Abstract
Adenosine-to-inosine (A-to-I) RNA editing entails the enzymatic deamination of adenosines to inosines by adenosine deaminases acting on RNA (ADARs). Dysregulated A-to-I editing has been implicated in various diseases, including cancers. However, the precise factors governing the A-to-I editing and their physiopathological implications remain as a long-standing question. Herein, we unravel that DEAH box helicase 9 (DHX9), at least partially dependent of its helicase activity, functions as a bidirectional regulator of A-to-I editing in cancer cells. Intriguingly, the ADAR substrate specificity determines the opposing effects of DHX9 on editing as DHX9 silencing preferentially represses editing of ADAR1-specific substrates, whereas augments ADAR2-specific substrate editing. Analysis of 11 cancer types from The Cancer Genome Atlas (TCGA) reveals a striking overexpression of DHX9 in tumors. Further, tumorigenicity studies demonstrate a helicase-dependent oncogenic role of DHX9 in cancer development. In sum, DHX9 constitutes a bidirectional regulatory mode in A-to-I editing, which is in part responsible for the dysregulated editome profile in cancer.
INTRODUCTION
Adenosine-to-inosine (A-to-I) RNA editing, a pivotal co- or post-transcriptional modification in eukaryotes, is catalyzed by adenosine deaminases acting on RNAs (ADARs) (1,2). The mammalian ADAR family comprises three structurally conserved members, ADAR1–3 (3,4). To date, only ADAR1 and ADAR2 have been reported to be catalytically active (5–7). More than a million A-to-I editing sites have been identified in the human transcriptome (8). This widespread enzymatic deamination of adenosines to inosines diversifies the transcriptome as general cellular machineries decode inosines as guanosines due to their structural similarity. A-to-I RNA editing has the potential to recode proteins (9,10), alter pre-mRNA splicing (11), mediate RNA interference (12,13), and affect the formation of ribonucleoprotein (RNP) complexes, transcript stability (14) and subcellular localization (15). Dysregulated expression of ADARs and A-to-I editing have been implicated in numerous diseases such as neurologic disorders and various cancers (16); however, the expression levels of ADARs are not always correlated with the editing frequency (17–20), indicating a multifaceted mode of regulation may be involved (20). It is therefore critical to elucidate the interwoven regulatory networks governing A-to-I editing. To this end, through conducting an unbiased screening for ADARs-interacting proteins using immunoprecipitation (IP) coupled with mass spectrometry (co-IP/MS), DEAH box helicase 9 (DHX9), also known as RNA helicase A (RHA) or nuclear DNA helicase II (NDH II), was identified as a ADARs-binding partner which forms a complex with ADAR1 and ADAR2 in the nucleus. Specific to its roles in RNA metabolism, DHX9 is known to be involved in translation (21), short interfering RNA (siRNA) (22) and circular RNA processing (23).
Although previous studies have reported that ADARs preferentially edit adenosines with certain 5′ and 3′ neighbouring nucleotides (24,25), the failure to identify conserved sequences suggests a more determining role of RNA structures in substrate specificity (26). The discovery of RNA helicases, an ubiquitous family of proteins that remodel RNA or RNP complexes in an energy-dependent manner (27), has prompted studies to investigate how helicases regulate cellular processes through structural remodeling. RNA helicases participate in nearly all aspects of RNA metabolism including splicing, translation, transcription and ribosome biogenesis (28). Preliminary evidence exists to demonstrate the participation of helicases in editing. Drosophila melanogaster homolog of DHX9, helicase maleless (Mle) has been suggested to coordinate two co-transcriptional processes, splicing and editing (29). Aberrant splicing of para transcripts was evident in Mlenapts background. In addition, although the editing process was partly dysregulated, the effects on editing were not as profound and the detailed regulatory mechanism adopted by the human DHX9 homolog in A-to-I editing regulation has not been thoroughly investigated. In our study, we uncovered a bidirectional regulator of A-to-I editing. More intriguingly, DHX9 exerts opposite regulatory effects dependent of the ADAR specificity of editing sites. Furthermore, our study provides fundamental mechanistic insights into how RNA helicase DHX9 regulates A-to-I editing, at least in part, through its helicase activities and its implications in cancer. We propose that DHX9 catalyzes active remodeling of the ADAR substrates into distinct structural signatures, exerting opposing regulatory effects which are dependent on the ADAR-specificity of editing sites. Moreover, we demonstrate the functional importance of DHX9 in tumorigenicity.
MATERIALS AND METHODS
Cell culture
Human embryonic kidney (HEK) 293T was grown in HyClone Dulbecco’s Modified Eagle Medium (DMEM; Thermo Scientific) supplemented with 10% fetal bovine serum (FBS; Thermo Scientific). SNU449 and EC109 cells were cultured in HyClone RPMI 1640 medium (Thermo Scientific) supplemented with 10% FBS. Unless otherwise stated, all the cell lines were incubated at 37°C, with 5% CO2.
GFP-trap and mass spectrometry
For identification of ADAR-interacting proteins, GFP-trap (Chromotrek) was used, as per manufacturer’s protocol, to co-immunoprecipitate GFP-tagged ADARs from transfected HEK 293T cells. Briefly, cells were lysed in 200 μl pre-chilled lysis buffer (10 mM Tris-hydrochloride (Tris–HCl) pH 7.5; 150 mM sodium chloride (NaCl); 0.5 mM ethylenediaminetetraacetic acid (EDTA); 0.5% Igepal-630; 1 × cOmplete protease inhibitor (Roche)). Clarified lysates were diluted with 300 μL pre-chilled washing buffer (10 mM Tris–HCl pH 7.5; 150 mM NaCl; 0.5 mM EDTA; 1 × cOmplete protease inhibitor (Roche)). Diluted lysates were incubated with 30 μl equilibrated GFP-Trap agarose beads (Chromotrek) for 1 h at 4°C under rotary agitation. The immunoprecipitate–bead complexes were washed with 1 ml washing buffer for 6 times. For elution, the immunoprecipitate-bead complexes were resuspended in 50 μl 2 × non-reducing sodium dodecyl sulfate (SDS) loading buffer (125 mM Tris–HCl, pH 6.8; 4% SDS; 20% glycerol; 0.01% bromophenol blue) and incubated at 95°C for 5 min.
The co-immunoprecipitate products were resolved on a 4–12% NuPage Novex Bis-Tris gel (Invitrogen). The gel was stained using the Colloidal Blue Staining Kit (Invitrogen) and digested with trypsin. Samples were analyzed on Thermo Scientific Easy-nLC coupled with Orbitrap XL. Survey full scan MS spectra (m/z 310–1400) were acquired with a resolution of r = 60 000, an AGC target of 1e6 and a maximum injection time of 700 ms. The 10 most intense peptide ions in each survey scan with an ion intensity of >2000 counts and a charge state ≥2 were isolated sequentially to a target value of 1e4 and fragmented in the linear ion trap by collisionally induced dissociation using a normalized collision energy of 35%. A dynamic exclusion was applied using a maximum exclusion list of 500 with repeat count of one, repeat and exclusion duration of 30 s. Peptide and protein quantification was performed with Scaffold using default settings. Database searches of MS data were performed using ipi.human.v3.68 fasta with tryptic specificity allowing maximum two missed cleavages, two labeled amino acids and an initial mass tolerance of 6 ppm for precursor ions and 0.5 Da for fragment ions. Cysteine carbamidomethylation was searched as a fixed modification, and N-acetylation and oxidized methionine were searched as variable modifications. Labeled arginine and lysine were specified as fixed or variable modifications, depending on the prior knowledge about the parent ion. Maximum false discovery rates were set to 0.01 for both protein and peptide. Proteins were considered identified when supported by at least one unique peptide with a minimum length of seven amino acids
Co-immunoprecipitation
EC109 cells were co-transfected with differentially tagged ADARs- and DHX9-encoding plasmids using jetPRIME (Polyplus). Transfected cells were harvested 24 h post-transfection. Cells were lysed using pre-chilled lysis buffer (50 mM Tris–HCl, pH 7.5; 150 mM NaCl; 0.1% Igepal-630; 1× EDTA-free cOmplete protease inhibitor (Roche)). All incubations were performed under rotary agitation. The lysates were first pre-cleared using 30 μl Dynabeads protein G (Invitrogen) at 4°C for 1 h. The pre-cleared lysates were then incubated with 5 μg of V5 antibody (Biorad MCA1360) at 4°C for 16 h. Subsequently, 50 μl of Dynabeads protein G (Invitrogen) was added and incubated at 4°C for 4 h. The immunoprecipitate–bead complexes were washed thrice with washing buffer (50 mM Tris–HCl, pH 7.5; 150 mM NaCl; 0.1% Igepal-630) at every interval of 5 min, on a rotating platform at 4°C. The immunoprecipitates were eluted using 50 μL 2 × non-reducing SDS loading buffer (125 mM Tris–HCl, pH 6.8; 4% SDS; 20% glycerol; 0.01% bromophenol blue).
The co-IP products were analyzed using Western blot. The following antibodies were used: mouse anti-V5 (1:5 000, 1 h; room temperature (RT); Biorad MCA1360), mouse anti-Flag-HRP (1:10 000, 1 h; RT; Sigma-aldrich A8592) and mouse anti-GFP (1:1 000, 2 h; RT; Santa Cruz sc-9996). For non-HRP conjugated primary antibodies, anti-mouse IgG-HRP (1:10 000, 1 h; RT; Santa Cruz sc-2005) was used for detection.
Immunofluorescence
Cells were cultured on coverslips for 24 h. Cells were washed with 1 × phosphate-buffered saline (PBS; 10 mM phosphate, 137 mM NaCl and 2.7 mM potassium chloride (KCl)) before fixation with 4% paraformaldehyde solution for 10 min at RT. Fixed cells were washed with 1 × PBS thrice, at every 5 min interval. Subsequently, cells were permeabilized with 0.5% Triton-X100 in 1 × PBS for 10 min at RT, followed by three washes with 1 × PBS at every 5 min interval. Serum-free protein block (Dako) was used and cells were blocked for 1 h at RT. Cells were blotted for 1 h at RT, with the following primary antibodies: rabbit anti-DHX9 (1:1 000; Abcam ab26271), mouse anti-ADAR1 (1:100; Abcam ab88574), mouse anti-ADAR2 (1:100; Sigma SAB1405426). Cells were washed thrice with 0.1% Tween-20 in 1 × PBS (PBST). Cells were co-stained in the dark with 4′,6-diamidino-2-phenylindole (DAPI; 0.2 µg/ml) and the following secondary antibodies for 30 min at RT: goat anti-mouse IgG-fluorescein conjugate (1: 1 000; Invitrogen #62–6511) and goat anti-rabbit IgG-rhodamine conjugate (1: 1 000; Invitrogen #31660); followed by three washes with PBST. The immunostained cells on the coverslips were mounted onto slides using SlowFade Gold antifade mountant (ThermoFisher Scientific). Immunostained cells were viewed using Olympus FV1000 confocal microscope.
Domain mapping
Serial deletion mutants of ADARs and DHX9 were cloned into pLenti6-V5 vector, in-frame with C-terminal V5 tag using the primers presented in ‘Supplementary Table S4’. For cloning of ADAR1 mutants, BamHI and XhoI restriction cut-sites were used. SacII and SpeI were used for cloning ADAR2 mutants. BamHI and SacII were used for cloning of DHX9 mutants. Respective deletion mutant-encoding plasmids were co-transfected into EC109 cells, together with the plasmids encoding its interacting partner. Co-IP was performed as per abovementioned.
Lentiviral shDHX9 vector construction and virus packaging
Tet-pLKO-puro was a gift from Dmitri Wiederschain (Addgene plasmid #21915). DHX9-targeting short hairpin RNAs (shRNAs) were designed using ‘The RNAi Consortium’ (Board Institute, USA). shDHX9 #1 (5′-GGGCTATATCCATCGAAATT-3′) and shDHX9 #2 (5′-GGTTCAGGTGGAAGGTTATAA-3′) were cloned into Tet-pLKO-puro vector at AgeI/EcoRI cut site, as previously described (30).
HEK293T cells were co-transfected with lentiviral packaging constructs and Tet-pLKO.1-puro construct using jetPRIME (Polyplus). Media were aspirated and replaced with fresh media at 24 h. Virus-containing media were collected 48 h post-transfection for subsequent transduction into EC109 and SNU449 cells.
DHX9 knockdown and protein analysis
Stably transduced EC109 cells were selected with 1 μg/ml puromycin (Invitrogen). DHX9 knockdown (KD) was induced with 2 μg/ml doxycycline (Dox) (Sigma-Aldrich) for 48 h. Cell lysates were lysed with NP-40 lysis buffer (50 mM Tris, pH 7.5; 150 mM NaCl; 1% Igepal-630; 1 × EDTA-free cOmplete protease inhibitor (Roche)). Protein lysates were quantified using Bradford assay (Biorad) and analyzed by western blots. The following antibodies were used: mouse DHX9 antibody (1:1 000, 2 h; RT; Abcam ab54593), mouse anti-ADAR1 (1:1 000, 2 h; RT; Abcam ab88574), mouse anti-ADAR2 (1:1 000, 2 h; RT; Sigma SAB1405426), mouse anti-actin (1:5 000, 1.5 h; RT; Abcam ab6276) and anti-mouse IgG-HRP (1:10 000, 1 h; RT; Santa Cruz sc-2005).
RNA sequencing and identification of editing events
A bioinformatics pipeline adapted from a previously published method (31) was used to identify RNA editing events (32). For each sample, raw reads were mapped to the reference human genome (hg19) and a splicing junction database generated from transcript annotations derived from UCSC, RefSeq, Ensembl and GENCODE (v19) by using Burrows–Wheeler Aligner with default parameters (bwa mem algorithm, v0.7.15-r1140) (33). To retain high quality data, polymerase chain reaction (PCR) duplicates were removed (samtools rmdup function, v1.4.1) (34) and the reads with mapping quality score <20 were discarded. Junction-mapped reads were then converted back to the genomic-based coordinates. An in-house perl script was utilized to call the variants from samtools pileup data and the sites with at least two supporting reads were retained. The candidate events were filtered by removing the single nucleotide polymorphisms (SNPs) reported in different cohorts (1000 Genomes Project (35), NHLBI GO Exome Sequencing Project (http://evs.gs.washington.edu/EVS/), dbSNP v138 (36)) and excluding the sites within the first six bases of the reads caused by imperfect priming of random hexamer during cDNA synthesis. For the sites not located in Alu elements, the candidates within the four bases of a splice junction on the intronic side, and those residing in the homopolymeric regions and in the simple repeats were all removed. Candidate variants located in the reads that map to the non-unique regions of the genome by using BLAST-like alignment tool (37) were also excluded. At last, only A-to-G editing sites based on the strand information from the strand-specific RNA-seq data were considered for all the downstream analyses. The same pipeline was applied for the cell lines and The Cancer Genome Atlas (TCGA) samples.
To identify high confidence editing events for particular analyses, the candidate sites were required to be supported by at least 20 reads and both shDHX9 KD treatments to result in at least 5% change in the editing level in the same direction.
Targeted sequencing
RNA-seq data were validated using targeted RT-PCR seq. The editing sites of interest were PCR amplified using gene-specific primers and pooled. Gene-specific primers were designed using previously published Perl script (38). ThruPLEX DNA-seq kit (Rubicon) was used as per manufacturer’s instructions, to add Illumina adaptors and indices onto the PCR amplicons. Briefly, 50 ng of each pool of PCR products were used for library construction and the libraries were amplified with five PCR cycles. Each pool of samples was uniquely indexed to allow for subsequent sample multiplexing. The libraries were size-selected for fragments ranging from 250 to 500 basepairs. Samples in equal molar amount were pooled to obtain final concentration of 12 pM and were spiked with 5% PhiX sequencing control V3 (Illumina). Sequencing was performed on MiSeq v3 1 × 150 bp (Illumina).
For targeted RT-PCR seq, a higher coverage filter (at least 100 reads) was applied. ADAR specificity of each site was assigned based on the following criteria: ADAR overexpression (OE) resulting in 5% or more increase in editing in the particular ADAR-overexpressing cell line, and having no editing change or change in opposite direction in the other ADAR-overexpressing cell line. Primer sequences are presented in ‘Supplementary Table S4’.
Rescue experiment
Using KAPA HiFi DNA polymerase (KAPA Biosystems) for site-directed PCR mutagenesis, shRNA-resistant DHX9 wild-type (DHX9WT) and helicase-deficient (DHX9K417R) cDNA sequences, with synonymous mutations introduced at shDHX9 #2 target sites, were individually cloned downstream of N-terminal Flag sequence in pPyCAGIP-Flag vector. Forward primer (5′-GAAGTGCAAGTAGAGGGCTACAACTACACTGGCATGGGAAATTCC-3′) and reverse primer (5′-GTTGTAGCCCTCTACTTGCACTTCACACATGAATTTCTGCCTGTT-3′) were used. K417R forward primer (5′-CTACTGGATGTGGGAGAACCACACAG-3′) and reverse primer (5′-CTGTGTGGTTCTCCCACATCCAGTAG-3′) were used to clone DHX9K417R by site-directed PCR mutagenesis. Day 1, EC109 cells were seeded on 10-cm plates at 80% confluency. Day 2, 10 μg of vector was transfected into EC109 cells using jetPRIME (Polyplus). The next day, transfected cells were replated on 15-cm plates, selected with puromycin (1 μg/ml; Invitrogen) and induced with Dox (2 μg/ml; Sigma-Aldrich). Two days post-induction (Day 5), cells were harvested.
Sanger sequencing
PCR products were purified using PCR purification kit (Qiagen) before being sent out to external company (1st Base, Singapore) for Sanger sequencing. Sequencing chromatograms were viewed using BioEdit software. The peak areas of adenosine and guanosine were calculated using ImageJ software and percentage of editing was expressed as peak area of guanosine over sum of peak area of adenosine and guanosine. Please refer to ‘Supplementary Table S4’ for the primer sequences.
RNA immunoprecipitation, RNA extraction and qRT-PCR
About 1 × 107 cells were used for RNA immunoprecipitation (RIP). Cell lysates were prepared with lysis buffer (50 mM Tris, pH 7.5; 150 mM NaCl; 1 mM EDTA; 1% Triton-X100; 1 × EDTA-free cOmplete protease inhibitor (Roche); 0.2U/μl SUPERase inhibitor (Ambion)). Two sets of 5% input were kept for protein and RNA analyses. For DHX9 RIP, lysates were pre-cleared with 3 μg rabbit IgG (Cell Signaling Technology #2729) and 30 μl Protein A Dynabeads (Invitrogen) on a rotating platform for 1 h at 4°C. Pre-cleared lysates were incubated with 5 μg rabbit IgG (Cell Signaling Technology #2729) or rabbit anti-DHX9 antibody (Abcam ab26271) at 4°C for overnight, on a rotating platform. Protein A Dynabeads (50 μl) were added to each tube and incubated at 4°C for 4 h on a rotating platform. The beads were washed thrice with 1 × TBS (50 mM Tris, pH 7.5; 150 mM NaCl). Ninety percent of the beads were used for RNA analysis. The remaining 10% was for western blot.
RNA extraction was done using RNeasy Mini kit (Qiagen) and cDNA was generated using Advantage reverse transcription kit (Clontech Laboratories), as per manufacturers’ protocols. GoTaq DNA polymerase (Promega) was used for qPCR. Enrichment was expressed as % input using the formulae: 100 × (2(-ΔCt[Normalized RIP])). Refer to ‘Supplementary Table S4’ for qPCR primers used.
Foci formation
Cells were seeded onto 6-well plates at a density of 1 × 103 cells per well. Spent media were replaced with fresh media containing 2 μg/ml of Dox and puromycin (1 μg/ml) every 3 days. Cells were stained with crystal violet solution (0.1% crystal violet, 25% methanol in water), after visible foci were formed.
Anchorage independent growth (soft agar assay)
For soft agar assay, 1 × 103 cells in 0.4% low-melting agarose were seeded on top of a solidified layer of 0.6% low-melting agarose in 6-well plates. Both the layers contained 2 μg/ml of Dox and puromycin (1 μg/ml). Cells were stained with crystal violet solution (0.005% crystal violet, 25% methanol in water), after visible colonies were formed.
For statistical analysis, GraphPad Prism version 7 for Windows (GraphPad Software, USA) was used to perform One-way Analysis of variance (ANOVA) followed by Tukey’s post-hoc test. Data were assumed to follow normal distribution.
In vivo tumorigenicity assay
To determine the in vivo tumorigenicity capabilities of cells, 2 × 106 cells were injected subcutaneously to the right and left flanks of 4–6 weeks old NOD scid gamma (NSG) mice. To induce DHX9 KD, mice were injected intraperitoneally with 600 μg Dox every other day. Tumor formation was monitored over a 3-week period and tumor volume was measured every other day once evident tumors were formed. Tumor volume was calculated using the formula: 0.5 × length × width × height. All animal experiments were approved by and performed in accordance with the Institutional Animal Care and Use Committees of National University of Singapore (NUS; Singapore). Independent Student’s t-test was performed on GraphPad Prism version 7 (GraphPad Software, USA) for Windows.
Tissue microarrays (TMAs) analysis
Tissue microarray (TMA) comprising a total 152 paired primary esophageal squamous cell carcinoma (ESCC) tumor and matched non-tumor (NT) tissues that were surgically removed and embedded in a paraffin block was obtained from Linzhou Cancer Hospital along with clinicopathological summaries between 2002 and 2004. None of these patients received preoperative chemotherapy and radiotherapy. All clinical samples used in this study involving human subjects were approved by the committees for ethical review of research at Zhengzhou University (Zhengzhou, China) and NUS (Singapore).
Immunohistochemical (IHC) staining and scoring
Sectioned TMA blocks (about 5 mm thick) were deparaffinized and rehydrated before immunohistochemical (IHC) staining. Endogenous peroxidase activity was inhibited with 3% hydrogen peroxide (Sigma-Aldrich) for 10 min at RT. For antigen retrieval, the slides were immersed in 10 mM citrate buffer (pH 6.0) and microwaved for 15 min. Non-specific binding was blocked with 5% normal goat serum (Invitrogen) in 1 × TBS for 10 min, followed by incubation with anti-DHX9 (1:1 000, Abcam ab26271) at 4°C overnight in a humidified chamber. The slides were sequentially incubated with biotinylated goat anti-rabbit IgG (1:100, Santa Cruz sc-2040) for 30 min at RT, followed by streptavidin-HRP conjugate for 30 min at RT. Isotope-matched human IgG was included as negative controls. For HRP enzymatic detection, 3,30-diaminobenzidine (DAB) substrate kit (Dako) was used, followed by counterstaining by Mayer’s hematoxylin. On the basis of staining intensities, the DHX9 immunoreactivity was scored as negative (0; total absence of staining), weak expression (1; faint staining in <50%, or moderate staining in <25% of tumor cells), moderate expression (2; moderate staining in > 25% to <75%, or strong staining in <25% of tumor cells) and strong expression (3; moderate staining in >75%, or strong staining in >25% of tumor cells).
DHX9 expression profiling in TCGA samples
RNA-seq data (fastq files) of 11 374 samples across 33 cancer types from TCGA were downloaded from dbGaP repository (2016). Each sample was processed as follows: raw reads were aligned to the reference human genome (hg19) by using STAR (v2.5.2a) (39). The gene expression quantification was performed by using featureCounts (v1.5.0-p3) (40) to obtain raw counts which were then normalized by dividing with the total number of uniquely mapped reads in the corresponding sample and multiplying by a factor of 100 million. The distribution of normalized expression levels for DHX9 was compared by using Wilcoxon rank sum test between the tumors and the normal samples, wherever available (24 out of 33 cancer types). For further analyses on RNA editing, 11 out of 24 cancer types, which had more than 5 normal samples and were shown to have significant DHX9 upregulation in the tumors compared to the normal samples were retained. A P-value less than 0.05 was regarded as significant up/down-regulation of DHX9 between the tumor and the normal samples.
Nuclear selective 2′-hydroxyl acylation by primer extension (SHAPE)-sequencing
Nuclear fractions of EC109-shScr and EC109-shDHX9 #2 cells were extracted using hypotonic buffer (10 mM Hepes pH 7.9; 1.5 mM magnesium chloride (MgCl2); 10 mM KCl; 0.1% Igepal CA-630; 0.5 mM dithiothreitol (DTT); 1 × EDTA-free cOmplete protease inhibitor (Roche)). Briefly, 1 × 107 intact nuclei were resuspended in 1 ml PBS. Appropriate amount of 0.5 M 2-methylnicotinic acid imidazolide (NAI) and anhydrous dimethyl sulfoxide (DMSO) was added to obtain a final concentration of 50 mM NAI or 10% DMSO (Sigma). The reaction was then incubated at 37°C for 15 min. A final concentration of 0.2 M DTT (Invitrogen) was used to quench the reaction. Nuclear pellets were washed once with pre-chilled 1 × PBS and pelleted at 1 000 × g, 4°C for 5 min. Nuclear RNA was extracted using RNeasy Mini kit (Qiagen). RNA samples prepared were sent to BGI, Hong Kong for library construction and sequencing. TruSeq stranded total RNA library preparation kit (Illumina) was used for library construction and libraries were sequencing on HiSeq4000 sequencing systems (Illumina) 2 × 100 bp, with two samples per lane.
The bioinformatics analysis of the samples was performed by using the default parameters published for icSHAPE pipeline (41). Briefly, after the removal of PCR duplicates, the reads were mapped to the human transcriptome by using bowtie2 (42). The transcript abundance was estimated by using the reads per kilobase of transcript per million values and the reverse transcription (RT) stops were calculated in each transcript. The background and the target RT stops from the control (DMSO) and the treated (NAI) samples, respectively, were normalized to calculate the enrichment reactivity scores for all the transcripts. These enrichment scores were further filtered to select the candidate transcripts with valid scores as well as high hit coverage and base density. The secondary structure of the representative substrates were then predicted guided by SHAPE reactivity scores using RNAfold tool from the ViennaRNA package 2.0 (43).
RESULTS
DHX9 interacts with ADARs in the nucleus
By conducting the co-IP/MS analysis, we uncovered 105 and 170 proteins that interact exclusively with ADAR1 and ADAR2 protein, respectively, and 238 proteins that bind to both ADAR1 and ADAR2 (Supplementary Figure S1 and Table S1). Among all candidate ADARs-binding proteins, 23 are helicases; 14 of the helicases interact with both ADAR members and 9 bind exclusively to ADAR2. DHX9, with the highest peptide sequence coverage among the helicases, was selected for further validation as a high confidence candidate. In agreement with the co-IP/MS data, GFP-tagged ADAR1 and ADAR2, but not GFP only, pulled down Flag-fused DHX9 in an ESCC cell line EC109 which has been used for an RNA editing study by our group previously (44), and reciprocally Flag-tagged ADAR1 and ADAR2 were detected in GFP-DHX9 IP products (Figure 1A). Both endogenous DHX9 and ADARs proteins were found to be expressed and co-localize in the nucleus of EC109 cells where the A-to-I editing of pre-mRNA transcripts occurs (Figure 1B and Supplementary Figure S2).
Figure 1.
DHX9 interacts with ADAR1 and ADAR2. (A) Reciprocal IP of ADARs and DHX9 in EC109 cells transfected simultaneously with 5 μg of ADAR1- or ADAR2- and 5 μg of DHX9-tagged with either GFP or Flag. GFP-tagged proteins were immunoprecipitated (IPed) using GFP-Trap system. Immunoblot analysis (IB) of GFP-pulldown products using antibodies against GFP and Flag shows specific binding between ADARs and DHX9. For each sample, 5% of the total cell lysate used in the IP reaction was loaded as an input control. (B) Confocal images of immunofluorescence staining of EC109 cells with antibodies against ADARs (green) or DHX9 (red). Nuclei were stained with DAPI (blue). Scale bars: 20 μm.
The domains involving the DHX9/ADARs interaction
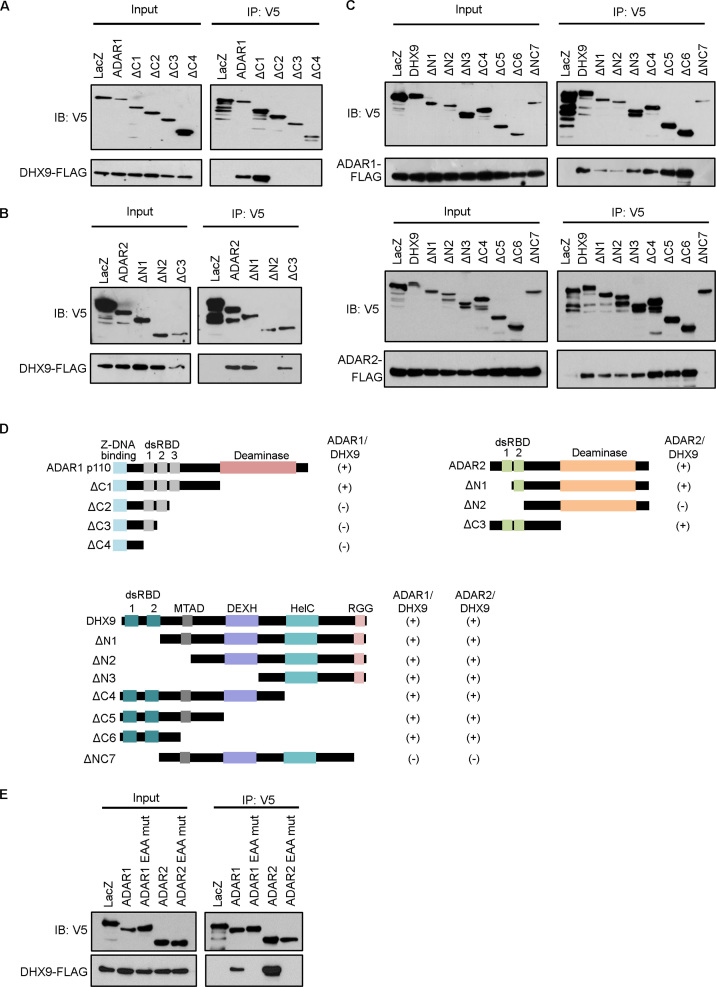
ADAR1p150 contains two Z-DNA binding domains (α and β), three double-stranded RNA binding domains (dsRBD1–3) and a C-terminal deaminase domain. The N-terminal Z-DNA binding domain α is absent in the predominant isoform, ADAR1p110. ADAR2 comprises two dsRBDs and a deaminase domain. DHX9 consists of a minimal transactivation domain and a helicase domain (HelC) flanked by two dsRBDs at N-terminus and a C-terminal ssRNA or ssDNA-binding domain containing arginine-glycine-glycine (RGG) repeats. To determine the interaction domain(s) that is involved in the ADARs–DHX9 interaction, serially truncated V5-tagged ADARs and DHX9 constructs were generated and used for co-IP analyses. These truncation constructs were co-transfected with the Flag-tagged partner construct in EC109 cells. The full-length ADAR1p110 (ADAR1-V5) and ΔC1-V5, but not ΔC2-V5, ΔC3-V5 and ΔC4-V5, pulled down Flag-DHX9 (Figure 2A). Similarly, Flag-DHX9 was detected in ADAR2-V5, ΔN1-V5 and ΔC3-V5 IP products (Figure 2B). Both Flag-ADAR1 and Flag-ADAR2 were pulled down by the full-length DHX9 (DHX9-V5) and all DHX9 truncation mutants except ΔNC7-V5 (Figure 2C). All these data suggest that the third dsRBD (dsRBD3) of ADAR1, the second dsRBD (dsRBD2) of ADAR2 and both dsRBDs and RGG of DHX9 are involved in the interaction of ADARs and DHX9 (Figure 2D).
Figure 2.
Domains involved in ADARs/DHX9 interaction. (A) Domain mapping of ADAR1 supporting ADAR1–DHX9 interaction. Lysates harvested from EC109 cells co-transfected with DHX9-Flag and ADAR1-V5 wild-type or truncation mutants were used for IP with anti-V5 antibodies. (B) Mapping of the ADAR2 domain required for ADAR2–DHX9 interaction. Lysates harvested from EC109 cells co-transfected with DHX9-Flag and ADAR2-V5 wild-type or truncation mutants were used for IP with anti-V5 antibodies. (C) Mapping of the DHX9 domain required for ADARs–DHX9 interaction. Lysates harvested from EC109 cells co-transfected with ADAR1-Flag or ADAR2-Flag and DHX9-V5 wild-type or truncation mutants were used for IP with anti-V5 antibodies. (D) Schematic diagrams of full-length ADAR1, ADAR2 and DHX9, with their truncation mutants. The extent of interaction is indicated as positive (+) and negative (−). dsRBD, double-stranded RNA binding domain; MTAD, minimal transactivation domain; HelC, helicase domain. (E) Co-IP of DHX9-V5 with wild-type or dsRNA-binding deficient (EAA) ADAR mutants. For each sample, 5% of the total cell lysates used in the IP reaction was loaded as an input control (Figure 2A–C and E).
The involvement of various RNA-binding associated domains in ADARs–DHX9 interaction prompted us to investigate whether the interaction is RNA dependent. ADAR mutants deficient in binding dsRNA were used to assess whether the loss of RNA binding in ADAR mutants affects interaction with Flag-DHX9. Point mutations introduced in the lysine-lysine-x-x-lysine (KKxxK) motifs within their dsRBDs, which make crucial contacts with RNA major groove, to glutamic acid-alanine-x-x-alanine (EAxxA) renders the EAA mutants incapable of binding dsRNA (45). The Flag-DHX9 was only pulled down by wild-type ADARs, and not the EAA mutants, suggesting that the interaction is RNA dependent (Figure 2E).
DHX9 reshapes global A-to-I RNA editing profiles
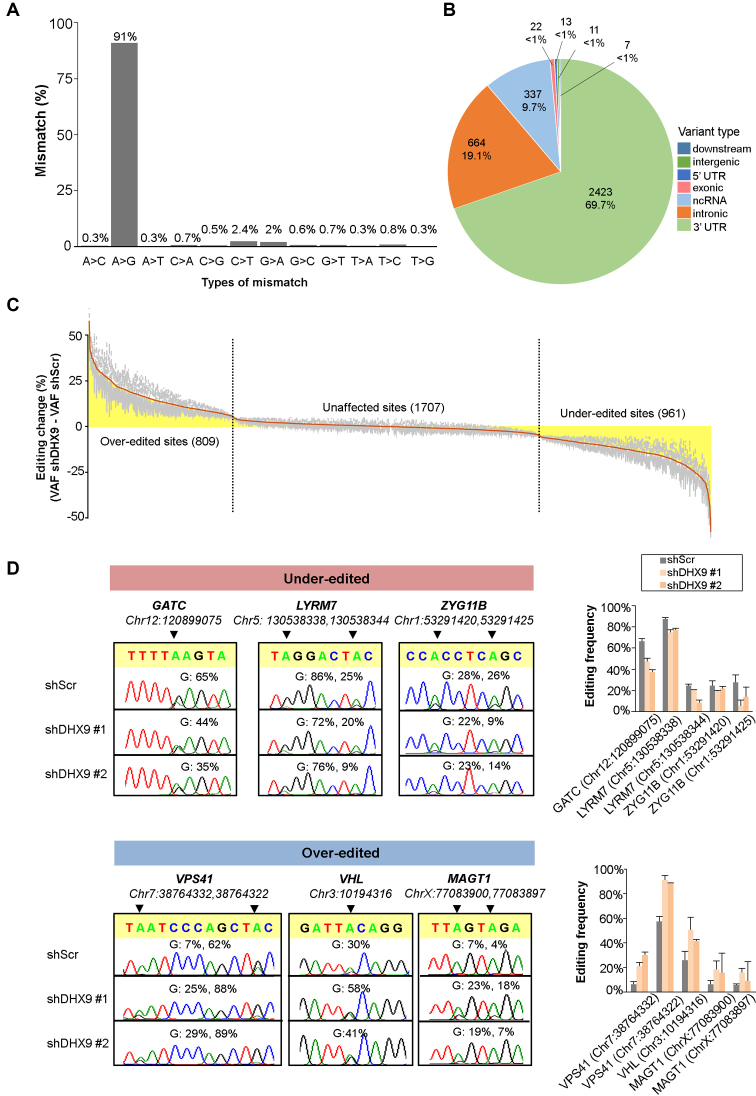
As demonstrated by our group previously, A-to-I RNA editing is dysregulated in ESCC and hepatocellular carcinoma (HCC) (44,46). In this study, RNA-seq analysis of EC109 cells reinforced the fact that A-to-I editing is the major contributor to nucleotide changes at transcriptome level and the vast majority of sites resides in 3′ untranslated regions (3′ UTRs), followed by intronic regions (Figure 3A and B). To investigate the regulatory role of DHX9 in A-to-I editing, we utilized a Dox-inducible KD system to silence DHX9 in EC109 cells. We first observed that both ADAR1 and ADAR2 were not affected by Dox dose-dependent KD of DHX9 (Supplementary Figure S3). As a result of DHX9 depletion (Supplementary Figure S4A), 1 770 sites (50.9%) out of the 3 477 high-confidence editing sites were affected (Figure 3C and Supplementary Table S2). Interestingly, DHX9 KD resulted in bidirectional changes in the A-to-I editing profiles, as reflected by two distinct subsets of editing sites, over-edited or under-edited (Figure 3C and D). We successfully validated 173 out of 224 (77.2%) randomly selected editing sites by targeted sequencing (targeted-seq) (Supplementary Figure S4C and Table S3). We further sequenced genomic DNA of EC109 cells to validate that these A-to-G nucleotide changes were introduced at the post-transcriptional level (Supplementary Figure S4D and Table S3). We observed no substantial difference in the distributions of genomic locations between over-edited and under-edited sites (Supplementary Figure S4E). Importantly, we observed similar effects of DHX9 depletion on editing in a HCC cell line SNU449, suggesting this DHX9-mediated bidirectional regulation of A-to-I editing is not likely to be specific to ESCC, but involved in different cancer types (Supplementary Figure S5 and Table S3). RIP-quantitative PCR (RIP-qPCR) of DHX9 confirmed DHX9 interacts with the ADAR substrates including GATC, LYRM7, ZYG11B, VPS41, VHL and MAGT1 (Supplementary Figure S6).
Figure 3.
DHX9 KD reshapes global A-to-I editing in a bidirectional manner. (A) Distribution of 12 types of nucleotide changes for the RNA editing events across the entire transcriptome of EC109 cells, as profiled by RNA-seq. (B) Distribution of A-to-I editing sites over annotated genomic regions. UTR, untranslated region; ncRNA, non-coding RNA. (C) RNA-seq analysis uncovers effects of DHX9 KD on global A-to-I editome. Changes in editing level are calculated by subtracting VAF of shDHX9 with VAF of shScr (VAFshDHX9-VAFshScr). Average editing changes induced by both shDHX9 #1 and #2 are represented by the orange line and shaded in yellow. Gray bars represent the standard error of the mean (s.e.m.). Events with equal or greater than 5% change in both KD samples and with a coverage of at least 20 reads are included in the analysis as the affected sites. VAF, variant allele frequency. (D) Sanger sequencing chromatograms illustrate editing of randomly selected over-edited and under-edited sites. Percentage represents the editing frequency calculated by taking the peak area of ‘G’ peak over sum of ‘A’ and ‘G’ peaks. Data represented as mean ± s.d. of technical triplicates from a representative of three independent experiments.
Editing substrate specificity determines the opposing effects of DHX9 on A-to-I editing
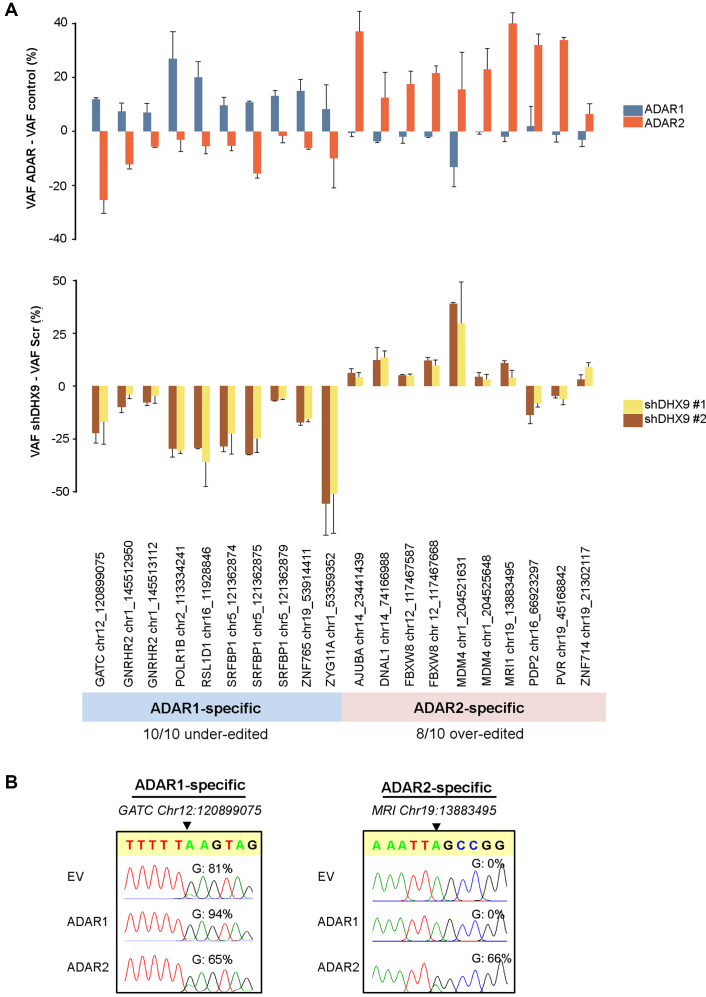
ADAR1 and ADAR2, with partial overlapping substrate specificity, are the primary enzymes catalyzing A-to-I editing in mammalian cells (47,48). To explore whether ADAR specificity of an editing site dictates the direction of regulation by DHX9, we first interrogated the ADAR substrate specificity of putative editing sites. We defined ‘ADAR1- or ADAR2-specific’ sites based on the following criteria: (i) greater than or equal to 5% increase in editing frequency of ‘ADAR1 or ADAR2-specific’ site upon either ADAR1 or ADAR2 overexpression (OE), respectively; and (ii) no increase in editing level of ‘ADAR1 or ADAR2-specific’ site upon ADAR2 or ADAR1 OE, respectively (Supplementary Figure S7). Besides, due to the lower efficiency of ADAR1 OE than that of ADAR2, one more criterion was applied for defining ‘ADAR2-specific’ sites; that is no more than 5% decrease in editing frequency after ADAR1 KD. Based on these criteria, we identified a total of 10 ADAR1-specific and 10 ADAR2-specific sites. Since ADAR1 and ADAR2 have the capability to form an inactive ADAR1/ADAR2 heterodimers that have distinct editing activities as their homodimeric counterparts (49,50), we did observe an antagonistic effect of ADAR OE on certain sites, such as an editing site (chr12:120899075) of GATC as well as a site (chr1:204521631) of MDM4 transcript (Figure 4A). We further examined the editing frequencies of these 20 specific sites in DHX9 KD cells. Strikingly, DHX9 KD resulted in 10 out of 10 (100%) ADAR1-specific sites to be under-edited. In contrast, 8 out of 10 (80%) ADAR2-specific sites were over-edited (Figure 4A and B), suggesting that the direction of change in editing caused by DHX9 KD is biased based on ADAR specificity of editing sites.
Figure 4.
Preferential repressive and stimulative roles of DHX9 dependent on ADAR specificity. (A) Editing frequency of sites determined using Targeted-seq. Upper panel: Bar charts showing effects of ADARs OE on the indicated editing sites in EC109 cells. Changes in editing level are calculated by subtracting VAF of ADAR1 or ADAR2 OE with VAF of empty vector control (VAFADAR-VAFcontrol); Lower panel: Bar charts showing effects of DHX9 KD on the indicated editing sites in EC109 cells. Changes in editing frequency are calculated by using VAFshDHX9-VAFshScr. Data represented as mean ± s.e.m. Experiment was performed with two biological replicates. VAF, variant allele frequency. (B) Representative Sanger sequencing chromatograms validate ADAR substrate specificity of two representative ADAR1- or ADAR2-specific sites. Percentage represents the editing frequency calculated by taking the peak area of ‘G’ peak over sum of ‘A’ and ‘G’ peaks.
Helicase activity of DHX9 is at least partially required for its A-to-I editing regulation
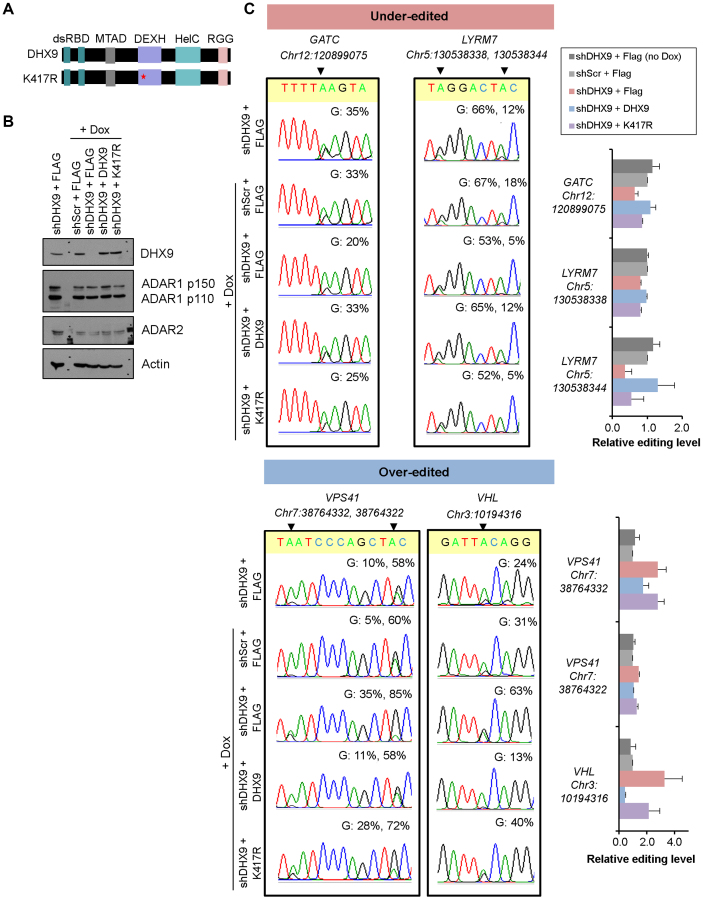
RNA helicases are involved in almost all aspects of RNA metabolism, orchestrating the various processes right from the transcription, processing, transport to decay of a given RNA (28). However, little is known about the function of helicases in modulating such RNA modification processes as A-to-I editing in mammals. In this study, to eliminate off-target effects of shRNAs against DHX9 (shDHX9) and more importantly investigate whether the helicase activity of DHX9 is required for A-to-I editing regulation, rescue assays were performed with the re-introduction of either wild-type DHX9 (DHX9WT) or helicase-deficient DHX9 (DHX9K417R) mutant, containing a lysine-to-arginine point mutation at residue 417 to abolish helicase activity (51), into DHX9 KD (EC109-shDHX9 #2) cells (Figure 5A). As expected, the DHX9WT successfully rescued the inhibitory or enhancing effect on editing caused by DHX9 KD (Figure 5B and C). In contrast, the DHX9K417R which is devoid of helicase activity was incapable of restoring the noted changes to the same extent as the DHX9WT, indicating the helicase activity is at least partially required for in A-to-I editing regulation (Figure 5B and C).
Figure 5.
DHX9 helicase activity is at least partially required for A-to-I editing regulation. (A) Schematic diagrams of wild-type DHX9 (DHX9WT) and helicase-deficient DHX9 (DHX9K417R). (B) Immunoblot analysis of DHX9 expression in the indicated group of cells. Inducible EC109 cells (EC109-shScr and EC109-shDHX9 #2) were transfected with Flag empty vector, Flag-tagged DHX9WT or DHX9K417R. −, in the absence of Dox; +, in the presence of Dox (2 μg/ml) for 48 h to turn on shRNA expression. Both EC109-shScr (+ Dox) and EC109-shDHX9 #2 (− Dox), transfected with Flag empty vector reflect basal editing level in the presence of endogenous DHX9. Dox-induced EC109-shDHX9 #2 cells were rescued with either Flag empty vector, Flag-DHX9WT or Flag-DHX9K417R. Actin as a loading control. (C) Sanger sequencing chromatograms showing the rescue effects of DHX9WT or DHX9K417R on representative sites affected by DHX9 depletion. Percentage represents the editing frequency calculated by taking the peak area of ‘G’ peak over sum of ‘A’ and ‘G’ peaks. Relative editing levels shown in the bar charts are expressed as relative to EC109-shScr (+ Dox), and represented as mean ± s.d. of technical triplicates from a representative of two independent experiments.
DHX9 is overexpressed in different cancer types and its helicase activity is functionally essential for cancer development
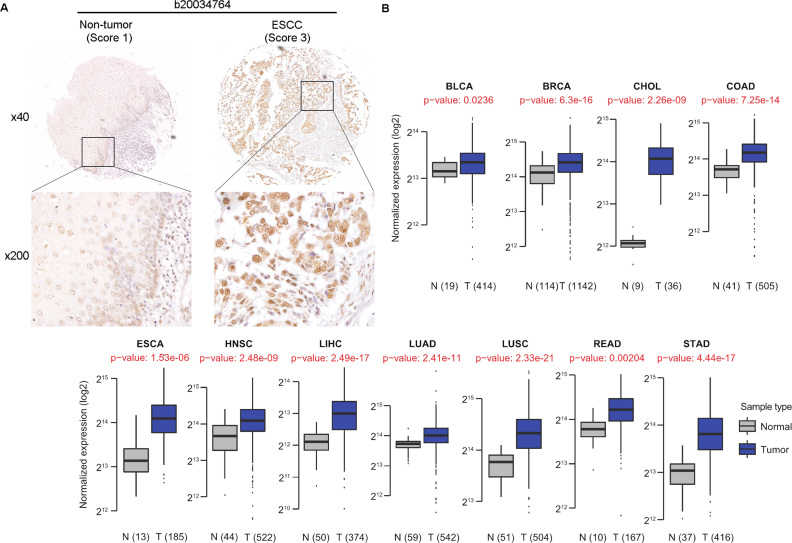
To dissect the functional importance of DHX9 during cancer development, we first examined the expression of DHX9 on a panel of TMAs consisting of 152 matched pairs of primary ESCC and NT esophagus specimens. Informative results of IHC staining were observed in 123 matched pairs of ESCC and NT specimens. We observed a differential nuclear expression of DHX9 between the primary ESCC and matched NT tissues. A detailed analysis of the IHC data revealed that DHX9 was overexpressed in 68.3% (84/123) of the analyzed ESCC tissues (Figure 6A). To further assess the clinical relevance of DHX9 in cancers, we analyzed DHX9 expression using the RNA-seq data from TCGA (52) and observed that DHX9 was significantly upregulated in tumor samples as compared to their corresponding NT tissues across 11 cancer types (P < 0.001, Wilcoxon signed rank test; Figure 6B).
Figure 6.
DHX9 is overexpressed in various cancer types. (A) A representative IHC staining of matched ESCC and NT specimens for DHX9 protein expression. Based on staining intensities, DHX9 immunoreactivities were scored as strong (3) and weak (1) expression in the primary ESCC and matched NT specimens, respectively. Magnified views (× 200) of the boxed regions are displayed in the lower panels. (B) Expression of DHX9 gene in 11 cancer types from TCGA. The number of normal (N) and tumor (T) samples compared are shown in brackets. Wilcoxon rank sum test was performed to compute the statistical significance. n.s. denotes no signficant difference (P > 0.05). BLCA: Bladder urothelial carcinoma; BRCA: Breast invasive carcinoma; CHOL: Cholangiocarcinoma; COAD: Colon adenocarcinoma; ESCA: Esophageal carcinoma; HNSC: Head and neck squamous cell carcinoma; LIHC: Liver hepatocellular carcinoma; LUAD: Lung adenocarcinoma; LUSC: Lung squamous cell carcinoma; READ: Rectum adenocarcinoma; STAD: Stomach adenocarcinoma.
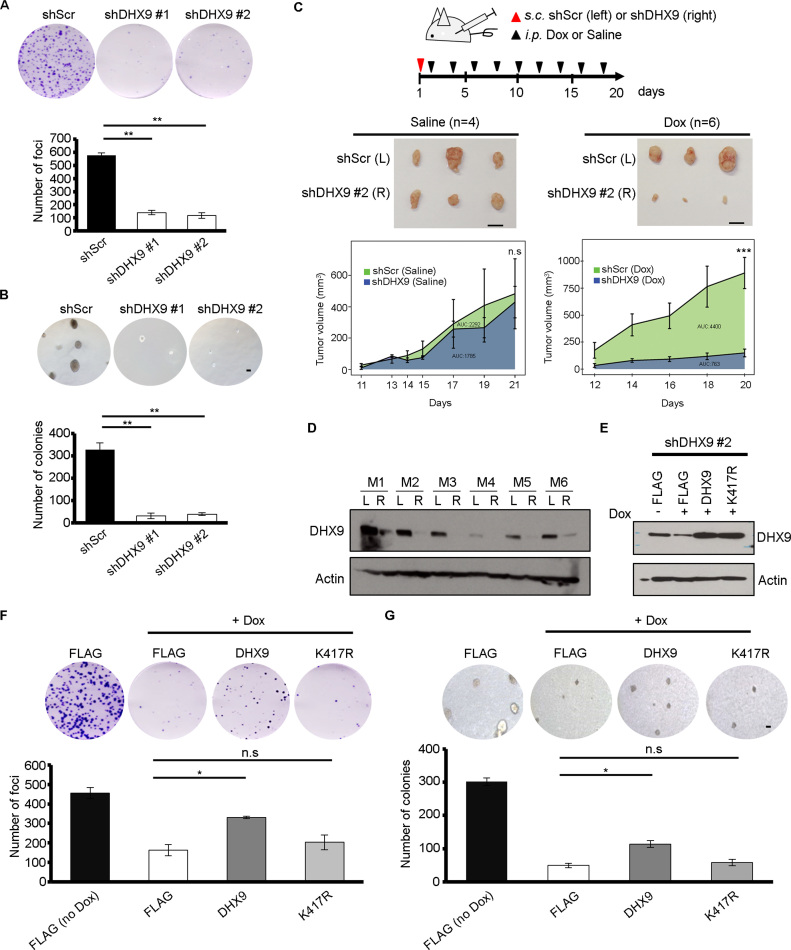
Further, we went on to assess the tumorigenic abilities of DHX9 KD EC109 cells in vitro and in vivo. DHX9 depletion by two shRNAs (shDHX9 #1 and shDHX9 #2) in EC109 cells dramatically reduced the frequency of foci formation, and the number of colonies formed in soft agar when compared to scrambled shRNA (shScr) (Figure 7A and B). In agreement with previous study, DHX9 suppression has deleterious effects on cell survival (53). Moreover, after subcutaneous (s.c.) injection of EC109 stable cells (shScr or shDHX9 #2) at day 1, mice received intraperitoneal (i.p.) injection of Dox or vehicle alone (saline) every 2 days during a 3-week observation period (Figure 7C; upper). Upon the injection of Dox, but not saline, xenograft tumors derived from shDHX9 #2 stable cells grew much slower than tumors derived from shScr stable cells (Figure 7C; middle and lower). Western blot analysis of DHX9 expression in xenograft tumor samples confirmed the persistent knockdown of DHX9 during the observation period (Figure 7D).
Figure 7.
DHX9 helicase activity is functionally essential for cancer development. (A) Quantification of foci formation in the indicated stable DHX9 KD (shDHX9 #1 and #2) and control (shScr) EC109 cells. **P < 0.01 and n.s denotes no significant difference. (B) Quantification of anchorage-independent colonies in the indicated stable DHX9 KD and control EC109 cells. **P < 0.01 and n.s denotes no significant difference. (C) Upper panel: Experimental design of in vivo tumorigenicity assay. Mice received i.p. injection of Dox or vehicle alone (saline) every 2 days during a 3-week observation period, post s.c. injection of the indicated stable DHX9 KD and control EC109 cells to the right (R) and left (L) flanks of mice, respectively. s.c., subcutaneous injection; i.p., intraperitoneal injection. Middle panel: Representative images of tumors derived from shDHX9 #2 or shScr cells in mice with i.p. injection of saline or Dox, at end point. Lower panel: Growth curves of tumors derived from the indicated cells in mice with i.p. injection of saline (n = 4) or Dox (n = 6), over a period of 3 weeks. Data are presented as area under the curve (AUC), in addition with the mean ± s.e.m. ***P < 0.001 determined by unpaired, two-tailed Student’s t-test. Scale bar: 1 cm. (D) Immunoblot analysis of DHX9 protein level in xenograft tumors derived from the indicated stable DHX9 KD (R, right flank) and control (L, left flank) EC109 cells in six mice (M1–6) with i.p. injection of Dox at end point. Actin as a loading control. (E) Immunoblot analysis of DHX9 protein level in shDHX9 #2 cells without or with the rescue by DHX9WT or DHX9K417R expression constructs. (F) Quantification of foci formation in shDHX9 #2 cells without or with the rescue of DHX9WT or DHX9K417R. *P < 0.05 and n.s. denotes no significant difference (P > 0.05). (G) Quantification of anchorage-independent colonies induced by shDHX9 #2 cells without or with the rescue of DHX9WT or DHX9K417R. *P < 0.05 and n.s. denotes no significant difference. All data are shown as mean ± s.d. of triplicate wells from a representative experiment of three independent experiments (Figure 7A, B, F and G), and statistical significance is determined by one-way ANOVA followed by Tukey’s post-hoc test.
Next, we examined whether the helicase activity is essential for the oncogenic capability of DHX9 by conducting functional rescue assays. In agreement, DHX9K417R failed to rescue the inhibitory effects of DHX9 KD on tumorigenicity while DHX9WT could partially rescue the tumorigenic effects (Figure 7E–G). Altogether, our findings demonstrate that helicase activity of DHX9 is required for its bidirectional regulation in A-to-I RNA editing, and is functionally essential for cancer development.
DISCUSSION
Although ADAR1 and ADAR2 deaminate all currently known A-to-I RNA editing sites, the poor correlation between ADAR levels and editing frequency has been observed in certain pathophysiological settings (17–20), raising the questions of how the editing activity of ADARs is regulated, and what are the additional cellular factors involved in A-to-I editing? In our screen for ADAR-interacting proteins, splicing-associated proteins such as U2 small nuclear RNA auxiliary factor (U2AF), heterogeneous RNPs and U4/U6 snRNPs were detected in the co-immunoprecipitates. In agreement, Raitskin et al. (54) reported that ADAR1 and ADAR2 are associated with splicing factors in large nuclear RNP particles. It is not surprising that we detected various splicing-associated proteins as splicing and A-to-I editing are spatially coordinated processes that occur co-transcriptionally and interdependently (55,56). These lend credence to our MS data.
In cells, RNA molecules are extensively bound by RNA binding proteins, and it remains elusive how ADAR enzymes gain access to bind and modify an editing site. Remodeling of RNPs and RNA structures, presumably regulated by various adenosine triphosphate-dependent RNA helicases, are therefore crucial for A-to-I editing. As reported previously, the overall editing efficiency of a substrate RNA may significantly be altered depending on the stability of dsRNA structure and/or splicing rate (29). The RNA helicases have been demonstrated to be pivotal players in RNA metabolism. However, there is paucity in knowledge on the regulatory mechanism adopted by RNA helicases in A-to-I editing. Here, in our screen for ADARs-interacting proteins, RNA helicase DHX9 was identified as a binding partner of both ADAR1 and ADAR2. The binding domain mapping revealed the requirement for dsRBDs in which the dsRBD3 of ADAR1, dsRBD2 of ADAR2 and both dsRBDs and RGG of DHX9 being responsible for ADARs–DHX9 interaction.
Further, an unbiased transcriptome-wide analysis of changes in A-to-I editing profiles by DHX9 unraveled that DHX9 plays a bidirectional role in the regulation of A-to-I editing. We robustly validated our DHX9 KD RNA-seq data by targeted-seq, covering 224 random sites and attaining a validation rate of 77.2%. The validated RNA-seq data illustrate the bidirectional regulatory role of DHX9 in A-to-I editing. This observation disputes the previously published in vitro study (57), in which recombinant DHX9, decreased editing of glutamate B receptor (GluR-B) transcripts, indicative of an antagonistic role of DHX9 in editing in vitro. One probable explanation for this discrepancy is that editing coordinates with other co-transcriptional processes, involving multiple proteins (57). The in vitro system may therefore not be able to recapitulate the coordinative effects exerted by multi-protein complexes in vivo. In a recent study, Aktas et al. (23) reported that DHX9 leads to translational inhibition of inverted Alu-containing reporters and severe RNA processing defects, notably major increase in circular RNA production. Co-depletion of DHX9 with ADAR1 enhanced circular RNA formation and dsRNA accumulation. In addition, 1 244 genes with 1 807 potential A-to-I RNA editing changes were found in DHX9 depleted cells; however, as these genes constitute only a small fraction of genes with splicing defects (4.5%) or expression changes (6%), the authors did not further their investigation in this aspect. Herein, we established a bidirectional regulatory role of DHX9 in A-to-I RNA editing. Intrigued by the bidirectional mode of regulation, we went on to investigate whether the substrate specificity of ADAR1 or ADAR2 determines the direction of regulation. Although various A-to-I editing databases are publicly available (58–60), none of these databases annotate the ADAR specificity of editing sites. In this study, we employed targeted-seq to assign the ADAR specificity to editing sites and deciphered 20 high-confidence ADAR1- or ADAR2-specific sites. Interestingly, we observed that DHX9 silencing preferentially represses editing of ADAR1-specific substrates, whereas augments editing of ADAR2-specific substrates.
We further attempted to decipher the mechanism driving the bidirectional regulatory effects. First, to investigate whether DHX9 affects binding of ADAR1 and ADAR2 to their substrates in an opposite manner, using RIP assays, we observed a general decrease in the binding affinity of both over-edited and under-edited substrates to ADAR1 and ADAR2 as a result of the Dox-induced DHX9 KD (data not shown). This refutes the possibility of DHX9 regulating A-to-I editing through affecting RNA substrate binding to ADARs. In concordance, ADAR2 binding and editing has been proposed to be separate events, where effective substrate binding does not result in efficient editing (61). Our findings demonstrate that DHX9 was found to be at least partially dependent on its helicase activity as revealed by the failure of helicase-deficient mutant DHX9K417R to rescue the editing phenotypes to the same extend as DHX9WT in DHX9 KD cells. This suggests that DHX9 may remodel the structures of ADAR substrates to exert differential regulatory effects. However, until now, factors defining the site selectivity and specificity of ADARs remain elusive. This gap in knowledge renders it challenging to address why DHX9 exhibits differential regulatory effects, depending on the ADAR specificity of the editing sites. Using selective 2′-hydroxyl acylation analysed by primer extension-sequencing (SHAPE-seq), we demonstrated that DHX9 actively remodeled structures of various RNA molecules (Supplementary Figure S8), but we failed to obtain structural information of transcripts-of-interest, which are the ADAR1- and ADAR2-specific sites regulated by DHX9. Without the distinct structural signatures of ADAR1- and ADAR2-specific sites being deciphered, it is currently impossible to overcome the technical limitations and make robust inferences based solely on our findings. Capitalizing on the dependence on helicase activity and active RNA structural remodeling activity of DHX9, we postulate that DHX9 remodels ADAR substrates into distinct structures; such that it augments and represses ADAR1- and ADAR2-specific sites, respectively.
Till date, the dysregulated expression of ADARs and A-to-I editing have been implicated in cancers (16). However, what are the precise factors governing the editing substrate selectivity and specificity remain as a long-standing question in this research area. It is intriguing to observe a preferential repressive or stimulative mode of regulation by DHX9, based on the ADAR substrate specificity. It has been reported that ADAR1 OE and ADAR2 downregulation in tumors, which enhances the editing rates of ADAR1 target transcripts (e.g. AZIN1 (9,44) and NEIL1 (62)) and impairs editing frequencies of ADAR2 substrates (e.g. COPA (46), GluA2 (63) and PODXL (32)), respectively, are involved in cancer development and progression. As a bidirectional regulator of A-to-I editing, DHX9 reshapes the editing profiles, which may further contribute to the dysregulation in editing and confer malignant transformation of cancer cells. Collectively, these findings highlight the distinct clinical implication of edited variants of ADAR1- and ADAR2-specific substrates. DHX9, through its bidirectional mode of A-to-I editing regulation, may therefore at least in part, be a critical mechanism in driving cancer development.
In sum, we elucidated the regulatory role of DHX9 in A-to-I editing. To our knowledge, our findings unravel the first bidirectional regulator in A-to-I editing. We propose that at least partially through its helicase activity, DHX9 catalyzes active remodeling of the ADAR substrates into distinct structural signatures, exerting opposing regulatory effects which are dependent on the ADAR-specificity of editing sites. Moreover, we demonstrated the functional importance of DHX9 in tumorigenicity.
DATA AVAILABILITY
The RNA-seq data are deposited in the following repository: Repository/DataBank Accession: GEO; Accession ID: GSE99789. Databank URL: http://www.ncbi.nlm.nih.gov/geo/. Bioinformatics code are available upon request.
Supplementary Material
ACKNOWLEDGEMENTS
We thank and acknowledge Prof. Xin-Yuan Guan (The University of Hong Kong, Hong Kong, China) and Dr Yanru Qin (the First Affiliated Hospital, Zhengzhou University, China) for providing ESCC TMAs, Dr Chen Zhi Xiong (National University of Singapore, Singapore) for providing DHX9 mutant constructs and Dr Kwok Chu Kit (City University of Hong Kong, Hong Kong) for his technical assistance in SHAPE experiment. We thank Dr Xavier Roca (Nanyang Technological University, Singapore) and Dr Sudhakar Jha for their critiques and insightful suggestions. The computational work for this article was partially performed on resources of National Supercomputing Centre, Singapore (https://www.nscc.sg).
Authors’ contribution: L.C. conceived and co-supervised the study with D.G.T. H.Q.H. and L.C. designed and performed the experiments. O.A. conducted all the bioinformatics analyses. H.Y. assisted in preliminary bioinformatics analyses. T.H.M.C., V.H.N., H.K. and J.S.L. assisted in the mouse work. L.Q. provided insightful suggestions. L.Q., J.H., D.J.T.T., S.J.T., Y.S. and F.B.M provided experimental materials. H.Q.H wrote the manuscript. O.A. and L.C. edited the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation Singapore; Singapore Ministry of Education under its Research Centres of Excellence initiative; NMRC Clinician Scientist- Individual Research Grant New Investigator Grant (CS-IRG NIG) [NMRC/CNIG/1117/2014]; NMRC Clinician Scientist-Individual Research Grant (CS-IRG) [NMRC/CIRG/1412/2014]; NUS Young Investigator Award (NUS YIA) [NUSYIA_FY14_P22]; NUS Start-up Fund [NUHSRO/2015/095/SU/01]; Singapore Ministry of Health’s National Medical Research Council under its Singapore Translational Research (STaR) Investigator Award; NIH/NCI Grant [R35CA197697]; Singapore Ministry of Education’s Tier 3 Grants [MOE2014-T3-1-006]. Funding for open access charge: Singapore Ministry of Education’s Tier 3 grants [MOE2014-T3-1-006].
Conflict of interest statement. None declared.
REFERENCES
- 1. Rodriguez J., Menet J.S., Rosbash M.. Nascent-seq indicates widespread cotranscriptional RNA editing in Drosophila. Mol. Cell. 2012; 47:27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang I.X., Core L.J., Kwak H., Brady L., Bruzel A., McDaniel L., Richards A.L., Wu M., Grunseich C., Lis J.T. et al. RNA-DNA differences are generated in human cells within seconds after RNA exits polymerase II. Cell Rep. 2014; 6:906–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bass B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002; 71:817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bass B.L., Nishikura K., Keller W., Seeburg P.H., Emeson R.B., O’Connell M.A., Samuel C.E., Herbert A.. A standardized nomenclature for adenosine deaminases that act on RNA. RNA. 1997; 3:947–949. [PMC free article] [PubMed] [Google Scholar]
- 5. Wagner R.W., Yoo C., Wrabetz L., Kamholz J., Buchhalter J., Hassan N.F., Khalili K., Kim S.U., Perussia B., McMorris F.A. et al. Double-stranded RNA unwinding and modifying activity is detected ubiquitously in primary tissues and cell lines. Mol. Cell. Biol. 1990; 10:5586–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. O’Connell M.A., Gerber A., Keller W.. Purification of human double-stranded RNA-specific editase 1 (hRED1) involved in editing of brain glutamate receptor B pre-mRNA. J. Biol. Chem. 1997; 272:473–478. [DOI] [PubMed] [Google Scholar]
- 7. Chen C.X., Cho D.S., Wang Q., Lai F., Carter K.C., Nishikura K.. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA. 2000; 6:755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bazak L., Haviv A., Barak M., Jacob-Hirsch J., Deng P., Zhang R., Isaacs F.J., Rechavi G., Li J.B., Eisenberg E. et al. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 2013; 24:365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen L., Li Y., Lin C.H., Chan T.H., Chow R.K., Song Y., Liu M., Yuan Y.F., Fu L., Kong K.L. et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat. Med. 2013; 19:209–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Paschen W., Schmitt J., Uto A.. RNA editing of glutamate receptor subunits GluR2, GluR5 and GluR6 in transient cerebral ischemia in the rat. J. Cereb. Blood Flow Metab. 1996; 16:548–556. [DOI] [PubMed] [Google Scholar]
- 11. Rueter S.M., Dawson T.R., Emeson R.B.. Regulation of alternative splicing by RNA editing. Nature. 1999; 399:75–80. [DOI] [PubMed] [Google Scholar]
- 12. Yang W., Chendrimada T.P., Wang Q., Higuchi M., Seeburg P.H., Shiekhattar R., Nishikura K.. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006; 13:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kawahara Y., Zinshteyn B., Sethupathy P., Iizasa H., Hatzigeorgiou A.G., Nishikura K.. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007; 315:1137–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Morita Y., Shibutani T., Nakanishi N., Nishikura K., Iwai S., Kuraoka I.. Human endonuclease V is a ribonuclease specific for inosine-containing RNA. Nat. Commun. 2013; 4:2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Z., Carmichael G.G.. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001; 106:465–475. [DOI] [PubMed] [Google Scholar]
- 16. Maas S., Kawahara Y., Tamburro K.M., Nishikura K.. A-to-I RNA editing and human disease. RNA Biol. 2006; 3:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lai F., Chen C.X., Lee V.M., Nishikura K.. Dramatic increase of the RNA editing for glutamate receptor subunits during terminal differentiation of clonal human neurons. J. Neurochem. 1997; 69:43–52. [DOI] [PubMed] [Google Scholar]
- 18. Maas S., Patt S., Schrey M., Rich A.. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:14687–14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Khermesh K., D’Erchia A.M., Barak M., Annese A., Wachtel C., Levanon E.Y., Picardi E., Eisenberg E.. Reduced levels of protein recoding by A-to-I RNA editing in Alzheimer's disease. RNA. 2016; 22:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu Y., Emeson R.B., Samuel C.E.. Serotonin-2C receptor pre-mRNA editing in rat brain and in vitro by splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase ADAR1. J. Biol. Chem. 1999; 274:18351–18358. [DOI] [PubMed] [Google Scholar]
- 21. Manojlovic Z., Stefanovic B.. A novel role of RNA helicase A in regulation of translation of type I collagen mRNAs. RNA. 2012; 18:321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Robb G.B., Rana T.M.. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol. Cell. 2007; 26:523–537. [DOI] [PubMed] [Google Scholar]
- 23. Aktas T., Avsar Ilik I., Maticzka D., Bhardwaj V., Pessoa Rodrigues C., Mittler G., Manke T., Backofen R., Akhtar A.. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017; 544:115–119. [DOI] [PubMed] [Google Scholar]
- 24. Polson A.G., Bass B.L.. Preferential selection of adenosines for modification by double-stranded RNA adenosine deaminase. EMBO J. 1994; 13:5701–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lehmann K.A., Bass B.L.. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry. 2000; 39:12875–12884. [DOI] [PubMed] [Google Scholar]
- 26. Lehmann K.A., Bass B.L.. The importance of internal loops within RNA substrates of ADAR1. J. Mol. Biol. 1999; 291:1–13. [DOI] [PubMed] [Google Scholar]
- 27. Singleton M.R., Dillingham M.S., Wigley D.B.. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007; 76:23–50. [DOI] [PubMed] [Google Scholar]
- 28. Jankowsky E. RNA helicases at work: binding and rearranging. Trends Biochem. Sci. 2011; 36:19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reenan R.A., Hanrahan C.J., Ganetzky B.. The mlenapts RNA helicase mutation in drosophila results in a splicing catastrophe of the para Na+ channel transcript in a region of RNA editing. Neuron. 2000; 25:139–149. [DOI] [PubMed] [Google Scholar]
- 30. Wiederschain D., Wee S., Chen L., Loo A., Yang G., Huang A., Chen Y., Caponigro G., Yao Y.M., Lengauer C. et al. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle. 2009; 8:498–504. [DOI] [PubMed] [Google Scholar]
- 31. Ramaswami G., Zhang R., Piskol R., Keegan L.P., Deng P., O’Connell M.A., Li J.B.. Identifying RNA editing sites using RNA sequencing data alone. Nat. Methods. 2013; 10:128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chan T.H., Qamra A., Tan K.T., Guo J., Yang H., Qi L., Lin J.S., Ng V.H., Song Y., Hong H. et al. ADAR-mediated RNA editing predicts progression and prognosis of gastric cancer. Gastroenterology. 2016; 151:637–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. The Genomes Project, C. A global reference for human genetic variation. Nature. 2015; 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K.. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001; 29:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kent W.J. BLAT–the BLAST-like alignment tool. Genome Res. 2002; 12:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang R., Li X., Ramaswami G., Smith K.S., Turecki G., Montgomery S.B., Li J.B.. Quantifying RNA allelic ratios by microfluidic multiplex PCR and sequencing. Nat. Methods. 2014; 11:51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 41. Flynn R.A., Zhang Q.C., Spitale R.C., Lee B., Mumbach M.R., Chang H.Y.. Transcriptome-wide interrogation of RNA secondary structure in living cells with icSHAPE. Nat. Protoc. 2016; 11:273–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lorenz R., Bernhart S.H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Qin Y.R., Qiao J.J., Chan T.H., Zhu Y.H., Li F.F., Liu H., Fei J., Li Y., Guan X.Y., Chen L.. Adenosine-to-inosine RNA editing mediated by ADARs in esophageal squamous cell carcinoma. Cancer Res. 2014; 74:840–851. [DOI] [PubMed] [Google Scholar]
- 45. Valente L., Nishikura K.. RNA binding-independent dimerization of adenosine deaminases acting on RNA and dominant negative effects of nonfunctional subunits on dimer functions. J. Biol. Chem. 2007; 282:16054–16061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chan T.H., Lin C.H., Qi L., Fei J., Li Y., Yong K.J., Liu M., Song Y., Chow R.K., Ng V.H. et al. A disrupted RNA editing balance mediated by ADARs (Adenosine DeAminases that act on RNA) in human hepatocellular carcinoma. Gut. 2014; 63:832–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang Isabel X., So E., Devlin James L., Zhao Y., Wu M., Cheung Vivian G.. ADAR regulates RNA editing, transcript stability, and gene expression. Cell Rep. 2013; 5:849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Riedmann E.M., Schopoff S., Hartner J.C., Jantsch M.F.. Specificity of ADAR-mediated RNA editing in newly identified targets. RNA. 2008; 14:1110–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chilibeck K.A., Wu T., Liang C., Schellenberg M.J., Gesner E.M., Lynch J.M., MacMillan A.M.. FRET analysis of in vivo dimerization by RNA-editing enzymes. J. Biol. Chem. 2006; 281:16530–16535. [DOI] [PubMed] [Google Scholar]
- 50. Gallo A., Keegan L.P., Ring G.M., O’Connell M.A.. An ADAR that edits transcripts encoding ion channel subunits functions as a dimer. EMBO J. 2003; 22:3421–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nakajima T., Uchida C., Anderson S.F., Lee C.G., Hurwitz J., Parvin J.D., Montminy M.. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997; 90:1107–1112. [DOI] [PubMed] [Google Scholar]
- 52. Cancer Genome Atlas Research, N. Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M.. The cancer genome atlas Pan-Cancer analysis project. Nat. Genet. 2013; 45:1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lee T., Paquet M., Larsson O., Pelletier J.. Tumor cell survival dependence on the DHX9 DExH-box helicase. Oncogene. 2016; 35:5093–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Raitskin O., Cho D.-S.C., Sperling J., Nishikura K., Sperling R.. RNA editing activity is associated with splicing factors in lnRNP particles: The nuclear pre-mRNA processing machinery. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:6571–6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Licht K., Kapoor U., Mayrhofer E., Jantsch M.F.. Adenosine to Inosine editing frequency controlled by splicing efficiency. Nucleic Acids Res. 2016; 44:6398–6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Solomon O., Oren S., Safran M., Deshet-Unger N., Akiva P., Jacob-Hirsch J., Cesarkas K., Kabesa R., Amariglio N., Unger R. et al. Global regulation of alternative splicing by adenosine deaminase acting on RNA (ADAR). RNA. 2013; 19:591–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bratt E.V.A., ÖHman M.. Coordination of editing and splicing of glutamate receptor pre-mRNA. RNA. 2003; 9:309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ramaswami G., Li J.B.. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 2014; 42:D109–D113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Picardi E., D’Erchia A.M., Lo Giudice C., Pesole G.. REDIportal: a comprehensive database of A-to-I RNA editing events in humans. Nucleic Acids Res. 2017; 45:D750–D757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kiran A., Baranov P.V.. DARNED: a DAtabase of RNa EDiting in humans. Bioinformatics. 2010; 26:1772–1776. [DOI] [PubMed] [Google Scholar]
- 61. Källman A.M., Sahlin M., Öhman M.. ADAR2 A→I editing: site selectivity and editing efficiency are separate events. Nucleic Acids Res. 2003; 31:4874–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Anadon C., Guil S., Simo-Riudalbas L., Moutinho C., Setien F., Martinez-Cardus A., Moran S., Villanueva A., Calaf M., Vidal A. et al. Gene amplification-associated overexpression of the RNA editing enzyme ADAR1 enhances human lung tumorigenesis. Oncogene. 2016; 35:4407–4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cenci C., Barzotti R., Galeano F., Corbelli S., Rota R., Massimi L., Di Rocco C., O’Connell M.A., Gallo A.. Down-regulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J. Biol. Chem. 2008; 283:7251–7260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data are deposited in the following repository: Repository/DataBank Accession: GEO; Accession ID: GSE99789. Databank URL: http://www.ncbi.nlm.nih.gov/geo/. Bioinformatics code are available upon request.