Figure 5.
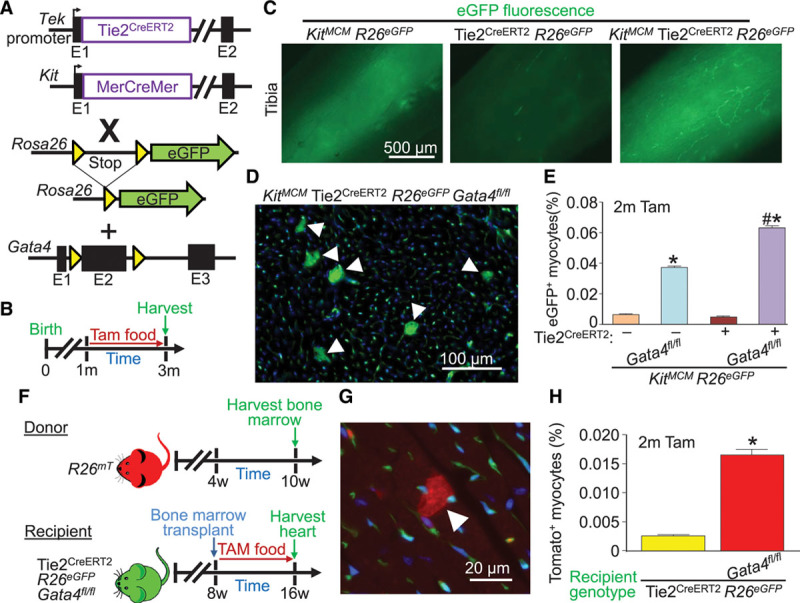
Global Tie2-lineage deletion of Gata4 augments appearance of Kit lineage– and bone marrow–labeled cardiomyocytes. A, Genetic models with the indicated alleles or transgenes in mice that were crossed. B, Experimental design of the protocol used in this figure. The Tie2 promoter from the Tek gene was used to drive the Cre-ERT2 cDNA. C, Whole-mount imaging of tibias from mice of the indicated genotypes of mice, highlighting labeled bone marrow cells (green) when the Kit lineage–tracing allele was used. D, Representative image showing accumulation of Kit allele–derived eGFP+ cardiomyocytes (arrowheads) in cardiac histological sections from the indicated mouse genotype 2 months after Gata4 deletion. E, Quantification of eGFP+ cardiomyocytes from KitMCM lineage tracing and KitMCM Tie2CreERT2 dual lineage tracing, with or without Gata4 deletion. Error bars are SEM, n=4 per group. *P<0.001 vs. KitMCM R26eGFP controls by 2-way ANOVA. #P<0.001 vs. dual KitMCM Tie2CreERT2 R26eGFP by 2-way ANOVA. F, Experimental design for bone marrow transplant of wild-type mTomato+ bone marrow into Tie2CreERT2 recipient mice with or without Gata4 deletion. G, Representative cardiac histological image of an mTomato+ cardiomyocyte (arrowhead) derived from bone marrow following bone marrow transplant from the mice shown in F. H, Quantification of mTomato+ cardiomyocytes from cardiac histological sections of recipient mice of the indicated genotypes following bone marrow transplant. Error bars are SEM, n=6 per group (10 sections per heart). *P<0.05 vs. Tie2CreERT2 R26eGFP recipient mice. eGFP indicates enhanced green fluorescent protein; and Tam, tamoxifen.
