Abstract
Baculovirus (BV) holds promise as a vector for anticancer gene delivery to combat the most common liver cancer–hepatocellular carcinoma (HCC). However, in vivo BV administration inevitably results in BV entry into non-HCC normal cells, leaky anticancer gene expression and possible toxicity. To improve the safety, we employed synthetic biology to engineer BV for transgene expression regulation. We first uncovered that miR-196a and miR-126 are exclusively expressed in HCC and normal cells, respectively, which allowed us to engineer a sensor based on distinct miRNA expression signature. We next assembled a synthetic switch by coupling the miRNA sensor and RNA binding protein L7Ae for translational repression, and incorporated the entire device into a single BV. The recombinant BV efficiently entered HCC and normal cells and enabled cis-acting transgene expression control, by turning OFF transgene expression in normal cells while switching ON transgene expression in HCC cells. Using pro-apoptotic hBax as the transgene, the switch-based BV selectively killed HCC cells in separate culture and mixed culture of HCC and normal cells. These data demonstrate the potential of synthetic switch-based BV to distinguish HCC and non-HCC normal cells for selective transgene expression control and killing of HCC cells.
INTRODUCTION
Hepatocellular carcinoma (HCC) is a primary malignancy of the liver and is the third leading cause of cancer deaths worldwide (1), hence entailing the need to develop effective therapeutic strategies. Baculovirus (BV) is an insect virus that is non-pathogenic to humans, yet BV also efficiently transduces a broad range of mammalian cells, including hepatocytes (2), HCC cells (3), endothelial cells (4) and stem cells (5). In transduced cells, BV confers transgene expression as long as the transgene is under the transcriptional control of an appropriate promoter. By delivering anticancer genes (6–10), BV is able to potently suppress tumor progression and thus holds promise for cancer gene therapy. Given these properties, we recently developed recombinant BVs to express miRNA (11) or long non-coding RNA (12) to combat HCC. Injection of these recombinant BVs into subcutaneous HCC tumors in mice suppressed the tumor progression. However, these studies used constitutive promoters to drive the transgene expression. In the clinical setting, in vivo administration of BV into patient's liver would inevitably transduce other adjacent normal cells (e.g. hepatocyte or vascular endothelial cells), resulting in leaky anticancer gene expression in normal cells and possible toxicity. Therefore, it is desirable to develop a recombinant BV that can control transgene expression exclusively in the target HCC cells.
Synthetic biology aspires to build functional biological devices to detect physiologically/pathologically relevant input signals so as to regulate output gene expression and cell behavior, using biomolecular components and genetic modules (13–18). Today researchers have designed a wide variety of synthetic switches and circuits to operate gene expression control in the transcription, translation and post-translation levels (19,20). Among these devices, trigger-inducible RNA riboswitches composed of a sensor (e.g. aptamer) and an actuator domain have been extensively developed to turn ON/OFF gene expression and can be wired to higher-order synthetic circuits (20–22). However, many of these riboswitches require adding an inducing ligand (20,22,23), thus making its in vivo applications in animals and humans more complicated. RNA switches can also be fabricated by appending miRNA complementary sequences at the 3′ untranslated region (UTR) of transgene to recognize intracellular miRNA and inhibit transgene expression (24). These miRNA-based sensing mechanisms have been exploited to turn ON/OFF gene expression for classifying cancer and non-cancer cells (25,26).
In addition, archaeal ribosomal protein L7Ae is an RNA-binding protein that binds with high affinity to RNA motif known as kink-turn (K-turn) and represses subsequent RNA translation (for review see (22)). The interaction of L7Ae:K-turn was first harnessed to construct an OFF switch to repress mRNA translation by inserting K-turn motifs at the 5′ UTR of a reporter gene (27). This translation repression property inspires the leverage of L7Ae:K-turn as a regulatory part for assembling synthetic switch (28–30), multi-input logic gate (31) and complex hierarchical synthetic circuits for applications such as cell classifier (32) and biocomputer (33).
To improve the safety profile of recombinant BV and selectively turn ON transgene expression in target HCC cells but not in normal cells, in this study we exploited the miRNA-based sensor and L7Ae:K-turn system to create a genetic switch, which senses the differential miRNA profiles in HCC and normal cells and responds with different outputs. We started from examining the miRNA expression profiles in HCC and normal cells and identified the ‘OncoMiR’ (the miRNA highly expressed in HCC cells but poorly expressed in normal cells) and ‘NormalMiR’ (the miRNA highly expressed in normal cells but poorly expressed in HCC cells). We next assembled a cis-acting genetic switch by coupling the L7Ae:K-turn translation repression system and miRNA sensors, and incorporated the entire genetic device into a single recombinant BV. We next evaluated whether the recombinant BV was able to discriminate HCC/normal cells and exclusively express the reporter gene (EBFP) in HCC cells. We also used the pro-apoptotic gene hBax as the transgene and evaluated whether the resultant BV was able to selectively kill HCC cells but not normal cells, in separate culture and mixed culture of HCC and normal cells.
MATERIALS AND METHODS
Cell culture
Insect cell Sf-9 was cultured using modified Grace's medium (Sigma) supplemented with 10% fetal bovine serum (FBS, Hyclone) at 27°C. Other mammalian cells were cultured in a 37°C, 5% CO2 incubator. HCC cells PLC/PRF/5 (designated as PLC thereafter), HepG2 and Hep3B were cultured using α-MEM medium (Gibco) containing 10% FBS. Mahlavu, Huh-7 and HEK293 were cultured in High glucose DMEM medium (Gibco) containing 10% FBS. Human umbilical vascular endothelial cells (HUVEC) were cultured in 1% gelatin-coated flasks using ECM medium (ScienCell Research Laboratories) supplemented with 5% FBS, 1% Penicillin-streptomycin and 1% Endothelial Cell Growth Supplement (ScienCell Research Laboratories).
Construction of recombinant BVs
miRNA sponge is the decoy sequence with miRNA binding sites capable of sequestering miRNAs. pBac-T2-CdE is a vector for BV construction (11), harboring CMV promoter, destabilized enhanced green fluorescent protein (d2EGFP), WPRE (woodchuck hepatitis virus posttranscriptional regulatory element, for mRNA stabilization) and poly A signal. Ten tandem repeats of miRNA sponge sequences derived from CXCR4 gene (not complementary to any known miRNA, used as scramble control) and 10 repeats of miRNA sponges targeting miR-196a-5p were synthesized (RNAi Core Facility, Academia Sinica, Taiwan) and cloned into pBac-T2-CdE at the 3′ UTR of d2EGFP to yield pBac-T2-Scramble and pBac-T2-s196, respectively. Eight tandem repeats of pre-miR-196a sequences were subcloned from pTA-miR196a (34) to pBac-T2-122 (11) at the 5′ UTR of d2EGFP to replace the original pre-miR-122 sequences and yielded pBac-T2-m196. The miRNA sponges and pre-miR-196a were driven by the CMV promoter and contained WPRE and poly A signal (PA) at the 3′ end.
To construct the synthetic switch, the genes encoding L7Ae, porcine teschovirus-1 2A (P2A) sequence and enhanced yellow fluorescent protein (EYFP) were PCR-amplified from pTK095 (Addgene (32)). The L7Ae-P2A-EYFP fragment was cloned into pBac-T2-s196 to remove d2EGFP and placed L7Ae-P2A-EYFP in between CMV promoter and miR-196a sponge to yield pBac-T2-L7Ae-s196. The miR-196 sponge was subsequently removed to yield pL7Ae/EYFP. Meanwhile, two repeats of K-turn (Kt) motif and the DNA sequence encoding enhanced blue fluorescent protein (EBFP) were PCR-amplified from pTK332 (Addgene (32)) and cloned into TA vector (Invitrogen). The DNA fragment encoding Kt-EBFP was cloned into pBac-T2-CdE to remove d2EGFP and yield pK-turn/EBFP. The entire cassette encoding CMV-Kt-EBFP-WPRE-PA was subcloned into pL7Ae/EYFP to yield pBac-T2-LKE.
To assemble the synthetic switch comprising the miRNA sensor and L7Ae:K-turn, hBax and 2 repeats of K-turn motif were PCR-amplified from hBax-C3-EGFP plasmid (Addgene) and pTK332 (Addgene), respectively, and cloned into TA vector. The gene fragment (Kt-hBax) was subsequently cloned into pBac-T2-CdE to replace d2EGFP and yielded pBac-T2-hBax. Next, 4 tandem repeats of miR-126 complementary sequences (4 × 126) were synthesized (Genescript) and cloned into pBac-T2-hBax at 3′ UTR of hBax. The fragment encoding Kt-hBax-4 × 126 was cloned into pBac-T2-L7Ae-s196 to yield pCirSB. Four tandem repeats of miR-196a complementary sequences (4 × 196) were synthesized (Genescript) and cloned into pCirSB to replace the miR-196a sponge and yielded pBac-T2-CirCB. Conversely, 4 tandem repeats of miR-126 complementary sequences were cloned into pK-turn/EBFP at the 3′ UTR of EBFP to yield pBac-T2-kt-EBFP-4 × 126. The cassette encoding Kt-EBFP-4 × 126 was cloned into pCirCB to replace Kt-hBax-4 × 126 in pCirCB to yield pBac-T2-CirCE.
pBac-T2-Scramble, pBac-T2-s196, pBac-T2-m196, pBac-T2-LKE, pBac-T2-CirCE and pBac-T2-CirCB plasmids were used to construct the corresponding recombinant BVs Bac-Scramble, Bac-s196, Bac-m196, LKE, CirCE and CirCB as described (11,35). The resultant BVs were amplified by infecting Sf-9 cells and the virus titer (pfu/ml) was determined as described (36).
BV transduction of mammalian cells
The BV transduction was performed as described (35–37). Briefly, cells were seeded to 6-well plates, cultured overnight and washed twice with phosphate-buffered saline (PBS, pH 7.4) prior to transduction. Depending on the multiplicity of infection (MOI), a certain volume of BV supernatant was diluted with fresh Grace's medium and then mixed with NaHCO3-free culture medium (e.g. DMEM or α-MEM) at a volumetric ratio of 1:4. The transduction was initiated by directly adding virus solution to the cells (750 μl/well), continued by gentle shaking on a rocking plate at room temperature for 6 h and the virus solution was replaced with fresh culture medium containing 3 mM sodium butyrate. After 18 h of incubation at 37°C (1 dpt), the cells were collected for analysis.
miRNA analysis by TaqMan® RT-qPCR and qPCR array
Human hepatocyte RNA and human liver tissue total RNA were purchased from ScienCell Research Laboratories and Clontech, respectively. For PLC, HUVEC and nude mice livers, total RNA was isolated using the Quick-RNA™ MiniPrep kit (Zymo Research).
For RT-qPCR miRNA analysis using TaqMan® microRNA assay, 10 ng of total RNA was reverse transcribed to cDNA using the TaqMan® microRNA reverse transcription kit (Applied Biosystems) and the StepOnePlus™ thermal cycler (Applied Biosystems) under the following conditions: 30 min at 16°C, 30 min at 42°C and 5 min at 85°C. The resultant cDNA was subjected to quantitative real-time PCR (qPCR) using TaqMan® Universal Mater Mix II (Applied Biosystems) and StepOnePlus™ with a single enzyme activation cycle of 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 60 s at 60°C. MiRNA expression levels were determined using the small nuclear RNA (snRNA) RNU6B as the reference gene and normalized to the results of a selected reference sample.
For miRNA array analyses, 0.5 μg of total RNA was reverse transcribed to cDNA using the QuantiMir Kit (System Biosciences) under the following conditions: 30 min at 37°C for polyadenylation; 5 min at 60°C for anchor dT adaptor annealing; 60 min at 42°C and 10 min at 95°C. The cDNA samples were subjected to qPCR on StepOnePlus™ using the Cancer MicroRNA qPCR Array Kit (System Biosciences) which comprises 95 miRNA-specific and U6-specific forward primers in a 96-well plate format and the 3′ universal reverse primer. SYBR Green qPCR master mix (Applied Biosystems) was added to a final concentration of 1×. The qPCR was performed under the following conditions: 2 min at 50°C, 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 60 s at 60°C. Expression levels of each mature miRNA were evaluated using U6 as the reference gene. The expression level of each miRNA in PLC cells was normalized to those in normal (HUVEC, hepatocyte, liver tissue) cells.
mRNA analysis
To analyze the L7Ae mRNA levels, total RNA was extracted using the Quick-RNA™ MiniPrep kit and reverse transcribed to cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) on StepOnePlus™ under the following conditions: 10 min at 25°C, 120 min at 37°C and 5 min at 85°C. The subsequent qPCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems) on StepOnePlus™ under the following conditions: a single enzyme activation cycle of 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 60 s at 60°C. 18S rRNA was used as the reference gene. qPCR primer sequences were designed as follows: L7Ae forward: AACGAGACGACAAAGGCTGT; L7Ae reverse: AAGGCTTCCAAGCTCCTTTC; 18s forward: AACCCGTTGAACCCCATT; 18s reverse: CCATCCAATCGGTAGTAGCG.
Spheroid formation assay
One day after BV transduction, cells were trypsinized, centrifuged and resuspended to 1 × 104 cells/ml in DMEM/F12 containing 10 ng/ml EGF (R&D Systems), 10 ng/ml FGF (R&D Systems) and 1× N2 supplement (Invitrogen). The cell suspension (100 μl/well) was added to the Ultra-Low Attachment 96-well plates (Corning) and cultured for 5 days. Spheroid formation was photographed with a microscope (Eclipse Ti2, Nikon) and the spheroid number was quantified using the built-in NIS-Elements software.
Reporter gene expression analysis
The EYFP and EBFP expressions were imaged using the fluorescence microscope (Eclipse Ti2, Nikon). To quantify the percentage of fluorescence-emitting cells and mean fluorescence intensity (arbitrary unit, a.u.), cells were trypsinized and analyzed using a flow cytometer (CytoFLEX, Beckman Coulter). The data were acquired from 10,000 events per sample with the following settings: 405 nm laser and 450/45 bandpass filter for EBFP as well as 488 nm laser and 525/40 nm bandpass filter for EYFP/EGFP. Data were analyzed using the CytoExpert software.
Cell death and apoptosis assays
PLC and HUVEC cells (3 × 105 cells/well) were separately seeded to 6-well plates, cultured overnight and mock-transduced or transduced with CirCB. The cell death was evaluated by microscopic observation at 1 dpt, followed by harvesting of all adherent and floating cells for trypan blue staining. The viable cell count was based on the staining and the viability was calculated.
For apoptosis analysis, all floating and adherent cells were collected at 1 dpt and resuspended in 2 ml culture medium for caspase-3 and -7 activity assay using Caspase-Glo 3/7 Assay kit (Promega). The cell suspension (100 μl) was seeded to 96-well plates and 100 μl Caspase-Glo solution was added to each well and incubated at room temperature for 30 min. The relative luminescence unit (RLU) of each sample was measured by the SpectraMax M2 microplate reader (Molecular Devices).
Evaluation of BV selectivity in PLC/HUVEC co-culture
PLC (1.5 × 105 cells/well) and HUVEC (1.5 × 105 cells/well) cells were co-seeded to 6-well plates, co-cultured overnight and were transduced with CirCE. At 1 dpt, the cells were immunostained with anti-CD31 antibody conjugated with PE-Cy7 (BD Biosciences) to characterize HUVEC cells. The HUVEC cells and EBFP expression in the mixed cell population were observed using the fluorescence microscope. The transduced co-cultured cells were also trypsinized, transferred to flow tubes and immunostained with anti-CD31 antibody conjugated with PE-Cy7 and anti-EGFR conjugated with AlexaFluor™ 647 (BD Bioscience). The immmunostained cells were subsequently analyzed by the flow cytometer (CytoFLEX) with the following settings: 405 nm laser and 450/45 bandpass filter for EBFP; 488 nm laser and 780/60 nm bandpass filter for PE-Cy7 as well as 638 nm laser and 660/20 nm bandpass filter for AlexaFluor™ 647. The CD31−/EGFR+ PLC cells and CD31+/EGFR− HUVEC cells were gated and the EBFP expression was quantified using the CytoExpert software.
Alternatively, the co-cultured PLC/HUVEC cells were mock-transduced or transduced with CirCB and the adherent cells were trypsinized at 1 dpt, transferred to flow tubes and immunostained with AlexaFluor™ 647-conjugated anti-EGFR antibody. The immunostained mixed cell population was analyzed by the flow cytometer for the percentage of EGFR+ PLC cells and EGFR− HUVEC cells.
Statistical analysis
All data were representative of at least 3 independent culture experiments. All quantitative data were analyzed using student's t-test and are expressed as means±standard deviations (SD). P < 0.05 was considered significant.
RESULTS
Involvement of miR-196a in HCC tumorigenicity
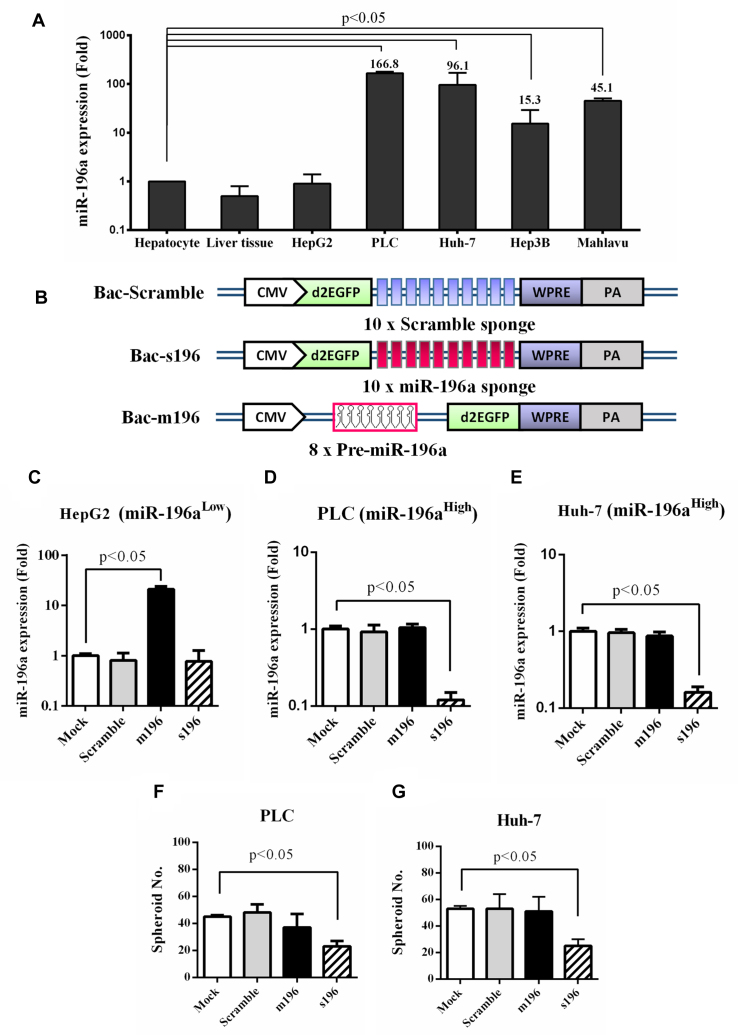
It was recently reported that miR-196a polymorphism is associated with HCC cancer risk (38) and recurrence (39). Therefore, we reasoned that miR-196a might be highly expressed in HCC cells and may serve as a novel target for sensing HCC cells. To assess this hypothesis, we first examined the miR-196a expression in several HCC cell lines and human liver tissue by RT-qPCR and normalized the data to that in human hepatocyte. Figure 1A depicts that miR-196a expression was aberrantly high in some tumorigenic HCC cells (e.g. PLC and Huh-7), but was low in the liver tissue and in the less tumorigenic HCC cell HepG2 (40).
Figure 1.
Involvement of miR-196a in HCC tumorigenicity. (A) MiR-196a expression in different HCC cells. (B) Recombinant BVs that expressed scramble sponge (Bac-Scramble), miR-196a sponge (Bac-s196) and pre-miR-196a (Bac-m196). (C–E) miR-196a levels in HepG2 (C), PLC (D) and Huh-7 (E) cells after BV transduction. (F–G) Spheroid formation in PLC (F) and Huh-7 (G) cells after BV transduction. The miR-196a levels in various HCC and normal cells were quantified by TaqMan® RT-qPCR and normalized to that in human hepatocytes. HCC cells were mock-transduced or transduced with recombinant BVs at MOI 200 and the miR-196a levels were analyzed by RT-qPCR at 1 dpt. The data represent means±SD of triplicated culture experiments.
To confirm the roles of miR-196a by modulating miR-196a levels, we constructed 3 recombinant BVs (Figure 1B): Bac-Scramble and Bac-s196 expressed 10 tandem repeats of scramble and miR-196a sponges (decoy targets of miRNA), respectively, while Bac-m196 expressed eight copies of miR-196a. The cells expressing low (HepG2) or high (PLC and Huh-7) miR-196a levels were separately transduced with the 3 recombinant BVs and analyzed by RT-qPCR (Figure 1C–E). In comparison with the mock-transduction control, Bac-Scramble barely disturbed miR-196a levels in all three cell types; Bac-m196 significantly enhanced miR-196a levels in HepG2 (Figure 1C) while Bac-s196 effectively knocked down miR-196a levels in PLC and Huh-7 (Figure 1D and E).
Sphere formation is an indicator of tumorigenicity (41). Both PLC and Huh-7 cells are highly tumorigenic and readily formed spheres whereas poorly tumorigenic HepG2 did not (Supplementary Figure S1), thus we chose PLC and Huh-7 cells to evaluate whether miR-196a knockdown suppressed sphere formation. Compared with Mock transduction, Bac-s196 transduction significantly (P < 0.05) suppressed the sphere formation of PLC and Huh-7 cells (Figure 1F–G and Supplementary Figure S1), confirming the correlation between miR-196a and tumorigenicity of PLC and Huh-7 cells. Bac-m196 transduction did not enhance the sphere formation (Figure 1F and G and Supplementary Figure S1) probably because miR-196a levels were sufficiently high such that Bac-m196 did not elevate miR-196a levels in PLC (Figure 1D) and Huh-7 (Figure 1E). Since PLC is a clinically relevant, tumorigenic HCC cell (42), expressed the highest level of miR-196a (≈166-fold versus hepatocyte, Figure 1A) and Bac-s196 transduction mitigated PLC’s ability to promote tubule formation (Supplementary Figure S2), an indicator of angiogenesis essential for tumor progression, we used PLC as the model HCC cell in subsequent experiments.
Identification of miRNAs suitable for distinguishing HCC and normal cells
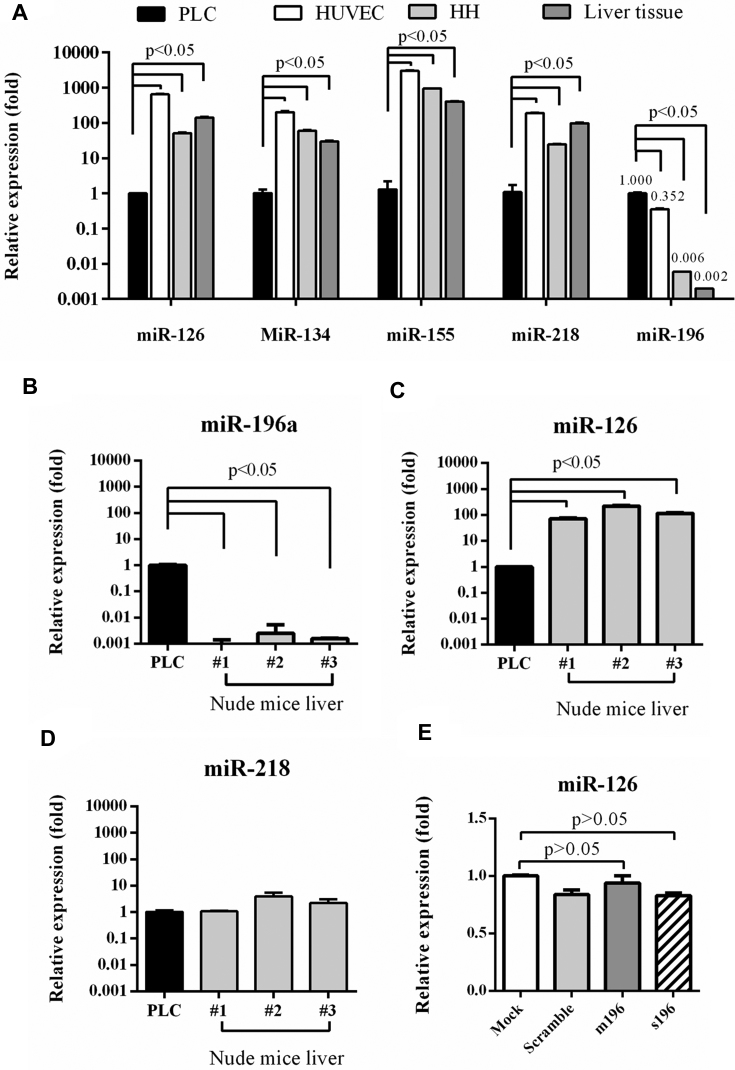
To classify HCC and normal cells, we screened more miRNAs that are specifically expressed in PLC (defined as ‘OncoMiR’) and in normal cells (defined as ‘NormalMiR’) by comparing miRNA expression profiles in PLC with those in human umbilical vascular endothelial cells (HUVEC), human hepatocytes and human liver tissue. MiRNA array analysis (Table 1) verified the high expression of miR-196a in PLC which was ≈7.8-fold that in HUVEC and >60-fold that in hepatocytes and in liver tissue. Table 1 also reveals a panel of miRNAs (e.g. miR-126, miR-134, miR-155 and miR-218) that was barely expressed in PLC. Such expression signature was further verified by RT-qPCR (Figure 2A), which showed that the expression levels of miR-126, miR-134, miR-155 and miR-218 in PLC were at least 10–1000 fold lower than those in HUVEC, human hepatocytes and liver tissue. Moreover, the miR-196a expression level in PLC was significantly higher (P < 0.05) than those in HUVEC (≈0.352-fold), hepatocyte (≈0.006-fold) and liver tissue (≈0.002-fold).
Table 1.
MicroRNA expression profile of PLC compared with human HUVEC, hepatocyte and liver tissue
| HUVEC | Hepatocyte | Liver | HUVEC | Hepatocyte | Liver | HUVEC | Hepatocyte | Liver | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| let7 | 0.25 | 0.12 | 0.06 | miR-153 | 0.26 | 1.34E+08 | 2.84E-11 | miR-200c | 1.00 | 0.02 | 0.06 |
| miR-7 | 0.25 | 0.25 | 0.26 | miR-154 | 0.01 | 0.01 | 0.03 | miR-202 | 0.25 | 1.01 | 0.99 |
| miR-92 | 0.25 | 0.06 | 0.25 | miR-155 | 0.00 | 0.00 | 0.00 | miR-203 | 0.49 | 0.03 | 0.06 |
| miR-93 | 0.25 | 0.06 | 0.50 | miR-15a | 0.12 | 0.50 | 131.08 | miR-204 | 0.13 | 0.06 | 0.03 |
| miR-9-1 | 126.28 | 8.34 | 8.10 | miR-15b | 0.25 | 0.25 | 0.50 | miR-205 | 0.25 | 0.07 | 0.12 |
| miR-101-1 | 0.12 | 0.52 | 0.06 | miR-16 | 0.25 | 528.07 | 0.13 | miR-206 | 0.25 | 0.07 | 1.02 |
| miR-103 | 0.25 | 0.25 | 0.25 | miR-17–3p | 0.50 | 0.51 | 1.00 | miR-21 | 0.12 | 0.02 | 1.99 |
| miR-106a | 0.25 | 2.00 | 0.51 | miR-17-5p | 0.25 | 0.02 | 0.24 | miR-210 | 1.01 | 0.50 | 1.00 |
| miR-106b | 0.24 | 0.25 | 0.99 | miR-181a | 0.12 | 0.12 | 0.25 | miR-214 | 0.50 | 0.13 | 0.12 |
| miR-107 | 0.50 | 2.02 | 0.12 | miR-181b | 0.12 | 0.02 | 0.12 | miR-215 | 7.87 | 1.04 | 0.00 |
| miR-10b | 0.01 | 0.50 | 0.06 | miR-181c | 0.49 | 0.06 | 0.50 | miR-372 | 0.25 | 0.14 | 0.25 |
| miR-1–1 | 0.02 | 0.52 | 0.25 | miR-181d | 0.06 | 0.06 | 1.00 | miR-373 | 0.25 | 0.02 | 0.25 |
| miR-122a | 3.95 | 0.00 | 0.00 | miR-183 | 30.84 | 2.03 | 31.72 | miR-218 | 0.01 | 0.00 | 0.00 |
| miR-125a | 0.12 | 0.25 | 0.13 | miR-185 | 0.50 | 1.01 | 0.25 | miR-219 | 0.25 | 0.55 | 0.01 |
| miR-125b | 0.01 | 0.02 | 0.01 | miR-186 | 1.98 | 2.17 | 0.99 | miR-22 | 0.12 | 0.25 | 0.00 |
| miR-126 | 0.00 | 0.00 | 0.00 | miR-188 | 0.50 | 0.26 | 0.25 | miR-488 | 0.51 | 0.03 | 0.00 |
| miR-128b | 0.99 | 0.14 | 0.50 | miR-18a | 0.25 | 0.12 | 3.98 | miR-221 | 0.06 | 1.00 | 0.00 |
| miR-132 | 0.49 | 0.06 | 0.51 | miR-190 | 0.25 | 0.02 | 0.25 | miR-222 | 0.03 | 3.97 | 0.00 |
| miR-133a | 0.51 | 1.03 | 0.00 | miR-191 | 0.49 | 0.25 | 0.00 | miR-223 | 0.51 | 0.00 | 0.00 |
| miR-134 | 0.03 | 0.00 | 0.00 | miR-192 | 62.29 | 0.50 | 0.00 | miR-224 | 0.12 | 0.13 | 0.00 |
| miR-135b | 0.25 | 0.02 | 0.00 | miR-194 | 126.60 | 0.51 | 0.00 | miR-23a | 0.25 | 0.06 | 0.12 |
| miR-136 | 0.03 | 0.03 | 0.00 | miR-195 | 0.12 | 0.50 | 0.00 | miR-24 | 0.25 | 0.06 | 0.12 |
| miR-137 | 0.00 | 0.03 | 0.03 | miR-196a | 7.79 | 64.47 | 63.97 | miR-25 | 0.25 | 0.02 | 0.25 |
| miR-140 | 0.12 | 0.24 | 0.25 | miR-197 | 0.25 | 4.05 | 0.00 | miR-26a | 0.50 | 0.13 | 0.03 |
| miR-141 | 0.49 | 0.26 | 0.24 | miR-198 | 0.49 | 0.03 | 0.00 | miR-26b | 0.99 | 15.83 | 0.06 |
| miR-142-3p | 0.25 | 0.02 | 0.01 | miR-199 | 0.06 | 0.06 | 0.00 | miR-27a+b | 0.25 | 260.76 | 0.13 |
| miR-143 | 2.03 | 0.06 | 0.00 | miR-30b | 0.49 | 4.04 | 0.00 | miR-30c | 0.12 | 130.92 | 0.06 |
| miR-145 | 0.23 | 0.01 | 0.00 | miR-19a+b | 0.24 | 8.03 | 0.00 | miR-29a+b+c | 0.25 | 0.02 | 0.25 |
| miR-146a | 0.24 | 0.13 | 0.25 | miR-95 | 0.99 | 0.06 | 0.12 | miR-30a-3p | 0.25 | 0.02 | 0.03 |
| miR-149 | 0.24 | 0.98 | 0.00 | miR-20a | 0.25 | 0.13 | 0.24 | miR-30a-5p | 0.25 | 132.84 | 0.06 |
| miR-150 | 0.03 | 0.02 | 0.00 | miR-200a | 0.22 | 0.01 | 0.03 | miR-296 | 0.25 | 0.51 | 0.99 |
| miR-151 | 0.12 | 0.12 | 0.00 | miR-200b | 0.25 | 0.02 | 0.06 | U6 snRNA | 1.00 | 1.00 | 1.00 |
The miRNA profiles were measured using the Cancer MicroRNA qPCR Array Kit. The expression level of each miRNA in PLC cells was normalized to those in normal (HUVEC, hepatocyte, liver tissue) cells.
miRNAs that were poorly expressed in PLC cells are shown in italics and blue (NormalMiR).
miRNAs that were highly expressed in PLC cells are bolded and shown in purple (OncoMiR).
Figure 2.
Identification of OncoMiR and NormalMiR. (A) Expression of miR-126, miR-134, miR-155, miR-218 and miR-196a in PLC, HUVEC, human hepatocyte and liver tissue. (B–D) miR-196a (B), miR-126 (C) and miR-218 (D) levels in mouse livers and PLC cells. (E) miR-126 levels in BV-transduced PLC cells. The miRNA levels were analyzed by RT-qPCR and normalized to those in PLC. The data represent means ± SD of at least three independent experiments.
Nude mouse is the potential xenograft tumor model for implantation of human PLC cells, thus we also analyzed the expression of miR-196a, miR-126 and miR-218 in nude mice livers. miR-134 and miR-155 levels were not analyzed due to difference in human and mouse sequences. The RT-qPCR analysis showed that in the livers of three nude mice miR-196a levels were tremendously lower (Figure 2B) while miR-126 levels were significantly higher (near 100-fold, Figure 2C) than those in PLC. miR-218 was also expressed in nude mice livers (Figure 2D) but to a lesser extent than miR-126.
These human and mouse data consistently proved that miR-196a expression was tremendously higher in PLC than in normal (HUVEC and liver) cells while miR-126 expression was remarkably higher in normal cells. Furthermore, transduction of PLC with recombinant BVs modulating miR-196a (Bac-m196 and Bac-s196) barely disturbed the miR-126 expression (Figure 2E). As such, miR-196a was selected as the ‘OncoMiR’ while miR-126 was chosen as the ‘NormalMiR’. Besides, hepatocytes are difficult to culture, thus HUVEC was chosen as the model normal cells.
Development of synthetic switch-based BV for selective transgene expression in HCC cells
To build a genetic device comprising both L7Ae:K-turn and OncoMiR/NormalMiR switch, we first constructed two plasmids, one co-expressing L7Ae and the reporter EYFP (enhanced yellow fluorescent protein) linked by a ribosome skipping P2A sequence while the other expressing the transgene EBFP (enhanced blue fluorescent protein) with two repeats of K-turn motifs at the 5′ UTR. Single transfection of HEK293 cells with each plasmid resulted in similar efficiency of EYFP and EBFP expression (Supplementary Figure S3). After co-transfection, EYFP expression remained robust but EBFP expression was markedly attenuated (Supplementary Figure S3), confirming the in trans inhibition of EBFP expression by L7Ae:K-turn.
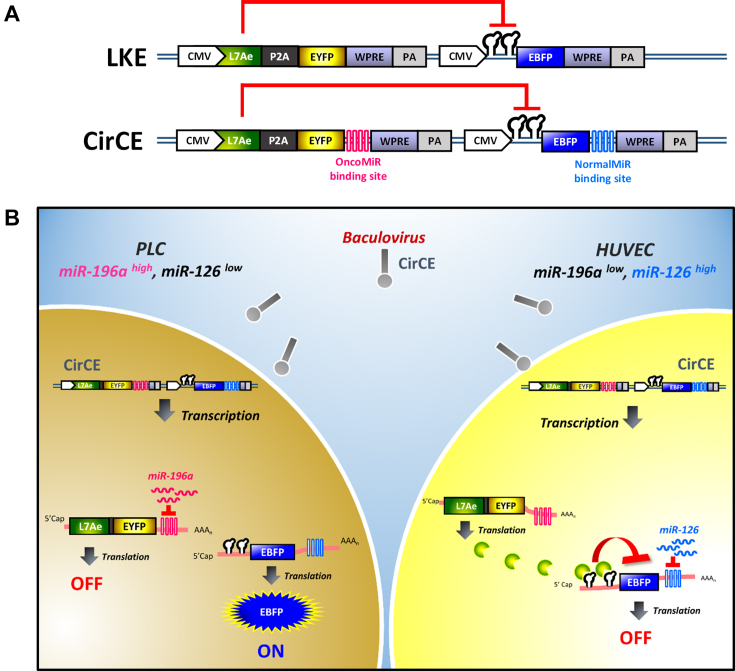
We next incorporated these two expression cassettes into a single recombinant BV LKE (Figure 3A), in which the first module co-expressed L7Ae and EYFP under the transcriptional control of CMV promoter while the second cassette expressed CMV-driven EBFP gene with K-turn motifs at the 5′ UTR. Here, EYFP served as the reporter to evaluate transduction efficiency while EBFP served as the reporter representing the transgene of interest. We envisioned that LKE transduction confers L7Ae and EYFP co-expression, yet EBFP expression would be repressed due to L7Ae binding to the K-turn motif. Exploiting the OncoMiR/NormalMiR as the ON/OFF switch, we added into LKE vector additional four tandem copies of miR-196a and miR-126 binding sites at the 3′ UTR of EYFP and EBFP genes, respectively, and built another recombinant BV CirCE (Figure 3A). CirCE transduction of PLC cells (high miR-196a, low miR-126) would turn ON EBFP expression due to the negligible miR-126 levels and high miR-196a levels that could turn OFF L7Ae translation (Figure 3B). In contrast, CirCE transduction of normal cells (low miR-196a, high miR-126) would turn ON L7Ae/EYFP expression due to the lack of miR-196a-mediated inhibition and turn OFF EBFP due to the miR-126 and L7Ae binding to the EBFP mRNA (Figure 3B).
Figure 3.
Recombinant BVs carrying the synthetic switch for transgene expression regulation. (A) Schematic illustration of recombinant BVs LKE and CirCE. CMV, cytomegalovirus immediate-early promoter. WPRE, woodchuck hepatitis virus posttranscriptional regulatory element. PA, poly A signal. (B) Schematic illustration of transgene expression induction (ON) and repression (OFF) in HCC and normal cells. CirCE transduction of PLC cells (high miR-196a, low miR-126) would turn ON EBFP expression due to the negligible miR-126 levels and high miR-196a levels that could turn OFF L7Ae translation. In contrast, CirCE transduction of HUVEC cells (low miR-196a, high miR-126) would turn ON L7Ae/EYFP expression due to the lack of miR-196a-mediated inhibition and turn OFF EBFP due to the miR-126 and L7Ae binding to the EBFP mRNA.
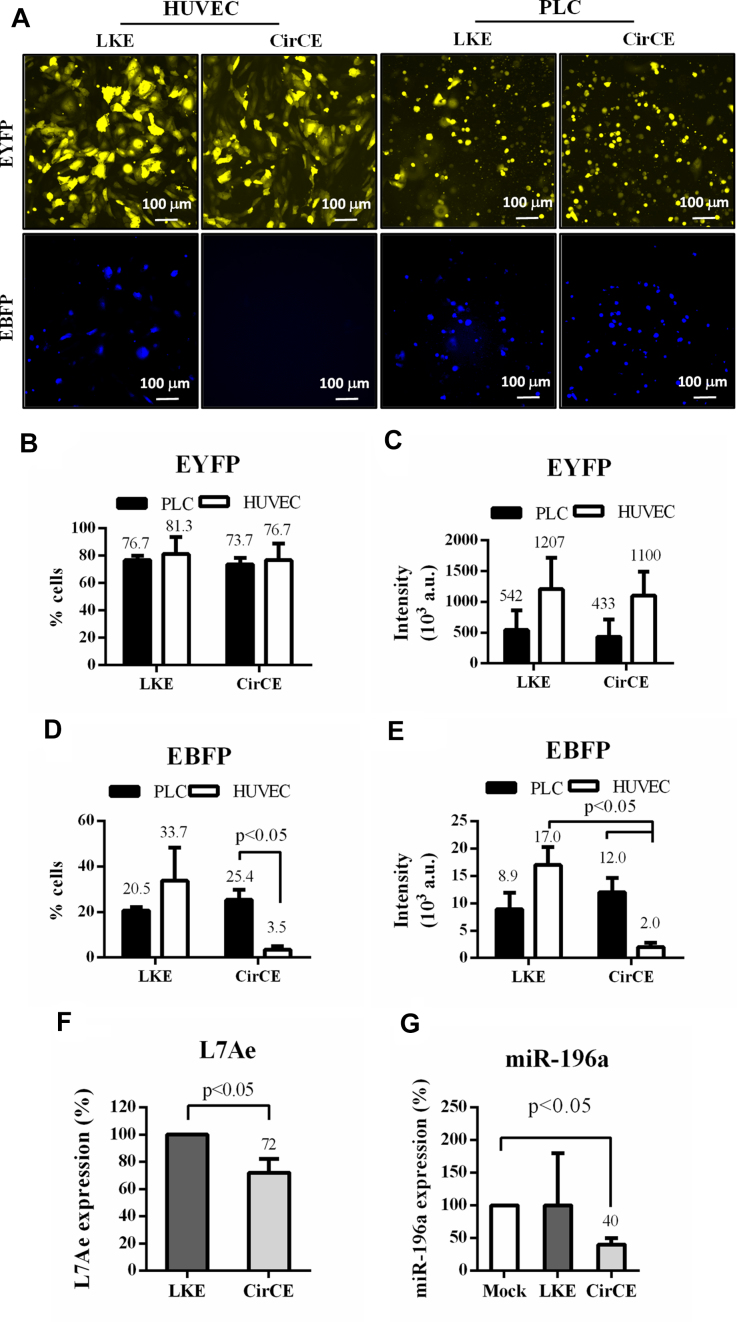
To validate the vector design, we transduced HUVEC and PLC cells separately and examined the reporter gene expression by fluorescence microscopy (Figure 4A) and flow cytometry (Figure 4B–E). LKE transduction gave rise to EYFP expression in ≈76.7% of PLC and ≈81.3% of HUVEC cells (Figure 4B) with high mean EYFP intensities (≈5.4 × 105 a.u. for PLC and ≈1.2 × 106 a.u. for HUVEC, Figure 4C), indicating effective transduction. However, LKE conferred EBFP expression in only ≈20.5% of PLC cells and ≈33.7% of HUVEC cells (Figure 4D) and the resultant mean EBFP intensities (≈8.9 × 103 a.u for PLC and ≈1.7 × 104 a.u. for HUVEC, Figure 4E) were significantly lower than corresponding EYFP intensities (Figure 4C), attesting that BV-carried L7Ae:K-turn effectively suppressed EBFP expression in PLC and HUVEC cells. With additional miR-126 binding sites at the 3′ UTR of EBFP, CirCE still conferred EBFP expression in ≈25.4% PLC cells with a mean intensity of 1.2 × 104 a.u., but gave rise to low EBFP expression in only ≈3.5% of HUVEC cells with a mean EBFP intensity of ≈2.0 × 103 a.u. (Figure 4D and E), proving that miR-126 in concert with L7Ae/K-turn enabled EBFP expression in PLC cells but efficiently turned OFF EBFP expression in normal cells. These data demonstrated that the synthetic switch-based CirCE was able to distinguish HCC and normal cells by selectively expressing transgene of interest in HCC cells.
Figure 4.
Selective transgene (EBFP) expression in HCC cells. (A) Fluorescence micrographs of cells after BV transduction. (B) Percentage of EYFP+ cells. (C) Mean EYFP intensity (×103 a.u.). (D) Percentage of EBFP+ cells. (E) Mean EBFP intensity (×103 a.u.). (F) L7Ae levels. (G) miR-196a levels. The HUVEC and PLC cells were transduced with LKE or CirCE at MOI 200 and examined by fluorescence microscopy or flow cytometry at 1 dpt. The L7Ae and miR-196a levels were analyzed by RT-qPCR at 1 dpt. The data represent means±SD of triplicated culture experiments.
Of note, CirCE transduction still conferred robust EYFP expression in ≈73.7% of PLC cells (Figure 4B) with a mean intensity of 4.3 × 105 a.u. (Figure 4C), which prompted us to suspect whether the miR-196a binding site in CirCE functioned properly in the miR-196a-rich PLC cells to inhibit L7Ae/EYFP expression. To evaluate this hypothesis, the L7Ae (Figure 4F) and miR-196a (Figure 4G) levels in transduced PLC cells were examined by RT-qPCR. When compared with the LKE transduction control, CirCE reduced ≈28% L7Ae level in PLC cells. Meanwhile, CirCE reduced ≈60% miR-196a levels as opposed to Mock transduction. Therefore, the miR-196a binding sites in CirCE indeed recruited miR-196a binding to the L7Ae-EYFP mRNA transcript to reduce L7Ae and miR-196a levels in PLC cells, although the degree of inhibition was suboptimal.
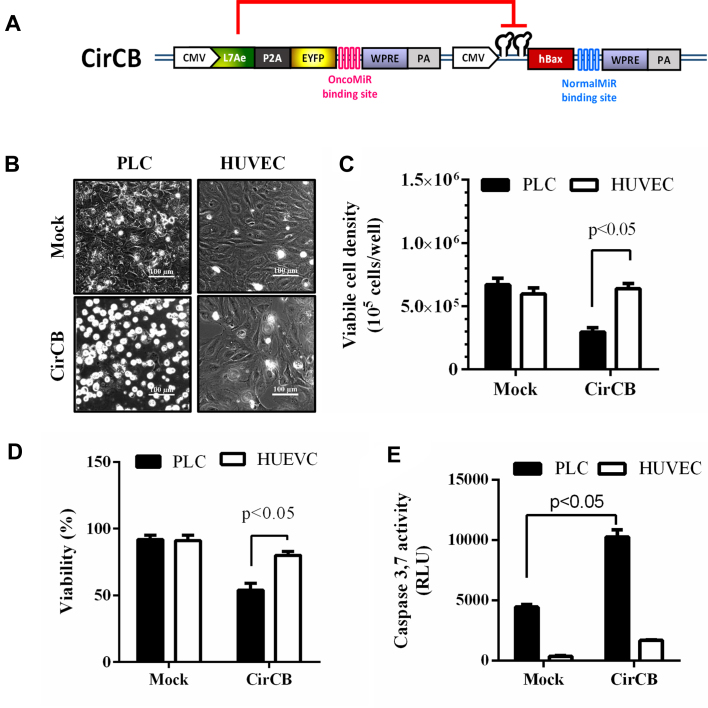
Synthetic-switch-based BV specifically killed HCC cells
To evaluate whether the synthetic switch-based BV was able to selectively kill HCC cells, we constructed a new recombinant BV CirCB which was similar to CirCE except that apoptosis-inducing gene hBax (13) was used as the transgene of interest (Figure 5A). The HCC (PLC) and normal (HUVEC) cells were seeded to 6-well plates (3 × 105 cells/well) and were mock-transduced or transduced with CirCB. The microscopic observation (Figure 5B) illustrated that CirCB transduction induced apparent cell floating and death for PLC cells, but not for HUVEC. All floating and adherent cells were further harvested for viable cell count and viability analysis. The mock-transduced PLC and HUVEC cells grew to ≈6.7 and 6.0 × 105 cells/well, respectively (Figure 5C), and viability remained >90% (Figure 5D), indicating the cell growth and health. The CirCB-transduced HUVEC cells also grew to ≈6.0 × 105 cells/well with a viability of ≈80%, attesting that CirCB transduction did not markedly damage normal cells. Notably, CirCB transduction of PLC cells significantly reduced the viable cell density to ≈3.3 × 105 cells/well (Figure 5C) and viability to 54% (Figure 5D). Since hBax is known to induce apoptosis through caspase 3 pathway, we further analyzed the apoptosis by luminescence-based Caspase 3,7 assay (Figure 5E). As shown, CirCB transduction of PLC cells elicited significantly more evident (P < 0.05) apoptosis than Mock transduction, but CirCB merely triggered a low degree of apoptosis in HUVEC cells. Figure 5 confirms that CirCB selectively induced apoptosis and death in HCC cells but provoked low cytotoxicity in normal cells.
Figure 5.
Selective killing of HCC cells in separate culture by the synthetic switch-based BV. (A) Schematic illustration of recombinant BV CirCB. CirCB was similar to CirCE except that the transgene was proapoptic hBax. (B) Cell morphology. (C) Viable cell density. (D) Viability. (E) Apoptosis induction. The HCC (PLC) and normal (HUVEC) cells were separately seeded to 6-well plates (3 × 105 cells/well) and were mock-transduced or transduced with CirCB. At 1 dpt, the cell morphology was observed under the microscope. All cells were then harvested and stained by trypan blue, followed by viable cell density and viability analyses. The cells were also subjected to luminescence-based Caspase 3,7 assay to quantify apoptosis. Higher relative luminescence units (RLU) indicate higher degrees of apoptosis. The data represent means±SD of triplicated culture experiments.
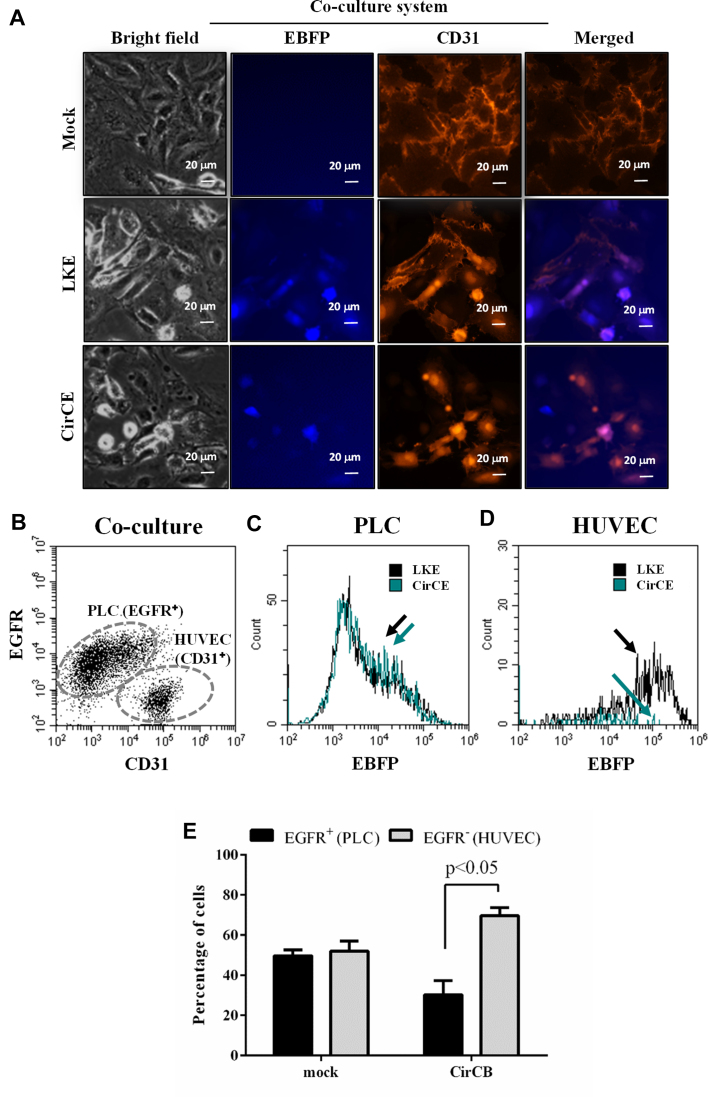
Selective transgene expression and killing in HCC cells in co-culture
To further evaluate whether the synthetic device was able to classify HCC cells in a population of different cells, we co-cultured HUVEC and PLC cells at a seeding density ratio of 1:1 in 6-well plates and transduced the cells with LKE or CirCE. Since HUVEC cells are CD31+/EGFR− while PLC cells are CD31−/EGFR+ (Supplementary Figure S4), we immunolabeled the co-cultured cells with anti-CD31 antibody to reveal CD31+ HUVEC cells and detected EBFP expression by a fluorescence microscope (Figure 6A). In the LKE-transduced HUVEC/PLC co-culture, EBFP expression was observed in CD31− PLC (blue) cells and CD31+ HUVEC (purple) cells. In the CirCE-transduced co-culture, EBFP expression was observed in the CD31− PLC (blue) cells and was barely observed in CD31+ HUVEC (orange) cells.
Figure 6.
Selective transgene expression and killing of HCC cells in PLC/HUVEC co-culture. (A) Fluorescence micrographs of co-cultured PLC and HUVEC cells. HUVEC and PLC cells were co-cultured at a seeding density ratio of 1:1 in 6-well plates (1.5 × 105 cells/well for each cell) and transduced with LKE or CirCE at MOI 200. The co-cultured cells were immunolabeled with anti-CD31 antibody and observed with a fluorescence microscope. (B–D) Immunofluorescence double labeling and flow cytometry analysis of BV-transduced co-cultured cells. HUVEC and PLC cells were co-cultured and transduced as in (A), immunostained with anti-EGFR and anti-CD31 antibodies, followed by flow cytometry analysis. The CD31−/EGFR+ PLC and CD31+/EGFR− HUVEC cells were gated (B) and the EBFP expression in PLC (c) and HUVEC (D) was elucidated. (E) Selective killing of HCC cells in co-culture by CirCB. HUVEC and PLC cells were co-cultured as in (A), mock-transduced or transduced with CirCB, immunostained with anti-EGFR antibody and analyzed by flow cytometry. The data are representative of at least 3 independent culture experiments.
To quantify the selective expression, the co-cultured cells were similarly transduced and subjected to immunolabeling with anti-EGFR and anti-CD31 antibodies, followed by flow cytometry analysis. Figure 6B confirmed that the immunofluorescence double labeling/flow cytometry was able to discriminate the co-cultured CD31−/EGFR+ PLC and CD31+/EGFR− HUVEC cells. Thus we further analyzed the EBFP expression in these PLC and HUVEC cell populations. Figure 6C shows that CirCE led to an EBFP expression profile similar to LKE in EGFR+/CD31− PLC cells. In CD31+/EGFR− HUVEC cells (Figure 6D), however, CirCE gave rise to evidently lower EBFP expression than LKE. These data indicated that CirCE conferred transgene (EBFP) expression more preferentially in PLC cells than in HUVEC cells in the mixed cell population.
To verify the selective killing, PLC and HUVEC cells were co-cultured by seeding at a 1:1 ratio, mock-transduced or transduced with CirCB, and immunolabeled with anti-EGFR antibody. Flow cytometry analysis of the mixed cell population (Figure 6E) depicted that after mock-transduction the percentages of EGFR+ PLC and EGFR− HUVEC cells were similar at ≈50%, but CirCB transduction resulted in significantly lower (P < 0.05) percentage of EGFR+ PLC cells (≈30%) than the percentage of EGFR− HUVEC cells (≈70%), indicating that CirCB transduction preferentially killed PLC cells in the mixed cell population. Figure 6 collectively demonstrates that the switch-based recombinant BV favorably conferred transgene (i.e. EBFP or hBax) expression and induced cell death in PLC cells within the mixed cell population.
DISCUSSION
The overriding objective of this study is to improve the safety profile of recombinant BV for clinical HCC cancer therapy, by incorporating a synthetic regulatory device comprising miRNA sensors and L7Ae:K-turn system for translational control of transgene expression in HCC cells. Although miRNA sensors based on distinct miRNA expression signature in different cells have been used for constructing genetic switches/circuits (for review see (20,24)), the majority of these studies simply used well-characterized miRNAs, rather than discovered new miRNA targets, for device fabrication. For instance, Xie et al. employed a panel of previously characterized miRNAs (miR-21, miR-17, miR-30a, miR-141, miR-142 and miR-146a) to classify HeLa and non-HeLa cell lines (25), while Hirosawa et al. used miR-21 and miR-302a-5p to discriminate HeLa and induced pluripotent stem cells, respectively (43).
Instead of using known miRNAs, hereby we uncovered miR-196a as an OncoMiR that is highly expressed in HCC cells but is poorly expressed in normal cells, and unveiled miR-126 as a novel NormalMiR that is expressed in normal cells but is barely expressed in HCC cells (Figures 1, 2 and Table 1). It has been shown that miR-196a targets a number of genes such as HOX-C8 (44) and HOX-B9 (45) and regulates angiogenesis (46) and PI3K/AKT signaling (47), which are critical to control cancer progression. miR-196a overexpressison is mostly found in cancer cells and is associated with the progression of different cancers including lung cancer (47), ovarian carcinoma (48), head and neck squamous cell carcinoma (45) and cervical cancer (49). However, overexpression and roles of miR-196a in other normal cells are rarely reported. Conversely, miR-126 regulates vascular integrity by repressing negative regulators of the VEGF pathway, including the Sprouty-related protein SPRED1 and phosphoinositol-3 kinase regulatory subunit 2 (50). These literature reports justify the section of miR-196a as the OncoMiR and miR-126 as the NormalMiR.
The OncoMiR/NormalMiR pair allowed us to engineer a miRNA sensing module coupled with the LA7e:K-turn system for incorporation into the recombinant BV (Figure 3). Although L7Ae:K-turn has been exploited to build regulatory switches and circuits (27–30,32,33), most previous studies demonstrate the conceptual design in model cancer cell lines (e.g. HeLa) and have yet to apply L7Ae:K-turn system to aggressive tumorigenic cancer cells (e.g. PLC (42)) and actual medical applications. Furthermore, previous L7Ae:K-turn-based genetic switches/circuits are encoded in plasmid DNA or synthetic RNA and are transfected into living cells by lipid-based reagents or nanoparticles (27,32,33). Despite the promise in controlling cell behaviors in vitro, transfection efficiency of liver cells is typically low (51), which constitutes a barrier for in vivo applications (e.g. liver cancer gene therapy) of these synthetic switches. Although other viral vectors such as lentivirus (52), adeno-associated viral vectors (53,54), adenoviruses (55,56), measles viruses (55) and alphaviruses (57) can carry gene switches to control gene expression, these viral vectors typically have limited cloning capacity. For instance, the cloning capacity of commonly used lentivirus and adeno-associated virus is 8 kb and 4.7 kb (58), respectively, which would restrict the size of transgene as well as the complexity and sophistication of the synthetic device.
In contrast, BV is able to transduce numerous cancer cells and stem cells at high efficiencies (35,59–62) and the large genome (≈134 kb) of BV allows it to accommodate exogenous gene cassettes of at least 38 kb (63). These attributes render BV an appealing vector to efficiently deliver complex gene switches and circuits. Indeed, the large cloning capacity allowed us to incorporate the whole device (≈7.3 kb) into a single BV for cis-acting regulation. Furthermore, the synthetic switch-based BV (CirCE) transduced HCC and HUVEC cells at efficiencies near 80% (Figure 4A–C) but was capable of distinguishing HCC and normal cells (Figure 4D and E), as judged from the significantly higher target transgene (EBFP) expression in PLC ( ≈ 25.4%, mean intensity ≈ 1.2 × 104 a.u.) than in HUVEC (≈ 3.5%, mean intensity ≈ 2.0 × 103 a.u.). These data confirmed that synthetic switch-regulated EBFP expression was selectively turned ON in HCC cells and OFF in normal cells.
In the current design, however, the magnitude of output (i.e. EBFP expression) was less than satisfactory in HCC cells, probably because the very strong CMV promoter transcribed too many L7Ae transcripts such that miR-196a binding was insufficient to tightly repress L7Ae in HCC cells (Figure 4F). This notion is evidenced by the high expression of EYFP in PLC cells (Figure 4A–C) which was linked to L7Ae by the P2A sequence. The leakiness of L7Ae resulted in binding to K-turn motif on the EBFP mRNA and alleviating the ON response of EBFP. This problem may be tackled by using a weaker promoter (e.g. EF-1α or PGK) to drive L7Ae and appending additional OncoMiR (e.g. miR-9-1 identified in Table 1) complementary sequences to the 3′ UTR to attenuate the L7Ae expression in HCC cells. Different L7Ae variants and modified K-turn motifs may also be used to tailor the binding affinity and hence the degree of inhibition (30). Alternatively, other RNA binding proteins such as bacteriophage coat protein MS2 (17,29,32,33), bacillus ribosomal protein S15 (29), Pumilio and FBF protein (64) and ribosomal protein L1 (65) may also be used in lieu of L7Ae.
Despite the suboptimal ON state of transgene, the switch-based BV still selectively triggered HCC cell apoptosis and death by using hBax as the transgene of interest (Figure 5). Importantly, the BV-carried regulatory switch was able to selectively express the transgene and kill HCC cells in the mixed populations of HCC and normal cells (Figure 6). These data collectively demonstrate the proof-of concept and implicate the potential of BV to deliver a synthetic switch for selective transgene expression and killing of HCC cells for safer HCC gene therapy. Future studies will be directed towards (i) improving the recombinant BV by removal of the EYFP reporter, (ii) more stringent reduction of L7Ae expression, (iii) use of more potent anti-HCC gene to augment the anticancer effect and (iv) in vivo application of the switch-based recombinant BV for the treatment of orthotopic HCC tumors in the liver.
Aside from HCC therapy, the device we engineered may be modified for the treatment of other cancers and self-restriction of transgene expression. For instance, CRISPR/Cas9 system has been widely utilized for programmable genome editing (66,67), but excessive Cas9 expression induces mutation at non-target genomic loci (‘off-target’ effect) (68) and triggers immunogenicity in vivo (69). Our device may be modified by linking Cas9 gene with L7Ae by a P2A sequence and adding a K-turn motif at the 5′ UTR of the transcript, so that the expressed L7Ae in turn provides a feedback negative regulation of L7Ae and Cas9 to self-restrict Cas9 expression and minimize the off-target effect.
In conclusion, we uncovered novel miRNA targets that are selectively expressed in HCC and normal cells, engineered a synthetic switch consisting of the miRNA sensor and L7Ae:K-turn actuator and incorporated the switch into recombinant BV. The synthetic switch-based recombinant BV distinguished HCC and normal cells by preferentially expressing the transgene and triggering HCC cell death in separate culture and co-culture. To our best knowledge, this is the first study exploiting the synthetic device-based BV to classify HCC and normal cells for selective induction of HCC cell death. Such design improves the safety profile and implicates the potential of this synthetic switch-based BV for future in vivo HCC gene therapy.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the RNAi Core Facility at Academia Sinica (Taiwan) for synthesizing the miRNA sponge sequences.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Veterans General Hospitals and University System of Taiwan Joint Research Program [VGHUST 105-G7-2-2, 106-G7-2-2, 107-G1-2-3, 105-G7-2-1, 106-G7-2-1]; Ministry of Science and Technology [MOST 106-2622-E-007-014-CC1, 106-2221-E-007-085-MY3, 107-3017-F-007-002 ]; Frontier Research Center on Fundamental and Applied Sciences of Matters, from the Featured Areas Research Center Program within the framwork of the Higher Education Sprout Project by the Ministry of Education [MOE 107QR001, 107Q2529E1], Taiwan. Funding for open access charge: VGHUST107-G1-2-3.
Conflict of interest statement. None declared.
REFERENCES
- 1. Balogh J., Victor D., Asham E.H., Burroughs S.G., Boktour M., Saharia A., Li X., Ghobrial R.M., Monsour H.P.. Hepatocellular carcinoma: a review. J. Hepatocell Carcinoma. 2016; 3:41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hofmann C., Sandig V., Jennings G., Rudolph M., Schlag P., Strauss M.. Efficient gene-transfer into human hepatocytes by baculovirus vectors. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:10099–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Delaney W.E., Miller T.G., Isom H.C.. Use of the hepatitis B virus recombinant baculovirus-HepG2 system to study the effects of (-)-beta-2′,3′-dideoxy-3′-thiacytidine on replication of hepatitis B virus and accumulation of covalently closed circular DNA. Antimicrob. Agents Chemother. 1999; 43:2017–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mansouri M., Bellon-Echeverria I., Rizk A., Ehsaei Z., Cianciolo Cosentino C., Silva C.S., Xie Y., Boyce F.M., Davis M.W., Neuhauss S.C.F. et al. . Highly efficient baculovirus-mediated multigene delivery in primary cells. Nat. Commun. 2016; 7:11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lin C.-Y., Lin K.-J., Kao C.-Y., Chen M.-C., Yen T.-Z., Lo W.-H., Chang Y.-H., Hu Y.-C.. The role of adipose-derived stem cells engineered with the persistently expressing hybrid baculovirus in the healing of massive bone defects. Biomaterials. 2011; 32:6505–6514. [DOI] [PubMed] [Google Scholar]
- 6. Wu C., Lin J., Hong M., Choudhury Y., Balani P., Leung D., Dang L.-H., Zhao Y., Zeng J., Wang S.. Combinatorial control of suicide gene expression by tissue-specific promoter and microRNA regulation for cancer therapy. Mol. Ther. 2009; 17:2058–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang J., Balasundaram G., Lo S.-L., Guang E.C.S., Xue J.M., Song J., Wan A.C.A., Ying J.Y., Wang S.. Microfibers fabricated by non-covalent assembly of peptide and DNA for viral vector encapsulation and cancer therapy. Adv. Mater. 2012; 24:3280–3284. [DOI] [PubMed] [Google Scholar]
- 8. Swift S.L., Rivera G.C., Dussupt V., Leadley R.M., Hudson L.C., Ma de Ridder C., Kraaij R., Burns J.E., Maitland N.J., Georgopoulos L.J.. Evaluating baculovirus as a vector for human prostate cancer gene therapy. PLos ONE. 2013; 8:e65557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Luo W.Y., Shih Y.S., Hung C.L., Lo K.W., Chiang C.S., Lo W.H., Huang S.F., Wang S.C., Yu C.F., Chien C.H. et al. . Development of the hybrid Sleeping Beauty-baculovirus vector for sustained gene expression and cancer therapy. Gene Ther. 2012; 19:844–851. [DOI] [PubMed] [Google Scholar]
- 10. Luo W.Y., Shih Y.S., Lo W.H., Chen H.R., Wang S.C., Wang C.H., Chien C.H., Chiang C.S., Chuang Y.J., Hu Y.C.. Baculovirus vectors for antiangiogenesis-based cancer gene therapy. Cancer Gene Ther. 2011; 18:637–645. [DOI] [PubMed] [Google Scholar]
- 11. Chen C.L., Wu J.C., Chen G.Y., Yuan P.H., Tseng Y.W., Li K.C., Hwang S.M., Hu Y.C.. Baculovirus-mediated miRNA regulation to suppress hepatocellular carcinoma tumorigenicity and metastasis. Mol. Ther. 2015; 23:79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen C.-L., Tseng Y.-W., Wu J.-C., Chen G.-Y., Lin K.-C., Hwang S.-M., Hu Y.-C.. Suppression of hepatocellular carcinoma by baculovirus-mediated expression of long non-coding RNA PTENP1 and MicroRNA regulation. Biomaterials. 2015; 44:71–81. [DOI] [PubMed] [Google Scholar]
- 13. Deans T.L., Cantor C.R., Collins J.J.. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell. 2007; 130:363–372. [DOI] [PubMed] [Google Scholar]
- 14. Way J.C., Collins J.J., Keasling J.D., Silver P.A.. Integrating biological redesign: Where snthetic biology came from and where it needs to go. Cell. 2014; 157:151–161. [DOI] [PubMed] [Google Scholar]
- 15. Smole A., Lainšček D., Bezeljak U., Horvat S., Jerala R.. A synthetic mammalian therapeutic gene circuit for sensing and suppressing inflammation. Mol. Ther. 2017; 25:102–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xue S., Yin J., Shao J., Yu Y., Yang L., Wang Y., Xie M., Fussenegger M., Ye H.. A synthetic-biology-inspired therapeutic strategy for targeting and treating hepatogenous diabetes. Mol. Ther. 2017; 25:443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Culler S.J., Hoff K.G., Smolke C.D.. Reprogramming cellular behavior with RNA controllers responsive to endogenous proteins. Science. 2010; 330:1251–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kemmer C., Gitzinger M., Baba Daoud-El, Djonov M., Stelling V., Fussenegger M.. Self-sufficient control of urate homeostasis in mice by a synthetic circuit. Nat. Biotechnol. 2010; 28:355–360. [DOI] [PubMed] [Google Scholar]
- 19. Ausländer S., Fussenegger M.. From gene switches to mammalian designer cells: present and future prospects. Trends Biotechnol. 2013; 31:155–168. [DOI] [PubMed] [Google Scholar]
- 20. Ausländer S., Fussenegger M.. Synthetic RNA-based switches for mammalian gene expression control. Curr. Opin. Biotechnol. 2017; 48:54–60. [DOI] [PubMed] [Google Scholar]
- 21. Kawasaki S., Fujita Y., Nagaike T., Tomita K., Saito H.. Synthetic mRNA devices that detect endogenous proteins and distinguish mammalian cells. Nucleic Acids Res. 2017; 45:e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Andries O., Kitada T., Bodner K., Sanders N.N., Weiss R.. Synthetic biology devices and circuits for RNA-based ‘smart vaccines’: a propositional review. Expert Rev. Vaccines. 2015; 14:313–331. [DOI] [PubMed] [Google Scholar]
- 23. Ganesan S.M., Falla A., Goldfless S.J., Nasamu A.S., Niles J.C.. Synthetic RNA–protein modules integrated with native translation mechanisms to control gene expression in malaria parasites. Nat. Commun. 2016; 7:10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peters G., Coussement P., Maertens J., Lammertyn J., De Mey M.. Putting RNA to work: translating RNA fundamentals into biotechnological engineering practice. Biotechnol. Adv. 2015; 33:1829–1844. [DOI] [PubMed] [Google Scholar]
- 25. Xie Z., Wroblewska L., Prochazka L., Weiss R., Benenson Y.. Multi-Input RNAi-based logic circuit for identification of specific cancer cells. Science. 2011; 333:1307–1311. [DOI] [PubMed] [Google Scholar]
- 26. Mohammadi P., Beerenwinkel N., Benenson Y.. Automated design of synthetic cell classifier circuits using a two-step optimization strategy. Cell Syst. 2017; 4:207–218. [DOI] [PubMed] [Google Scholar]
- 27. Saito H., Kobayashi T., Hara T., Fujita Y., Hayashi K., Furushima R., Inoue T.. Synthetic translational regulation by an L7Ae–kink-turn RNP switch. Nat. Chem. Biol. 2010; 6:71–78. [DOI] [PubMed] [Google Scholar]
- 28. Saito H., Fujita Y., Kashida S., Hayashi K., Inoue T.. Synthetic human cell fate regulation by protein-driven RNA switches. Nat. Commun. 2011; 2:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Endo K., Stapleton J.A., Hayashi K., Saito H., Inoue T.. Quantitative and simultaneous translational control of distinct mammalian mRNAs. Nucleic Acids Res. 2013; 41:e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stapleton J.A., Endo K., Fujita Y., Hayashi K., Takinoue M., Saito H., Inoue T.. Feedback control of protein expression in mammalian cells by tunable synthetic translational inhibition. ACS Syn. Biol. 2012; 1:83–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Auslander S., Stucheli P., Rehm C., Auslander D., Hartig J.S., Fussenegger M.. A general design strategy for protein-responsive riboswitches in mammalian cells. Nat. Methods. 2014; 11:1154–1160. [DOI] [PubMed] [Google Scholar]
- 32. Wroblewska L., Kitada T., Endo K., Siciliano V., Stillo B., Saito H., Weiss R.. Mammalian synthetic circuits with RNA binding proteins for RNA-only delivery. Nat. Biotechnol. 2015; 33:839–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ausländer S., Ausländer D., Müller M., Wieland M., Fussenegger M.. Programmable single-cell mammalian biocomputers. Nature. 2012; 487:123. [DOI] [PubMed] [Google Scholar]
- 34. Liao Y.-H., Chang Y.-H., Sung L.-Y., Li K.-C., Yeh C.-L., Yen T.-C., Hwang S.-M., Lin K.-J., Hu Y.-C.. Osteogenic differentiation of adipose-derived stem cells and calvarial defect repair using baculovirus-mediated co-expression of BMP-2 and miR-148b. Biomaterials. 2014; 35:4901–4910. [DOI] [PubMed] [Google Scholar]
- 35. Sung L.-Y., Chen C.-L., Lin S.-Y., Li K.-C., Yeh C.-L., Chen G.-Y., Lin C.-Y., Hu Y.-C.. Efficient gene delivery into cell lines and stem cells using baculovirus. Nat. Protoc. 2014; 9:1882–1899. [DOI] [PubMed] [Google Scholar]
- 36. Sung L.-Y., Chen C.-L., Lin S.-Y., Hwang S.-M., Lu C.-H., Li K.-C., Lan A.-S., Hu Y.-C.. Enhanced and prolonged baculovirus-mediated expression by incorporating recombinase system and in cis elements: a comparative study. Nucleic Acids Res. 2013; 41:e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lo S.-C., Li K.-C., Chang Y.-H., Hsu M.-N., Sung L.-Y., Vu T.A., Hu Y.-C.. Enhanced critical-size calvarial bone healing by ASCs engineered with Cre/loxP-based hybrid baculovirus. Biomaterials. 2017; 124:1–11. [DOI] [PubMed] [Google Scholar]
- 38. Tian T., Wang M., Zhu W., Dai Z.-M., Lin S., Yang P.-T., Liu X.-H., Liu K., Zhu Y.-Y., Zheng Y. et al. . MiR-146a and miR-196a-2 polymorphisms are associated with hepatitis virus-related hepatocellular cancer risk: a meta-analysis. Aging. 2017; 9:381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xu X., Ling Q., Wang J., Xie H., Wei X., Lu D., Hu Q., Zhang X., Wu L., Zhou L. et al. . Donor miR-196a-2 polymorphism is associated with hepatocellular carcinoma recurrence after liver transplantation in a Han Chinese population. Int, J. Cancer. 2016; 138:620–629. [DOI] [PubMed] [Google Scholar]
- 40. Qiu G.H., Xie X., Xu F., Shi X., Wang Y., Deng L.. Distinctive pharmacological differences between liver cancer cell lines HepG2 and Hep3B. Cytotechnology. 2015; 67:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ishiguro T., Ohata H., Sato A., Yamawaki K., Enomoto T., Okamoto K.. Tumor-derived spheroids: Relevance to cancer stem cells and clinical applications. Cancer Sci. 2017; 108:283–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu L., Cao Y., Chen C., Zhang X., McNabola A., Wilkie D., Wilhelm S., Lynch M., Carter C.. Sorafenib blocks the RAF/MEK/ERK pathway, inhibits tumor angiogenesis, and induces tumor cell apoptosis in hepatocellular carcinoma model PLC/PRF/5. Cancer Res. 2006; 66:11851–11858. [DOI] [PubMed] [Google Scholar]
- 43. Hirosawa M., Fujita Y., Parr Callum J.C., Hayashi K., Kashida S., Hotta A., Woltjen K., Saito H.. Cell-type-specific genome editing with a microRNA-responsive CRISPR–Cas9 switch. Nucleic Acids Res. 2017; 45:e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mueller D.W., Bosserhoff A.K.. MicroRNA miR-196a controls melanoma-associated genes by regulating HOX-C8 expression. Int. J. Cancer. 2011; 129:1064–1074. [DOI] [PubMed] [Google Scholar]
- 45. Darda L., Hakami F., Morgan R., Murdoch C., Lambert D.W., Hunter K.D.. The role of HOXB9 and miR-196a in head and neck squamous cell carcinoma. PLos One. 2015; 10:e0122285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Suárez Y., Sessa W.C.. microRNAs as novel regulators of angiogenesis. Cir. Res. 2009; 104:442–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Guerriero I., D’Angelo D., Pallante P., Santos M., Scrima M., Malanga D., De Marco C., Weisz A., Laudanna C., Ceccarelli M. et al. . Analysis of miRNA profiles identified miR-196a as a crucial mediator of aberrant PI3K/AKT signaling in lung cancer cells. Oncotarget. 2017; 8:19172–19191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fan Y., Fan J., Huang L., Ye M., Huang Z., Wang Y., Li Q., Huang J.. Increased expression of microRNA-196a predicts poor prognosis in human ovarian carcinoma. Int. J. Clin. Exp. Pathol. 2015; 8:4132–4137. [PMC free article] [PubMed] [Google Scholar]
- 49. Villegas-Ruiz V., Juárez-Méndez S., Pérez-González O.A., Arreola H., Paniagua-García L., Parra-Melquiadez M., Peralta-Rodríguez R., López-Romero R., Monroy-García A., Mantilla-Morales A. et al. . Heterogeneity of microRNAs expression in cervical cancer cells: over-expression of miR-196a. Int. J. Clin. Exp. Pathol. 2014; 7:1389–1401. [PMC free article] [PubMed] [Google Scholar]
- 50. Fish J.E., Santoro M.M., Morton S.U., Yu S., Yeh R.-F., Wythe J.D., Ivey K.N., Bruneau B.G., Stainier D.Y.R., Srivastava D.. miR-126 regulates angiogenic signaling and vascular integrity. Dev. Cell. 2008; 15:272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kotterman M.A., Schaffer D.V.. Engineering adeno-associated viruses for clinical gene therapy. Nat. Rev. Genet. 2014; 15:445–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Brown B.D., Cantore A., Annoni A., Sergi L.S., Lombardo A., Valle Della. A microRNA-regulated lentiviral vector mediates stable correction of hemophilia B mice. Blood. 2007; 110:4144–4152. [DOI] [PubMed] [Google Scholar]
- 53. Strobel B., Klauser B., Hartig J.S., Lamla T., Gantner F., Kreuz S.. Riboswitch-mediated attenuation of transgene cytotoxicity increases adeno-associated virus vector yields in HEK-293 cells. Mol. Ther. 2015; 23:1582–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dastor M., Schreiber J., Prochazka L., Angelici B., Kleinert J., Klebba I., Doshi J., Shen L., Benenson Y.. A workflow for in vivo evaluation of candidate inputs and outputs for cell classifier gene circuits. ACS Synth. Biol. 2018; 16:474–489. [DOI] [PubMed] [Google Scholar]
- 55. Ketzer P., Kaufmann J.K., Engelhardt S., Bossow S., von Kalle C., Hartig J.S., Ungerechts G., Nettelbeck D.M.. Artificial riboswitches for gene expression and replication control of DNA and RNA viruses. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E554–E562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ketzer P., Haas S.F., Engelhardt S., Hartig J.S., Nettelbeck D.M.. Synthetic riboswitches for external regulation of genes transferred by replication-deficient and oncolytic adenoviruses. Nucleic Acids Res. 2012; 40:e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bell C.L., Yu D., Smolke C.D., Geall A.J., Beard C.W., Mason P.W.. Control of alphavirus-based gene expression using engineered riboswitches. Virology. 2015; 483:302–311. [DOI] [PubMed] [Google Scholar]
- 58. Borel F., Kay M.A., Mueller C.. Recombinant AAV as a platform for translating the therapeutic potential of RNA interference. Mol. Ther. 2014; 22:692–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Airenne K.J., Hu Y.-C., Kost T.A., Smith R.H., Kotin R.M., Ono C., Matsuura Y., Wang S., Yla-Herttuala S.. Baculovirus: an insect-derived vector for diverse gene transfer applications. Mol. Ther. 2013; 21:739–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen C.-Y., Lin C.-Y., Chen G.-Y., Hu Y.-C.. Baculovirus as a gene delivery vector: recent understandings of molecular alterations in transduced cells and latest applications. Biotechnol. Adv. 2011; 29:618–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li K.-C., Chang Y.-H., Yeh C.-L., Hu Y.-C.. Healing of osteoporotic bone defects by baculovirus-engineered bone marrow-derived MSCs expressing microRNA sponges. Biomaterials. 2016; 74:155–166. [DOI] [PubMed] [Google Scholar]
- 62. Hsu M.-N., Liao H.-T., Li K.-C., Chen H.-H., Yen T.-C., Makarevich P., Parfyonova Y., Hu Y.-C.. Adipose-derived stem cell sheets functionalized by hybrid baculovirus for prolonged GDNF expression and improved nerve regeneration. Biomaterials. 2017; 140:189–200. [DOI] [PubMed] [Google Scholar]
- 63. Cheshenko N., Krougliak N., Eisensmith R.C., Krougliak V.A.. A novel system for the production of fully deleted adenovirus vectors that does not require helper adenovirus. Gene Ther. 2001; 8:846–854. [DOI] [PubMed] [Google Scholar]
- 64. Abil Z., Zhao H.. Engineering reprogrammable RNA-binding proteins for study and manipulation of the transcriptome. Mol. BioSyst. 2015; 11:2658–2665. [DOI] [PubMed] [Google Scholar]
- 65. Ohno H., Inoue T.. Designed Regular Tetragon-Shaped RNA–Protein Complexes with Ribosomal Protein L1 for Bionanotechnology and Synthetic Biology. ACS Nano. 2015; 9:4950–4956. [DOI] [PubMed] [Google Scholar]
- 66. Wright A.V., Nunez J.K., Doudna J.A.. Biology and applications of CRISPR systems: harnessing nature's toolbox for genome engineering. Cell. 2016; 164:29–44. [DOI] [PubMed] [Google Scholar]
- 67. Maeder M.L., Gersbach C.A.. Genome-editing technologies for gene and cell therapy. Mol. Ther. 2016; 24:430–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D.. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013; 31:822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chew W.L., Tabebordbar M., Cheng J.K.W., Mali P., Wu E.Y., Ng A.H.M., Zhu K., Wagers A.J., Church G.M.. A multifunctional AAV–CRISPR–Cas9 and its host response. Nat. Methods. 2016; 13:868–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.