Abstract
The possible role of C-X-C chemokine receptor type 4 (CXCR4)/CXCR7 signaling pathway in pre-eclampsia (PE) was investigated to study the expression of CXCR4/CXCR7 in serum of PE patients and its correlation and relationship with PE. Twenty patients with mild PE and 40 cases with severe PE enrolled into the PE group and 60 cases of the normal pregnancy group during the same period were selected as the objects of study. The changes in expression of serum CXCR4 messenger ribonucleic acid (mRNA) and CXCR7 mRNA were detected by reverse transcription-quantitative polymerase chain reaction (RT-qPCR), and CXCR4/CXCR7 protein concentration in serum was determined by enzyme-linked immunosorbent assay (ELISA). The pregnancy termination time of gravid was earlier in the severe PE group than that in the normal pregnancy group (P<0.05); the mean arterial pressure (MAP), systolic pressure, diastolic pressure were higher in patients with mild and severe PE than those in the normal pregnancy group (P<0.05). The results of RT-qPCR showed that the mRNA expression of serum CXCR4 and CXCR7 in PE patients were distinctly higher than those in the normal pregnancy group (P<0.05). The expression level of CXCR4 mRNA was positively correlated with that of CXCR7 mRNA (r=0.567, P=0.02). The results of ELISA displayed that the content of CXCR4/CXCR7 in serum of patients with PE was remarkably higher than that in the normal pregnancy group (P<0.05); the expression of serum CXCR4/CXCR7 in patients with severe PE was higher than in those with mild PE (P<0.05). The expression level of serum CXCR4 protein was positively correlated with that of CXCR7 protein (r=0.563, P=0.01). The expression level of CXCR4/CXCR7 may be closely related to the formation of PE.
Keywords: CXCR4, CXCR7, signaling pathway, pre-eclampsia, correlation
Introduction
Pre-eclampsia (PE) (1–3) is a unique and multisystem-related disease in pregnant women, of whom the incidence rate is approximately 7%. It is clinically characterized (4,5) by endothelial dysfunction, platelet aggregation abnormality and hypertension proteinuria, which can also cause slow growth and development in fetus. PE is an important cause of fetal death, affecting many families. The current research on the mechanism of PE is not mature, and two theories have been recognized: i) Abnormal immune mechanism (6), which means that abnormal immunity of pregnant women and fetus can induce PE and ii) injury of vascular endothelial cells (7) means that PE is a vascular disease caused by oxidative stress due to placental ischemia-reperfusion injury. Therefore, there are more studies on oxidative stress in PE patients.
A study indicated that (8) PE occurs in women during pregnancy due to a variety of regulatory molecules secreted into the blood of pregnant women, which is caused by the abnormality of trophoblast infiltration leading to the incorrect implantation of placenta in the uterus thus resulting in the persistent lack of nutrients such as O2. C-X-C chemokine receptor type 4 (CXCR4)/CXCR7 (9,10) is an important signaling pathway during pregnancy, which is highly expressed in the placenta. It is involved in embryogenesis and implantation of placenta; moreover, they can also form a dipolymer, aggregate β-inhibitory protein and activate its related signaling transduction pathways. The study indicates that CXCR4 and CXCR7 combined with CXCL12 can regulate signaling pathways such as phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT)/ fructooligosaccharide (FOS) and Ser/Thr kinase (Raf)/ methyl ethyl ketone (MEK)/extracellular signal-regulated kinase 1 (ERK1) that participate in the differentiation of trophoblast cells and formation of placenta. Therefore, we suspect PE of pregnant women is related to the abnormal expression of CXCR4/CCR7.
The present study investigated the correlation of CXCR4/CXCR7 signaling pathway with PE through detecting the expression levels of CXCR4 and CXCR7 in the serum of women.
Patients and methods
Clinical data
Twenty patients with mild PE and 40 cases with severe PE enrolled into PE group and 60 cases of normal pregnancy group during the same period admitted to Wenzhou People's Hospital from January 2015 to January 2017 were selected. The changes in expression of serum CXCR4 messenger ribonucleic acid (mRNA) and CXCR7 mRNA were detected by reverse transcription-polymerase chain reaction (RT-PCR), and CXCR4/CXCR7 protein concentration in the serum of women during pregnancy was determined by enzyme-linked immunosorbent assay (ELISA). Patients were diagnosed as mild and severe PE accordingly (11). i) Mild PE: after 20 weeks of gestation, BP ≥140/90 mmHg; urinary protein ≥0.3 g/24 h or random urinary protein (+); symptoms such as upper abdominal discomfort and headache. ii) Severe PE:BP ≥160/110 mmHg; urinary protein ≥2.0 g/24 h or random urinary protein (++); Serum creatinine >1,061 µmol/l; platelet <100 × 109/l; microangiopathy hemolysis (elevated LDH); elevated serum ALT or AST; persistent headache or other neurological or visual disorders; persistent upper abdominal discomfort. The 24 h urinary protein quantification was tested by routine urine test and defined as: 0.15–0.5 g was microalbuminuria (±), 0.5–1 g was mild proteinuria (+), and 1–4 g was moderate proteinuria (++), >4 g was severe proteinuria (+++). The pregnant women enrolled in this study had no organic inflammation, systemic infection, hypertension or diabetes, and all of them were delivered by caesarean section. Pre-analytical routine including Systolic pressure, Diastolic pressure and Random proteinuria were performed. The study was approved by the Ethics Committee of Wenzhou People's Hospital (Wenzhou, China) and informed consent was signed by the patients or their families.
Extraction of ribonucleic acid
Whole blood samples of the patients were collected in the morning fasting state. The sample was tested in 2 h to obtain serum by centrifugation at 2,500 × g for 10 min. The serum total RNA was extracted by TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the instructions provided by Invitrogen; Thermo Fisher Scientific, Inc. The concentration and purity of the extracted RNA were analyzed by an ultraviolet spectrophotometer (Hitachi, Tokyo, Japan), and the integrity of RNA was analyzed by 3% agarose gel electrophoresis.
Synthesis of complementary deoxyribonucleic acid (cDNA)
The RNA was reversely transcribed into cDNA, referring to the TaqMan® MicroRNA Reverse Transcription kit instructions (Thermo Fisher Scientific, Inc.). The reaction included 37°C for 45 min and 95°C for 5 min. The production was preserved at −20°C.
RT-qPCR
The reaction system was 25 µl in total, including pre-denaturation at 95°C for 5 min, denaturation at 95°C for 30 sec, annealing at 60°C for 45 sec, extension at 72°C for 3 min, a total of 35 cycles, finally extension at 72°C for 5 min. The production of PCR was preserved at 4°C. PCR was carried out by ABI Prism 7900 PCR instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.). CXCR4 mRNA: upstream primer 5′-GAGTCGATGCTGATCCCAAT-3′ and downstream primer 5′-GGCAGGTGAAAGGGATGTAG-3′; CXCR7 mRNA: upstream primer 5′-CCTACGTGGTGGTCTTCCTT-3′ and downstream primer 5′-GGCAGGTGAAAGGGATGTAG-3′. U6 was used as the internal reference of reaction (upstream primer: 5′-CTCGCTTCGGCAGCACA-3; downstream primer: 5′-AACGCTTCACGAATTTGCGT-3′) (Guangzhou Shangeng Biological Technology Co., Ltd., Guangzhou, China). A 3-well was duplicated for all samples, and the result was analyzed by 2−∆∆Cq (12). The specimen was stored for use within 7 days.
Detection of CXCR4/CXCR7 protein
The protein levels of CXCR4 and CXCR7 were detected according to the instructions of ELISA kit (Nanjing Jin Yibai Biological Technology Co., Ltd., Nanjing, China), including the collection of serum specimen (preservation at −20°C, the specimen was stored for use within 7 days), preparation of reagents and standards, and production of standard curves. The optical density (OD) value was measured at 450 nm.
Statistical analysis
The data were analyzed by Statistical Product and Service Solutions (SPSS) 19.0 software (IBM Corp., Armonk, NY, USA). Enumeration data were analyzed by Chi-square test, and measurement data are expressed by (mean ± SD). Comparison between groups was done using One-way ANOVA test followed by SNK test as its post hoc test. The correlation of expression level of CXCR4 with CXCR7 was analyzed by Pearson's correlation analysis. The correlation of expression level of CXCR4/CXCR7 with PE was analyzed by Cox regression. P<0.05 was considered to indicate a statistically significant difference.
Results
General data
There was no difference in age and number of fetus between the PE group and normal pregnancy group. All the subjects enrolled in this study were delivered by caesarean section (Table I).
Table I.
Comparison of general data between the PE group and normal pregnancy group.
| General data | PE group (n=60) | Normal pregnancy group (n=60) | P-value |
|---|---|---|---|
| Age (years) | 27.6±4.1 | 28.9±4.8 | 0.798 |
| Pregnancy termination of time (week) | 37.4±2.1 | 39.8±2.5 | 0.657 |
| Delivery mode | Caesarean section | Caesarean section | |
| Infant weight (Kg) | 3.1±0.5 | 3.3±0.5 | 0.742 |
| Mild PE patient (n) | 20 | ||
| Severe PE patients (n) | 40 | ||
| Number of fetus | 0.546 | ||
| Woman with single birth (n) | 58 (96.67%) | 59 (98.33%) | |
| Woman with twins (n) | 2 (3.33%) | 1 (1.67%) | |
| Systolic pressure (mmHg) | 156.52±11.35 | 107.69±11.87 | 0.021 |
| Diastolic pressure (mmHg) | 94.44±7.47 | 67.80±8.95 | 0.034 |
| Random proteinuria | +/++ | −/± |
There was no difference in pregnancy termination time between the mild PE patients and normal pregnancy group (37.9±1.8 vs. 39.8±2.5) (P>0.05). The pregnancy termination time of patients with severe PE was earlier than that in the normal pregnancy group (36.9±2.4 vs. 39.8±2.5) (P<0.05).
The systolic pressure, diastolic pressure and mean arterial pressure (MAP) were higher in patients with mild and severe PE than those in the normal pregnancy group (P<0.05). There was a difference in infant weight between the mild and severe PE patients and normal pregnancy group (P>0.05). The differences in systolic and diastolic pressure, MAP and infant weight between patients with mild and severe PE were not detected (P>0.05) (Table II).
Table II.
Comparisons of clinical features between the normal pregnancy group and PE group.
| Clinical features | Normal pregnancy group (i) | Mild PE patient (ii) | Severe PE patient (iii) |
|---|---|---|---|
| Systolic pressure (mmHg) | 113.4±9.8 | 137.8±11.2 | 155.2±11.9 |
| Diastolic pressure (mmHg) | 72.7±7.2 | 91.3±9.7 | 101.2±9.2 |
| MAP (mmHg) | 86.2±7.6 | 106.5±9.4 | 117.9±8.7 |
| Infant weight (kg) | 3.3±0.5 | 3.4±0.4 | 2.7±0.6 |
| i vs. ii | i vs. iii | ii vs. iii | |
| Systolic pressure | P=0.045 | P=0.024 | P=0.267 |
| Diastolic pressure | P=0.037 | P=0.023 | P=0.358 |
| MAP | P=0.034 | P=0.021 | P=0.432 |
| Infant weight | P=0.876 | P=0.056 | P=0.051 |
The results of RT-qPCR showed that the expression levels of CXCR4 mRNA and CXCR7 mRNA in the serum of mild and severe PE patients were distinctly higher than those in the normal pregnancy group (P<0.05). There were no statistically significant differences in serum CXCR4 mRNA and CXCR7 mRNA between patients with severe and mild PE (P>0.05) (Table III).
Table III.
Comparison of expression levels of CXCR4 mRNA and CXCR7 mRNA.
| mRNA | Normal pregnancy group (i) | Mild PE patients (ii) | Severe PE patients (iii) |
|---|---|---|---|
| CXCR4 mRNA | 0.319±0.121 | 0.697±0.134 | 0.740±0.142 |
| CXCR7 mRNA | 0.542±0.079 | 0.791±0.155 | 0.798±0.134 |
| i vs. ii | i vs. iii | ii vs. iii | |
| CXCR4 mRNA | 0.024 | 0.017 | 0.728 |
| CXCR7 mRNA | 0.031 | 0.032 | 0.987 |
The results of ELISA displayed that the protein content of CXCR4/CXCR7 in the serum of patients with mild and severe PE was remarkably higher than that in the normal pregnancy group (P<0.05). The expression level of CXCR4/CXCR7 protein in the serum of patients with severe PE was higher than that in those with mild PE (P<0.05) (Table IV).
Table IV.
Comparison of protein expression levels between CXCR4 and CXCR7 (ng/ml).
| Protein | Normal pregnancy group (i) | Mild PE patients (ii) | Severe PE patients (iii) |
|---|---|---|---|
| CXCR4 protein | 11.083±2.445 | 18.275±7.582 | 25.765±4.660 |
| CXCR7 protein | 3.048±0.673 | 4.468±1.130 | 5.707±1.576 |
| i vs. ii | i vs. iii | ii vs. iii | |
| CXCR4 protein | 0.036 | 0.021 | 0.029 |
| CXCR7 protein | 0.044 | 0.032 | 0.040 |
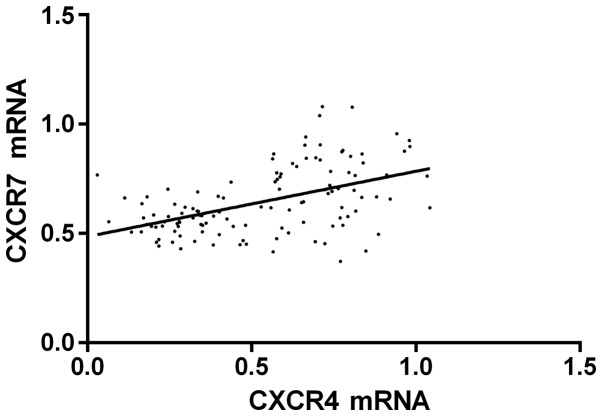
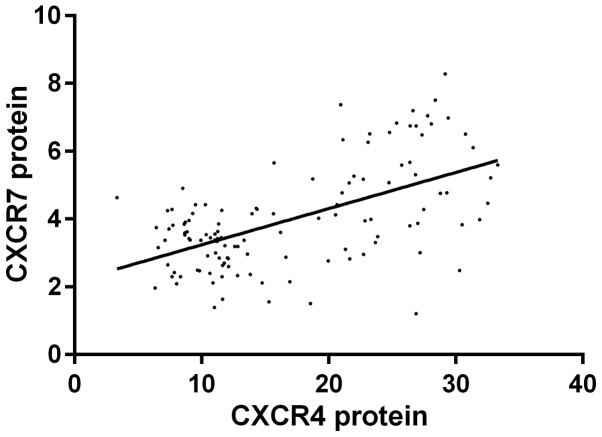
Pearson's correlation analysis indicated that the expression level of CXCR4 mRNA was positively correlated with CXCR7 mRNA (r=0.501, P<0.001); the expression level of CXCR4 protein was positively correlated with CXCR7 protein (r=0.572, P<0.001); COX regression analysis revealed that CXCR4/CXCR7 was related to the formation of PE, and 95% confidence intervals (CI) were [2.124 (1.557–4.479), P=0.002; 2.315 (1.231–3.524), P=0.002] (Figs. 1 and 2).
Figure 1.
Logistic regression analysis between CXCR4 mRNA and CXCR7 mRNA. r=0.485, P<0.001.
Figure 2.
Logistic regression analysis between CXCR4 protein and CXCR7 protein. r=0.568, P<0.001.
Discussion
The present study investigated the correlation of CXCR4/CXCR7 signaling pathway with PE through detecting the expression levels of CXCR4 and CXCR7 in the serum of 120 women during pregnancy. The results showed that the expression levels of CXCR4 mRNA and CXCR7 mRNA in the serum of PE patients were distinctly higher than those in the normal pregnancy group (P<0.05), and the expression levels of proteins were higher in the PE patients than those in the normal pregnancy group. Thus, it could be indicated that CXCR4/CXCR7 was related to the formation of PE, and the correlation coefficient was (r=0.563, P=0.01). The results of Karakus et al (13) indicated that CXCR4 polymorphism was related to the development of PE. The study by Lu et al (9) also demonstrated that CXCR4 and CXCR7 were associated with trophoblast apoptosis, which may be related to the formation and development of severe PE. Their results were consistent with our results, further confirming the correlation of CXCR4/CXCR7 with PE.
The result of this study indicated that there was no difference in the expression of CXCR4/CXCR7 between mild and severe PE patients, so we cannot confirm whether CXCR4/CXCR7 has a correlation with progression of PE, which may be related to the small number of the selected samples.
The human placenta is the most important fetal development organ during pregnancy, regulating the dynamic gene expression associated with placental and fetal development. The balanced expression of various genes in the placenta can maintain pregnancy, including fetal development. However, the abnormal expression of genes in the placenta can cause obstetric diseases such as PE, which has a great adverse effect on fetal growth and survival (14). PE accounts for 2.7% of all fetal perinatal death factors, 12% of fetal intrauterine growth restriction factors and 19% of preterm birth factors (15). Therefore, the prevention, diagnosis and treatment of PE are very important issues.
The study by Bramham et al (16) displayed that plasma growth factor (PlGF) may help to guide clinical decisions for admission and delivery in PE patients. The research by Stepan et al (17) also verified that utilization of the soluble fms-like tyrosine kinase-1 (sFlt-1)/PlGF ratio may help optimize care through ameliorating the management of women suspected of PE. In our results, CXCR4/CXCR7 may play a supporting role in the diagnosis of PE. At present, the more popular and effective research on the prevention and treatment of PE is low-dose aspirin treatment in pregnant women with high risk of PE, which may reduce the risk of PE (18,19). A limitation of the present study is that the reference gene U6 may differ from the CXCR4/7 mRNAs in processing and stability. The stability of different mRNA structures is different (20).
In conclusion, the expression level of CXCR4/CXCR7 may be closely related to the formation of PE, which seriously affects the quality of life of infants. Attention should be paid to prevention, diagnosis and treatment of PE by pregnant women and clinicians.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
ZZ analyzed and interpreted the patient data, and was a major contributor in writing the manuscript. HZ performed the experiments. JZ participated in the experiments and the design of the study. HC participated in the analysis and discussion of the data. XC was a major contributor in designing the methods. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Wenzhou People's Hospital (Wenzhou, China). Signed informed consents were obtained from the patients or their families.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chen Q, Sousa JD, Snowise S, Chamley L, Stone P. Reduction in the severity of early onset severe preeclampsia during gestation may be associated with changes in endothelial cell activation: A pathological case report. Hypertens Pregnancy. 2016;35:32–41. doi: 10.3109/10641955.2015.1100309. [DOI] [PubMed] [Google Scholar]
- 2.McGinnis R, Steinthorsdottir V, Williams NO, Thorleifsson G, Shooter S, Hjartardottir S, Bumpstead S, Stefansdottir L, Hildyard L, Sigurdsson JK, et al. FINNPEC Consortium; GOPEC Consortium: Variants in the fetal genome near FLT1 are associated with risk of preeclampsia. Nat Genet. 2017;49:1255–1260. doi: 10.1038/ng.3895. [DOI] [PubMed] [Google Scholar]
- 3.Li XL, Chen TT, Dong X, Gou WL, Lau S, Stone P, Chen Q. Early onset preeclampsia in subsequent pregnancies correlates with early onset preeclampsia in first pregnancy. Eur J Obstet Gynecol Reprod Biol. 2014;177:94–99. doi: 10.1016/j.ejogrb.2014.03.043. [DOI] [PubMed] [Google Scholar]
- 4.Mol BWJ, Roberts CT, Thangaratinam S, Magee LA, de Groot CJM, Hofmeyr GJ. Pre-eclampsia. Lancet. 2016;387:999–1011. doi: 10.1016/S0140-6736(15)00070-7. [DOI] [PubMed] [Google Scholar]
- 5.Rana S, Karumanchi SA, Lindheimer MD. Angiogenic factors in diagnosis, management, and research in preeclampsia. Hypertension. 2014;63:198–202. doi: 10.1161/HYPERTENSIONAHA.113.02293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levron Y, Dviri M, Segol I, Yerushalmi GM, Hourvitz A, Orvieto R, Mazaki-Tovi S, Yinon Y. The ‘immunologic theory’ of preeclampsia revisited: a lesson from donor oocyte gestations. Am J Obstet Gynecol. 2014;211(383):e1–e5. doi: 10.1016/j.ajog.2014.03.044. [DOI] [PubMed] [Google Scholar]
- 7.Meeme A, Buga GA, Mammen M, Namugowa A. Endothelial dysfunction and arterial stiffness in pre-eclampsia demonstrated by the EndoPAT method. Cardiovasc J Afr. 2017;28:23–29. doi: 10.5830/CVJA-2016-047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaiworapongsa T, Chaemsaithong P, Yeo L, Romero R. Pre-eclampsia part 1: Current understanding of its pathophysiology. Nat Rev Nephrol. 2014;10:466–480. doi: 10.1038/nrneph.2014.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu J, Zhou WH, Ren L, Zhang YZ. CXCR4, CXCR7, and CXCL12 are associated with trophoblastic cells apoptosis and linked to pathophysiology of severe preeclampsia. Exp Mol Pathol. 2016;100:184–191. doi: 10.1016/j.yexmp.2015.12.013. [DOI] [PubMed] [Google Scholar]
- 10.van Zuylen WJ, Ford CE, Wong DD, Rawlinson WD. Human cytomegalovirus modulates expression of noncanonical Wnt receptor ROR2 to alter trophoblast migration. J Virol. 2015;90:1108–1115. doi: 10.1128/JVI.02588-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.August P, Sibai BM. Preeclampsia: Clinical Features and Diagnosis. In: Lockwood CJ, Barss VA, editors. UpToDate. Waltham, MA: 2015. [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. METHODS. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Karakus S, Bagci B, Bagci G, Sancakdar E, Yildiz C, Akkar O, Cetin A. SDF-1/CXCL12 and CXCR4 gene variants, and elevated serum SDF-1 levels are associated with preeclampsia. Hypertens Pregnancy. 2017;36:124–130. doi: 10.1080/10641955.2016.1250904. [DOI] [PubMed] [Google Scholar]
- 14.Myatt L, Redman CW, Staff AC, Hansson S, Wilson ML, Laivuori H, Poston L, Roberts JM. Global Pregnancy CoLaboratory: Strategy for standardization of preeclampsia research study design. Hypertension. 2014;63:1293–1301. doi: 10.1161/HYPERTENSIONAHA.113.02664. [DOI] [PubMed] [Google Scholar]
- 15.Dodd JM, O'Brien C, Grivell RM. Preventing pre-eclampsia - are dietary factors the key? BMC Med. 2014;12:176. doi: 10.1186/s12916-014-0176-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bramham K, Seed PT, Lightstone L, Nelson-Piercy C, Gill C, Webster P, Poston L, Chappell LC. Diagnostic and predictive biomarkers for pre-eclampsia in patients with established hypertension and chronic kidney disease. Kidney Int. 2016;89:874–885. doi: 10.1016/j.kint.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 17.Stepan H, Herraiz I, Schlembach D, Verlohren S, Brennecke S, Chantraine F, Klein E, Lapaire O, Llurba E, Ramoni A, et al. Implementation of the sFlt-1/PlGF ratio for prediction and diagnosis of pre-eclampsia in singleton pregnancy: Implications for clinical practice. Ultrasound Obstet Gynecol. 2015;45:241–246. doi: 10.1002/uog.14799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Käehne LV, Lundin ICR. Treatment with low-dose acetylsalicylic acid can reduce risk of pre-eclampsia in high-risk pregnant women. Ugeskr Laeger. 2017;179:2017. (In Danish) [PubMed] [Google Scholar]
- 19.Rolnik D, Wright D, Poon L, O'Gorman N, Syngelaki A, de Paco Matallana C, Akolekar R, Cicero S, Janga D, Singh M, et al. OC01. 04: Aspirin versus placebo in pregnancies at high-risk of preterm pre-eclampsia: A multicentre, double-blind, placebo-controlled trial. Ultrasound Obstet Gynecol. 2017;50:1–47. doi: 10.1002/uog.17573. [DOI] [Google Scholar]
- 20.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.