Abstract
In contrast to GNRA tetraloop receptors that are common in RNA, receptors for the more thermostable UNCG loops have remained elusive for almost three decades. An analysis of all RNA structures with resolution ≤3.0 Å from the PDB allowed us to identify three previously unnoticed receptors for UNCG and GNRA tetraloops that adopt a common UNCG fold, named ‘Z-turn’ in agreement with our previously published nomenclature. These receptors recognize the solvent accessible second Z-turn nucleotide in different but specific ways. Two receptors participating in a complex network of tertiary interactions are associated with the rRNA UUCG and GAAA Z-turns capping helices H62 and H35a in rRNA large subunits. Structural comparison of fully assembled ribosomes and comparative sequence analysis of >6500 rRNA sequences helped us recognize that these motifs are almost universally conserved in rRNA, where they may contribute to organize the large subunit around the subdomain-IV four-way junction. The third UCCG receptor was identified in a rRNA/protein construct crystallized at acidic pH. These three non-redundant Z-turn receptors are relevant for our understanding of the assembly of rRNA and other long-non-coding RNAs, as well as for the design of novel folding motifs for synthetic biology.
INTRODUCTION
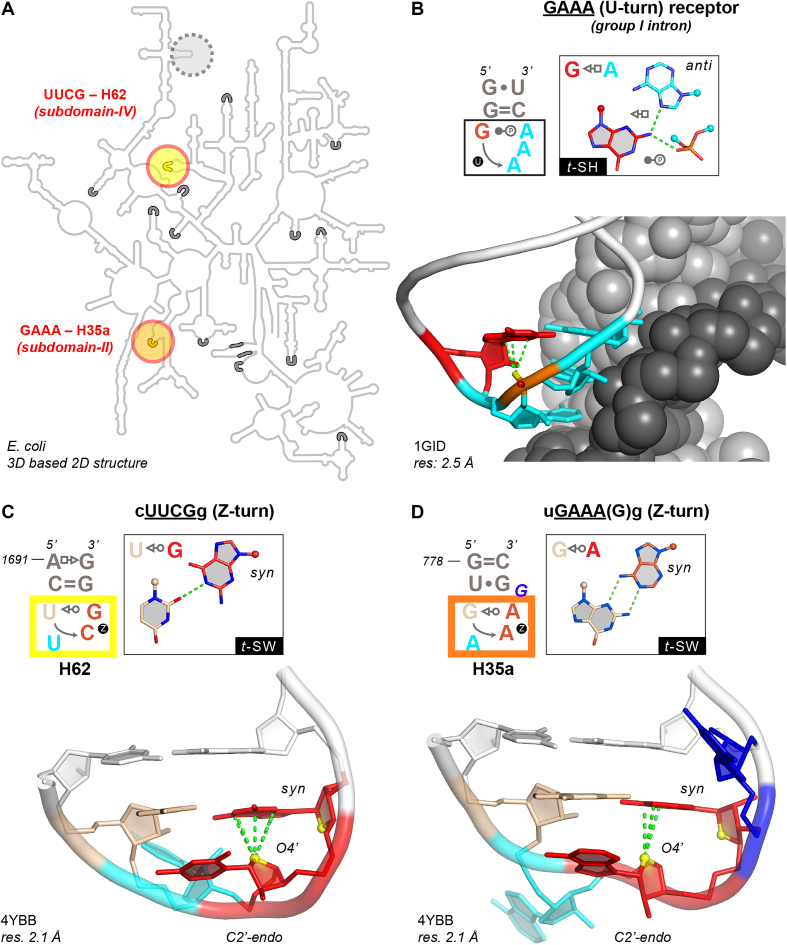
RNA architectures are formed by long-range tertiary interaction networks involving secondary structure elements such as tetraloops (1). Among those, GNRAs—N = any nucleotide, R = A or G—are more common than UNCGs (e.g. ≈14 versus ≈2 in the Escherichia coli rRNA large subunit or LSU; Figure 1A). A rRNA secondary structure search established that the cUUCGg loop is only third in frequency after the cGAAAg and cGUGAg loops, and that GNRA and UNCG sequences represent respectively 82% and 11% of the total count of inferred tetraloops (2) (note that capital and lower-case letters correspond to loop and stem nucleotides, respectively. At the time, these observations were surprising since UNCG loops were measured in vitro to be thermodynamically more stable than GNRAs (3) and, therefore, thought to be more prone to form stabilizing tertiary (3D) interactions.
Figure 1.
UUCG/GAAA Z-turn folds and GNRA U-turn receptor. (A) E. coli LSU 2D structure (44) with circled H62 (yellow) and H35a (orange) Z-turns and 14 identified U-turns (grey segments); a third non-conserved Z-turn among rRNA structures is circled in grey (see Supplementary Figure S1). (B) Structure of a U-turn bound to its receptor (69). Colors: first G in red; stacked phosphate in orange; stacked adenines in cyan; double helical receptor strands in light and dark grey. (C, D) H62 cUUCGg and H35a uGAAA(G)g Z-turns with the numbering of PDBid 4YBB (32,70). Colors: CpG and ApA Z-steps in red; Z-step O4’ atoms in yellow; solvent accessible second nucleotide in cyan, bulged G in blue; closing base pair nucleotides in white. (B–D) Oxygen/nucleobase stacking distances ≤3.5 Å in green.
To rationalize this discrepancy, it was proposed that tetraloop frequencies correlate better to the loop potential to establish 3D interactions than to their thermodynamic stability (1,4,5). Indeed, GNRAs were recognized to be central to the folding of large RNAs and RNPs like group I introns (6–8), group II introns (9), ribonuclease P (10–12) and ribosomes (13,14), as a result of their ability to form a characteristic U-turn (15) that allows the Watson–Crick edges of the three stacked nucleobases to interact with double helical receptors (Figure 1B). Hence, GNRAs became a widely used motif in the design of constructs for RNA crystallization (16–18). Furthermore, synthetic GNRA receptor variants were selected in vitro to expand the growing biotechnology toolbox of 3D motifs (1,19–21). Receptors for UNAC and GANC loops with exposed base-edges were also reported (22–24).
On the other hand, cognate receptors for the more thermostable UNCG loops have remained elusive (24). Thus, these loops were tagged as ‘loners’ and were deemed unimportant for forming long-range tertiary interactions (25). As a result, UNCG loops are nowadays merely considered as stable caps for hairpin stems, nucleation sites for RNA folding and protein-binding sites (26–29). Quite understandably, the different abilities of GNRA and UNCG loops to form long-range RNA/RNA contacts originate from their specific 3D structures.
To better appreciate the peculiarities of UNCG loops (Figure 1C), we recently extended the description of their structural signature by establishing that their CpG step adopts a characteristic and up to now under-appreciated Z-DNA/RNA conformation—see method section and (30). Thus, by analogy to the better-known GNRA U-turns (Figure 1B) (15,31), these folds were named ‘Z-turns’ (32). We could establish that Z-turn folds are not limited to UNCG sequences, as they are also accessible to GNRA sequences through a rare U-to-Z transition (Figure 1D). Subsequently, we and others demonstrated that a large array of sequences including YNNG (Y = U/C) and GNNA can fold as Z-turns (32–34) as summarized in (32). An important characteristic of these Z-turns is related to their solvent accessible second nucleobase, which has a Watson–Crick edge that can participate in base specific tertiary contacts (Figure 1C, D). Such interaction types are rare and were never described in a loop/receptor framework—see below and (35,36).
Here, we ascertain through a survey of PDB structures with resolutions ≤3.0 Å that, although infrequent when compared to U-turn/receptors, Z-turn/receptors do exist in natural and synthetic RNA constructs, where they fulfill structural and functional roles. Within the present PDB structures, we identified three non-ambiguous and distinct Z-turn receptors, their rarity hinting to why they remained unnoticed for so long. Among those, two Z-turn receptors in rRNA are located within the intricate structural context provided by the LSU subdomain-IV four-way junction (Supplementary Figure S1). These loop/receptor systems involve the subdomain-IV cUU2CGg H62 and the subdomain-II uGA2AA(G)g H35a loops (Figure 1A) and take specific advantage of the solvent accessible second nucleobase of the Z-turns that are marked by a superscript in these E. coli loop sequences (Supplementary Figure S1)—note that bulged residues not belonging to the Z-turn tetranucleotide sequences are in parenthesis (Figure 1D). The third Z-turn receptor requires acidic pH crystallisation conditions but may nonetheless be of value for the design of RNA nanosystems such as pH dependent sensors.
Overall, we believe this work to be relevant to the current interest in RNA structure prediction, in particular for long non-coding RNAs. These RNAs may contain conserved tetraloops that might undergo U-to-Z transitions, interact with complex RNA architectures such as junctions, adopt transient/alternative structures upon protein binding, and approach the nucleotide length of LSU particles (37–40). This suggest that these RNAs may fold as complex 3D structures, although this remains a subject of debate (41). Further, the characterization of these Z-turn receptors could stimulate the discovery of atypical folding principles and open perspectives in the design of novel motifs relevant to synthetic biology systems (42,43).
MATERIALS AND METHODS
Throughout, we use the nucleotide numbering scheme of the E. coli 2D structure (44) along with the related 2.1 Å resolution X-ray structure (PDBid: 4YBB) as a reference unless otherwise specified. To localize potential Z-turn receptors, we first searched for Z-turn motifs in the pool of PDB X-ray and cryo-EM structures with resolution ≤3.0 Å (32). These motifs are defined as tetranucleotide folds comprising 1–4 nucleobase-nucleobase H-bond(s) and a 3–4 oxygen–π, also named O4’–π, stacking contact (30,32,45) (Figure 1C, D), the 3–4 nucleotides forming a Z-step similar to those observed in Z-DNA/RNA. The distance between the O4’ atom and the nucleobase plane has to be ≤3.5 Å. In addition, the projection of the O4’ atom on the nucleobase plane must lie within the nucleobase aromatic cycle. A polygon-offset of 0.5 Å was used to account for 3D structure determination inaccuracies. These values were calculated by the DSSR tool package (46). Typically, the fourth nucleotide of these turns adopts a syn conformation as in the UNCG loops embedding a CpG Z-step. Although rare variants with the fourth nucleobase in anti, called Zanti-turns, do exist (32), they were not considered in this study—see for example the E. coli cGC2145CAg with the adenine in anti. Note that this loop embeds atoms with B-factors above 80 Å2 and displays weak electron density as shown by the RNABricks2 web service (47). The corresponding ‘e-density’ score is associated either with weak signal (below 1.0 sigma on average) or real-space correlation coefficients value below 0.7. Alike, a cUA1535:DACGg ‘Z-turn’ is capping H59 in subdomain-III. However, this loop does not respect the O4’–π, stacking contact criteria we listed above and displays atoms with B-factors ≥80 Å2 (Supplementary Figure S1; Figure 1A).
In this ensemble, we then searched Z-turns that establish tertiary RNA/RNA contacts with at least one of their four nucleotides and found the three non-redundant instances described in the result section. Given that we based our search on structures with resolution ≤3.0 Å, it remains possible that some Z-turn motifs appearing in low resolution structures were missed. However, these strict criteria allowed us to be confident in the motifs we describe. For naming base pairs, the nomenclature developed by Leontis and Westhof was used (48).
For analyzing rRNA large subunit (LSU) structures that gather two of the Z-turn receptors we found, we collected a sample of LSU structures covering all species present in the PDB and selected the best resolution X-ray/cryo-EM models (Supplementary Table S1), ending up with 30 structures of resolutions ranging from 2.1 to 9.0 Å (Supplementary Table S2). For crystallographic structures with multiple asymmetric units, only the ribosome with the lowest average B-factors in the asymmetric unit was considered; in 4YBB this corresponds to 23S rRNA chain DA and 16S rRNA chain AA. In 4YBB, the sequences of the two tetraloops bound to the Z-turn receptors are cUUCGg (H62) and uGAAA(G)g (H35a)—note that for 2D and 3D structures, helix but not residue numberings are conserved among all rRNAs. To cope with these differences, Supplementary Table S2 provides residue number and chain label correspondences for all surveyed LSUs. We disregarded the mitochondrial B. taurus and L. tarentolae structures that have resolutions not better than 10 Å, along with a few other LSUs for which atomic details are missing (Supplementary Table S1). Since we are aware that most of the LSU 3D structures were obtained by molecular replacement and homology modeling, we visually inspected all X-ray and cryo-EM density maps associated with the Supplementary Table S2 structures to verify that the Z-turns were appropriately modeled. For redundancy criteria, please see reference (32).
For sequence alignments, the SILVA_128_LSURef_tax_silva_trunc.fast file containing 154 297 rRNA LSU sequences was downloaded from the SILVA website (49). The SILVA taxonomy (50) was extracted from the FASTA sequence headers. The file was parsed to keep only the first sequence for a given organism and, consequently, was downsized to 7848 sequences. Then, this file was split in three smaller files for archaea, bacteria and eukarya cytoplasmic LSUs. We excluded organelle LSUs for which the limited number of available sequences appears significantly less homogeneous and, therefore, more problematic to analyze. Inspecting such sequences would have little impact on the characterization of UNCG and related Z-turn receptors. At the beginning of each of these files, one or two reference sequences (from structures with available 3D data, see Supplementary Table S2) were added: H. marismortui to archaea (209 sequences), E. coli and T. thermophilus to bacteria (3735 sequences), S. cerevisiae and H. sapiens to eukarya (2341 sequences). LSU sequences were aligned with SSU-ALIGN (51), using a custom-built LSU template as reference for the alignments. This template was generated by SSU-build, a module of SSU-ALIGN, using the sequence family seeds RF02540 (LSU_rRNA_archaea, 92 organisms), RF02541 (LSU_rRNA_bacteria, 102 organisms) and RF02543 (LSU_rRNA_eukarya, 89 organisms) extracted from the Rfam.seed file (available on: ftp://ftp.ebi.ac.uk/pub/databases/Rfam/CURRENT/). These sequence seeds are part of the Rfam 12.2 release (http://rfam.xfam.org/, January 2017) (52) and were originally derived from the Comparative RNA Web (CRW) site (53). The resulting alignments were analyzed with UGENE (54).
RESULTS
Only three non-ambiguous and non-redundant Z-turn receptors in solved RNA structures
To assess the remarkable low rate of occurrence of Z-turn receptors, we searched all RNA X-ray and cryo-EM structures in the PDB with resolutions ≤3.0 Å for Z-turns participating in tertiary interactions. We considered only tertiary RNA/RNA contacts involving one or more of the four tetraloop nucleotides and excluded loops not meeting Z-turn geometric features (see Materials and Methods) or displaying nucleotide atoms with B-factors >80 Å2, to discount potentially overfitted data and increase confidence in the retained motifs. Further, we excluded proximity contacts and considered that a Z-turn receptor must establish at least one base specific interaction with the attached Z-turn. With these criteria, we characterized only three non-redundant Z-turn/receptors in the available pool of PDB structures: two in rRNA that involve the H62 and H35a Z-turns interacting with the subdomain-IV four-way junction and one in an RNA/protein construct crystallized at acidic pH, along with a few outliers. Even if RNA/RNA contacts involving Z-turns are rare but not unusual, we found that base specific interactions are limited to these three examples.
Structure and sequence conservation of the H62 and H35a Z-turns in rRNA LSUs
To assess the significance of the limited number of non-redundant Z-turn receptors, we analyzed first their conservation in rRNA through an inspection of all best resolution X-ray and cryo-EM LSU structures from archaea, bacteria, eukarya and chloroplasts/mitochondria (Supplementary Tables S1 and S2). We considered that a high degree of recurrence of these motifs is required for a conserved structural and functional potential. Therefore, for each organism and organelle for which LSU 3D structures are available, we first confirmed through visual inspection that in every the subdomain-IV H62 UUCG and subdomain-II H35a GAAA sequences that we found associated with Z-turn receptors—as well as rare tetraloop variants (Supplementary Figure S2)—were modeled as Z-turns. To obtain confidence in these rRNA structures which were often obtained through molecular replacement or homology modeling, we visually inspected all related experimental X-ray and cryo-EM density maps and found that they unambiguously matched the modeled Z-turns. Although we deliberately included in our sample some poor resolution structures (>4 Å; see Supplementary Table S2), it was possible in all instances to confirm the presence of a Z-turn given that, even at low-resolution, Z-turn and U-turn densities are sufficiently distinctive to avoid misidentifications (see Materials and Method section). In addition, we assessed that all H62 UUCG loops are closed by a cis-Watson–Crick c=g or g=c pair (Supplementary Table S2) while, as inferred from 3D structure analysis (Supplementary Table S2; Supplementary Figure S3), the H35a closing pair is mainly a wobble u•g pair or its isosteric c•a+ counterpart (55–57),
Next, we extended the views gained through LSU visualization by analyzing the sequence conservation of these hairpins with a focus on cytoplasmic rRNA (see Materials and Method section). For H62, the UUCG sequence dominates in all life domains, followed by the Z-turn-compatible UUAG, UUUG and CUUG sequences (Table 1), resulting in a strict conservation of the second U (≈99%). Typically, the first nucleotide of the loop is a pyrimidine (U/C) and loop-closing c=g pairs largely exceed g=c pairs (Table 1) as generally observed for tetraloops (58,59). This stringent sequence and structure conservation of the H62 Z-turn, of their closing pair and of their stem length (60) suggest strong selective pressure associated with the need for conservation of higher-order tertiary interactions.
Table 1.
Sequence variations for the cUUCGg H62 and uGAAA(G)g H35a Z-turns. In total, 6285 sequences were analyzed: 3735 bacteria, 209 archaea and 2341 eukarya
| Bacteria | Archaea | Eukarya | |
|---|---|---|---|
| H62 sequences | |||
| UUCG | 92% | 79% | 62% |
| UUUG | 2% | 14% | 15% |
| UUAG | 5% | 4% | <1% |
| CUUG | <1% | 1% | 20% |
| Total | >99% | 98% | >97% |
| Closing pair | |||
| c=g | 93% | 84% | 98% |
| g=c | 7% | 16% | 1% |
| Total | ≈100% | ≈100% | ≈99% |
| H35a sequences | |||
| GAAA | 99% | 71% | 98% |
| AAAA | <1% | 17% | <1% |
| GAUA | <1% | 1% | ≈0% |
| Total | >99% | 89% | >98% |
| Fifth bulged base | |||
| G | 71% | 81% | 99% |
| A | 5% | 12% | ≈0% |
| U | 24% | 6% | ≈0% |
| C | ≈0% | 0% | 0% |
| Total | ≈100% | >99% | >99% |
| Closing pair | |||
| u•g | 82% | 62% | 0% |
| c•a+ | 1% | 17% | 99% |
| u-a | <1% | 12% | <1% |
| a-u | 10% | 0% | 0% |
| u•u | 6% | 4% | 0% |
| Total | ≈100% | ≈95% | >99% |
The calculated percentages exclude gaps in sequence alignments that are limited to ≈4% for eukarya H35a and between 0 to <2% otherwise.
The H62 UUCG Z-turn receptor
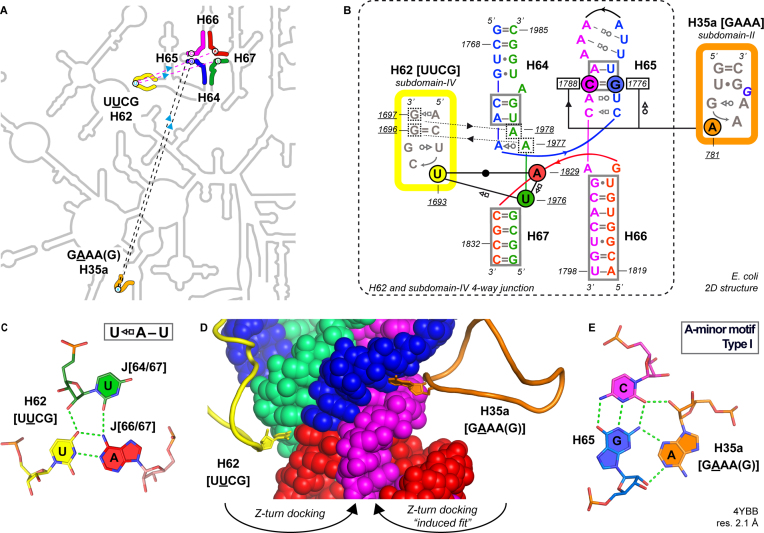
To verify the involvement of the H62 Z-turn in tertiary interactions, we inspected 28 cytoplasmic LSU structures—note that H62 is absent in mitochondrial rRNA with the exception of mitochondrial yeast rRNA (Supplementary Table S2; (61))—and observed that this Z-turn establishes a conserved tertiary interaction with the 4-way junction of subdomain-IV (Figure 2A, B). The solvent accessible second uridine of the UU2CG Z-turn wedges into a notch to form a U2–A pair with A1829 from the J66/67 joining region (Figure 2C). This recurrent tertiary contact reveals that the core of the four-way junction acts as a UUCG receptor (Figure 2B, D). Comparative sequence analysis point to a ≈100% conservation of the U2 and AJ66/67 residues suggesting that the corresponding U2-AJ66/67 base pair is strictly conserved in all life domains. Moreover, given the occurrence of various Z-turn-compatible loop sequences (UU2CG, UU2AG, UU2UG and CU2UG; Table 1), this LSU-embedded receptor may bind any ‘YU2NG’ sequence. For now, besides UUCG, only a single structure of a UUAG(U) Z-turn was visualized in the mitochondrial S. cerevisiae ribosome (PDBid: 3J6B; Supplementary Figure S2A).
Figure 2.
H62 and H35a Z-turn receptors in the rRNA subdomain-IV four-way junction. (A) Zoom on the E. coli LSU 2D structure (see also Figure 1A; Supplementary Figure S1) highlighting H62 (yellow) and H35a (orange) Z-turns and their receptors within the subdomain-IV four-way junction (light blue arrow: docking directionality). (B) 2D structure of the four-way junction and the UUCG/GAAA Z-turns (48,71); cis-WW stem pairs of the junction are boxed; the two non-specific four-way junction to H62 UUCG hairpin interactions are marked by dashed lines; the residue numbers of the H65 C=G pair forming a A-minor base triple with H35a are boxed. (C) Base triple associated with the H62 UUCG Z-turn receptor. (D) CPK view highlighting the insertion of the H62 (yellow) and H35a (orange) Z-turn second nucleotides in four-way junction notches. (E) A-minor base triple associated with the H35a GAAA Z-turn receptor.
Within this loop/receptor motif, the U2–A pair is part of a [UH62–AJ66/67]•UJ64/67 base triple (Figure 2B, C). In a few instances, the third U is replaced by a C, a G or is not at contact distance from the U–A pair suggesting weaker phylogenetic constraints on its nature. Therefore, it can be inferred that the U2–A pair is the main H62 UU2CG structural determinant of this Z-turn receptor system. In addition, several contacts involving the Z-turn stem and the four-way junction are noted (Supplementary Figure S4) underscoring a tight loop/receptor complementarity.
The H35a GAAA(G) Z-turn receptor
Interestingly, we found that the LSU subdomain-IV four-way junction embeds a second receptor, which is specific to the H35a uGAAA(G)g Z-turn. It shares substantial similarities with the H62 UUCG receptor (Figure 2B). In both cases, the specificity of the receptor is ensured by the formation of a tertiary interaction involving the solvent accessible second nucleotide of the loop. What is characteristic of the H35a GA2AA Z-turn receptor is the ‘type I’ A-minor base triple (62) that involves a C=G pair of the H65 stem (Figure 2B, E). This [C=G]•A2 base triple is conserved to ≈100% in all three life domains, and the tetraloop sequence is either GA2AA (≈99% in bacteria, ≈98% in eukarya; ≈70% in archaea; Table 1), GA2CA (mitochondrial H. sapiens; PDBid: 5OOM; Supplementary Table S2), or AA2AA (17% corresponding to 33 sequences in archaea; for a model structure of a AA2AA Z-turn, see Supplementary Figure S5).
In contrast to the c=g closing pair of the H62 cUUCGg/receptor system (Supplementary Figure S4), in H35a the closing pair does not contact the receptor. Instead, two non-specific contacts are formed between the third and fourth nucleotides of the GAA3A4 sequence and the 2′-hydroxyl groups of the U–A pair in H65 (Figure 2B). The absence of contacts between the GAAA receptor and the Z-turn closing pairs may explain the greater sequence variability of these pairs (Table 1). We also visualized potential contacts established by the bulged fifth nucleotide. Although it is mainly a G (Table 1), it is not systematically involved in tertiary interactions and no conserved interactions are seen across species.
A third Z-turn receptor needs acidic conditions
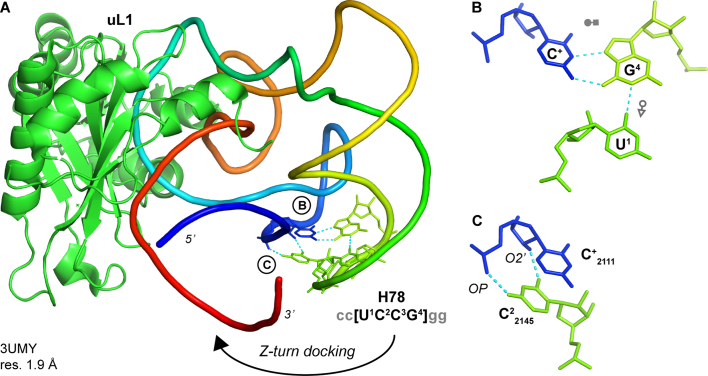
In our structural sample of RNA structures with resolution ≤3.0 Å, we characterized only one other motif that qualifies as a Z-turn receptor (PDBid: 3UMY; Figure 3).This motif was identified in several crystal structures of a ribosomal T. thermophilus H78 fragment in complex with native and mutant uL1 r-proteins (63,64). The single stranded receptor is separated by 30 nucleotides from a UC2CG Z-turn and involves a protonated cytosine (C+2111). The loop/receptor interactions are highly specific since they require the presence of a C2 and forbid the occurrence of other nucleotides. Yet, since this H78 UC2145CG loop does not adopt a Z-turn in the full-length T. thermophilus LSUs (Figure 3B), it is probable that this rare UCCG receptor is specific to crystal forms obtained at non-physiological 5.6–6.0 pH (63,64) and, therefore, not relevant to functional ribosomes. However, this specific design may be relevant to synthetic biology (see Discussion).
Figure 3.
UCCG receptor in a crystal structure of a rRNA/r-protein fragment at a 5.6–6.0 pH (63,64). (A) The RNA fragment is colored according to increasing residue numbers from blue (5′) to red (3′); the uL1 r-protein is green. The H78 UCCG Z-turn along with the receptor C+ residues are shown as sticks; closing pairs are not shown for clarity. The circled letters refer to the interactions shown in the right panels. The docking direction is marked by an arrow. (B) Base triple that involves a C+ residue. (C) Nucleobase C2 (green) to backbone C (blue) contacts.
Z-turn receptor outliers involving A-minor motifs
In addition, a conserved interaction between a cUA344CGg Z-turn (h14) and a gGA160AAc U-turn (h8) has been reported in small rRNA subunits (35,65). This interaction involves the formation of an A-minor motif between the second nucleotide of the GA2AA U-turn and the closing c=g pair of the Z-turn. Therefore, it corresponds to a GAAA loop interacting with a GAAA receptor closed by a UACG Z-turn that, in our views, does not qualify as a Z-turn receptor since the RNA/RNA contacts involve only the Z-turn stem (PDBid: 4YBB; Supplementary Figure S6A, B). We also noticed a crystal lattice contact (PDBid: 5BO3; Supplementary Figure S6C, D) involving two symmetry related gUACGc Z-turns, where each of the solvent accessible ‘A’ nucleobase establishes an A-minor interaction with the closing g=c pair of the adjacent turn (36). Here also, we consider that this self-recognition contact does not qualify as a Z-turn receptor.
DISCUSSION
We were able to establish that Z-turn receptors exist in RNA and are significant for rRNA structures. The two Z-turn receptors that we identified in rRNA require the large structural context provided by the subdomain-IV four-way junction. This architecture contrasts with the simpler double stranded structures of the more common GNRA receptor (Figure 1B). Most likely, the subdomain-IV structural complexity explains why Z-turn receptors remained elusive and why efforts to select them in vitro —typically from a pool of sequences that cannot exceed a few dozen nucleotides in length—have been unsuccessful (20).
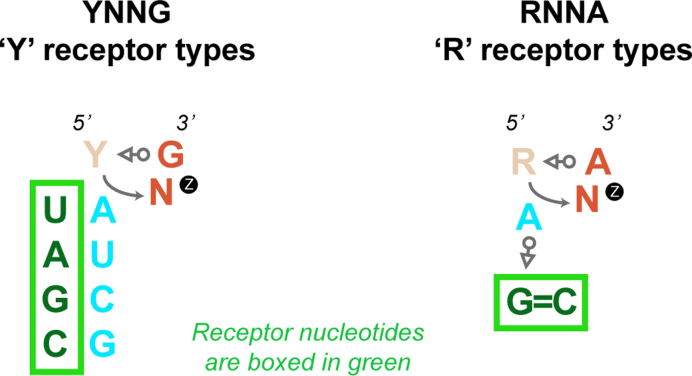
Our data assert that the LSU Z-turn receptors belong to one of two subtypes: those that accommodate turns starting with a pyrimidine (Y = U/C) and display a ‘YU2NG’ consensus sequence, and those that accommodate turns starting with a purine (R = G/A) and display a ‘RA2NA’ consensus sequence. The first subtype can be categorized as a pyrimidine or ‘Y’ receptor associated with sequences adopting stable Z-turns like UUCG loops closed by a trans–Sugar/Watson–Crick pair (t–S/W; Figure 1C). The proposed docking mechanism for such receptors is relatively straightforward. For instance, there is little doubt that the H62 UU2CG loop conserves its Z-turn once formed—probably very early after transcription—and must await the completion of the folding of the subdomain-IV four-way junction before attaching to the receptor first through its closing pair (Supplementary Figure S4) and second through the formation of the specific A-U2 pair (see arrow, Figure 2D). The “docking” or folding mechanism for the UCCG Z-turn at acidic conditions is probably very similar (Figure 3).
The second subtype is a purine or ‘R’ receptor that is associated with GNRA-like sequences for which a Z-turn is not anticipated (32). Thus, an ‘induced fit’ docking mechanism involving a U-to-Z transition needs to be envisaged contrasting with the simpler docking mechanism proposed for H62 (see arrows, Figure 2D). A self-evident structural difference between these two ‘Y’ and ‘R’ receptors is the nature of the base-specific interactions involving the second loop nucleotide. In the ‘Y’ subtype, the solvent accessible residue forms a U2-A Watson–Crick pair, while in the ‘R’ subtype, the A2 residue establishes an A-minor interaction to a C=G pair. Interestingly, we did not find any notable covariations regarding the ‘R’ and the ‘Y’ type receptors involving the solvent accessible second nucleobase of the Z-turn, although exceptions might emerge in the future. This underlines again the observed structural conservation of the loop and its receptor in rRNA. However, strictly speaking, there is no obvious reason to avoid covariations at the UH62-AJ66/67 pair for the ‘Y’ receptor—except for the need to form a base triple with UJ64/67 (Figure 2C)—while only a limited set of covariations at the H35a level involving a A-minor motif could be envisaged given the restricted base pairing possibilities of such base triples. Potential covariations associated with the ‘Y’ and ‘R’ rRNA receptor types are summarized in Figure 4 and suggest that, outside rRNA, Z-turn receptors and associated covariations may be integrated in the design of novel folds pertinent for synthetic biology. Of course, a few other combinations could be envisaged such as: the ‘A’ of a UACG Z-turn could form a A-minor triple with a C=G pair as for a ‘R’ receptor, or the second ‘A’ of a GANA Z-turn could be involved in a Watson–Crick pair as for a ‘Y’ receptor, therefore, opening a large array of Z-turn/receptor combinations. Yet, some interrogations remain that we could not fully address. Why are these H62 and H35a motifs so highly conserved in rRNA and why don’t we observe a YNNG instead of a GAAA loop at the end of H35a? More surprisingly, why are covariations almost inexistent in the present rRNA sequence dataset?
Figure 4.
Schematic representation of the ‘Y’ and ‘R’ Z-turn receptor types as observed in rRNA and possible covariations (see also Figure 2). The ‘Y’ receptor involves a Watson–Crick base pair between the second nucleotide of the Z-turn (cyan) and a complementary base pair of the receptor in green (Figure 2C). The ‘R’ receptor involves the formation of a A-minor motif between the second nucleotide of the Z-turn (cyan) and a G=C pair in green (Figure 2D). For ‘R’ compared to the ‘Y’ receptors, it is more difficult to propose meaningful covariations.
On a methodological note, our conservative approach for locating Z-turns may have led to the exclusion of a few Z-turns that might emerge as valid once better resolution structures will become available. However, it is worth noting that, aside applying resolution criteria, we did not consider the E. coli H59 UA1535CG Z-turn of the LSU subdomain-III (Figure 1A; Supplementary Figure S1) given that neither its fold, sequence nor stem length are conserved in available rRNA structures and that it adopts a deformed Z-turn structure that does not form RNA/RNA contacts, underlying the versatility of rRNA structures in non-conserved regions. Therewith, we are confident that Z-turn receptors are much less frequent than U-turn receptors in the RNA world.
We anticipate that the so far exclusive presence of natural Z-turn receptors in rRNA may reflect the currently limited size of large RNAs and RNPs in structural databases. With ramping structural biology efforts towards atomic details of large RNPs and considering the length of many non-coding RNAs and their roles as transient scaffolds for protein binding, Z-turn receptors may emerge in other biologically relevant systems. After all, UNCGs and other loops prone to adopt a Z-turn (32) are conserved among non-coding RNAs for example in disease-causing bacteria (37), in fungi—e.g., SART1 (66)—in plants (40) and in viral RNA (67,68).
Although long non-coding RNAs do not always comprise four-way junctions, the example of the uL1 complex suggests that, in the future, other artificial constructs may embed Z-turn receptors. Surely, additional Z-turn recognition schemes relevant to synthetic biology remain to be found. From there on, as witnessed for GNRA U-turns (1), establishing new Z-turn receptor designs is only a matter of time.
Supplementary Material
ACKNOWLEDGEMENTS
L.D., Q.V. and P.A. wish to thank Prof. Eric Westhof and Dr Pascale Romby for their support. We also wish to thank Dr Eric Nawrocki and Prof. Sean Eddy for their kind help regarding SSU-ALIGN, as well as Dr John A. Hammond, Dr Daniel Eiler, Prof. Jeff Kieft, Prof. Norm Pace and Prof. Tom Cech for their feedback on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
French National Research Agency [ANR-15-CE11-0021-01].
Conflict of interest statement. None declared.
REFERENCES
- 1. Fiore J.L., Nesbitt D.J.. An RNA folding motif: GNRA tetraloop-receptor interactions. Q. Rev. Biophys. 2013; 46:223–264. [DOI] [PubMed] [Google Scholar]
- 2. Sheehy J.P., Davis A.R., Znosko B.M.. Thermodynamic characterization of naturally occurring RNA tetraloops. RNA. 2010; 16:417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Antao V.P., Tinoco I.. Thermodynamic parameters for loop formation in RNA and DNA hairpin tetraloops. Nucleic Acids Res. 1992; 20:819–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. SantaLucia J. Jr., Kierzek R., Turner D.H.. Context dependence of hydrogen bond free energy revealed by substitutions in an RNA hairpin. Science. 1992; 256:217–219. [DOI] [PubMed] [Google Scholar]
- 5. Wu L., Chai D.G., Fraser M.E., Zimmerly S.. Structural variation and uniformity among tetraloop-receptor interactions and other loop-helix interactions in RNA crystal structures. PLoS One. 2012; 7:e49225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Michel F., Westhof E.. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 1990; 216:585–610. [DOI] [PubMed] [Google Scholar]
- 7. Jaeger L., Michel F., Westhof E.. Involvement of a GNRA tetraloop in long-range RNA tertiary interactions. J. Mol. Biol. 1994; 236:1271–1276. [DOI] [PubMed] [Google Scholar]
- 8. Costa M., Michel F.. Frequent use of the same tertiary motif by self-folding RNAs. EMBO J. 1995; 14:1276–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Toor N., Keating K.S., Taylor S.D., Pyle A.M.. Crystal structure of a self-spliced group II intron. Science. 2008; 320:77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brown J.W., Nolan J.M., Haas E.S., Rubio M.A.T., Major F., Pace N.R.. Comparative analysis of ribonuclease P RNA using gene sequences from natural microbial populations reveals tertiary structural elements. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:3001–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Massire C., Jaeger L., Westhof E.. Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J. Mol. Biol. 1998; 279:773–793. [DOI] [PubMed] [Google Scholar]
- 12. Torres-Larios A., Swinger K.K., Pan T., Mondragon A.. Structure of ribonuclease P - a universal ribozyme. Curr. Opin. Struc. Biol. 2006; 16:327–335. [DOI] [PubMed] [Google Scholar]
- 13. Ban N., Nissen P., Hansen J., Moore P.B., Steitz T.A.. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000; 289:905–920. [DOI] [PubMed] [Google Scholar]
- 14. Noller H.F. RNA structure: reading the ribosome. Science. 2005; 309:1508–1514. [DOI] [PubMed] [Google Scholar]
- 15. Jucker F.M., Pardi A.. GNRA tetraloops make a U-turn. RNA. 1995; 1:219–222. [PMC free article] [PubMed] [Google Scholar]
- 16. Reyes F.E., Garst A.D., Batey R.T.. Strategies in RNA crystallography. Methods Enzymol. 2009; 469:119–139. [DOI] [PubMed] [Google Scholar]
- 17. Reiter N.J., Osterman A., Torres-Larios A., Swinger K.K., Pan T., Mondragon A.. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature. 2010; 468:784–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Coonrod L.A., Lohman J.R., Berglund J.A.. Utilizing the GAAA tetraloop/receptor to facilitate crystal packing and determination of the structure of a CUG RNA helix. Biochemistry. 2012; 51:8330–8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Geary C., Baudrey S., Jaeger L.. Comprehensive features of natural and in vitro selected GNRA tetraloop-binding receptors. Nucleic Acids Res. 2008; 36:1138–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ishikawa J., Fujita Y., Maeda Y., Furuta H., Ikawa Y.. GNRA/receptor interacting modules: versatile modular units for natural and artificial RNA architectures. Methods. 2011; 54:226–238. [DOI] [PubMed] [Google Scholar]
- 21. Tanaka T., Furuta H., Ikawa Y.. Guo P. Natural selection and structural polymorphism of RNA 3D structures involving GNRA loops and their receptor motifs. 2013; CRC Press; 109–120. [Google Scholar]
- 22. Zhao Q., Huang H.C., Nagaswamy U., Xia Y., Gao X., Fox G.E.. UNAC tetraloops: to what extent do they mimic GNRA tetraloops. Biopolymers. 2012; 97:617–628. [DOI] [PubMed] [Google Scholar]
- 23. Keating K.S., Toor N., Pyle A.M.. The GANC tetraloop: a novel motif in the group IIC intron structure. J. Mol. Biol. 2008; 383:475–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ishikawa J., Furuta H., Ikawa Y.. An in vitro-selected RNA receptor for the GAAC loop: modular receptor for non-GNRA-type tetraloop. Nucleic Acids Res. 2013; 41:3748–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hall K.B. Mighty tiny. RNA. 2015; 21:630–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ennifar E., Nikulin A., Tishchenko S., Serganov A., Nevskaya N., Garber M., Ehresmann B., Ehresmann C., Nikonov S., Dumas P.. The crystal structure of UUCG tetraloop. J. Mol. Biol. 2000; 304:35–42. [DOI] [PubMed] [Google Scholar]
- 27. Butcher S.E., Pyle A.M.. The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks. Acc. Chem. Res. 2011; 44:1302–1311. [DOI] [PubMed] [Google Scholar]
- 28. Thapar R., Denmon A.P., Nikonowicz E.P.. Recognition modes of RNA tetraloops and tetraloop-like motifs by RNA-binding proteins. Wiley Interdiscip. Rev. RNA. 2014; 5:49–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cheong H., Kim N., Cheong C.. RNA structure: tetraloops. ELS. 2015; Chichester: John Wiley & Sons, ltd. [Google Scholar]
- 30. D’Ascenzo L., Leonarski F., Vicens Q., Auffinger P.. ‘Z-DNA like’ fragments in RNA: a recurring structural motif with implications for folding, RNA/protein recognition and immune response. Nucleic Acids Res. 2016; 44:5944–5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Auffinger P., Westhof E.. An extended structural signature for the tRNA anticodon loop. RNA. 2001; 7:334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. D’Ascenzo L., Leonarski F., Vicens Q., Auffinger P.. Revisiting GNRA and UNCG folds: U-turns versus Z-turns in RNA hairpin loops. RNA. 2017; 23:259–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Du Z., Yu J., Andino R., James T.L.. Extending the family of UNCG-like tetraloop motifs: NMR structure of a CACG tetraloop from coxsackievirus B3. Biochemistry. 2003; 42:4373–4383. [DOI] [PubMed] [Google Scholar]
- 34. Bottaro S., Lindorff-Larsen K.. Mapping the universe of RNA tetraloop folds. Biophys. J. 2017; 113:257–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Proctor D.J., Schaak J.E., Bevilacqua J.M., Falzone C.J., Bevilacqua P.C.. Isolation and characterization of a family of stable RNA tetraloops with the motif YNMG that participate in tertiary interactions. Biochemistry. 2002; 41:12062–12075. [DOI] [PubMed] [Google Scholar]
- 36. Fernandez-Millan P., Autour A., Ennifar E., Westhof E., Ryckelynck M.. Crystal structure and fluorescence properties of the iSpinach aptamer in complex with DFHBI. RNA. 2017; 23:1788–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lindgreen S., Umu S.U., Lai A.S., Eldai H., Liu W., McGimpsey S., Wheeler N.E., Biggs P.J., Thomson N.R., Barquist L. et al. . Robust identification of noncoding RNA from transcriptomes requires phylogenetically-informed sampling. PLoS Comput. Biol. 2014; 10:e1003907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Morris K.V., Mattick J.S.. The rise of regulatory RNA. Nat. Rev. Genet. 2014; 15:423–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Somarowthu S., Legiewicz M., Chillon I., Marcia M., Liu F., Pyle A.M.. HOTAIR forms an intricate and modular secondary structure. Mol. Cell. 2015; 58:353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hawkes E.J., Hennelly S.P., Novikova I.V., Irwin J.A., Dean C., Sanbonmatsu K.Y.. COOLAIR antisense RNAs form evolutionarily conserved elaborate secondary structures. Cell Rep. 2016; 16:3087–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rivas E., Clements J., Eddy S.R.. A statistical test for conserved RNA structure shows lack of evidence for structure in lncRNAs. Nat. Methods. 2017; 14:45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Grabow W.W., Jaeger L.. RNA self-assembly and RNA nanotechnology. Acc. Chem. Res. 2014; 47:1871–1880. [DOI] [PubMed] [Google Scholar]
- 43. Geary C., Chworos A., Verzemnieks E., Voss N.R., Jaeger L.. Composing RNA nanostructures from a syntax of RNA structural modules. Nano Lett. 2017; 17:7095–7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Petrov A.S., Bernier C.R., Hershkovits E., Xue Y.Z., Waterbury C.C., Hsiao C.L., Stepanov V.G., Gaucher E.A., Grover M.A., Harvey S.C. et al. . Secondary structure and domain architecture of the 23S and 5S rRNAs. Nucleic Acids Res. 2013; 41:7522–7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chawla M., Chermak E., Zhang Q., Bujnicki J.M., Oliva R., Cavallo L.. Occurrence and stability of lone pair-π stacking interactions between ribose and nucleobases in functional RNAs. Nucleic Acids Res. 2017; 45:11019–11032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lu X.J., Bussemaker H.J., Olson W.K.. DSSR: an integrated software tool for dissecting the spatial structure of RNA. Nucleic Acids Res. 2015; 43:e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chojnowski G., Walen T., Bujnicki J.M.. RNA Bricks-a database of RNA 3D motifs and their interactions. Nucleic Acids Res. 2014; 42:D123–D131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Leontis N.B., Stombaugh J., Westhof E.. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002; 30:3497–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glockner F.O.. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013; 41:D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yilmaz P., Parfrey L.W., Yarza P., Gerken J., Pruesse E., Quast C., Schweer T., Peplies J., Ludwig W., Glockner F.O.. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014; 42:D643–D648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nawrocki N.E. Structural RNA homology search and alignment using covariance models. 2009; Saint Louis: Washington University. [Google Scholar]
- 52. Nawrocki E.P., Burge S.W., Bateman A., Daub J., Eberhardt R.Y., Eddy S.R., Floden E.W., Gardner P.P., Jones T.A., Tate J. et al. . Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 2015; 43:D130–D137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cannone J.J., Subramanian S., Schnare M.N., Collett J.R., D'Souza L.M., Du Y., Feng B., Lin N., Madabusi L.V., Muller K.M. et al. . The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002; 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Okonechnikov K., Golosova O., Fursov M., team U.. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012; 28:1166–1167. [DOI] [PubMed] [Google Scholar]
- 55. Masquida B., Westhof E.. On the wobble GoU and related pairs. RNA. 2000; 6:9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hesslein A.E., Katunin V.I., Beringer M., Kosek A.B., Rodnina M.V., Strobel S.A.. Exploration of the conserved A+C wobble pair within the ribosomal peptidyl transferase center using affinity purified mutant ribosomes. Nucleic Acids Res. 2004; 32:3760–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Siegfried N.A., O’Hare B., Bevilacqua P.C.. Driving forces for nucleic acid pK(a) shifting in an A(+).C wobble: effects of helix position, temperature, and ionic strength. Biochemistry. 2010; 49:3225–3236. [DOI] [PubMed] [Google Scholar]
- 58. Bevilacqua P.C., Blose J.M.. Structures, kinetics, thermodynamics, and biological functions of RNA hairpins. Annu. Rev. Phys. Chem. 2008; 59:79–103. [DOI] [PubMed] [Google Scholar]
- 59. Vanegas P.L., Horwitz T.S., Znosko B.M.. Effects of non-nearest neighbors on the thermodynamic stability of RNA GNRA hairpin tetraloops. Biochemistry. 2012; 51:2192–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hedenstierna K.O., Siefert J.L., Fox G.E., Murgola E.J.. Co-conservation of rRNA tetraloop sequences and helix length suggests involvement of the tetraloops in higher-order interactions. Biochimie. 2000; 82:221–227. [DOI] [PubMed] [Google Scholar]
- 61. Greber B.J., Ban N.. Structure and function of the mitochondrial ribosome. Annu. Rev. Biochem. 2016; 85:103–132. [DOI] [PubMed] [Google Scholar]
- 62. Nissen P., Ippolito J.A., Ban N., Moore P.B., Steitz T.A.. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:4899–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tishchenko S., Gabdulkhakov A., Nevskaya N., Sarskikh A., Kostareva O., Nikonova E., Sycheva A., Moshkovskii S., Garber M., Nikonov S.. High-resolution crystal structure of the isolated ribosomal L1 stalk. Acta Cryst. 2012; D68:1051–1057. [DOI] [PubMed] [Google Scholar]
- 64. Tishchenko S., Kostareva O., Gabdulkhakov A., Mikhaylina A., Nikonova E., Nevskaya N., Sarskikh A., Piendl W., Garber M., Nikonov S.. Protein-RNA affinity of ribosomal protein L1 mutants does not correlate with the number of intermolecular interactions. Acta Cryst. 2015; D71:376–386. [DOI] [PubMed] [Google Scholar]
- 65. Sahu B., Khade P.K., Joseph S.. Functional replacement of two highly conserved tetraloops in the bacterial ribosome. Biochemistry. 2012; 51:7618–7626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li S., Breaker R.R.. Identification of 15 candidate structured noncoding RNA motifs in fungi by comparative genomics. BMC Genomics. 2017; 18:785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Prostova M.A., Gmyl A.P., Bakhmutov D.V., Shishova A.A., Khitrina E.V., Kolesnikova M.S., Serebryakova M.V., Isaeva O.V., Agol V.I.. Mutational robustness and resilience of a replicative cis-element of RNA virus: Promiscuity, limitations, relevance. RNA Biol. 2015; 12:1338–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Prostova M.A., Deviatkin A.A., Tcelykh I.O., Lukashev A.N., Gmyl A.P.. Independent evolution of tetraloop in enterovirus oriL replicative element and its putative binding partners in virus protein 3C. Peerj. 2017; 5:e3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cate J.H., Gooding A.R., Podell E., Zhou K.H., Golden B.L., Kundrot C.E., Cech T.R., Doudna J.A.. Crystal structure of a group I ribozyme domain - Principles of RNA packing. Science. 1996; 273:1678–1685. [DOI] [PubMed] [Google Scholar]
- 70. Noeske J., Wasserman M.R., Terry D.S., Altman R.B., Blanchard S.C., Cate J.H.. High-resolution structure of the Escherichia coli ribosome. Nat. Struct. Mol. Biol. 2015; 22:336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Laing C., Schlick T.. Analysis of four-way junctions in RNA structures. J. Mol. Biol. 2009; 390:547–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.