Abstract
R-loops are three-stranded RNA:DNA hybrid structures essential for many normal and pathobiological processes. Previously, we generated a quantitative R-loop forming sequence (RLFS) model, quantitative model of R-loop-forming sequences (QmRLFS) and predicted ∼660 000 RLFSs; most of them located in genes and gene-flanking regions, G-rich regions and disease-associated genomic loci in the human genome. Here, we conducted a comprehensive comparative analysis of these RLFSs using experimental data and demonstrated the high performance of QmRLFS predictions on the nucleotide and genome scales. The preferential co-localization of RLFS with promoters, U1 splice sites, gene ends, enhancers and non-B DNA structures, such as G-quadruplexes, provides evidence for the mechanical linkage between DNA tertiary structures, transcription initiation and R-loops in critical regulatory genome regions. We introduced and characterized an abundant class of reverse-forward RLFS clusters highly enriched in non-B DNA structures, which localized to promoters, gene ends and enhancers. The RLFS co-localization with promoters and transcriptionally active enhancers suggested new models for in cis and in trans regulation by RNA:DNA hybrids of transcription initiation and formation of 3D-chromatin loops. Overall, this study provides a rationale for the discovery and characterization of the non-B DNA regulatory structures involved in the formation of the RNA:DNA interactome as the basis for an emerging quantitative R-loop biology and pathobiology.
INTRODUCTION
The R-loop is a co-transcriptionally formed three-stranded hybrid nucleic acid structure, which consists of two components: a nascent RNA transcript segment with its DNA template and a fragment of a displaced non-template DNA in a single-stranded conformation. In vitro, the RNA pairs with one of the two DNA strands in the region of homology to form an R loop in which one element is an RNA:DNA duplex and the other is single-stranded DNA (ssDNA) (1). Such RNA:DNA heteroduplex formation is mostly determined by nucleic acid sequence and has its highest stability in the case of poly-purine RNA binding to a poly-pyrimidine DNA-template (2). In vitro evidence of R-loop formation co-transcriptionally was shown using negatively supercoiled DNA plasmids of Escherichia coli mutants without active topoisomerase I (3,4). The R-loop originates behind the moving RNA polymerase, and the effects of DNA supercoiling, translation, DNA topoisomerases and RNase H on R-loop formation have been elucidated (3–5).
Studies of eukaryotic RNA:DNA hybrids under physiological conditions began on human B-cell heavy chain immunoglobulin switch (Ig S) regions, where R-loops have been detected and cleaved by the nucleotide excision repair nucleases, contributing to the class switch recombination and antibody diversity (6–8). Further studies of the Ig S region sequences established the high importance of a few short guanine clusters (called R-loop initiation zone, RIZ) and a further G-rich DNA segment (called R-loop elongation zone REZ) in the non-template DNA strand as the determinants of the initiation of the R-loop formation and its stabilization in the Ig S regions, respectively (9–15).
The effect of R-loop formation on transcription elongation and genetic stability was first uncovered in yeast with impaired Pol II transcription elongation bearing mutations in a THO/TREX complex (16). R-loops provide a substrate for an activation-induced cytosine deaminase (9), induce instability at the expanded tri- and hexanucleotide repeats (17–21) and cause single- and double-stranded DNA breaks (20–30), transcription interference and DNA replication block (26–31). R-loop formation is associated with neurodegenerative diseases, including amyotrophic lateral sclerosis (20,21), frontotemporal dementia (20,21), non-ketotic hyperglycinemia (32,33), spinocerebellar ataxia type 1 (SCA1), myotonic dystrophy (DM1), and fragile X type A syndrome [(20,21,30,31,34), reviewed in (35–39)]. Proteins that prevent R-loop formation are ASF/SF2 splicing factor (40) and BRCA2 (41), whereas senataxin (42) and capping enzyme (43) resolve and promote R-loop formation, respectively.
While R-loops were first described in 1976 (1) and were for many years associated with only a few specific genes, in recent years, our understanding of their critical function and prevalence in the genomes has advanced, revolutionizing the field. Recently, to detect R-loops, genome-wide experimental approaches using immune-detection of RNA:DNA hybrids (44) and computational prediction structural models have been developed (32,33,45–51). RNA:DNA hybrid immunoprecipitation (44) with S9.6 antibodies using either cDNA or ssDNA can be supplemented by high-throughput sequencing, resulting in DRIP-seq (RNA:DNA immunoprecipitation following high-throughput sequencing) (45–47), strand-specific modification of DRIP-seq method, called DRIPc-seq (RNA:DNA immunoprecipitation followed by RNA purification and conversion to cDNA, coupled to high-throughput sequencing) (47) and RDIP-seq (RNA:DNA immunoprecipitation following high-throughput sequencing) (48). S1-DRIP-seq (S1 nuclease DNA:RNA immunoprecipitation with deep sequencing) has been developed for mapping hybrid-prone regions in budding yeast Saccharomyces cerevisiae (49). Consistent with in silico predictions (32,33,50), DRIP-seq, DRIPc-seq and RDIP-seq sequencing methods identified thousands of RNA:DNA hybrid peak regions non-uniformly distributed in human and mouse genomes and highlighted the associations of R-loops with DNA methylation status, open chromatin and promoter- and enhancer-like chromatin signatures (45–48). Along the yeast genome, multiple ‘R-loop hot-spots’ associated with highly expressed Pol II transcribed genes, ribosomal genes, telomeres and transposons have been reported (49). In yeast, nematodes, and mammalian cells, R-loop formation is also positively correlated with H3S10P, a histone modification associated mainly with mitotic chromatin condensation (51).
However, to date, the structures of DNA regions, which can form R-loops when transcribed, the regions number and localization of the region boundaries at the genome scale and the mechanistic model(s) of R-loop formation remain unresolved. Therefore, computational prediction and systematic characterization of the genomic regions responsible for RNA:DNA hybridization is essential to better understand the features and mechanisms of R-loop formation and might provide insights into their functions under normal cellular conditions, as well as their pathological and evolutionary roles.
To address these questions, we have previously developed a quantitative model of R-loop-forming sequences (QmRLFS) (32,50). This structural model was based on the observations that guanine-rich natural and artificial RNA sequences can form more stable double-stranded hybrids with cytosine-rich ssDNA than reverse-complementary DNA sequences (2,6,12–15). In vivo, such ‘heteromerous’ conformations can be generated by the hybridization of the C-rich DNA template strand with the nascent RNA. In (33,50), using the literature data regarding structure, size and strands of available experimentally defined RLFSs and thermodynamic characteristics of RNA:DNA hybridization, we assumed that linked G-rich RIZ and REZ DNA sequences represented general components of the major R-loop forming sequences (RLFS) in mammalian genomes. We proposed the quantitative model of RLFS (QmRLFS) that has predicted strand-specific chromosome coordinates of the putative RNA:DNA hybrids and R-loops at the gene and genome scales (32,50). In the human genome, the QmRLFS model has predicted R-loops in 664 774 regions (33,50).
The in vitro data based the in silico RLFS model were preferentially predicted in most of the protein-coding genes with a preferred location at the proximal promoter region of the human and other organisms genes (32,50). Consistency between the predicted RLFS genome coordinates and the available in vitro and in vivo RNA:DNA hybridization data sets has ranged between 84 and 91% (32,33,50). We developed a database to predict the RLFSs, their lengths, strands and boundaries. We also mapped the localizations of RLFSs in gene body and in the human genome as well as genes and genomes of several other organisms [(32,33), http://rloop.bii.a-star.edu.sg/?pg2=stats]. Recently, R-loop database (DB) has been updated; in particular, it includes publicly available genome-wide RNA:DNA hybrid/R-loop formation datasets for the humans and mice (32,33,50). This DB provides a unique resource for integrative analyses, design of experiments and finding solutions related to diverse problems of R-loop biology.
Here, we conduct a comprehensive statistical, structural and functional analysis of the RLFSs and its clusters and compare their characteristics with experimental RNA:DNA hybrid/R-loop datasets at the gene and genome scales in the human genome.
MATERIALS AND METHODS
Annotation of genes and RLFSs
In our study, we used GENCODE release 24 human gene and transcript annotations mapped to the NCBI build GRCh37 primary assembly (hg19) (52). In several analyses carried out by QmRLFS (http://rloop.bii.a-star.edu.sg/?pg=qmrlfs-finder) (50) and R-loop DB (http://rloop.bii.a-star.edu.sg/) (33), we specified and used the human assembly GRCh37 in the Ensembl Release 75 gene annotation system (https://www.ensembl.org/info/data/mysql.html). The GENCODE gene set presents a full merge between HAVANA manual annotation and Ensembl automatic annotation. For studied genes and gene biotypes, both systems provided very similar annotations and statistics. We referred to the annotation in the contexts of the analysis (see also Supplementary Materials: Annotation of genes, gene type and regions).
DRIP-seq, DRIPc-seq, RDIP-seq, CpG islands, RLFS, SkewR and G4-quadruplex data
Raw datasets from DNA:RNA immune-precipitation followed by sequencing (DRIP-seq) experiments (45,46) performed with human pluripotent NT2 (or Ntra2) cells were downloaded from the NCBI Sequence Read Archive (https://trace.ddbj.nig.ac.jp/DRASearch) under the accession numbers SRR393964 and SRP020088, respectively. In the DRIP-seq data of one study (45), FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) found an unexpectedly high fluctuation of base proportions at various read positions and a modest proportion (∼20%) of mapped reads to the genome (2 M uniquely mapped reads from 12 M reads in total), indicating a low-quality of sequence data. Due to these and other facts (Supplementary Materials: DRIP-based datasets, their qualitative and quantitative characteristics; Supplementary Tables S1 and 2), we excluded the DRIP-seq dataset (45) from our further analysis. The raw sequence data in (46) followed the quality control criteria, and after our mapping of the significant DRIP-seq peak regions, we used the processed data in our further analyze (Supplementary Materials: DRIP-based datasets, their qualitative and quantitative characteristics; Supplementary Tables S1 and 2).
DRIP-seq data for NT2 (47) and K562 cells (47), DRIPc-seq data for NT2 cells (47) and RDIP-seq data for IMR90 and HEK293T cells (48) were downloaded from the NCBI Gene Expression Omnibus (GEO) data repository and processed for our analysis (see Supplementary Materials: DRIP-based data sets, their qualitative and quantitative characteristics, Supplementary Table S1). Details are given in Supplementary Materials: Identification of the genes, transcription start site (TSS)-proximal and transcription end site (TES)-proximal regions; CpG islands, RLFS, SkewR, and G4-quadruplex data.
CAGE-seq data analysis
Cap analysis of gene expression (CAGE)-seq data in .ctss file format (mapped, un-normalized data in a single base-pair resolution format) for 32 cell lines and cell types (whole cell fraction) was retrieved from the FANTOM5 data repository using CAGEr R package (53) (Supplementary Materials: CAGE-seq data analysis; CAGE clusters in unidirectional and divergent gene promoters). The numbers of identified CAGE-clusters for all cell types used in the study are given in Supplementary Tables S3 and 4.
U1 snRNP sites and putative polyadenylation signal (PAS) sequence identification
To analyze genome-wide associations of RLFSs with pre-mRNA splicing events, including intronic cleavage and polyadenylation regulating gene expression, we carried out a motif search for U1 snRNP splice sites (or U1 sites) and putative polyadenylation (poly-A) signals (PAS) (AATAAA/ATTAAA) in the Find Individual Motif Occurrences (FIMO) software from MEME-Suite toolkit (54). The details of the analysis and datasets supporting this analysis are presented in the ‘Materials and Methods’ section and Supplementary Materials: U1 snRNP sites and PASs identification) and Supplementary Table S10.
Enhancer data
Cell-type-specific expression data of enhancers and their coordinates (‘robust’ set, N = 32 693) were obtained from the Human Transcribed Enhancer Atlas [(55), http://enhancer.binf.ku.dk/enhancers.php]. Due to a very narrow width of enhancer regions (293 bp on average), an additional 500 bases were added for calculation of the numbers of intersecting RLFSs.
Statistical methods and software
R programming language version 3.2.3 was mainly used for data analysis. Gene ontology enrichment was performed with the topGO R package (version 2.22.0) (56). Custom scripts were written in R and bash. Bedtools v2.17.0 software was used to intersect genomic intervals and peak regions. Cytel Studio-9 software (Cytel Software Corporation) and Statistica 7.1 (StatSoft, Inc.) were used for statistical testing. SigmaPlot-11 software (CyStat Software, Inc.) was used for data analysis and the graphical presentation of our results.
RESULTS
R-loop formation structure distributions in the human genome, genes and gene regions
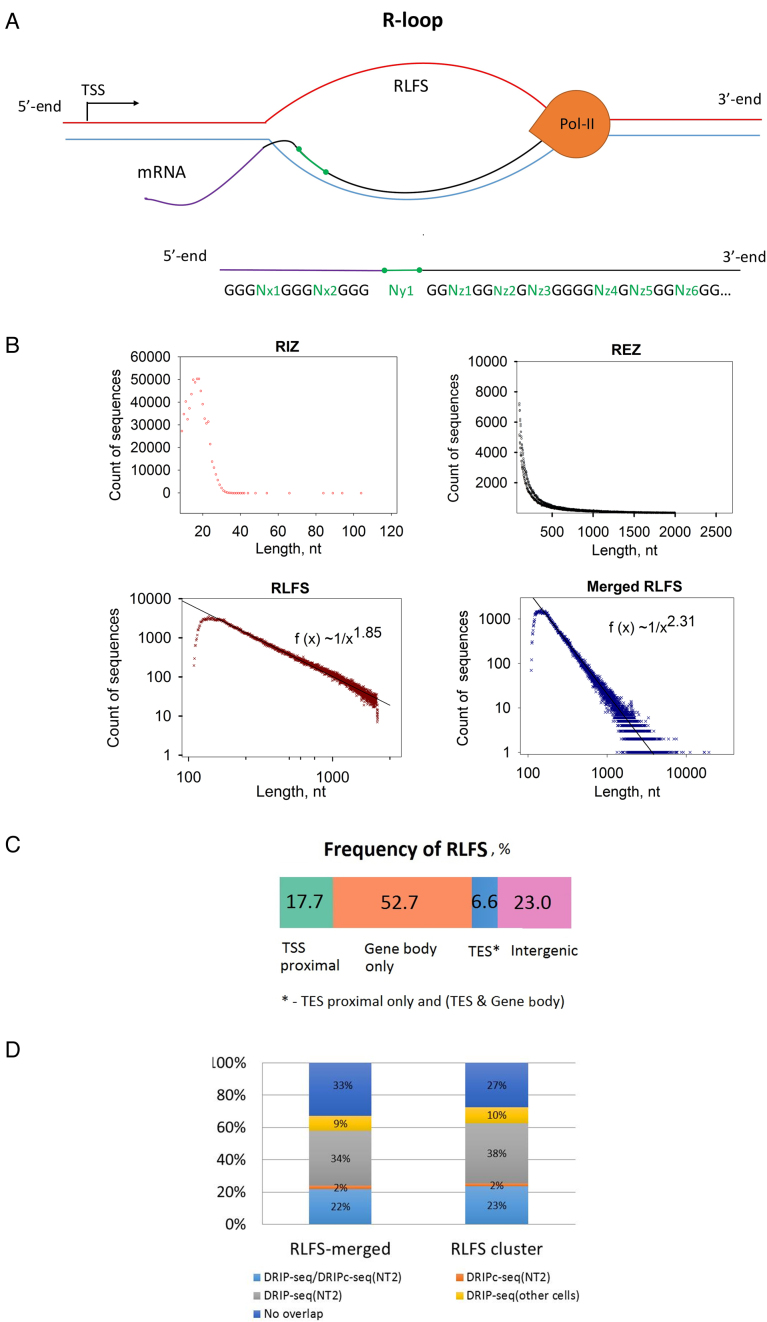
Figure 1A shows a schema of a transcriptional R-loop (top panel) and a structure of the RLFS, including the RIZ, linker and RIZ sequences (bottom panel), quantified by QmRLFS (32,50). In the human genome, QmRLFS-finder predicted a strand-specific localization of 664,791 RLFSs. In the human genome and other genomes, RLFSs are often organized in the overlapped or grouped RLFSs (32,33,50), which often overlapped with or embedded to CpG islands and G-rich regions (32,38,57). If the RLFSs on the same stand are overlapped by at least one nucleotide, we call such a sequence subset an RLFS cluster (32,33,50). The RLFS clusters formed unique sequences. When no further RLFSs overlapped the long merged sequence, this sequence, called a merged RLFS region, had maximum length and was mapped and counted.
Figure 1.
Statistical distributions and characteristics of the RLFS and RLFS's structures. (A) Structural model of RLFS: short G-cluster-rich region thought to be responsible for the initiation of R-loop formation (RIZ), structurally non-specified short linker (linker) and linker downstream of long high-/moderate-G-density region (called R-loop elongation zone or REZ). REZ could provide for RNA:DNA hybrid/R-loop stabilization. For detailed quantitative characteristics of the QmRLFS model, see (33,50). (B) The length frequency distribution of the RIZ, REZ, RLFS and merged RLFSs. Power law-like function tails on the right of the RLFS and merged RLFS length distributions fit data well (goodness of fit linear regression; P < 0.001). (C) The distribution (%) of RLFSs in ‘gene body’, ‘TSS-proximal’, ‘TES-proximal’ and ‘intergenic’ genome regions (Supplementary Materials: Identification of the genes, TSS-proximal and TES-proximal regions). (D) Merged RLFS and clustered RLFS regions overlapped with DRIP-Seq and RDIP-Seq peak regions defined in (45–48) in promoters (−1 kb; +2 kb from the TSS), TES (+2 kb; −1 kb from the annotated TES), gene bodies (excluding 2 kb from 5′ and 3′ gene ends) and outside of annotated genes (with 2 kb added to 5′ and 3′ gene ends). All genes longer than 4.5 kb were considered (N = 17 889 genes).
QmRLFS consolidated the RLFSs onto 229 816 non-overlapped RLFS-merged regions (33). By our estimates, ∼2.33% of the chromosome length in the 3326 Mbp human genome is covered by RLFS-merged G-rich regions. R-loop DB provides the Ensembl-based annotation of RLFS-positive annotated genes and their proximal regulatory regions. This database includes 64 102 genes mapped on the human chromosomes: ∼76% (511 651/664 791) RLFSs are co-localized within gene body regions, including 2 kb upstream and 2 kb downstream gene-flanking regions (33). Approximately 4.4% of the total genes’ spans includes the merged RLFS regions. An RLFS that is localized in any genic region(s) is considered a gene-associated RLFS. We observed that in most gene-associated RLFS regions, the RLFSs are organized in overlapping regions, forming in total 169 222 merged RLFS regions. The total number of merged (and clustered) RLFS regions are similar between the positive and negative strands of the chromosomes (Supplementary Table S6). However, within gene regions and the ±2 kb gene proximity regions, the merged RLFS regions and the RLFS clusters are preferentially localized in the gene sense (non-template) strand of double-helical DNA, with respective frequencies 0.621 and 0.628, suggesting their evolution and functional involvement.
Figure 1B shows the sequence length frequency distribution functions of the RIZ, REZ, RLFS and merged RLFS regions. All these experimental functions are unimodal and skewed to the right. However, the functions and their parameters are different. For instance, the most frequent (mode) sequence length of RIZ, RLFS and merged RLFS regions is 18, 139 and 148 nt, respectively (Supplementary Table S6). The QmRLFS-defined minimal and maximal length of RLFS (109 and 2079 nt) and merged RLFS regions (110 and 19 366 nt) provides a genome-wide mapping of almost all expected RLFSs. Figure 1B shows that ∼95% of the human genome RLFSs are included in the QmRLFS-predicted sequences. Our estimates consist of the length data observed in vitro and in vivo case studies for stable RNA:DNA hybrid/R-loop lengths (10–13,15,18,57). We characterize the underlying distribution functions of the sequence length of the RIZ, REZ, RLFS and merged RLFS regions. Using working definitions of the genome regions of the TSS-proximal, gene body and TES-proximal regions (Supplementary Materials: Identification of the genes, TSS-proximal and TES-proximal regions), we found that in the human genome, 75.4% of RLFSs are assigned to these three types of RLFS-positive regions, providing 17.7, 52.7 and 6.6% of the RLFSs, accordingly (Figure 1C and Supplementary Table S7A). Additionally, our comparative analysis of the number of RLFSs in the transcripts suggests the higher transcriptional activity associated with the long RLFSs, including (i) downstream TSS, gene body and upstream TES, (ii) downstream TSS and gene body, and (iii) gene body and upstream TES (Supplementary Table S7B). However, transcriptional activity was negatively associated with RLFSs belonging to the gene body only (Supplementary Table S7B).
We found that RLFSs are significantly overlapped with the peak regions defined in genome-wide RNA:DNA hybrid/R-loop detections (46–48) (Figure 1D; Supplementary Figures S1D and 2). To characterize these associations in the TTS-proximal, TES-proximal and gene body regions (Supplementary Materials: Identification of the genes, TSS-proximal and TES-proximal regions), we used protein-coding genes longer than 2 kb, which corresponded to 93.1% of all annotated protein-coding genes (Supplementary Figure S1A). In the regions of the genes annotated by GENCODE, we found that the sequence length frequency functions of the significant peak regions (SPRs) exhibit a right-skewed unimodal form and overlap with the RLFS regions (Supplementary Figure S1). Assessing the significance of the overlap between peak regions of the experimental datasets from (45–48) and RLFSs by the sequence reshuffling test (Supplementary Materials: Test of non-randomness of RLFS allocation) (Figure 1D and Supplementary Figure S3) demonstrated that many thousands of SPRs were RLFS-positive in the gene-associated and intergenic regions. In the promoter regions, the percentage of RLFS-positive peak regions was most reproducible (varied between 74 and 92%) (Supplementary Figure S3B). Supplementary Figure S3C and D demonstrates a certain similarity of the general patterns of RLFS, DRIP-seq and DRIPc-seq distributions for NT2 cells in the vicinity of the TSS and TES regions. However, certain dissimilarities between RLFS, DRIP-seq and DRIPc-seq data were also observed.
In total, the DRIP-seq and DRIPc-seq experimental data (peak region length) collectively overlapped with 67% (70 344/105 127) of merged RLFS regions and 73% (41 267/56 884) of cluster RLFS regions, predicted on the gene strand within the gene body regions and the ±2 kb TTS and TES proximal flanking regions.
Supplementary Figure S4 shows the experimental datasets in a chromosome 19 region, including 10 genes and their transcribed forms. This is a representative locus demonstrating differences and similarities between the peak regions defined by DRIP-seq, DRIPc-seq and RDIP-seq in the studied cell lines (nine datasets). Supplementary Figure S4 also includes two RNA-seq datasets for NT2 cells and RPLS regions. This figure shows modest similarity and remarkable variations between publications (45–48). In particular, RDIP-seq peak regions (48) were much shorter (and less intensive) than the peak regions defined by DRIP-seq, DRIPc-seq (47). In contrast, a vast majority RLFS regions overlapped with or were embedded in one or more SPRs across the experiments. These and our multiple other findings suggest the inter-publication data variations. These findings also suggest that the RLFSs could be used for unbiased verification of the experimentally defined RNA:DNA hybrid/R-loop regions within and between datasets and suggest a potential for discovering new RLFSs, RNA:DNA hybrids/R-loops. In the next sections, we will carry out detailed comparative analysis of the RLFS predictions with RNA:DNA hybrid/R-loop experimental data.
Identification of variations and leading technical factors affecting similarity and reproducibility of the genome-wide RNA:DNA hybrid/R-loop datasets
Recently, two research groups have developed immunoprecipitation (IP)-based RNA:DNA hybrid detection protocols and provided genome-wide data indicative of R-loops in the human genome (45–48). The datasets were obtained from four cell lines (NTERA2 (NT2), K562, IMR90 and HEK293T) using DRIP-seq (45) (NT2), improved DRIP-seq (NT2, K562, (46,47)), DRIPc-seq (NT2, (46)) and RDIP-seq (IMR90, and HEK293T, (48)), distinguished by their protocols and peak calling results. All these methods rely on the high affinity of the S9.6 monoclonal antibody to the RNA:DNA hybrid-specific immune-precipitation (44). However, the studies (45–48) differed in sample preparation, immunoprecipitation treatment procedures, the specific sequencing library construction protocols and the library sizes. For instance, Supplementary Figures S1–S3A show significant variations of the total number of SPRs mapped on the human reference genome over the sequence library sizes in (45) versus (46) versus (47) versus (48). In (45), DRIP-seq raw data were of low quality (Supplementary Figure S1), and the peak regions and the peak region intensity values were poorly reproducible in other DRIP-based libraries (46,47). Furthermore, in comparison to data reported in (45), the number of re-analyzed SPRs (Supplementary Figure S1) was much smaller than in other DRIP-based libraries (46,47) (‘Materials and Methods’ section; Supplementary Materials: DRIP-seq based datasets, their qualitative and quantitative characteristics). We have excluded the DRIP-seq dataset (45) from our further analysis.
The re-analyzed sequence dataset quality and the alignment results reported in (46) were acceptable for the peak calling analysis (Supplementary Figure S2). However, we found that the total number of SPRs in the dataset from (46) (n = 4181 peak regions) was ∼10–20 times smaller than that from other datasets (47) (Supplementary Figure S3A). In comparison with (47), the peak region signal intensity values were also systematically smaller. These facts can be explained by the relatively low sensitivity of the datasets in (46) in comparison to other DRIP-seq datasets.
Regardless of the reported consistency between datasets for distinct cell lines (NT2 and K562 in (47) or IMR90 and HEK293T in (48)), we observed significant variations between datasets (47,48)), as well as essential differences in the experimental methods, developed in (47) and (48) (Supplementary Figures S1B–3 and S5A). For instance, in contrast to RLFS and DRIP-seq and DRIPc-seq data, the RDIP-seq libraries derived from two very different cell types (IMR90, HEK293) are represented by similar and considerably low total number of the SPRs. The frequency distributions of the peaks occurred at the proximity of TSS and TES were also similar to each other (Supplementary Figure S5). However, they were remarkably differed from the other datasets. Next, the RNA:DNA hybrid/R-loop region boundaries defined by RDIP-seq (48) were much closer to the RLFS peak region boundaries predicted by QmRLFS and GC skew score (Supplementary Figures S1, 6 and Table S8). The RLFS allocation relative to the genome-wide detected datasets by sequence reshuffling analysis provided quantified support for these findings (Supplementary Figure S2). On the other hand, the DRIP-seq and DRIPc-seq SPR intensity on average were much higher and more frequent in the critical gene regions (TSS, TES) compared to the RDIP-seq SPRs, indicating the higher sensitivity of DRIP-seq and DRIPc-seq. Additionally, DRIPc-seq peak region signals were strand-specific, and this property was significantly concordant with the RLFS coordinates. Detailed analysis of genome-wide methods’ biases and associated data variations are presented in Supplementary Figures S1–7 and Table S8, and Supplementary Materials: Data variation analysis; Mapping variations affect gene and genome scale peak region distributions and sequencing technology biases.
Across the studied datasets, only 369 peak regions were commonly identified (called ‘reproducible peak regions’, RPR, Supplementary Table S9). This number corresponded to <10% of the peak regions reported by each study. The 289 genes were associated with the RPRs. A significant fraction of the RPRs were relatively long and overlapped, and they belonged to the TSS-TES proximal regions, TSS-gene body and TES-gene body regions (Supplementary Table S9B and Figure S7A). Of the reproducible peak regions, 30.6 and 10.8% RPRs were in TSS-proximal and TES-proximal regions, respectively (Supplementary Figure S7A), and 38.8 and 19.8% were in gene body and intergenic regions, respectively. The TSS-proximal and TES-proximal gene regions were more highly enriched by RLFSs in comparison to all gene bodies (Figure 1C), and the RPR lengths were similar to the lengths of the RDIP-seq peak region and the RLFS regions (Supplementary Table S8). The RPR genes were involved in regulation of transcription, from Pol II binding, protein binding, RNA metabolism, transcriptional and translational regulation, RNA splicing, cellular response to DNA damage stimulus, nucleolus, and nuclear lumen localization, and several other biological processes, molecular functions and cellular compartments (Supplementary Figure S7B and Table S9C–E). These characteristics are in agreement with the RNA:DNA hybrid/R-loop computational prediction models and previous experimental results (32–33,45–48,50).
Comparison of the performance of genome-wide experimental and computational methods
In vivo R-loop detection methods, such as non-denaturing bisulfite footprinting and DRIP-qPCR, can provide the most direct and precise evidence of R-loop formation by capturing ssDNA and RNA:DNA hybrid components of the R-loop structure, respectively (9,45). Here, we used publicly available experimentally defined (non-genome-wide) RNA:DNA hybrid/R-loop mapping data (32,33,35,38,50) as a reference to compare the performance of the genome-wide experimental data and computational predictions.
Supplementary Table S10 summarizes the collected the reference R-loop datasets and our results. To account for various factors that influence R-loop formation, we combined the information from both in vitro and in vivo studies. In addition, we performed DRIP-qPCR experiments to examine the FOXO1 gene QmRLFS-predicted R-loops in SKOV3 ovarian and MDA-MB-436 breast cancer cells (Supplementary Figure S8, Supplementary Materials: DNA-RNA immunoprecipitation assay; PCR analysis from the DRIP assay) and included the DRIP-qPCR mapping results in the reference dataset. We identified the boundaries and lengths of the promoter, gene body and TES regions (Supplementary Figure S9A) and estimated the specificity, sensitivity and accuracy of the DRIP-seq datasets for NT2 cells and RLFS models (Supplementary Figure S9B; Supplementary Materials: Sensitivity, specificity, accuracy and balance accuracy). Comparison of the overlapped DRIP-seq (46,47), DRIP-seq and the DRIPc-seq peak regions (47), and the QmRLFS prediction regions (50) with the reference regions showed 90, 77.8, 89 and 89% specificity, respectively (Supplementary Figure S9B; Supplementary Materials: Evaluation of performance of computational R-loop prediction methods and experimental data). The sensitivity (which determined by the library sizes) was 45% for the DRIP-seq in (46,47), 84% for DRIP-seq in (47) and 72% for DRIPc-seq in (47). For QmRLFS, the estimated sensitivity was 72%. The overall accuracy was 58% for DRIP-seq (46,47), 76.5% for DRIP-seq (47) and 82.4% for DRIPc-seq (47) datasets (Supplementary Figure S9B). The accuracy of QmRLFS (50) was 76.5%. The Kendal tau correlation coefficients between the binary scores of the 34 reference and studied datasets were significant, with comparable correlation coefficient values of r = 0.41 for DRIP-seq in (46), r = 0.54 for DRIPc-seq in (47), r = 0.58 for DRIP-seq in (47) and r = 0.54 for RLFS (all at P < 0.001). These results demonstrate similar performance of DRIP-seq, DRIPc-seq (47) and QmRLFS.
Supplementary Figure S6A shows that the lengths of a significant fraction of the DRIP-seq and DRIPc-seq SPRs are considerably longer than the lengths of the RDIP-seq data and QmRLFS predicted regions. Supplementary Figure S6B shows that in contrast to merged and clustered RLFSs, the experimentally defined SPRs lack the GC-skewed pattern significance vs random sequence subset control. These results are reproduced within the promoter, gene body, gene end and nearest neighbor terminal-promoter regions (Supplementary Figure S6C and D).
Supplementary Figure S6E shows the comparative analysis of the experimental and predicted profiles of genome-wide IP-based and computational results in the FMR1-related region as an example of the method's performance. An R-loop region has been previously defined at the end of the first exon end and start of the first intron of this gene (18). The R-loop is associated with a CpG island (Supplementary Figure S6E). Supplementary Figure S6E shows the differences and similarities between the localizations as well as the frequency distribution profiles of the peak regions defined by DRIP-seq, DRIPc-seq and RLFS in the FMR1 gene body and its downstream proximal promoter region. In contrast to DRIPc-seq, QmRLFS (i) accurately identifies the boundaries of the experimentally defined R-loop region (18) and the DRIP-seq SPR (47) in the first exon–first intron region but (ii) does not identify any other RLFSs within the FMR1 locus, which were detected by DRIP-seq and DRIPc-seq (long regions with false-positive signals).
Note that in total and across the gene expression groups, the balance accuracy was the highest for QmRLFS (74%) and was 60% for SkewR and 71.1% for CpG island index (CGI) (Supplementary Figure S10). A detailed analysis of the performance and comparisons of the CGI, SkewR and QmRLFS methods for the genes, the gene expression levels and the proximal gene regions are presented in Supplementary Data: Evaluation and comparing the performance of CGI, SkewR and QmRLFS methods.
RLFSs constitute a substantial fraction of the transcribed human genome, being a discriminative feature between protein-coding genes, non-coding RNA genes and pseudogenes
A major fraction of gene types in annotation systems are represented by the genes encoding for proteins, non-coding RNA genes and pseudogenes (52,58). Our identification of RLFS loci in all 20 014 GENCODE-annotated protein-coding genes, antisense long non-coding transcribed loci (which are often associated with an antisense direction of protein-coding gene promoters) (n = 5564), lincRNAs (n = 7674) and pseudogenes (n = 14 501), including ±2 kb genic proximal flanking regions, showed that 79, 68, 58 and 28% of them were RLFS-positive, respectively (Supplementary Figure S11). We also analyzed the pseudogenes that can contained introns, which are thought to be produced by gene duplication (unprocessed pseudogenes), and the pseudogenes that lacked introns, which are thought to arise from reverse transcription of mRNA followed by reinsertion of the cDNA into the genome (processed pseudogenes). Our analysis of these types of pseudogenes revealed that the unprocessed (n = 2612) and processed pseudogenes (n = 10 283) were distinguished by RLFS: 36 and 24% of them, respectively, were RLFS-positive unprocessed and processed pseudogenes, respectively. Furthermore, we found high enrichment of RLFS in the transcribed pseudogene loci: 74% of transcribed unprocessed pseudogenes, but only 48% of processed pseudogenes, were RLFS-positive.
Considering only promoter regions, we then asked what was the frequency distribution of RLFS occurrences in TSS proximal of different types of genes, as these genes are subject to different transcriptional regulatory programs and differ in GC content (46) and, potentially, R-loop formation (32,33,50). Supplementary Figure S11A shows the essential variation of the percentage of the RLFS-positive in the (−500/+1000) bp interval around a TSS for the genes of different categories: protein-coding, lincRNA, antisense genes and pseudogenes. Supplementary Figure S11B shows the distributions of RLFSs, CGI and GC-content in the promoter regions of the protein-codding, lincRNA, antisense genes and pseudogenes. Only the GENCODE genes longer than 3 kb are shown in Supplementary Figure S11.
Together, these results demonstrate that the RLFSs and associated RNA-DNA hybrids and R-loops are common transcriptional regulation genome components whose frequency decreased in the order protein-coding genes→antisense lncRNA→lincRNA→ pseudogenes, suggesting key roles of the RLFSs in primate genome regulation and evolution.
RLFSs and their cluster allocations correlate with CAGE clusters and transcription initiation rate and predict transcriptional directionality
To further evaluate the quantitative associations of RLFSs with transcription initiation loci and their expression levels, we used CAGE sequencing data (53) (see ‘Materials and Methods’ section and Supplementary Materials: CAGE-seq data analysis, and Supplementary Table S3). The results showed that RLFSs are co-localized with 24–46% of all cellular TSSs defined by CAGE signals (called CAGE clusters, see ‘Materials and Methods’ section and Supplementary Materials: CAGE-seq data analysis; CAGE clusters in unidirectional and divergent gene promoters) across many cell types (Supplementary Table S3). In all datasets, the intensity of RLFS-positive CAGE cluster signals was higher than those of RLFS-negative CAGE cluster signals (Supplementary Table S3).
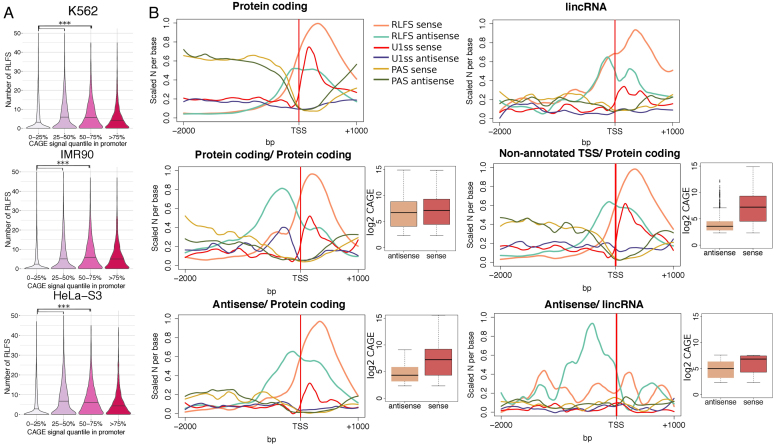
Furthermore, considering CAGE-based TSSs located in annotated gene promoters, 73–82% of these TSSs were closely associated with RLFSs (Supplementary Table S4). The number of RLFSs per promoter correlated with CAGE signal intensity (Figure 2). RLFSs were significantly predominant in promoters of moderately and highly expressed genes compared to lower-expressed genes (0–25% quantile range) (Figure 2A, P < 1.0e-10), suggesting a role of the TSS-proximal RLFS clusters (and merged RLFS regions) in the activation and switching-on of gene functions. We observed that in uni-directionally and bi-directionally transcribed gene promoters, RLFS provides the strand-specific prediction of R-loop formation (Supporting Materials: CAGE clusters in unidirectional and divergent promoters; Figure 2B). Merged RLFSs were preferentially located on the sense DNA strand, with a maximum RLFS density at ∼300–350 nt downstream of the TSSs of stand-alone protein-coding, lncRNA and non-annotated genes (Figure 2B). These results are consistent with the experimental studies of RNA:DNA hybrids/R-loops regardless different cell types (46,59,60).
Figure 2.
RLFS boundaries correlate with TSSs and transcription directionality. (A) Distributions of the numbers of RLFSs at the proximity of promoter regions. To define unidirectional promoters, −500, +1 kb regions of annotated gene TSSs without intersecting TSSs on the opposite strand were considered (N = 52 900), CAGE clusters were defined as described in the ‘Materials and Methods’ section (CAGE-Seq data analysis); promoters were divided into four classes by the CAGE clusters signal intensity (0–25, 25–50, 50–75, 75–100 percentiles). For each cell line, the promoters with a single CAGE cluster were selected, and the numbers of overlapping RLFSs per promoter region were calculated. The black line on violin plot denotes median of the distribution; RLFSs were significantly enriched in promoters of moderately expressed genes (50–75% of CAGE signal intensity) compared to low (0–25%) and low-moderately expressed (25–50%) (P-value < 2.2e-16 by one-sided Wilcoxon rank sum test). (B) RLFS, U1 and PAS motif distributions on the sense and antisense DNA strands in promoters of stand-alone protein-coding genes (N = 4793), lincRNAs (N = 194) and divergent gene pairs: protein-coding/protein-coding (N = 522), protein-coding/antisense transcripts (N = 204), protein-coding/non-annotated transcripts (overlapping with a CAGE cluster on the antisense strand, N = 954) and lincRNA/antisense transcripts (N = 36). Promoters were classified as described in the ‘Materials and Methods’ section (defining unidirectional and divergent gene promoters). The sequence/signal count densities were scaled per maximum number considering sequences/signals from both sense and antisense strands. Red and brown box plots illustrate sequence/signal distributions of the total number of CAGE clusters on the sense and antisense strands downstream (1 kb) and upstream (2 kb) of the annotated TSS, respectively.
In protein-coding/protein-coding divergent gene pairs, RLFSs were almost equally and symmetrically distributed around TSS regions on positive and negative DNA strands. However, on the negative strand, the maximum value of the RLFS density was located ∼300 nt relative to the TTS, while on the positive strand, the maximum value was located ∼250 nt relative to the TTS. In the protein-coding/non-coding RNA divergent gene pairs, the RLFS density functions on the negative and positive strands have different shapes and span different regions (Figure 2B).
In protein-coding genes, these regions formed distinct density patterns: a unimodal frequency distribution with an almost symmetrical shape with a maximum value at a distance of ∼300 nt relatively to TTS. For antisense gene pairs, the frequency function was bimodal, with two maximum values: at 260–280 nt downstream of the TSS of the coding gene and at 200 nt upstream of the protein-coding gene TSS. Furthermore, the distribution changed across the gene types and was associated with their expression level (Figure 2B). We also observed significant differences between the RLFS densities at the TSS and gene expression patterns in non-annotated TSS versus protein-coding regions and antisense-lncRNA pairs (Figure 2B). Notably, only in the protein-coding gene pairs was the expression of sense and antisense genes relatively high, and there were not many differences in their averages across the genes.
The specification of transcription orientation can be controlled by multiple factors, one of which might be R-loops. Indeed, according to the QmRLFS model (Figure 1A), the order of RIZ and REZ sequences of RLFS determined the transcriptional directionality. Furthermore, the RLFS in proximal TSS or/and TES form the RLFS clusters with the same directionality, alone or in combination with DRIPc-seq peak region signals (47), provided even stronger transcriptional directionality.
G-rich U1 sites and U1-PAS signals axis patterns associated RLFS
The observed associations of the RLFS, RNA–DNA hybrids and R-loops with check-points of RNA-pol transcription sites suggested an involvement of these nucleic acid structures in splicing, elongation and isoform diversity of the multi-exon genes (32). Our R-loop DB data analysis showed a high frequency of the co-occurrences of the exon-intron junction regions with the RLFS, RNA:DNA hybrids/R-loop regions, suggesting that such structures play a role in the regulation of U1 sites and polyadenylation signals (PASs). We observed that the frequency distribution of guanine occurrences in the distinct 10-nt U1 site sequences found on 5′end downstream of the genes is skewed, and guanine-rich U1 sites are highly abundant (Supplementary Figure S12 and Table S11). Some 75% (212/283) of distinct 10-nt U1 site sequences included four to seven guanines; such splicing donor sites represent 76.6% (1 219 490/1 592 914) of U1 sites at the human genome scale. ANOVA (Statistica 7) showed high confidence (P < 0.000001) and a positive correlation (r = 0.374) between the number of guanines in the distinct U1 site sequences and the number of occurrences of the U1 sites in the human genome (Supplementary Figure S12). The G-rich U1 donor sites (with 4–7 guanines) tended to be more abundant than best matched U1 donor sites (Supplementary Table S11B). For instance, the most G-rich U1 donor site (GGAG|GTGAGG) was found in 36 510 gene regions, while the perfect matched U1 site (CCAG|GTAAGT) was found only in 5043 gene regions. These findings suggest that a large fraction of U1 sites are G rich (and strongly C poor). In TTS proximal downstream regions (mostly in the firset exon–first intron junctions), the association between U1 sites and RLFS regions was most notable. Supplementary Figure S8A shows DRIP-PCR results supporting this point. It shows the QmRLFS-predicted DNA segments and the DRIP-PCR-detected RNA:DNA hybrids/R-loops that included the U1 splice site of the FOXO1 gene.
The putative PASs at the 5′ ends are U1 site-associated asymmetric sequence determinants, forming a U1-PAS axis, involved in promoter-proximal mRNA cleavage and polyadenylation (61). In general, on the sense (non-template) strand, the frequency of distributions of the RLFS and PAS at the 5′ ends had distinct sequence structures, and their sites were mutually exclusive. We observed such patterns for divergent protein-coding gene pairs and different types of divergent genes, including lincRNAs (Figure 3B). However, such patterns were not significant on the antisense strand. Consistent with CAGE data, Figure 3 shows that inclusion of U1 sites in RLFS regions and exclusion of PAS is a common genome architecture hosting RLFSs. This allows for us to predict the transcript initiation and diversity regions, transcriptional direction(s) and alternative isoforms.
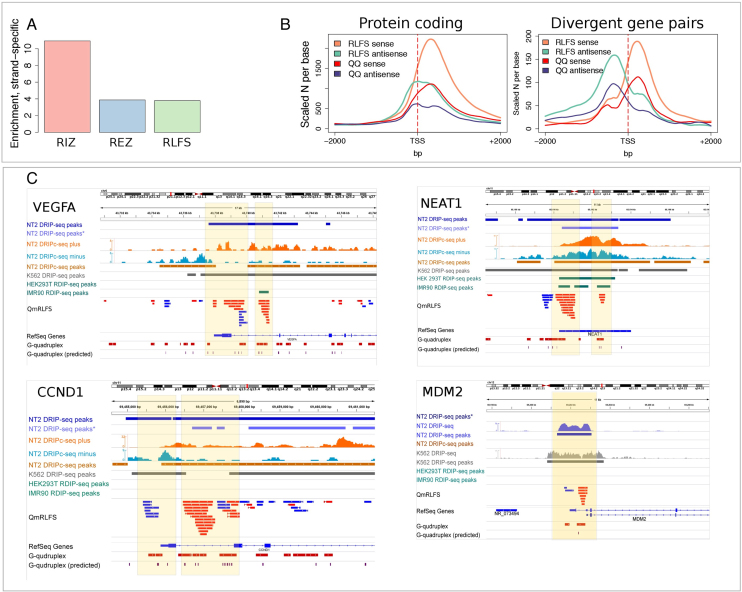
Figure 3.
RLFSs are co-enriched with experimental G4 sequences genome-wide. (A) Enrichment of experimental G4 structures on the sense RLFS strand. Enrichment was calculated as a ratio of number of the G4-positive merged RLFS sequences (RIZ, REZ or entire RLFS with at least one G4 on the sense strand) to the number of the G4-positive merged RLFS sequences (RIZ, REZ or entire RLFS with at least one G4, respectively) found on the same genome double strand position on the antisense strand. The strand orientation was defined by RLFS strand. (B) Distributions of RLFSs and experimental G4s in the proximal promoters (around TSS) of stand-alone protein-coding genes and the protein-coding/protein-coding divergent gene pairs. RLFSs were merged in a strand-specific manner to provide the same scaling with non-overlapping G4. (C) Genome browser shots showing RNA:DNA hybrids/R-loops and G4s in VEGFA, NEAT1, CCND1 and MDM2 gene promoters. Asterisk for DRIP-seq data denotes data from (46). Detailed descriptions of the maps and associations are presented in Supplementary Materials: examples of the RLFSs highly enriched with G repeats strand-specific G4-quadruplexes.
Together, the observed distribution patterns suggest roles for RLFSs, RNA:DNA hybrids and R-loops in a general model of a U1-PAS axis (61) as the RNA processing initiators and the spliceosome complex recruitment sites.
Genome-wide high level of structural integrity of the RLFSs, RNA:DNA hybrids, R-loops and G4 quadruplexes
RNA:DNA hybrids/R-loops represent a non-canonical three-strand nucleic acid structure located often in CpG islands and G-rich regions. The G4 motif is a sequence that can form a quadruplex structure (57,62). For a few genes, it has been reported that co-transcriptionally formed stable RNA:DNA hybrids on the template DNA strands could accompany computationally predicted G4 motif(s)-rich regions on the non-template DNA strand, called G-loops (57,62–64). However, simultaneous genome-wide detected G4 motifs with RLFSs and precise mutual localization RNA–DNA hybrids/R-loops have not been reported. The genome-wide structural classification of the experimental G-loops has not yet been studied.
The QmRLFS-predicted RLFSs can be considered strand-specific G-rich and G-repeat-rich sequences. However, the RLFS ssDNA may not contain canonical or non-canonical G4 sequences or G4 motifs on the same strand. Based on the QmRLFS model, the RIZ of RLFS should contain two or more short G-tracks, separated by short non-G nucleotides (e.g. GGGNxGGGNxGGG and GGGGGNxGGGG). When additional one or two G-tracts are present in an RIZ DNA sequences, G4 may be formed. REZ sequences are G-rich and longer than RIZ or linker (32,50) (Figure 1).
First, we focused on the structural associations of RIZ, REZ and entire RLFSs with the canonical (predicted) G4s and the G4-rich structures experimentally defined and mapped genome-wide (65). In total, we re-mapped the 410 924 experimentally defined G4-rich regions in the human genome (hg19). The proportions of canonical G4s overlapping with these experimental G4s were quite close to what was reported by Chambers et al. (65) (e.g. 73% of canonical G4s overlapped with experimental G4-PDS).
Second, we mapped these G4 structures onto RIZ and REZ sequences of RLFSs. We observed that 75% of individual RLFSs intersecting with protein-coding genes (308 163 of 410 924) contained at least one experimentally defined G4 structure, and 77.8% of these G4 structures were located in RIZ, while the remaining structures were located in REZ, in the QmRLFS model. Interestingly, relative to the template (transcribed) DNA strand, a >10.9-fold enrichment of G4 structures located on the non-template (non-transcribed) strand was observed in RIZ, and an ∼4-fold enrichment of G4 structures was observed in REZ (Figure 3A). Importantly, the G4 density distribution form across TSS regions was almost mirror symmetrical and was embedded to the RLFS density distribution (Figure 3B). The both distribution functions are strongly correlate in unidirectional and divergent proximal promoter regions (Figure 3B). Considering estimates of the proportions of RLFS-positive promoter regions in different gene types (Figures 1C and 2), these findings predict that at least 56% of protein-coding genes, 38% of antisense lncRNA genes and 23% of lncRNA genes include G4-positive RLFS complexes, potentially could form G-loop-like conformations.
Third, using R-loop DB tools, we found that the G4-rich sequences were frequently included in the RLFSs of human and other mammalians. We found such combined non-B DNA structure regions preferentially within or nearby the evolutionarily conserved gene regulatory regions of the mammalian genomes, particularly within or nearby CpG and the proximal promoter regions of transcribtion regulatory genes.
Figure 3C (left bottom panel) provides the examples of multiple G4-rich RLFSs supported by RNA-DNA hybrids/R-loop and other datasests. The G4 formation in the vascular endothelial growth factor (VEGF-alpha) proximal promoter region was previously reported (66,67), and subsequent studies revealed the 5′ RNA G-quadruplex structure that is also essential for IRES-mediated translation initiation and ribosome recruitment to its mRNA (68,69). We observed notable consistency between QmRLFS, DRIP-seq and DRIPc-seq data in the VEGF-alpha proximal promoter region of NT2 and K-562 cells (Figure 3C). We found the co-localization and the strand-specific direction of QmRLFSs, RNA:DNA hybrids and G4-rich regions (experimental and computationally predicted) not only in the VEGF-alpha proximal promoter region but also in introns, indicating bidirectional transcriptional initiation sites.
Using R-loop DB, we found many interesting examples of the co-localization of the RLFSs and their clusters with the other signals essential for control of the non-coding regulatory genes. Figure 3C (right bottom panel) shows the lncRNA gene NEAT1 as another representative example. NEAT1 RNA is an essential architectural component of paraspeckle nuclear bodies in mammals (33). It is associated with cancer, neurologic and other diseases. Figure 3C shows that experimentally defined RNA:DNA hybrid-predicted G4s and G4-rich regions (70) were included in the complex genome architecture predicted by the RLFS cluster. The prediction of RLFSs in the proximal promoter region of this single exon gene was consistent with the results of all DRIP- and RDIP-based experimental data (which were included in the R-loop database (33)). Figure 3C (bottom panels) shows the G4-RLFS structures and several regulatory sequences in the loci of other two genes, CCND1 and MDM2, essential in the cell cycle, apoptosis pathways and diseases. We also observed G4, RLFS and DRIP-PCR co-localization at the beginning of the 1st intron (and other introns) of the FOXO1 gene (Supplementary Figure S8). We refer to Supplementary Materials: Examples of the RFLSs highly enriched with G repeats strand-specific G4-quadruplexes (Figure 3C), which contains an additional analysis of the RFLSs highly enriched with G repeats and strand-specific G4-quadruplexes.
The G4-positive RLFSs, evolutionarily conserved in human and mouse, showed a high enrichment of the RNA-Pol II promoter, positive regulation of gene expression, chromatin modification, transcription regulation activity and transcription factor activity GO terms.
Thus, our results demonstrate a high level of structural integrity of the RLFSs, RNA:DNA hybrids, R-loops and G4 regions and suggest the roles of the G4-positive RLFS in evolution, biological complexity and diversity, transcription, metabolic pathways, cell growth, differentiation and diseases.
Reverse-forward RLFS pairs with bidirectional R-loop formation patterns
Analysis of the RLFSs and experimental RNA:DNA hybrid profiles using R-loopDB tools revealed that the RLFSs are often organized as the bidirectional RLFS clusters (Figure 4A and B), whose members ranged from a few to several dozen RLFSs preferentially localized at TSS proximity regions (Supplementary Figure S8 and Figure 4C). To quantify these structures, we identified all paired reverse-forward RLFS loci (PRLs) as the RLFSs separated by at most 500 bp and combined these pairs by merging individual RLFSs (shared between the pairs) located on the same DNA strand (Figure 4A, Supplementary materials: Identification of reverse-forward RLFS loci). The corresponding schematic sequence models of a PRL are shown in the top panel of Figure 4B, and the examples of the PRL in the proximal promoter and U1 site region of FOXO1 gene are presented in the bottom panel of Figure 4A. In total, in the human genome we identified 24 296 PRLs. The IDs of the identified PRLs, the number of RLFSs in the PRLs, their genome coordinates, sequence spans and data referring to embedded/overlapped G4s based on DRIPc-seq are presented in Supplementary Table S12.
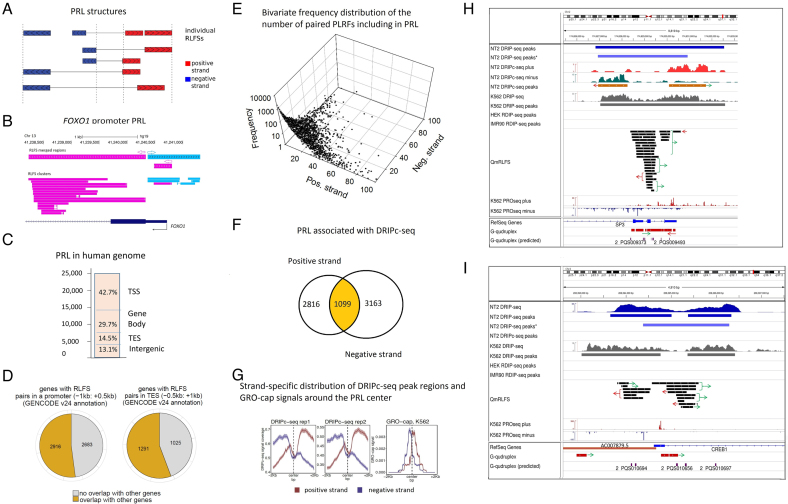
Figure 4.
Analysis of the paired reversed-forward RLFS loci (PRLs). (A) A schema for the identification of neighbor-paired RLFS loci on the forward and reverse DNA strands. The center of a PRL was defined as the middle point between the rightmost reverse-strand RLFS and the leftmost forward-strand RLFS in each pair. The model assumed that most of the pairs would be functional within such a sequence span and in the distal region approximately corresponding to two nucleosome spans. (B). PRL structure in the FOXO1 region, including the promoter, exon 1 and the 1st intron 5′ splice site. (C) The genome-wide RPL distribution (N = 24 296). TSS, TES, gene body and intergenic regions; they were defined similarly to Figure 2B. (D) The frequencies of the PRLs co-localized with the singleton (orphan) genes and the PRLs co-localized with the gene clusters defined at TSS- and TES- proximity regions. The left panel shows the numbers of genes with RPL with and without localization of other genes at the promoter proximity. The right panel shows the numbers of the genes with RPL with and without localization of other genes at the TES proximity. (E) The bivariate distribution of the number of RLFSs included in the PRL set, observed on the positive and negative DNA strands. For each strand, the power law-like frequency distribution that fit well by the Kolmogorov–Waring function has a long tail on the right side (71). This function specifies many sequence types and families, including RLFS (32,71). (F) Strand-specific DRIPc-seq peak regions density functions (NT2 cells) (47) are associated with PRL regions and asymmetrically localized on the PRL flanks. Strand-specific densities of DRIPc-seq peak regions (replicates 1 and 2) and GRO-cap (right) signals are distributed around the PRL center. The results are shown for the positive and negative DNA strands (depicted in red and blue, respectively). (G) Strand-specific distribution of DRIPc-seq peak regions and GRO-cap signals around the PRL center. The left and central panels show the density of DRIPc-seq peak region for two experimental replicates. The right panel shows the densities of the GRO-cap signals region around the PRL center. The results obtained from the genomes of K562 cells (72). (H) Co-localization analysis of the PRL defined within the SP3 gene promoter region. Experimental data integrated via UCSC genome browser tracks (done via R-loop DB tools (33)), including the RNA:DNA hybrid/R-loop profiles (DRIP-based experiments) and the experimental G4-rich region datasets downloaded from the GSE63874 NCBI GEO data repository. Computationally predicted canonical G4s and non-B DNA structures downloaded from the non-B DNA database https://nonb-abcc.ncifcrf.gov. (I) Co-localization analysis of the PRL predicted within the CREB1 gene promoter region. All tracks are the same as in panel H.
We observed that 86.9% (19 811/24 296) of PRLs were predominantly localized in gene-proximal or gene body regions (Figure 4C); the remaining 13.1% of PRLs were distributed in distal intergene regions. The proportion of non-paired RLFS-merged regions in gene-proximal or gene body regions was significantly smaller (∼60%). The difference between the two proportions was 21.35% (the 95% confidence interval of the difference between the two proportions was 20.84 to 21.86%, with P < E-10). Furthermore, in the TSS-proximal regions, the fraction of RLFSs in PRLs was twice that in the whole RLFS set (P < 1E-10 by the difference of two proportions test). Notably, among the 5602 PRL-positive TSS gene regions, 44.3% of events were associated with a single gene (i.e. not bidirectional or unidirectional neighboring genes) (Figure 4D). We observed a similar proportion (47.95%) among the 2316 PRL-positive TES regions.
The frequency distribution of the number of RLFSs in PRLs is skewed right, toward the rare abundant-RLFS-count events and could be quantified by the Kolmogorov-Waring distribution (71) (Figure 4E). A steady state birth-death stochastic process leading to that distribution, assumes preferential selection of the “fitted” regulatory elements (e.g., RLFSs) in gene strand in the course of genome evolution. A relatively small number of PRLs formed the highly enriched clusters of RLFSs (Supplementary Table S12), including a dozen RLFSs, which we called ‘super PRLs’. Interestingly, when the number of RLFSs in a PRLs becomes larger, the number of RLFSs in the gene sense strand increases (Figure 4E) suggesting a role in gene functional specialization.
Figure 4F and G provides several basic characteristics of the PRLs supported by DRIP-seq, DRIPc-seq and GRO-cap experimental datasets. Importantly, the DRIPc-seq peak regions overlapped with or localized at 29.1% (7078/24 296) of PRLs (P < E-10). A total of 39.8% (2816/7078) of the 7078 PRLs were associated with the DRIPc-seq peak regions on the positive strand, and 44.7% (3163/7078) of the PRLs were associated with the DRIPc-seq peak regions on the negative strand. Additionally, 15.5% (1099/7078) of the PRLs were associated with the DRIPc-seq peak regions detected on both strands (Figure 4F). We observed that 93.2% (1024/1099) of the PRLs were localized in gene-proximal or gene body regions, suggesting the positive evolution selection and biological significance of the RNA:DNA hybrids/R-loops specified by this PRL subset (Supplementary Table S12).
Figure 4G shows the strand-specific DRIPc-seq peak region density functions (for NT2 cells) (47) that are associated with PRL regions and asymmetrically localized on the PRL flanks. Strand-specific densities of DRIPc-seq peak regions (replicates 1 and 2) and GRO-cap protocol (72) signals are distributed around the PRL center (right panel of Figure 4F). GRO-cap captures TSSs for both stable and unstable transcripts, which allowed us conduct detailed associations between the transcripts with promoter signals. Our analyses suggest the functional activity of the PRL RLFSs on the reverse and direct strands. Using a nuclear run-on protocol called GRO-cap capturing TSSs for both stable and unstable transcripts, we analyzed the RLFS at the enhancers in the human cells K562 reported in (72). Figure 4G (right panel) shows the density of GRO-cap signals for K562 cells. These results differentiate the strand-specific GRO-cap signals relative to PRL position and indicates that PRL play important roles in early phase of transcription initiation.
The results of the RLFS mapping analysis also showed that PRLs are often localized in the alternative TSS, splice variants and different gene regulatory signals, including G4s, DNA methylation sites, genome segmentation and DNase I–hypersensitive regions (Supplementary Matherials: ChIP-seq and DNAase-data). We observed PRLs at proximal nascent transcript initiation sites and bidirectional or unidirectional promoter regions.
Figure 4H and I shows two examples of the co-localization analysis of PRLs with promoter regulatory regions of transcription factor genes, including the RNA:DNA hybrid/R-loop profiles, G4s and the precision nuclear run-on sequencing (PRO-Seq) signals mapping the Pol II active sites. PRO-Seq mapped the Pol II pausing sites. Figure 4H shows the customized USCS tracks of these sites in SP3 promoter region containing three mRNA isoforms annotated in Ref-seq (and seven isoforms annotated in Ensembl Transcript DB (Build 75), data not shown). Figure 4H shows that the Pol II pausing sites (observed as the highest peaks) are included the PRLs. Importantly, the experimentally defined G4-rich regions and the predicted G4s are highly abundant in the PRLs on both stands of promoter regions; 43% of PRLs include the experimentally defined and predicted G4-rich regions. We did not identify the G4-rich regions in 13.7% of PRLs (Supplementary Table S12). Supplementary Figure S13 shows high-density, diverse non-B DNA structures and RLFSs in the alternative start regions of SP3. Multiple experimentally supported G4-rich regions and computationally predicted G4s associated with RLFS cluster and RNA:DNA hybrid/R-loop data maps are observed. Such complex architectures were observed via R-loop DB tools. These findings suggest an integrity and a co-evolution of the PRL, G4-rich and other non-B DNA sequences in transcription factor gene promoter regions.
The gene CREB1 encodes a transcription factor of the leucine zipper family of DNA-binding proteins. Figure 4I shows a predicted PRL in the CREB1 promoter region with the activity strongly supported by DRIP-seq datasets. The opposite strand of the CREB1 proximal TSS encodes the antisense noncoding RNA gene AC007879.5 (ENSG00000223725), which encodes two transcripts overlapping the 5′ end of CREB1. Figure 4I shows a strand-specific co-localization of the RLFS belonging to a PRL with PRO-seq sites determining the Pol II active sites and Pol II pausing sites on the DNA strands. As in the case of SP3, we also found that the RLFS cluster regions are highly abundant with G4s, G4-rich and other non-B DNA regions. These findings suggest a novel PRL-mediated regulatory mechanism(s) of the gene expression controlling a functional activity of divergent antisense gene pairs.
Intergenic RLFSs are often co-located with transcribed enhancers that form RNA:DNA hybrids
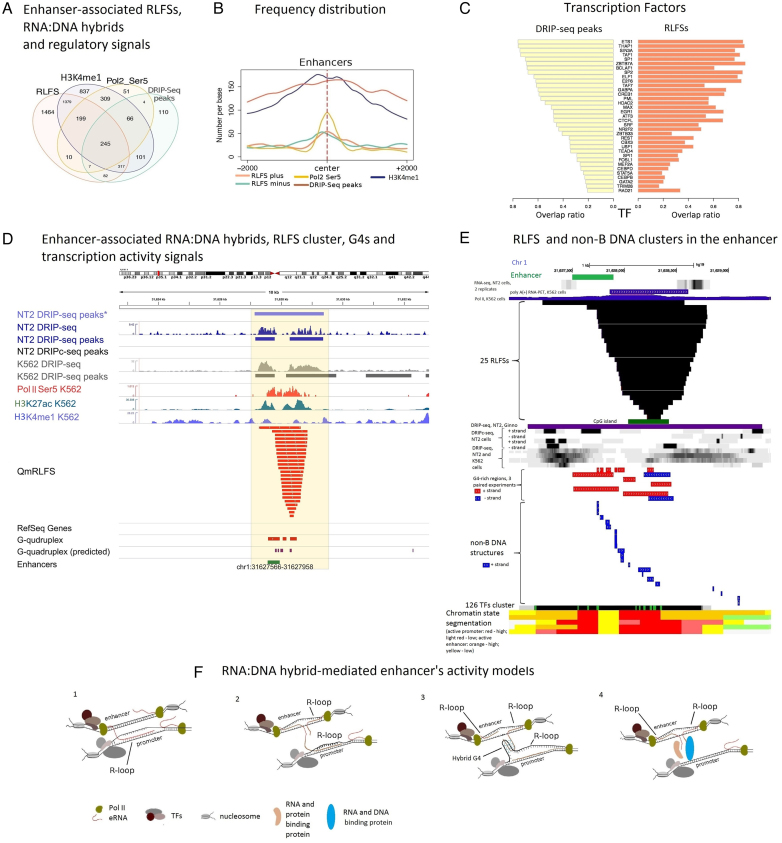
A number of recent genome-wide studies indicated that mammalian enhancers represent transcriptionally active units that produce relatively short non-coding RNA molecules, called ‘eRNAs’, which are typically capped and can be polyadenylated or not (72–74). The eRNA expression level was directly associated with the time- and tissue-specific activity of enhancers (Supplementary Figure S14B). Transcriptional activity at enhancers may result in nucleic acid structures including R-loops (38), although the prevalence of R-loops at enhancers and exact boundary mapping of RLFSs, and RLFS-associated RNA:DNA hybrids/R-loops in eRNA transcript units still require further genome-wide evaluation.
Here, we analyzed 32 693 transcribed human enhancers (55) for the presence of RLFSs (‘Materials and Methods’ section: ‘Enhancers data’). QmRLFS models predicted R-loop formation in 9998 (30.6%) of these enhancers. The RLFS-positive enhancers located at least 2 kb away (called extragenic enhancers) from the annotated genes consist of 37% (3703/9998) of the enhancers; ∼25% of these enhancers were localized at most 0.5 kb away from the RLFSs. The enrichment analysis indicated the high significance of a co-localization of the QmRLFS sequences DRIPc-seq regions (P < 0.001). Specifically, using DRIP-seq peak region data for K562 cells (47), we observed that 9.3% (932/9998) of enhancers were DRIP-seq-positive, with consensus regions between QmRLFS models and DRIP-seq data consisting of 651 enhancer regions, which are 70% (651/932) of experimentally defined RNA:DNA hybrid/R-loop events in K562 cells. Data on the associations of the RLFS-positive enhancers within H3K27ac, H3K4me1 and Pol II binding and DRIP-seq peak regions detected in the K562 cell genome are shown in Supplementary Table S13A.
To explore whether the experimentally defined RNA:DNA hybrid/R-loop formation occurred in transcribed enhancers, we used the ENCODE project data repository for 562 cells (75). We used ChiP-seq H3K4Me1 histone mark data and the data of initiating Pol II (Ser5 CTD modification), CAGE enhancers expression data across 809 samples from (55,76) and DRIP-seq data for the K562 cell line from (47). Our results indicate that the enhancer-associated RLFSs (and their merged regions and clusters) co-localized with experimentally defined RNA:DNA hybrid-forming genome regions (defined by DIPseq), the H3K4Me1 histone mark, and initiating Pol II data on hundreds of transcribed enhancer sites (Figure 5A). The common subset on Venn diagram included 245 RLFSs accociated with intergenic enhansers (Figure 5A). Importantly, the RLFS nucleotide density function in proximal regions of the 245 intergenic enhancers is high abundant (RLFS plus vs RLFS minus), unimodal and almost symmetrical relatively to enhancer's center (Figure 5B).
Figure 5.
Characteristics of the RLFSs associated with transcribed enhancers. (A) Co-localization analysis of the RLFS, H3K4Me1, Pol II Ser5 and DRIP-seq peak regions for K562 cells within transcribed intergenic enhancer regions. Only the enhancers located at least 2 kb away from the annotated genes were considered for the analysis. (B) The distributions of the common RLFS (N = 245), H3K4Me1, Pol II Ser5 and DRIP-seq peak region sequences around enhancer centers. (C) Similarity of the transcription factor overlap ratio values in the DRIP-seq and RLFS regions co-localized with the enhancers. (D) Co-localization analysis of the enhancer-associated RNA:DNA hybrids, RLFS clusters, G4s and transcription activity signals with the enhancer G-rich region. Asterisk for DRIP-seq data denotes data from (46). Other signal profiles are drawn based on the datasets described in ‘Materials and Methods’ section. (E) RLFS and non-B DNA sequence clusters in the e-NKAIN1 enhancer. Data visualization and co-localization analysis was done via integration of the USCS genome browser and R-loop database tracks. Experimental G4-rich region datasets were downloaded from GSE63874 NCBI GEO data repository. Characterization of the structural and functional statuses of the enhancer region was described in ‘Materials and Methods’ section. Computationally predicted canonical G4s and non-B DNA structures were downloaded from non-B DNA database https://nonb-abcc.ncifcrf.gov. Types of 22 non-B DNA sequences are the following (from the left to the right): G4 motif, mirror repeat, short tandem repeat, G4 motif, mirror repeat, G4 motif, direct repeat, short tandem repeat, mirror repeat, G4 motif, G4 motif, mirror repeat, direct repeat, inversed repeat, mirror repeat, inversed repeat, G4 motif, direct repeat, inversed repeat, short tandem repeat, mirror repeat, short tandem repeat. (F) The structural models of R-loop involvement in promoter–enhancer interactions. 1. A nascent eRNA displaces non-template ssDNA in a transcribing gene promoter region and links the active enhancer to the transcribed gene. 2. Two bi-directionally transcribed nascent eRNAs form the enhancer-associated R-loops (eR-loops), leading to a local stabilization of the active enhancer that helps the nascent eRNA to form a non-canonical DNA:RNA hybrid in transcribed gene promoter (e.g. DNA:e-RNA-DNA triplex, e-RNA-mediated R-loop) in trans. 3. A nascent e-RNA displaces a non-template ssDNA near the enhancer, stabilizes an eRNA-mediated R-loop in cis and, via Hoogsteen binding (forming a hybrid G4), links to a non-template ssDNA in an R-loop conformation of a gene promoter in trans. 4. eRNA-protein-DNA complex (or protein-mediated DNA binding) interaction in trans.
Figure 5A shows that a vast majority of the enhancer-associated DRIP-seq peak regions in K562 cells are RLFS positive. The RLFS nucleotides are distributed along the enhancer regions, having maximal density around the center of the enhancer, corresponding to the maximum of the initiating Pol II peak density, DRIP-seq peak region localization and H3K4me1 signals (Figure 5B). Supplementary Figure S14A shows the results of clustering analysis of CAGE, H3K4Me1, H3K4me3, H3K36me3, H3K79me2, H3K27ac, H3K9ac, DNase, and Pol II (Ser5 and Ser2 CTD modification) data mapped within and around merged RLFS regions. These signals form 3 ordered clusters: (H3K27ac, H3K9ac, DNaseI, H3K4me2 and H3K4me3), (GAGE, Pol II (Ser5, Ser2 CTD modification)) and (H3K36me3, H3K4me1 and H3K27Me3). These results in combination with the results of Figure 5A-B suggest structural and functional roles of RLFSs (and RNA-DNA hybrids/R-loops) in the binding of Pol II with transcription initiation sites and histone modification signals marking open enhanser's chromatin. Additionally, Figure 5C shows the similarity of the transcription factor overlap ratio values between the DRIP-seq and RLFS regions co-localized with the enhancers (Supplementary Materials: ChiP-Seq and DNase-Seq data).
Figure 5D shows an example of an enhancer region mapped upstream of NKAIN1 gene. The gene start is 24.6 kb downstream of the enhancer end (defined by FANTOM5). NKAIN is a member of the mammalian protein family with similarity to Drosophila Nkain and interacts with the beta subunit of Na, K-ATPase (77). NKAIN1 could be considered a ‘typical’ enhancer-neighboring gene of neuronal cells. We called the enhancer e-NKAIN1. Supplementary Table S13B provides the sequence data of e-NKAIN1. The table includes the sequences and annotation of the 25 RLFSs formed the RLFS cluster associated with e-NKAIN1. DRIP-seq and DRIP-c seq signals (47) suggest R-loops associated with RLFS clusters. The Pol II activity and enhancer marks (H3K27ac, H3K4me1) suggest the transcriptional activity of the RLFS-positive enhancers. We found that the predicted G4 sequences (78) and experimentally defined G4-rich regions (65) are also included in the region of interest (Figure 5D). Figure 5E provides additional experimental data characterizing the structural and functional status of the e-NKAIN1 region. Figure 5E shows the transcription activity of the enhancer loci (duplicated experiments) in the NT2 ((47), RNA-seq data) and K562 cells (Pol II ChIA-PET signals from ENCODE; Sample ID: GSM970213). Figure 5E shows RLFS cluster and DRIP-seq data from K562 and NT2 cells (including replicated data) without and with RNase H1 treatment (preventing RNA:DNA formation (46,47)). The RLFS and DRIP-seq regions are co-localized with TF binding regions (ChIP-seq data for 161 TFs), as determined by the ENCODE project and USCS CpG region span. We detected the chromatin open state of the region with the USCS tracks for the nine active promoters and enhancer marks defined jointly across nine cell types by a multivariate hidden Markov model. We extend our model of the active enhancer via mapping results of the experimental G4-rich regions and computationally predicted G4s and diverse types of non-B DNA sequences ((65,70,78); http://ttsmi.bii.a-star.edu.sg/; https://nonb-abcc.ncifcrf.gov).
In total, the results of Figure 5D and E suggests that the e-NKAIN1 enhancer region is cell type-specific and transcriptionally active and that not only RLFS-mediated RNA:DNA hybrids/R-loops but also abundant G4s and other non-canonical (non-B DNA) sequences localize there and probably form secondary and more complex spatio-temporal distant structures on both DNA strands.
Thus, RLFS co-localization and integrative data analyses suggest the plausible structural and functional involvement of the RNA:DNA hybrids/R-loops in the structural polymorphism of the nucleic acids conformations and dynamic formation of high-order chromatin structures occurring via R-loop-mediated 3-D formation of (partially known) functional regulatory signals. In such scenarios, our extended models include RNA–DNA complex-binding proteins and ncRNAs, and assume the involvement of diverse non-B DNA conformations and RNA–DNA–protein complexes acting in cis and in trans.
DISCUSSION
This study is the first to characterize the underlying distribution functions of the sequence length of the RIZ, REZ, RLFS and merged and clustered RLFS regions in mammalian genome. The present study provides evidence that RLFSs
can high accurately and reproducibly identify strand-specific localization of RNA:DNA hybrids/R-loops at the single-nucleotide and genome levels;
are significantly predominant in the promoters of moderately and highly expressed genes compared to lower-expressed genes;
are closely overlapped with 73–82% of CAGE-positive TSSs (considering CAGE-positive TSSs located in annotated gene promoters), suggesting a role of the TSS-proximal RLFSs in the switching-on/-off of gene functions and alternative promoters;
are often organized in overlapped RLFS and RLFS clusters. The number and place of these structures near TSS, TES and enhancer's regions is correlated with key transcription, splicing, epigenetic and other key regulatory signals;
are common structural and functional components of RNA:DNA hybrids/R-loops formed in association with G4s and other non-canonical nucleic acid conformations in the critical genome and gene-regulatory regions in protein-coding, bidirectional and long ncRNA genes and in distant (non-genic) regulatory sites (e.g. enhancers);
are often organized as bidirectional RLFS clusters enriched by non-B DNA structures, whose members range from a few to several dozen RLFSs and are commonly localized at promoter, gene ends and enhancer regions;
offer structural insights into the RNA:DNA interactome, playing roles in in cis and in trans transcriptional regulation, gene type classification, genome complexity and transcriptome diversity, and structural high-order chromatin organization and dynamics;
can play essential roles in transcription activation and Pol II pausing in most coding genes and a significant fraction of non-protein-coding genes, transcribed loci (lncRNAs) and transcribed unprocessed pseudogenes; and
provide a novel structural categorization of the gene types and pseudogenes, including protein-coding genes, antisense lncRNAs, lincRNAs and unprocessed and processed pseudogenes.
We found that more than 75% of RLFSs are localized in annotated gene loci or their closest boundaries upstream TSS and downstream TES. In average across human genome, 22% of the RLFSs are present in TSS-proximal regions, and 47.7% in gene bodies, whereas ∼6% of the RLFSs are found close to the TES.
Using a reference dataset and QmRLFS predictions for genome regions, we evaluated the inherent technical biases and variations of each genome-wide RNA:DNA hybrid/R-loop method analyzed in this study. We found biases in the SPR mapping. The SPR localization boundary were often different relatively to CpG, R-Skew and GC, RLFS positive regions. The SPR distribution profiles over the whole genome and gene-regulatory regions were variated across the methods. For example, the DRIPc-seq, DRIP-seq and RDIP-seq SPR profiles in gene-associated proximal regions are often underrepresented with SPRs in the GC-rich and CpG-positive promoter regions. However, the SPRs in DRIP-seq and DRIPc-seq libraries are over-represented in the intragenic regions. These facts are reflected in relatively high frequency of long span RNA:DNA hybrids/R-loop regions and the boundaries expansion associated with sequencing depth in DRIP-seq and DRIPc-seq libraries (47). These finding were supported by comparative analysis with DRIP-seq data (46), SPR set data, defined by non-immunoprecipitation methods and RLFSs.
The RDIP-seq, DRIP-seq and DRIPc-seq SPR under-representation at most proximal promoters and the 1st exon–1st intron regions can be explained by the well-known bias of the Illumina sequencing technology read undercounting (80,81) that is primarily affected by the GC-rich content of the actual DNA fragments in the sequence library.
Recently, Halasz et al. (59) thoroughly evaluated several factors in the experimental procedure of DRIP-seq. The authors found that the genome fragmentation method in DRIP-seq and DRIPc-seq (restriction enzyme digestion) led to the over-representation of lengthy DRIP SPR fragments over coding genes/ORFs, and this bias was enhanced in the first exons of multi exon genes. This prevents the assignment of a precise biological function to a significant fraction of RNA:DNA hybrids/R-loops. These data agree with our statistical estimates and gene level observations using R-loop DB and support our conclusions regarding the unexpectedly long lengths of DRIP-seq and DRIPc-seq SPRs and the missing data in promoter-proximal gene regions determining RNA:DNA hybrid/R-loop formation.
In summary, we conclude that essential protocol differences, technical biases, the scarcity of currently available data and the lack of biological replicates in genome-wide studies contribute to the observed variations within and between the studies and datasets. In this context, to identify precise detection method and map the R-loop ssDNA and complementary nascent RNA sequences using long-read and single-molecule sequencing technologies could be encouraged. At last, we identified the 289 RPRs-associated genes that provide confidence and reproducible GO characteristics of the RNA:DNA hybrid/R-loop positive genes and could be used as R-loop positive references.
In mammalian and many other high-eukaryotic genomes, RNA:DNA hybrids/R-loops are predominant in promoters, where these structures are associated with high CpG island abundance, GC content and GC skew, and in transcription termination loci, where, conversely, these structures are primarily depleted in GC content (47). However, recent results in plants (82) and yeast (49) suggest alternatives or complementary sequence models of R-loop formation, which might require different approaches for their computational prediction and experimental identification. Although current studies have primarily focused on deciphering R-loop formation in GC-rich and relatively long regions of transcribed genes, much remains to be explored about R-loop formation in other DNA sequences (for example in trinucleotide repeats loci (21)), where yet unknown molecular mechanisms might be predictive of R-loop formation in vivo.
Our analyses showed that the RLFSs broadly include experimentally detected G4 motif-rich DNAs in a strand-specific manner (in non-template strand). The G4 motif-rich DNA regions are preferentially co-localized with RIZ structures of RLFSs.
It has been hypothesized that once formed in an R-loop, G4 may play a role in stabilization of the RNA:DNA hybrid (57,62). Additionally, G4 structures formed on non-template DNA strands behind the elongating RNA polymerase complex may lead to polymerase pausing and initiation of R-loop formation, which under some circumstances leads to DNA breaks, mutations and genome instability (27,35–39,57,62). Our findings suggest that the G4-rich and other non-B DNA structures could modulate RNA:DNA hybridization/R-loop formation and organize collectively diverse complex structural-functional regulatory modules involving in many regulatory and pathological processes in cells and multicellular organisms.
The R-loop structure includes G4 in (anti-parallel or parallel) conformations that mostly included in the non-template single strand and could form complex conformations (62–64). Inverse repeats can adopt hairpin structures, which are formed by a fold in a single strand of DNA. Homo (purine/pyrimidine tracks) may fold into several types of intramolecular triplexes (78,79). We propose that in cis-acting nascent RNAs may favor triplex and many other non-canonical conformation and in association with RLFSs and R-loops provide diverse RNA:DNA hybrid regulatory conformations and interactions. Due to structural and thermodynamic characteristics, these non-canonical structures could be included in the sets of the extend RLFS structures and R-loop models. Figure 6 shows probable structural RNA:DNA hybrid/R-loop models in which the RLFS region includes triplexes, G4s and other non-canonical nucleic acid structures (Supplementary Materials: Extended R-loop models including RNA:DNA hybrids and alternative non-canonical nucleic acid structures). Depending on the sequence context and external factors, the models could provide for positive and negative cooperation interactions between the structural modules of such complex genome architectures. Verification of these models can be performed.
Figure 6.
The extended R-loop models including RNA:DNA hybrids and alternative non-canonical nucleic acid structures that often co-localize and form stable conformations during nascent transcription process. The proposed models include (top left) G4, (top right) G4, triplex and hairpin, (central left) G4, triplex, two hairpins (on the positive and negative strands, respectively) and an i-motif structure, (central right) intra-molecule DNA and DNA:RNA hybrid G4s, (bottom left) intra-molecule DNA G4 and DNA:RNA triplex on the positive strand and (bottom right) the duplicated R-loops formed by a single nascent RNA. Detail description of the models A-F see in Supplementary Materials: Extended R-loop models including RNA:DNA hybrids and alternative non-canonical nucleic acid structures.
Interestingly, the G4 structures include not only canonical G4 motifs but also other repeat G-rich motifs, such as those with longer loops, steam loop or bulge formation sequences in G4-rich regions (62–66,83). For instance, according to computational modeling and in vitro validation studies (84), the duplex stem-loop-containing quadruplex formation motifs in the human genome are highly enriched and preferentially mapped within G4-rich regions in the TSS region. The duplex stem-loop secondary structures are diverse and could be thermodynamically more stable than the G4s (85). The associations of the duplex stem-loop-containing quadruplex with the R-loop may play important biological roles and may be used as highly specific diagnostic and therapeutic targets.
We introduced and characterized a new class of the RLFS clusters, called PRLs, whose members range from a few to a dozen RLFSs, preferentially localized at the proximity of bidirectional and unidirectional TSSs and often including alternative promoter regions. In total, we identified 24 296 PRL in the human genome. Our results suggest that the PRL-positive regions in head-to-head antisense gene pair may promote concordant bi-directional transcriptional activation (86,90). We observed that PRLs are also enriched at TES proximity regions.
Our results suggest that PRLs include a widespread subset of non-B DNA-rich structures. Importantly, the DRIPc-seq peak regions (which are strand specific) overlapped with or located at 29.1% of PRL. In total, our findings suggest the important roles of PRL and their integrity with G4-rich and other non-B DNA structures in gene expression, transcription profiles diversity and diverse cellular functions. RLFS, G4s, steam loop and bulge formation sequences in G4-rich regions could be involved in binding and recruitment RNA- and DNA-binding proteins. Indeed, recent studies have suggested that G4s and RNA:DNA hybrids can cooperate in both positive and negative manners. They may influence to chromatin modification status, metabolic process, genome stability and recruitment of the key regulatory proteins and ncRNAs to specific DNA sites (20,21,57,83,87–89). Enzymes such as DHX9 and Pif1 helicase can unwind G4 DNA and/or resolve RNA:DNA hybrids (38,63,64,83,91). As a result, RNA degradation and export factors may lead to RNA:DNA hybrid resolution and suppress R-loop and G4 accumulation. By unwinding these non-B DNA structures, DHX9 and Pif1 may significantly contribute to transcriptional activation, resolving the pol II–DNA-pol conflicts and maintenance of genome stability.
The application of DRIP-seq and DRIPc-seq methods in human and mouse cells has enabled the identification of RNA:DNA hybrids/R-loops associated with enhancer- and insulator-like chromatin state signatures (47). RNA:DNA hybrids/R-loops can be associated with an open chromatin state, the activity of Pol II, the H3K4me1 and H3K27ac marks, and certain transcription factor binding sites associated with enhancers (38,47,48). The results obtained using RLFS are consistent with these observations. Moreover, we identified RNA:DNA hybrids in 30% (9998) of the annotated enhancers; of these, 3703 enhancers in the extragenic 2-kb gene regions were RLFS positive. These results could be used as a new resource for future studies of the RLFS-and RNA:DNA hybrid-positive enhances.
Our results demonstrate that the R-loop-positive extragenic enhancer regions include not only RLFS-determined RNA:DNA hybrids/R-loops but also high-order DNA conformations, RNA–DNA hybrids and regulatory signals acting in cis and in trans. These findings suggest a novel level of biological complexity and regulation functions of the R-loop-positive enhances and new perspectives for highly specific therapeutic targeting.
However, the precise mechanisms of the R-looping processes and G4s are mostly unknown. We propose four RLFS-involved mechanisms of the intergenic enhancer occurring near the gene promoter (Figure 5F). The 1st model proposes that the R-loops could be formed via nascent eRNA and may act as a trans-regulatory sequence directly forming RNA:DNA hybrids with a GC-skewed C-rich strands in distant open gene promoter regions. The second model proposes that RNA:DNA hybrids/R-loops are initially formed within an enhancer region and subsequently the nascent eRNA forms R-loop in the promoter region in trans. The third model proposes the eRNA:promoter DNA hybrid G4 (or G4s), which may be a new and important structural element of the high-order DNA-RNA interactome. Our fourth model proposes that an eRNA, forming an R-loop within an enhancer region, could in trans facilitate a recognition of single-stranded displaced DNA by proteins mediating site-specific interactions between an enhancer and an active gene promoter region. In particular, model 4 suggests that RNA-binding and/or RNA- and DNA-binding proteins could be involved. All these models suggest a formation of the R-loop-mediated polynucleosome RNA:DNA hybrids and proteins at 3-D structures as candidates of the underlying components of the dynamic organization of an RNA-mediated nucleosome scaffold.
We showed that the RLFS density function is positively correlated with the splicing donor signal disposition and negatively correlated with putative PAS at the 5′ ends of different gene types and divergent gene pairs, confirming the functional and evolutionarily important association between R-loops, gene origin and RNA splicing. Our findings support the hypothesis that R-loops are involved in transcriptional U1-PAS axis gene specification, gene origin from divergent gene pairs and the evolution of transcriptome complexity, at least in humans and other mammals (61,92).
Notably, the use of the predictions of specific RLFSs, RNA:DNA hybrids, R-loops and more complex RNA:DNA structures should facilitate the physical isolation of specific genes or genome R-loop regions and develop new types of CRISPR/Cas9 RLFS-specific gene editing and genome engendering. The datasets and theoretical models developed in this study may provide a useful starting point for future applications of RLFSs and R-loops in biology and medicine.
In summary, we conclude that computationally identified RLFSs are high-confidence, common, strand-specific DNA elements playing mechanistic roles in the formation and regulation of a co-transcriptional RNA:DNA interactome. Our results and models provide comprehensive, experimentally testable information for further quantitative validation. Non-canonical DNA structures such as G4s, G4-rich sequences, triplexes and repeats are common RLFS-associated conformations, which collectively constitute a structural basis for transcriptional control, chromatin organization and dynamics, biogenesis, and multiple cellular functions. These structures could form diverse modules acting in in cis and in trans regulation on the gene, genome and transcriptome scales, providing under some circumstances a mechanistic basis for the R-loop-mediated functions in normal and disease-associated RNA:DNA interactions. We identified a class of experimentally supported RLFS cluster regions, called reverse-forward RLFS paired loci, which are preferentially localized at bi-directionally and uni-directionally transcribed gene promoter and enhancer regions and enriched with G4-rich and other non-B DNA sequence structures. Our results suggest critical roles of RLFSs in transcription initiation regulation, promoter switching, splicing, 3D structural chromatin organization and dynamics, pervasive anti-sense transcription, cellular responses to environmental stimuli, proliferation, epigenetic factors, differentiation and many diseases. Applications of our R-loop database and interactive computational tools could accelerate future methods development and studies of emerging R-loop biology and pathobiology.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge the efforts of the editor and reviewers in their critical assessment and improvement of our work. V.B. thanks the SIPGA/A*STAR program for financial support of his internship at BII/A*STAR.
Authors’ contribution: V.A.K. conceived the concept, motivations and objectives for this study. V.A.K., P.J. and V.B. performed statistical tests, data analysis, provided interpretation of results and wrote method’s sections and initial version of the manuscript. Y.S.P. carried out validation experiments. T.W. and P.J. generated RLFS data and together with V.A.K. and V.B. integrated these data with other datasets. V.A.K. wrote the final version of the manuscript. All authors have read and approved the final manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Agency for Science, Technology and Research (A*STAR), Singapore. Funding for open access charge: Bioinformatics Institute/A-STAR.
Conflict of interest statement. None declared.
REFERENCES
- 1. White R.L., Hogness D.S.. R loop mapping of the 18S and 28S sequences in the long and short repeating units of Drosophila melanogaster rDNA. Cell. 1977; 10:177–192. [DOI] [PubMed] [Google Scholar]
- 2. Ratmeyer L., Vinayak R., Zhong Y.Y., Zon G., Wilson W.D.. Sequence specific thermodynamic and structural properties for DNA.RNA Duplexes. Biochemistry. 1994; 33:5298–5304. [DOI] [PubMed] [Google Scholar]
- 3. Drolet M., Phoenix P., Menzel R., Masse E., Liu L.F., Crouch R.J.. Overexpression of RNase H partially complements the growth defect of an Escherichia coli delta topA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:3526–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Massé E., Drolet M.. Escherichia coli DNA topoisomerase I inhibits R-loop formation by relaxing transcription-induced negative supercoiling. J. Biol. Chem. 1999; 274:16659–16664. [DOI] [PubMed] [Google Scholar]
- 5. Broccoli S., Rallu F., Sanscartier P., Cerritelli S.M., Crouch R.J., Drolet M.. Effects of RNA polymerase modifications on transcription-induced negative supercoiling and associated R-loop formation. Mol. Microbiol. 2004; 52:1769–1779. [DOI] [PubMed] [Google Scholar]
- 6. Reaban M.E., Lebowitz J., Griffin J.A.. Transcription induces the formation of a stable RNA.DNA hybrid in the immunoglobulin alpha switch region. J. Biol. Chem. 1994; 269:21850–21857. [PubMed] [Google Scholar]
- 7. Daniels G.A., Lieber M.R.. RNA: DNA complex formation upon transcription of immunoglobulin switch regions: implications for the mechanism and regulation of class switch recombination. Nucleic. Acids. Res. 1995; 23:5006–5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tian M., Alt F.W.. Transcription-induced cleavage of immunoglobulin switch regions by nucleotide excision repair nucleases in vitro. J. Biol. Chem. 2000; 275:24163–24172. [DOI] [PubMed] [Google Scholar]
- 9. Yu K., Chedin F., Hsieh C.-L., Wilson T.E., Lieber M.R.. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 2003; 4:442–451. [DOI] [PubMed] [Google Scholar]
- 10. Huang F.T., Yu K., Hsieh C.L., Lieber M.R.. Downstream boundary of chromosomal R-loops at murine switch regions: implications for the mechanism of class switch recombination. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:5030–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yu K., Roy D., Huang F.T., Lieber M.R.. Detection and structural analysis of R-loops. Methods Enzymol. 2006; 409:316–329. [DOI] [PubMed] [Google Scholar]
- 12. Roy D., Yu K., Lieber M.R.. Mechanism of R-Loop formation at immunoglobulin class switch sequences. Mol. Cell. Biol. 2008; 28:50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roy D., Lieber M.R.. G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas high G density without clustering is sufficient thereafter. Mol. Cell. Biol. 2009; 29:3124–3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gyi J.I., Conn G.L., Lane A.N., Brown T.. Comparison of the thermodynamic stabilities and solution conformations of DNA·RNA hybrids containing purine-rich and pyrimidine-rich strands with DNA and RNA duplexes. Biochemistry. 1996; 35:12538–12548. [DOI] [PubMed] [Google Scholar]
- 15. Zhang Z.Z., Pannunzio N.R., Hsieh C.-L., Yu K., Lieber M.R.. The role of G-density in switch region repeats for immunoglobulin class switch recombination. Nucleic. Acids Res. 2014; 42:13186–13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huertas P., Aguilera A.. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell. 2003; 12:711–721. [DOI] [PubMed] [Google Scholar]
- 17. Lin Y., Dent S.Y.R., Wilson J.H., Wells R.D., Napierala M.. R loops stimulate genetic instability of CTG.CAG repeats. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:692–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Loomis E.W., Sanz L.A., Chédin F., Hagerman P.J.. Transcription-associated R-loop formation across the human FMR1 CGG-repeat region. PLoS Genet. 2014; 10:e1004294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Groh M., Lufino M.M.P., Wade-Martins R., Gromak N.. R-loops associated with triplet repeat expansions promote gene silencing in friedreich ataxia and fragile X syndrome. PLoS Genet. 2014; 10:e1004318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haeusler A.R., Donnelly C.J., Periz G., Simko E.A.J., Shaw P.G., Kim M.-S., Maragakis N.J., Troncoso J.C., Pandey A., Sattler R. et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014; 507:195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reddy K., Schmidt M.H.M., Geist J.M., Thakkar N.P., Panigrahi G.B., Wang Y.-H., Pearson C.E.. Processing of double-R-loops in (CAG)·(CTG) and C9orf72 (GGGGCC)·(GGCCCC) repeats causes instability. Nucleic Acids Res. 2014; 42:10473–10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li X., Manley J.L.. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005; 122:365–378. [DOI] [PubMed] [Google Scholar]
- 23. Stirling P.C., Chan Y.A., Minaker S.W., Aristizabal M.J., Barrett I., Sipahimalani P., Kobor M.S., Hieter P.. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev. 2012; 26:163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wahba L., Amon J.D., Koshland D., Vuica-Ross M.. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol. Cell. 2011; 44:978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sollier J., Stork C.T., García-Rubio M.L., Paulsen R.D., Aguilera A., Cimprich K.A.. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol. Cell. 2014; 56:777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wellinger R.E., Prado F., Aguilera A.. Replication fork progression is impaired by transcription in hyperrecombinant yeast cells lacking a functional THO complex. Mol. Cell. Biol. 2006; 26:3327–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tuduri S., Crabbé L., Conti C., Tourrière H., Holtgreve-Grez H., Jauch A., Pantesco V., De Vos J., Thomas A., Theillet C. et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol. 2009; 11:1315–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Helmrich A., Ballarino M., Tora L.. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell. 2011; 44:966–977. [DOI] [PubMed] [Google Scholar]
- 29. Gan W., Guan Z., Liu J., Gui T., Shen K., Manley J.L., Li X.. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 2011; 25:2041–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sordet O., Redon C.E., Guirouilh-Barbat J., Smith S., Solier S., Douarre C., Conti C., Nakamura A.J., Das B.B., Nicolas E. et al. Ataxia telangiectasia mutated activation by transcription- and topoisomerase I-induced DNA double-strand breaks. EMBO Rep. 2009; 10:887–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Colak D., Zaninovic N., Cohen M.S., Rosenwaks Z., Yang W.Y., Gerhardt J., Disney M.D., Jaffrey S.R.. Promoter-bound trinucleotide repeat mRNA drives epigenetic silencing in fragile X syndrome. Science. 2014; 343:1002–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wongsurawat T., Jenjaroenpun P., Kwoh C.K., Kuznetsov V.. Quantitative model of R-loop forming structures reveals a novel level of RNA-DNA interactome complexity. Nucleic Acids Res. 2012; 40:e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jenjaroenpun P., Wongsurawat T., Sutheeworapong S., Kuznetsov V.A.. R-loopDB: a database for R-loop forming sequences (RLFS) and R-loops. Nucleic Acids Res. 2016; 45:D119–D127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yeo A.J., Becherel O.J., Luff J.E., Cullen J.K., Wongsurawat T., Jenjaroenpoon P., Kuznetsov V.A., McKinnon P.J., Lavin M.F.. R-loops in proliferating cells but not in the brain: implications for AOA2 and other autosomal recessive ataxias. PLoS One. 2014; 9:e90219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Aguilera A., García-Muse T.. R loops: from transcription byproducts to threats to genome stability. Mol. Cell. 2012; 46:115–124. [DOI] [PubMed] [Google Scholar]
- 36. Skourti-Stathaki K., Proudfoot N.J.. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014; 28:1384–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hamperl S., Cimprich K.A.. The contribution of co-transcriptional RNA:DNA hybrid structures to DNA damage and genome instability. DNA Repair (Amst.). 2014; 19:84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Santos-Pereira J.M., Aguilera A.. R loops: new modulators of genome dynamics and function. Nat. Rev. Genet. 2015; 16:583–597. [DOI] [PubMed] [Google Scholar]
- 39. Costantino L., Koshland D.. The Yin and Yang of R-loop biology. Curr. Opin. Cell Biol. 2015; 34:39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li X., Manley J.L.. New talents for an old acquaintance: the SR protein splicing factor ASF/SF2 functions in the maintenance of genome stability. Cell Cycle. 2005; 4:1706–1708. [DOI] [PubMed] [Google Scholar]
- 41. Bhatia V., Barroso S.I., García-Rubio M.L., Tumini E., Herrera-Moyano E., Aguilera A.. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014; 511:362–365. [DOI] [PubMed] [Google Scholar]
- 42. Skourti-Stathaki K., Proudfoot N.J., Gromak N.. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell. 2011; 42:794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kaneko S., Chu C., Shatkin A.J., Manley J.L.. Human capping enzyme promotes formation of transcriptional R loops in vitro. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:17620–17625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Boguslawski S.J., Smith D.E., Michalak M.A., Mickelson K.E., Yehle C.O., Patterson W.L., Carrico R.J.. Characterization of monoclonal antibody to DNA RNA and its application to immunodetection of hybrids. J. Immunol. Methods. 1986; 89:123–130. [DOI] [PubMed] [Google Scholar]
- 45. Ginno P.A., Lott P.L., Christensen H.C., Korf I., Chédin F.. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell. 2012; 45:814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ginno P.A., Lim Y.W., Lott P.L., Korf I., Chedin F.. GC skew at the 5′ and 3′ ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res. 2013; 23:1590–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sanz L.A., Hartono S.R., Lim Y.W., Steyaert S., Rajpurkar A., Ginno P.A., Xu X., Chédin F.. Prevalent, dynamic, and conserved R-loop structures associate with specific epigenomic signatures in mammals. Mol. Cell. 2016; 63:167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nadel J., Athanasiadou R., Lemetre C., Wijetunga N.A., Ó Broin P., Sato H., Zhang Z., Jeddeloh J., Montagna C., Golden A. et al. RNA:DNA hybrids in the human genome have distinctive nucleotide characteristics, chromatin composition, and transcriptional relationships. Epigenet. Chromatin. 2015; 8:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wahba L., Costantino L., Tan F.J., Zimmer A., Koshland D.. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev. 2016; 30:1327–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jenjaroenpun P., Wongsurawat T., Yenamandra S.P., Kuznetsov V.A.. QmRLFS-finder: a model, web server and stand-alone tool for prediction and analysis of R-loop forming sequences. Nucleic Acids Res. 2015; 43:W527–W534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Castellano-Pozo M., Santos-Pereira J.M., Rondón A.G., Barroso S., Andújar E., Pérez-Alegre M., García-Muse T., Aguilera A.. R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol. Cell. 2013; 52:583–590. [DOI] [PubMed] [Google Scholar]
- 52. Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012; 22:1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Haberle V., Forrest A.R.R., Hayashizaki Y., Carninci P., Lenhard B.. CAGEr: precise TSS data retrieval and high-resolution promoterome mining for integrative analyses. Nucleic Acids Res. 2015; 43:e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Grant C.E., Bailey T.L., Noble W.S.. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011; 27:1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T. et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014; 507:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Alexa A., Rahnenfuhrer J.. topGO: enrichment analysis for Gene Ontology. R package version 2.22.0. 2010. [Google Scholar]
- 57. Duquette M.L., Huber M.D., Maizels N.. G-rich proto-oncogenes are targeted for genomic instability in B-cell lymphomas. Cancer Res. 2007; 67:2586–2594. [DOI] [PubMed] [Google Scholar]
- 58. Kung J.T.Y., Colognori D., Lee J.T.. Long noncoding RNAs: past, present, and future. Genetics. 2013; 193:651–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Halász L., Karányi Z., Boros-Oláh B., Kuik-Rózsa T., Sipos É., Nagy É., Mosolygó-L Á., Mázló A., Rajnavölgyi É., Halmos G. et al. RNA-DNA hybrid (R-loop) immunoprecipitation mapping: an analytical workflow to evaluate inherent biases. Genome Res. 2017; 27:1063–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang X., Chiang H.C., Wang Y., Zhang C., Smith S., Zhao X., Nair S.J., Michalek J., Jatoi I., Lautner M. et al. Attenuation of RNA polymerase II pausing mitigates BRCA1-associated R-loop accumulation and tumorigenesis. Nat. Commun. 2017; 8:15908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Almada A.E., Wu X., Kriz A.J., Burge C.B., Sharp P.A.. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013; 499:360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Maizels N., Gray L.T.. The G4 genome. PLoS Genet. 2013; 9:e1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhou R., Zhang J., Bochman M.L., Zakian V.A., Ha T.. Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. Elife. 2014; 3:e02190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Paeschke K., Bochman M.L., Garcia P.D., Cejka P., Friedman K.L., Kowalczykowski S.C., Zakian V.A.. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013; 497:458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chambers V.S., Marsico G., Boutell J.M., Di Antonio M., Smith G.P., Balasubramanian S.. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015; 33:877–881. [DOI] [PubMed] [Google Scholar]
- 66. Sun D., Guo K., Rusche J.J., Hurley L.H.. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005; 33:6070–6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sun D., Guo K., Shin Y.J.. Evidence of the formation of G-quadruplex structures in the promoter region of the human vascular endothelial growth factor gene. Nucleic Acids Res. 2011; 39:1256–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Morris M.J., Negishi Y., Pazsint C., Schonhoft J.D., Basu S.. An RNA G-quadruplex is essential for cap-independent translation initiation in human VEGF IRES. J. Am. Chem. Soc. 2010; 132:17831–17839. [DOI] [PubMed] [Google Scholar]
- 69. Bhattacharyya D., Diamond P., Basu S.. An independently folding RNA G-quadruplex domain directly recruits the 40S ribosomal subunit. Biochemistry. 2015; 54:1879–1885. [DOI] [PubMed] [Google Scholar]
- 70. Kwok C.K., Marsico G., Sahakyan A.B., Chambers V.S., Balasubramanian S.. rG4-seq reveals widespread formation of G-quadruplex structures in the human transcriptome. Nat. Methods. 2016; 13:841–844. [DOI] [PubMed] [Google Scholar]
- 71. Kuznetsov V.A. Tatarinova TV, Nikolsky Y. Mathematical modeling of avidity distribution and estimating general binding properties of transcription factors from genome-wide binding profiles. Biological Networks and Pathway Analysis. 2017; NY: Springer; 193–276. [DOI] [PubMed] [Google Scholar]
- 72. Core L.J., Martins A.L., Danko C.G., Waters C.T., Siepel A., Lis J.T.. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat. Genet. 2014; 46:1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kim T.-K., Hemberg M., Gray J.M., Costa A.M., Bear D.M., Wu J., Harmin D.A., Laptewicz M., Barbara-Haley K., Kuersten S. et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010; 465:182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lam M.T.Y., Li W., Rosenfeld M.G., Glass C.K.. Enhancer RNAs and regulated transcriptional programs. Trends Biochem. Sci. 2014; 39:170–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. The ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. The FANTOM Consortium and the RIKEN PMI and CLST (DGT) A promoter-level mammalian expression atlas. Nature. 2014; 507:462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gorokhova S., Bibert S., Geering K., Heintz N.. A novel family of transmembrane proteins interacting with β subunits of the Na,K-ATPase. Hum. Mol. Genet. 2007; 16:2394–2410. [DOI] [PubMed] [Google Scholar]
- 78. Jenjaroenpun P., Chew C.S., Yong T.P., Choowongkomon K., Thammasorn W., Kuznetsov V.A.. The TTSMI database: a catalog of triplex target DNA sites associated with genes and regulatory elements in the human genome. Nucleic Acids Res. 2015; 43:D110–D116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Cer R.Z., Donohue D.E., Mudunuri U.S., Temiz N.A., Loss M.A., Starner N.J., Halusa G.N., Volfovsky N., Yi M., Luke B.T. et al. Non-B DB v2.0: a database of predicted non-B DNA-forming motifs and its associated tools. Nucleic Acids.Res. 2013; 41:D94–D100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Benjamini Y., Speed T.P.. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res. 2012; 40:e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. DaCosta J.M., Sorenson M.D.. Amplification biases and consistent recovery of loci in a double-digest RAD-seq protocol. PLoS One. 2014; 9:e106713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sun Q., Csorba T., Skourti-Stathaki K., Proudfoot N.J., Dean C.. R-loop stabilization represses antisense transcription at the arabidopsis FLC locus. Science. 2013; 340:619–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Dolinnaya N.G., Ogloblina A.M., Yakubovskaya M.G.. Structure, properties, and biological relevance of the DNA and RNA G-quadruplexes: overview 50 years after their discovery. Biochemistry. 2016; 81:1602–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lim K.W., Jenjaroenpun P., Low Z.J., Khong Z.J., Ng Y.S., Kuznetsov V.A., Phan A.T.. Duplex stem-loop-containing quadruplex motifs in the human genome: a combined genomic and structural study. Nucleic Acids Res. 2015; 43:5630–5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lim K.W., Nguyen T.Q.N., Phan A.T.. Joining of multiple duplex stems at a single quadruplex loop. J. Am. Chem. Soc. 2014; 136:17969–17973. [DOI] [PubMed] [Google Scholar]
- 86. Grinchuk O.V., Jenjaroenpun P., Orlov Y.L., Zhou J., Kuznetsov V.A.. Integrative analysis of the human cis -antisense gene pairs, miRNAs and their transcription regulation patterns. Nucleic Acids Res. 2009; 38:534–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Chen P.B., Chen H.V., Acharya D., Rando O.J., Fazzio T.G.. R loops regulate promoter-proximal chromatin architecture and cellular differentiation. Nat. Struct. Mol. Biol. 2015; 22:999–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Shrestha P., Xiao S., Dhakal S., Tan Z., Mao H.. Nascent RNA transcripts facilitate the formation of G-quadruplexes. Nucleic Acids Res. 2014; 42:7236–7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Salvi J.S., Chan J.N.Y., Szafranski K., Liu T.T., Wu J.D., Olsen J.B., Khanam N., Poon B.P.K., Emili A., Mekhail K.. Roles for Pbp1 and caloric restriction in genome and lifespan maintenance via suppression of RNA-DNA Hybrids. Dev. Cell. 2014; 30:177–191. [DOI] [PubMed] [Google Scholar]
- 90. Boque-Sastre R., Soler M., Oliveira-Mateos C., Portela A., Moutinho C., Sayols S., Villanueva A., Esteller M., Guil S.. Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:5785–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Chakraborty P., Grosse F.. Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair (Amst.). 2011; 10:654–665. [DOI] [PubMed] [Google Scholar]
- 92. Wu X., Sharp P.A.. Divergent transcription: a driving force for new gene origination. Cell. 2013; 155:990–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.