Abstract
It is becoming increasingly clear that chromosome organization plays an important role in gene regulation. High-resolution methods such as 4C, Capture-C and promoter capture Hi-C (PCHiC) enable the study of chromatin loops such as those formed between promoters and enhancers or CTCF/cohesin binding sites. An important aspect of 4C/Capture-C/PCHiC analyses is the reliable identification of chromatin loops, preferably not based on visual inspection of a DNA contact profile, but on reproducible statistical analysis that robustly scores interaction peaks in the non-uniform contact background. Here, we present peakC, an R package for the analysis of 4C/Capture-C/PCHiC data. We generated 4C data for 13 viewpoints in two tissues in at least triplicate to test our methods. We developed a non-parametric peak caller based on rank-products. Sampling analysis shows that not read depth but template quality is the most important determinant of success in 4C experiments. By performing peak calling on single experiments we show that the peak calling results are similar to the replicate experiments, but that false positive rates are significantly reduced by performing replicates. Our software is user-friendly and enables robust peak calling for one-vs-all chromosome capture experiments. peakC is available at: https://github.com/deWitLab/peakC.
INTRODUCTION
The 3D organization of the genome plays an important role in the regulation of genes. Many examples have been described of enhancer sequences that activate gene expression over large distances. One of the most extreme cases is a Shh enhancer, which regulates its target gene at a distance of ∼1 Mb (1–3). In contrast to activating promoter–enhancer loops, insulated neighborhoods prevent the spurious activation of neighboring genes by a distal super-enhancer (4), in line with their proposed role as insulator sequences (5). To measure the organization of the 3D genome, various adaptations of the original 3C (6) method have been developed (4C (7)/5C (8)/Hi-C (9)/Capture-C (10)/promoter Capture Hi-C (11)). 4C, Capture-C and promoter capture Hi-C (PCHiC) are so-called one-vs-all methods, which enable the measurement of contact frequencies of a single locus (the ‘viewpoint’) in the case of 4C and Capture-C or multiple single loci in the case of PCHiC with the entire genome in-depth and at high resolution.
In previous work we have presented strategies for the identification of far-cis interactions (generally > 5 Mb from the viewpoint) (12,13). However, the strongest regulatory interactions occur in near-cis regions, in the form of preferential chromatin loops, for instance between promoters and enhancers or between convergently oriented CTCF sites (14,15). To discover these loops, here we focus on identifying significant interactions, apparent as peaks in 4C/Capture-C data. To this end we have defined a number of characteristics a contact peak should adhere to. First, a loop or contact peak should be identified in replicate experiments. Second, when the genomic region of a peak identified in a 4C experiment is used as a viewpoint in a subsequent 4C experiment, it should identify the original viewpoint; a characteristic we term reciprocity. Finally, because of the nature of ‘C’-methods, neighboring fragments co-migrate in the cross-linking step and therefore peaks cannot be interpreted at the single fragment level.
A crucial first step in the identification of peaks is to properly model the background distribution, which is non-trivial for two reasons. First, the background in a 4C/Capture-C experiment is highly non-uniform, with the ‘random’ contact frequency decreasing with an increase in the distance from the viewpoint. In addition, the background distribution can differ substantially depending on where in the genome the viewpoint is chosen. For instance, if a viewpoint is close to the border of a topologically associating domain (TAD) there will be higher contact frequencies with loci inside the TAD compared to outside the TAD, leading to a strong skew in the distribution of 4C coverage. We (16) and others (17) have previously used monotonic regression to empirically model the background distribution. The FourCSeq analysis method (17) uses a transformation of the data and subsequent variance stabilizing normalization prior to fitting the regression model and the identification of peaks. We developed peakC, a novel non-parametric peak caller for 4C, Capture-C and PCHiC data based on monotonic regression and rank-product based statistics. Our 4C-Seq peak calling dataset enables us to put a lower bound on the number of reads required for robust peak calling and show that template complexity is a more important determinant for success in a 4C experiment than read depth. Furthermore, we show that peakC can be applied to Capture-C data resulting in robust kilobase resolution peak calling and to PCHiC data enabling the analysis of thousands of genomic regions in parallel.
MATERIALS AND METHODS
44C dataset
E14 ES cells (IB10 cells) were cultured in BRL-conditioned Dulbecco's Modified Eagle Medium (DMEM with High Glucose, GlutaMAX™, pyruvate; Life Technologies) supplemented with 10% fetal calf serum (FCS), non-essential amino acids (NEAA) (Life Technologies), 1000 U/ml leukemia inhibitory factor (LIF) and 2-mercaptoethanol. We isolated fetal livers from e14.5 embryos by dissection and obtained single cell suspensions by filtration through a cell strainer (BD biosciences). 4C was performed as described previously (18) using DpnII as a first restriction enzyme and Csp6I as a second restriction enzyme. For mESCs we generated three biological replicates from individual dishes (∼10 × 106 cells). For fetal livers, biological replicates were generated from individual embryos from a single pregnant mouse. We used barcoded primers to distinguish between replicates and tissues. 4C PCR amplicons were sequenced on a HiSeq 2500.
Sequencing reads consist of a viewpoint specific sequence which is equal to the forward primer that was used in the 4C PCR and the sequence that was ligated to the viewpoint fragment. The primer sequence is used to identify the reads that belong to a specific viewpoint. After splitting the reads into viewpoint specific fastq files, the primer sequence, excluding the restriction site (i.e. GATC for DpnII) is trimmed from the reads. The trimmed reads are subsequently mapped to the genome (mm9) with bowtie2 (19) using standard settings. Reads with a mapping quality of 1 or higher are retained. Next, we apply three filters for the mapped reads: whether (i) they overlap with a restriction fragment end, (ii) whether they are unique in the genome and (3) whether they are ‘blind’ fragments or not. We will briefly explain these filters below.
Before any 4C mapping is done we perform an in silico digestion of the reference genome (in our case mm9) using the first and second restriction enzyme (in our case DpnII/MboI and Csp6I, respectively). In the 3C ligation step, every restriction fragment has two restriction fragment ends (or fragment ends), that can ligate to the viewpoint fragment. Fragment ends are therefore the natural highest resolution of 4C experiments (12,20). The in silico fragment ends are mapped back to the reference genome, from which we get the position of the original fragment end and whether the fragment end is unique in the genome. We remove non-unique fragment ends from our dataset, because filtering non-uniquely mapping reads would otherwise lead to an excess of zeros in the data at these fragment ends. Finally, a special class of fragments that are ligated to the viewpoint fragment, but that do not contain a restriction site for the secondary restriction enzyme (i.e. DpnII-DpnII fragments or so-called ‘blind’ fragments (18)) were also removed from the dataset, because they give a systematically lower level of signal. The analyses described in this work are focused on the intrachromosomal (‘cis’) interactions and we performed read depth normalization across experiments by simple scaling of mapped reads to 1 million mapped reads in cis. The data have been deposited to GEO under accession number GSE105177.
External data
Capture-C data was taken from (10) and (21) and was downloaded from GEO (accession GSE67959 and GSE97867, respectively). Erythroblast PHiC data was taken from (11) and was downloaded from https://osf.io/u8tzp/.
External ChIPseq datasets were downloaded for CTCF and H3K27ac in mESC (16,22) and Ter119+ cells (21,23).
peakC was developed in R and is provided as a package on github: https://github.com/deWitLab/peakC. The package enables peak calling for both single and replicate experiments. The reading functions of peakC perform the normalization of the data (see above) and provide the user with some quality characteristics (for instance the number of captured fragments within the 100 kb flanking the viewpoint). For the analysis a subset of the data is chosen (for instance 1 Mb up- and downstream of the viewpoint). We urge the user to exercise caution is selecting the size of the genomic window. We have observed a slight tendency for a higher probability of false positive identification of peaks at more proximal sites (data not shown). All the statistical analyses implemented in peakC that we describe below are performed on this subset of the data flanking the viewpoint.
Identification of spatial contacts in replicate experiments
The main assumption in our 4C analysis is that the contact frequency decreases monotonically with the distance. We use monotonic regression to model the background contact frequency. Because the background contact frequency can differ between the regions upstream and downstream of the viewpoint (see Figure 1A) we model these independently. In order to identify regions that are significantly contacted by the viewpoint we have developed a statistical framework that enables the analysis of replicate 4C experiments from the same viewpoint. For every experiment a background model is calculated. Next, we calculate both the ratio (R) and the difference (Δ) between the running mean of the observed fragment end coverage and the expected background coverage for this fragment end. The number of fragment ends in the running mean can be set using the parameter wSize. For 4C experiments we typically set this value to 21, for the Capture-C data we have set wSize to 11. Our peak calling consists of a two-step process. First, we determine fragment ends whose coverage significantly exceeds background coverage using a rank-product statistic based on the Δ scores from multiple replicate experiments. Second, we apply an empirical post-hoc effect size threshold filter based on the R scores. Both will be discussed below.
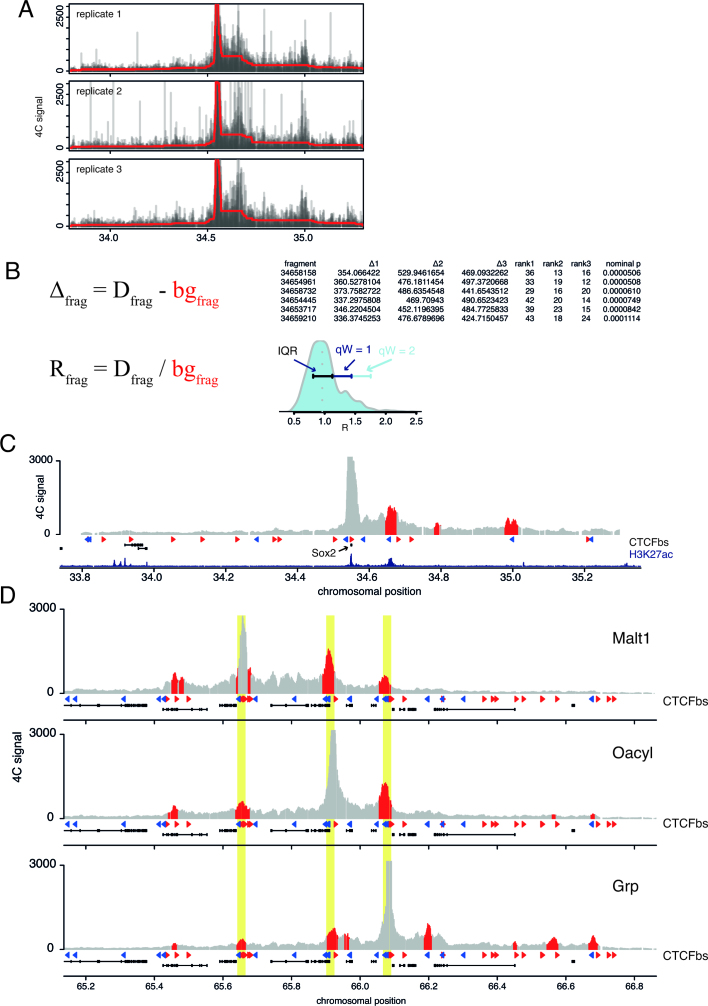
Figure 1.
Monotonic regression procedure for peak calling in 4C data. (A) 4C profiles for the Sox2 locus in mouse ESCs are shown. Read coverage to individual fragments normalized to 1 million intrachromosomal reads is shown for three replicates. red line shows the background distribution estimated from a monotonic regression analysis for the up- and downstream regions separately. (B) Summary of the parameters that are used for the identification of peaks. Δfrag is calculated as the difference between the coverage over the fragment (Dfrag) and the estimated background value for that fragment (bgfrag). For every fragment three Δfrag values are determined (one for every replicate) and the corresponding rank within each replicate. The ranks are multiplied and transformed into a P-value (see Materials and Methods). For the second threshold the ratio (Rfrag) between Dfragand bgfrag is calculated. The threshold value is set as Q3 + qRw x IQR (Q3 is the third quartile and IQR the Interquartile Range). By varying the qRw threshold can be set more or less stringent. The data is based on the three replicates shown in (A). (C) Combined 4C profile of the replicates in (A). Red and blue triangles show the orientation of CTCF binding sites. Levels of the active enhancer mark H3K27ac from mESCs (22) are shown in darkblue. (D) Combined sextuplicate 4C profiles and peak calls for reciprocal viewpoints. Viewpoint names indicate the closest gene. Red and blue rectangles show the orientation of CTCF binding sites.
For combining multiple replicate experiments we make use of the rank-product (24). Originally developed for the identification of differentially expressed genes, rank products are an intuitive, non-parametric way of modeling variation observed across independent experiments. Within each experiment we rank the Δ scores (from high to low), giving us a rank for every fragment in each experiment. We combine experiments by calculating the product of the ranks for every fragment. For significance testing, the observed rank products are compared to their sampling distribution under a permutation null model (25). A simple approximation to this distribution, which facilitates the computation of P-values, is provided by Koziol (26): This is implementend in the statistical programming language R as follows: 1 – pgamma(–log(r/(n + 1)k), k, scale = 1), where r is the rank-product, n is the number of fragments and k is the number of samples. Although, an exact calculation of the P-value has been proposed by Eisinga et al. (27), for larger samples this is computationally prohibitive. A Benjamini and Hochberg multiple testing procedure (28) is then applied with the aim to control for the false discovery rate (‘fdr’). We use the implementation in R by the function p.adjust for this and control the fdr at level alphaFDR; a parameter whose value we typically set to 0.1.
Next, we calculate the average R of the replicate experiments and determine the quartiles and interquartile range (IQR) of the distribution of R. Let Q3 be the third quartile, then the threshold for R is set as Q3+ qWr * IQR (Figure 1B). qWr can be set as a parameter in peakC to filter results based on effect size and is typically set to 1, after which the data-adaptive threshold is calculated using the formula.
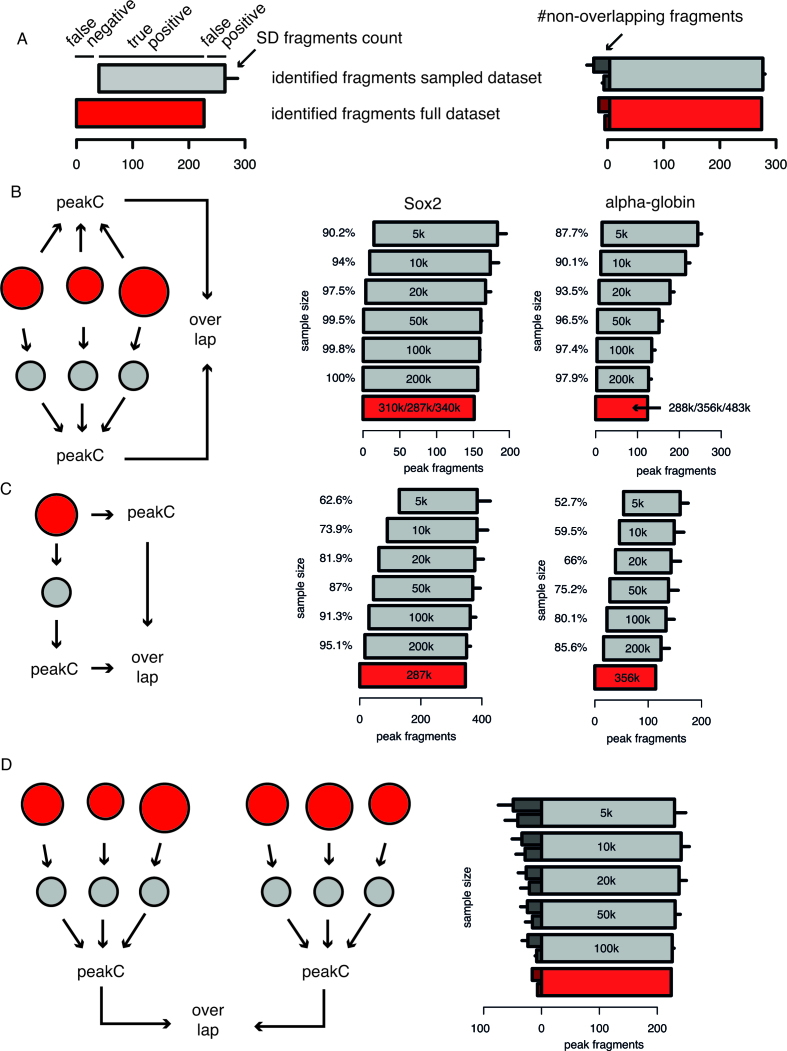
In order to estimate the effect of the sequencing depth of 4C experiments on the reproducibility of the peak calling we performed an in silico analysis where we repeatedly subsampled the 4C data and recalculated the significant interactions. All the fragment ends within a running mean window containing at least one significant fragment end are called as significant fragments. We treat the fragments called in the full data set as the ‘truth’ and calculate the number of true positive, false positive and false negative fragments based upon the overlap between the significant fragments in subsampled datasets and those from the full dataset.
Identification of peaks in single experiments
The approach we follow to identify peaks in a single experiment is also based on monotonic regression. However, since single experiments do not allow us to model the effect of random variation on the 4C coverage, rather than calculating a rank-product based P-value we determine an empirical threshold analogous to the effect size filter for R scores. Fragments are selected when the Δ score is above Q3Δ + qWd * IQR and the R score of the same fragment is greater than Q3R + qWr * IQR. Typical values that seem to work well across our dataset are qWd set to 1.5 and qWr set to 1. The computational analysis that guides the selection of ‘optimal’ values for these parameters, i.e. values that result in high reproducibility of peaks in replicate experiments, will be discussed in the Results section.
FourCSeq analysis
FourCSeq analysis was performed as described in the vignette (https://bioconductor.org/packages/release/bioc/vignettes/FourCSeq/inst/doc/FourCSeq.pdf). In the calls to the FourCSeq analysis function getZScores, we used the following parameters: fitFun = distFitMonotone, removeZeros = TRUE, minCount = 0 and minDist = 20 000. In the parameter sweep we varied two parameters: zScoreThresh and fdrTresh. The z-score was varied between 1.96 and 50 and the FDR between 0.1 and 0.001.
RESULTS
To test our peak calling software, we generated a set of high-resolution 4C datasets. We prepared biological triplicate 4C templates in two mouse tissues, embryonic stem cells (ESCs) and fetal liver. On each of these six templates we assessed the contact profile of 10 different viewpoints, resulting in 60 individual 4C datasets. Eight of these 10 viewpoints were from CTCF binding sites close to TAD borders or loop anchors. The remaining two viewpoints were designed close to the promoter of the mESC-specific gene Sox2 and the fetal liver specific gene α-globin. The tissue-specific expression of these genes is associated with differential chromatin architecture (Supplementary Figure S1A), presumably because the (super)-enhancer-promoter interactions guide the tissue-specific expression. In addition to this collection of data we generated six templates in ES cells for three different viewpoints that showed reciprocal interactions between CTCF binding sites. Read counts and other statistics can be found in Supplementary Table S1. All of the viewpoints showed a certain degree of distal looping. We used these data to evaluate our peak calling procedure.
We sought to develop a robust non-parametric method for the identification of contact peaks in the 4C data. Contact peaks are genomic regions that in replicate experiments show a statistically significant increase in contact frequency over the expected background. We would like to emphasize that contact peaks are different from contact frequency. We acknowledge that contact frequency can be an important measure for promoter–enhancer communication, however, our aim is to identify peaks (or preferential loops), rather than regions with high contact frequency per se. In some ways measuring of contact frequency is easier as it is directly related to the coverage in a 4C experiment. Identification of contact peaks is less trivial as it requires the correct estimation of the background model. peakC fits the background independently for every replicate using monotonic regression (29) and estimates the decay trend in 4C contacts. Monotonic (or isotonic) regression calculates a flexible regression model under the constraint that the dependent variable can only increase or decrease with an increase in the independent variable. This is a reasonable assumption for a chromatin fiber because in a random, unrestrained, polymer model the contact frequency is expected to decrease exponentially with increasing distance for an ensemble of polymers (30). Figure 1A shows the raw 4C data for the promoter of the Sox2 gene in mESCs. The red line shows the estimated background contact frequency. This example clearly indicates why it is important to calculate the background model for up- and downstream sequences independently: contact profiles of individual sites can be highly asymmetric. Ignoring this could lead to either over- or underestimation of the background model. In the next step of the algorithm the difference (Δ) and ratio (R) between the fragment coverage and the predicted fragment coverage is calculated and a statistical analysis is performed to identify significant fragments (see Methods for further details, Figure 1B). As an example, Figure 1C shows the result of the peak calling for the Sox2 locus in mESCs. The most prevalent interaction is the one with the super-enhancer (2,16), but an additional interaction with a very distal convergently oriented CTCF site is also identified. peakC can thus faithfully identify promoter–enhancer interactions and CTCF-mediated chromatin loops.
Because performing replicate experiments is not always possible, for instance due to limited source material, we have also developed a slightly modified version of our peak calling method for single template 4C experiments. The single template peak calling algorithm is based on double thresholding, i.e. on the distributions of both Δ and R (see Materials and Methods for a detailed explanation). In Supplementary Figure S1b we show the results of statistical analysis with peakC on a triplicate experiment. In Supplementary Figure S1C, the results of application of peakC to the individual replicates separately is shown. Although most of the called peaks are shared between the combined and the single experiments, clearly there are regions that are identified in only one of the replicates. Although we do not have a ground truth for 4C peaks, we reason that it is likely that regions identified exclusively in one of three replicate 4C experiments result from random variation in 4C ligation and amplification frequencies and hence constitute false positive peak calls. Below, we will therefore analyze and compare the results from peak calling in single and replicate 4C experiments in more detail.
An important way to validate a 4C peak is to determine reciprocity. For a peak to represent a specific interaction or loop, it should also be appreciable in the reciprocal 4C experiment where the interacting locus is chosen as viewpoint. Therefore, to further validate peakC we designed viewpoints at sites identified as peaks by peakC in our first set of 4C experiments and performed 4C PCRs on six separate 4C templates. Figure 1D shows that for every interacting site for which we performed 4C, we were able to identify the original viewpoint as a contact peak, emphasizing the robustness of our peak calling method.
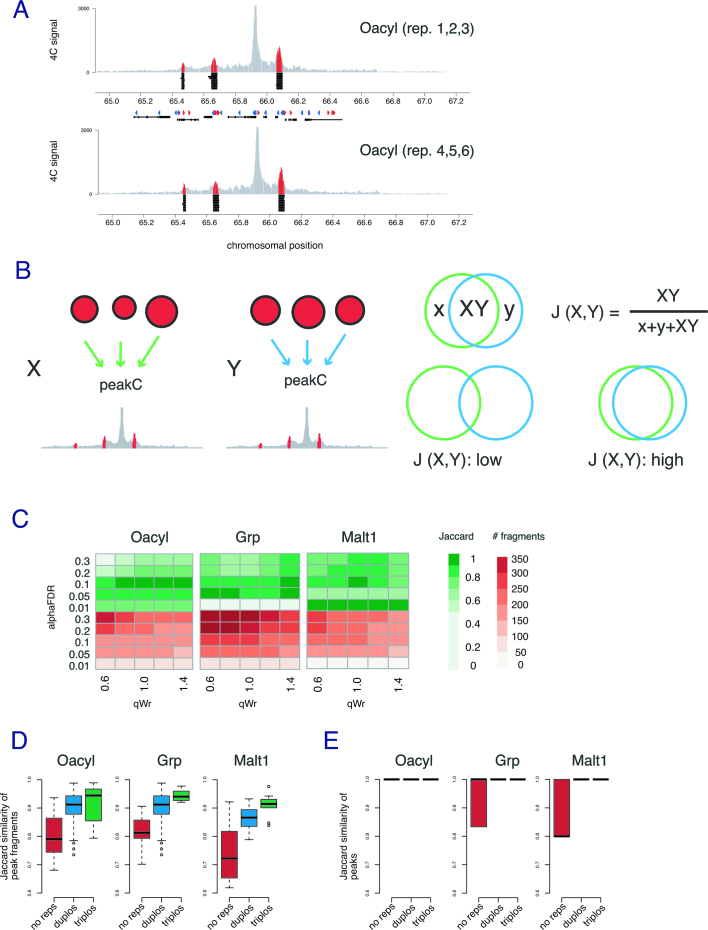
Peak sizes and their degree of overlap between replicates depend on the significance threshold parameters alphaFDR and qWr (see Methods). To optimize these parameters and assess the importance of using replicate templates in a 4C experiment, we divided the dataset into non-overlapping pairs of three replicates (n = 10). As shown in Figure 2A, at a given combination of thresholds peakC consistently finds for all 20 sets of triplicates three contact peaks: they do however differ somewhat in their start and end position across the sets. We subsequently systematically varied the threshold parameters, performed peak calling and determined their similarity by calculating the Jaccard similarity coefficient (i.e. the intersection divided by the union) of the restriction fragments called under these conditions (Figure 2B). The optimal parameters we thus selected were alphaFDR = 0.1 and qWr = 1 (Figure 2C) (for further details, see methods). When reducing the number of replicates per set to two or even single experiments, the average Jaccard similarity coefficient drops correspondingly (Figure 2D). Importantly, we find this trend for all three viewpoints, which suggests that robust peak calling benefits from the inclusion of replicate experiments. We repeated this analysis, but this time calculating overlap between peak regions (defined as collapsed sets of consecutive significant peak fragments) where two peaks overlap when there is an overlap of at least one fragment (Figure 2E). This analysis reveals that peakC is very consistent in identifying broader regions with increased contacts and that when only a single 4C template is available, more regions are being called that cannot be reproduced in independent 4C experiments.
Figure 2.
Replicate experiments show high level of reproducibility for contact peak calls. (A) Plots of Oacyl 4C profiles (4C coverage per 1 million of mapped reads) in two independent sets of triplicate 4C experiments. Genomic regions indicated in black underneath the peak regions in red represent the peaks called in all 20 different (non-overlapping) sets of triplicates (10 in each panel). Red and blue triangles show the orientation of CTCF binding sites. (B) Schematic showing how to compute the Jaccard coefficient to measure the overlap of peak fragments identified by peakC in independent sets. The panel on the left shows 2 independent sets (labeled X and Y) of triplicate 4C experiments. The peak fragments identified are represented in the Venn diagram by a green (set X) and a blue (set Y) circle, and partitioned into subsets labeled x, XY and y representing fragments found exclusively in set X, the overlap of fragments found in set X and set Y and fragments found exclusively in set Y respectively. The formula shows how to calculate the corresponding Jaccard coefficient. (C) Heatmaps of average Jaccard coefficients (green) and average number of peak fragments identified (red) with peakC in independent sets of triplicates for different values of the significance thresholds alphaFDR and qRw for three viewpoints. (D) Boxplots of Jaccard coefficients of independent sets of 4C experiments for optimal values of the significance thresholds alphaFDR and qRw for the sets of triplicate 4C experiments for three different viewpoints. e) Box plots of Jaccard coefficients for comparisons on the level of peaks (sets of collapsed consecutive peak fragments) rather than individual fragments.
In order to further benchmark our method, we compared the peak calls from peakC to peak calls from FourCseq, an alternative 4C analysis method. We first analyzed a viewpoint from the original FourCseq publication (31), that studied the interaction profile of cis-regulatory modules in Drosophila embryos. Viewpoint CRM_4319 shows a stable loop across three different tissue/timepoints (whole embryo 3–4 h, whole embryo 6–8 h and mesoderm 6–8 h) (Supplementary Figure S2A). Both FourCseq and peakC consistently identify this loop across the tissues. However, in the regions between the loop and the viewpoint there seems to be more discrepancy between the interaction calls, with peakC being seemingly better at identifying the visually apparent peaks and preventing identification of seemingly spurious peaks. In order to objectively measure reproducibility, we used our set of six templates to compare two sets of three templates that were processed under the exact same conditions. We first performed a parameter sweep for FourCSeq, like we did for peakC to obtain the parameters that give the most optimal reproducibility. Our first observation was that the FDR threshold affects neither the reproducibility nor the number of called fragments very much (Supplementary Figure S2B). The z-score threshold does affect the number of called fragments, however, the reproducibility as measured by the Jaccard index is not strongly affected. Only at very high z-score thresholds (i.e. >50) for data from the Grp viewpoint FourCSeq gives good reproducibility, but this is for a very small number of fragments (∼7). In Supplementary Figure S2C we show the effect of varying the z-score threshold for the Oacyl viewpoint (see Figure 2A for comparison to peakC). It is important to note that the individual fragments identified by FourCSeq show a low reproducibility, suggesting a high rate of false positives. We chose rational thresholds for FourCSeq (i.e. FDR < 0.05 and z-score > 7.13) and determined the reproducibility between the various sets of replicates. Supplementary Figure S2D shows that the Jaccard index for FourCSeq for almost every comparison is <50%, with Oacyl and Malt1 having Jaccard indices between 40% and 45%. peakC peak calling on the other hand, on average results in Jaccard indices >90%. From these comparisons we conclude that peakC is more conservative in its peak calls than FourCseq, but that the called peaks are more reproducible. In the Discussion we examine how the fundamental differences between the two methods can lead to differences in the results.
Our 4C dataset and peak calling procedure offers an opportunity to assess the effect of 4C data quality on the reproducibility of 4C results. We investigated the minimum amount of reads required for reproducible peak calling by subsampling analysis. We iteratively (100x) and randomly sampled 200k, 100k, 50k, 20k, 10k and 5k reads from our datasets and performed peak calling. To estimate the effect of sequencing depth on performance we each time calculate the number of fragments that are shared between the original dataset and the sampled dataset (see Figure 3A for explanation). Figure 3B shows the results for two tissue-specific genes (Sox2 in mESC and α-globin (Hba1) in fetal liver). When we systematically perform the subsampling analysis for our 10 viewpoints, we find that with only 20k intrachromosomal reads per experiment we still are able to identify >90% of the original peak fragments in 15/20 4C experiments, with the majority of the 20 experiments identifying > 95% of the original fragments. Note also that downsampling does not necessarily come at the cost of a strongly increased false positive rate (Figure 3, Supplementary Figures S3–S4).
Figure 3.
Downsampling analysis shows lower bound for read depth in 4C experiments. (A) Explanatory figure for the interpretation of (B) and (C) (left) and (D) right. (B) Barplots show the effect of downsampling on replicate experiments. Replicate 4C datasets were downsampled to a fixed number of reads (shown inside the bars). Note that the number of reads per replicate is shown. peakC analysis was performed on the subsampled dataset and the result was compared to the full dataset. Results for Sox2 in mESCs and α-globin in fetal livers are shown. (C) Similar analysis as in (B) but with single experiments instead of replicate experiments. One of the three replicates was chosen for the analysis from the Sox2 and α-globin datasets. (D) Barplots show the effect of downsampling on reproducibility. A sextuplicate 4C dataset is randomly split into two sets of triplicate experiments. Both sets are subsequently subsampled to a fixed amount of reads (number of reads per replicate is shown inside the bars). After peakC analysis, the overlap in the fragments called as peaks is plotted.
We also performed the downsampling analysis on the single experiments. As expected downsampling with single experiments leads to higher levels of false positives and false negatives, however the effect is rather modest. It is important to note here that this analysis shows that discrepancy in regions that were identified as peaks across the single experiments, were persistent through repeated downsampling (Supplementary Figure S5B). This means that deeper sequencing of 4C amplicons will not result in more reliable contact peak calling. Rather, based on these analyses we conclude that the inclusion of an additional 4C template is a far more effective method to reduce false positive peak calls.
To further emphasize the importance of adding replicate template experiments, we determined the impact of downsampling on two sets of replicate experiments. To this end we downsampled 6 replicate experiments and divided them into two groups of three replicates and compared the identified contact peaks between the two experiments. As is clear from Figure 3D downsampling to only 20k intrachromosomal reads per replicate results in only limited numbers of false positive and false negative contact peak fragments. Summarizing, these sampling results show that (i) our peak calling algorithm with replicates is very robust between two sets of replicate experiments, (ii) that only a very limited number of reads is necessary for reproducible peak calling and (iii) the inclusion of replicate templates reduces false positive contact peak calls.
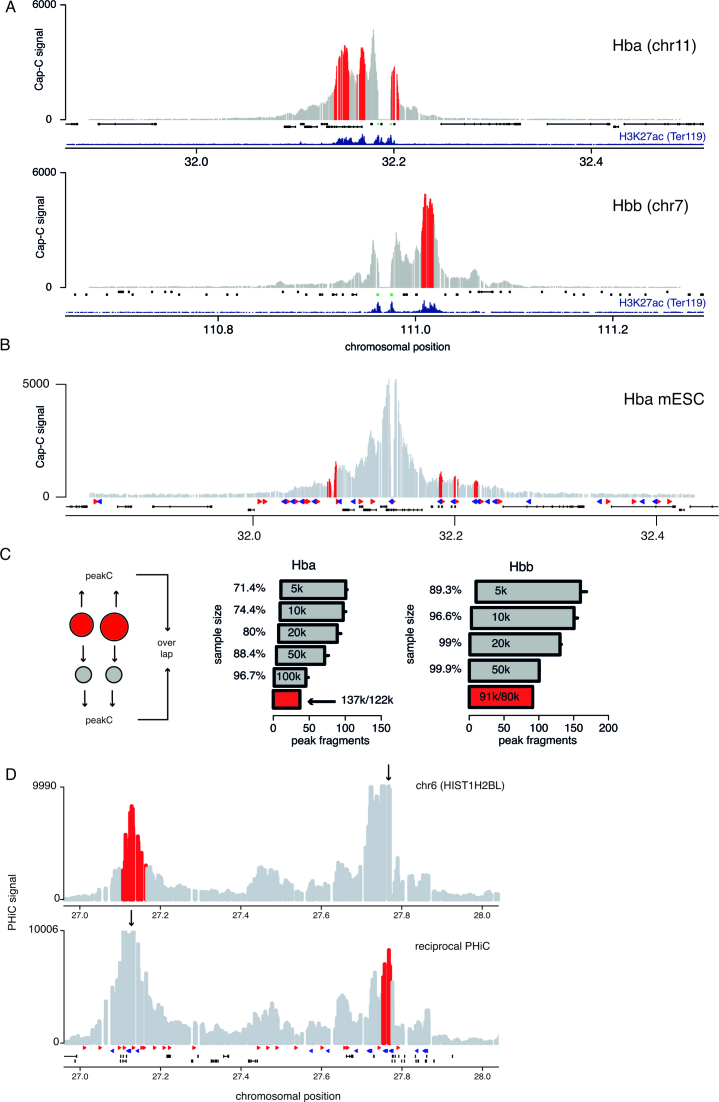
As an alternative to the inverse PCR-based 4C, one can also use a hybridization capture strategy to identify ligated fragments. Because the so-called Capture-C strategy (10,32) is also a one-vs-all strategy like 4C, peakC can be used to identify interactions from these data. Figure 4A shows examples from a multiplexed capture strategy (10) for the α-globin (Hba) and β-globin (Hbb) genes in murine ter119+ cells. peakC clearly identifies the super-enhancers known as the locus control regions that regulate these genes as interactions. In Figure 4B, we show an analysis of a Capture-C experiment in mESCs, which shows a more subtle interaction pattern. From a CTCF binding site in the α-globin region we identify three consecutive CTCF sites as interaction partners, even though the interactions are much less pronounced in mESCs compared to ter119+ cells. These results show the sensitivity of combining Capture-C with the peakC analysis method. By performing a downsampling analysis we can assess the amount of reads at which the signal starts to break down (Figure 4C). The results suggest that decreasing the amount of reads in a Capture-C experiment does not generate a large number of false negatives, but that the number of false positives does increase. It is important to note that, because Capture-C is less sensitive to spurious fragment amplification, smaller window sizes can be used for the identification of interaction regions; a window size of five still gives significant interactions, which brings the resolution in the kilobase range.
Figure 4.
peakC enables the identification of peaks in Capture-C data. (A) peakC output for Capture-C data from the α-globin (Hba, top) and β-globin locus (Hbb, bottom) in mouse erythroid cells. Capture-C data was taken from (10). α-globin and β-globin genes are marked in green. Below the Capture-C data the H3K27ac ChIPseq signal (23) is plotted in darkblue. (B) peakC output for Capture-C data from a CTCF site in mESCs in the α-globin locus. Capture-C data was taken from (21). CTCF data was taken from (16). (C) Downsampling analysis similar to Figure 2B) for the Capture-C datasets shown in A). (D) peakC output for a selected locus from PCHiC data from human erythroblasts.
By using large capture libraries, it is possible to investigate many different sites in parallel, albeit with less depth per viewpoint or bait. We used peakC to perform peak calling on a promoter capture Hi-C (PCHiC) dataset that was generated for human erythroblasts to investigate the interactome of over 30k promoters (11). Whereas the previously analyzed 4C/Capture-C templates were generated with a 4-bp restriction enzyme (i.e. DpnII/MboI), it is important to note that the PCHiC template was generated generated with a 6-bp restriction enzyme (i.e. HindIII), which lowers the resolution of the analysis. Figure 4D shows an example of peakC analysis on these PCHiC data for one of the H2B genes on chromosome 6. We used the reciprocal capture HiC dataset to perform peakC analysis from the site that was identified in the promoter capture. For the reciprocal capture experiment peakC identifies the original promoter as a significant interaction. Comparison with CTCF binding data from lymphoblasts (33) shows that this loop is formed between two convergently oriented CTCF sites. These results show that peakC is a general-purpose analysis toolkit for one-vs-all 3D genome methods.
DISCUSSION
Identifying interactions in chromosome capture data is a non-trivial task because of the non-uniform distribution of the background. Previous methods have tried to model this distribution by averaging over multiple ‘viewpoints’ (34) or by calculating a genome-wide average (35,36). This enables the identification of regions with significantly increased contact frequency compared to a genome-wide average. The downside of these methods is that this can lead to either under- or overestimation of the number of contacts depending on the genomic location of the region. For instance, when a viewpoint is located close to a TAD boundary the distribution of captures (or interactions) is heavily skewed (37), an observation which was crucial to the identification of TADs. Furthermore, if only one or a few viewpoints are available for study, it is not possible to calculate a genome-wide or multi-viewpoint average.
Recent developments in high-resolution Hi-C have led to the identification of preferential chromatin loops, many of which overlap with TAD boundaries (14). This higher resolution also resulted in a different method for the identification of loops, rather than selecting regions that show an increase over an inferred background model, regions were selected that show an increase in signal both upstream and downstream of the loop anchors (14), a choice that was mostly enabled by the increase in resolution and sequencing throughput. Our previous work on CTCF loops followed a similar rationale (16). In the current work, we have extended our previously described peak calling algorithm to also include replicate experiments. The use of replicates also enables the calculation of test statistics and the identification of significantly contacted regions. In order to not have to rely on arbitrary thresholds, inclusion of replicate experiments is therefore strongly advocated. Our results show that sequencing depth should not be a limiting factor for performing these.
Because it is not always feasible to include replicates in experiments, especially when a large number of genomic sites is under investigation, we have also developed a robust method for identifying peaks from single 4C experiments. Many of the peaks we have identified in our replicate experiment were also identified in our single experiments, often in all three replicates individually. However, because the chance of identifying false positive peaks is substantial, the peaks identified in single experiments should be thoroughly scrutinized.
peakC is not the only method for the identification of interactions in 4C data. We performed side-by-side comparison of peakC to FourCseq. Using default and very commonly used significance thresholds, FourCseq seems to call more significant interactions. We show here that the overlap between the fragments that are significantly identified by FourCSeq in two independent sets of replicates is generally <50%, whereas for peakC this number is generally >90%. What differences in the methods could account for these differences in reproducibility? Although both methods rely on monotonic regression for the estimation of the background model, FourCseq uses a smooth monotone fit on the variance-stabilizing transformed data and peakC uses pool-adjacent-violators algorithm (PAVA) isotonic regression on the untransformed data. A more fundamental difference is that, peakC analyses windows of fragments whereas FourCseq performs single fragment analysis. We believe that interpretation of signals from single fragments should be performed with extreme caution. We have previously shown that the amplification of a fragment can be dependent on sequence characteristics, such as length, of said fragment (18). We further advocate the averaging of signals of multiple fragments to acknowledge that neighbor fragments inevitably co-migrate when loops are formed. Finally, the inverse 4C PCR may result in non-systematic amplification of DNA fragments. In our opinion, because peakC takes these characteristics of the 4C method into account we find that peakC shows a higher reproducibility between sets of replicate experiments.
In conclusion, peakC is a lightweight R package that enables conservative but robust loop calling and thereby facilitates the systematic analysis of 4C, Capture-C and PCHiC experiments.
DATA AVAILABILITY
The data have been deposited to GEO under accession number GSE105177. peakC is available at: https://github.com/deWitLab/peakC.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Adrien Melquiond and Valerio Bianchi for mapping 4C data and members of the de Laat and de Wit labs for discussion.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Research Council (ERC) [StG 637587 HAP-PHEN to E.d.W.];Netherlands Organisation for Scientific Research (NWO) [NWO-VICI 724.012.003 to W.d.L.]; Oncode Institute which is partly financed by the Dutch Cancer Society. Funding for open access charge: ERC.
Conflict of interest statement. None declared.
REFERENCES
- 1. Tolhuis B., Palstra R.J., Splinter E., Grosveld F., de Laat W.. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell. 2002; 10:1453–1465. [DOI] [PubMed] [Google Scholar]
- 2. Li Y., Rivera C.M., Ishii H., Jin F., Selvaraj S., Lee A.Y., Dixon J.R., Ren B.. CRISPR reveals a distal super-enhancer required for Sox2 expression in mouse embryonic stem cells. PLoS One. 2014; 9:e114485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lettice L.A., Horikoshi T., Heaney S.J.H., van Baren M.J., van der Linde H.C., Breedveld G.J., Joosse M., Akarsu N., Oostra B.A., Endo N. et al. Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:7548–7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dowen J.M., Fan Z.P., Hnisz D., Ren G., Abraham B.J., Zhang L.N., Weintraub A.S., Schuijers J., Lee T.I., Zhao K. et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014; 159:374–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chung J.H., Whiteley M., Felsenfeld G.. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993; 74:505–514. [DOI] [PubMed] [Google Scholar]
- 6. Dekker J., Rippe K., Dekker M., Kleckner N.. Capturing chromosome conformation. Science. 2002; 295:1306–1311. [DOI] [PubMed] [Google Scholar]
- 7. Simonis M., Klous P., Splinter E., Moshkin Y., Willemsen R., de Wit E., van Steensel B., de Laat W.. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet. 2006; 38:1348–1354. [DOI] [PubMed] [Google Scholar]
- 8. Dostie J., Richmond T.A., Arnaout R.A., Selzer R.R., Lee W.L., Honan T.A., Rubio E.D., Krumm A., Lamb J., Nusbaum C. et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006; 16:1299–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009; 326:289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Davies J.O.J., Telenius J.M., McGowan S.J., Roberts N.A., Taylor S., Higgs D.R., Hughes J.R.. Multiplexed analysis of chromosome conformation at vastly improved sensitivity. Nat. Methods. 2015; 13:74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Javierre B.M., Sewitz S., Cairns J., Wingett S.W., Várnai C., Thiecke M.J., Freire-Pritchett P., Spivakov M., Fraser P., Burren O.S. et al. Lineage-Specific genome architecture links enhancers and Non-coding disease variants to target gene promoters. Cell. 2016; 167:1369–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Splinter E., de Wit E., van de Werken H.J.G., Klous P., de Laat W.. Determining long-range chromatin interactions for selected genomic sites using 4C-seq technology: from fixation to computation. Methods. 2012; 58:221–230. [DOI] [PubMed] [Google Scholar]
- 13. Splinter E., de Wit E., Nora E.P., Klous P., van de Werken H.J.G., Zhu Y., Kaaij L.J.T., van Ijcken W., Gribnau J., Heard E. et al. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev. 2011; 25:1371–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rao S.S.P., Huntley M.H., Durand N.C., Stamenova E.K.. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014; 159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vietri Rudan M., Barrington C., Henderson S., Ernst C., Odom D.T., Tanay A., Hadjur S.. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 2015; 10:1297–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de Wit E., Vos E.S.M., Holwerda S.J.B., Valdes-Quezada C., Verstegen M.J.A.M., Teunissen H., Splinter E., Wijchers P.J., Krijger P.H.L., de Laat W.. CTCF binding polarity determines chromatin looping. Mol. Cell. 2015; 60:676–684. [DOI] [PubMed] [Google Scholar]
- 17. Klein F.A., Pakozdi T., Anders S., Ghavi-Helm Y., Furlong E.E.M., Huber W.. FourCSeq: analysis of 4C sequencing data. Bioinformatics. 2015; 31:3085–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van de Werken H.J.G., Landan G., Holwerda S.J.B., Hoichman M., Klous P., Chachik R., Splinter E., Valdes-Quezada C., Oz Y., Bouwman B.A.M. et al. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat. Methods. 2012; 9:969–972. [DOI] [PubMed] [Google Scholar]
- 19. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Van De Werken H.J.G., De Vree P.J.P., Splinter E., Holwerda S.J.B., Klous P., De Wit E., De Laat W.. 4C technology: protocols and data analysis. Methods Enzymol. 2012; 513:89–112. [DOI] [PubMed] [Google Scholar]
- 21. Hanssen L.L.P., Kassouf M.T., Oudelaar A.M., Biggs D., Preece C., Downes D.J., Gosden M., Sharpe J.A., Sloane-Stanley J.A., Hughes J.R. et al. Tissue-specific CTCF-cohesin-mediated chromatin architecture delimits enhancer interactions and function in vivo. Nat. Cell Biol. 2017; 19:952–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kowalczyk M.S., Hughes J.R., Garrick D., Lynch M.D., Sharpe J.A., Sloane-Stanley J.A., McGowan S.J., De Gobbi M., Hosseini M., Vernimmen D. et al. Intragenic enhancers act as alternative promoters. Mol. Cell. 2012; 45:447–458. [DOI] [PubMed] [Google Scholar]
- 24. Breitling R., Armengaud P., Amtmann A., Herzyk P.. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004; 573:83–92. [DOI] [PubMed] [Google Scholar]
- 25. Breitling R., Armengaud P., Amtmann A., Herzyk P.. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004; 573:83–92. [DOI] [PubMed] [Google Scholar]
- 26. Koziol J.A. Comments on the rank product method for analyzing replicated experiments. FEBS Lett. 2010; 584:941–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Eisinga R., Breitling R., Heskes T.. The exact probability distribution of the rank product statistics for replicated experiments. FEBS Lett. 2013; 587:677–682. [DOI] [PubMed] [Google Scholar]
- 28. Benjamini Y., Hochberg Y.. Controlling the false discovery Rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995; 57:289–300. [Google Scholar]
- 29. de Leeuw J., Hornik K., Mair P.. Isotone optimization in R: Pool-Adjacent-Violators algorithm (PAVA) and active set methods. J. Stat. Softw. 2009; 32:1–24. [Google Scholar]
- 30. Rippe K. Making contacts on a nucleic acid polymer. Trends Biochem. Sci. 2001; 26:733–740. [DOI] [PubMed] [Google Scholar]
- 31. Ghavi-Helm Y., Klein F.A., Pakozdi T., Ciglar L., Noordermeer D., Huber W., Furlong E.E.M.. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 2014; 512:96–100. [DOI] [PubMed] [Google Scholar]
- 32. Hughes J.R., Roberts N., McGowan S., Hay D., Giannoulatou E., Lynch M., De Gobbi M., Taylor S., Gibbons R., Higgs D.R.. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat. Genet. 2014; 46:205–212. [DOI] [PubMed] [Google Scholar]
- 33. Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2013; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sanyal A., Lajoie B.R., Jain G., Dekker J.. The long-range interaction landscape of gene promoters. Nature. 2012; 489:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jin F., Li Y., Dixon J.R., Selvaraj S., Ye Z., Lee A.Y., Yen C.-A., Schmitt A.D., Espinoza C.A., Ren B.. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013; 503:290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ay F., Bailey T.L., Noble W.S.. Statistical confidence estimation for Hi-C data reveals regulatory chromatin contacts. Genome Res. 2014; 24:999–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B.. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012; 485:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data have been deposited to GEO under accession number GSE105177. peakC is available at: https://github.com/deWitLab/peakC.