Fig. 5.
HLA-associated adapted variants enriched in Saskatchewan are preferentially located within provincial sequence clusters.
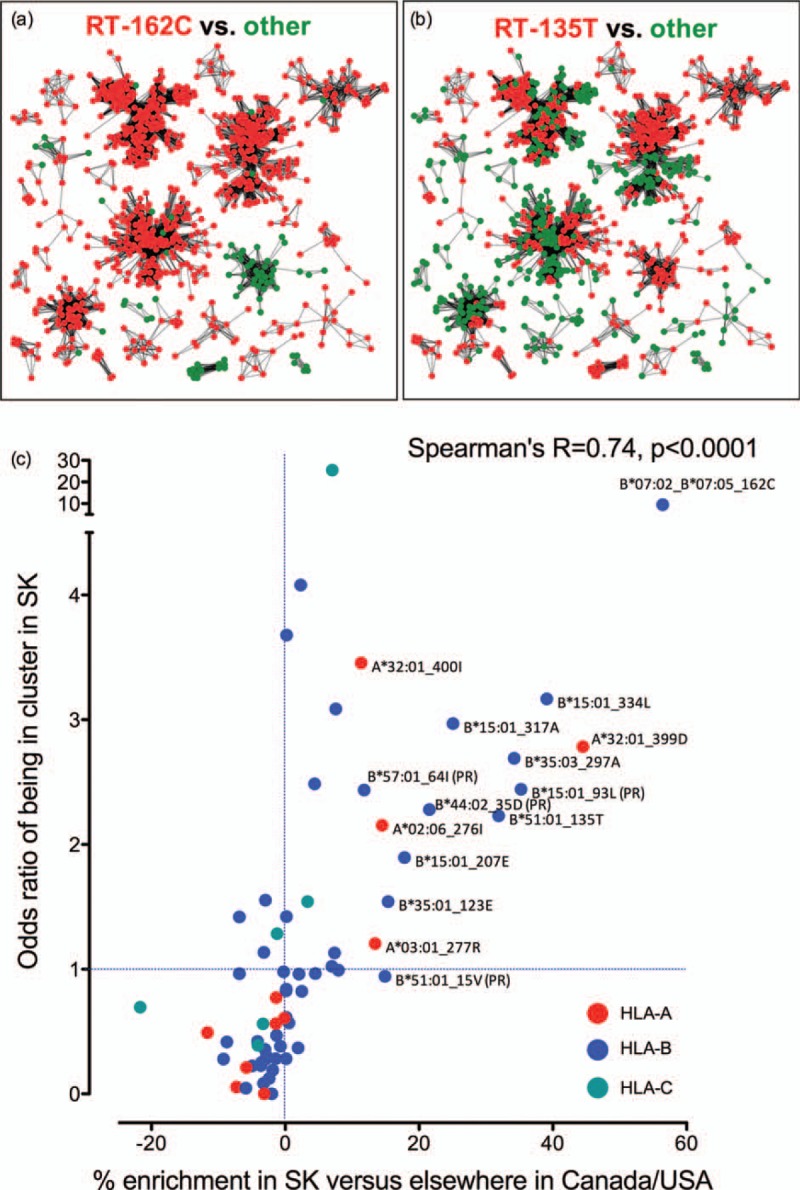
Panel a: Distribution of the B∗07 : 02/05-associated RT-S162C adaptation within major Saskatchewan sequence clusters. Sequences identified as being in a cluster of minimum size 5 were graphed such that each node represented an HIV sequence from a unique individual, and edges represent patristic (tip-to-tip) distances separating them. Sequences harbouring 162C are red, all others are green. Panel b: Distribution of the B∗51 : 01-associated RT-I135T adaptation within major Saskatchewan sequence clusters. Sequences harbouring 135T in red, all others are green. Panel c: Each dot represents one of the 70 HLA-associated adapted variants investigated in the present study, coloured by the HLA locus restricting it (HLA-A: red; HLA-B: blue and HLA-C: teal). For each mutation, its absolute % enrichment in Saskatchewan vs. CA/US (that is, the Δ of the linked values shown in Figures 2b–d) is plotted alongside its odds ratio of being located in an HIV sequence residing in a cluster in Saskatchewan, revealing a highly significant correlation between these factors. Mutations that are more than 10% enriched in Saskatchewan versus CA/US are labeled with their identity and HLA restriction. Results indicate that HLA-associated adapted variants enriched in Saskatchewan are preferentially located within sequence clusters.
