) or absence (red bars) of REST. Data are represented as the mean values of the percentage of input signal ± S.E.M. (n = 3 independent experiments). Control IgG for both conditions are represented by grey and white bars. (F) ChIP-qPCR experiments showing the enrichment of REST and EHMT2 at regulated promoters, in presence (black bars) or absence (blue bars) of DDX17/DDX5. Data are represented as the mean values ± S.E.M. (n = 3 independent experiments) of the fold increase in ChIP signal relative to the background signal (control IgG). (G) ChIP-qPCR experiments showing the DDX17/DDX5-independent binding of REST at the LHFPL4 promoter that is regulated only by REST. Details as in E.
We next used public chromatin immunoprecipitation (ChIP)-seq datasets from ENCODE and ChIP enrichment analysis (ChEA) to look for enriched transcription factor binding sites at the promoter of genes that are specifically activated in the double REST+DDX5/DDX17 knockdown condition. This revealed a strong enrichment of REST binding sites, as expected (Figure 5D, Supplementary Table S2). Accordingly, ChIP-qPCR experiments showed that REST bound to the REST/DDX17/DDX5-regulated promoters (Figure 5E, upper panel). DDX17 was also enriched at these promoters and its binding was inhibited by the silencing of REST (Figure 5E, lower panel). As a control, we detected some DDX17 binding on a non-regulated intergenic region that was not reduced upon REST silencing (Figure 5E, Neg.). Note that we could not determine whether DDX5 is also recruited to REST-targeted promoters as we did not succeed in performing ChIP with this factor in SH-SY5Y cells. Collectively, these observations suggest that DDX17 is recruited on REST-target promoters in a REST-dependent manner.
Interestingly, we observed that DDX17/DDX5 silencing reduced REST binding to its target promoters (Figure 5F, left panel). Supporting the idea that DDX17/DDX5 are required for the assembly of a REST-dependent complex on repressed genes, the silencing of both RNA helicases also reduced the binding of EHMT2, a global REST corepressor (Figure 5F, right panel) (9,60).
Finally, we also looked at REST and DDX17 binding to the promoter of the LHFPL4 gene, which was regulated only by REST and not by DDX17/DDX5 (Supplementary Figure S4C and Supplementary Table S1). DDX17 bound weakly to this promoter, but in a REST-independent manner (Figure 5E, LHFPL4 panel). In addition, REST interacted with the LHFPL4 promoter in a DDX17/DDX5-independent manner (Figure 5G), in agreement with our previous observation that not all REST target genes are coregulated by DDX17/DDX5 (Supplementary Table S1).
Altogether, these experiments demonstrated that DDX17 (and possibly DDX5) is a bona fide transcriptional coregulator of REST and suggested that the combined loss of REST and DDX17 contributes to efficiently derepress a number or neuronal genes during neuronal differentiation.
DISCUSSION
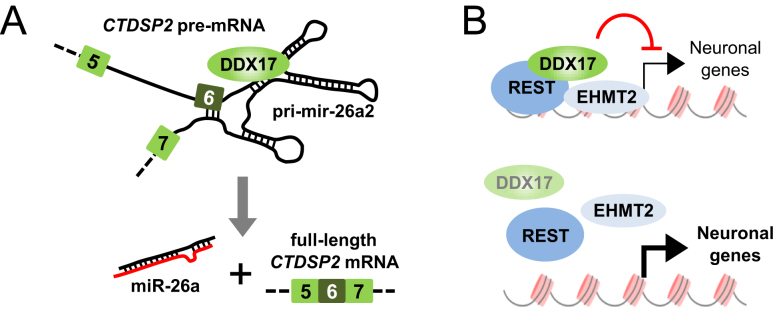
In this study, we uncovered a novel major function of RNA helicases in the REST gene circuitry that regulates the early stages of neuronal differentiation (Figure 6). First, DDX17 and DDX5 co-repress a number of REST target genes, including genes that are induced during neuronal differentiation and that have neuron-associated functions (Figures 4–5). Second, DDX5 and DDX17 control the processing of miR-26a2 precursor and are required for the biogenesis of other miRNAs during differentiation, which are known to target members of the REST complex, including REST itself and DDX17 (Figures 1–3 and Supplementary Figure S2). This likely contributes to the induction of REST-repressed neuronal genes.
Figure 6.
Model for the dual function of DDX17 in neural precursors. (A) Upon induction of differentiation, the CTDSP2 gene produces primary transcripts that can be correctly processed only in presence of DDX17 and DDX5 RNA helicases. This allows the production of mature miR-26a that is a direct regulator of DDX17 expression. (B) DDX17 associates with REST to some of its target promoters, along with EHMT2 which is known to promote a repressive chromatin environment. Upon silencing of DDX17/DDX5 genes, the binding of REST to promoters is weaker, and a subset of DDX17/DDX5- and REST-repressed neuronal genes are activated.
So far the activity of DDX5 and DDX17 in transcription regulation has been mostly studied in the context of the coactivation of various transcription factors (29). However, both proteins were shown to inhibit the transcription of a few transfected reporters by recruiting histone deacetylase 1 (61). To the best of our knowledge, this report is the first demonstration that DDX17 or DDX17 coregulates the activity of a major transcription repressor at a genome-wide scale. DDX17 binds in a REST-dependent manner to the promoter of their repressed genes (Figure 5), indicating that DDX17 is recruited on promoters by transcriptional repressors. Interestingly, we observed a reduced binding of REST and its cofactor EHMT2 in the absence of DDX5/DDX17 (Figure 5F), suggesting that these RNA helicases may stabilize the association of the whole REST complex to their target promoter. At this stage it remains to determine whether this is due to a cooperative binding of REST and DDX17 to DNA, or because the RNA helicase promotes a chromatin environment favourable for REST binding, or to another reason.
One hypothesis is that RNA helicases contribute, perhaps with long noncoding RNAs (lncRNA), to correctly position the transcription factor and its associated proteins at its target promoter, as scaffolding elements that would then allow the local modification of chromatin. Supporting this idea, the Rmrp lncRNA functionally mediates the interaction between DDX5 and RORγt (30), and another lncRNA, MeXis, was recently shown to control the binding of both DDX17 and liver X receptor to the promoter of their target gene, where it locally modified the chromatin architecture of this gene (62). As REST interacts with various lncRNAs that are thought to promote the assembly of chromatin modifying complexes to its target genes (63,64), it will be important to determine whether the REST-associated function of DDX17 is linked to these (or other) lncRNAs, and whether this may participate to the establishment of repressive chromatin marks on their target promoters.
The processing of pri-miRNAs by the microprocessor relies on the precise positioning of Drosha and DGCR8 onto the precursor stem–loop, forming a complex of which stoichiometry and structure have been revealed recently (52,65). Microprocessor assembly and pri-miRNA processing are regulated by many factors, which includes sequence elements (66–68), lncRNA (69), RNA binding proteins (70), as well as DDX5 and DDX17 RNA helicases (32–36). DDX17 has been shown to bind its target pri-miRNAs in a sequence-specific manner, either at the level of the stem-loop, or immediately downstream (34,71). Yet, how RNA helicases precisely cooperate with the Microprocessor is unclear.
Our results on the regulation of pri-mir-26a2 processing help to clarify this relationship. We propose that an important function of DDX5 and DDX17 is to ensure the proper folding of the pri-miRNA stem-loop by destabilizing competing RNA structures that likely impair the assembly of Microprocessor. Interestingly, those RNA helicases regulate alternative splicing via a similar mechanism. DDX5 has been shown to destabilize inhibitory structures around the 5′ splice site of alternative exons in the H-Ras and Tau transcripts (40,41), and our previous results indicated this could be a common feature of many DDX5/DDX17-regulated exons (26).
A majority of miRNAs is located within protein-coding or non-coding genes (reviewed in (53)). Splicing and intronic pri-miRNA cleavage were initially described as two independent events that have little effect on each other (72,73), at least for miRNAs that have a pure intronic localization (the situation is more complex for miRNAs overlapping splice sites and for mirtrons). This model was recently challenged as it was shown that the knockdown of splicing factors has a more specific and negative impact on the expression of intronic miRNAs compared to intergenic miRNAs (74). The results presented here may help to resolve this controversy. Our work suggests that intronic regulatory sequences can simultaneously affect the processing of an intronic miRNA precursor and the splicing of its host intron, but that both processes are mechanistically uncoupled. Indeed, the silencing of RNASEN (Drosha) altered pri-mir-26a2 maturation without affecting intron 6 splicing (Figure 2H and I). An interesting hypothesis is that the processing of some pri-miRNAs is connected to the splicing of their host intron when their shared intronic environment is unfavorably structured, requiring the presence of helicases such as DDX5 or DDX17. Note that we cannot rule out that splicing of the host intron is a first step to allow pri-miR-26a2 processing, but, we could not test this possibility as we failed to force CTDSP2 exon 6 skipping using specific antisense RNA or by knocking down the 70K subunit of U1 snRNP.
In addition to control miR-26a expression, DDX5 and DDX17 appear to be required for the biogenesis of other retinoic acid-induced miRNAs (Figure 1C), including miR-9 and miR-124, two major regulators of brain development (13,75). Apart from miR-26a, our data do not allow at this stage to know whether all altered miRNAs are direct targets of DDX5 and/or DDX17, although evidence for direct regulation has been provided elsewhere, in different settings, for miR-132 and miR-181b (26,76). It is interesting to underline that miR-9, miR-124 and miR-26a/b have a broad impact on the expression of several components of the REST complex. For example miR-9 reduces the expression of REST and DDX17 (Figure 3 and Supplementary Figure S2) (15–17), while miR-124 targets CoREST (18). Moreover miR-26a targets its own CTDSP2 transcript (Supplementary Figure S2C and (21), which codes for an RNA polymerase II CTD phosphatase that is a functional partner of REST in non-neuronal cells (20). We now show that miR-26a/b directly downregulate DDX17 and to a lesser extent REST (Figure 3 and Supplementary Figure S2). Our data suggest that like in zebrafish (21), miR-26a/b have a positive influence on human neuronal differentiation (Supplementary Figure S2F), even if further experiments will be required to fully address this point.
This cocktail of miRNAs is essential for neurogenesis, as evidenced by their numerous mRNA targets in the mouse brain (54) and by their effect on the conversion of non-neuronal cells to neurons (55–57). Their coordinated targeting of the REST complex contributes to the inactivation of this complex. Taken individually, each of those miRNAs does not necessarily have a very strong effect on their target. However, as each miRNA targets several components of the REST complex, and REST complex components are often targeted by at least two different miRNAs, this supports a model in which different miRNA-related pathways efficiently converge toward the downregulation of the REST complex. Targeting multiple components from the same complex may be necessary to reach a threshold level of activity under which the expression of the targeted genes will be significantly affected. This may be crucial during neuronal differentiation, since the timing of gene expression must be precisely controlled from cell cycle exit to final neuronal maturation (19). In agreement with this hypothesis, we showed that REST silencing on its own has little effect on gene expression compared to the silencing of both REST and DDX7/DDX5 (Figure 4). The coordinated targeting of several REST complex members may thus be necessary to promote the timely expression of neuronal genes.
Collectively, the complex interplay between RNA helicases, miRNAs and the REST complex is likely important to guarantee the robustness of this genetic circuit and the orchestration of the highly organized succession of events that shape the transcriptome of differentiating neuronal cells. DDX17, which is required both for the activity of REST and for the expression of miRNAs that downregulate REST complex components, is at the heart of mechanisms that control cell fate decisions.
DATA AVAILABILITY
Raw RNA-Seq datasets have been deposited on NCBI Gene Expression Omnibus (GEO) under the accession number GSE98871.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank our past and present colleagues from the team for their daily help and comments throughout the course of this project. This work was initiated at the Cancer Research Center of Lyon (CRCL) and concluded at LBMC. The authors are grateful to their colleagues from these two institutes for inspiring discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Agence Nationale pour la Recherche [ANR-16-CE12-0009-01]; Fondation ARC [PGA120140200853]; Institut National du Cancer [2014-156]; Ligue contre le Cancer (Comités du Rhône et de la Loire); AFM-Téléthon and Association Hubert Gouin ‘Enfance et Cancer’. M.P.L., S.T. and C.B.-P. received fellowships respectively from the Fondation ARC (post-doctoral fellowship), AFM-Téléthon (doctoral fellowship) and the Région Rhône-Alpes (doctoral fellowship). G.G. was supported by the Agence Nationale pour la Recherche. Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ebert M.S., Sharp P.A.. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012; 149:515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ernsberger U. Regulation of gene expression during early neuronal differentiation: evidence for patterns conserved across neuron populations and vertebrate classes. Cell Tissue Res. 2012; 348:1–27. [DOI] [PubMed] [Google Scholar]
- 3. Ninkovic J., Gotz M.. How to make neurons–thoughts on the molecular logic of neurogenesis in the central nervous system. Cell Tissue Res. 2015; 359:5–16. [DOI] [PubMed] [Google Scholar]
- 4. Chong J.A., Tapia-Ramirez J., Kim S., Toledo-Aral J.J., Zheng Y., Boutros M.C., Altshuller Y.M., Frohman M.A., Kraner S.D., Mandel G.. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995; 80:949–957. [DOI] [PubMed] [Google Scholar]
- 5. Schoenherr C.J., Anderson D.J.. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995; 267:1360–1363. [DOI] [PubMed] [Google Scholar]
- 6. Chen Z.F., Paquette A.J., Anderson D.J.. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat. Genet. 1998; 20:136–142. [DOI] [PubMed] [Google Scholar]
- 7. Andres M.E., Burger C., Peral-Rubio M.J., Battaglioli E., Anderson M.E., Grimes J., Dallman J., Ballas N., Mandel G.. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:9873–9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lunyak V.V., Burgess R., Prefontaine G.G., Nelson C., Sze S.H., Chenoweth J., Schwartz P., Pevzner P.A., Glass C., Mandel G. et al. . Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002; 298:1747–1752. [DOI] [PubMed] [Google Scholar]
- 9. Roopra A., Qazi R., Schoenike B., Daley T.J., Morrison J.F.. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol. Cell. 2004; 14:727–738. [DOI] [PubMed] [Google Scholar]
- 10. Ballas N., Grunseich C., Lu D.D., Speh J.C., Mandel G.. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005; 121:645–657. [DOI] [PubMed] [Google Scholar]
- 11. Qureshi I.A., Gokhan S., Mehler M.F.. REST and CoREST are transcriptional and epigenetic regulators of seminal neural fate decisions. Cell Cycle. 2010; 9:4477–4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Conaco C., Otto S., Han J.J., Mandel G.. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:2422–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abernathy D.G., Yoo A.S.. MicroRNA-dependent genetic networks during neural development. Cell Tissue Res. 2015; 359:179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Visvanathan J., Lee S., Lee B., Lee J.W., Lee S.K.. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007; 21:744–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Packer A.N., Xing Y., Harper S.Q., Jones L., Davidson B.L.. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J. Neurosci. 2008; 28:14341–14346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Laneve P., Gioia U., Andriotto A., Moretti F., Bozzoni I., Caffarelli E.. A minicircuitry involving REST and CREB controls miR-9-2 expression during human neuronal differentiation. Nucleic Acids Res. 2010; 38:6895–6905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Giusti S.A., Vogl A.M., Brockmann M.M., Vercelli C.A., Rein M.L., Trumbach D., Wurst W., Cazalla D., Stein V., Deussing J.M.. MicroRNA-9 controls dendritic development by targeting REST. eLife. 2014; 3:doi:10.7554/eLife.02755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Volvert M.L., Prevot P.P., Close P., Laguesse S., Pirotte S., Hemphill J., Rogister F., Kruzy N., Sacheli R., Moonen G. et al. . MicroRNA targeting of CoREST controls polarization of migrating cortical neurons. Cell Rep. 2014; 7:1168–1183. [DOI] [PubMed] [Google Scholar]
- 19. Nechiporuk T., McGann J., Mullendorff K., Hsieh J., Wurst W., Floss T., Mandel G.. The REST remodeling complex protects genomic integrity during embryonic neurogenesis. eLife. 2016; 5:e09584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yeo M., Lee S.K., Lee B., Ruiz E.C., Pfaff S.L., Gill G.N.. Small CTD phosphatases function in silencing neuronal gene expression. Science. 2005; 307:596–600. [DOI] [PubMed] [Google Scholar]
- 21. Dill H., Linder B., Fehr A., Fischer U.. Intronic miR-26b controls neuronal differentiation by repressing its host transcript, ctdsp2. Genes Dev. 2012; 26:25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Linder P., Jankowsky E.. From unwinding to clamping - the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 2011; 12:505–516. [DOI] [PubMed] [Google Scholar]
- 23. Jarmoskaite I., Russell R.. RNA helicase proteins as chaperones and remodelers. Annu. Rev. Biochem. 2014; 83:697–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bourgeois C.F., Mortreux F., Auboeuf D.. The multiple functions of RNA helicases as drivers and regulators of gene expression. Nat. Rev. Mol. Cell biology. 2016; 17:426–438. [DOI] [PubMed] [Google Scholar]
- 25. Fuller-Pace F.V., Moore H.C.. RNA helicases p68 and p72: multifunctional proteins with important implications for cancer development. Future Oncol. 2011; 7:239–251. [DOI] [PubMed] [Google Scholar]
- 26. Dardenne E., Polay Espinoza M., Fattet L., Germann S., Lambert M.P., Neil H., Zonta E., Mortada H., Gratadou L., Deygas M. et al. . RNA helicases DDX5 and DDX17 dynamically orchestrate transcription, miRNA, and splicing programs in cell differentiation. Cell Rep. 2014; 7:1900–1913. [DOI] [PubMed] [Google Scholar]
- 27. Li H., Lai P., Jia J., Song Y., Xia Q., Huang K., He N., Ping W., Chen J., Yang Z. et al. . RNA helicase DDX5 inhibits reprogramming to pluripotency by miRNA-based repression of RYBP and its PRC1-dependent and -independent functions. Cell Stem Cell. 2017; 20:462–477. [DOI] [PubMed] [Google Scholar]
- 28. Bourgeois C.F., Auboeuf D.. The RNA helicase DDX5 is a reprogramming roadblock. Stem Cell Invest. 2017; 4:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fuller-Pace F.V. The DEAD box proteins DDX5 (p68) and DDX17 (p72): multi-tasking transcriptional regulators. Biochim. Biophys. Acta. 2013; 1829:756–763. [DOI] [PubMed] [Google Scholar]
- 30. Huang W., Thomas B., Flynn R.A., Gavzy S.J., Wu L., Kim S.V., Hall J.A., Miraldi E.R., Ng C.P., Rigo F. et al. . DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature. 2015; 528:517–522. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31. Caretti G., Schiltz R.L., Dilworth F.J., Di Padova M., Zhao P., Ogryzko V., Fuller-Pace F.V., Hoffman E.P., Tapscott S.J., Sartorelli V.. The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev. Cell. 2006; 11:547–560. [DOI] [PubMed] [Google Scholar]
- 32. Suzuki H.I., Yamagata K., Sugimoto K., Iwamoto T., Kato S., Miyazono K.. Modulation of microRNA processing by p53. Nature. 2009; 460:529–533. [DOI] [PubMed] [Google Scholar]
- 33. Hong S., Noh H., Chen H., Padia R., Pan Z.K., Su S.B., Jing Q., Ding H.F., Huang S.. Signaling by p38 MAPK stimulates nuclear localization of the microprocessor component p68 for processing of selected primary microRNAs. Sci. Signal. 2013; 6:ra16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mori M., Triboulet R., Mohseni M., Schlegelmilch K., Shrestha K., Camargo F.D., Gregory R.I.. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell. 2014; 156:893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moy R.H., Cole B.S., Yasunaga A., Gold B., Shankarling G., Varble A., Molleston J.M., tenOever B.R., Lynch K.W., Cherry S.. Stem-loop recognition by DDX17 facilitates miRNA processing and antiviral defense. Cell. 2014; 158:764–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Motino O., Frances D.E., Mayoral R., Castro-Sanchez L., Fernandez-Velasco M., Bosca L., Garcia-Monzon C., Brea R., Casado M., Agra N. et al. . Regulation of microRNA 183 by cyclooxygenase 2 in liver Is DEAD-Box Helicase p68 (DDX5) dependent: role in insulin signaling. Mol. Cell. Biol. 2015; 35:2554–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Honig A., Auboeuf D., Parker M.M., O’Malley B.W., Berget S.M.. Regulation of alternative splicing by the ATP-dependent DEAD-box RNA helicase p72. Mol. Cell. Biol. 2002; 22:5698–5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guil S., Gattoni R., Carrascal M., Abian J., Stevenin J., Bach-Elias M.. Roles of hnRNP A1, SR proteins, and p68 helicase in c-H-ras alternative splicing regulation. Mol. Cell. Biol. 2003; 23:2927–2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lin C., Yang L., Yang J.J., Huang Y., Liu Z.R.. ATPase/helicase activities of p68 RNA helicase are required for pre-mRNA splicing but not for assembly of the spliceosome. Mol. Cell. Biol. 2005; 25:7484–7493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Camats M., Guil S., Kokolo M., Bach-Elias M.. P68 RNA helicase (DDX5) alters activity of cis- and trans-acting factors of the alternative splicing of H-Ras. PLoS One. 2008; 3:e2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kar A., Fushimi K., Zhou X., Ray P., Shi C., Chen X., Liu Z., Chen S., Wu J.Y.. RNA helicase p68 (DDX5) regulates tau exon 10 splicing by modulating a stem-loop structure at the 5′ splice site. Mol. Cell. Biol. 2011; 31:1812–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dardenne E., Pierredon S., Driouch K., Gratadou L., Lacroix-Triki M., Espinoza M.P., Zonta E., Germann S., Mortada H., Villemin J.P. et al. . Splicing switch of an epigenetic regulator by RNA helicases promotes tumor-cell invasiveness. Nat. Struct. Mol. Biol. 2012; 19:1139–1146. [DOI] [PubMed] [Google Scholar]
- 43. Samaan S., Tranchevent L.C., Dardenne E., Polay Espinoza M., Zonta E., Germann S., Gratadou L., Dutertre M., Auboeuf D.. The Ddx5 and Ddx17 RNA helicases are cornerstones in the complex regulatory array of steroid hormone-signaling pathways. Nucleic Acids Res. 2014; 42:2197–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Combaret V., Turc-Carel C., Thiesse P., Rebillard A.C., Frappaz D., Haus O., Philip T., Favrot M.C.. Sensitive detection of numerical and structural aberrations of chromosome 1 in neuroblastoma by interphase fluorescence in situ hybridization. Comparison with restriction fragment length polymorphism and conventional cytogenetic analyses. Int. J. Cancer. 1995; 61:185–191. [DOI] [PubMed] [Google Scholar]
- 45. Herbette M., Mercier M.G., Michal F., Cluet D., Burny C., Yvert G., Robert V.J., Palladino F.. The C. elegans SET-2/SET1 histone H3 Lys4 (H3K4) methyltransferase preserves genome stability in the germline. DNA Repair (Amst.). 2017; 57:139–150. [DOI] [PubMed] [Google Scholar]
- 46. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma’ayan A.. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013; 14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stallings R.L., Foley N.H., Bray I.M., Das S., Buckley P.G.. MicroRNA and DNA methylation alterations mediating retinoic acid induced neuroblastoma cell differentiation. Semin. Cancer Biol. 2011; 21:283–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cochella L., Hobert O.. Diverse functions of microRNAs in nervous system development. Curr. Top. Dev. Biol. 2012; 99:115–143. [DOI] [PubMed] [Google Scholar]
- 51. Quick-Cleveland J., Jacob J.P., Weitz S.H., Shoffner G., Senturia R., Guo F.. The DGCR8 RNA-binding heme domain recognizes primary microRNAs by clamping the hairpin. Cell Rep. 2014; 7:1994–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kwon S.C., Nguyen T.A., Choi Y.G., Jo M.H., Hohng S., Kim V.N., Woo J.S.. Structure of human DROSHA. Cell. 2016; 164:81–90. [DOI] [PubMed] [Google Scholar]
- 53. Mattioli C., Pianigiani G., Pagani F.. Cross talk between spliceosome and microprocessor defines the fate of pre-mRNA. Wiley Interdiscipl. Rev. RNA. 2014; 5:647–658. [DOI] [PubMed] [Google Scholar]
- 54. Chi S.W., Zang J.B., Mele A., Darnell R.B.. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009; 460:479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yoo A.S., Crabtree G.R.. ATP-dependent chromatin remodeling in neural development. Curr. Opin. Neurobiol. 2009; 19:120–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Xue Y., Ouyang K., Huang J., Zhou Y., Ouyang H., Li H., Wang G., Wu Q., Wei C., Bi Y. et al. . Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2013; 152:82–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xue Y., Qian H., Hu J., Zhou B., Zhou Y., Hu X., Karakhanyan A., Pang Z., Fu X.D.. Sequential regulatory loops as key gatekeepers for neuronal reprogramming in human cells. Nat. Neurosci. 2016; 19:807–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rockowitz S., Lien W.H., Pedrosa E., Wei G., Lin M., Zhao K., Lachman H.M., Fuchs E., Zheng D.. Comparison of REST cistromes across human cell types reveals common and context-specific functions. PLoS Comput. Biol. 2014; 10:e1003671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dietrich N., Lerdrup M., Landt E., Agrawal-Singh S., Bak M., Tommerup N., Rappsilber J., Sodersten E., Hansen K.. REST-mediated recruitment of polycomb repressor complexes in mammalian cells. PLoS Genet. 2012; 8:e1002494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zheng D., Zhao K., Mehler M.F.. Profiling RE1/REST-mediated histone modifications in the human genome. Genome Biol. 2009; 10:R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wilson B.J., Bates G.J., Nicol S.M., Gregory D.J., Perkins N.D., Fuller-Pace F.V.. The p68 and p72 DEAD box RNA helicases interact with HDAC1 and repress transcription in a promoter-specific manner. BMC Mol. Biol. 2004; 5:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sallam T., Jones M., Thomas B.J., Wu X., Gilliland T., Qian K., Eskin A., Casero D., Zhang Z., Sandhu J. et al. . Transcriptional regulation of macrophage cholesterol efflux and atherogenesis by a long noncoding RNA. Nat. Med. 2018; 24:304–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y.. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010; 329:689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ng S.Y., Johnson R., Stanton L.W.. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012; 31:522–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nguyen T.A., Jo M.H., Choi Y.G., Park J., Kwon S.C., Hohng S., Kim V.N., Woo J.S.. Functional anatomy of the human microprocessor. Cell. 2015; 161:1374–1387. [DOI] [PubMed] [Google Scholar]
- 66. Auyeung V.C., Ulitsky I., McGeary S.E., Bartel D.P.. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013; 152:844–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Du P., Wang L., Sliz P., Gregory R.I.. A biogenesis step upstream of microprocessor controls miR-17-92 expression. Cell. 2015; 162:885–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang F., Song W., Zhao H., Ma Y., Li Y., Zhai D., Pi J., Si Y., Xu J., Dong L. et al. . The RNA-binding protein QKI5 regulates primary miR-124-1 processing via a distal RNA motif during erythropoiesis. Cell Res. 2017; 27:416–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Liz J., Portela A., Soler M., Gomez A., Ling H., Michlewski G., Calin G.A., Guil S., Esteller M.. Regulation of pri-miRNA processing by a long noncoding RNA transcribed from an ultraconserved region. Mol. Cell. 2014; 55:138–147. [DOI] [PubMed] [Google Scholar]
- 70. Ha M., Kim V.N.. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014; 15:509–524. [DOI] [PubMed] [Google Scholar]
- 71. Moy R.H., Cherry S.. DDX17: structured RNA recognition drives diverse outputs. Cell Cycle. 2014; 13:3467–3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kim Y.K., Kim V.N.. Processing of intronic microRNAs. EMBO J. 2007; 26:775–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Morlando M., Ballarino M., Gromak N., Pagano F., Bozzoni I., Proudfoot N.J.. Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol. 2008; 15:902–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Janas M.M., Khaled M., Schubert S., Bernstein J.G., Golan D., Veguilla R.A., Fisher D.E., Shomron N., Levy C., Novina C.D.. Feed-forward microprocessing and splicing activities at a microRNA-containing intron. PLoS Genet. 2011; 7:e1002330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Coolen M., Katz S., Bally-Cuif L.. miR-9: a versatile regulator of neurogenesis. Front. Cell Neurosci. 2013; 7:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Remenyi J., Bajan S., Fuller-Pace F.V., Arthur J.S., Hutvagner G.. The loop structure and the RNA helicase p72/DDX17 influence the processing efficiency of the mice miR-132. Scientific Rep. 2016; 6:22848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Geissler V., Altmeyer S., Stein B., Uhlmann-Schiffler H., Stahl H.. The RNA helicase Ddx5/p68 binds to hUpf3 and enhances NMD of Ddx17/p72 and Smg5 mRNA. Nucleic Acids Res. 2013; 41:7875–7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mallinjoud P., Villemin J.P., Mortada H., Polay Espinoza M., Desmet F.O., Samaan S., Chautard E., Tranchevent L.C., Auboeuf D.. Endothelial, epithelial, and fibroblast cells exhibit specific splicing programs independently of their tissue of origin. Genome Res. 2014; 24:511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw RNA-Seq datasets have been deposited on NCBI Gene Expression Omnibus (GEO) under the accession number GSE98871.