Abstract
CK2 is an essential protein kinase implicated in various cellular processes. In this study, we address a potential role of this kinase in chromatin modulations associated with transcription. We found that CK2 depletion from yeast cells leads to replication-independent increase of histone H3K56 acetylation and global activation of H3 turnover in coding regions. This suggests a positive role of CK2 in maintenance/recycling of the histone H3/H4 tetramers during transcription. Interestingly, strand-specific RNA-seq analyses show that CK2 inhibits global cryptic promoters driving both sense and antisense transcription. This further indicates a role of CK2 in the modulation of chromatin during transcription. Next, we showed that CK2 interacts with the major histone chaperone Spt6, and phosphorylates it in vivo and in vitro. CK2 phosphorylation of Spt6 is required for its cellular levels, for the suppression of histone H3 turnover and for the inhibition of spurious transcription. Finally, we showed that CK2 and Spt6 phosphorylation sites are important to various transcriptional responses suggesting that cryptic intragenic and antisense transcript production are associated with a defective adaptation to environmental cues. Altogether, our data indicate that CK2 mediated phosphorylation of Spt6 regulates chromatin dynamics associated with transcription, and prevents aberrant transcription.
INTRODUCTION
All DNA-related processes within the cell such as replication, transcription and repair occur in the context of chromatin. The basic unit of chromatin, the nucleosome, consists of 147 base pairs of DNA wrapped around a histone octamer (1). To make DNA accessible to the various macromolecules involved in the processes listed above, cells need to modulate chromatin structure. Different factors can alter chromatin structure; these include histone modifiers, ATP-dependent chromatin remodelers, histone chaperones and histone variants. Modulation of chromatin structure during transcription elongation has been the focus of several studies [reviewed in (2,3)]. Diverse sets of factors have been identified to play important roles in altering chromatin structure during transcription elongation, e.g. Isw1, Isw2, SWI/SNF, Chd1, Spt6, the FACT complex (Spt16-Pob3), Set2, Set1 and the Paf1 complex [reviewed in (3–5)].
Nucleosomes are a major obstacle to transcription elongation (6). Cells have developed intricate mechanisms involving histone chaperones (HCs) to deal with this challenge. HCs promote disassembly of nucleosomes in front of RNAP II and reassembly in its wake [reviewed in (7–9)]. Chromatin reassembly in the wake of transcription is related to the important issue of dealing with the consequences of RNAP II passage (10,11). The cell must re-establish chromatin structure to repress a large number of sites that should not allow transcription initiation (12–14). The consequences of this spurious transcription are not known. However, it may interfere with many important processes, such as gene regulation, self-renewal of stem cells, genome stability and viral infections (15–18). HCs play an important role in the regulation of spurious transcription presumably by reassembling nucleosomes in the wake of RNAP II. Interestingly, recent studies showed that HCs involved in yeast spurious transcription regulation, such as Spt2 and Spt6, use the histone H3/H4 molecules displaced by transcription and recycle them to reassemble nucleosomes following RNAP II passage (19,20). This function is of great importance because it blocks the incorporation of newly synthesized histones that are highly acetylated (21,22) which may contribute to the maintenance of histone marks in transcribed regions. Acetylated histones tend to favor initiation from cryptic promoters located within coding regions. Furthermore, in addition to blocking spurious transcription, nucleosome reassembly mediated by HCs, such as Spt6, prevents wrongful localization of the variant histone H2A.Z in transcribed units (23).
Spt6 is a major conserved H3/H4 HC, essential for the repression of spurious transcription and the maintenance of nucleosomes (12,13,23–27). Many questions regarding the function and regulation of Spt6 remain without answers. Interestingly, we have found that Spt6 interacts with the HC Spt2 and that this interaction is regulated by Casein Kinase 2 (CK2) through the phosphorylation of Spt2 (28). CK2 is an essential serine/threonine kinase involved in many cellular processes [reviewed in (29,30)]. CK2 is commonly deregulated in cancer, and several studies suggest a link between CK2 dysfunction and epigenetic modulations in favor of the tumorigenesis process (31,32). Moreover, it has been shown in different organisms that CK2 interacts with many factors involved in chromatin modulations and may regulate their functions. Studies in Saccharomyces cerevisiae (33–35), Plasmodium falciparum (36), Drosophila (37), and mammals (38) strongly suggest a conserved and extensive role of CK2 in chromatin modulations. However, this role and the concerned molecular mechanisms remain elusive. Importantly, as mentioned previously, we have found that CK2 interacts with Spt2, phosphorylates it and thereby disrupts its interaction with Spt6 (28). We found that this regulation plays a role in the association of Spt2 with coding regions and into the function of this HC in the repression of spurious transcription in yeast. In addition to the link between CK2, Spt2 and Spt6, this kinase has been directly involved in interactions with other factors that are implicated in chromatin modulations during transcription elongation. Indeed, CK2 cooperates with the FACT complex to phosphorylate the PAF complex and may regulate indirectly the level of H2B ubiquitylation in coding regions (33,39). Proteomic analyses show that it also co-purifies with Spt4/Spt5, FACT and other elongation related factors (35,40). Together, these observations strongly suggest a potential role of CK2 in chromatin dynamics during transcription elongation.
In this study, we have specifically addressed the role of CK2 in chromatin dynamics associated with transcription. Using ChIP and ChIP-seq approaches, we have found that CK2 regulates histone H3 dynamics outside of replication, indicating a role in nucleosomes turnover associated with transcription. In cells depleted of CK2 activity, newly synthesized histone H3 are incorporated in transcribed regions at a higher rate than what should be expected in normal cells. As a consequence, cryptic sense and antisense transcripts levels are increased in these cells, indicating that chromatin refolding in transcribed units is deficient. We next addressed the question of how CK2 regulates this process. We reasoned that CK2 could regulate the function of key factors that are involved in chromatin dynamics associated with transcription. Importantly, we found that it interacts and phosphorylates the HC Spt6, which plays a major role in chromatin refolding during transcription elongation (12,13,20,23,26). Furthermore, we identified the CK2 phosphorylation sites in Spt6 and showed that mutation of these sites affects the function of this protein. Interestingly, our global analyses of transcripts by RNA-seq indicate that CK2 phosphorylation sites are involved in the regulation of cryptic sense and antisense transcription. Moreover, comparison between CK2 and Spt6 mutant data sets clearly shows that both regulate the levels of similar cryptic transcripts suggesting an overlapping function. Surprisingly, we found that Spt6 cellular levels are directly controlled by CK2 and their restauration bypass the need for normal CK2 activity for the repression of cryptic transcription. Finally, we show that CK2 and Spt6 phosphorylation are required for the dynamic response to environmental changes and various stresses. Taken together, our data show that CK2 regulates the dynamics of chromatin in transcribed regions by modulating Spt6 stability.
MATERIALS AND METHODS
Saccharomyces cerevisiae strains and plasmids
All strains used in this study are isogenic to S288C (41) and are listed in Supplementary Table S1. Strains were constructed by standard genetic methods, either by crosses or by transformation. Yeast cells expressing different SPT6 alleles were constructed by transformation of SPT6 plasmid containing the indicated alleles in SPT6/Δspt6 diploid strains followed by tetrad dissection. SPT6-FLAG, CKA1-13MYC, CKA2-13MYC, IWS1-13MYC, CKB2-TAP, alleles were constructed by integrating the DNA encoding the particular epitope at the 3′-end of the respective gene (42,43). The cka1Δ::KANMX6, spt6Δ::KANMX6 and bar1Δ::NatMX4 alleles were constructed by replacing the open reading frame with KANMX6 or NatMX4 selection marker (42,44). The point mutation in the CKA2 allele (D225N) was introduced as described in (45) with NatMX6 as a selection marker. The plasmids were constructed using standard molecular biology techniques. 6-His fusion plasmids were constructed by insertion of PCR amplified fragments into the appropriate sites of the pet15b vectors (Novagen). The pCC11 SPT6 WT plasmid is described elsewhere (46). Plasmids expressing Spt6 were generated by mutagenesis of pCC11 using Quick Change Multi Site-Directed Mutagenesis Kit from Agilent Technologies following the manufacturer's protocol. All mutations were verified by sequencing. FLAG-SPT6 was amplified from genomic DNA and inserted in the YCp50 plasmid. Plasmid YCp has been described elsewhere (47). Growth of different strains was monitored by spot tests on the indicated medium
Western blot and antibodies
Proteins were extracted in the presence of trichloroacetic acid, separated on 8–15% SDS-PAGE gels, and transferred to nitrocellulose membrane. Membranes were incubated with anti p-Ser5 (Millipore clone 3E8, 04-1572), anti p-Ser2 (Millipore clone 3E10, 04-1571), anti Rpb1 (8WG16 Covance, MMS-126R), anti Pgk1 (Invitrogen 459250) anti-H3 (Abcam Ab1791), anti-H3K36me3 (Abcam Ab9050), anti-Flag (Sigma F3165), anti-Myc (Covance MMS-150R), anti-Tap (Open Biosystems CAB1001) and anti His (Clontech 631212). Ponceau red staining and Pgk1 signal were used to determine equal loading.
Protein purification and in vitro phosphorylation assay
Recombinant 6His-tagged proteins were expressed in Escherichia coli BL21 bacteria and purified with Ni2+-nitrilotriacetic acid (NTA)-agarose (Qiagen) according to the manufacturer's protocol. Tandem affinity purification (TAP) of Ckb2 was done as described previously (43). In vitro phosphorylation of recombinant proteins was done as described previously (28). His-tagged recombinant proteins (1 μg) were incubated for 30 min at 30°C with CK2 purified from yeast in kinase buffer (final concentration of 80 mM NaCl–KCl, 25 mM Tris–HCl [pH 8], 10 mM MgCl2, 1 mM DTT, 50 μM cold ATP and 1 μCi [γ-32P]ATP). Samples were run on 10% SDS-PAGE gels, blotted onto nitrocellulose, dried and exposed to film.
Immunoprecipitation experiments
Co-immunoprecipitation assays were performed as described previously (48). Briefly, 10 μl of anti-Flag M2 agarose beads were incubated overnight at 4°C with yeast whole-cell extract (WCE) (5 mg of total protein) in binding buffer (20 mM HEPES [pH 7.5], 300 mM NaCl, 10% glycerol, 0.1% NP-40, 2 μg/ml of leupeptin and pepstatin, 5 μg/ml of aprotinin and 1 mM phenylmethylsulfonyl fluoride [PMSF]). Beads were washed three times with the binding buffer. Bound proteins were analyzed by Western blotting. For His pulldown assay, 300 ng of recombinant 6xHis-Spt6 coupled to Ni-NTA agar beads were incubated with an equal amount of Ckb2-TAP purified from yeast in pulldown buffer (20 mM Hepes (pH 7.5), 150 mM NaCl, 10% glycerol, 100 μg/ml BSA, 0.5 mM DTT, 0.1% NP-40 and 2 μg/ml each of leupeptin and pepstatin, 5 μg/ml aprotinin) for 3 h at 4°C. Beads were washed three times with pulldown buffer and bound proteins were analyzed by western blot with an antibody against TAP-tag.
Protein stability assay
Cells were grown to mid-log phase in synthetic medium with 2% glucose as the carbon source and reinoculated in 50 ml fresh media with 0.003% SDS at OD600 0.5 for 3 h. Then, cultures were shifted to 37°C, cycloheximide (Sigma, 7698) was added to a final concentration of 40 μg/ml to stop protein synthesis and cells were harvested at different time points. Cell lysates were prepared in lysis buffer and protein concentration was determined using the Bradford assay. Proteins were analyzed by SDS-PAGE and western blotting using the same amount of total proteins for each time point.
Transcriptional response assays
For galactose induction, strains were grown in YP 2% raffinose, to an OD600 of 0.8, G1 arrested with α factor and shifted to 37°C for 30 min. Then, galactose was added to a final concentration of 2% and cells were harvest at the indicated time. For amino-acid starvation, cells were grown in synthetic media lacking histidine (SC-HIS) to an OD600 of 0.8 and shifted or not to 37°C for 30 min. Then, 3-aminotriazole (3AT, Sigma A8056) was added to the final concentration of 40 mM and cells were harvest after 1 h of treatment.
Chromatin immunopurification (ChIP)
ChIP experiments were performed as described previously (21). Efficient G1 arrest (at least 95%) of yeast cells was achieved by adding 500 ng/ml α factor for 2–3 h. Immunoprecipitation of Rpb1 was performed using 2–3 μg of the 8WG16 anti-CTD antibody per immunoprecipitation (Covance, MMS-126R). Antibodies against p-Ser5 (3E8, 04-1572) and p-Ser2 CTD (3E10, 04-1571) were purchased from Millipore and ChIP was performed using 2 μg per immunoprecipitation. The immunoprecipitations of H3 and H3K56ac were done with 0.5 μg of anti-H3 antibody per immunoprecipitation (Abcam, 1791) and 0.2 μg of anti-H3K56 antibody per immunoprecipitation (Millipore, 07-677-I), respectively. Anti H3K36me3 (Abcam, 9050) were used at 0.5 μg per immunoprecipitations. Finally, the Flag ChIPs were done using 10 μl of anti-Flag agarose beads per immunoprecipitation (Sigma, A2220), and Myc ChIPs were done using 3–5 μg of anti-Myc 9E10 per immunoprecipitation (Covance monoclonal antibodies, MMS-150R). ChIP experiments were analysed by qPCR with oligonucleotides listed in Supplementary Table S2. All ChIP experiments were done at least in triplicates; P value < 0.05 (*), P value < 0.01 (**) and P value < 0.001 (***).
RNA analyses
Total RNA was isolated using the hot-phenol method. In northern blot analyses, 20 to 40 μg of RNA were separated on a 1% agarose formaldehyde–MOPS gel and transferred to a nylon membrane. The FLO8, SCR1, DDC1, SPB4, PHO84 and YML122C probes were amplified by PCR and radiolabeled by random priming. For RT-quantitative PCR (qPCR) quantifications, cDNAs were generated using the Invitrogene M-MLV reverse transcriptase kit and their levels were measured by real-time PCR using LightCycler 480 Sybr green I master kit purchased from Roche. Sequences and detailed protocols used in RT-qPCR are available in online supplementary file. All RT-qPCR experiments were done at least in triplicates; P value < 0.05 (*), P value < 0.01 (**) and P value < 0.001 (***) are indicated. RNA-seq experiments were performed following TruSeq stranded total RNA illumina protocol and Illumina next generation sequencing was performed as 50 base pairs single-end reads (TruSeq was performed at the McGill University and Génome Québec Innovation Centre, Montréal, Canada).
RNA-seq data processing
The sequenced reads from each sample were preprocessed with Trimmomatic (49) and aligned with bwa (50) to S. cerevisiae genome assembly R64-1-1 (GCA_000146045.2). The number of reads for each exon annotated in Ensembl Release 77 was calculated for both the sense and antisense strands. Differential expression of gene was assessed using DESeq (51). The subset of reads falling within the 5′-most and 3′-most 10% of each exon were also calculated. To avoid confounding effects from non-cryptic antisense transcription, overlapping genes were excluded from further analyses. Furthermore, to reduce the amount of noise from inactive genes, genes with fewer than 20 reads in their 5′ region or 10 reads in their 3′ region were also excluded (2670 genes in total). For each gene, the following metrics were calculated: antisense ratio = log2(readsantisense/readssense), antisense enrichment = antisense ratioCK2/Spt6 – antisense ratioWT, three prime ratio = log2(reads3′/reads5′) and three prime enrichment = three prime ratioCK2/Spt6 – three prime ratioWT. Additionally, using samtools (52), all libraries were split into sense and antisense reads according to the underlying genes. Coverage of all genes over a hundred equal-sized bins were then calculated using the bam Coverage and compute Matrix utilities from the deeptools suite (53). The average coverage over all genes from the TSS to the TES were then normalized into reads per million (RPM) and plotted for each library.
ChIP-Seq analyses
Illumina next generation sequencing was performed as 50 base pairs single-end reads at the McGill University and Génome Québec Innovation Centre, Montréal, Canada. The sequenced reads were aligned to the S. cerevisiae genome assembly R64-1-1 (GCA_000146045.2) using bowtie v0.12.8 (54) (Coverage was calculated using bedtools (http://bedtools.readthedocs.io/) (55) and was normalized into reads per million (RPM). Metagenes plotting the average coverage for every gene going from transcription start site (TSS) to transcription end site (TES) were then produced using the ngs.plot tool version 2.08 (56). Briefly, we used the ngs.plot.r function with the sarCer3 database, the gene body option and a window of 1000 bp around the gene.
RESULTS
CK2 regulates replication-independent H3K56 acetylation in transcribed regions
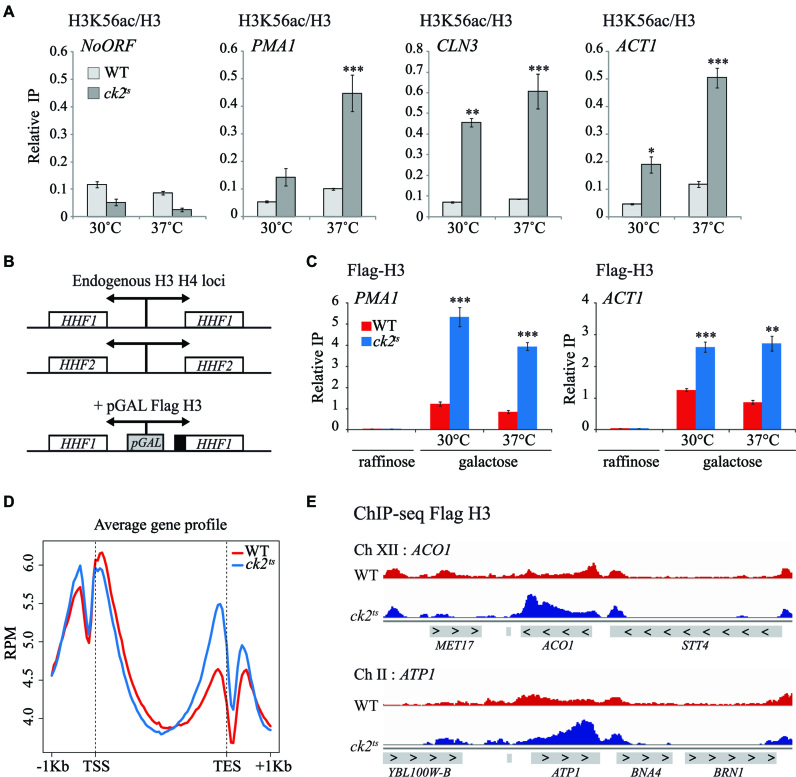
Several reports involved CK2 functionally and physically with chromatin regulators including Spt2 and Spt6 histone chaperones (28,34,35,38). Importantly, we and others found that Spt2 and Spt6 maintain the histone H3/H4 tetramer in coding regions by repressing replication-independent H3 exchange and turnover at several genes (19,20,57). Whether CK2 is implicated in histone H3 dynamics is not known. In order to address this question, we analyzed, in vivo, the role of CK2 in the regulation of H3/H4 exchange in G1 arrested cells. Because histone H3/H4 turnover is tightly associated with H3K56ac levels outside of S-phase (21), we first assessed by ChIP assays the level of H3K56ac in G1 arrested cells in wild-type or ck2ts mutant. As shown in Figure 1A, depletion of CK2 activity is associated with higher levels of new H3K56ac in the coding regions of transcribed genes PMA1, CLN3 and ACT1. In contrast, no such effect was observed in the non-transcribed intergenic region of chromosome V. Thus, our data suggest that CK2 controls the histone H3 exchange.
Figure 1.
Histone H3 turnover is increased in ck2ts mutant at coding regions of genes. (A) CK2 is required for the repression of replication-independent H3K56ac incorporation. ChIP assays assessing the H3K56 acetylation level in WT and ck2ts. Cells were G1-arrested and subsequently grown for 2 h at 30 or 37°C. The values shown represent the average and standard errors of three independent experiments measuring H3K56ac levels (IP/Input) relative to histone H3 occupancy (IP/Input). Coding regions of different genes were tested and NoORF is a non-transcribed control locus of chromosome V. (*) P value < 0.05; (**) P value < 0.01; (***) P value < 0.001. (B) Schematic representation of Flag-H3 inducible construction in YAN1104 and YAN1105 strains. A galactose-inducible form of H3, inserted in URA3 locus, was fused to Flag epitope and co-expressed with histone H4. (C) CK2 is required for the replication-independent repression of the new H3 deposition. ChIP experiment of newly synthesized Flag-H3 in WT and ck2ts. Cells were grown at 30°C in YP-Raffinose, arrested in G1, and then Galactose was added to induce Flag-H3 synthesis for 1 h at 30 or 37°C. The values shown represent the average and standard errors of three independent experiments measuring Flag-H3 (IP/Input) in coding regions of two transcribed genes. (*) P value < 0.05; (**) P value < 0.01; (***) P value < 0.001. (D) CK2 represses global replication-independent incorporation of newly synthesized H3 in the 3′end of genes. Metagene analysis of ChIP-seq data showing newly synthesized Flag-H3 signal of WT and ck2ts strains aligned over transcribed regions of genes. Transcription start site (TSS) and transcription end site (TES) are shown. (E) A view of Flag-H3 ChIP-seq data at two genomic regions of chromosomes XII and II illustrating increased Flag-H3 in ck2ts.
CK2 prevents global histone exchange in 3′end of genes independently of H3K36me3
We tested directly the H3 exchange (Figure 1B and C) by measuring the level of newly synthesized histone Flag-H3 incorporation at various locations in G1 arrested cells as described previously (21). Upon galactose induction in G1 arrested cells, Flag-H3 is synthesized and incorporated in the genome, mainly within the 5′ and 3′ transcribed regions and not in the coding regions of genes (21,58). ChIP-qPCR assays showed that similarly to H3K56ac, depletion of CK2 resulted in higher incorporation of new Flag-H3 into coding regions of active genes (Figure 1C). This observation confirms that CK2 prevents histone exchange in the coding regions of PMA1, ACT1 and CLN3 genes. Then, we analyzed the global exchange in wild-type and ck2ts by Flag-H3 ChIP-seq in G1 arrested cells. Metagene analyses of replication-independent Flag-H3 incorporation indicated that CK2 inhibition led to increased levels of new H3 association (Figure 1D). This experiment was performed twice with similar results as shown in Supplementary Figure S1A. Interestingly, this effect was globally localized in 3′ end of the coding regions of genes as further illustrated in Figure 1E showing two different regions containing ATP1 and ACO1 genes. Importantly, previous studies indicated that histone H3 exchange in 3′end of genes is regulated by H3K36 tri-methylation (H3K36me3) mediated by Set2 (22). Our observation suggested the possibility that CK2 could modulate the H3 exchange by regulating directly or indirectly H3K36me3. To address this possibility, we analyzed the effect of CK2 depletion on the global level of H3K36me3 in yeast cells. As indicated in Supplementary Figure S1B, the level of H3K36me3 in restrictive conditions was similar to that of the wild-type. Moreover, H3K36me3 ChIP assays at various genes indicated that CK2 is not involved in the modulation of this histone mark (Supplementary Figure S1C). Thus, our data indicate that CK2 effect on histone H3 dynamics is independent of H3K36me3.
CK2 inhibition affects global redistribution and phosphorylation of RNAP II
CK2’s role in histone H3 dynamics outside of S-phase may suggest a global involvement of this kinase on transcription by RNAP II. Because replication-independent histone H3 exchange is correlated with transcription by RNAP II (21,58), it is possible that the H3 dynamics observed in ck2ts mutants is the result of a change in RNAP II level, distribution or activity. To address this question, we analyzed the association of RNAP II by Rpb1 ChIP-seq in wild-type and ck2ts cells in the same conditions described for Flag-H3 experiments. The reads obtained after Rpb1 ChIP-seq were aligned for each strain on the transcription start site (Supplementary Figure S2A). In contrast to new H3 deposition, we did not observe a consistent change in the global Rpb1 distribution upon CK2 loss. We next grouped the genes with respect to their RNAP II level and analyzed the effect of CK2 on Rpb1 distribution. As shown, in all groups, CK2 depletion had no effect on the pattern of Rpb1 distribution (Supplementary Figure S2B). We also tested the phosphorylation level of cellular Rpb1 in presence or absence of CK2 activity. Interestingly, CK2 depletion led to global decrease in the phosphorylation of CTD-Serine 2 (Supplementary Figure S2C). This suggests that CK2 may affect RNAP II activity.
CK2 inhibits antisense transcription
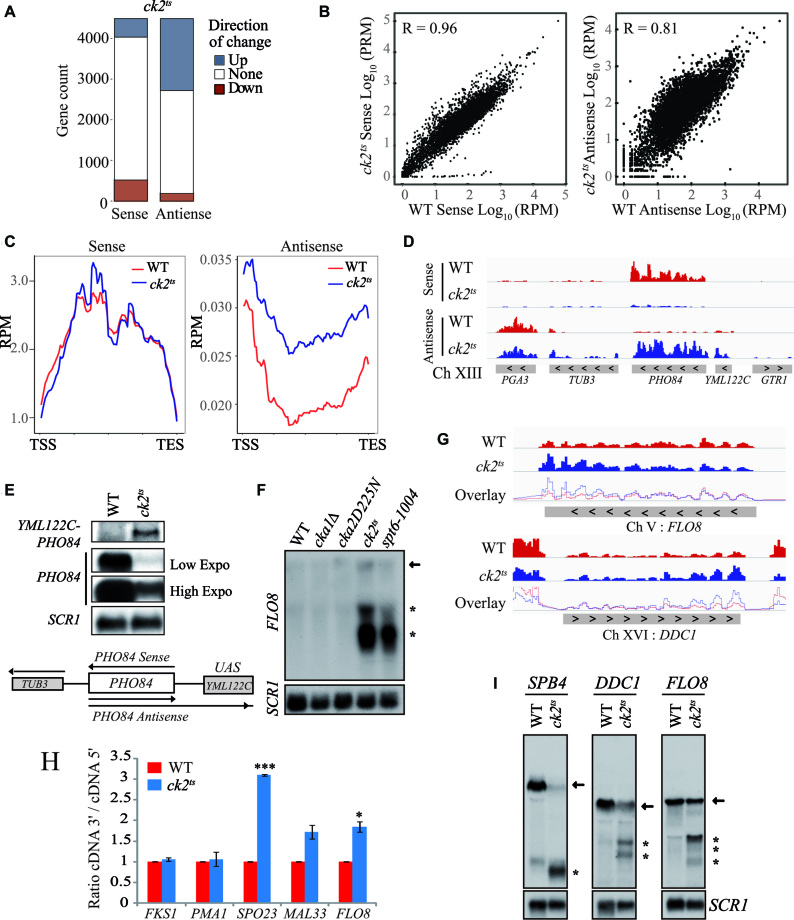
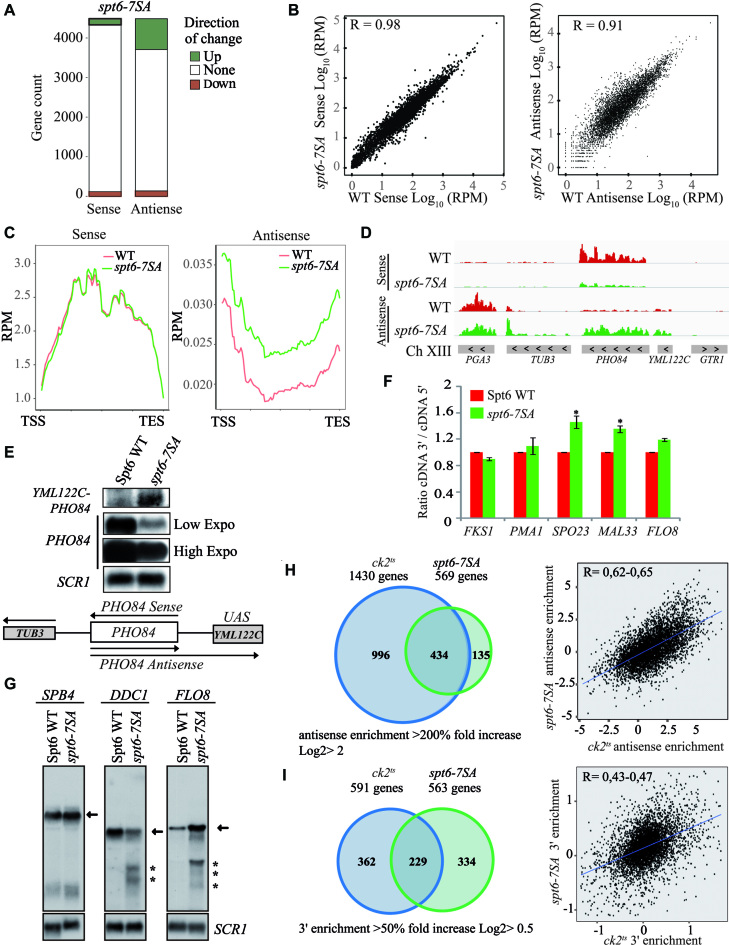
The role of CK2 in histone H3 dynamics, RNAP II distribution and phosphorylation suggests a potential global involvement of this kinase in the regulation of transcription. To identify a possible role of CK2 on transcripts levels, we performed strand-specific RNA-seq comparing WT to ck2ts at restrictive temperature. Strikingly, a CK2-depletion led to an increase of antisense transcripts levels in a large number of genes (Figure 2A). The genes for which the antisense transcript levels were up are overrepresented. The number of genes showing more antisense transcripts is nine times higher than those having antisense transcript levels down. This ration up/down of 9:1 is much higher to what could be expected when we compare it with the gene count of up/down sense ratio that is approximately of 1:1 (P value of 10e–70). Next, we compared wild-type and ck2ts RNA-seq datasets. Wild-type and ck2ts sense transcripts showed high level of similarity. However, the Pearson correlation between WT antisense transcripts and ck2ts was significantly reduced (Figure 2B). This effect to antisense transcripts levels was further confirmed by metagene analysis aligning sense and antisense transcripts to the transcription unit (Figure 2C; Supplementary Figure S3). Antisense enrichment relative to WT has been calculated for each gene in ck2ts mutant, and 1430 genes showed at least 2-fold increase in their antisense ratio. This experiment was performed twice with similar results each time and the correlation between the two experiments is excellent (r = 0.98) as shown in Supplementary Figure S4. To validate the RNA-seq data regarding the altered antisense transcription, we analyzed a locus located in chromosome XIII containing PHO84 and YML122C genes (Figure 2D). PHO84 antisense transcription produces a long transcript that extend into YML122C (15,59). We conducted Northern blot analyses of PHO84 and YML122C in WT and ck2ts cells. In the conditions of our experiment, YML122C is not transcriptionally active and no sense or antisense transcripts from this gene were observed in WT cells. However, depletion of CK2 resulted in the production of a long antisense transcript (PHO84-YML122C) starting in PHO84 and extending into YML122C (Figure 2E). The observation of the transcript in ck2ts cells confirms the RNA-seq data obtained for this locus and further indicates that CK2 inhibits antisense transcription.
Figure 2.
CK2 regulates transcriptional accuracy. (A) CK2 is required for the repression of antisense transcription in a high number of genes. A representation of genes count affected by at least two-fold either positively (blue) or negatively (red) in ck2ts for sense or antisense transcripts levels. (B) The ck2ts antisense transcripts are less similar to those of wild-type (WT) strain. WT and ck2ts sense (left panel) or antisense (right panel) log10 RPM scores were plotted for each transcription unit. The Pearson correlation (R value) was calculated and is shown in each panel. (C) CK2 represses global antisense transcription. Metagene analysis showing WT and ck2ts RNA-seq RPM signals of the sense (left panel) or antisense (right panel) strands aligned over transcribed regions of genes. Transcription start site (TSS) and transcription end site (TES) are shown. (D) A view of PHO84 locus (chromosome XIII) sense and antisense strands transcripts data obtained by RNA-seq in WT (red) or ck2ts (blue). (E) CK2 represses antisense transcription at the PHO84 locus. Northern blot analyses of PHO84-YML122C locus transcripts using different probes. PHO84 probe was used to identify PHO84 sense transcripts while YML122C probe was used specifically to follow the long antisense transcript that starts in PHO84 and extends into YML122C. As shown, this transcript is only observed in ck2ts. (F) CK2 is required for the suppression of spurious transcription from the cryptic promoter of FLO8 gene. Total RNA was isolated from WT, cka1Δ, cka2D225N, ck2ts and spt6-1004 cells grown at 30°C and shifted to 37°C for 2 h. The probe for the northern analysis was generated against 3′-end of FLO8, and SCR1 was used as a loading control. The arrow indicates full-length FLO8 RNA transcripts, and the asterisk indicates FLO8 short transcripts resulting from cryptic initiation. (G) CK2 mutation leads to 3′enrichment of sense transcripts at FLO8 and DDC1. The sense strands transcripts data were obtained by sequencing total RNA of WT (red) or ck2ts (blue). The overlay of WT and ck2ts signals illustrates the 3′ RNA enrichment in ck2ts mutants. (H) CK2 mutation leads to 3′enrichment of sense transcripts at SPO23, MAL33 and FLO8. In this experiment, transcripts levels of the indicated genes were analyzed by RT-qPCR. The 5′ and 3′ regions of each gene were amplified and the values shown are 3′/5′ ratios representing the average and standard errors of three independent experiments. (*) P value < 0.05; (***) P value < 0.001. (I) CK2 is required for the repression of spurious transcription at DDC1 and SPB4. Total RNA was isolated from cells after two hours of growth at 37°C and analysed by Northern Blot with probes specific for 3′-end of FLO8, DDC1 and SPB4. SCR1 was used as a loading control. Arrows indicate full-length RNA transcripts, and asterisks indicate short transcripts.
CK2 represses sense spurious transcription from intragenic cryptic promoters
Recent studies highlighted the role of many chromatin remodelers and HCs in the repression of cryptic transcription and established a clear link between chromatin structure regulation in the transcription unit and the repression of both antisense and sense cryptic transcription from within coding regions (13,23,27,60). Because CK2 regulates antisense transcription similar to chromatin regulators, we wanted to know if CK2 depletion was also associated with sense spurious transcription from known cryptic promoters. For that, we first analyzed FLO8 model gene containing well characterized sense cryptic promoters in its coding region (12,26). FLO8 cryptic promoters are activated when chromatin is not properly refolded after RNAP II passage (12,26,61). Northern blot analyses were performed on total RNA isolated from wild-type, ck2ts and spt6-1004 cells as control (Figure 2F). These analyses showed high levels of FLO8 short cryptic transcripts in spt6-1004 cells grown at restrictive temperature, indicating the activation of the sense cryptic promoters. Interestingly, in the ck2ts cells, we observed strong signals indicating the presence of short cryptic transcripts. Thus, CK2 plays a crucial role in the repression of spurious transcription from FLO8 cryptic promoters and contributes in the maintenance of proper chromatin structure in its coding region. We next analyzed our RNA-seq data to look for a potential global role of CK2 in the regulation of intragenic spurious transcription. Previous studies showed that derepression of sense cryptic intragenic transcription results in the enrichment of transcripts initiated in the 3′end of genes leading to a global alteration in the 3′/5′ transcripts ratio signal (12,27). To address the role of CK2 in cryptic transcription, we generated a ratio of 3′ over 5′ RNA-seq sense signal to obtain a value for each gene. These values were highly reproducible as shown by the pairwise correlations between each replicate (Supplementary Figure S5). When comparing WT and ck2ts 3′/5′ ratios, we identified 591 genes in ck2ts with 50% increase (Supplementary Table S3). Representative genes with altered ratio or 3′ enrichment relative to WT are shown in Figure 2G. To validate our results, we used different approaches. First, we assessed the 3′/5′ expression ratio by RT-qPCR at two other genes exhibiting, in our RNA-seq data, a substantial 3′ enrichment in ck2ts, SPO23 and MAL33. FLO8 is used as positive 3′enrichment control, PMA1 and FKS1 as negative 3′enrichment controls (Figure 2H). As expected for FLO8, SPO23 and MAL33, CK2 depletion led to an alteration in the 3′/5′ transcript ratio suggesting that this kinase represses cryptic transcription from within these genes. As a second approach to further confirm our analyses, we tested spurious transcription by Northern blot probing specific genes. Our RNA-seq data predicted spurious initiation at DDC1 and SPB4 in ck2ts mutant. Interestingly, several spurious cryptic transcripts produced from these genes were identified in previous studies (12,27). Using 3′end specific probes, we found clear bands indicating short transcripts initiated from cryptic promoters of FLO8, DDC1 and SPB4 (Figure 2I). These observations further confirm that CK2 is essential for the repression of spurious transcription from cryptic promoters located within the coding regions of genes.
CK2 interacts with and phosphorylates the transcription elongation associated histone chaperone Spt6
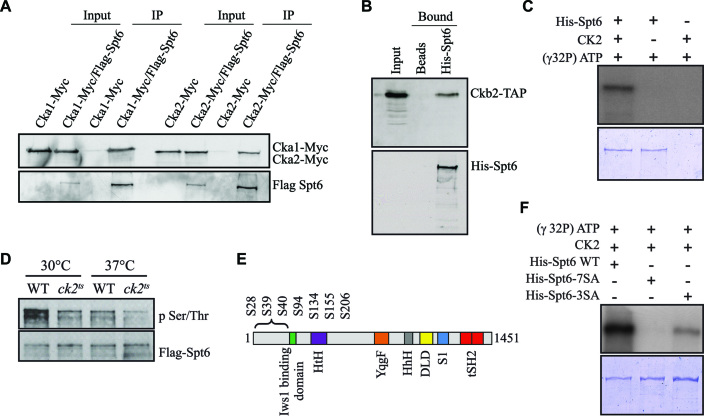
Our data indicate a global role of CK2 in the modulation of chromatin structure during transcription elongation. We wanted to address how CK2 regulates this process. Several observations initially suggested that CK2 might fulfill this function at least in part by regulating Spt6 activity. First, Spt6 is one of the most important chromatin organizer in the coding regions of genes throughout evolution (13,16,62). Second, we established that CK2 regulates the Spt6-Spt2 interaction during elongation (28). Third, peptides from the various subunits of CK2 have been identified by mass spectrometry in tandem affinity-purified Spt6 samples (35,40). Fourth, Spt6 was found phosphorylated at three CK2 consensus motifs (35). To investigate the functional relationship between CK2 and Spt6, we first assessed whether this kinase stably interacts with Spt6 in vivo. For this purpose, we purified FLAG-Spt6 from yeast strains containing two epitope-tagged CK2 subunits. CK2 subunits Cka1-Myc and Cka2-Myc co-purified with FLAG-Spt6 (Figure 3A). To test if this interaction was direct, we performed a His pulldown assay using purified 6xHis-Spt6. Ckb2 purified from yeast by TAP was incubated with 6xHis-Spt6 coupled with Ni-NTA agarose. As shown in Figure 3B, Ckb2-TAP was pulldown by 6xHis-Spt6 indicating a direct interaction between the histone chaperone and the kinase.
Figure 3.
CK2 interacts with Spt6 and phosphorylates it in vitro and in vivo. (A) Spt6 interacts with different subunits of CK2 in vivo. Flag-Spt6 was immunopurified from strains expressing Cka1-Myc or Cka2-Myc. The purified samples were analyzed by western blot with antibodies against the Myc and Flag epitopes. A purification control was obtained using strains expressing untagged Spt6. (B) Spt6 interacts directly with CK2 in vitro. A His pulldown assay was performed using equal amounts of Ckb2-TAP purified from yeast and recombinant 6xHis-Spt6. Ni-NTA agar beads alone served as a control. (C) CK2 phosphorylates Spt6. In vitro phosphorylation assay of recombinant 6xHis-Spt6 with CK2 kinase purified from yeast. (D) Spt6 is phosphorylated in vivo in a CK2-dependent manner. Flag-Spt6 was immunopurified from WT and ck2ts strains grown at 30°C or at 37°C for 2 h. The purified samples were analyzed by western blot with antibodies against phosphoserine/phosphothreonine or Flag epitope. (E) Schematic representation of various domains of Spt6: potential CK2 target sites (S28, S39, S40, S94, S134, S155, S206) are all found in the N-terminal region. (F) The N-terminal phosphosites of Spt6 (S28, S39, S40, S94, S134, S155, S206) are required for the phosphorylation by yeast CK2. In vitro phosphorylation assay of recombinant 6xHis-Spt6 WT, and non-phosphorylatable mutants (6xHis-Spt6-3SA and -7SA) using CK2 kinase purified from yeast.
We next determined whether Spt6 was directly phosphorylated by CK2 using an in vitro phosphorylation assay with recombinant 6xHis-Spt6 and tandem affinity-purified Ckb2 from yeast. CK2 was able to incorporate [γ]-32Pi into Spt6 (Figure 3C). This indicates that Spt6 is a direct substrate for CK2. Interestingly, it has been previously shown that Spt6 is phosphorylated in vivo at various CK2 consensus motifs (35,63). To investigate if Spt6 is modified by CK2 in vivo, we immunopurified FLAG-Spt6 from WT and ck2ts yeast strains and analyzed the purified material by Western blot with an antibody against phospho-serine/threonine. As shown in Figure 3D, Spt6 was recognized by the anti-phospho-serine/threonine in WT extracts, further confirming that Spt6 is indeed a phosphoprotein in vivo. This signal was significantly altered in the ck2ts mutant at the restrictive temperature, suggesting an important role for CK2 in the regulation of the phosphorylation state of Spt6 in vivo. Taken together, our data indicate that CK2 interacts directly with Spt6 and phosphorylates this elongation factor both in vitro and in vivo.
Next, we aimed to identify the sites in Spt6 that are phosphorylated by CK2. Previously, three potential CK2 phosphorylation sites (S94, S134 and S206) in Spt6 were shown to be modified in vivo (35). We analyzed by mass spectrometry the phosphorylation profile of Spt6 and confirmed that at least two sites (S94, S134) were indeed modified in vivo (Supplementary Figure S6). Interestingly, our mass spectrometry analyses allowed the identification of at least one additional site in N-terminal region of Spt6: S155 (Supplementary Figure S6). Moreover, five CK2 phosphorylation target sites have been reported for Spt6 in vivo (63,64). Notably, all the above-mentioned phosphorylation sites are located in the N-terminal region of Spt6. Furthermore, using the NetPhos 3.1 server, we predicted a total of 7 ‘high score’ potential CK2 target sites in the N-terminal region of Spt6 (Figure 3E). We therefore elected to mutate either the three initially identified, or all seven potential CK2 target sites to alanine residues to produce and collect the resulting recombinant protein. Then, we subjected the two mutant proteins to in vitro phosphorylation by purified CK2 from yeast. The Spt6 mutant in which the three initial CK2 target sites had been eliminated, exhibited a reduced but residual level of phosphorylation, while no [γ]-32Pi was incorporated into a mutant Spt6 protein in which all seven CK2 potential target sites had been eliminated, indicating that these sites include all CK2 phosphorylation sites used in vitro (Figure 3F).
CK2 Phosphorylation in Spt6 are important for its chromatin function
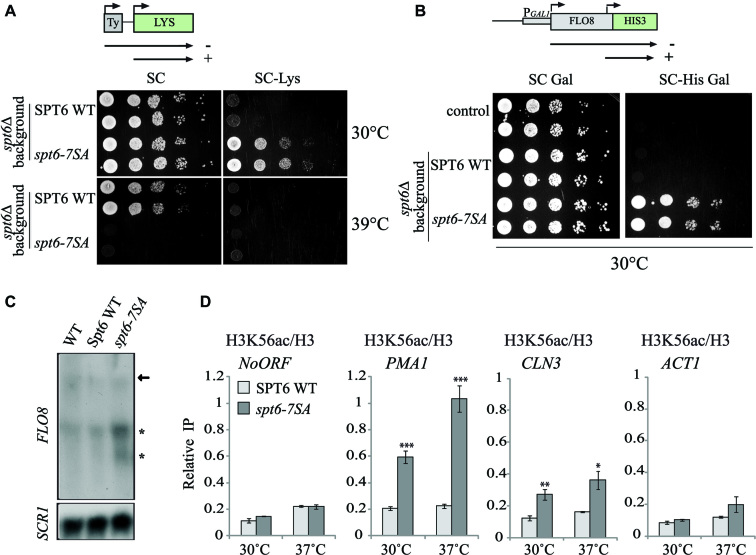
To analyze the potential function of the identified CK2 target sites, we constructed strains expressing Spt6-7SA mutant form and probed a set of phenotypes associated with Spt6 function. One aspect of the SPT phenotype is a defect in Spt6 function that suppresses δ insertion mutations in the promoter of the LYS2 genes. Thus, a WT strain with a lys2-128δ allele is unable to grow in the absence of lysine. A defect in Spt6 function suppresses this phenotype and the strain is no longer auxotrophic for lysine (46 ). As shown in Figure 4A, cells expressing spt6-7SA mutant exhibited thermosensitivity, and an spt- phenotype (loss of lysine auxotrophy) on SC-Lys. An spt- phenotype indicates defect in Spt6 function associated with chromatin organization (65). Defective Spt6 function is also associated with spurious transcription from cryptic promoters within the coding regions of transcriptionally active genes (12). We analyzed the consequence of the expression of non phosphorylatable Spt6 mutant on the repression of intragenic cryptic promoter at FLO8 using the reporter system pGAL1-FLO8-HIS3. The expression from the FLO8 cryptic promoter, as measured by growth in the absence of histidine, indicates spurious transcription (Figure 4B), reflecting a defect in chromatin refolding during transcription. We also analyzed cryptic transcription at FLO8 by Northern blot and detected short cryptic transcripts in yeast expressing spt6-7SA (Figure 4C). This result shows that CK2 target sites found in the N-terminal region of Spt6 are important for Spt6 chromatin functions. Finally, we assessed by ChIP assays the level of H3K56ac in G1 arrested cells in WT or spt6-7SA mutant (Figure 4D). Importantly, the inhibition of Spt6 phosphorylation by CK2 was associated with higher specific incorporation of new H3K56ac in the coding regions of transcribed genes (non-transcribed intergenic region of chromosome V was used as negative control). Our data strongly suggest that CK2-dependant phosphorylation of the Spt6 N-terminal region is involved in the modulation of histone H3 dynamics during transcription. Taken together, these data show that CK2 phosphorylation sites of the Spt6 N-terminal region are required for Spt6 chromatin functions.
Figure 4.
Phosphorylation by CK2 regulates Spt6 chromatin function. (A) Mutation of CK2 phosphosites in Spt6 leads to spt- phenotype. SPT6 WT and spt6-7SA (Serine to Alanine) strains containing the Lys2-128δ SPT reporter allele were serially diluted and spotted on synthetic complete (SC) or SC medium lacking lysine (SC-Lys) at 30 or 39°C. The spt6-7SA allele is thermosensitive and has an spt- phenotype. (B) The spt6-7SA is not able to suppress spurious transcription from FLO8 cryptic promoter. WT and spt6Δ strains expressing SPT6 WT or spt6-7SA containing pGAL1-FLO8-HIS3 spurious transcription reporter were serially diluted and spotted on synthetic complete (SC) medium, or SC medium lacking histidine (SC-His) with Galactose as a carbon source. The spt6-7SA allele growth in absence of histidine indicates intragenic cryptic initiation within FLO8 coding region. (C) The spt6-7SA mutant is associated with cryptic transcription. Cells expressing either SPT6 WT or spt6-7SA were grown in YPD at 37°C. Total RNA was isolated and analyzed by Northern blot with a probe specific for 3′-end of FLO8. SCR1 served as loading control. (D) Replication-independent histone H3K56ac level is increased at coding regions of genes in spt6-7SA mutant. ChIP assays assessing the H3K56 acetylation level in G1-arrested cells expressing Spt6 or the mutated Spt6-7SA version. Cells were G1-arrested and subsequently grown for 2 h at 30 or 37°C. The values shown represent the average and standard errors of three independent experiments measuring H3K56ac levels (IP/Input) relative to histone H3 occupancy (IP/Input). Coding regions of different genes were tested and NoORF is a non-transcribed control locus of chromosome V. (*) P value < 0.05; (**) P value < 0.01; (***) P value<0.001.
Spt6 phosphorylation sites play an essential role in the global repression of antisense and cryptic transcription
To characterize further the role CK2-dependent Spt6 phosphorylation in transcription at high resolution, we performed strand-specific RNA-seq in mutant comparing WT to spt6-7SA. Approximately the same number of genes for which the sense transcripts levels were affected either positively or negatively (Figure 5A). Strikingly, spt6-7SA mutation led to the increase of antisense transcription in a significantly higher number of genes (P value < 10e–70, Ratio UP/Down 6:1). This is consistent with what we observed for the ck2ts mutant and suggests that Spt6 phosphorylation sites are required for the repression of antisense transcription. We next analyzed the data by comparing WT and spt6-7SA sense and antisense transcripts. Also, similar to what have been found in ck2ts cells, the Pearson correlation between WT and spt6-7SA data sets is reduced for the antisense transcripts (Figure 5B). The metagene analyses aligning sense and antisense transcripts to transcription start site further confirmed that the most striking effect of spt6-7SA mutation is to antisense transcription (Figure 5C and Supplementary Figure S7). Next, we confirmed our global data by testing antisense transcription in a specific locus. As for ck2ts, the RNA-seq analysis showed that spt6-7SA allele is associated with a significant induction of antisense transcription in PHO84 locus (Figure 5D). We probed for the gene YML122C located upstream of PHO84. This gene is not transcriptionally active, no sense or antisense transcripts are observed unless antisense transcript initiates from PHO84 coding region and extends into YML122C (Figure 5E). In spt6-7SA allele, a probe located in YML122C revealed a long antisense transcript (PHO84-YML122C) starting in PHO84 and extending into YML122C (Figure 5E). Hence, these data indicate that CK2 phosphorylation sites in Spt6 are required for the repression of antisense transcription in yeast cells.
Figure 5.
Spt6 phosphorylation regulates cryptic intragenic and antisense transcription. (A) Spt6 is required for the repression of antisense transcription in a high number of genes. A representation of genes count affected by at least two-fold either positively (green) or negatively (red) in spt6-7SA for sense or antisense transcripts levels. (B) The spt6-7SA antisense transcripts are less correlated with those of the wild-type (WT) strain. WT sense (left panel) or antisense (right panel) log10 RPM scores for each transcription unit were plotted against spt6-7SA datasets. The Pearson correlation (R value) was calculated and is shown in each panel. (C) CK2 phosphorylation sites in SPT6 are required for the global repression of antisense transcription. Metagene analysis showing WT and spt6-7SA RPM RNA-seq signals of the sense (left panel) or antisense (right panel) strands aligned over transcribed regions of genes. Transcription start site (TSS) and transcription end site (TES) are shown. A higher level of antisense transcription is observed in spt6-7SA. (D) Spt6 phosphosites are essential to the repression of antisense transcription in PHO84 locus. A view of PHO84 locus (chromosome XIII) sense and antisense strands transcripts data obtained by RNA-seq in WT (red) or spt6-7SA (green). (E) The CK2 phosphorylated residues play a crucial role in the repression of antisense transcription at PHO84 locus. This observation was shown here by Northern blot analyses of PHO84-YML122C locus transcripts using different probes. PHO84 probe was used to identify PHO84 sense transcripts while YML122C probe was used specifically to follow the long antisense transcript that starts in PHO84 and extends into YML122C. (F) Mutation of CK2 phosphorylated residues in Spt6 leads to 3′enrichment of sense transcripts at SPO23, MAL33 and FLO8. In this experiment, transcripts levels of the indicated genes were analyzed by RT-qPCR. The 5′ and 3′ regions of each gene were amplified and the values shown are 3′/5′ ratios representing the average and standard errors of three independent experiments. (*) P value < 0.05. (G) CK2-dependent phosphorylation of Spt6 is required for the repression of spurious transcription at DDC1 and SPB4. Total RNA was isolated from cells after two hours of growth at 37 and analysed by northern blot with probes specific for 3′-end of FLO8, DDC1, and SPB4. SCR1 was used as a loading control. Arrows indicate full-length RNA transcripts, and asterisks indicate short transcripts. (H) Significant overlap between ck2ts and spt6-7SA datasets for the genes with an increased antisense transcripts levels. The Venn diagram compares the number of genes with antisense enrichment in ck2ts and spt6-7SA mutants. The right panel is a correlation plot of antisense enrichment between ck2ts and spt6-7SA mutants. The ck2ts Antisense/Sense ratio relative to WT for each gene was plotted against spt6-7SA Antisense/Sense ratio relative to WT. The Pearson correlation coefficient (R value) was calculated and shows a positive and significant correlation between the two ratios. (I) Significant number of genes with predicted intragenic cryptic transcripts, identified by 3′enrichmnent, in ck2ts overlap with those of spt6-7SA mutants. The Venn diagram compares the number of genes with 3′ enrichment in ck2ts and spt6-7SA mutants. The right panel is a correlation plot of 3′ enrichment between ck2ts and spt6-7SA mutants. The 3′ennrichment of ck2ts for each gene was plotted against that of spt6-7SA. The Pearson correlation coefficient (R value) was calculated and shows a positive and significant correlation between the two data sets regarding cryptic transcripts.
Spt6 is involved in the repression of global spurious transcription from cryptic promoters (12). As shown in Figure 4, mutations of CK2 phosphorylation sites lead to the derepression of the FLO8 cryptic promoter suggesting that CK2 regulation of Spt6 may be involved in global control of sense cryptic transcripts level. To analyze the global role of CK2 regulation on cryptic transcripts, we measured the 3′/5′ ratios in the spt6-7SA mutant for all genes and compared them to that of wild-type cells. In spt6-7SA, we found 563 genes with increased ratio suggesting that CK2 phosphorylation sites play an important role in the repression of cryptic transcription (Figure 5I, Supplementary Table S3). These data are reproducible as attested by the correlation shown in Supplementary Figure S8. To validate these results, we measured the 3′/5′ ratio by RT-qPCR at 3 genes, FLO8, SPO23 and MAL33, exhibiting 3′ enrichment in spt6-7SA mutant RNA-seq signals (Figure 5F). Interestingly, as predicted by our RNA-seq analyses, spt6-7SA mutation is associated with a significant increase of 3′/5′ transcripts ratio at SPO23 or MAL33 and a slight one at FLO8 (Figure 5F). This effect is not observed in the negative controls PMA1 and FKS1, confirming that CK2 regulation of Spt6 controls the cryptic transcripts levels at several genes identified by our analyses. Moreover, our RNA-seq 3′/5′ ratios data also predicted the existence of cryptic transcripts produced from within DDC1 3′end but not in SPB4 3′ end region. This was surprising because SPB4 is a common model gene used in several studies of cryptic transcription and has been found to be affected in ck2ts cells (12). To further confirm our analyses, we probed the transcripts of DDC1 and SPB4 by Northern blot (Figure 5G). We observed clear bands indicating short transcripts initiated from 3′end cryptic promoters of FLO8 and DDC1, but not for SPB4 as predicted by our analyses. Therefore, these data suggest significant but not complete overlap between ck2ts and spt6-7SA RNA-seq data sets. Together our observations indicate that CK2 phosphorylation of Spt6 is required for the repression of spurious transcription from cryptic promoters located within the coding regions of genes.
Spt6 phosphorylation mutants regulate cryptic sense and antisense transcription at promoters that are also affected by CK2
As suggested by our Northern blot experiments there are overlaps between ck2ts and spt6-7SA but also some differences as shown for SPB4. We wanted to address this question globally and know if the mutation of Spt6 phosphorylation sites affects cryptic sense or antisense transcription at the same sites as CK2. We first compared ck2ts and spt6-7SA RNA-seq data sets for antisense transcripts and found positive and significant overlap between the two data sets (Figure 5H left panel and Supplementary Figure S9A). Moreover, we found positive correlation between the two data sets (Figure 5H right panel). Interestingly, we also observed the same type of relations between the two sets of data regarding the cryptic transcripts identified by the 3′/5′ ratio (Figure 5I and Supplementary Figure S9B). The overlap was much higher than what would be expected by chance with P values less than 8.529784e-67 (Figure 5H and I; Supplementary Figure S9). Remarkably, 76% (434 out 569) of the genes showing an increase of antisense transcription in spt6-7SA also produce higher antisense transcripts in ck2ts (Figure 5H). Together, these data establish a strong link between Spt6 phosphorylation and the global regulation of transcription accuracy by CK2. However, it must be noted that the group of CK2 genes producing pervasive antisense transcripts extends well beyond those affected in spt6-7SA. This suggests that, in addition to CK2–Spt6 pathway, this kinase may regulate additional sites independently of Spt6.
CK2 phosphorylation sites are required for the stability of Spt6
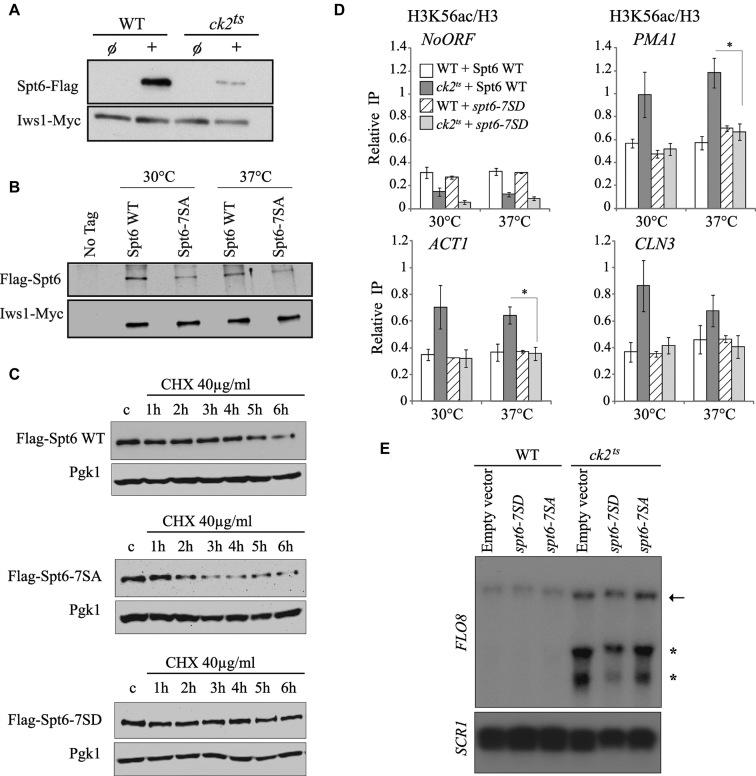
The essential kinase CK2 regulates transcription accuracy possibly by modulating the activity of chromatin factors involved in the re-setting of the chromatin structure in the path of transcription. We have found that CK2 interacts and phosphorylates Spt6 which plays a major role in the refolding of nucleosomes in the body of genes. Our global data indicate that CK2 represses spurious transcription at many genes that are also affected by the loss of its phosphorylation sites in Spt6. These observations suggest that CK2 may influence, at least in part, the accuracy of transcription by phosphorylating Spt6 and thereby modulating its activity. Interestingly, by studying the effect of CK2 on Spt6/Iws1 complex, we observed that the level of the complex is significantly dependent on CK2 phosphorylation of Spt6 in vitro (data not shown). This effect is not observed in vivo (data not shown). However, we observed that CK2 depletion has a significant effect on total Spt6 protein level as indicated by Western blots shown in Figure 6A. This finding suggests a mechanism involving CK2 that regulates the cellular level of Spt6. We tested this by assessing the level of Spt6 in spt6-7SA cells. As shown in Figure 6B, mutation of CK2 phosphosites is clearly associated with a decreased level of Spt6. Interestingly, reducing the level of Spt6 just by 50% results in an impaired function, as shown in Supplementary Figure S10, supporting previous reports (46,65). The reduction of Spt6 level could be the consequence of either a defect in the production of the protein or an impaired stability. Because we did not observe a significant and a reproducible effect of CK2 on the production of SPT6 transcripts by RT-QPCR (data not shown), we decided to test the second possibility by asking whether CK2 phosphorylation sites contributes to Spt6 stability. For that, Spt6 level was analyzed in wild-type and CK2 phosphosites mutant spt6-7SA cells grown in presence of cycloheximide, a protein synthesis inhibitor. As shown in Figure 6C and Supplementary Figure S11, Spt6–7SA protein level decay was significantly faster than Spt6 WT decay. The half -life of Spt6-7SA was around two hours while that of Spt6 WT is approximately four hours. Importantly, expressing a version of Spt6 mimicking CK2 phosphorylation stabilized the protein and no significant decay was observed after six hours of cycloheximide treatment. This result indicates that CK2 phosphorylation sites play a major role in the maintenance of normal Spt6 level.
Figure 6.
CK2 phosphorylation sites are critical for Spt6 stability and chromatin functions. (A) CK2 mutant is associated with a decrease of Spt6 cellular level. Flag-Spt6 and Iws1-Myc proteins were analyzed by western blots on total extracts from wild type (WT) or ck2ts heat-shocked cells using antibodies against Flag and Myc epitopes. (B) Mutation in CK2-dependent phosphosites leads to a decrease in Spt6 protein level. Flag-Spt6 and Iws1-Myc proteins were analyzed by western blots on chromatin extracts of cells expressing Flag-Spt6 or Flag-Spt6-7SA. (C) CK2-dependent phosphorylation of Spt6 is required for its stability. Flag-Spt6, Flag-Spt6-7SA (non-phosphrylatable) and Flag-Spt6-7SD (phosphomimic) proteins were analyzed by western blots on total extracts from heat-shocked cells treated for the time indicated with cycloheximide (CHX). Pgk1 was used as a loading control. Mutation of Spt6 phosphosites to alanine reduces its half-life while the change toward aspartic acid residues stabilizes this protein. (D) Spt6 phosphomimic mutant spt6-7SD suppresses the increased H3K56ac deposition observed in ck2ts mutant. ChIP assays assessing the H3K56 acetylation level in WT and ck2ts expressing Spt6 or Spt6-7SD. Cells were G1-arrested and subsequently grown for 1 h at 30 or 37°C. The values shown represent the average and standard errors of three independent experiments measuring H3K56ac levels (IP/Input) relative to histone H3 occupancy (IP/Input). Coding regions of different genes were tested and NoORF is a non-transcribed control locus of chromosome V. (*) P value < 0.05. (E) Spt6 phosphomimic mutant spt6-7SD suppresses the cryptic transcription in ck2ts at FLO8 gene. Cells expressing either Spt6-7SD or Spt6-7SA were grown in YPD at 30°C and then shifted 37°C for two hours. Total RNA was extracted and analyzed by northern blot using a specific FLO8 3′ end probe. SCR1 is a loading control.
Restoring Spt6 cellular level in CK2 depleted cells suppresses H3K56ac incorporation and spurious transcription
Our later findings regarding Spt6 stability bring about an interesting possibility involving CK2 regulation of histone H3 dynamics. Indeed, one can assume that if CK2 regulation modulates histone H3 dynamics by maintaining Spt6 cellular levels, then mimicking CK2 phosphorylation of Spt6 could affect this dynamic. To address this possibility, we analyzed the incorporation of new H3 in CK2 phosphosites mutants at the coding regions of several genes. The level of H3K56ac, a surrogate of new H3 incorporation, was assessed by ChIP assays in wild-type or ck2ts G1 arrested cells expressing either Spt6 WT or Spt6-7SD (Figure 6D). Interestingly, when the ck2ts cells express a wild-type Spt6, the H3K56ac level is increased at the coding regions of active genes at both permissive and restrictive temperatures. This shows higher replication-independent incorporation of newly synthesized H3 in ck2ts and indicates that in these regions, H3/H4 tetramers that are disrupted by elongating RNAP II are not recovered. Instead, in these locations, new H3/H4 tetramers are used to reassemble nucleosomes. Importantly, when ck2ts cells express a version of Spt6 that mimics CK2 phosphorylation, a clear suppression of the new H3 incorporation was observed at the coding regions of active genes. This indicates that CK2 function in the regulation of replication-independent H3/H4 dynamics is mediated by the phosphorylation of Spt6 and suggests that stable level of this factor plays a key role in this process. To further study the effect of Spt6 phosphorylation state on CK2 chromatin function, we asked if the suppression of histone H3 incorporation observed in ck2ts cells expressing a version mimicking CK2 phosphorylation is associated with a change in the spurious transcription. To this end, we analyzed by Northern blot the transcripts of FLO8 gene. As shown in Figure 6E, in restrictive conditions, short transcripts are clearly observed in ck2ts cells expressing Spt6 WT or Spt6-7SA. Interestingly, ck2ts cells expressing Spt6-7SD display significantly less FLO8 short transcripts indicating that a stable level of Spt6 suppresses spurious transcription from cryptic promoters in cells that are depleted in CK2 activity. Together, our findings demonstrate that CK2’s function in suppression of spurious transcription is at least in part dependent on the regulation of Spt6 stability through its direct phosphorylation.
CK2 and Spt6 phosphorylation sites are important for the efficient transcriptional response to environmental signals and stresses
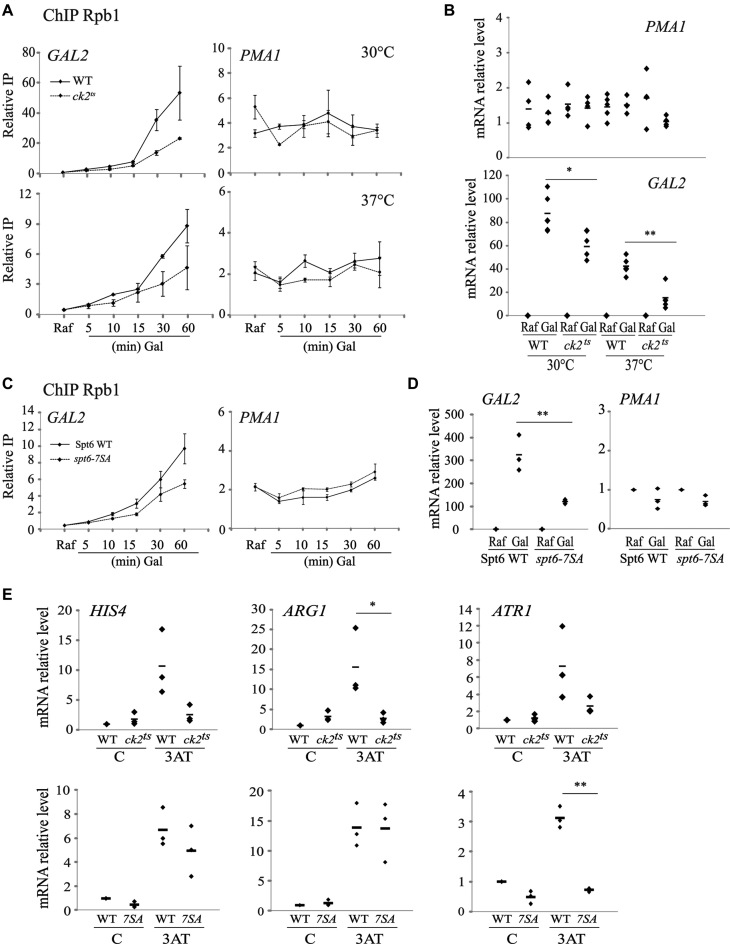
CK2 has a widespread role in chromatin dynamics and the control of transcription accuracy without impacting transcripts levels of most protein-encoding genes. Indeed, in standard conditions used in our study, the important production of spurious sense and antisense transcripts in CK2 depleted cells has only limited effects on gene expression. This surprising observation is however not restricted to CK2 and many chromatin modifiers have limited impact on gene expression (66). Interestingly, elegant studies showed that chromatin regulators have greater effect on the dynamics of gene expression than on the steady state transcription (66). Thus, it is possible that, similar to chromatin regulators, CK2 would modify the dynamics of transcriptional response. To test this, we first asked whether CK2 has a role in the dynamics of galactose induction at galactose responsive genes. In Figure 7A, a time-course of Rpb1 (a subunit of RNAP II) recruitment to GAL2 gene is shown in both wild-type and ck2ts at permissive and restrictive temperatures. Interestingly, at both conditions, we observed a clear reduction of Rpb1 recruitment at GAL2 while no such effect was observed at the constitutive gene PMA1. This defect was associated with a clear reduction of the GAL2 mRNA production, indicating that CK2 affects the cellular response to carbon source shift (Figure 7B). Next, we wanted to know if CK2 phosphorylation sites in Spt6 are important for Rpb1 recruitment and GAL2 transcriptional induction. Importantly, similar to ck2ts mutant, the recruitment of Rpb1 and the induction of GAL2 induction are both impaired in spt6-7SA (Figure 7C and D). We further tested other transcriptional responses as histidine starvation by treating WT or mutant cells with 3-aminotriazole (3AT), a competitive inhibitor of His3. Both ck2ts and spt6-7SA mutations affected significantly this response regarding mRNA levels of different metabolic genes activated by amino-acid starvation such as HIS4, ARG1 and ATR1 (Figure 7E). We conclude that CK2 and the phosphorylation of Spt6 are required for the dynamic adaptation of cells to environmental changes and stresses.
Figure 7.
CK2 and its Spt6 phosphorylation sites are required for transcriptional response. (A) Rpb1 recruitment to galactose-inducible gene is affected by CK2 depletion. ChIP assays analysing Rpb1 recruitment to galactose inducible GAL2 or to constitutive PMA1 genes. Cells from WT or ck2ts strains were grown in raffinose to mid-log phase, galactose was added and cultures were shifted to the indicated temperatures (30°C: permissive or 37°C: non-permissive). The values shown represent the average and standard errors of three independent experiments measuring Rpb1 relative levels (IP/Input). (B) CK2 is required for the transcript level induction of the galactose responsive gene GAL2. RT-QPCR assessing transcripts levels of GAL2 and PMA1 upon galactose induction in WT and ck2ts. Total RNAs, extracted from cells treated as described in A, were analysed by RT-qPCR. (C) Rpb1 recruitment to galactose-inducible gene is affected by mutations of CK2 phosphosites in Spt6. ChIP assays analysing Rpb1 recruitment to galactose inducible GAL2 or to constitutive PMA1 genes. Cells expressing Spt6 or Spt6-7SA mutated version were grown in raffinose to mid-log phase and then galactose was added for the indicated time. The values shown represent the average and standard errors of three independent experiments measuring Rpb1 relative levels (IP/Input). (D) Spt6 phosphorylation sites are required for the transcript level induction of the galactose responsive gene GAL2. RT-QPCR assessing transcripts levels of GAL2 and PMA1 upon galactose induction in cells expressing WT Spt6 or Spt6-7SA. Total RNAs, extracted from cells treated as described in C, were analysed by RT-qPCR. (E) CK2 and its phosphorylation sites in Spt6 are required for the transcriptional response to histidine starvation. WT, ck2ts or cells expressing spt6-7SA were grown in SC-His medium, heat shocked at 37°C for 30 min and then treated with 40 mM 3-aminotriazole (3AT) for 1 h to induce amino acid starvation response. Total RNA was extracted, used to produce cDNA and quantified by QPCR at HIS4, ARG1 and ATR1 genes. For all RT-QPCR, the relative level is the ratio of the indicated RNA to the SCR1 transcript level. (*) P value < 0.05; (**) P value < 0.01; (***) P value < 0.001.
DISCUSSION
Proper refolding of chromatin during the course of transcription is tightly regulated. Defects in chromatin refolding have been linked to spurious transcription from cryptic promoters within the coding regions of different genes (12,14,22,23). Cryptic transcription is a widespread phenomenon in yeast, with many genes displaying such aberrant event. Importantly, it has also been reported in mammals (18,67). Different factors can suppress cryptic transcription, including histones, regulators of histone genes, chromatin remodeling factors, transcription elongation factors and histone chaperones (12,23,68). CK2’s subunits have been associated with some of these factors. Indeed, subunits of CK2 have been found to be associated with different factors that modulate chromatin structure during transcription elongation (35,39,40). Previous studies suggested that CK2 may regulate chromatin structure (33,34,69). However, an exact mechanism of how such regulation operates remained largely elusive. Here we show, for the first time, clear and direct evidence of CK2’s involvement in this process. Our data further support a physical and a functional link between CK2 and an essential HC, indicating a specific role of CK2 in chromatin structure dynamics. Furthermore, we identified a potential mechanism of how CK2 may regulate chromatin modulations associated with transcription elongation. By regulating this chromatin process, CK2 contributes directly to the suppression of cryptic intragenic sense and antisense transcription.
CK2 controls the nature of nucleosomes in the 3′end of transcribed regions
During transcription elongation, nucleosomes are unfolded to allow the progression of the transcription machinery along DNA (3). Upon RNAP II passage, nucleosomes are refolded back, mainly by mechanisms that preserve the histone H3/H4 tetramers. These mechanisms involve histone H3/H4 chaperones such as Spt2 and Spt6 (19,20,57). In addition to nucleosomes refolding, histones are further modified by enzymes, including Set2, which incorporates methyl groups on lysine 36 of histone H3 (70). This modification is of great importance because it directs the deacetylation of histone H3/H4 tails and thereby stabilizes nucleosomes (22,70). Together, reassembly of nucleosomes and chemical modifications of histones are required for the repression of spurious transcription and the focusing of RNAP II on real promoters leading to production of transcripts translated into proteins. Our data show a key role of CK2 in chromatin remodeling in 3′end of coding regions. First, we found that CK2 activity is required for the control of H3K56 acetylation level in transcribed regions of several genes (Figure 1A). Second, we showed that new histone H3 incorporation in coding regions, a surrogate of nucleosomes turnover in these locations (5), is also tightly and globally controlled by CK2 (Figure 1C). Third, the high level of histone H3 exchange in CK2 depleted cells is specifically located in the 3′end regions of genes (Figure 1D). Thus, CK2 controls the nature of nucleosomes that are refolded back after the passage of RNAP II and this has important consequences. Indeed, a marked increase of replication-independent histone H3/H4 exchange means that those nucleosomes have different marks and may therefore display different properties. A high level of H3K56ac outside of S-phase indicates not only a higher histone H3/H4 turnover but also a higher general acetylation level of histone H3/H4 (22,70). As discussed above, acetylation of H3/H4 in coding region has a major impact on the role of the chromatin structure as barrier against RNAP II wrongful association in these locations. Our work shows that CK2 is required for the control of H3K56ac levels in coding regions and may influence indirectly other histone acetylation sites that could be important for the inhibition of cryptic promoters located within coding regions.
One important observation regarding CK2 and chromatin modulations, is that the high level of new histone H3 incorporation we observed in CK2 depleted cells is located in the 3′end regions of genes. These regions are typically enriched in histone H3K36me3 mark, which is known to play a major role in the inhibition of various cryptic promoters, and is involved in the control of histone H3 exchange at these locations (22). In addition, similar to a set2 mutant, ck2ts did not affect the global levels of histones in coding regions (data not shown). Furthermore, Spt6/Iws1 complex is a key regulator of H3K36me3 and Set2 activity (62). Thus, CK2 has similarity towards Set2 on histone H3 exchange in 3′ of coding regions, H3K56ac levels, histone H3 occupancy, and Spt6 function. Consequently, one could easily imagine that CK2 may influence chromatin modulations through the direct or indirect control of H3K36 methylation. Our data discard this possibility. First, in ck2ts cells, we did not observe an effect on H3K36me3 at all genes tested (Supplementary Figure S1C and data not shown). Second, the global level of these modifications was not significantly different in CK2 depleted cells from that of wild-type cells (Supplementary Figure S1B). Third, the recruitment of Set2 enzyme was directly assessed by ChIP assays in ck2ts cells and no significant change has been observed (data not shown). Therefore, we conclude that CK2 controls the chromatin dynamics in the coding regions through one or more pathway(s) and none of these include Set2 and H3K36me3.
CK2-mediated phosphorylation of Spt6 modulates chromatin in transcribed regions
Several new observations made in this work point toward a direct regulation of one of the main histone chaperone involved in chromatin modulations during transcription elongation. Indeed, we found that Spt6 is phosphorylated on several residues by CK2 both in vivo and in vitro (Figure 3). These phosphosites have an important impact on the function of Spt6 and their mutation leads to defects in the suppression of Ty transcription (Figure 4). In addition, they are required for the control of H3K56ac levels and recycling of H3/H4 tetramers in coding regions. Consequently, the Spt6 version mutated in CK2 phosphosites cannot achieve the repression of spurious transcription, as shown by reporter assay, Northern blot and RNA-seq data (Figures 4 and 5). Importantly, we demonstrated that mimicking CK2 phosphorylation results in rescue of ck2ts phenotypes on both histone H3K56ac and spurious transcription (Figure 6C and D). Thus, we feel confident that a significant part of CK2 regulation proceeds through Spt6 phosphorylation. This is especially the case for histone H3 dynamics, given the fact that the expression of an Spt6 version mimicking CK2 phosphorylation in ck2ts mutants leads to a complete repression of histone H3K56ac increased incorporation in the tested coding regions (Figure 6D). However, CK2 control of cryptic sense and antisense transcription extends beyond the role of Spt6. Indeed, despite good overlap between ck2ts and spt6-7SA data sets regarding genes producing intragenic or antisense transcripts, differences do exist. This is well illustrated by the comparison of ck2ts and spt6-7SA genes that were affected for antisense transcripts. We observed that almost 80% of genes affected in spt6-7SA also produce antisense transcripts in ck2ts. However, CK2 mutant affects many more genes and only 32% are common with spt6-7SA. Several possibilities could explain these observations. In our experimental conditions, ck2tscould simply have indirect effects that would result in more drastic phenotypes. Alternatively, CK2 could regulate different factor(s) that would modulate directly the initiation from different cryptic sense or antisense promoters. Interestingly, CK2 has been directly linked with FACT and PAF elongation complexes and H2B ubiquitylation (33,35,39) which were associated with spurious transcription and chromatin reassembly (71,72). The existence of such an alternative pathway is supported by the partial suppression of CK2 spurious transcription when Spt6-7SD is expressed.
Finally, we should mention that Spt6 partner, Iws1, is a potential target of CK2 (35). In addition to Spt6, CK2 may regulate the other subunit of the Spt6/Iws1 complex and this regulation could have a significant role in the overall control of transcription accuracy. Thus, it would be interesting to test if CK2 modulates Iws1 and how this potential regulation affects the chromatin function of Spt6/Iws1complex and spurious transcription. In any ways, CK2 regulates Spt6 function by maintaining its cellular levels. This has direct consequences on histone H3/H4 dynamics in coding region and thereby transcription accuracy.
CK2 may help RNAP II to focus on real promoters and therefore allows optimal cell response to environmental challenges
In cells depleted of CK2 activity, we measured high levels of cryptic sense and antisense transcripts (Figure 2). It is likely that this spurious transcription would have a major impact on bona fide genes transcription. Indeed, an increasing number of studies in various organisms show that non-coding RNA and their transcription directly and indirectly regulate the transcription of genes (15,17,73,74). Our data do not allow us to draw clear conclusions on steady state transcripts levels of genes (Figure 2C). One could expect that transcription from a cryptic promoter driving either sense or antisense transcription would impact and compete with the normal canonical promoter. We assessed this possibility by analyzing the link between changes in the transcription of bona fide genes and the presence of spurious transcription in these genes. Unfortunately, we did not observe a significant correlation between the normal transcript variation and production of spurious transcription (data not shown). Importantly, this is consistent with previous observations made on spt6-1004 mutant cells in which massive spurious transcription has been observed (12,27). In this mutant, no significant correlation between spurious transcription and transcription from canonical promoters has been reported (27). It was proposed that absence of obvious link may be caused by spt6-1004s' drastic effects on chromatin structure including massive loss of nucleosomes (25). Importantly, in our experimental conditions, no such effect has been observed in CK2 depleted cells and histone H3 levels taken as an assessment of nucleosomes occupancy was not changed (data not shown). Thus, in the light of our data, it seems unlikely that mutant with less drastic effect would allow us to uncover or analyze better a potential relationship between spurious and canonical transcription. Hence, the question of the spurious transcription effect on the bona fide gene expression remains unanswered. Two possibilities explaining the transcriptional role of CK2 may be discussed. First, cryptic transcription could have no impact on gene expression. This would be consistent with many observations regarding the phenotypes of mutants that produce spurious transcripts without a significant outcome on transcription from regular promoters. Indeed, mutants of the HCs Spt2 or the methyltransferase Set2 are associated with cryptic transcription that has virtually no impact on their growth fitness or even global gene expression (75,76). However, it is important to note that despite their importance, many chromatin factors mutants have mild effects on global gene expression. Presumably, this is due to homeostatic mechanisms that compensate for the loss of chromatin regulators (66).
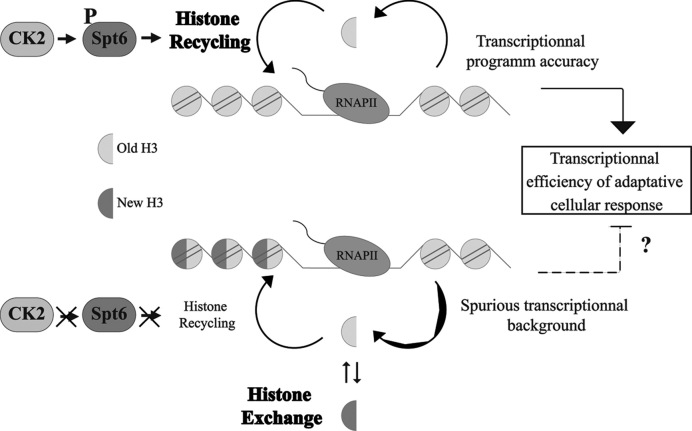
Interestingly, looking at model genes, such as PHO5, suggest a second possibility that may explain the potential transcription role of CK2. Indeed, in steady state standard conditions, PHO5 transcription is not generally affected by the loss of many chromatin factors. This is however not the case when cells are challenged to produce rapidly the Pho5 alkaline phosphatase (77,78). Therefore, in the case of CK2, we reasoned that CK2 repression of spurious transcription may be important for the response to environmental challenges. As shown in our study, all stresses conditions tested in ck2ts mutant led to the observation of a defect in the transcriptional response (Figure 7). Moreover, this was also observed in the Spt6 phosphosite mutant (Figure 7). Our observation is consistent with elegant global studies showing that chromatin factors have only mild effects on steady state transcription, but are severely impaired in transcriptional stress response (66). Without excluding other possibilities, such as a role of CK2/Spt6 phosphorylation in the transcriptional induction itself, a disturbed chromatin dynamics and the induction of spurious transcription may affect transcriptional response. In summary, our study shows a new role of CK2 in the regulation of histone H3/H4 dynamics through the phosphorylation of the HC Spt6. We propose a mechanism (see model Figure 8) where CK2 phosphorylates the Spt6 N-terminal domain and these modifications stabilize Spt6. Consequently, this HC recycles H3/H4 tetramers and nucleosomes are adequately refolded back. Finally, we suggest that CK2 modulates other substrate that collaborate to the inhibition of spurious transcription. Future studies focused on these factors should give us a full picture on how this conserved kinase controls the accuracy of transcription.
Figure 8.
Mechanism of chromatin regulation by CK2. In presence of CK2 activity, Spt6 is phosphorylated and its level is maintained. This allows the proper recycling of histones H3/H4 tetramers and ensures the accuracy of transcription by repressing spurious transcription. Altogether, these functions could lead to the efficiency of the transcriptional response.
DATA AVAILABILITY
The genomic datasets from this study have been deposited in NCBI’s Gene Expression Omnibus (GEO) database under accession number GSE109080.
Supplementary Material
ACKNOWLEDGEMENTS
We thank S. Bilodeau and S. Hussein for the critical review of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
CIHR [PJT-148780]; A. Nourani holds a Canadian Chair research. Funding for open access charge: CIHR.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kornberg R.D. Structure of chromatin. Ann. Rev. Biochem. 1977; 46:931–954. [DOI] [PubMed] [Google Scholar]
- 2. Selth L.A., Sigurdsson S., Svejstrup J.Q.. Transcript elongation by RNA polymerase II. Ann. Rev. Biochem. 2010; 79:271–293. [DOI] [PubMed] [Google Scholar]
- 3. Venkatesh S., Workman J.L.. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 2015; 16:178. [DOI] [PubMed] [Google Scholar]
- 4. Kulaeva O.I., Hsieh F.K., Chang H.W., Luse D.S., Studitsky V.M.. Mechanism of transcription through a nucleosome by RNA polymerase II. Biochim. Biophys. Acta. 2013; 1829:76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rando O.J., Winston F.. Chromatin and transcription in yeast. Genetics. 2012; 190:351–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gaykalova D.A., Kulaeva O.I., Volokh O., Shaytan A.K., Hsieh F.-K., Kirpichnikov M.P., Sokolova O.S., Studitsky V.M.. Structural analysis of nucleosomal barrier to transcription. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E5787–E5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Avvakumov N., Nourani A., Côté J.. Histone chaperones: modulators of chromatin marks. Mol. Cell. 2011; 41:502–514. [DOI] [PubMed] [Google Scholar]
- 8. Gurard-Levin Z.A., Quivy J.-P., Almouzni G.. Histone Chaperones: Assisting histone traffic and nucleosome dynamics. Annu. Rev. Biochem. 2014; 83:487–517. [DOI] [PubMed] [Google Scholar]
- 9. Hammond C.M., Strømme C.B., Huang H., Patel D.J., Groth A.. Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell Biol. 2017; 18:141–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kireeva M.L., Walter W., Tchernajenko V., Bondarenko V., Kashlev M., Studitsky V.M.. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol. Cell. 2002; 9:541–552. [DOI] [PubMed] [Google Scholar]
- 11. Studitsky V.M., Walter W., Kireeva M., Kashlev M., Felsenfeld G.. Chromatin remodeling by RNA polymerases. Trends Biochem. Sci. 2004; 29:127–135. [DOI] [PubMed] [Google Scholar]
- 12. Cheung V., Chua G., Batada N.N., Landry C.R., Michnick S.W., Hughes T.R., Winston F.. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 2008; 6:2550–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. DeGennaro C.M., Alver B.H., Marguerat S., Stepanova E., Davis C.P., Bahler J., Park P.J., Winston F.. Spt6 regulates intragenic and antisense transcription, nucleosome positioning, and histone modifications genome-wide in fission yeast. Mol. Cell. Biol. 2013; 33:4779–4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shetty A., Kallgren S.P., Demel C., Maier K.C., Spatt D., Alver B.H., Cramer P., Park P.J., Winston F.. Spt5 plays vital roles in the control of sense and antisense transcription elongation. Mol. Cell. 2017; 66:77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Camblong J., Beyrouthy N., Guffanti E., Schlaepfer G., Steinmetz L.M., Stutz F.. Trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes Dev. 2009; 23:1534–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gérard A., Ségéral E., Naughtin M., Abdouni A., Charmeteau B., Cheynier R., Rain J.-C., Emiliani S.. The integrase cofactor LEDGF/p75 associates with Iws1 and Spt6 for postintegration silencing of HIV-1 gene expression in latently infected cells. Cell Host Microbe. 2015; 17:107–117. [DOI] [PubMed] [Google Scholar]
- 17. Lenstra T.L., Coulon A., Chow C.C., Larson D.R., Lenstra T.L., Coulon A., Chow C.C., Larson D.R.. Single-Molecule imaging reveals a switch between spurious and functional ncRNA transcription article single-molecule imaging reveals a switch between spurious and functional ncRNA transcription. Mol. Cell. 2015; 60:597–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xie L., Pelz C., Wang W., Bashar A., Varlamova O., Shadle S., Impey S.. KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. EMBO J. 2011; 30:1473–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen S., Rufiange A., Huang H., Rajashankar K.R., Nourani A., Patel D.J.. Structure-function studies of histone H3/H4 tetramer maintenance during transcription by chaperone Spt2. Genes Dev. 2015; 29:1326–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kato H., Okazaki K., Iida T., Nakayama J.-I., Murakami Y., Urano T.. Spt6 prevents transcription-coupled loss of posttranslationally modified histone H3. Scientific Rep. 2013; 3:2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rufiange A., Jacques P.É., Bhat W., Robert F., Nourani A.. Genome-Wide Replication-Independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell. 2007; 27:393–405. [DOI] [PubMed] [Google Scholar]
- 22. Venkatesh S., Smolle M., Li H., Gogol M.M., Saint M., Kumar S., Natarajan K., Workman J.L.. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature. 2012; 489:452–455. [DOI] [PubMed] [Google Scholar]
- 23. Jeronimo C., Watanabe S., Kaplan C.D., Peterson C.L., Robert F.. The histone chaperones FACT and Spt6 restrict H2A.Z from intragenic locations. Mol. Cell. 2015; 58:1113–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bortvin A., Winston F.. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science. 1996; 272:1473–1476. [DOI] [PubMed] [Google Scholar]
- 25. Ivanovska I., Jacques P.-É., Rando O.J., Robert F., Winston F.. Control of chromatin structure by spt6: different consequences in coding and regulatory regions. Mol. Cell. Biol. 2011; 31:531–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kaplan C.D., Laprade L., Winston F.. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003; 301:1096–1099. [DOI] [PubMed] [Google Scholar]
- 27. Uwimana N., Collin P., Jeronimo C., Haibe-Kains B., Robert F.. Bidirectional terminators in Saccharomyces cerevisiae prevent cryptic transcription from invading neighboring genes. Nucleic Acids Res. 2017; 45:6417–6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bhat W., Boutin G., Rufiange A., Nourani A.. Casein kinase 2 associates with the yeast chromatin reassembly factor Spt2/Sin1 to regulate its function in the repression of spurious transcription. Mol. Cell. Biol. 2013; 33:4198–4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meggio F., Pinna L.a. One-thousand-and-one substrates of protein kinase CK2. FASEB J. 2003; 17:349–368. [DOI] [PubMed] [Google Scholar]
- 30. St-Denis N.A., Litchfield D.W.. From birth to death: The role of protein kinase CK2 in the regulation of cell proliferation and survival. Cell. Mol. Life Sci. 2009; 66:1817–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Deplus R., Blanchon L., Rajavelu A., Boukaba A., Defrance M., Luciani J., Rothé F., Dedeurwaerder S., Denis H., Brinkman A.B. et al. . Regulation of DNA methylation patterns by CK2-mediated phosphorylation of Dnmt3a. Cell Rep. 2014; 8:743–753. [DOI] [PubMed] [Google Scholar]
- 32. Wu S.Y., Lee A.Y., Lai H.T., Zhang H., Chiang C.M.. Phospho switch triggers brd4 chromatin binding and activator recruitment for gene-specific targeting. Mol. Cell. 2013; 49:843–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Basnet H., Su X.B., Tan Y., Meisenhelder J., Merkurjev D., Ohgi K.A., Hunter T., Pillus L., Rosenfeld M.G.. Tyrosine phosphorylation of histone H2A by CK2 regulates transcriptional elongation. Nature. 2014; 516:267–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Calvert M.E.K., Keck K.M., Ptak C., Shabanowitz J., Hunt D.F., Pemberton L.F.. Phosphorylation by casein kinase 2 regulates Nap1 localization and function. Mol. Cell. Biol. 2008; 28:1313–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Krogan N.J., Kim M., Ahn S.H., Zhong G., Kobor M.S., Cagney G., Emili A., Shilatifard A., Buratowski S., Greenblatt J.F.. RNA polymerase II elongation factors of saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 2002; 22:6979–6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dastidar E.G., Dayer G., Holland Z.M., Dorin-Semblat D., Claes A., Chene A., Sharma A., Hamelin R., Moniatte M., Lopez-Rubio J.-J. et al. . Involvement of plasmodium falciparum protein kinase CK2 in the chromatin assembly pathway. BMC Biol. 2012; 10:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bouazoune K., Brehm A.. dMi-2 chromatin binding and remodeling activities are regulated by dCK2 phosphorylation. J. Biol. Chem. 2005; 280:41912–41920. [DOI] [PubMed] [Google Scholar]
- 38. Li Y., Keller D.M., Scott J.D., Lu H.. CK2 phosphorylates SSRP1 and inhibits its DNA-binding activity. J. Biol. Chem. 2005; 280:11869–11875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bedard L.G., Dronamraju R., Kerschner J.L., Hunter G.O., Axley E.D., Boyd A.K., Strahl B.D., Mosley A.L.. Quantitative analysis of dynamic protein interactions during transcription reveals a role for casein kinase II in Polymerase-associated Factor (PAF) complex phosphorylation and regulation of histone H2B monoubiquitylation. J. Biol. Chem. 2016; 291:13410–13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gavin A.-C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.-M., Cruciat C.-M. et al. . Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002; 415:141–147. [DOI] [PubMed] [Google Scholar]
- 41. Winston F., Dollard C., Ricupero‐Hovasse S.L.. Construction of a set of convenient saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995; 11:53–55. [DOI] [PubMed] [Google Scholar]
- 42. Longtine M.S., McKenzie A., Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R.. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998; 14:953–961. [DOI] [PubMed] [Google Scholar]
- 43. Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Séraphin B.. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001; 24:218–229. [DOI] [PubMed] [Google Scholar]
- 44. Goldstein A.L., McCusker J.H.. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae - Goldstein - 1999 - Yeast - Wiley Online Library. Yeast. 1999; 15:1541–1553. [DOI] [PubMed] [Google Scholar]
- 45. Kitazono A.A., Tobe B.T.D., Kalton H., Diamant N., Kron S.J.. Marker-fusion PCR for one-step mutagenesis of essential genes in yeast. Yeast. 2002; 19:141–149. [DOI] [PubMed] [Google Scholar]
- 46. Clark-Adams C.D., Winston F.. The SPT6 gene is essential for growth and is required for delta-mediated transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 1987; 7:679–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rose M.D., Novick P., Thomas J.H., Botstein D., Fink G.R.. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987; 60:237–243. [DOI] [PubMed] [Google Scholar]
- 48. Nourani A., Doyon Y., Utley R.T., Lane W.S., Côté J., Allard P.. Role of an ING1 growth regulator in transcriptional activation and targeted histone acetylation by the NuA4 complex role of an ING1 growth regulator in transcriptional activation and targeted histone acetylation by the NuA4 complex. Mol. Cell. Biol. 2001; 21:7629–7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Anders S., Huber W.. Differential expression analysis for sequence count data. Genome Biol. 2010; 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shen L., Shao N., Liu X., Nestler E.. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014; 15:284–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ferrari P., Strubin M.. Uncoupling histone turnover from transcription-associated histone H3 modifications. Nucleic Acids Res. 2015; 43:3972–3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dion M.F., Kaplan T., Kim M., Buratowski S., Friedman N., Rando O.J.. Dynamics of Replication-Independent histone turnover in budding yeast. Science. 2007; 315:1405–1408. [DOI] [PubMed] [Google Scholar]
- 59. Camblong J., Iglesias N., Fickentscher C., Dieppois G., Stutz F.. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007; 131:706–717. [DOI] [PubMed] [Google Scholar]
- 60. van Bakel H., Tsui K., Gebbia M., Mnaimneh S., Hughes T.R., Nislow C.. A compendium of nucleosome and transcript profiles reveals determinants of chromatin architecture and transcription. PLoS Genet. 2013; 9:e1003479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Carrozza M.J., Li B., Florens L., Suganuma T., Swanson S.K., Lee K.K., Shia W.-J., Anderson S., Yates J., Washburn M.P. et al. . Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005; 123:581–592. [DOI] [PubMed] [Google Scholar]
- 62. Yoh S.M., Lucas J.S., Jones K.a. The Iws1: Spt6: CTD complex controls cotranscriptional mRNA The Iws1: Spt6: CTD complex controls cotranscriptional mRNA biosynthesis and HYPB / Setd2-mediated histone H3K36 methylation. Genes Dev. 2008; 3422–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Albuquerque C.P., Smolka M.B., Payne S.H., Bafna V., Eng J., Zhou H.. A multidimensional chromatography technology for In-depth phosphoproteome analysis. Mol. Cell. Proteomics. 2008; 7:1389–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Li X., Gerber S.A., Rudner A.D., Beausoleil S.A., Haas W., Villén J., Elias J.E., Gygi S.P.. Large-Scale phosphorylation analysis of α-Factor-Arrested saccharomyces cerevisiae. J. Proteome Res. 2007; 6:1190–1197. [DOI] [PubMed] [Google Scholar]
- 65. McCullough L., Connell Z., Petersen C., Formosa T.. The abundant histone chaperones Spt6 and FACT collaborate to assemble, inspect, and maintain chromatin structure in saccharomyces cerevisiae. Genetics. 2015; 201:1030–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Weiner A., Chen H.V., Liu C.L., Rahat A., Klien A., Soares L., Gudipati M., Pfeffner J., Regev A., Buratowski S. et al. . Systematic dissection of roles for chromatin regulators in a yeast stress response. PLoS Biol. 2012; 10:e1001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hainer S.J., Gu W., Carone B.R., Landry B.D., Rando O.J., Mello C.C., Fazzio T.G.. Suppression of pervasive noncoding transcription in embryonic stem cells by esBAF. Genes Dev. 2015; 29:362–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Smolle M., Venkatesh S., Gogol M.M., Li H., Zhang Y., Florens L., Washburn M.P., Workman J.L.. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat. Struct. Mol. Biol. 2012; 19:884–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Barz T. Genome-wide expression screens indicate a global role for protein kinase CK2 in chromatin remodeling. J. Cell Sci. 2003; 116:1563–1577. [DOI] [PubMed] [Google Scholar]
- 70. Li B., Gogol M., Carey M., Pattenden S.G., Seidel C., Workman J.L.. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev. 2007; 21:1422–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chu Y., Simic R., Warner M.H., Arndt K.M., Prelich G.. Regulation of histone modification and cryptic transcription by the Bur1 and Paf1 complexes. EMBO J. 2007; 26:4646–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Fleming A.B., Kao C.-F., Hillyer C., Pikaart M., Osley M.A.. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol. Cell. 2008; 31:57–66. [DOI] [PubMed] [Google Scholar]
- 73. Martens J.A., Wu P.Y.J., Winston F.. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 2005; 19:2695–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wilusz J.E., Sunwoo H., Spector D.L.. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009; 23:1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Nourani A., Robert F., Winston F.. Evidence that Spt2/Sin1, an HMG-like factor, plays roles in transcription elongation, chromatin structure, and genome stability in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006; 26:1496–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Venkatesh S., Li H., Gogol M.M., Workman J.L., Workman J.L.. Selective suppression of antisense transcription by Set2-mediated H3K36 methylation. Nat. Commun. 2016; 7:13610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Korber P., Barbaric S., Luckenbach T., Schmid A., Schermer U.J., Blaschke D., Hörz W.. The histone chaperone Asf1 increases the rate of histone eviction at the yeast PHO5 and PHO8 promoters. J. Biol. Chem. 2006; 281:5539–5545. [DOI] [PubMed] [Google Scholar]
- 78. Williams S.K., Truong D., Tyler J.K.. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:9000–9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genomic datasets from this study have been deposited in NCBI’s Gene Expression Omnibus (GEO) database under accession number GSE109080.