Abstract
Telomerase adds telomeric repeats to chromosome ends by processive copying of a template within the telomerase RNA bound to telomerase reverse transcriptase. Telomerase RNAs have single-stranded regions that separate the template from a 5′ stem and 3′ pseudoknot, and mammals gained additional stem P2a.1 separating the template from the pseudoknot. Using human telomerase, we show that the length of template 3′-flanking single-stranded RNA is a determinant of repeat addition processivity whereas template 5′-flanking single-stranded RNA and P2a.1 are critical for activity but not processivity. In comparison, requirements for the template sequence itself are confounding: different substitutions of the same position have strikingly different consequences, from improved processivity and activity to complete inactivation. We discovered that some altered-template sequences stabilize an alternative RNA conformation that precludes the pseudoknot by base-pairing of one pseudoknot strand to the template 3′ end. Using mutations to reduce over-stability of the alternative conformation, we restore high activity and processivity to otherwise inactive altered-template telomerase ribonucleoproteins. In cells, over-stabilization or destabilization of the alternative state severely inhibited biogenesis of active telomerase. Our findings delineate roles for human telomerase RNA template-flanking regions, establish a biologically relevant pseudoknot-alternative RNA conformation, and expand the repertoire of human telomerase repeat synthesis.
INTRODUCTION
Telomerase is a ribonucleoprotein (RNP) reverse transcriptase specialized for restoring the telomeric repeats eroded from chromosome ends during genome replication (1). Although telomerase is active in every cell division of single-celled organisms, multicellular animals including humans largely silence telomerase after early embryogenesis, with complete repression or only tightly regulated transient expression in human somatic cells (2,3). This repression of telomerase contributes to tumor suppression by limiting the proliferative capacity of any individual cell lineage. Remarkably, ∼90% of human cancers have reactivated telomerase (4,5). On the other hand, even slight deficiencies of normal human telomerase activation during early development lead to disease by premature exhaustion of telomere-limited proliferation. The spectrum of disorders associated with compromised telomerase function includes pulmonary fibrosis, aplastic anemia, dyskeratosis congenita, and other tissue renewal failures (6). The life-challenging consequences of aberrantly high or aberrantly low human telomerase function make this enzyme a promising therapeutic target.
Active telomerase RNP can be assembled by heterologous expression of the two catalytic core components: telomerase reverse transcriptase (TERT) and telomerase RNA (TER) (7). TERTs generally have four domains: an N-terminal (TEN) domain, high affinity RNA-binding domain (TRBD), reverse transcriptase domain (RT), and C-terminal extension (CTE) analogous to a polymerase thumb (8). The latter three domains, TRBD-RT-CTE, have a ring-like architecture established by TRBD-CTE interaction, thereby encircling a central cavity with the active site (9). The TEN domain is separated from the TRBD-RT-CTE (henceforth TERT ring) by a largely disordered linker, but it is stably positioned above the active-site face of TERT ring near the CTE (10). By an unknown mechanism, TEN domain stabilizes the initially short DNA-RNA duplex formed at the template 3′ end in the active site for productive elongation (8). In comparison to TERT, TER is highly divergent in sequence and secondary structure, ranging from ∼150–200 nucleotides (nt) in ciliates to ∼320–540 nt in vertebrates and ∼930 to >2000 nt in yeasts. Nonetheless, TER does possess functionally conserved motifs revealed by mutagenesis and complementation approaches (11). These motifs include a template, varying in length and reverse-transcription fidelity; a pseudoknot (PK) with characteristic base-triple pairings, variably spaced from the template 3′ end; and stem–loop/stem-junction motif folded separately from the template/PK (t/PK) that interacts with TERT and can be required for RNP activity (8,11). In human telomerase RNA (hTR), the t/PK and the stem–loop/stem-junction conserved region (CR) 4/5 combined are necessary and sufficient for telomerase reconstitution in cell extract or in cells (12–15). In this work, we use a minimized functional hTR (hTRmin) with t/PK connected to CR4/5 by a short linker (12,13). The hTRmin-TERT RNP is nearly indistinguishable from endogenous hTR-TERT RNP in catalytic activity and is biologically functional for telomere maintenance (12,13).
To regenerate its internal template, unlike other DNA polymerases, telomerase must release single-stranded rather than duplexed DNA product (16). Product release from the template can occur without release from the enzyme, allowing realignment of the product 3′ end with the template 3′ end for processive synthesis of another repeat (repeat addition processivity, or RAP). Some disease-associated human TERT mutations affect RAP (17–19), and RAP defects from TERT mutations, hTR mutations, or small molecule telomerase inhibitors compromise telomere maintenance or the telomere length set-point of homeostasis (20,21). The generally accepted model for a telomerase catalytic cycle of RAP begins with pairing of a primer 3′ end to the template 3′ alignment region that is a partial duplication of the residues that are involved in templating repeat synthesis. The initially short duplex engages the active site and is extended until the template 5′ boundary, with some duplex dissociation at the template 3′ end (20). In human telomerase, the template 5′ boundary is enforced in part by template 5′ end spacing from a 5′-flanking stem (hTR P1) (22), as typical of other telomerases (11), but also in large part by the sequence of the product–template duplex (12,22–25). Then, a proposed opening of the active site cavity would allow full duplex unpairing (26,27). The TERT ring could retain interactions with the DNA strand of template duplex upon its dissociation to single-stranded DNA (12,20), while the released template realigns to a ‘default’ positioning of the template 3′ end near the active site. If new duplex of the template and product 3′ ends is captured productively in the active site, telomerase commences another round of repeat synthesis.
Despite many previous studies of internal template use by telomerase, an open question remains: what are the RNA structure and/or protein interaction(s) that govern the positioning of unpaired template 3′ end in the active site, prior to duplex formation? Ciliate TERs contain a shared Template Recognition Element (TRE) sequence 3′ of the template, in the single-stranded segment connecting template to the PK. This TRE motif, on an RNA oligonucleotide also containing the template, can direct trans-template use by the Tetrahymena telomerase active site (28). As would be expected for a motif involved in template positioning, mutations in the ciliate Tetrahymena TRE compromise nucleotide addition at the 3′ end of the template and consequently compromise RAP (29,30). In both ciliate and human telomerase holoenzymes, the single-stranded template 3′-flanking region threads from the inner active site cavity of the TERT ring to its outside surface near the TEN domain (27,31,32). However, unlike the ciliate TERs, vertebrate TERs lack sequence conservation 5′ or 3′ of the template (33,34). Again unlike telomerase from Tetrahymena, trans-template use by human telomerase does not require an RNA oligonucleotide to contain template 3′-flanking sequence, although this sequence is stimulatory (12,35). Furthermore, in mammalian TERs, the template-flanking 3′ region gained stem P2a.1, which has functional and structural association to the TERT TEN domain (12).
Here, we characterize the functions and functional interplay of human telomerase template 3′-flanking single-stranded RNA, P2a.1, template 5′-flanking single-stranded RNA and the template sequence itself. Initial studies were performed using the activity-sufficient hTRmin added in excess to rabbit reticulocyte lysate (RRL) reactions that translate TERT in vitro. Conclusions from this reconstitution system were extended to cellular assembly conditions, using transient transfection to co-express hTRmin and TERT. Generally, template 3′-flanking single-stranded RNA deletions or insertions compromised RAP, whereas loss of P2a.1 or changes in the template 5′-flanking region did not alter RAP but did alter activity level. In addition, we discovered an alternative conformation of the t/PK, which can be deleteriously over-stabilized by introducing a template substitution that creates only a single additional alternative-conformation base pair. Because an alternative fold of TERT-free TER has been evident for telomerase from the ciliate Tetrahymena (36,37), a PK-alternative conformation of telomerase RNA could be universal. We adopted a strategy of adjusting the stability of the alternative fold independently of the active t/PK conformation, by which we rescued catalytic activity on several altered-template sequences. We show that disfavoring the alternative t/PK fold has no impact on RNA-TERT interaction but compromises activity reconstitution in cells. Our results yield new insights about the similarities and differences in mechanisms that guide TER and TERT co-folding and internal template use across evolutionarily divergent telomerase enzymes.
MATERIALS AND METHODS
Telomerase reconstitution in RRL
Wild-type (WT) and mutant hTRmin RNAs were transcribed from linearized plasmids using T7 RNA polymerase and purified by denaturing gel electrophoresis. TERT was expressed using the TNT-T7 coupled reticulocyte lysate system according to manufacturer's instructions (Promega) in reactions containing 40 ng/μl 3xFLAG-TERT pCITE expression construct and a vast molar excess (100 ng/μl) of purified hTRmin. RRL reactions for TERT expression and RNP assembly were incubated at 30°C for 3.5 h. TERT-hTR interaction is primarily determined by CR4/5, which was unaltered in the hTR variants studied in this work.
Telomerase reconstitution in cells
For RNP assembly in cells, WT and mutant hTRmin expression constructs were cloned in a pBS vector backbone with the U3 small nucleolar RNA promoter, a 5′ leader sequence (LhTRmin Leader C), and a MALAT1 triplex motif followed by an RNaseP cleavage site; this expression context generates biologically stable and functional hTRmin telomerase by 3′ processing of a non-polyadenylated precursor to the triplex motif 3′ boundary (13). Human U2OS cells were transiently transfected using calcium phosphate for expression of human TERT N-terminally tagged with tandem Protein A domains and triple FLAG peptide epitope (ZZF-TERT) from a pcDNA3.1 expression construct (13) and the hTRmin from pBS constructs. Cells were harvested after 48 hours and lysed by 3 freeze-thaw cycles in hypotonic lysis buffer (HLB: 20 mM HEPES at pH 8.0, 2 mM MgCl2, 0.2 mM EGTA, 10% glycerol, 0.1% NP-40, 1 mM DTT, 0.1 mM PMSF and Sigma protease inhibitor cocktail). NaCl was then added to 400 mM final concentration, and the whole-cell extract was cleared by centrifugation. Samples were normalized to 1.0–1.5 mg/ml by Bradford assay using BSA standards.
Normalized extract samples were used in equivalent amounts as inputs for northern blots and immunoblots. For immunopurification with FLAG M2 monoclonal antibody resin (Sigma-Aldrich), inputs were diluted to bring NaCl concentration to 150 mM. Samples were bound to FLAG resin by end-over-end rotation at 4°C for 2 h, and then washed three times with HLB containing 150 mM NaCl. After the final wash, the immunopurified (IP) samples were split into three tubes, and residual buffer was removed using a syringe. IP samples for RNA and protein analyses were boiled at 70°C for 5 min. IP samples for activity assays were used directly, without elution.
Telomerase activity assays
Activity assays were performed in 20 μl reaction mixtures containing either 4 μl RRL-reconstituted telomerase or cellular RNP immobilized on FLAG resin, diluted into activity assay buffer (50 mM Tris-acetate at pH 8.0, 3 mM MgCl2, 1 mM EGTA, 1 mM spermidine, 5 mM DTT and 5% glycerol). Samples were incubated with 500 nM of T21-GTTAGG primer for WT hTRmin or primers complementary to hTRmin altered-template sequence with the same repeat permutation. Primer extension reactions were initiated by adding 5 μM dGTP spiked with 0.1 μM α-32P-dGTP (3000 Ci/mmol, 10 mCi/ml, Perkin-Elmer), 250 μM dATP, 250 μM dTTP and 250 μM dCTP, unless otherwise mentioned. Reaction mixtures were incubated at 30°C for 1 h. Products were extracted using phenol–chloroform–isoamyl alcohol, precipitated with ethanol, and resolved on 12% polyacrylamide/7 M urea/0.6× Tris borate–EDTA gels. Gels were dried and exposed to phosphorimager screens. Products were visualized by scanning on Typhoon (GE healthcare) and subsequently analyzed using ImageJ software (NIH).
Northern blots
RNA samples were resolved by denaturing PAGE and transferred to Hybond N+ nylon membrane (GE healthcare), UV-crosslinked and incubated in Church's buffer (1% BSA, 1 mM EDTA, 0.5 M NaP04 at pH 7.2, 7% SDS) with 15% formamide at 50°C for membrane blocking. 32P-end-labeled probes complementary to regions in the hTR 3′ of P2 (5′-TAGAATGAACGGTGGAAGGCGGCAGGCCGAGGCT-3′) and hTR CR4/5 (5′-TCGCGGTGGCAGTGGGTGCCTCCGGAGAAGCCC-3′) were added and incubated at 50°C overnight. Membranes were washed three times with 1× SSC + 0.1% SDS at 50°C and exposed on phosphorimager screens. Signal was visualized by scanning on Typhoon (GE healthcare) and subsequently analyzed using ImageJ software (NIH).
Immunoblots
Protein samples were resolved on 4–12% SDS-PAGE gels and transferred to nitrocellulose membrane. Membranes were blocked in 4% non-fat milk (Carnation) in TBS buffer (150 mM NaCl, 50 mM Tris at pH 7.5) and then incubated with primary antibodies at 4°C overnight. FLAG-tagged TERT was detected using mouse anti-FLAG (1:4000, F1804, Sigma), and the internal control tubulin was detected using mouse anti-tubulin (1:500, DM1A, Calbiochem). After primary antibody incubation, membranes were washed extensively in TBS then incubated with goat anti-mouse IR 800 secondary antibody (1:3000, A-14, Santa Cruz Biotechnology) in 4% non-fat milk in TBS buffer for 1 h at room temperature. After extensive washing in TBS, blots were scanned using a LI-COR Odyssey imager (LI-COR Biosciences).
RESULTS
The length of template 3′-flanking single-stranded RNA is critical for high RAP
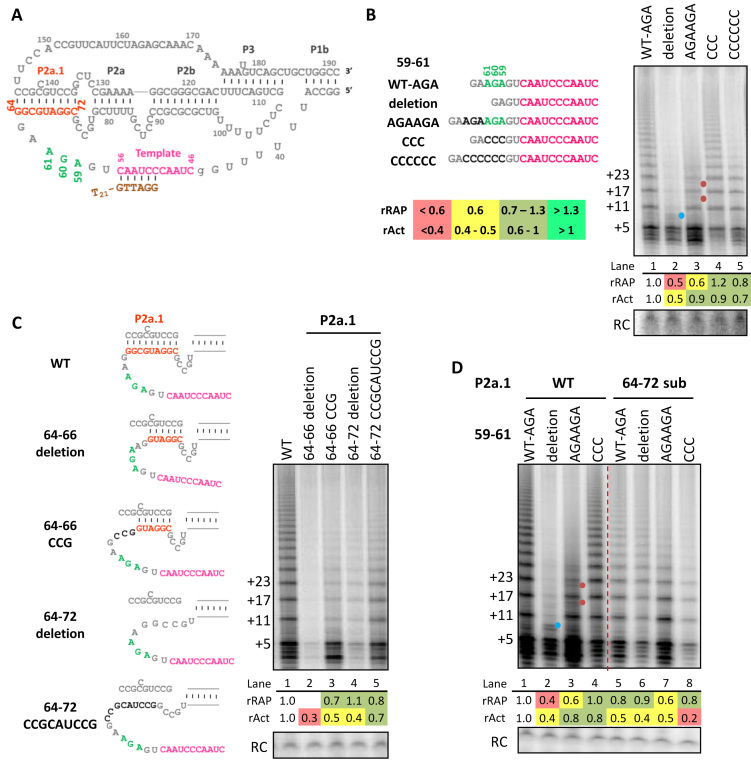
The absence of a phylogenetically conserved sequence motif 3′ of the vertebrate TER template suggests the possibility that vertebrate telomerases rely more on template base-pairing with DNA than on single-stranded template 3′ end positioning in the active site to support efficient initiation of repeat synthesis. On the other hand, the vertebrate template 3′-flanking region could have unrecognized sequence determinants of template 3′ end positioning. We therefore investigated whether the hTR template 3′-flanking region contributes to RAP. The Tetrahymena TER TRE UCU55-57 is spaced from the template residues encoding the TT of the telomeric repeat by 3 nt, with the equivalent sequence in hTR corresponding to AGA59-61 (Figure 1A and Supplementary Figure S1). The Tetrahymena TRE is followed by a substantial amount of single-stranded RNA, whereas hTR has at most 2 single-stranded nt on either side of AGA59-61 between the template and P2a.1 (Supplementary Figure S1). We used RRL reconstitution of TERT with hTRmin having the substitution U45g to the immediate 5′ of the template to be able to preclude template 5′ boundary bypass in activity assays without dCTP (Figure 1A). This substitution did not substantially alter telomerase activity level or RAP (Supplementary Figure S2, lanes 1–2 and 17–18). All further hTRmin mutations were made in the U45g background (simplified to WT hereafter). Telomerase activity was assayed by direct extension of primer T21-GTTAGG with radiolabeled dNTP mixtures. This primer has a telomeric repeat 3′-end permutation with unique alignment to the template, and its 5′ poly-thymidine tail improves binding to telomerase without possibility for primer folding into quadruplex structures (12). A typical product ladder reflected complete first-repeat synthesis by addition of 5 nt (+5) and subsequent 6-nt repeat additions (Figure 1B, product lengths indicated at left). To test whether sequence of the template 3′-flanking region affects overall activity or RAP, we made single, double and triple nt substitutions of AGA59–61. None of these had a dramatic impact on activity or RAP (Figure 1B, lane 4; Supplementary Figure S3).
Figure 1.
The template 3′-flanking single-stranded region length and P2a.1 stem differentially influence RAP and activity. (A) Diagram of the hTR t/PK with the proximal part of paired stem P1. Numbering indicates position within full-length hTR. Template is in pink, the 3′ template-flanking AGA59–61 is in green, and the bottom (5′) strand of P2a.1 in orange. Also shown is template alignment of the T21-GTTAGG primer used for activity assays in brown. Activity assays of RRL-reconstituted hTRmin mutants are shown with (B) template 3′-flanking 59–61 region length variations, (C) P2a.1 deletions or substitutions and (D) 59–61 and P2a.1 double mutations. Schematics for mutants are shown with mutated bases in darker gray. Assays were carried out using primer T21-GTTAGG in presence of all four dNTPs. The number of nucleotides added to the primer is indicated to the left. Here and in subsequent figure panels, the product precipitation recovery control (RC) was cropped from the faster-migrating region of the same gel. Blue dot indicates products from template 5′ boundary bypass, while red dot indicates products dissociated or stalled before complete repeat synthesis. Relative RAP (rRAP) values were calculated by dividing the collective intensities of products ≥2 repeats by the intensity of first-repeat products then normalizing the ratio to wild-type (WT) hTRmin enzyme. Relative activity (rAct) values were calculated by normalizing the collective intensities of all products to WT hTRmin enzyme. Values of rRAP and rAct are color-coded according to the key provided in (B) and Supplementary Figure S2, with severe defects indicated in red, moderate defects in yellow, levels in similar range as WT in light green, and improvements over WT in bright green. Similar results were obtained from two to four independent experimental replicates.
We next made several insertion or deletion (indel) mutants. In comparison to AGA59-61CCC substitution, we tested AGA59-61 deletion, AGA duplication, or AGA replacement by 6 cytosines (Figure 1B). All of the 59–61 indel changes reduced RAP (Figure 1B). We monitored changes in RAP by dividing the collective intensities of products ≥2 repeats by the initial repeat intensity then normalizing this ratio to the WT enzyme, to obtain rRAP. Also we evaluated changes in activity by normalizing collective intensities of total products to WT hTRmin enzyme to obtain rAct. The vast excess of hTRmin used for RRL reconstitution of human telomerase, combined with retention of the wild-type hTR CR4/5 motif that determines affinity for TERT, reduces the possibility of TERT interaction differences across the hTRmin sequence variants studied in this work. A severe RAP defect was observed with the deletion mutant, which had an additional product doublet between +5 and +11 (Figure 1B, lane 2, blue dot) that disappeared in reactions lacking dCTP (Supplementary Figure S2, note the absence of the lane 21 blue-dot product in lane 5). These findings indicate that shorter template 3′-flanking single-stranded RNA length favored template 5′ boundary bypass (dCTP-dependent products) and also compromised RAP independent of template 5′ boundary bypass (in reactions with or without dCTP).
Increasing the length of template 3′-flanking single-stranded RNA by AGA duplication also reduced RAP (Figure 1B, lanes 3 and 5). Unlike AGA deletion, template 3′-flanking region increases in length by insertion of AGA did not induce template 5′ boundary bypass, because inclusion or exclusion of dCTP from the activity assays did not affect the product profile (Supplementary Figure S2). However, AGA duplication did result in minor additional products, likely from premature dissociation before the enzyme reached the template 5′ boundary, because these products were detected in reactions with and without dCTP for any hTRmin enzyme with template 3′-flanking single-stranded region expansion (Figure 1B, lane 3, red dots; these products are more evident in Supplementary Figure S2, lanes 6–8 and 22–24). Curiously, combined mutation and expansion of the template 3′-flanking region by AGA replacement with CCCCCC had less impact on RAP than did expansions that retained an AGA (Figure 1B, compare lane 5 to lane 3; Supplementary Figure S2, lanes 6–9 and 22–25). Overall, we conclude that the length of the template 3′-flanking single-stranded region is more important for high RAP than its native sequence.
To investigate the mechanism by which the template 3′-flanking region length influences RAP, we tested whether template 3′-flanking region mutations compromised enzyme turnover using a pulse-chase primer extension protocol (Supplementary Figure S4). Telomerase was bound to a 27-nt primer and radiolabeled dNTP mix was added to initiate the initial primer extension. After 10 min, 100-fold molar excess of an 18-nt chase primer was added and DNA synthesis was allowed to continue, removing aliquots at regular time intervals. Changing the length of template 3′-flanking single-stranded RNA did not impede enzyme turnover, indicating that the reduced-RAP enzymes dissociate from product DNA rather than stall with bound product. Thus, RAP defects from changing the length of template 3′-flanking region do not reflect failure to dissociate the template–product duplex.
Stem P2a.1 improves human telomerase template use
Mammalian TERs acquired stem P2a.1 in place of some template 3′-flanking single-stranded RNA (Supplementary Figure S1). To investigate the significance of P2a.1 in human telomerase, we deleted or substituted different lengths of the P2a.1 bottom strand (5′ strand) with intact P2a.1 top-strand sequence, thereby partially or completely disrupting P2a.1 (Figure 1C, left). The larger substitution and deletion were less deleterious than the smaller substitution and deletion, and both substitution mutants (Figure 1C, lanes 3 and 5) showed better activity than the corresponding deletion mutants (lanes 2 and 4). Enzymes with the larger substitution or deletion of the bottom strand of P2a.1 had RAP close to normal. These results prompt the conclusion that P2a.1 is not critical for human telomerase RAP but is stimulatory for activity. Also, if P2a.1 is present, a full helical turn of duplex rather than a half-turn is optimal.
Curiously, compared to length changes at template 3′-flanking AGA59-61 (Figure 1B), length changes introduced by disruption of P2a.1 had little impact on RAP (Figure 1C). In an attempt to understand this disparity, we combined the entire P2a.1 bottom-strand substitution of residues 64–72 with changes to AGA59-61 (Figure 1D). When AGA59-61 deletion was combined with P2a.1 disruption, we observed a substantial gain in RAP compared to the 59–61 deletion mutant alone (Figure 1D, compare lane 2 to lane 6), along with loss of minor products from template 5′ boundary bypass (lane 2, blue dot). Also, combining AGA duplication with P2a.1 disruption suppressed the prematurely dissociated products detected with AGA duplication alone (Figure 1D, compare lane 3 with red-dot products to lane 7). Thus, unpairing P2a.1 compensated for changes in length of template 3′-flanking single-stranded RNA to allow high RAP. In comparison, combining the AGA59-61CCC sequence substitution with P2a.1 disruption was synthetically deleterious for activity (Figure 1D, compare lane 8 to lanes 4 and 5). This activity defect suggests that the P2a.1 stem and 59–61 sequence have mutually reinforcing roles in allowing efficient template use by the active site.
Changes to template 5′-flanking single-stranded RNA influence overall activity more than RAP
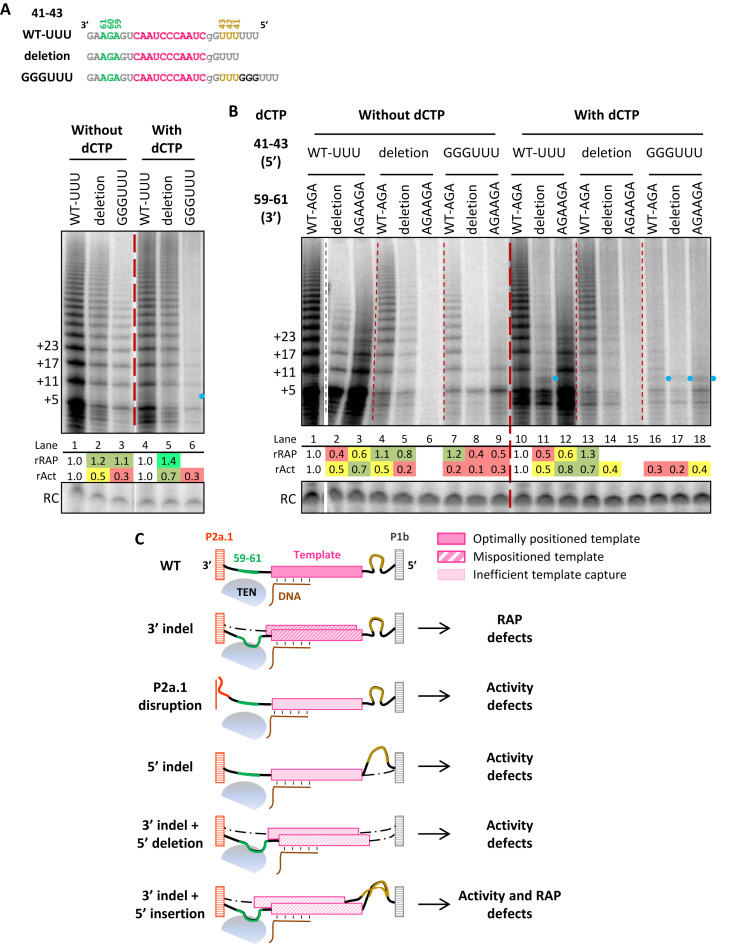
In general, vertebrate TERs have a pyrimidine-rich single-stranded sequence between stem P1 and the template 5′ end, with the exception of 5′-truncated rodent TERs lacking P1 (33). This template 5′-flanking single-stranded region is a defined length in Tetrahymena TER (Supplementary Figure S1), determinant of the template 5′ boundary and RAP (38). To probe whether vertebrate TER template 5′-flanking single-stranded RNA length has an influence on RAP, we assayed hTRmin mutants with a deletion of the 5′ flanking UUU41-43 or with a triplet guanosine insertion to generate GGGUUU (Figure 2A). Unlike the RAP inhibition by template 3′-flanking 59–61 deletion or duplication, template 5′-flanking 41–43UUU deletion or expansion reduced activity without inhibition of RAP in reactions lacking dCTP (Figure 2A, lanes 1–3). However, allowing template 5′ boundary bypass by inclusion of dCTP resulted in compromised RAP from template 5′-flanking region expansion (Figure 2A, lanes 1–6) along with the appearance of template 5′-boundary bypass product (lane 6, blue dot). This finding is consistent with previous studies of the role of the template 5′-flanking P1 in setting the template 5′ boundary (11,22).
Figure 2.
The template 5′-flanking region length contributes to activity and defining the template 5′-boundary. (A) Activity assay of hTRmin 41–43 indel mutants reconstituted in RRL. Diagrams of template 5′-flanking region length changes are shown with template in pink, AGA59–61 in green, UUU41–43 in yellow, and added sequence in darker gray. The assays were carried out using primer T21-GTTAGG in presence of all 4 dNTPs or omitting dCTP. (B) Activity assay of 59–61 and 41–43 combination mutants reconstituted in RRL in presence of all 4 dNTPs or omitting dCTP. Blue dots indicate products from template 5′ boundary bypass observed only in the presence of dCTP. Values of rRAP and rAct were calculated as described in Figure 1. Similar results were obtained from 2 independent experimental replicates. (C) Model for activity and RAP defects induced by length changes in the template 5′- and 3′-flanking single-stranded RNA regions and P2a.1 stem disruption. Schematic shows template position post-translocation with TEN domain of TERT near the 3′ end of template-product hybrid.
We also assayed combinations of template 5′-flanking indels and template 3′-flanking indels. Of interest, combined deletion of 41–43 and 59–61 preserved RAP (Figure 2B, lane 5). Combining template 5′-flanking 41–43 expansion with template 3′-flanking 59–61 deletion or duplication was deleterious for activity (Figure 2B, lanes 1–3 versus 7–9) and for template 5′-flanking sequence insertion caused template boundary bypass in reactions with dCTP (Figure 2B, lanes 16–18, blue dots). Activity became undetectable with the ‘template shift’ combination of 41–43 deletion and 59–61 AGA duplication (Figure 2B, lane 6). Altogether, our results suggest that template positioning relies on a balance of constraints from the 5′ and 3′ flanking elements (Figure 2C). RAP defects imposed by changing the length of template 3′-flanking single-stranded RNA could reflect more heterogeneous template positioning prior to DNA engagement (Figure 2C, mispositioned template). The roles of P2a.1 pairing and template 5′-flanking single-stranded region length could instead be to improve activity by favoring template threading through the active site cavity; without P2a.1 or proper length of the 5′ flanking region, a lower fraction of RNP would have template within reach of the active site (Figure 2C, inefficient template capture). Deletion but not expansion of the template 5′-flanking single-stranded RNA region could at least partially offset the loss of RAP from template 3′ indels, but no combination showed improved activity (Figure 2C).
Human telomerase has sequence-dependent tolerance for template changes
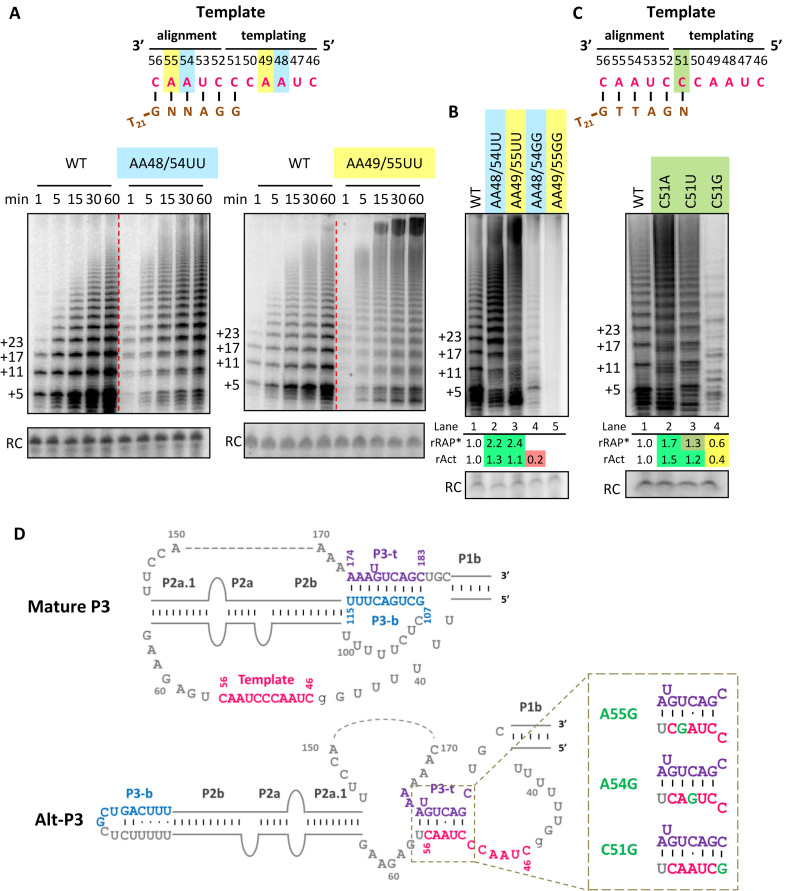
We next sought to test whether template sequence itself matters for template positioning in the active site. Human telomerase template sequence is known to influence the template 5′ boundary, as well as the rate and RAP of processive repeat synthesis, through sequence-specific RNA-DNA duplex recognition (12,23,24,39,40). Addition of product TT across from template AA48-49, followed by 2 nt of additional synthesis, gives a signal to ‘pause.’ This pausing improves template 5′ boundary fidelity but also reduces the rate of next-repeat synthesis, with the consequence that mutant repeats templated by altered AA48-49 are synthesized with higher rate and thus longer product lengths of high RAP (23,24).
Using hTRmin for RRL reconstitution, we recapitulated results from previous studies using other forms of hTR (39) in assays of paired alignment- and templating-region mutations AA48/54UU or AA49/55UU (Figure 3A). Enzymes assembled by both mutants of hTRmin show an increased rate of repeat addition, especially with the AA49/55UU template substitution: at a 5 min time point, AA48/54UU enzyme had added ∼18 detectable repeats and AA49/55UU enzyme >20 repeats compared to ∼9 repeats added by WT enzyme (Figure 3A). Compared to WT, AA48/54UU and AA49/55UU enzymes had greater RAP. As expected (23), the Tetra-U mutant hTRmin with all four template-region adenosines substituted to uridines also supported robust activity, but under our assay conditions, slightly less RAP than WT hTRmin enzyme (Supplementary Figure S5). We also compared activity of hTRmin template mutants assembled with TERT ring lacking the TEN domain (Supplementary Figure S5A, bottom panel). Curiously, even TERT without a TEN domain can support a low level of RAP upon abrogation of the template signal for synthesis pausing (AA48–49). For subsequent experiments with altered template hTRmin (Figures 3–6, Supplementary Figures S5 and S9), we quantified RAP by dividing the collective intensities of products ≥10 repeats by the collective intensities of products <10 repeats then normalizing the ratio to WT hTRmin enzyme, to obtain rRAP* (compared to a cutoff of ≥2 for rRAP previously; Figures 1 and 2, Supplementary Figures S2 and S3). This change reflects the need to discriminate opposite mutation impacts: the loss of RAP from template-flanking region mutation versus the increase in RAP by mutation of the template sequence itself. The color-coding for comparisons of rRAP* remained the same as used for rRAP (Figure 1B).
Figure 3.
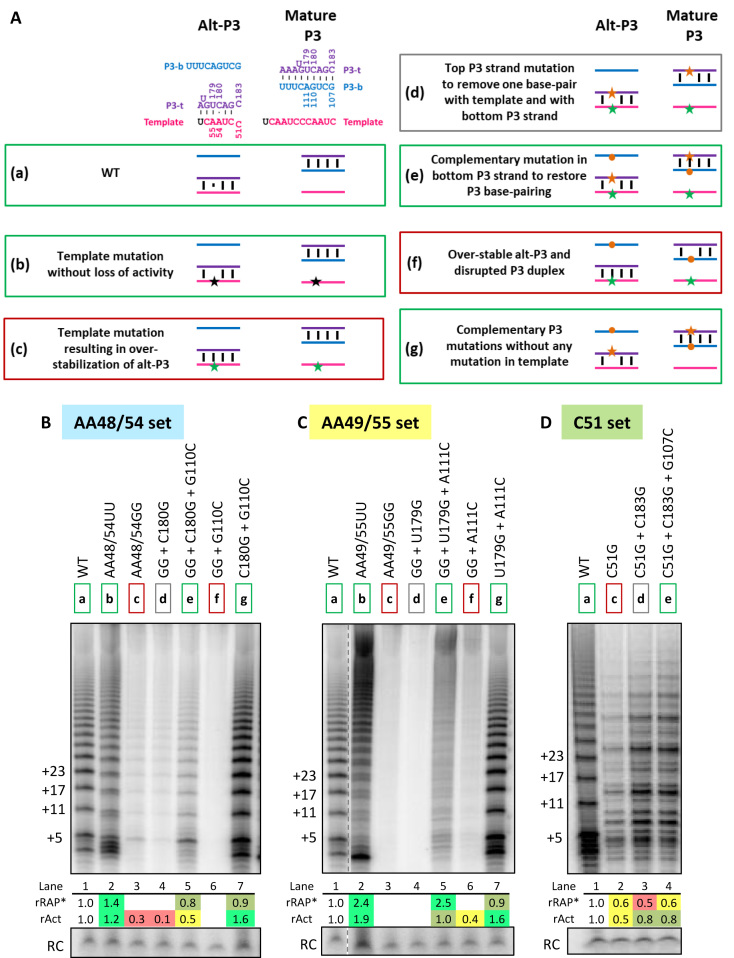
Template mutations at the same position have different impact on activity and RAP of hTRmin telomerase assembled in vitro. (A) Activity assay of hTRmin mutants with A to U template mutations reconstituted in RRL, assayed over a time course in reactions omitting dCTP. Template sequence is shown at top, with shading indicating matching alignment and templating positions that pair to the same position of a telomeric repeat. Primer is shown in brown; N indicates a base complementary to the altered template. (B) Comparison of enzyme activity for templates with a pair of A mutated to U versus G, assayed in the presence of all dNTPs. (C) Activity assay of enzymes with mutations at the C51 template position, with an illustration at top parallel to (A). rRAP* values were calculated by dividing the collective intensities of products ≥10 repeats by the collective intensities of products <10 repeats and then normalizing the ratio to WT hTRmin enzyme. rAct values were calculated as described in Figure 1. For (B) and (C), similar results were obtained from two to three independent experimental replicates. (D) Two structures of the hTR t/PK with coloring to indicate the template (pink) and P3 strands top (P3-t, purple) and bottom (P3-b, blue). Active ‘Mature P3’ conformation has a template accessible for pairing with substrate, whereas Alt-P3 does not (adapted from (36)). At right, altered template sequences predicted to additionally stabilize alt-P3 are shown with the template mutation in green.
Figure 6.
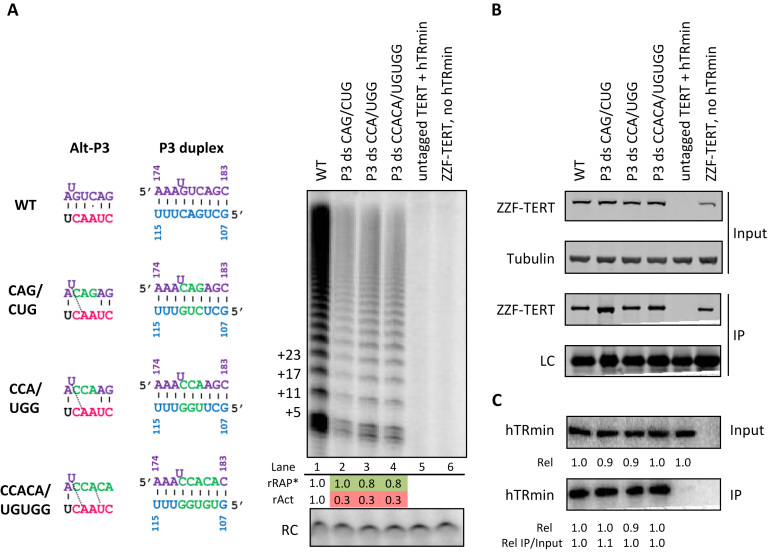
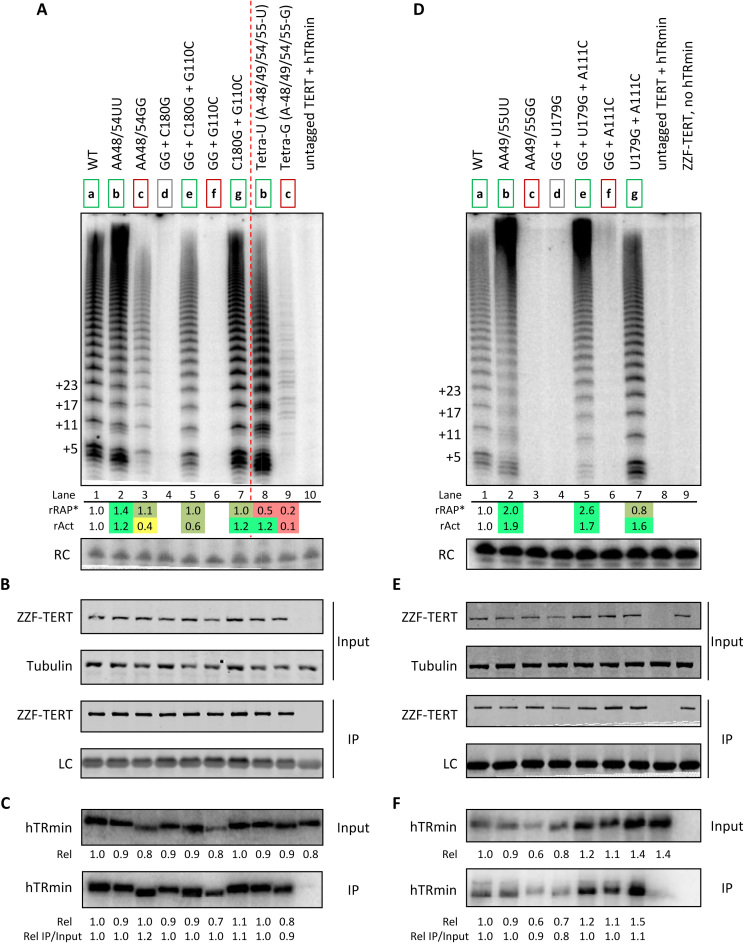
Alt-P3 may promote but is not essential for active telomerase assembly in in cells. (A) Activity assay, (B) immunoblot and (C) northern blot to follow assembly and activity of P3 duplex-swap hTRmin mutations co-expressed with ZZF-tagged TERT in U20S cells, paralleling the experiments described in Figure 5. Values for rRAP* and rAct were calculated as described in Figure 3. Similar results were obtained from two independent experimental replicates.
Unexpectedly, in contrast to the AA48/54UU and AA49/55UU mutants, AA48/54GG and AA49/55GG hTRmin reconstitutions showed little to no activity (Figure 3B). Similarly, the hTRmin Tetra-G mutant with all template adenosines substituted to guanosines was unable to reconstitute active RNP (Supplementary Figure S5B). In parallel, we tested substitutions of template position C51, using template-matched T21-GTTAGN primer so that the initial dNTP to be incorporated was the same across the altered templates in the first round of repeat synthesis (Figure 3C, schematic). We found that although C51A and C51U were tolerated for template use, C51G was not and showed reduced activity and RAP with dramatically altered product repeat addition ladder (Figure 3C). These observations raised the prospect that template ability to be positioned in the active site depends on template sequence itself. However, a different hypothesis that ultimately prevailed derives from the detection of an alternative pairing of Tetrahymena PK stem 3, in which the template is base-paired to what in active TER is the 3′ (top) strand of stem 3 (Supplementary Figure S1) (36,37). Tetrahymena TER alternative stem 3 alternative conformation (stem 3-alt) is favored in full-length TER without TERT. A putative hTR alternative P3 (alt-P3) could form by pairing part of the template to the 3′ (top) strand of active hTR P3 (Supplementary Figure S1) (36). This putative hTR alt-P3 would be predicted to have less thermodynamic stability than the P3-containing mature t/PK fold, which has been confirmed to predominate bulk hTR in vitro and in vivo (41). However, if biological context stabilizes alt-P3 and/or favors its accumulation as a kinetic intermediate in PK folding, a small amount of additional alt-P3 stabilization by template sequence changes could prevent the active-enzyme t/PK structure from forming (Figure 3D).
Alternative conformations of hTR t/PK explain template sequence requirements for activity
To test whether altered-template over-stabilization of an hTR alt-P3 conformation can account for the activity defects of some template mutants, we designed a strategy to rescue template use by the AA48/54GG, AA49/55GG and C51G hTRmin RNPs by eliminating their increase in stability of alt-P3 (Figure 4A, Supplementary Figures S6–S8). We mutated a P3 top-strand base such that it could no longer pair with the mutated template (change from state (c) to (d), Figure 4A). However, the mutated base could no longer form P3, which is essential for RNP activity (Supplementary Figure S9A). Hence, we next made a compensitory mutation in the P3 bottom strand to restore P3, thereby affecting alt-P3 stability without disruption of P3 (change from state (d) to state (e), Figure 4A). For comparison, we also made the combination of over-stable alt-P3 and disrupted P3 (state (f), Figure 4A), and as a control we tested the compensatory change in P3 top and bottom strands with no template mutation (state (g)).
Figure 4.
Over-stabilization of alt-P3 inhibits the activity of telomerase assembled in vitro. (A) Compensatory mutation strategy to rescue template mutations with over-stabilized alt-P3. (a) Alt-P3 (left) and Mature P3 (right) states of WT hTRmin with the strand coloring used throughout the schematics. Numbering for the bases mutated in this assay is indicated. (b) Template mutation without loss of activity (black star). (c) Template mutation that would over-stabilize alt-P3 state (green star). (d) Top P3 strand mutation (orange star) to remove one base-pair with template and with bottom P3 strand. (e) Complementary mutation in bottom P3 strand (orange circle) to restore P3 duplex base-pairing. (f) Mutations in template and bottom P3 strand. (g) Mutations in top and bottom strands of P3 without any template mutation. Positive control and successful rescue combinations predicted to have high catalytic activity are boxed green, those with little to no activity are boxed in red, and others are boxed in gray. (B–D) Activity assays for rescue of mutation at different template positions: (B) AA48/54, (C) AA49/55 and (D) C51. Shading for these template mutation sets is same as in Figure 3. The hTRmin mutants were reconstituted in RRL and assayed with a template-specific primer in the presence of all dNTPs. Mutations are indicated with the corresponding color-coded boxed ‘a–g’ states. Values for rRAP* and rAct were calculated as described in Figure 3. Similar results were obtained from 2 independent experimental replicates.
As described above, compared to the template AA48/54UU substitution, the template AA48/54GG substitution was strongly inhibitory (Figure 4B, lanes 1–3). The AA48/54GG mutations would stabilize alt-P3 by forming a G:C base-pair of 54G and C180 (Figure 3D, lower right). Disruption of the additional alt-P3 pairing of 54G by P3 top-strand mutation C180G did not rescue activity, but if the P3 base-pair was restored with G110C, activity rescue was observed (Figure 4B, lanes 3–5). Similarly, we rescued the entirely inactive AA49/55GG template (Figure 4C, lanes 1–3) that could form an alt-P3 G:U wobble base-pair between 55G and U179 (Figure 3D). Disruption of 55G alt-P3 base-pairing to P3 top strand with the mutation U179G did not restore activity, but with the additional mutation A111C to restore P3, activity improved dramatically, albeit with a notable smearing of the product ladder indicative of changes in limiting steps of repeat synthesis (Figure 4C, lanes 3–5). Like the AA49/55UU enzyme, the AA49/55GG enzyme had the increased RAP resulting from abrogation of the synthesis pause encoded in the product–template duplex sequence (24). As expected, the combination of over-stable alt-P3 and disrupted P3 was deleterious for activity, whereas compensatory change of P3 top and bottom strands alone was not (Figure 4B and C, lanes 6–7).
We used a similar mutagenesis strategy to test whether the low activity of enzyme with template C51G could be improved. C51G would over-stabilize alt-P3 by adding an additional base pair with C183 (Figure 3D, Supplementary Figure S8). Reverting the increase in alt-P3 stability with the P3 top strand mutation C183G (Supplementary Figure S8) gave a significant increase in activity, although still with the altered product profile indicative of changes in limiting steps of repeat synthesis (Figure 4D, lanes 1–3). Curiously, there was no further gain in activity from adding the G107C mutation to restore P3 (Figure 4D, lanes 3–4; Supplementary Figure S8). The P3 top-strand mutation C183G lies at the edge of the P3 duplex near P1, so P3 could form without the terminal base-pair. Overall, our results support the existence of an alt-P3 conformation and show that human telomerase template use can be expanded to new sequences by precluding over-stability of alt-P3.
Cellular telomerase reconstitution supports the importance of a fine-tuned stability of alt-P3
Human telomerase reconstitution in RRL requires a vast excess of purified hTRmin, with the RNA pre-folded and added prior to TERT synthesis. These assembly conditions could desensitize the detection of roles for alternative RNA structures as intermediates in hTR folding to its mature, functional conformation. Therefore, to extend our studies of a role for alternative RNA conformation, we reconstituted telomerase by transient co-expression of ZZF-TERT and WT or mutant hTRmin in telomerase-negative human U20S cells. Cellular telomerase RNP containing hTRmin is fully functional for telomere maintenance (13). TERT was affinity purified from cell extract using FLAG antibody resin, and the bound material was tested for catalytic activity, TERT, and hTRmin (Figure 5 sets A, B, C or D, E, F). We initially compared the template-only mutations tested by RRL reconstitution, with similar activity assay results: the 2-nt template mutations AA48/54GG and AA49/55GG inhibited activity while AA48/54UU and AA49/55UU did not (Figure 5A and D, lanes 1–3). The 4-nt Tetra-U and Tetra-G mutations of all template adenosines also altered activity in parallel with the RRL reconstitution results (Figure 5A, lanes 8–9; Supplementary Figure S5B).
Figure 5.
Over-stabilization of alt-P3 decreases active telomerase assembly in cells. (A) Activity of ZZF-tagged TERT RNP reconstituted in U20S cells with the AA48/54 set and Tetra-U/Tetra-G set of hTRmin template mutations. Cellular ZZF-TERT RNP was immunopurified (IP) and assayed directly on FLAG beads using template-specific primers in presence of all dNTPs. A background control expressed untagged TERT with WT (lane 10). Mutations are indicated with their corresponding ‘a-g’ states described for Figure 3. Values for rRAP* and rAct were calculated as described in Figure 3. (B) Immunoblot detection of ZZF-TERT in cell extracts and after immunopurification. Tubulin was used as the extract loading control for input samples and the FLAG-antibody light chain was non-specifically detected as a loading control for IP samples. (C) Northern blot detection of the overexpressed hTRmin in the cell extracts and after immunopurification. Normalized hTRmin RNA levels for the input and IP samples are shown relative to WT. (D–F) Experiments paralleling A–C for the AA49/55 set of hTRmin template mutations. Two background controls were included (lanes 8–9). Similar results were obtained from two independent experimental replicates.
Remarkably, the loss of activity by cell-reconstituted telomerase with template AA48/54GG or AA49/55GG was rescued by P3 top- and bottom-strand mutations that offset the gain in alt-P3 stability (Figure 5A and D, lane 5). No activity was detected for enzymes with template substitutions and disrupted P3, whereas enzymes with the P3 top- and bottom-strand compensatory mutations alone had robust activity (Figure 5A and D, lanes 4, 6–7). As expected, quantified copurification of hTRmin with TERT demonstrated that each t/PK-mutant RNA retained TERT interaction (Figure 5B, C and E, F), consistent with a primary role for hTR CR4/5 in hTR-TERT interaction (15). Paralleling the RRL assays (Figure 4B and C), enzyme assembled in cells by hTRmin with the compensatory P3-strand mutations U179G + A111C was more active than WT enzyme (Figures 4C and 5D, compare lanes 1 and 7) despite partial destabilization of alt-P3 (Supplementary Figure S7g). These experiments suggest a deleterious impact of alt-P3 over-stability.
To address whether alt-P3 has a role in active RNP biogenesis, we tested combinations of 3 or 5 nt P3 top- and bottom-strand mutations that are compensatory for P3 but should eliminate alt-P3 (Figure 6A, left). Despite intact P3, these mutations greatly reduced active RNP assembly in cells (Figure 6A). Quantification of hTRmin co-purified with TERT showed that each t/PK-mutant RNA retained a wild-type level of TERT interaction (Figure 6B and C), independent of the consequence of mutations on enzyme activity. Combined with the robust activity of enzyme with partial destabilization of alt-P3 by compensatory P3-strand mutations U179G + A111C (Figure 5D, lane 7), we suggest that alt-P3 impact is by competition for mature P3 pairing rather than as a specific structure. This is consistent with a lack of requirement for alt-P3 in RRL reconstitution (Supplementary Figure S9B and C), which in addition to a vast excess of hTRmin also obliges numerous folding chaperones to generate the active RNP (7,8). Thus, the role of alternative hTR structure formation is most consistent with function as an intermediate in cellular co-folding of hTR with TERT (Figure 7).
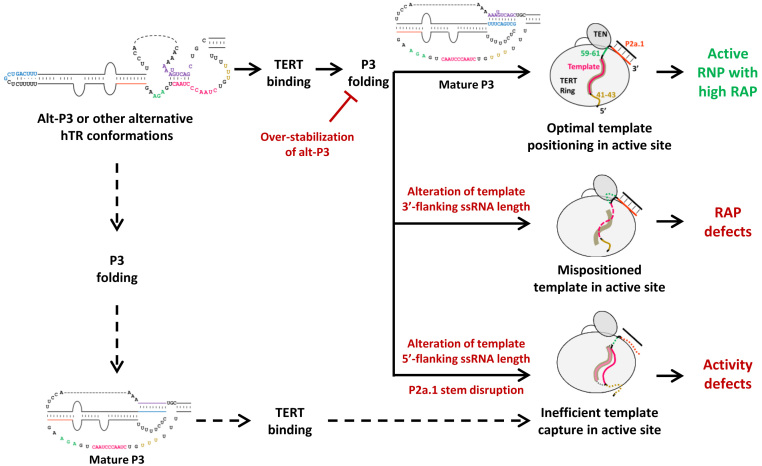
Figure 7.
Model for human telomerase activity and RAP dependence on hTR folding in the t/PK domain, TERT interaction, and template threading. The shaded groove in illustrations at right indicates the productive threading of template across the active site cavity; ss, single-stranded.
DISCUSSION
In this study, we defined RNA sequence and structure determinants of the human telomerase template positioning important for template use and RAP. Unlike template 5′ boundary recognition, which in human telomerase involves sequence-specific readout of the product–template duplex (24,40), template 3′ end positioning requires guidance for the path of single-stranded RNA. We showed that template 3′-flanking RNA influences repeat synthesis, with distinct requirements for RAP versus activity overall. Template 5′ flanking RNA length influences RAP less directly through enforcing the template 5′ boundary, since loss of RAP can be suppressed by preventing template 5′ boundary bypass. In contrast, loss of activity from template 5′ flanking RNA length changes cannot be rescued. Activity requirements for P2a.1 and template sequence were also explored. A surprising role for the template region was uncovered in our studies of template sequence changes: the template 3′ end can base-pair with a strand of P3 to form a duplex with stability limited for active RNP biogenesis. Together these findings bring new insights about how sequence and structure within the TER t/PK domain enable and constrain activity and RAP.
Template positioning relies on contributions from template 3′- and 5′-flanking regions and P2a.1
Paradigms for TER motif contributions to template positioning emerged from initial studies using the Tetrahymena enzyme. In ciliates, the conserved template 3′-flanking TRE motif contributes specifically to template positioning and 3′ end use in the active site (28,29). Here we show that for hTR, template 3′-flanking single-stranded RNA length is more important than sequence. This conclusion meshes with the conservation of this single-stranded region length in mammals with P2a.1, including in rodents with pyrimidine- rather than purine-rich template 3′-flanking sequence and no template 5′-flanking P1 (none differ from hTR by more than 2 nt of length, see (34)).
The template 3′-flanking single-stranded RNA threads from template in the active site cavity (the template 5′ end being placed there by TRBD anchoring off its 5′ side) to an exit path over the CTE and around the TEN domain, ending at the TEN domain surface that abuts the junction of template 3′-flanking sequence and P2a.1 (32). Human telomerase P2a.1 is rigidly fixed in tertiary structure as the terminus of a long, bent duplex extending from the PK. A much shorter mimic of P2a.1 containing a 3 bp stem can be formed in medaka fish Oryzias latipes TER, hinting at an elaboration of P2a.1 through vertebrate evolution (42). We suggest that TERT could constrain template 3′-flanking single-stranded RNA in a manner that accounts for the length specificity evident in our studies above, consistent with human telomerase holoenzyme structure (32) and a yeast TEN domain model for its function as a product–template duplex-splitter at the template 3′ end (43). This interaction could provide some constraint to support RAP in the absence of P2a.1.
In contrast to the RAP requirement for a specific length of template 3′-flanking single-stranded RNA connecting template to P2a.1, template 5′-flanking single-stranded RNA length had more impact on activity overall. Thus, template 3′- and 5′-flanking regions have distinct contributions to template positioning in the active site cavity (Figures 2C and 7). We propose that while the fixed length of the template 3′-flanking region plays a role in accurately repositioning the template during processive repeat synthesis, length of the template 5′-flanking region influences the overall probability of template capture in the active site cavity. Increased length of template 5′-flanking single-stranded RNA allows template 5′ boundary bypass to some extent in human telomerase (22), but to greater extent in ciliate telomerases that define the template 5′ end by a steric impediment of RNA duplex and/or bound TERT (38,44). Here, we found that altering the length of 3′-flanking single-stranded RNA, in the presence of P2a.1, also encouraged template 5′ boundary bypass. That mutations of template 3′-flanking sequence increased template 5′ boundary bypass is consistent with results from trans-template oligonucleotide assays of template-less hTRmin RNP, which found that inclusion of wild-type template 3′-flanking sequence on the template oligonucleotide improved activity and decreased template 5′ boundary bypass (12). These findings suggest the possibility that the template 5′-flanking and 3′-flanking single-stranded regions contribute to duplex handling, reflected in template 5′ boundary fidelity, as well as to the positioning of single-stranded template, reflected in overall activity and RAP defects even without template 5′ boundary bypass.
Interestingly, while an insertion in the hTR template 3′-flanking region or a deletion in the template 5′-flanking region individually had little impact on activity, the combination of these two indels as a template ‘shift’ in location between P1 and P2a.1 dramatically reduced telomerase activity. A less dramatic aggravation was observed in activity assays of Tetrahymena telomerase with TERs bearing deletions in the template 3′-flanking TRE and insertions in the template 5′-flanking single-stranded region (45). Because the human and Tetrahymena enzymes differ in their template 5′ boundary definition mechanisms (11), evolutionary conservation of template 5′-flanking regions would not be expected. Nonetheless, there is a general dependence of template use on the combination of template 3′-flanking and template 5′-flanking single-stranded RNA lengths.
Template sequence influences the catalytic cycle
Mutagenesis of hTR and TERT in previous studies revealed that telomerase synthesis rate and RAP can vary independently (39,46). Both rate and RAP increase with mutation of hTR template A49, which when paired with product acts as a signal to stop synthesis at the template 5′ boundary (24). Pairing of the same product position to template 3′ end A55 slows incorporation of the first nt of the next repeat (23). Our results extend these findings, showing increased RAP for enzymes reconstituted in RRL or in cells with hTRmin harboring an AA49/55UU or AA49/55GG template (the latter in the background of additional U179G and A111C). We also observed a RAP increase for enzymes reconstituted in RRL or in cells with hTRmin harboring the AA48/54UU template. These findings are notable in the consistency of increased RAP for enzymes synthesizing diverse repeat sequences: GGTTAG (WT), GGATAG (49U), GGCTAG (49G) or GGTAAG (48U). Thus, the wild-type enzyme may have evolved under selection to constrain the rate of sequential repeat synthesis, perhaps to sensitize the enzyme for regulation at telomeres.
Active RNP folding occurs via a putative intermediate
The PK forms in the bulk of hTR synthesized in vitro or in vivo, with or without TERT (41). However, NMR and FRET studies indicate that the hTR t/PK can sample alternative conformations (47–51). Compared to human, the Tetrahymena TER PK is smaller and less stably folded, making it logical that PK folding in full-length TER depends on TER association to TERT (37,52,53). Recombinant RNA with just the Tetrahymena PK sequence does fold as a PK (10,36,52). However, if template sequence is included adjacent to the PK, the t/PK instead folds as a long hairpin (36,37). This alternative conformation (Supplementary Figure S1) has been proposed to be an intermediate in the formation of active RNP, with melting of stem 3-alt and formation of mature stem 3 only upon TERT binding. Step-wise TER folding could be necessary to allow the template and flanking single-stranded RNA to thread across the full width of the TERT ring on the face with the active site cavity, followed by PK formation to clasp the template into activity-permissive positioning (31,36). Deleterious over-stabilization of Tetrahymena TER stem 3-alt would explain the otherwise unaccountably severe loss of activity imposed by TER C62G mutation in RRL-assembled RNP, although this severity was not evident for cell-assembled RNP (29,30). The biological impact of Tetrahymena TER alt-P3 destabilization remains to be investigated by future studies.
Given the high thermodynamic stability of the hTR PK compared to its Tetrahymena counterpart, it was not obvious that hTR would have a functionally relevant alt-P3. Also, it is unintuitive that a minimally base-paired structure such as proposed for hTR alt-P3 previously (36) and here could, with a single additional base-pair, present a major challenge for folding of active RNP. Nonetheless, our results support these conclusions. Formation of alt-P3 state appears more important in cells than in RRL for assembly of active RNP, which could be due to the RRL excess of hTRmin, abundance of chaperones, and/or co-translational rather than post-translational TERT-hTR assembly, among other differences. We cannot exclude the possibility that this difference in the impact of alt-P3 elements relates to an RNA folding difference that we have not anticipated. To account for how alt-P3 can be relevant for active RNP assembly in cells, we suggest that an alt-P3-specific feature of hTR t/PK sequence or structure is recognized by a biogenesis factor, whose binding would favor the otherwise frail base-pairing of alt-P3 and stabilize this conformation as a productive intermediate of active RNP assembly. Alt-P3 conformation and any biogenesis factors accompanying the alt-P3 state of hTR could also be important to protect the template from modification or cleavage prior to its protection by TERT. Curiously, mouse TER can form a much longer alt-P3 duplex than hTR (Supplementary Figure S1), which could compensate for lack of P1 to protect the TER 5′ end prior to its association with TERT. The benefit of a stronger alt-P3 pairing in 5′-truncated mouse TER would account for the template alignment-region A to G mutation that reduces mouse telomerase RAP (54).
We suggest that the nascent hTR transcript initially folds as alt-P3 but with time folds to the more thermodynamically stable PK. RNA remodeling from alt-P3 to P3 could be promoted by TERT interaction or occur with kinetics independent of TERT. If TERT binds CR4/5 prior to P3 formation, subsequent PK formation generates an active RNP (Figure 7, top). However, if the PK folds before TERT binds CR4/5, PK formation precludes template threading across the full width of the TERT ring above the active site cavity, producing inactive RNP (Figure 7, dashed lines). We suggest that productive template threading also relies on features of template-flanking single-stranded regions and P2a.1, which places the 3′ end of template 3′-flanking single stranded RNA at a specific surface of the TERT TEN domain (32). If premature PK formation places the template in position(s) inaccessible to the active site in a fraction of the RNP population, that fraction would be inactive, reflected as loss of RNP activity overall (i.e., a lower percentage of the RNP population has template ‘captured’ in the active site cavity; Figure 7). On the other hand, if the entire population of RNP has template in the active site cavity but less favorable template 3′ end positioning prior to DNA pairing, the activity defect manifests as lower RAP (Figure 7).
It remains an open question whether hTR adopts a diversity of t/PK folding intermediates other than alt-P3, any number of which could be sampled for interaction by TERT but only some of which could undergo successful folding to active RNP. As precedent, the t/PK domain of Saccharomyces cerevisiae TER has two conformations with alternative stem pairing (55) as does the Tetrahymena TER t/PK (36,37). The evidence presented in this work for an alt-P3 hTR conformation extends previous studies in yeast and ciliate systems that together suggest a potentially fundamental principle for co-folding of TER and TERT. By developing mutagenesis strategies that preserve a finely balanced stability of alt-P3 while changing template sequence, we substantially expanded the template repertoire that can be used by human telomerase. This accomplishment facilitates the use of altered-template telomerases to make defined changes in telomeric repeat sequence at chromosome ends, which has revealed telomere protection requirements and mechanisms for telomere length homeostasis (56–60).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank R.A. Wu and H. Upton for initial guidance with RRL assays, and J. Vogan and X. Zhang for initial guidance with cellular assays. The authors also thank R.A. Wu, H. Upton and T.H.D. Nguyen for constructive comments on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM054198, HL079585 to K.C.]. Funding for open access charge: National Institutes of Health [GM054198, HL079585 to K.C.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Blackburn E.H., Greider C.W., Szostak J.W.. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat. Med. 2006; 12:1133–1138. [DOI] [PubMed] [Google Scholar]
- 2. Aubert G. Telomere dynamics and aging. Prog. Mol. Biol. Transl. Sci. 2014; 125:89–111. [DOI] [PubMed] [Google Scholar]
- 3. Aubert G., Lansdorp P.M.. Telomeres and aging. Physiol Rev. 2008; 88:557–579. [DOI] [PubMed] [Google Scholar]
- 4. Shay J.W., Wright W.E.. Role of telomeres and telomerase in cancer. Semin. Cancer Biol. 2011; 21:349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shay J.W. Role of telomeres and telomerase in aging and cancer. Cancer Discov. 2016; 6:584–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Armanios M., Blackburn E.H.. The telomere syndromes. Nat. Rev. Genet. 2012; 13:693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Weinrich S.L., Pruzan R., Ma L., Ouellette M., Tesmer V.M., Holt S.E., Bodnar A.G., Lichsteiner S., Kim N.W., Trager J.B. et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 1997; 17:498–502. [DOI] [PubMed] [Google Scholar]
- 8. Wu R.A., Upton H.E., Vogan J.M., Collins K.. Telomerase mechanism of telomere synthesis. Annu. Rev. Biochem. 2017; 86:439–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mitchell M., Gillis A., Futahashi M., Fujiwara H., Skordalakes E.. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat. Struct. Mol. Biol. 2010; 17:513–518. [DOI] [PubMed] [Google Scholar]
- 10. Jiang J., Miracco E.J., Hong K., Eckert B., Chan H., Cash D.D., Min B., Zhou Z.H., Collins K., Feigon J.. The architecture of Tetrahymena telomerase holoenzyme. Nature. 2013; 496:187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Podlevsky J.D., Chen J.J.. Evolutionary perspectives of telomerase RNA structure and function. RNA Biol. 2016; 13:720–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu R.A., Collins K.. Human telomerase specialization for repeat synthesis by unique handling of primer-template duplex. EMBO J. 2014; 33:921–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vogan J.M., Zhang X., Youmans D.T., Regalado S.G., Johnson J.Z., Hockemeyer D., Collins K.. Minimized human telomerase maintains telomeres and resolves endogenous roles of H/ACA proteins, TCAB1, and Cajal bodies. eLife. 2016; 5:e18221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tesmer V.M., Ford L.P., Holt S.E., Frank B.C., Yi X., Aisner D.L., Ouellette M., Shay J.W., Wright W.E.. Two inactive fragments of the integral RNA cooperate to assemble active telomerase with the human protein catalytic subunit (hTERT) in vitro. Mol. Cell. Biol. 1999; 19:6207–6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mitchell J.R., Collins K.. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase in vivo and in vitro. Mol. Cell. 2000; 6:361–371. [DOI] [PubMed] [Google Scholar]
- 16. Collins K. Single-stranded DNA repeat synthesis by telomerase. Curr. Opin. Chem. Biol. 2011; 15:643–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Robart A.R., Collins K.. Investigation of human telomerase holoenzyme assembly, activity, and processivity using disease-linked subunit variants. J. Biol. Chem. 2010; 285:4375–4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alder J.K., Cogan J.D., Brown A.F., Anderson C.J., Lawson W.E., Lansdorp P.M., Phillips J.A. 3rd, Loyd J.E., Chen J.J., Armanios M.. Ancestral mutation in telomerase causes defects in repeat addition processivity and manifests as familial pulmonary fibrosis. PLoS Genet. 2011; 7:e1001352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zaug A.J., Crary S.M., Jesse Fioravanti M., Campbell K., Cech T.R.. Many disease-associated variants of hTERT retain high telomerase enzymatic activity. Nucleic Acids Res. 2013; 41:8969–8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu R.A., Tam J., Collins K.. DNA-binding determinants and cellular thresholds for human telomerase repeat addition processivity. EMBO J. 2017; 36:1908–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pascolo E., Wenz C., Lingner J., Hauel N., Priepke H., Kauffmann I., Garin-Chesa P., Rettig W.J., Damm K., Schnapp A.. Mechanism of human telomerase inhibition by BIBR1532, a synthetic, non-nucleosidic drug candidate. J. Biol. Chem. 2002; 277:15566–15572. [DOI] [PubMed] [Google Scholar]
- 22. Chen J.L., Greider C.W.. Template boundary definition in mammalian telomerase. Genes Dev. 2003; 17:2747–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen Y., Podlevsky J.D., Logeswaran D., Chen J.J.. A single nucleotide incorporation step limits human telomerase repeat addition activity. EMBO J. 2018; 37:e97953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brown A.F., Podlevsky J.D., Qi X., Chen Y., Xie M., Chen J.J.. A self-regulating template in human telomerase. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:11311–11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moriarty T.J., Marie-Egyptienne D.T., Autexier C.. Regulation of 5′ template usage and incorporation of noncognate nucleotides by human telomerase. RNA. 2005; 11:1448–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang W., Lee Y.S.. A DNA-hairpin model for repeat-addition processivity in telomere synthesis. Nat. Struct. Mol. Biol. 2015; 22:844–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chan H., Wang Y., Feigon J.. Progress in human and Tetrahymena telomerase structure determination. Annu. Rev. Biophys. 2017; 46:199–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miller M.C., Collins K.. Telomerase recognizes its template by using an adjacent RNA motif. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:6585–6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cunningham D.D., Collins K.. Biological and biochemical functions of RNA in the Tetrahymena telomerase holoenzyme. Mol. Cell. Biol. 2005; 25:4442–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eckert B., Collins K.. Roles of telomerase reverse transcriptase N-terminal domain in assembly and activity of Tetrahymena telomerase holoenzyme. J. Biol. Chem. 2012; 287:12805–12814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jiang J., Chan H., Cash D.D., Miracco E.J., Ogorzalek Loo R.R., Upton H.E., Cascio D., O’Brien Johnson R., Collins K., Loo J.A. et al. Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions. Science. 2015; 350:aab4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nguyen T., Tam J., Wu R., Greber B., Toso D., Nogales E., Collins K.. Cryo-EM structure of substrate-bound human telomerase holoenzyme. Nature. 2018; 557:190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen J.L., Blasco M.A., Greider C.W.. Secondary structure of vertebrate telomerase RNA. Cell. 2000; 100:503–514. [DOI] [PubMed] [Google Scholar]
- 34. Podlevsky J.D., Bley C.J., Omana R.V., Qi X., Chen J.J.. The telomerase database. Nucleic Acids Res. 2008; 36:D339–D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Qi X., Xie M., Brown A.F., Bley C.J., Podlevsky J.D., Chen J.J.. RNA/DNA hybrid binding affinity determines telomerase template-translocation efficiency. EMBO J. 2012; 31:150–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cash D.D., Feigon J.. Structure and folding of the Tetrahymena telomerase RNA pseudoknot. Nucleic Acids Res. 2017; 45:482–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cole D.I., Legassie J.D., Bonifacio L.N., Sekaran V.G., Ding F., Dokholyan N.V., Jarstfer M.B.. New models of Tetrahymena telomerase RNA from experimentally derived constraints and modeling. J. Am. Chem. Soc. 2012; 134:20070–20080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Autexier C., Greider C.W.. Boundary elements of the Tetrahymena telomerase RNA template and alignment domains. Genes Dev. 1995; 9:2227–2239. [DOI] [PubMed] [Google Scholar]
- 39. Drosopoulos W.C., Direnzo R., Prasad V.R.. Human telomerase RNA template sequence is a determinant of telomere repeat extension rate. J. Biol. Chem. 2005; 280:32801–32810. [DOI] [PubMed] [Google Scholar]
- 40. Wu R.A., Collins K.. Sequence specificity of human telomerase. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:11234–11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zemora G., Handl S., Waldsich C.. Human telomerase reverse transcriptase binds to a pre-organized hTR in vivo exposing its template. Nucleic Acids Res. 2016; 44:413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang Y., Yesselman J.D., Zhang Q., Kang M., Feigon J.. Structural conservation in the template/pseudoknot domain of vertebrate telomerase RNA from teleost fish to human. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:e5125–e5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Petrova O.A., Mantsyzov A.B., Rodina E.V., Efimov S.V., Hackenberg C., Hakanpaa J., Klochkov V.V., Lebedev A.A., Chugunova A.A., Malyavko A.N. et al. Structure and function of the N-terminal domain of the yeast telomerase reverse transcriptase. Nucleic Acids Res. 2018; 46:1525–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lai C.K., Miller M.C., Collins K.. Template boundary definition in Tetrahymena telomerase. Genes Dev. 2002; 16:415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Berman A.J., Akiyama B.M., Stone M.D., Cech T.R.. The RNA accordion model for template positioning by telomerase RNA during telomeric DNA synthesis. Nat. Struct. Mol. Biol. 2011; 18:1371–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xie M., Podlevsky J.D., Qi X., Bley C.J., Chen J.J.. A novel motif in telomerase reverse transcriptase regulates telomere repeat addition rate and processivity. Nucleic Acids Res. 2010; 38:1982–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hengesbach M., Kim N.K., Feigon J., Stone M.D.. Single-molecule FRET reveals the folding dynamics of the human telomerase RNA pseudoknot domain. Angew. Chem. Int. Ed. Engl. 2012; 51:5876–5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Parks J.W., Kappel K., Das R., Stone M.D.. Single-molecule FRET-Rosetta reveals RNA structural rearrangements during human telomerase catalysis. RNA. 2017; 23:175–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Theimer C.A., Finger L.D., Trantirek L., Feigon J.. Mutations linked to dyskeratosis congenita cause changes in the structural equilibrium in telomerase RNA. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:449–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Theimer C.A., Blois C.A., Feigon J.. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol. Cell. 2005; 17:671–682. [DOI] [PubMed] [Google Scholar]
- 51. Kim N.K., Zhang Q., Zhou J., Theimer C.A., Peterson R.D., Feigon J.. Solution structure and dynamics of the wild-type pseudoknot of human telomerase RNA. J. Mol. Biol. 2008; 384:1249–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mihalusova M., Wu J.Y., Zhuang X.. Functional importance of telomerase pseudoknot revealed by single-molecule analysis. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:20339–20344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sperger J.M., Cech T.R.. A stem–loop of Tetrahymena telomerase RNA distant from the template potentiates RNA folding and telomerase activity. Biochemistry. 2001; 40:7005–7016. [DOI] [PubMed] [Google Scholar]
- 54. Chen J.L., Greider C.W.. Determinants in mammalian telomerase RNA that mediate enzyme processivity and cross-species incompatibility. EMBO J. 2003; 22:304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu F., Kim Y., Cruickshank C., Theimer C.A.. Thermodynamic characterization of the Saccharomyces cerevisiae telomerase RNA pseudoknot domain in vitro. RNA. 2012; 18:973–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hanish J.P., Yanowitz J.L., de Lange T.. Stringent sequence requirements for the formation of human telomeres. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:8861–8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kirk K.E., Harmon B.P., Reichardt I.K., Sedat J.W., Blackburn E.H.. Block in anaphase chromosome separation caused by a telomerase template mutation. Science. 1997; 275:1478–1481. [DOI] [PubMed] [Google Scholar]
- 58. Stohr B.A., Xu L., Blackburn E.H.. The terminal telomeric DNA sequence determines the mechanism of dysfunctional telomere fusion. Mol. Cell. 2010; 39:307–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Diolaiti M.E., Cimini B.A., Kageyama R., Charles F.A., Stohr B.A.. In situ visualization of telomere elongation patterns in human cells. Nucleic Acids Res. 2013; 41:e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li S., Rosenberg J.E., Donjacour A.A., Botchkina I.L., Hom Y.K., Cunha G.R., Blackburn E.H.. Rapid inhibition of cancer cell growth induced by lentiviral delivery and expression of mutant-template telomerase RNA and anti-telomerase short-interfering RNA. Cancer Res. 2004; 64:4833–4840. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.