Abstract
Linker histone H1 has a key role in maintaining higher order chromatin structure and genome stability, but how H1 functions in these processes is elusive. Here, we report that acetylation of lysine 85 (K85) within the H1 globular domain is a critical post-translational modification that regulates chromatin organization. H1K85 is dynamically acetylated by the acetyltransferase PCAF in response to DNA damage, and this effect is counterbalanced by the histone deacetylase HDAC1. Notably, an acetylation-mimic mutation of H1K85 (H1K85Q) alters H1 binding to the nucleosome and leads to condensed chromatin as a result of increased H1 binding to core histones. In addition, H1K85 acetylation promotes heterochromatin protein 1 (HP1) recruitment to facilitate chromatin compaction. Consequently, H1K85 mutation leads to genomic instability and decreased cell survival upon DNA damage. Together, our data suggest a novel model whereby H1K85 acetylation regulates chromatin structure and preserves chromosome integrity upon DNA damage.
INTRODUCTION
Chromatin structure and genome integrity is preserved through highly organized cellular machineries, including linker histone H1. In mammalian cells, H1 consists of a family of >10 isoforms that redundantly regulate chromatin organization (1,2). Triple knockout of three of these H1 isoforms in murine cells causes ∼50% total H1 loss and general chromatin structural aberrations, but only affects the expression of a limited number of genes (3). In Drosophila, H1 prevents genome instability whereas deletion of H1 increases R-loop accumulation and sister-chromatid exchange (4). H1 is also essential for in vitro reconstitution of 30-nm chromatin fibers, which is critical to forming higher order chromatin structure (5). These data indicate that H1 has a key role in maintaining higher order chromatin structure.
Mammalian H1 has a tripartite structure consisting of a short N-terminal domain, a highly conserved globular domain and a long unstructured C-terminal domain (6). The mechanisms as to how H1 binds chromatin are still evolving, but it is now generally accepted that both the globular and C-terminal domains contribute to binding H1 to the nucleosome and maintaining chromatin condensation and higher order 30-nm chromatin structure (1). The H1 globular domain is critical for its dynamic binding to the nucleosome dyad and linker DNA (7–12). Deletion or disruption of specific residues within the globular domain can alter the binding affinity or binding mode of H1 to chromatin (9,12–16). For example, mutating arginine 54 (R54) to an alanine or lysine impairs H1 binding to nucleosomes and results in global chromatin decompaction (16). In murine cells, mutating several lysine residues to alanine, including lysine 85, leads to decreased H1 binding affinity to chromatin (9). At last, mutating Drosophila H1 lysine 95, which is homologous to human lysine 85 (referred to as H1K85 hereafter), strongly reduces H1 binding to nucleosomes in vitro (12). These reports support that the H1 globular domain, especially H1K85, is important in regulating H1 dynamics and chromatin structure. The underlying mechanisms and biological relevance of this regulation in vivo need further investigation.
Histone post-translational modifications (PTMs) are crucial for regulating chromatin structure and genome stability as dysregulated histone PTMs can cause cellular disorders including cancer (17,18). Although the functional link between core histone modifications and genome stability is well established, modifications of linker histone are also critical to preserve genome integrity (19). For example, deacetylation of H1K26 by SIRT1 results in enriched H1 on chromatin and formation of facultative heterochromatin (20). H1 facilitates the recruitment of heterochromatin protein 1 (HP1) to promote heterochromatin formation, but H1 phosphorylation disrupts this interaction and leads to disassembly of higher order chromatin structure (21–23). Furthermore, peptidylarginine deiminase 4 (PADI4)-mediated citrullination (the conversion of arginine to citrulline) within the H1 globular domain during cellular reprogramming leads to chromatin decondensation (16). These studies underlie the importance of H1 PTMs in regulating genome condensation and stability, but how H1 modifications (especially to its globular domain) regulate chromatin structure is unclear.
As well as its role in packaging and preserving genetic information, chromatin structure is extensively reorganized and remodeled during the DNA damage response (DDR) and DNA repair (24,25). Acetylation of histone H4, which destabilizes higher order chromatin structure and allows DNA repair factors to access damaged chromatin, is essential in the DDR and DNA repair (26,27). Mechanistically, histone acetylation modulates chromatin structure by altering histone–DNA electrostatic charges and recruiting remodeling factors and complexes (28,29). This ‘access–repair–restore’ model further illustrates how histone modifications and chromatin remodeling machineries regulate chromatin accessibility and organization to promote DNA repair (30). Dynamic acetylation of core histones, which is balanced by histone deacetylases (HDACs) and acetyltransferases (HATs), is crucial for chromatin remodeling and maintaining genome integrity (31). How H1 acetylation is dynamically regulated in response to DNA damage and whether it is involved in the modulation of chromatin structure is largely unknown.
In this study, we engineered and expressed different H1K85 mutations to investigate whether acetylation is important for H1 to regulate chromatin structure. We found that a K85 acetylation-mimic mutation (H1K85Q) leads to increased H1 nucleosome binding and condensed chromatin structure by upregulating the interaction between H1 and core histones. H1K85 acetylation (referred to as H1K85ac hereafter) is dynamically regulated in response to DNA damage, which is balanced by the HAT p300/CBP-associated factor (PCAF) and HDAC1. Most importantly, H1K85ac facilitates the recruitment of heterochromatin protein 1 (HP1). This study thus reveals a novel mechanism by which acetylation of a single H1 site within the globular domain modulates DNA damage-induced chromatin dynamics.
MATERIALS AND METHODS
Cell culture, antibodies and reagents
HCT116, HeLa, HEK293T, A549, H1299, Calu3 and MDA-MB-231 cells were cultured in McCoy’s 5A medium or DMEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin in a 37°C incubator with a 5% CO2 and humidified atmosphere. The antibodies used in this study include: anti-H1 and anti-pan-H3-ac from Active Motif; anti-H1.4, anti-β-actin and anti-Biotin from Santa Cruz; anti-H1K85ac and anti-H1K63ac from PTM Biolabs; anti-H3K14ac, anti-H3K9ac, anti-H4K16ac, anti-histone H3, anti-histone H4 from Abcam; anti-PCAF, anti-phospho-H2AX (S139), anti-H2A, anti-HP1α, anti-SMC1, anti-SUV39H1, anti-pan-acetyl-lysine, anti-HDAC1, anti-HDAC2, anti-HDAC3 and anti-HDAC8 from Cell Signaling; anti-HP1β and anti-HP1γ from GeneTex; anti-FLAG from Sigma; anti-GST from APPLYGEN; anti-GFP from MBL. Adriamycin, etoposide, camptothecin (CPT), isopropy-β-D-thiogalactoside (IPTG), trichostatin A and nicotinamide were purchased from Sigma; anacardic acid was purchased from Selleck.
Plasmid and RNAi
H1.2 and H1.4 cDNAs were cloned into p3 × FLAG-CMV-10, EGFP-C2, pET28b and pGEX-4T3. p300, hMOF, TIP60, HDAC1, HDAC3 and HDAC8 cDNAs were cloned into p3 × FLAG-CMV-10. FLAG-PCAF, GST-PCAF and GST-PCAF-HAT2 (amino acids 352–658) were gifts from Dr Ye Zhang (Chinese Academy of Medical Sciences). FLAG-HDAC2 was a gift from Dr. Jiadong Wang (Peking University Health Science Center). All mutations were generated using a site-directed mutagenesis kit (Vazyme). The following siRNAs were used to silence target genes by transient transfection with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions:
HDAC1: 5′-UGGCCAUCCUGGAACUGCUAAAGUA-3′;
HDAC2: 5′-GAAGAUCCAGACAAGAGAAUUUCUA-3′;
HDAC3: 5′-CGGGAUGGCAUUGAUGACCAGAGUU-3′;
HDAC8: 5′-GACGGAAAUUUGAGCGUAUUCUCUA-3′.
Establishment of stable cell lines
shRNA knockdown
Stable HDAC1 knockdown cells were generated using Lipofectamine 2000 tranfection of shRNA constructs with the HDAC1 siRNA sequences cloned into a pGPH1/Hygro vector (GenePharma).
CRISPR-Cas9 gene editing
Knockout cells were generated via Lipofectamine 2000 transfection of sgRNAs cloned into a pCRISPR-LvSG03 vector (GeneCopoeia). The protocol followed was as previously described (32).
SDS-PAGE and immunoblotting
Protein quantification, sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting procedures were performed as previously described (33). Equal amounts of proteins were sized fractionated on a 6–15% SDS-PAGE gel.
Histone acid extraction
Histone was extracted as previously described (34). Briefly, cell pellets were lysed in hypotonic buffer, re-suspended in 0.4 N sulfuric acid and precipitated by trichloroacetic acid. The histone pellet was washed with acetone and dissolved in ddH2O.
Co-IP
Cell pellets were lysed in lysis buffer (20 mM Tris·HCl pH 8.0, 137 mM NaCl, 10% glycerol, 2 mM ethylenediaminetetraacetic acid (EDTA), 1% protease inhibitor cocktail, 1% NP-40) for 30 min on ice. The supernatant was collected after centrifugation and incubated with the indicated antibodies at 4°C rotating overnight. Protein G or A Sepharose slurry (GE healthcare) was added and incubated with the samples for 3 h at 4°C. The beads were washed and boiled in 2 × protein sample buffer at 100°C for 5 min before SDS-PAGE and immunoblotting.
Real-time RT-PCR assay
Total RNA was extracted in TRIzol (Invitrogen) and precipitated in ethanol. cDNA was then synthesized using a HiScript cDNA Synthesis Kit (Vazyme). The relative expression of Sat-2 and GAPDH were measured by real-time polymerase chain reaction (PCR) with the following primers: Sat-2, 5′-CATCGAATGGAAATGAAAGGAGTC-3′ (sense) and 5′-ACCATTGGATGATTGCAGTCAA-3′ (antisense); GAPDH, 5′-CAGCAAGAGCACAAGAGGAA-3′ (sense), 5′-CCCCTCTTCAAGGGGTCTAC-3′ (antisense).
FISH
Cells were synchronized into metaphase by treatment with colchicine (0.5 μM) for 2.5 h. Chromosomes were spread following incubation with a centromere-specific PNA probe (Panagene) and then observed with a fluorescent microscope.
ChIP and ChIP-seq
ChIP assay was performed as previously described (34). Briefly, cells were cross-linked with 1% formaldehyde and lysed in lysis buffer (1% SDS, 5 mM EDTA, 50 mM Tris·HCl pH 8.0, 1 mM phenylmethanesulfonyl fluoride (PMSF), 1% protease inhibitor cocktail) for 10 min on ice. The lysates were sonicated on ice and then centrifuged for 10 min at 12 000 rpm at 4°C. The supernatant was collected and precleared in dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris·HCl pH 8.0) with protein G or A Sepharose and salmon DNA (Thermo Fisher) for 2 h at 4°C. The precleared samples were immunoprecipitated with indicated antibodies at 4°C overnight. Protein G or A Sepharose and salmon sperm DNA were added and incubated for 3 h at 4°C with rotation. The beads were washed, eluted and heated at 65°C overnight to reverse the cross-links. DNA was purified using a PCR Cleaning Kit (Macherey-Nagel) and real-time PCR was performed using the following primers: 5′-TCTTCTTCAAGGACGACGGCAACT-3′ (sense) and 5′-TTGTAGTTGTACTCCAGCTTGTGC-3′ (antisense); genomic control: 5′-CCTGAGCAAAGACCCCAA-3′ (sense) and 5′-TTACTTGTACAGCTCG-3′ (antisense). For ChIP-seq, the lysates were sonicated to generate DNA fragments of 200–500 bp. Purified DNA was analyzed by deep sequencing (BGI).
SILAC labeling and quantitative MS
The experiment was performed as previously described (35). Briefly, HeLa cells were grown in medium with the light or heavy isotope of U-C6 (12C6 or 13C6) separately for more than six generations. The experimental group was treated with adriamycin (1 μM) for 12 h. The cells were then lysed and mixed for quantitative mass spectrometry (MS) analysis (PTM Biolabs).
In vitro acetylation assay
PCAF and other acetyltransferases were purified and incubated with substrates in acetylation buffer (50 mM Tris·HCl, pH 8.0, 50 mM NaCl, 4 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol (DTT), and 10% glycerol) with or without acetyl-CoA (5 mM) for 1 h at 30°C. The reactions were stopped by adding 5 × protein sample buffer and the samples were boiled at 100°C for 5 min before SDS-PAGE and immunoblotting.
In vitro deacetylation assay
HDACs were purified and incubated with histones extracted from TSA-treated cells in deacetylation buffer (10 mM Tris·HCl pH 8.0, 10 mM NaCl, 10% glycerol) for 1 h at 30°C. The reactions were stopped by adding 5 × protein sample buffer and the samples were boiled at 100°C for 5 min before SDS-PAGE and immunoblotting.
Laser micro-irradiation
Cells were grown on a glass-bottomed dish and irradiated with a 365 nm pulsed nitrogen UV laser (16 Hz pulse, 41% laser output) generated using a Micropoint System (Andor). The cells were either fixed immediately in 4% paraformaldehyde or cultured at 37°C for the indicated times.
Immunofluorescence microscopy
Cells were fixed in 4% paraformaldehyde and then permeabilized with Triton X-100. After blocking with 0.8% bovine serum albumin (BSA) and incubating with the indicated primary antibodies at 4°C rotating overnight, the samples were washed with 0.8% BSA and incubated with secondary antibodies (conjugated to FITC/TRITC) for 2 h at room temperature in the dark. After washing and embedding in DAPI, the samples were observed under an Olympus FV1000-IX81 confocal microscope.
FRAP
Live-cell microscopy was performed at 37°C with 5% CO2 under a Leica SP5 inverted confocal microscope (488 nm argon laser line, 63 × objective). Images of the selected regions of interest (ROI: 2 × 2 mm) were captured and quantified (2 s intervals for 180 s) following the bleach. The fluorescence signals measured in the ROI were normalized to the pre-bleach signals.
MNase sensitivity assay
Cell pellets were lysed in buffer A (10 mM HEPES pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M Sucrose, 10% Glycerol, 1mM DTT, 0.1% Triton X-100) for 8 min on ice. The nuclei were pelleted and digested with 10 U/ml MNase (Takara) in digestion buffer (10 mM Tris·HCl pH 7.5, 1 mM NaCl, 3 mM MgCl2, 1 mM CaCl2) for 3 min at 37°C. Genomic DNA was purified using a DNA purification kit (QIAGEN) and separated by 1.2% agarose gel electrophoresis. DNA bands were stained with ethidium bromide and visualized under a Gel Doc XR+ system (Bio-Rad).
Sedimentation experiments
Sedimentation experiments were performed as previously described (5). Briefly, HIS-tagged WT H1.4, H1.4K85Q or H1.4K85R were separately added to the reconstituted nucleosome arrays. Sedimentation experiments were performed using a Beckman Coulter ProteomeLab XL-I using a 4-hole An-60Ti rotor. The data were analyzed and the sedimentation coefficients were calculated. The average sedimentation coefficients were determined at the boundary midpoint.
Colony formation assay
Cells were cultured in 6-well plates for 24 h and then treated with or without etoposide (10 μM) for 2 h. The cells were then washed five times with PBS and re-cultured in fresh medium for 2 weeks. Cells were stained with crystal violet and the number of colonies consisting of >50 cells were counted.
Statistics
Data were analyzed by Student’s t-test. A P < 0.05 was considered statistically significant (NS, P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001). At least three independent experiments were performed in all cases.
RESULTS
H1K85Q acetylation-mimic mutation promotes chromatin condensation
Although H1 lysine 85 (H1K85) may regulate chromatin condensation, how it is involved in this process remains largely unknown. Sequence alignment of different linker histone H1 isoforms and across different species showed that the globular domain (and K85) is highly conserved (Supplementary Figure S1A and B), suggesting that this site may have important, evolutionarily conserved functions. To investigate how H1K85 is involved in chromatin organization and H1 dynamics, we mutated H1K85 to prevent its possible modification by replacing lysine with an arginine (K85R), or to mimic acetylation by replacing lysine with glutamine (K85Q).
First, we performed an analytical ultracentrifugation assay on nucleosome arrays reconstituted with K85Q, K85R or wild-type (WT) H1 proteins, which directly reflects the condensation of 30-nm chromatin fiber in vitro. Surprisingly, we found that the sedimentation coefficient of the nucleosome arrays containing WT or K85R were similar but considerably smaller (by ∼3S) than the H1K85Q nucleosome array (Figure 1A and B). The ultracentrifugation experiments were repeated with an increased ratio of H1 to nucleosome, which produced similar results (Figure 1C), suggesting that H1K85Q increases the level of condensed 30-nm chromatin structure formation in vitro.
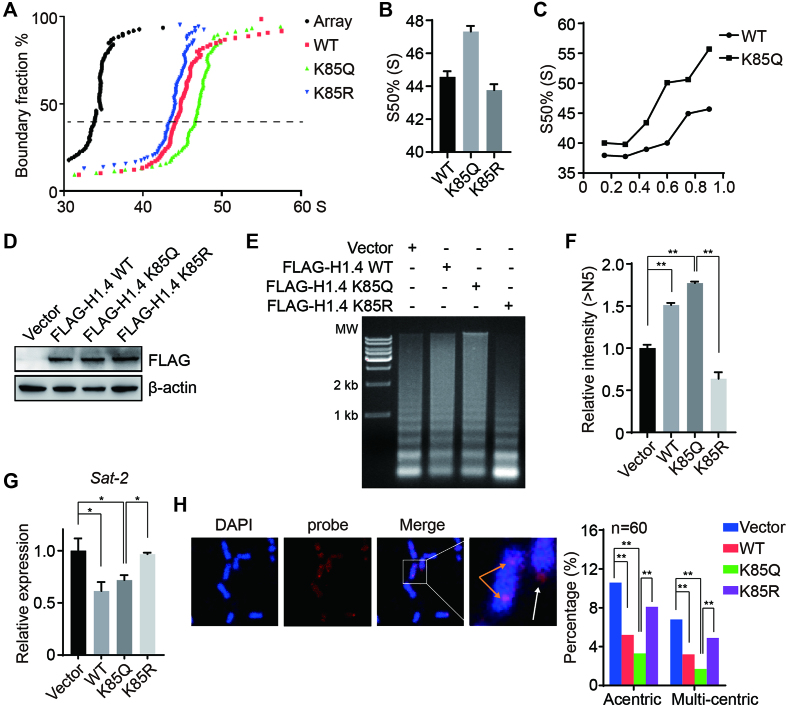
Figure 1.
Acetylation-mimic H1K85 mutations cause chromatin condensation. (A) Recombinant WT, K85Q or K85R HIS-tagged H1.4 was added to the reconstituted nucleosome array and subjected to in vitro analytical ultracentrifugation assay. A nucleosome array without H1.4 was used as a control. (B) Values from (A) at a boundary fraction of 50%. Data represent the means ± SD. (C) Sedimentation coefficient of the nucleosome array containing an increased ratio of H1 to nucleosome at a 50% boundary fraction. (D) H1.4 KO HeLa cells were stably-transfected with WT, K85Q and K85R H1.4 plasmids and whole cell lysates were analyzed by immunoblotting. (E) H1.4 KO HeLa cells were transfected with the indicated plasmids and chromatin fractions were extracted and analyzed by micrococcal nuclease (MNase) sensitivity assay. (F) Quantifications of lane signal intensities (upper bands, >N5) in (D). All data represent the means ± SD. (G) Sat-2 relative expression was measured by real-time PCR in H1.4 KO HeLa cells stably-transfected with WT, K85Q or K85R H1.4 plasmids. (H) H1.4 KO HeLa cells were stably transfected with the indicated plasmids and subjected to FISH. Metaphase chromosomes were labeled using a pan-centromere probe (red). Chromosomes were counterstained with DAPI (blue). The white arrow indicates acentric chromosomes, and the orange arrow indicates multiple-centric chromosomes. The centromere aberrances were statistically analyzed (n = 50).
To determine the role of the H1K85Q mutation in maintaining higher order chromatin structure in vivo, we stably transfected an empty vector, WT, K85Q or K85R H1.4 expression constructs into H1.4 knockout (KO) HeLa cells (Figure 1D). Expression of WT or mutant H1.4 plasmids in HeLa cells showed that they could be efficiently incorporated onto chromatin (Supplementary Figure S1C). Here, we found that K85Q cells were more resistant to micrococcal nuclease (MNase) digestion than WT or K85R cells (Figure 1E and F), indicative of a more condensed chromatin structure in K85Q cells. To verify whether the H1K85Q mutation leads to abnormal genome stability and integrity, we measured the transcript levels of centromeric satellite repeats Sat-2, which is a marker of heterochromatin structure. Consistent with previous results (36), over-expression of WT H1.4 led to decreased Sat-2 expression (Figure 1G) that was comparable with K85Q cells. Conversely, the K85R mutation did not repress Sat-2 expression (Figure 1G), suggesting that K85Q and WT H1 compact chromatin, but H1K85R may loosen chromatin. Fluorescence in situ hybridization (FISH) using human-specific pan-centromere peptide nucleic acid probes confirmed the structural chromosomal changes. Here, vector-only or K85R cells displayed more severe centromere defects than WT and K85Q cells (Figure 1H). Together, these data unexpectedly reveal that an acetylation-mimic mutation of H1K85 (H1K85Q) compacts higher order chromatin structure both in vitro and in vivo.
H1K85Q acetylation-mimic mutation alters H1 dynamics and promotes H1 binding to core histones
We next dissected how the H1K85Q mutation alters chromatin structure. As H1 regulates chromatin structure through its dynamic binding to chromatin, we proposed that the H1K85 mutation may alter the H1 binding dynamics. To verify this hypothesis, we used fluorescence recovery after photobleaching (FRAP) to measure the dynamic binding of H1 to chromatin. GFP-tagged WT, K85Q or K85R H1.4 expression constructs were transfected into H1.4 KO cells; these mutations had no detectable influence on the expression or subcellular localization of H1.4 (Supplementary Figure S1D). Surprisingly, WT H1.4 and H1.4K85R exhibited similar recovery kinetics whereas the recovery speed of H1.4K85Q was significantly slower than either H1.4K85R or WT H1.4 (Figure 2A). Similar results were obtained by FRAP for GFP-tagged H1.2 and its corresponding mutants (Figure 2B).
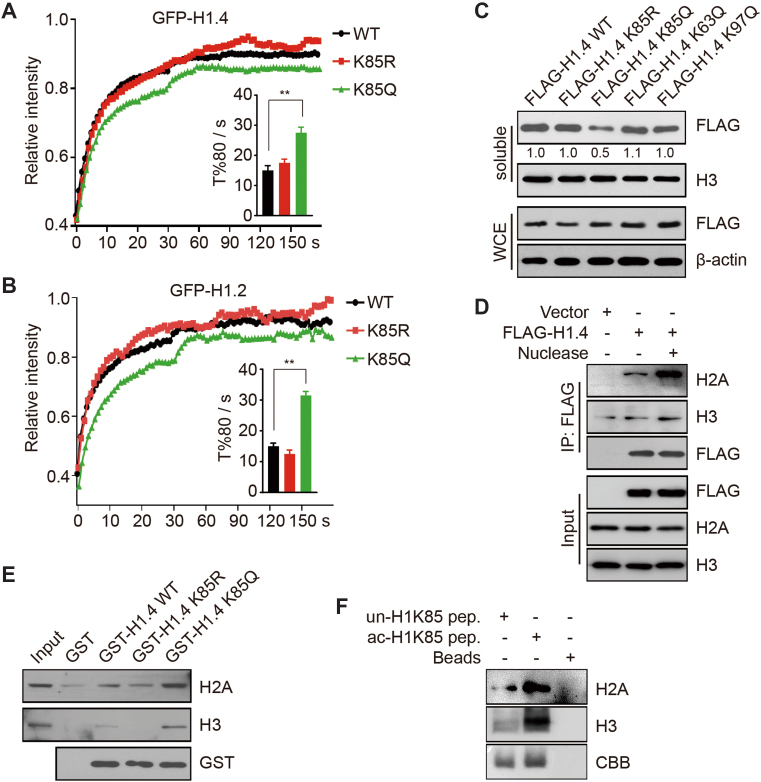
Figure 2.
Acetylation-mimic H1K85 mutation alters H1 dynamics and promotes H1 binding to core histones. (A and B) HCT116 cells were transfected with WT, K85R and K85Q GFP-H1.4 or GFP-H1.2 and subjected to FRAP. T%80 indicates the time when the fluorescent signals recovered to 80% of the original intensity after bleaching. All data represent the means ± SD. (C) HCT116 cells were transfected with the indicated plasmids and chromatin-bound proteins were extracted by salt extraction. Whole cell extracts and soluble proteins were analyzed by immunoblotting. The relative band intensity from three independent immunoblots are shown, relative to FLAG-H1.4 WT which was normalized to 1. (D) HCT116 cells were transfected with FLAG-H1.4 or an empty-vector. Cell extracts were immunoprecipitated using FLAG-conjugated M2 beads with or without nuclease and then analyzed by immunoblotting. (E) WT, K85R and K85Q GST-H1.4 or GST alone were incubated with recombinant H3 or H2A and analyzed by in vitro binding assay. (F) Biotin-tagged unmodified K85 or acetylated K85 peptides were incubated with recombinant H3 or H2A and analyzed by in vitro binding assay.
To confirm these results, we performed a salt extraction assay using high concentration sodium chloride to measure the binding dynamics of WT and H1 mutants. Here, proteins with higher chromatin binding affinity tend to be less readily extracted into the soluble fraction. Consequently, FLAG-tagged H1.4K85Q was extracted ∼2-fold less than FLAG-tagged WT, K85R and other mutant expression constructs (Figure 2C), which further suggests that K85Q increases H1 binding affinity to chromatin. These data indicate that the acetylation-mimic mutant H1K85Q decreases H1 mobility and increases H1 affinity to chromatin in vivo.
Histone acetylation decreases DNA–histone interactions. As such, we hypothesized that H1K85Q may promote H1 binding to chromatin by facilitating H1 interaction with core histones. To prove this hypothesis, we performed a co-IP (co-immunoprecipitation) assay to detect the interaction between H1 and core histones. The interaction between H1.4 and core histones H3 and H2A markedly increased when the nucleosomal DNA was digested by nuclease (Figure 2D); these data indicate that the association between H1 and core histones increases when H1 detaches from DNA. To further confirm that H1 directly interacts with core histones instead of nucleosomal DNA, we performed an in vitro pull-down assay using H1 mutants and recombinant core histones. While H1 interacted with core histones, H1.4K85Q exhibited stronger binding than either WT H1.4 or H1.4K85R (Figure 2E and Supplementary Figure S1E). At last, we performed a peptide pull-down assay using biotin-tagged unmodified or acetylated H1K85 peptides and found that acetylated H1K85 peptide bound more strongly to H3 and H2A than unmodified peptide (Figure 2F). Together, these data suggest that the acetylation-mimic mutant H1K85Q regulates H1 dynamic binding to chromatin by promoting the interaction between H1 and core histones.
H1K85 acetylation is dynamically regulated in response to DNA damage
To explore the biological functions of the H1K85Q mutation in vivo, we postulated that H1K85 may be regulated by acetylation in human cells. To test this hypothesis, we generated H1K85ac or H1K63ac (histone H1 lysine 63 acetylation)-specific antibodies for comparison, as H1K63 has minimal effects on H1 dynamics and chromatin structure (12). Slot blot and peptide competition assay demonstrated that these antibodies were specific (Supplementary Figure S2A and B). Similarly, H1K85ac or H1K63ac antibodies could not recognize K85 or K63 mutants, respectively (Supplementary Figure S2C and D), further confirming the specificity of these two antibodies. In addition, we verified that different H1 variants were acetylated at K85 site (Supplementary Figure S2E), suggesting that H1K85ac is a ubiquitous, but not variant-specific acetylation mark. By immunoblotting, we detected H1K85ac in various human cancer cell lines and mouse tissues (Figure 3A and Supplementary Figure S2F). We then investigated whether H1K85ac was involved in chromatin-based processes, such as transcription. Chromatin immunoprecipitation-sequencing (ChIP-seq) of HCT116 cells showed that H1K85ac largely bound intergenic regions (53%) and gene bodies (39%), whereas only 7% H1K85ac bound regions 2 kb upstream of the transcription start sites (Supplementary Figure S2G). As the proportion of H1K85ac bound to potential promoter regions was not significantly different to that of the random peaks (∼6%), these data suggest that H1K85ac may not be mainly involved in general transcriptional regulation. By comparison, ∼40% H1K34ac is enriched at promoters, and thus is proposed to be an active transcriptional regulator (37).
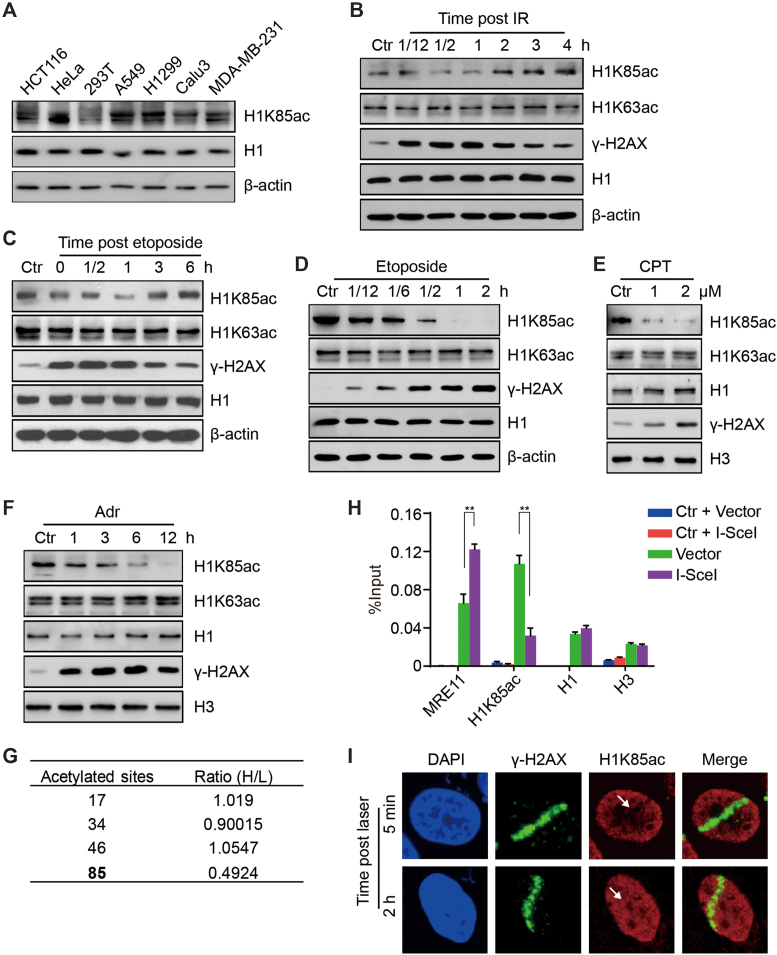
Figure 3.
Acetylation of H1K85 is dynamically regulated in response to DNA damage. (A) Protein from different cancer cell lines was extracted and whole cell lysates were analyzed by immunoblotting. (B and C) HCT116 cells were treated with 10 Gy IR or etoposide (40 μM) for 1 h and released for the indicated times and whole cell lysates were analyzed by immunoblotting. (D) HCT116 cells were treated with etoposide (40 μM) for the indicated times and whole cell lysates were analyzed by immunoblotting. (E and F) HCT116 cells were treated with CPT for 12 h at the indicated concentrations or adriamycin (Adr, 1 μM) for the indicated times and histones were analyzed by immunoblotting. (G) H1 acetylation sites identified by SILAC. The relative ratio of the adriamycin treated group (H) to the control group (L) is shown. (H) DR-GFP U2OS cells were transfected with I-SceI endonuclease or an empty vector and subjected to ChIP with the indicated antibodies. Ctr indicates a specific genomic locus distal from the I-SceI cut site and was used as a negative control. MRE11 was used as a positive control. All data represent the means ± SD. (I) HCT116 cells were micro-irradiated and analyzed by immunofluorescence at 5 min or 2 h post-irradiation. The white arrow indicates the irradiation path.
To investigate whether H1K85ac is involved in the cellular response to DNA damage, we treated cells with ionizing radiation (IR). Intriguingly, H1K85ac, but not H1K63ac, immediately reduced after IR exposure and they reached the lowest levels 1 h post IR (Figure 3B and Supplementary Figure S3A). H1K85ac gradually restored to normal levels after 1 h post IR, which may indicate completion of DNA repair (Figure 3B and Supplementary Figure S3A). HeLa cells were also treated with etoposide (a topoisomerase II inhibitor and DNA damage inducer) and then released to drug-free medium for different time intervals. Here, H1K85ac exhibited the same patterns of dynamic change as observed with IR (Figure 3C). In addition, persistent exposure to etoposide resulted in reduced H1K85ac levels in a time-dependent and dose-dependent manner (Figure 3D and Supplementary Figure S3B). DNA damage induced by other genotoxic reagents, CPT and adriamycin (Adr), also led to decreased H1K85ac levels (Figure 3E and F). At last, stable isotope labeling with amino acids in cell culture (SILAC) followed by quantitative MS of HeLa cells treated with adriamycin (Supplementary Figure S3C) showed that H1K85ac was markedly reduced after adriamycin treatment, whereas acetylation of many other H1 lysine residues remained virtually unchanged (Figure 3G). More importantly, MS did not detect other types of modifications at H1K85 site, suggesting that H1K85ac is specific (Supplementary Figure S3D).
To further confirm the reduced H1K85ac levels on chromatin, we used a U2OS DR-GFP (direct repeat green fluorescent protein) cell line and the endonuclease I-SecI to induce a targeted, single DNA damage site. ChIP analysis around the I-SecI site showed that H1K85ac enrichment decreased upon induction of DNA damage by I-SecI, whereas the levels of total H1 and core histones remained largely unaltered (Figure 3H). We also exposed HeLa cells to laser micro-irradiation and analyzed H1K85ac by immunofluorescence. Here, H1K85ac immediately decreased in the DNA damage path 5 min after micro-irradiation and recovered 2 h later (Figure 3I). In addition, we explored whether H1K85ac could be directly involved in regulating DDR and repair. We showed that overexpression of FLAG-tagged WT H1.4 or its K85 mutants did not markedly influence DNA damage-induced Ataxia-telangiectasia mutated (ATM) activation or loading of early DDR factors, such as ATM or the KU complex (Supplementary Figure S3E and F). Interestingly, overexpressing WT H1.4 did not affect cellular activities of homologous recombination (HR) and non-homologous end joining (NHEJ)-mediated DNA repair, whereas overexpression of H1.4 K85 mutants led to decreased HR and NHEJ efficiencies (Supplementary Figure S3G and H). Altogether, these data show that H1K85ac is dynamically regulated in response to DNA damage and that persistent DNA damage reduces H1K85ac. This effect may be associated with chromatin relaxation and DNA repair after DNA damage.
PCAF acetylates H1K85 acetylation in vivo and in vitro
Next, we sought to identify the HATs responsible for H1K85ac. We transfected several typical HATs, including p300, MOF, PCAF and TIP60, into HCT116 cells and found that H1K85ac was specifically up-regulated in the presence of over-expressed PCAF, a GNAT family member (Figure 4A and Supplementary Figure S4A). Overexpression of another GNAT family member, GCN5, showed little effects in H1K85ac levels (Supplementary Figure S4B). To confirm PCAF-dependent regulation of H1K85ac, we separately transfected WT and enzyme activity dead (E570A/D610A, ED) PCAF into HCT116 cells and found that WT PCAF increased H1K85ac whereas ED PCAF had no effect (Figure 4B). Anacardic acid, a specific PCAF inhibitor, markedly downregulated H1K85ac compared to control, thus further supporting that PCAF is a specific H1K85 acetyltransferase (Supplementary Figure S4C).
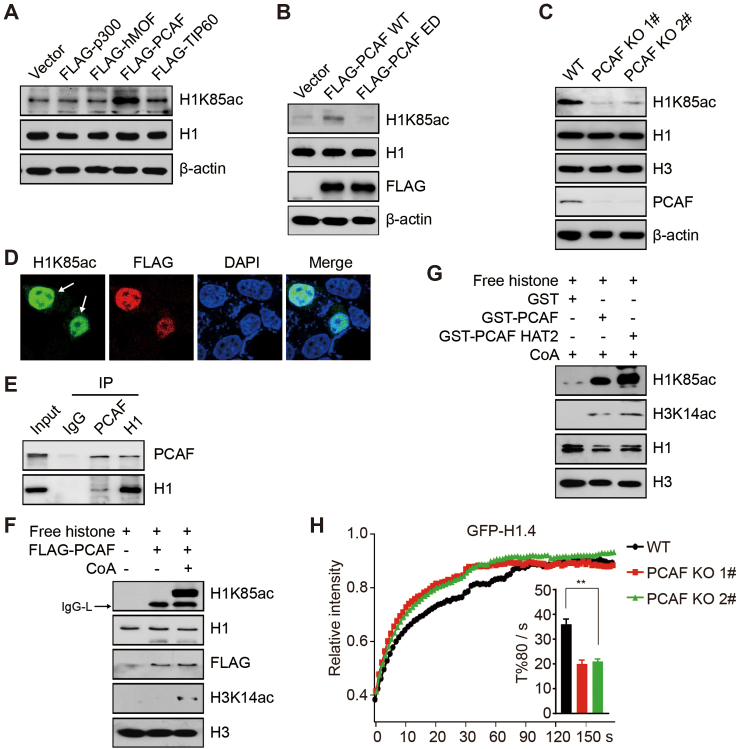
Figure 4.
PCAF acetylates H1K85 acetylation in vivo and in vitro. (A and B) HCT116 cells were transfected with the indicated plasmids and whole cell lysates were analyzed by immunoblotting. (C) WT and PCAF KO (1# and 2#) HCT116 cells were analyzed by immunoblotting. (D) PCAF KO (1#) cells were transfected with FLAG-PCAF and analyzed by immunofluorescence. The arrows indicate PCAF KO cells with overexpressed FLAG-PCAF plasmids. (E) HCT116 cells lysates were immunoprecipitated using an anti-IgG, anti-PCAF or anti-H1 antibody and analyzed by immunoblotting. (F) FLAG-PCAF was purified from HEK293T cells and subjected to in vitro acetylation assay with or without acetyl-CoA using free histones as substrates. IgG-L indicates the light chain of IgG. (G) GST, GST-PCAF or GST-PCAF HAT2 was purified from Escherichia coli and subjected to in vitro acetylation assay using free histones as substrates. (H) PCAF KO (1#, 2#) and WT cells were transfected with GFP-H1.4 and subjected to FRAP analysis. All data represent the means ± SD.
We next generated PCAF KO HCT116 cells using clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 technology (Supplementary Figure S4D). Here, H1K85ac levels were significantly reduced in these KO cells (Figure 4C). We then reintroduced a FLAG-tagged PCAF plasmid into the PCAF KO cells and found by immunofluorescent assay that H1K85ac signal intensity increased upon re-introduction of PCAF (Figure 4D). A co-IP assay using endogenous antibodies or ectopically expressed FLAG-tagged PCAF confirmed an interaction between PCAF and histone H1 in vivo (Figure 4E and Supplementary Figure S4E). Moreover, we found that PCAF is gradually recruited to chromatin in response to DNA damage (Supplementary Figure S4F), indicating that recovery of H1K85ac is dependent on PCAF.
We then established an in vitro acetylation assay to verify whether PCAF is truly responsible for H1K85ac. Here, both recombinant or immunoprecipitated full-length PCAF could catalyze H1K85ac in the presence of acetyl-coenzyme A (Figure 4F and Supplementary Figure S4G). The catalytic core of PCAF that contains a specific HAT2 domain (amino acids 352–658) was sufficient to acetylate H1K85ac in vitro (Figure 4G). Other HATs, including p300 and TIP60 exhibited low activity toward H1K85ac (Supplementary Figure S4H and I). At last, FRAP demonstrated that H1 bound less tightly to chromatin in PCAF KO cells than in WT cells (Figure 4H and Supplementary Figure S4J). Taken together, these results suggest that PCAF is the predominant HAT to acetylate H1K85.
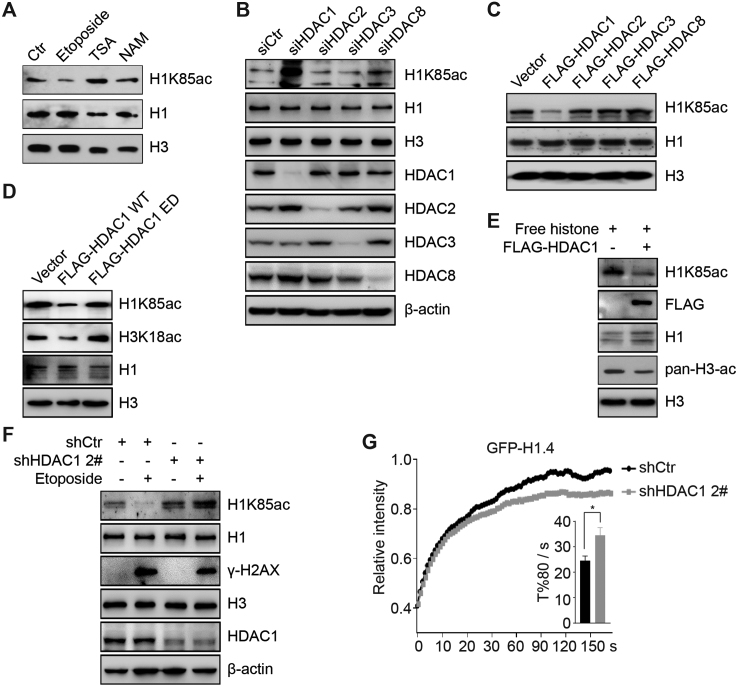
HDAC1 deacetylates H1K85 acetylation in vivo and in vitro
To explore how H1K85ac is reduced in the early response to DNA damage, we aimed to identify the responsible H1K85 deacetylase. Cellular exposure to Trichostatin A (a broad class I and II HDAC inhibitor) but not nicotinamide (a class III HDAC inhibitor) dramatically induced H1K85ac, indicating that H1K85ac might be regulated by class I or II but not class III HDACs (Sirtuins) (Figure 5A). To further support this hypothesis, we individually knocked down the nucleic class I HDACs, including HDAC1, HDAC2, HDAC3 and HDAC8, using siRNA in HCT116 cells. Knockdown of HDAC1, but not HDAC2, 3 or 8, resulted in a significant increase in H1K85ac levels compared to control (Figure 5B). Meanwhile, over-expression of HDAC1 led to decreased H1K85ac levels, whereas over-expression of other class I HDACs had minimal effects (Figure 5C).
Figure 5.
HDAC1 deacetylates H1K85 in vivo and in vitro. (A) HCT116 cells were treated with etoposide (40 μM) for 2 h, TSA (1 μM) for 12 h or NAM (5 mM) for 12 h. Histone was extracted and analyzed by immunoblotting. (B) HCT116 cells were transfected with the indicated siRNAs and whole cell extracts were analyzed by immunoblotting. (C and D) HCT116 cells were transfected with the indicated plasmids and histone was analyzed by immunoblotting. (E) FLAG-HDAC1 was purified from HEK293T cells and subjected to in vitro deacetylation assay using free histones as substrates. The reaction was analyzed by immunoblotting. (F) Stable HDAC1 knockdown (shHDAC1, 2#) or control cells (shCtr) were treated with etoposide and whole cell extracts were analyzed by immunoblotting. (G) Stable HDAC1 knockdown (shHDAC1, 2#) or control cells (shCtr) were transfected with GFP-H1.4 and analyzed by FRAP. All data represent the means ± SD.
We then generated an enzyme activity dead (H141A, ED) HDAC1 construct, to further identify the relationship between HDAC1 and H1K85ac. Over-expression of WT HDAC1 reduced H1K85ac levels compared to ED HDAC1, which had a minimal effect (Figure 5D). In addition, an in vitro deacetylation assay to verify the direct role of HDAC1 in deacetylating H1K85ac showed that HDAC1 potently deacetylated H1K85ac in vitro (Figure 5E). Other class I HDACs, although displayed strong activity toward histone H3, were unable to deacetylate H1K85ac in vitro (Supplementary Figure S5A–C).
At last, H1K85ac reduction in response to DNA damage was abrogated in HDAC1 stable knockdown HCT116 (shHDAC1) cells (Figure 5F and Supplementary Figure S5D), suggesting that HDAC1 helps regulate H1K85ac reduction in response to DNA damage. These results were confirmed by FRAP, whereby the mobility of GFP-tagged H1 decreased in shHDAC1 compared to control cells (Figure 5G). Together, these data suggest that HDAC1 deacetylates H1K85ac in vivo and in vitro.
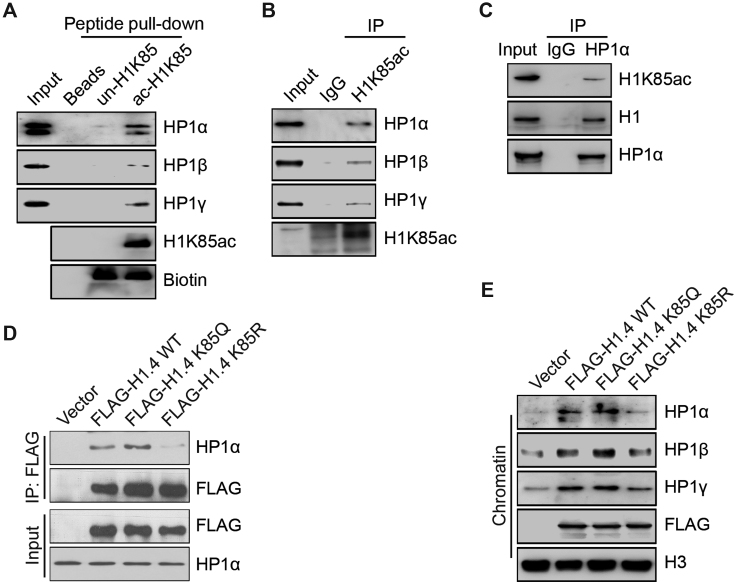
H1K85 acetylation promotes the recruitment of heterochromatin protein 1 (HP1)
To better understand how H1K85ac promotes chromatin condensation in vivo, we studied whether H1K85ac influences the recruitment of other chromatin regulators. Peptide pull-down of biotin-labeled acetylated H1K85 peptides followed by MS identified HP1 as a specific interacting protein with acetylated H1K85 (Supplementary Figure S6A). Consistently, MS also identified core histones to specifically interact with acetylated H1K85 peptides (Supplementary Figure S6A), and acetylated H1K85 interacted with all HP1 isoforms, including HP1α, HP1β and HP1γ in vitro (Figure 6A). Next, we confirmed these findings by co-IP assays in cancer cells using H1K85ac-specific and HP1α antibodies (Figure 6B and C). Other chromatin condensing factors, such as SUV39H1 and SMC1 (structural maintenance of chromosomes protein 1), did not interact with H1K85ac (Supplementary Figure S6B). These results suggest that H1K85ac may specifically assist HP1 recruitment to chromatin.
Figure 6.
H1K85 acetylation promotes the recruitment of heterochromatin protein 1 (HP1). (A) Chromatin-bound proteins of HCT116 cells were extracted and incubated with biotin-tagged unmodified K85, acetylated K85 peptide or beads alone and analyzed by in vitro peptide pull-down assay. (B and C) Chromatin-bound proteins of HCT116 cells were extracted and then immunoprecipitated using the indicated antibodies for co-IP and then analyzed by immunoblotting. (D) WT, K85Q and K85R FLAG-H1.4 or empty-vector alone were transfected into HeLa cells. Cell extracts were immunoprecipitated using FLAG-conjugated M2 beads and then analyzed by immunoblotting. (E) WT, K85Q and K85R FLAG-H1.4 or empty-vector were transfected into HeLa cells. Chromatin-bound proteins were extracted and analyzed by immunoblotting.
To test whether H1K85ac functions through HP1 to regulate chromatin structure, we performed a co-IP with WT, K85Q or K85R H1.4 expression constructs to assess the interaction between H1 and HP1. Here, the interaction between HP1 and K85Q H1 was stronger than the interactions between HP1 and WT or K85R H1 (Figure 6D). In addition, chromatin fractionation demonstrated that H1 over-expression promoted HP1 enrichment onto chromatin: K85Q H1 was more effective at promoting HP1 than WT H1, whereas K85R H1 repressed HP1 recruitment (Figure 6E). The chromatin levels of SUV39H1 and SMC1 were unaffected by over-expression of H1 or its mutants (Supplementary Figure S6C). Together, these results suggest that H1K85ac recruits HP1 and H1K85ac dynamics are associated with the dynamic mobilization of HP1 upon DNA damage.
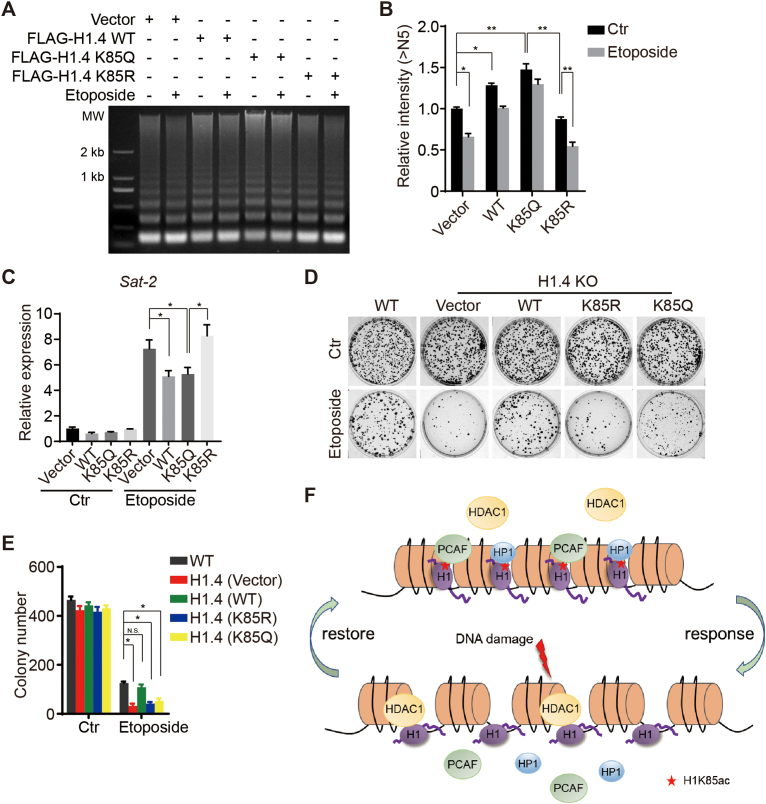
H1K85 acetylation regulates genome stability and cell survival in response to DNA damage
To further explore the biological relevance of H1K85ac and its dynamics in response to DNA damage in cancer cells, we investigated whether H1K85ac participates in the regulation of dynamic chromatin condensation upon DNA damage. To verify this hypothesis, we performed an MNase sensitivity assay and found that DNA damage-induced chromatin decondensation was markedly repressed in H1.4 KO cells reintroduced with K85Q H1, compared to more relaxed chromatin upon DNA damage in cells reintroduced with K85R H1 (Figure 7A and B). In addition, we performed real-time PCR assays to evaluate the Sat-2 expression levels upon DNA damage. Here, Sat-2 levels in H1.4 WT and K85Q cells were significantly decreased upon receipt of DNA damaging stimuli (Figure 7C), indicating that the relaxing of heterochromatin after DNA damage was impeded in K85Q H1 reintroduced cells. Together, these data indicate that H1K85ac leads to chromatin condensation, and decreasing H1K85ac results in chromatin decompaction in response to DNA damage.
Figure 7.
H1K85 acetylation regulates genome stability and cell survival in response to DNA damage. (A) HCT116 cells were transfected with the indicated plasmids and treated with etoposide (40 μM) for 2 h. Cells were extracted and analyzed by MNase sensitivity assay. (B) Quantifications of lane signal intensity (upper bands, >N5) in (A). All data represent the means ± SD. (C) Relative expression of Sat-2 in H1.4 KO cells stably-transfected with vector, WT, K85Q or K85R H1.4 after etoposide treatment (20 μM) for 4 h, determined by real-time PCR. (D and E) HCT116 cells were stably transfected with the indicated plasmids and subjected to colony formation assay after etoposide treatment (10 μM) for 2 h. All data represent the means ± SD. (F) Model of H1K85ac-mediated regulation of chromatin structure in the context of DNA damage. H1K85ac is balanced by PCAF and HDAC1. H1K85ac compacts chromatin by tethering H1 to core histones and recruiting HP1. In response to DNA damage, H1K85ac rapidly decreases, leading to reduced H1 binding affinity to chromatin and reduced enrichment of HP1, resulting in chromatin decondensation. Upon completion of DNA repair, H1K85ac levels increase and permit the restoration of chromatin structure.
Failure to create an open chromatin environment (i.e. persistently condensed chromatin) or restore a condensed silencing chromatin state (i.e. persistently relaxed chromatin) after DNA repair results in genome instability and cell death (38). Interestingly, we found that survival of H1.4 KO cells exposed to DNA damage stress was impaired but reintroduction of WT H1.4 could rescue this defect (Figure 7D and E). This finding indicates the critical functions of H1.4 in regulating genome stability. However, stable transfection of either H1.4K85R or H1.4K85Q failed to restore defective survival in H1.4 KO cells upon DNA damage (Figure 7D and E), suggesting that both reduction and restoration of H1K85ac is crucial in regulating DNA damage-induced alteration of chromatin structure. As both the K85R and K85Q mutation block the dynamic changes of H1K85ac, these results demonstrate that H1K85ac is a key regulator of genome stability during the DDR and repair.
DISCUSSION
In this study, we demonstrate that the acetylation of H1K85 is a critical regulator of chromatin structure. H1K85ac is balanced by PCAF and HDAC1, which mediate chromatin dynamics in response to DNA damage. Therefore, we propose a model whereby H1K85ac facilitates H1 binding to core histones and HP1 recruitment to promote chromatin condensation, whereas DNA damage-induced dynamic changes of H1K85ac permits chromatin relaxation and restoration to ensure genome stability (Figure 7F).
The H1 globular domain is a pivotal determinant of H1 binding to chromatin. Although the C-terminal domain was previously considered a primary factor, it has been accepted that the globular domain and C terminus cooperate together to regulate H1 binding to chromatin (1). Mutation of a single site within the globular domain of H1 can markedly affect its binding affinity to chromatin. In this study, both in vitro and in vivo assays demonstrated that a K85Q mutation decreased H1 mobility and condensed chromatin to a higher degree than a K85R mutation or WT H1. Mutating two other lysine residues within the globular domain, K63 or K97, had a minimal effect on H1 binding, indicating the specificity of K85 perhaps due to its distinct structural localization or dynamic modification status. However, some in vivo experiments also showed that K85Q mutation condensed chromatin as much as WT H1, whereas the K85R mutation caused chromatin loosening. This effect may be explained by the differential abundance of endogenous H1K85ac under different circumstances and the distinct localization of H1K85ac in specific genomic loci, such as the centromeric regions.
Histone modifications are key regulators of chromatin structure. For example, histone H3 lysine 9 trimethylation (H3K9me3) by SUV39H1 recruits a repressive complex containing HP1 and marks the formation of heterochromatin (39–41). Histone H4 lysine 16 acetylation (H4K16ac) inhibits the formation of higher order chromatin structure in vitro (27). In this study, we identified H1K85ac as a critical factor in regulating genome stability and chromatin structure. Differing from the traditional view that acetylation neutralizes the positive charge of histones and leads to decreased histone–DNA binding and chromatin relaxation (28,42), we found that H1K85ac condenses higher order chromatin structure. As H1 is not located within the octamer and exhibits complex binding modes to the nucleosome, we proposed that H1K85ac may increase H1 binding to chromatin or other regulatory factors, hence resulting in chromatin compaction. We proved this hypothesis by showing that the K85Q mutation and acetylated H1K85 peptides promoted the interaction between H1, core histones and HP1. These results are further supported by the notion that nucleosome stability is not only regulated by histone–DNA interactions, but also by histone–histone and histone–chaperone interactions (17). For instance, H3K56 acetylation increases the binding affinity of the histone chaperone complex chromatin assembly factor 1 (Caf1), which promotes histone deposition and nucleosome assembly (43). Although H1 interacts with SUV39H1 and promotes its recruitment in Drosophila (44), our assays did not find that H1K85ac was involved in SUV39H1 recruitment. Interestingly, previous studies have shown that H1-mediated HP1 recruitment is influenced by a modification to the H1 N terminus, such as lysine 26 (K26) methylation or serine 27 (S27) phosphorylation (22). Here, we provide the first evidence that HP1 enrichment onto chromatin is also regulated by modification of the H1 globular domain, suggesting that both the H1 N terminus and globular domain contribute to HP1 recruitment.
In this study, we also report that H1K85ac exhibits differential dynamic patterns when exposed to IR or other DNA damaging reagents. A plausible explanation for this effect is that IR-mediated damage allows the cells to recover and repair once the exposure is complete, whereas chemical reagents can cause persistent DNA damage. We demonstrated this point by showing equivalent dynamics of H1K85ac between IR treatment and withdrawal of etoposide. In addition, these H1K85ac dynamics are consistent with DNA damage-induced chromatin mobilization. The classic ‘access–repair–restore’ model implies that chromatin is actively decondensed upon DNA damage to facilitate the recruitment of DNA repair factors, after which the chromatin is restored (30,45). Dynamic histone modifications, such as H4K16ac and H3K9me3, are key factors controlling chromatin structure in the DDR and repair (46). We are the first to show that dynamic H1 acetylation is an additional, critical regulator of this process. Moreover, we show that H1K85ac condenses chromatin, whereas K85Q and K85R mutations abrogate H1K85ac dynamics. Therefore, both the K85R mutation (which could not stabilize chromatin organization as WT H1) and the K85Q mutation (which failed to relax chromatin fibers) can lead to impaired cell survival in response to DNA damage. Our results are further supported by the theory that failure to establish an accessible chromatin environment or to restore chromatin compaction leads to defective chromatin functions, such as transcription (47,48).
H1 isoforms share significant sequence homology, especially within the conserved globular domain (49). Therefore, it is difficult to delete or mutate a specific H1 site in human cells, unlike in Drosophila or yeast that expresses a single H1 variant. In addition, the structural model of H1 binding to the nucleosome is different in Drosophila, chicken or yeast compared to human cells (5,12,13,50). Moreover, H1.4 and H1.5, which are almost identical H1 variants, are proposed to bind to the nucleosomes in distinct manners (5,51,52). These reports suggest that H1–nucleosome binding patterns are complicated in human cells. We showed that H1K85 is highly conserved and H1K85ac is present in almost all H1 variants. Mutation of K85 also affected the chromatin binding of different H1 variants. Furthermore, we did not detect other types of modification at H1K85, strengthening the specificity of H1K85ac. Therefore, it is reasonable to propose that H1K85ac is a conserved H1 modification but not variant-specific. To characterize the biological functions of H1K85ac, we used H1.4 for deletion and mutation analysis, which is one of the most abundant H1 variants in cancer cells and has the strongest chromatin-condensing effects (53). The fact that H1.4 binds off the nucleosome dyad and does not interact with linker DNA in any specific manner supports our model that H1K85ac promotes H1 interaction with core histones (5).
HDAC1 and PCAF regulate the acetylation status of core histones and various non-histone proteins, but their association with H1 has not been previously reported. We found that H1K85ac is specifically regulated by HDAC1 and PCAF in vivo and in vitro. Although HDAC1 and HDAC2 shared significant sequence homology, our results showed that only HDAC1 is responsible for H1K85 deacetylation. This specificity may be explained by their different structural organization, binding partners or genomic localization. For example, HDAC1 and HDAC2 exhibit distinct genomic distribution patterns (54). HDAC1 and HDAC2 also show unique functions in stem cell differentiation or memory formation (55,56). In addition, HDAC1 is instantly recruited to chromatin upon DNA damage (57), consistent with our observation that H1K85 undergoes rapid deacetylation in response to DNA damage. We also showed that GCN5, a family member of PCAF, could not acetylate H1K85ac, which may be due to their diverse complex formation with different subunits (58,59). Interestingly, we found that PCAF is gradually recruited to chromatin during DNA damage repair, which is supported by a recent report that PCAF is recruited to UV-induced DNA damage sites (60). These data suggest that HDAC1 and PCAF are responsible for DNA damage-induced H1K85ac dynamics.
This study presents a novel model by which acetylation of lysine 85 within the globular domain of H1 promotes the condensation of higher order chromatin structure, which acts to preserve genome stability and cell survival. Histone modifications and histone regulators are promising targets to treat human cancers (61,62). Although dedicated studies are still warranted for further demonstration, our findings may provide the first mechanistic insight to target H1 acetylation as a cancer therapy.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Guohong Li (Chinese Academy of Sciences, China) for helping the in vitro 30-nm chromatin assays and Dr. Yawen Bai (National Institute of Health, USA) for critical comments on the manuscript. The authors also appreciate Dr. Ye Zhang (Chinese Academy of Medical Sciences, China), Dr. Jianyuan Luo (Peking University, China), Dr. Jiadong Wang (Peking University, China) and Dr. Zhijie Chang (Tsinghua University, China) for sharing the HAT and HDAC plasmids. Finally, the authors thank Dr. Jessica Tamanini (Shenzhen University) for proofreading the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key R&D Program of China [2017YFA0503900]; National Natural Science Foundation of China [81621063, 81530074, 31570812, 81720108027]; National Institutes of Health of USA [DK071900, CA129325, CA178765 to R.G.R]; Shenzhen Municipal Commission of Science and Technology Innovation [JCYJ20160427104855100, JCYJ20170818092450901]. Funding for open access charge: National Key R&D Program of China [2017YFA0503900].
Conflict of interest statement. None declared.
REFERENCES
- 1. Fyodorov D.V., Zhou B.R., Skoultchi A.I., Bai Y.. Emerging roles of linker histones in regulating chromatin structure and function. Nat. Rev. Mol. Cell Biol. 2018; 19:192–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hergeth S.P., Schneider R.. The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle. EMBO Rep. 2015; 16:1439–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fan Y., Nikitina T., Zhao J., Fleury T.J., Bhattacharyya R., Bouhassira E.E., Stein A., Woodcock C.L., Skoultchi A.I.. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005; 123:1199–1212. [DOI] [PubMed] [Google Scholar]
- 4. Bayona-Feliu A., Casas-Lamesa A., Reina O., Bernues J., Azorin F.. Linker histone H1 prevents R-loop accumulation and genome instability in heterochromatin. Nat. Commun. 2017; 8:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Song F., Chen P., Sun D., Wang M., Dong L., Liang D., Xu R.M., Zhu P., Li G.. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 2014; 344:376–380. [DOI] [PubMed] [Google Scholar]
- 6. Allan J., Hartman P.G., Crane-Robinson C., Aviles F.X.. The structure of histone H1 and its location in chromatin. Nature. 1980; 288:675–679. [DOI] [PubMed] [Google Scholar]
- 7. Goytisolo F.A., Gerchman S.E., Yu X., Rees C., Graziano V., Ramakrishnan V., Thomas J.O.. Identification of two DNA-binding sites on the globular domain of histone H5. EMBO J. 1996; 15:3421–3429. [PMC free article] [PubMed] [Google Scholar]
- 8. Duggan M.M., Thomas J.O.. Two DNA-binding sites on the globular domain of histone H5 are required for binding to both bulk and 5 S reconstituted nucleosomes. J. Mol. Biol. 2000; 304:21–33. [DOI] [PubMed] [Google Scholar]
- 9. Brown D.T., Izard T., Misteli T.. Mapping the interaction surface of linker histone H1(0) with the nucleosome of native chromatin in vivo. Nat. Struct. Mol. Biol. 2006; 13:250–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Catez F., Ueda T., Bustin M.. Determinants of histone H1 mobility and chromatin binding in living cells. Nat. Struct. Mol. Biol. 2006; 13:305–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stasevich T.J., Mueller F., Brown D.T., McNally J.G.. Dissecting the binding mechanism of the linker histone in live cells: an integrated FRAP analysis. EMBO J. 2010; 29:1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou B.R., Feng H., Kato H., Dai L., Yang Y., Zhou Y., Bai Y.. Structural insights into the histone H1-nucleosome complex. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:19390–19395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou B.R., Jiang J., Feng H., Ghirlando R., Xiao T.S., Bai Y.. Structural mechanisms of nucleosome recognition by linker histones. Mol. Cell. 2015; 59:628–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou B.R., Feng H., Ghirlando R., Li S., Schwieters C.D., Bai Y.. A small number of residues can determine if linker histones are bound on or off dyad in the chromatosome. J. Mol. Biol. 2016; 428:3948–3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Okosun J., Bodor C., Wang J., Araf S., Yang C.Y., Pan C., Boller S., Cittaro D., Bozek M., Iqbal S. et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet. 2014; 46:176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Christophorou M.A., Castelo-Branco G., Halley-Stott R.P., Oliveira C.S., Loos R., Radzisheuskaya A., Mowen K.A., Bertone P., Silva J.C., Zernicka-Goetz M. et al. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature. 2014; 507:104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tessarz P., Kouzarides T.. Histone core modifications regulating nucleosome structure and dynamics. Nat. Rev. Mol. Cell Biol. 2014; 15:703–708. [DOI] [PubMed] [Google Scholar]
- 18. Waldmann T., Schneider R.. Targeting histone modifications–epigenetics in cancer. Curr. Opin. Cell Biol. 2013; 25:184–189. [DOI] [PubMed] [Google Scholar]
- 19. Izzo A., Schneider R.. The role of linker histone H1 modifications in the regulation of gene expression and chromatin dynamics. Biochim. Biophys. Acta. 2016; 1859:486–495. [DOI] [PubMed] [Google Scholar]
- 20. Vaquero A., Scher M., Lee D., Erdjument-Bromage H., Tempst P., Reinberg D.. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell. 2004; 16:93–105. [DOI] [PubMed] [Google Scholar]
- 21. Nielsen A.L., Oulad-Abdelghani M., Ortiz J.A., Remboutsika E., Chambon P., Losson R.. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell. 2001; 7:729–739. [DOI] [PubMed] [Google Scholar]
- 22. Daujat S., Zeissler U., Waldmann T., Happel N., Schneider R.. HP1 binds specifically to Lys26-methylated histone H1.4, whereas simultaneous Ser27 phosphorylation blocks HP1 binding. J. Biol. Chem. 2005; 280:38090–38095. [DOI] [PubMed] [Google Scholar]
- 23. Hale T.K., Contreras A., Morrison A.J., Herrera R.E.. Phosphorylation of the linker histone H1 by CDK regulates its binding to HP1alpha. Mol. Cell. 2006; 22:693–699. [DOI] [PubMed] [Google Scholar]
- 24. Price B.D., D’Andrea A.D.. Chromatin remodeling at DNA double-strand breaks. Cell. 2013; 152:1344–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hauer M.H., Gasser S.M.. Chromatin and nucleosome dynamics in DNA damage and repair. Genes Dev. 2017; 31:2204–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bird A.W., Yu D.Y., Pray-Grant M.G., Qiu Q., Harmon K.E., Megee P.C., Grant P.A., Smith M.M., Christman M.F.. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002; 419:411–415. [DOI] [PubMed] [Google Scholar]
- 27. Shogren-Knaak M., Ishii H., Sun J.M., Pazin M.J., Davie J.R., Peterson C.L.. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006; 311:844–847. [DOI] [PubMed] [Google Scholar]
- 28. Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997; 389:349–352. [DOI] [PubMed] [Google Scholar]
- 29. Bannister A.J., Kouzarides T.. Regulation of chromatin by histone modifications. Cell Res. 2011; 21:381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Soria G., Polo S.E., Almouzni G.. Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol. Cell. 2012; 46:722–734. [DOI] [PubMed] [Google Scholar]
- 31. Gong F., Miller K.M.. Mammalian DNA repair: HATs and HDACs make their mark through histone acetylation. Mutat. Res. 2013; 750:23–30. [DOI] [PubMed] [Google Scholar]
- 32. Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu W.G., Hileman T., Ke Y., Wang P., Lu S., Duan W., Dai Z., Tong T., Villalona-Calero M.A., Plass C. et al. 5-aza-2′-deoxycytidine activates the p53/p21Waf1/Cip1 pathway to inhibit cell proliferation. J. Biol. Chem. 2004; 279:15161–15166. [DOI] [PubMed] [Google Scholar]
- 34. Cao L.L., Wei F., Du Y., Song B., Wang D., Shen C., Lu X., Cao Z., Yang Q., Gao Y. et al. ATM-mediated KDM2A phosphorylation is required for the DNA damage repair. Oncogene. 2016; 35:301–313. [DOI] [PubMed] [Google Scholar]
- 35. Chen Y., Colak G., Zhao Y.. SILAC-based quantification of Sirt1-responsive lysine acetylome. Methods Mol. Biol. 2013; 1077:105–120. [DOI] [PubMed] [Google Scholar]
- 36. Izquierdo-Bouldstridge A., Bustillos A., Bonet-Costa C., Aribau-Miralbes P., Garcia-Gomis D., Dabad M., Esteve-Codina A., Pascual-Reguant L., Peiro S., Esteller M. et al. Histone H1 depletion triggers an interferon response in cancer cells via activation of heterochromatic repeats. Nucleic Acids Res. 2017; 45:11622–11642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kamieniarz K., Izzo A., Dundr M., Tropberger P., Ozretic L., Kirfel J., Scheer E., Tropel P., Wisniewski J.R., Tora L. et al. A dual role of linker histone H1.4 Lys 34 acetylation in transcriptional activation. Genes Dev. 2012; 26:797–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang D., Zhou J., Liu X., Lu D., Shen C., Du Y., Wei F.Z., Song B., Lu X., Yu Y. et al. Methylation of SUV39H1 by SET7/9 results in heterochromatin relaxation and genome instability. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:5516–5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nakayama J., Rice J.C., Strahl B.D., Allis C.D., Grewal S.I.. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001; 292:110–113. [DOI] [PubMed] [Google Scholar]
- 40. Cheutin T., McNairn A.J., Jenuwein T., Gilbert D.M., Singh P.B., Misteli T.. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003; 299:721–725. [DOI] [PubMed] [Google Scholar]
- 41. Lachner M., O’Carroll D., Rea S., Mechtler K., Jenuwein T.. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001; 410:116–120. [DOI] [PubMed] [Google Scholar]
- 42. Strahl B.D., Allis C.D.. The language of covalent histone modifications. Nature. 2000; 403:41–45. [DOI] [PubMed] [Google Scholar]
- 43. Li Q., Zhou H., Wurtele H., Davies B., Horazdovsky B., Verreault A., Zhang Z.. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008; 134:244–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lu X., Wontakal S.N., Kavi H., Kim B.J., Guzzardo P.M., Emelyanov A.V., Xu N., Hannon G.J., Zavadil J., Fyodorov D.V. et al. Drosophila H1 regulates the genetic activity of heterochromatin by recruitment of Su(var)3-9. Science. 2013; 340:78–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Polo S.E., Almouzni G.. Chromatin dynamics after DNA damage: the legacy of the access-repair-restore model. DNA Repair (Amst.). 2015; 36:114–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sulli G., Di Micco R., d’Adda di Fagagna F.. Crosstalk between chromatin state and DNA damage response in cellular senescence and cancer. Nat. Rev. Cancer. 2012; 12:709–720. [DOI] [PubMed] [Google Scholar]
- 47. Adam S., Polo S.E., Almouzni G.. Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell. 2013; 155:94–106. [DOI] [PubMed] [Google Scholar]
- 48. Dinant C., Ampatziadis-Michailidis G., Lans H., Tresini M., Lagarou A., Grosbart M., Theil A.F., van Cappellen W.A., Kimura H., Bartek J. et al. Enhanced chromatin dynamics by FACT promotes transcriptional restart after UV-induced DNA damage. Mol. Cell. 2013; 51:469–479. [DOI] [PubMed] [Google Scholar]
- 49. Harshman S.W., Young N.L., Parthun M.R., Freitas M.A.. H1 histones: current perspectives and challenges. Nucleic Acids Res. 2013; 41:9593–9609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schafer G., Smith E.M., Patterton H.G.. The Saccharomyces cerevisiae linker histone Hho1p, with two globular domains, can simultaneously bind to two four-way junction DNA molecules. Biochemistry. 2005; 44:16766–16775. [DOI] [PubMed] [Google Scholar]
- 51. Syed S.H., Goutte-Gattat D., Becker N., Meyer S., Shukla M.S., Hayes J.J., Everaers R., Angelov D., Bednar J., Dimitrov S.. Single-base resolution mapping of H1-nucleosome interactions and 3D organization of the nucleosome. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:9620–9625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bednar J., Garcia-Saez I., Boopathi R., Cutter A.R., Papai G., Reymer A., Syed S.H., Lone I.N., Tonchev O., Crucifix C. et al. Structure and dynamics of a 197 bp nucleosome in complex with linker histone H1. Mol. Cell. 2017; 66:384–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Happel N., Doenecke D.. Histone H1 and its isoforms: contribution to chromatin structure and function. Gene. 2009; 431:1–12. [DOI] [PubMed] [Google Scholar]
- 54. Wang Z., Zang C., Cui K., Schones D.E., Barski A., Peng W., Zhao K.. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009; 138:1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Guan J.S., Haggarty S.J., Giacometti E., Dannenberg J.H., Joseph N., Gao J., Nieland T.J., Zhou Y., Wang X., Mazitschek R. et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009; 459:55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dovey O.M., Foster C.T., Cowley S.M.. Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:8242–8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Miller K.M., Tjeertes J.V., Coates J., Legube G., Polo S.E., Britton S., Jackson S.P.. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol. 2010; 17:1144–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yang X.J. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 2004; 32:959–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lee K.K., Workman J.L.. Histone acetyltransferase complexes: one size doesn’t fit all. Nat. Rev. Mol. Cell Biol. 2007; 8:284–295. [DOI] [PubMed] [Google Scholar]
- 60. Zhao M., Geng R., Guo X., Yuan R., Zhou X., Zhong Y., Huo Y., Zhou M., Shen Q., Li Y. et al. PCAF/GCN5-mediated acetylation of RPA1 promotes nucleotide excision repair. Cell Rep. 2017; 20:1997–2009. [DOI] [PubMed] [Google Scholar]
- 61. Li Z., Zhu W.G.. Targeting histone deacetylases for cancer therapy: from molecular mechanisms to clinical implications. Int. J. Biol. Sci. 2014; 10:757–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Li Y., Li Z., Zhu W.G.. Molecular mechanisms of epigenetic regulators as activatable targets in cancer theranostics. Curr. Med. Chem. 2017; doi:10.2174/0929867324666170921101947. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.