Abstract
Background
The transient middle cerebral artery occlusion (tMCAO) model is used for studying the molecular mechanisms of ischemic damage and neuroprotection. Numerous studies have demonstrated the role of individual genes and associated signaling pathways in the pathogenesis of ischemic stroke. Here, the tMCAO model was used to investigate the genome-wide response of the transcriptome of rat brain tissues to the damaging effect of ischemia and subsequent reperfusion.
Results
Magnetic resonance imaging and histological examination showed that the model of focal ischemia based on endovascular occlusion of the right middle cerebral artery for 90 min using a monofilament, followed by restoration of the blood flow, led to reproducible localization of ischemic damage in the subcortical structures of the brain. High-throughput RNA sequencing (RNA-Seq) revealed the presence of differentially expressed genes (DEGs) in subcortical structures of rat brains resulting from hemisphere damage by ischemia after tMCAO, as well as in the corresponding parts of the brains of sham-operated animals. Real-time reverse transcription polymerase chain reaction expression analysis of 20 genes confirmed the RNA-Seq results. We identified 469 and 1939 genes that exhibited changes in expression of > 1.5-fold at 4.5 and 24 h after tMCAO, respectively. Interestingly, we found 2741 and 752 DEGs under ischemia–reperfusion and sham-operation conditions at 24 h vs. 4.5 h after tMCAO, respectively. The activation of a large number of genes involved in inflammatory, immune and stress responses, apoptosis, ribosome function, DNA replication and other processes was observed in ischemia–reperfusion conditions. Simultaneously, massive down-regulation of the mRNA levels of genes involved in the functioning of neurotransmitter systems was recorded. A Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis showed that dozens of signaling pathways were associated with DEGs in ischemia–reperfusion conditions.
Conclusions
The data obtained revealed a global profile of gene expression in the rat brain sub-cortex under tMCAO conditions that can be used to identify potential therapeutic targets in the development of new strategies for the prevention and treatment of ischemic stroke.
Electronic supplementary material
The online version of this article (10.1186/s12864-018-5039-5) contains supplementary material, which is available to authorized users.
Keywords: tMCAO, RNA-Seq, Gene expression, mRNA, Neurotransmission, Inflammation
Background
Among the cerebrovascular diseases of the brain, ischemic stroke remains one of the leading causes of mortality and disability [1–3]. This disease occurs as a result of a critical reduction in blood flow in the brain tissue, which leads to the death of neurons and glial cells, accompanied by massive inflammation. The models of cerebral ischemia in laboratory animals are used to understand the processes underlying the damage and recovery of neurological functions after cerebral ischemic injury, as well as to study the protective properties of drugs [4–10]. The main strengths and weaknesses of the most commonly used animal models of acute ischemic stroke have been described in a number of review articles [11–13].
The model of transient middle cerebral artery occlusion (tMCAO) is widely used for the development of neuroprotective therapeutic approaches. tMCAO is based on temporal artery occlusion and subsequent restoration of blood flow. According to Howells et al., the model that was created using an intravascular filament was used in 42.2% of 2582 neuroprotection experiments [13]. The tMCAO model reflects events that occur in ischemic stroke in humans in the treatment of thrombolytic agents [14, 15]. The results of clinical studies indicate that, currently, thrombolysis is one of the most effective and affordable methods of treatment of ischemic stroke [16, 17]. Concomitantly, it is known that reperfusion after thrombolysis can enhance the damage caused by ischemia [18]. The brain damage caused by ischemia–reperfusion (IR) leads to the disruption of the functioning of multiple genes [19–21]. However, at present, most studies have analysed the features of the expression of a small number of genes associated only with one or a few signaling pathways [5, 22–25].
In the present study, we used high-throughput mRNA sequencing (RNA-Seq) to study the genome-wide response of the transcriptome to the damaging effect of IR in a tMCAO model based on endovascular artery occlusion (90 min) and subsequent reperfusion. The tMCAO model created using this duration and method of occlusion is one of the most commonly used models of rat cerebral ischemia [26–28]. We identified hundreds of differentially expressed genes (DEGs) and their functional associations in the rat brain sub-cortex, which included necrotic and penumbra areas under the tMCAO model conditions. These new data indicate a complex spatial-temporal genome regulation of brain cells under IR that can be used to identify potential therapeutic targets in the development of new strategies for the prevention and treatment of ischemic stroke.
Results
Characterization of ischemia using magnetic resonance imaging (MRI)
Using the diffusion-weighted imaging (DWI) and T2-weighted imaging (T2 WI) modes of magnetic resonance imaging (MRI), we detected the location and volume of ischemic foci in animals after tMCAO. A typical the diffusion-weighted imaging (DWI) of the formation of ischemic injury areas with a subcortical (cerebral subcortex) and hemispheric (cerebral subcortex plus cortex) localization in the brain of rats at 4.5 and 24 h after tMCAO is shown in Fig. 1. Among the 29 animals that were subjected to tMCAO and decapitated at 24 h (IR24 group), 25 and 4 rats exhibited a subcortical and hemispheric localization of the focus, respectively, at 3 h after operation. However, 24 h after occlusion, the number of animals with a subcortical localization decreased to 18 and the number of animals with a hemispheric localization increased to 11. Among the 10 animals that were subjected to tMCAO and decapitated at 4.5 h (IR4.5 group), 8 and 2 rats exhibited a subcortical and hemispheric localization of the focus, respectively. The statistical indicators (the median (Me) and the lower (LQ) and upper (UQ) quartiles for the 25 and 75 percentile interval) of animals from IR24 and IR4.5 groups are shown in Table 1 and Additional file 1, respectively.
Fig. 1.
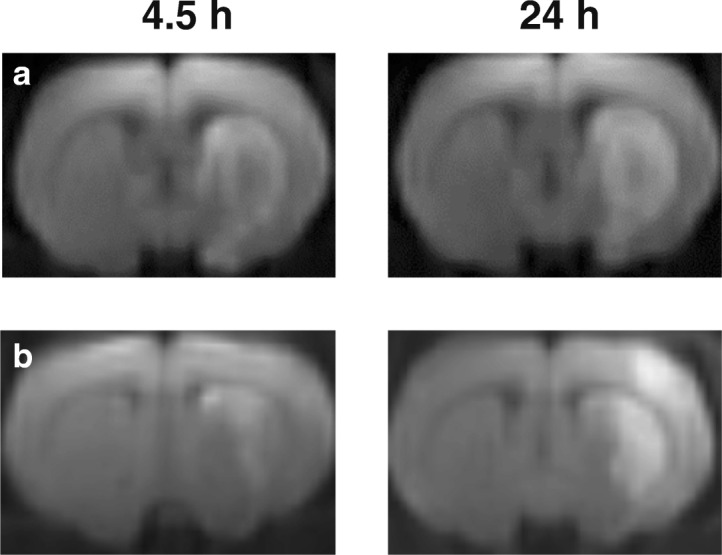
Characterization of ischemia using magnetic resonance imaging. A typical diffusion-weighted imaging (DWI) of the formation of ischemic injury areas with a subcortical localization both 4.5 and 24 h after tMCAO (a), as well as subcortical and hemispheric localization at 4.5 and 24 h after tMCAO, respectively (b) in the rat brain
Table 1.
Characterization of ischemia using magnetic resonance imaging
| Time after operation (h) | Type of ischemic focus localization | Number of observations | Volume (mm3) Me [LQ;UQ] | |
|---|---|---|---|---|
| DWI | T2 WI | |||
| 3 | subcortical | 25 | 41.86 [37.97; 53.66] | 19.5 [10.93; 28.7] |
| hemispheric | 4 | 173.72 [130.31; 208.9] | 93.18 [84.37; 95.73] | |
| 24 | subcortical | 18 | 46.1 [41.6; 56.92] | 40.96 [36.06; 51.11] |
| hemispheric | 11 | 178.2 [119.01; 201.7] | 142.48 [91.72; 173.09] | |
Statistical indicators (the median (Me) and the lower (LQ) and upper (UQ) quartiles for the 25 and 75 percentile interval) for 29 animals at 3 and 24 h after tMCAO are shown
Morphology of brain tissues in the experimental groups
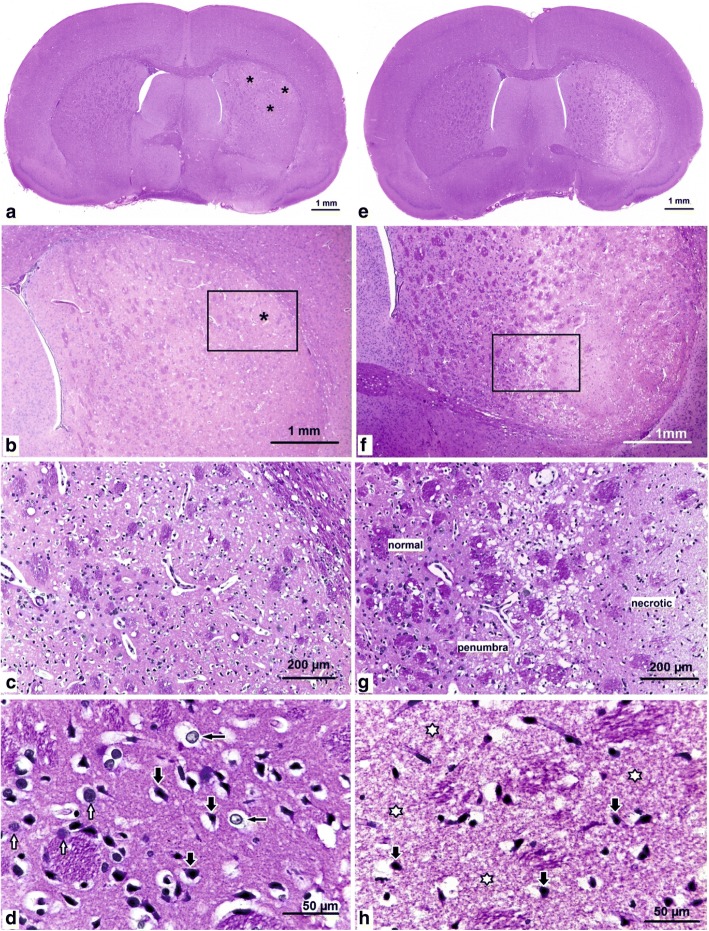
A typical image of the formation of ischemic injury areas with a subcortical localization in the brain of rats at 4.5 and 24 h after tMCAO is shown in Fig. 2. Focal ischemic lesions of the brain tissue in the form of oedema and foci of “enlightenment” were found in the dorsolateral regions of the subcortical region of the right hemisphere at 4.5 h after tMCAO. The most pronounced changes were observed at − 0.3 mm from the bregma (Fig. 2a, b). Concomitantly, the area of the developing ischemic damage covered a significant area, from the caudoputamen to the corpus callosum and outer capsule. Microscopic examination revealed a drop in the lumen of the capillaries and their emptying, pronounced perivascular oedema, sparsity and bleaching of the neuropil caused by oedema and vacuolization and the appearance of numerous hyperchromic neurons with peri-cellular oedema (Fig. 2c). According to pathomorphological criteria, a significant fraction of neurocytes was in a state of hypoxic damage (reduction of basophilia of chromatin in the nuclei, oedema and homogenization of the cytoplasm) and death (pyknosis and destruction of nuclei and cytoplasmic lysis) (Fig. 2d). The pronounced dystrophic changes observed in the pericarion of most neurons were not detected in the medial region of the caudoputamen outside the zones of the visualized focal ischemia.
Fig. 2.
Photomicrographs of haematoxylin and eosin-stained sections of the rat brain in tMCAO model conditions. a–d 4.5 h after tMCAO. e–h 24 h after tMCAO. a, e Coronal rat brain sections at the level of − 0.3 mm from the bregma. Asterisks indicate the damaged area involving the caudoputamen nucleus of the right hemisphere. b–d High-magnification images of the zone of ischemic injury in the dorsolateral region of the caudoputamen. c Area of panel (b) marked with a rectangle. d Hypoxic damage to neurons, with pyknotic nuclei and peri-cellular oedema indicated in the ischemic zone (thick black arrows); decrease of nuclear basophilia in the neurons (thin black arrows); single intact neurons (white arrows). f–h Infarction zone within the caudoputamen at higher magnification. g Area of panel (f) marked with a rectangle. The change zone: from normal through to ischemic-injured (penumbra) to necrotic tissue. h Ischemic necrosis of the brain tissue in the central core of an infarct destruction of the neuropil (white asterisks); dead “pyknotic” neurons (thick black arrows)
The developmental regions of the subcortical infarction zone were observed in the lateral caudoputamen region, from + 1.7 to − 3.3 mm from the bregma, at 24 h after tMCAO. In the range from − 0.3 to − 2.0 mm from the bregma, the infarction zone had an elongated shape in the dorsoventral direction and captured most of the caudoputamen lateral region (Fig. 2e), with distinct visualization of the penumbra and necrosis zones (Fig. 2f, g). The core of the infarction was represented by completely necrotized neural tissue with destruction of all elements of the neuropil and dead “pyknotic” neurons (Fig. 2h). The neurons, both with attributes of ischemic damage and single neurocytes without pronounced pathological changes, were located in the penumbra, the size of which exceeded that of the necrotic zone. The pathological changes were not detected in the medial region of the caudoputamen. In sham-operated animals (SH), the histological pattern of the capillary network and the morphology of neurons in the brain cortex, subcortical centres and the intermediate brain corresponded to the norm.
RNA-Seq analysis of gene expression profiles in tMCAO model conditions
Using RNA-Seq, the expression level of 17,352 genes was determined in rat brain sub-cortex under tMCAO conditions. Sham-operated animals were used as a control to analyse the mRNA levels of DEGs in the rat sub-cortex after tMCAO at 4.5 h (IR4.5 vs. SH4.5) and 24 h after operation (IR24 vs. SH24), respectively. In addition, there were DEGs under sham-operation conditions at 24 vs. 4.5 h after surgical procedure (SH24 vs. SH4.5) and under ischemia–reperfusion conditions at 24 vs. 4.5 h after tMCAO (IR24 vs. IR4.5).
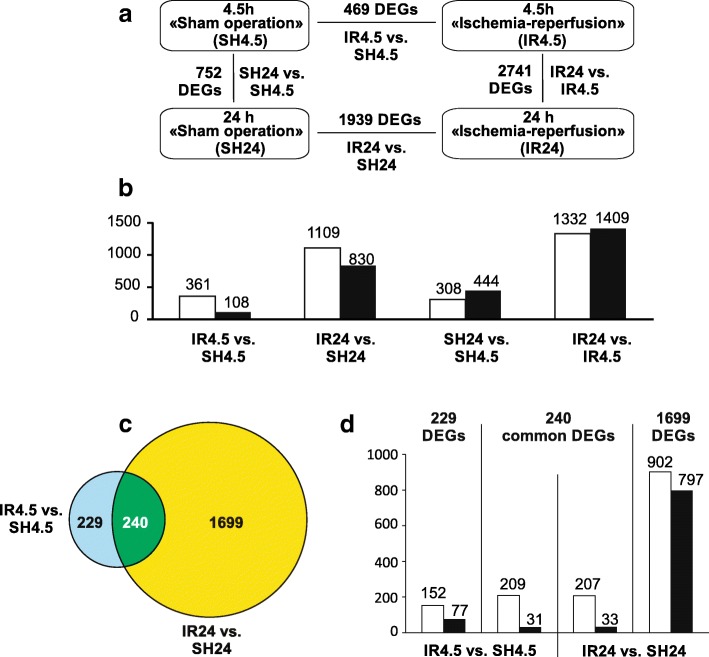
The number of DEGs in the rat brain sub-cortex in the animal groups studied is shown in the Fig. 3a. There were 469 and 1939 DEGs in IR4.5 vs. SH4.5 and IR24 vs. SH24, respectively. Concomitantly, the number of up-regulated DEGs was greater than the number of down-regulated DEGs at both time points (Fig. 3b). There were 2741 DEGs in IR24 vs. IR4.5. This was the maximum number of DEGs observed between the animal groups studied (Fig. 3b). In particular, we detected an approximately equal number of up-regulated (1332) and down-regulated (1409) DEGs. A similar analysis found 752 DEGs in response to sham operation in SH24 vs. SH4.5, 308 DEGs were up-regulated and 444 down-regulated in this group.
Fig. 3.
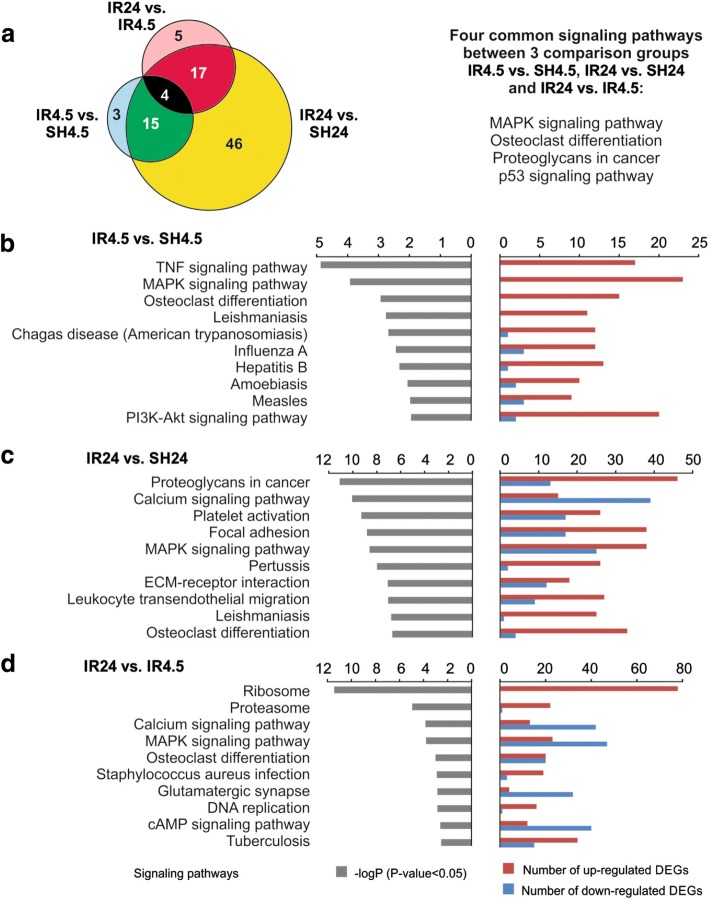
RNA-Seq analysis of differentially expressed genes (DEGs) in tMCAO model conditions. There are RNA-Seq results in IR4.5 vs. SH4.5, IR24 vs. SH24, IR24 vs. IR4.5, SH24 vs. SH4.5. a Schematic comparison of the results obtained for the groups. b The numbers of up- and down-regulated DEGs are indicated by white and black bars, respectively. The numbers placed above the bars indicate the number of DEGs. c The numbers of DEGs overlapped in two pairwise comparisons: IR4.5 vs. SH4.5 and IR24 vs. SH24. d Diagrams showing DEGs that lie within the section of sets on the Venn diagrams (c). The numbers of up- and down-regulated DEGs are indicated by white and black bars, respectively. The numbers placed above the bars indicate the number of DEGs. Genes with a change in expression > 1.5-fold compared with the baseline value and with a Padj < 0.05 were selected for analysis
There were 229 genes (152 up- and 77 down-regulated) that changed their expression in IR4.5 vs. SH4.5, but didn’t change it in IR24 vs. SH24 (Fig. 3c). Simultaneously, there were 1699 genes (902 up- and 797 down-regulated) that changed their expression in IR24 vs. SH24, but didn’t change it in IR4.5 vs. SH4.5 (Fig. 3c). All genes predominantly have binding and catalytic activity, but the former DEGs additionally have transcription factor (Npas4, Fosl2, Fosb), cytokine (Ccl2, Ccl3, Il1a, Il1b), hormone (Oxt, Avp) and transporter (Sec4a5, Kcnj13, Abcg2) activity, whereas the latter predominantly have cytokine (Ccl6, Cx3cl1, Il16, Nrtn, Tnfsf9) and growth factor (S100a4, Tbrg1, Fgf2) activity.
We detected 240 common DEGs in the rat sub-cortex between IR4.5 vs. SH4.5 and IR24 vs. SH24 (Fig. 3c). The expression of more than 86% of these DEGs was increased (Fig. 3d). It should be noted that IR had the opposite effect on several genes in IR4.5 vs. SH4.5 and IR24 vs. SH24. In particular, we found 17 up-regulated DEGs (Nr4a1, Nr4a2, Nr4a3, Egr2, Egr3, Egr4 and others) and 15 down-regulated DEGs (Bst2, Mx2, Ifi27, Aldh1a2, Xdh and others) in IR4.5 vs. SH4.5, that exhibited a reversion in the direction of expression in IR24 vs. SH24 (see Additional file 2). The former DEGs predominantly have ligand-activated sequence-specific DNA-binding RNA polymerase II transcription factor activity, whereas the latter predominantly have hydrolase and oxidoreductase activity, as well as receptor-binding and hormone activities.
There were 396 common DEGs in rat sub-cortex between IR24 vs. IR4.5 and SH24 vs. SH4.5 (see Additional file 3: Figure S1A). In particular, 132 DEGs were up-regulated and 264 down-regulated under the sham–operation condition, and 143 DEGs were up-regulated and 253 down-regulated after the development of ischemia–reperfusion damage (see Additional file 3: Figure S1B).
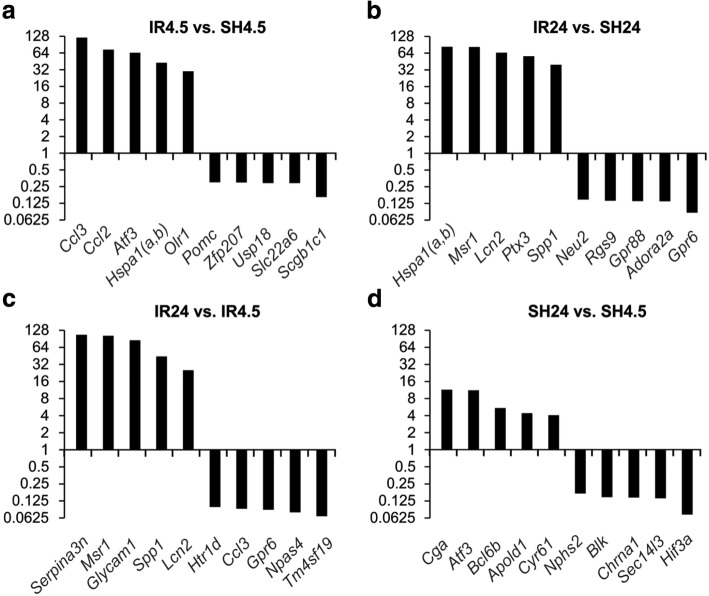
The top five genes in each of the comparison groups (i.e. those that exhibited the greatest fold change in expression in the subcortical structures of the rat brain) are presented in Fig. 4. The table shows the up-regulation of the mRNA levels of the Ccl3 and Ccl2 chemokine genes (> 120-fold and > 70-fold) and those of the transcription factor Atf3 gene (> 64-fold) at 4.5 h after tMCAO vs. sham operation (Fig. 4a). The mRNA levels of the heat-shock protein Hspa1(a,b) gene and Msr1 gene, which encodes the macrophage scavenger receptor 1, were increased by > 80-fold in IR24 vs. SH24 (Fig. 4b).
Fig. 4.
The top five DEGs of each comparison group. There are genes with the greatest fold change in expression in the subcortical structures of the rat brain, as assessed using RNA-Seq. In the comparison of data groups IR4.5 vs. SH4.5 (a), IR24 vs. SH24 (b), IR24 vs. IR4.5 (c) and SH24 vs. SH4.5 (d). Differences were considered statistically significant at the probability of Padj < 0.05
The expression of the Serpina3n gene, which encodes a serine (or cysteine) peptidase inhibitor, clade A, member 3 N, as well as that of the Msr1 gene, was increased by > 100-fold in IR24 vs. IR4.5 (Fig. 4c). The fold change of DEGs in SH24 vs. SH4.5 was less than under IR (Fig. 4d).
Verification of the RNA-Seq results using real-time reverse transcription polymerase chain reaction (RT–PCR)
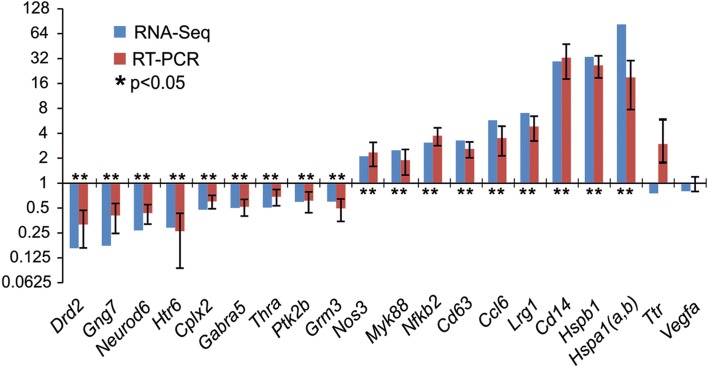
Real-time reverse transcription polymerase chain reaction (RT–PCR) analysis of the expression of 20 genes was used to verify the RNA-Seq results (Fig. 5). mRNA isolated from the subcortical brain structures of nine rats at 24 h after tMCAO and of nine sham-operated rats was used. Both methods of analysis identified three top genes with a maximum positive expression change (Hspa1(a,b), Hspb1 and Cd14) and three top genes with a maximum negative expression change (Drd2, Gng7 and Neurod6). Moreover, both methods identified genes with moderately significant changes in expression levels: up-regulated genes (Ccl6, Cd63, Nfkb2, Myd88 and Nos3) and down-regulated genes (Grm3, Ptk2b, Thra, Cplx2, Gabra5 and Htr6). By contrast, significant results regarding the differential expression of the Ttr and Vegfa genes were not obtained using the two methods (Fig. 5). Thus, the real-time RT–PCR results confirmed the RNA-Seq data sufficiently; the slight variations in gene expression detected by these methods may have been caused by differences in the methodology and statistical processing of data used in each case (Fig. 5).
Fig. 5.
Real-time reverse transcription polymerase chain reaction (RT-PCR) verification of the RNA-Seq results. Data for comparison between the “ischemia–reperfusion” and “sham–operation” groups at 24 h after operation (IR24 vs. SH24) are shown. Two reference genes Gapdh and Rpl3 were used to normalize PCR-results. In each group, there were at least 9 rats. Eighteen genes that change the expression of more than 1.5-fold from the baseline value and whose P-value lower 0.05, as well as two other genes were selected for analysis
Biological processes and functional categories of the proteins encoded by the DEGs identified under the ischemia–reperfusion condition
Analysis of the Gene Ontology database using the PANTHER tool allowed the identification of the biological processes associated with the DEGs identified in the rat brain sub-cortex under the tMCAO model condition. Table 2 shows that the biological processes associated with the DEGs could be divided into 13 functional categories for all comparison groups. The top five categories associated with the largest number of genes included the cellular process, the metabolic process, biological regulation, response to stimuli and localization. The proteins encoded by DEGs were also classified according to their molecular functions. The classification of DEGs according to the molecular functions of their encoded proteins revealed that the largest number of DEGs belonged to the categories of binding, catalytic and transporter activity, as well as signal-transducer and receptor activity (Table 3). The functional categories of the proteins encoded by the DEGs were identified using the David program (see Additional file 4). The damaging effect of IR at 4.5 h after tMCAO affected the expression levels of DEGs associated with 34 functional categories. The total number of functional categories associated with the effects of IR at 24 h after tMCAO exceeded 80 (see Additional file 4), while only six functional categories (phosphoproteins, developmental proteins, biological rhythms, disulphide bonding, glycoproteins and angiogenesis) were associated with SH.
Table 2.
Analysis of the biological processes associated with the DEGs using the PANTHER tool
| Biological Processes | IR4.5 vs. SH4.5 | IR24 vs. SH24 | IR24 vs. IR4.5 | SH24 vs. SH4.5 |
|---|---|---|---|---|
| cellular process (GO:0009987) | 189 | 675 | 920 | 245 |
| metabolic process (GO:0008152) | 136 | 450 | 661 | 168 |
| biological regulation (GO:0065007) | 73 | 257 | 326 | 93 |
| response to stimulus (GO:0050896) | 72 | 226 | 275 | 73 |
| localization (GO:0051179) | 37 | 154 | 192 | 49 |
| cellular component organization or biogenesis (GO:0071840) | 34 | 151 | 205 | 50 |
| developmental process (GO:0032502) | 32 | 139 | 164 | 45 |
| multicellular organismal process (GO:0032501) | 19 | 122 | 167 | 37 |
| immune system process (GO:0002376) | 21 | 59 | 51 | 14 |
| biological adhesion (GO:0022610) | 15 | 44 | 40 | 10 |
| locomotion (GO:0040011) | 15 | 37 | 43 | 9 |
| reproduction (GO:0000003) | 1 | 10 | 12 | 5 |
| rhythmic process (GO:0048511) | 2 | 1 | 3 | 3 |
Associative analysis for all comparison groups was carried out according to the Gene Ontology (GO) database
Table 3.
Analysis of the molecular functions associated with the DEGs using the PANTHER tool
| Molecular Functions | IR4.5 vs. SH4.5 | IR24 vs. SH24 | IR24 vs. IR4.5 | SH24 vs. SH4.5 |
|---|---|---|---|---|
| antioxidant activity (GO:0016209) | 0 | 6 | 6 | 2 |
| binding (GO:0005488) | 122 | 409 | 530 | 150 |
| catalytic activity (GO:0003824) | 99 | 384 | 463 | 124 |
| receptor activity (GO:0004872) | 18 | 77 | 104 | 19 |
| signal transducer activity (GO:0004871) | 23 | 71 | 89 | 25 |
| structural molecule activity (GO:0005198) | 8 | 50 | 90 | 13 |
| transporter activity (GO:0005215) | 24 | 90 | 140 | 44 |
| channel regulator activity (GO:0016247) | 0 | 2 | 2 | 0 |
| translation regulator activity (GO:0045182) | 0 | 7 | 10 | 0 |
Associative analysis for all comparison groups was carried out according to the Gene Ontology (GO) database
Fifteen functional categories of the proteins encoded by DEGs under IR are presented in Table 4. These were chosen because the expression of the DEGs that encoded these proteins was predominantly changed in each of the categories unidirectionally. Accordingly, the number of up- and down-regulated DEGs is also indicated in Table 4. A detailed analysis of the DEGs detected in each of the comparison groups is presented in Additional files 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19. It should be noted that there were up-regulated DEGs associated with the inflammatory, immune and stress responses, as well as with apoptosis and regulation of transcription, in the first hours after the restoration of blood flow in the subcortical structures of the rat brain (see Additional files 5, 6, 10, 11, 16 and 18). Both the number of DEGs associated with the above categories and their level of expression were increased significantly at 24 h after tMCAO compared with the sham-operation conditions. However, a large number of down-regulated DEGs were associated with the transcription process (see Additional file 18). Moreover, dozens of down-regulated DEGs were associated with the functioning of neurotransmitter systems in IR24 vs. SH24 (see Additional files 12 and 17).
Table 4.
Analysis of the functional categories of the proteins encoded by DEGs
| Functional Categories | IR4.5 vs. SH4.5 | IR24 vs. SH24 | IR24 vs. IR4.5 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Up | Down | Padj | Up | Down | Padj | Up | Down | Padj | |
| Innate immunity | 10 | 3 | 5.04E-04 | 35 | 3 | 2.17E-09 | 30 | 10 | 9.38E-07 |
| Inflammatory response | 12 | 0 | 5.89E-05 | 16 | 2 | 6.39E-03 | 10 | 10 | 3.73E-02 |
| Stress response | 7 | 0 | 2.80E-03 | 12 | 0 | 1.70E-02 | |||
| Differentiation | 14 | 3 | 3.38E-02 | 29 | 24 | 1.62E-03 | 31 | 42 | 1.58E-04 |
| Cell adhesion | 12 | 4 | 2.20E-02 | 29 | 23 | 7.52E-05 | 28 | 30 | 6.19E-03 |
| Transcription regulation | 35 | 5 | 3.54E-04 | 67 | 48 | 1.77E-04 | 51 | 108 | 9.26E-06 |
| DNA replication | 14 | 0 | 4.54E-03 | 16 | 4 | 1.31E-04 | |||
| Adaptive immunity | 13 | 4 | 5.51E-05 | 11 | 6 | 2.20E-03 | |||
| Apoptosis | 38 | 10 | 3.38E-04 | 41 | 14 | 9.35E-03 | |||
| Synapse | 2 | 58 | 4.10E-07 | 3 | 84 | 1.19E-11 | |||
| Ion channel | 10 | 43 | 8.05E-05 | 17 | 71 | 5.76E-12 | |||
| Proteasome | 21 | 1 | 1.30E-06 | ||||||
| Ribosomal protein | 73 | 4 | 8.77E-11 | ||||||
| Translation regulation | 10 | 3 | 4.80E-02 | ||||||
| Neurogenesis | 9 | 23 | 1.30E-04 | 7 | 29 | 2.17E-03 | |||
Associative analysis of DEGs for comparison groups under ischemia–reperfusion conditions was carried out according to the DAVID database. The number of up-regulated (Up) and down-regulated (Down) DEGs in the condition of the tMCAO model, as well as the P-values adjusted using the Benjamini–Hochberg procedure (Padj), are shown
The number of DEGs identified in IR24 vs. IR4.5 exceeded that detected in IR24 vs. SH24. In particular, we detected the up-regulation of DEGs that encode proteins associated with the functional categories of DNA replication, proteasome, translation regulation and ribosomal function in IR24 vs. IR4.5 (see Additional files 9, 14, 15 and 19). It should be noted that we detected dozens of down-regulated DEGs that encoded proteins involved in the functioning of neurotransmitter systems and in the regulation of transcription, cell adhesion, differentiation and neurogenesis (see Additional files 7, 8, 12, 13 and 18). Moreover, we identified single DEGs that were associated with the stress response, neurogenesis, transmission of nerve impulses and other processes in the subcortical structures of the rat brain in SH24 vs. SH4.5 (see Additional files 12, 13, 16 and 18).
Signaling pathways associated with DEGs identified under the ischemia–reperfusion conditions
Using DAVID v6.8, a Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was used to annotate the functions of the DEGs identified in the rat brain sub-cortex under the IR (see Additional file 20). We found 22 and 82 signaling pathways associated with DEGs in IR4.5 vs. SH4.5 and IR24 vs. SH24, respectively. The former included predominantly signaling pathways involved in the inflammatory response, while the latter included signaling pathways involved in inflammation and neurotransmission. It should be noted that we identified 26 signaling pathways (involved in inflammation, neurosignaling, the formation and functioning of ribosomes, the functioning of proteases, DNA replication and other processes) that were associated with DEGs in the subcortical structure of the rat brain in IR24 vs. IR4.5 (see Additional file 20). A comparative analysis of the signaling pathways identified showed that four signaling pathways that participate in the inflammatory response (MAPK, proteoglycans in cancer, osteoclast differentiation and the p53 signaling pathway) were common to all groups and reflected the effects of IR (Fig. 6a, see Additional file 20). In addition, 15 other common signaling pathways were identified between IR4.5 vs. SH4.5 and IR24 vs. SH24, and 17 common signaling pathways were identified between IR24 vs. IR4.5 and IR24 vs. SH24 groups (Fig. 6a, see Additional file 20). The most significant signaling pathways associated with DEGs in the groups studied are shown in Fig. 6b, c and d. Several signaling pathways involved in the inflammatory response (PI3K-Akt, TNF and others) were associated with DEGs in the rat brain sub-cortex at 4.5 h after tMCAO (Fig. 6b, see Additional file 20). The activation of DEGs associated with focal adhesion, leukocyte trans-endothelial migration, proteoglycan in cancer and other signaling pathways was observed at 24 h after tMCAO (Fig. 6c, see Additional file 20). A comparative analysis of the DEGs detected under the two conditions (IR24 vs. IR4.5 and IR24 vs. SH24) revealed signaling pathways involved in the inflammatory response. However, calcium signaling, glutamatergic synapse, cAMP signaling and other neurosignaling pathways were predominantly associated with DEGs that were down-regulated (Fig. 6d, see Additional file 20). It should be noted that signaling pathways of the ribosome, the proteasome, DNA replication and purine metabolism were predominantly associated with DEGs that were up-regulated in the rat brain sub-cortex in IR24 vs. IR4.5 (see Additional file 20). It should be noted that no reliable association between the DEGs and any signaling pathway was identified in SH24 vs. SH4.5.
Fig. 6.
KEGG database analyses of the signaling pathway annotations for differentially expressed genes (DEGs). a Venn diagrams showing common and unique signaling pathways associated with DEGs in IR4.5 vs. SH4.5, IR24 vs. SH24, IR24 vs. IR4.5, SH24 vs. SH4.5. The numbers in the sectors indicate the number of signaling pathway annotations for DEGs. b, c, d The most significant signaling pathways and corresponding P-values adjusted using the Benjamini–Hochberg procedure, as well as the number of up- and down-regulated DEGs in IR4.5 vs. SH4.5 (b), IR24 vs. SH24 (c), IR24 vs. IR4.5 (d). Genes with a change in expression > 1.5-fold compared with the baseline value and with a P-value < 0.05 were selected for analysis
Discussion
Experimental animal models are of great importance for understanding the mechanisms of damage and survival of neurons after ischemic brain injury, as well as for the development of new strategies of neuroprotection. We adapted the tMCAO model, which is used in the development of neuroprotective approaches, for the genome-wide transcriptome analysis of gene expression profiles in the rat brain sub-cortex under conditions of cerebral IR. The main objective of the study was the identification of DEGs, biological processes and signaling pathways associated with the response of brain cells to IR.
MRI and pathomorphological results showed that the specific tMCAO model used in the present study (mode and time of occlusion and reperfusion) allowed the induction of a focal ischemic injury with a predominant subcortical localization. Unlike postmortem triphenyl tetrazolium chloride (TTC) staining, which is used often to localize the area of ischemia [20, 29–31], MRI allowed us to localize the emerging ischemic damage in each animal in vivo and to observe the dynamics of its development (Fig. 1). Animals with acute transient impairment of the cerebral circulation manifested their primary lesions as ischemia of the nervous tissues against the background of vascular disturbances. The standard haematoxylin and eosin (HE) staining allowed the verification of changes in the infarct area at the cellular level (Fig. 2).The microscopic changes in the blood supply zone of the right MCA were characterized by the formation of loci of hypoxic damage of neurons with varying severity at 4.5 h after occlusion. Classical zones of formation of an ischemic infarct with deep destructive changes and death of nerve cells in necrotic zones and the large penumbra region were observed at 24 h after occlusion. Thus, MRI and histological examination indicated that the tMCAO model with the characteristics used in the present study led to ischemic injuries with a reproducible localization and volume of ischemic injury, including both the necrosis zone and penumbra. Consequently, the tMCAO model implemented here can be used both for studying the mechanisms of ischemic injury and for testing neuroprotective drugs.
It is well known that cerebral ischemia causes a cascade of biochemical and transcriptome changes in brain tissues [20, 32]. Numerous studies have indicated that early-response genes (c-fos and c-jun) [33] and zinc finger genes trigger cell proliferation and differentiation [34, 35], while genes that encode heat-shock proteins, are involved in the inflammatory response and cytoskeleton organization [36] and others are predominantly activated after the onset of ischemia. It has been shown that reperfusion after ischemia causes additional damage in brain cells, including destruction of the endothelial cells of the microvascular brain, disturbance of the blood–brain barrier, accumulation of excess oxygen radicals and activation of apoptosis [10, 18, 37–39]. Cerebral ischemic injury caused by artery occlusion in combination with reperfusion leads to numerous disorders of the regulation of mRNAs involved in the cerebral ischemic response [19–21].
We performed a genome-wide analysis of the transcriptome via RNA-Seq in the subcortical structures of the rat brain that contained the ischemic focus and the penumbra region under the conditions of the tMCAO model. Dozens of DEGs were identified using high-throughput RNA-Seq. An analysis of the expression of 20 genes by real-time RT–PCR confirmed the RNA-Seq results. The use of two time points (4.5 and 24 h) for the tMCAO model significantly increased the number of previously known genes involved in the early and late response. We found that 469 and 1939 genes exhibited changes in expression > 1.5-fold at 4.5 and 24 h after tMCAO, respectively. It should be noted that these genes were up-regulated predominantly under the tMCAO model conditions. Among these, we found genes involved in the inflammatory, immune and stress response, as well as in apoptosis and the regulation of transcription (see Additional files 6, 11, 12, 17 and 18). Under IR, the mRNA level of genes encoding chemokines (Ccl2, Ccl3), heat-shock proteins (Hspa1(a,b)), the macrophage scavenger receptor 1 (Msr1), the secreted phosphoprotein 1 (Spp1) and the suppressor of cytokine signaling 3 (Socs3), as well as other genes encoding proteins involved in inflammatory and immune responses, was increased by > 10-fold (Fig. 4). The significant activation of gene expression observed here may indicate the response of living cells in the penumbra to the effects of damage.
Studies with several MRI experiments in rat models of tMCAO demonstrated that stroke-induced infarct evolved into an area of injury, maximal at 24 h [40, 41]. In numerous studies, the molecular mechanisms of IR damage and the effects of neuroprotective agents on the infarct area were studied at 24 h, which is considered the most appropriate time point [19, 20, 27, 42–44]. Wang et al. studied the molecular mechanism of IR pathogenesis using RNA-Seq at 24 h after tMCAO in the hippocampus of rats. Those authors detected 182 DEGs, most of which were up-regulated [20]. A GO analysis showed that these DEGs were mainly associated with inflammation, stress, immune response, glucose metabolism, apoptosis [20]. Our analysis of gene expression under tMCAO conditions using RNA-Seq confirmed these results. However, we found that a > 10-fold greater number of DEGs were associated with a larger number of functional categories and signaling pathways at 24 h after tMCAO. We studied a region of the rat brain that was much larger and included not only the hippocampus but also other subcortical structures that were characterized apparently by a large variety of surviving cells in the peri-infarct region.
We found that 82 signaling pathways were associated with DEGs in IR24 vs. SH24 (see Additional file 20). Among them there were NF-kappa B, TNF, Pathways in cancer, Phagosome, HIF-1, VEGF, Chemokine and other signaling pathways. The significance of the inflammatory signaling pathways (Platelet activation, Focal adhesion, MAPK and others) was increased at 24 h compared with 4.5 h after tMCAO (Fig. 6b, c). In addition, the number of both up-regulated and down-regulated DEGs was increased. It should be noted that among the genes that were down-regulated at 24 h after tMCAO, we found genes involved in the regulation of transcription and the functioning of the neurosignaling pathway (glutamatergic synapse, cholinergic synapse, dopaminergic synapse, calcium signaling pathway and others) (see Additional files 12 and 17).
The analysis of differential gene expression in the rat brain sub-cortex in IR24 vs. IR4.5 and SH24 vs. SH4.5 led to the elucidation of the response of the transcriptome to the development of ischemic injury and to sham operation. It should be noted that the largest number of DEGs (2741) was associated with 26 signaling pathways in IR24 vs. IR4.5. In IR24 vs. IR4.5 we found a significant activation of the expression of genes involved in biosynthetic cell systems (ribosome, proteasome, DNA replication and purine metabolism functional categories), which was not detected in IR4.5 vs. SH4.5 and IR24 vs. SH24 (Fig. 6d, see Additional file 20). The effect obtained indicated a large-scale reorganization of nucleic acid and protein biosynthesis that is apparently related to the adaptive response of brain cells to the damage caused by IR. Concomitantly, between the time points of 4.5 to 24 h after tMCAO, an overwhelming effect of IR on the expression of hundreds of genes involved in the regulation of transcription, cell adhesion, differentiation, neurogenesis and the neurosignaling system was observed (see Additional files 7, 8, 12, 13, 17 and 18). Our results clearly showed that the down-regulation of neurosignaling activity was the consequence of the development of ischemia–reperfusion damage in the tMCAO model. This massive down-regulation of DEGs may be related to the death of cells in the necrotic region or reflect the response of cells of the penumbra to the IR.
The effect of sham operation on gene expression profile in SH24 vs. SH4.5 was highlighted by the differential expression of 752 genes, which included single genes involved in the stress response (Hspe1), neurogenesis and differentiation (Neurod6), in the response to inflammation (Cd14, Tlr7) and in several other processes (see Additional files 8, 10, 13 and 16). It should be noted that in SH24 vs. SH4.5 no reliable association was identified between the DEGs and any signaling pathway. However, about 400 common DEGs were found between the SH24 vs. SH4.5 and IR24 vs. IR4.5, at that the direction of their expression change coincided in both cases predominantly. Therefore, it is necessary to consider the contribution of the SH effect when analysing the pathological effect of IR on gene expression in the tMCAO model.
Conclusions
Thus, the study of the transcriptome profile of cells in the subcortical structures of the brain under tMCAO conditions led to the identification of DEGs that encode proteins that participate in various functional categories, biological processes and signaling pathways, via which brain cells respond to IR. Our results revealed the activation of a large number of genes involved in inflammation, the immune response, apoptosis and the stress response. Simultaneously, under IR, a massive down-regulation of genes that ensure the functioning of neurotransmitter systems was observed. The response of the transcriptome to SH in the condition of the tMCAO model detected here should be considered in studies of the genetic mechanisms underlying the regulation of the response of brain cells to the damaging effects of IR. Therefore, analysis of the transcriptome is one of the most important approaches in a comprehensive study of the effects of the development of ischemic damage. Undoubtedly, this will form the scientific basis of new medical technologies and the development of highly effective drugs to treat this condition.
Methods
Animals
White rats of the Wistar line (weight, 200–250 g) were obtained from the Experimental Radiology sector in A. Tsyb Medical Radiological Research Center, Obninsk, Russian Federation. The animals were maintained on a 12 h light/dark cycle at a temperature of 22–24 °C, with free access to food and water. The animals were divided into the “sham operation” (SH) and “ischemia–reperfusion” (IR) groups.
Transient cerebral ischemia rat model
The transient cerebral ischemia rat model was induced by endovascular occlusion of the right middle cerebral artery using a monofilament (Doccol Corporation, USA) for 90 min, and subsequent reperfusion for 3 or 22.5 h using the method of Koizumi et al., [45] with modifications. The rats were decapitated at 4.5 or 24 h after tMCAO (group “IR4.5” or “IR24”, respectively). Prior to the surgical procedure, rats were anesthetized using 3% isoflurane; the anaesthesia was maintained using 1.5–2% isoflurane and the EZ–7000 small animal anaesthesia system (E-Z Anesthesia, USA). The sham-operated rats (groups “SH4.5” and “SH24”, respectively) were subjected to a similar surgical procedure under anaesthesia (neck incision and separation of the bifurcation), but without tMCAO. Each experimental group consisted of at least fifteen animals. The subcortical structures were separated from the rat brain. Tissues were placed in RNAlater solution for 24 h at 0 °C and then stored at − 70 °C.
MRI
The MRI study of the characteristics and size of the ischemic injury of rat brains was carried out using small animal 7 T ClinScan tomograph (Bruker BioSpin, USA). The standard protocol included the following modes: DWI with mapping of the apparent diffusion coefficient for assessing acute ischemic damage, and T2 WI in the transversal plane. Magnetic resonance angiography (3D-TOF MRA) was used for visualization of the main arteries and control of the recanalization. Quantitative assessment of the volume of the infarction focus was performed using the ImageJ software package (Wayne Rasband, National Institute of Mental Health, Bethesda, MD, USA). MRI was performed immediately before decapitation in rats from the IR4.5 group. In rats from the IR24 group, MRI was performed twice: at 3 h after tMCAO and immediately before decapitation.
Histological examination of rat brains
Tissue samples of rat brains from each group (IR4.5, IR24, SH4.5 and SH24; n = 3–4) were immersed in Buin’s fluid for 24 h and washed with 70% ethanol. Tissue samples were dehydrated and embedded in Histomix® (BioVitrum, Russia). Tissue sectioning was performed with the orientation of two tissue blocks for subsequent excision into coronary sections at the level from − 4.0 to − 0.5 and from − 0.5 to + 5 mm from the bregma. Sections with a thickness of 5–6 μm obtained through 0.5–1 mm on a microtome (Leica RM2235, Germany) were stained with haematoxylin and eosin (BioVitrum) after dewaxing. Histological specimens were examined under a microscope (Leica DM 1000) with a micrograph to digital camera (Leica ICC50 HD). Morphological analysis was performed with allowance for normal and pathological central nervous system variants [46–48]. Stereotactic mapping of the damaged zones and accurate determination of the level of sections were performed according to an atlas of the rat brain.
RNA isolation
Total RNA from the subcortex, was isolated using TRIzol reagent (Invitrogen, Thermo Fisher Scientific) and acid guanidinium thiocyanate–phenol–chloroform extraction [49]. The isolated RNA was treated with deoxyribonuclease I (DNase I) (Thermo Fisher Scientific) in the presence of RiboLock ribonuclease (RNase) inhibitor (Thermo Fisher Scientific), according to the manufacturer’s recommended protocol. Deproteinization was performed using a 1:1 phenol:chloroform mixture. The isolated RNA was precipitated with sodium acetate (3.0 M, pH 5.2) and ethanol. RNA integrity was checked using capillary electrophoresis (Experion, BioRad, USA).
RNA-Seq
Total RNA isolated from the subcortical structures of the brain, including the lesion focus, was used in this experiment. The RNA-Seq experiment was conducted with the participation of ZAO Genoanalytika, Russia. For RNA-Seq, the polyA fraction of the total RNA was obtained using the oligoT magnetic beads of the Dynabeads® mRNA Purification Kit (Ambion, USA). cDNA (DNA complementary to RNA) libraries were prepared using the NEBNext® mRNA Library Prep Reagent Set (NEB, USA). The concentration of cDNA libraries was measured using Qbit 2.0 and the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, USA). The length distribution of library fragments was determined using the Agilent High Sensitivity DNA Kit (Agilent, USA). Sequencing was carried out using an Illumina HiSeq 1500 instrument. At least 10 million reads (1/50 nt) were generated.
RNA-Seq data analysis
Four variants of the comparisons of RNA-Seq results were used to identify DEGs under tMCAO model conditions. Two variants showed DEGs under ischemia–reperfusion after tMCAO at 4.5 h vs. sham operation at 4.5 h after surgical procedure (IR4.5 vs. SH4.5) and under ischemia–reperfusion after tMCAO at 24 h vs. sham operation at 24 h after surgical procedure (IR24 vs. SH24). In addition, there were DEGs. Two other variants identified DEGs under sham-operation conditions at 24 vs. 4.5 h after surgical procedure (SH24 vs. SH4.5) and under ischemia–reperfusion conditions at 24 vs. 4.5 h after tMCAO (IR24 vs. IR4.5). Each of the comparison groups (IR4.5, IR24, SH4.5 and SH24) included three animals. To obtain the most representative results, the RNA-Seq analysis was performed using brain samples of rats with a subcortical localization of the ischemic focus exclusively. All genes were annotated on the NCBI Reference Sequence database. The levels of gene expression were measured as fragments per kilobase per million reads using the Cuffdiff program. Only genes that exhibited changes in expression > 1.5-fold and had a P-values adjusted using the Benjamini–Hochberg procedure lower 0.05 (Padj < 0.05) were considered.
cDNA synthesis
cDNA synthesis was conducted in 20 μl of reaction mixture containing 2 mg of RNA using the reagents of a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) in accordance with the manufacturer’s instructions. Oligo (dT)18 primers were used to analyse mRNA.
Real-time RT–PCR
The 25 μl PCR mixture contained 2 μl of 0.2× reverse transcriptase reaction sample, forward and reverse primers (5 pmol each), 5 μl of 5× reaction mixture (Evrogen Joint Stock Company) including PCR buffer, Taq DNA polymerase, deoxyribonucleoside triphosphates (dNTP) and the intercalating dye SYBR Green I. Primers specific to the genes studied were selected using OLIGO Primer Analysis Software version 6.31 and were synthesized by the Evrogen Joint Stock Company (see Additional file 21). The amplification of cDNAs was performed using a StepOnePlus Real-Time PCR System (Applied Biosystems, USA) in the following mode: stage 1 (denaturation), 95 °C, 10 min; stage 2 (amplification with fluorescence measured), 95 °C, 1 min; 65 °C, 1 min; 72 °C, 1 min (40 cycles).
Data analysis of real-time RT–PCR and statistics
Two reference genes Gapdh and Rpl3 were used to normalize the cDNA samples [50]. Calculations were performed using BestKeeper, version 1 [51] and Relative Expression Software Tool (REST) 2005 software [52]. The manual at the site ‘REST.-gene-quantification.info’ (http://www.gene-quantification.de/rest-2009.html) was used to evaluate expression target genes relative to the expression levels of the reference genes. The values were calculated as EfCt(ref)/EfCt(tar), where Ef is the PCR efficiency, Ct(tar) is the average threshold cycle (Ct) of the target gene, Ct(ref) is the average Ct of the reference gene, and EfCt(ref) is the geometric average EfCt of the reference genes. PCR efficiencies were assessed using the amplification of a series of standard dilutions of cDNAs and computed using REST software [52]. The efficiency values for all PCR reactions were in the range 1.83 to 2.08 (see Additional file 21). At least 9 animals were included in each comparison group. When comparing data groups, statistically significant differences were considered with the probability p < 0.05. Additional calculations were performed using Microsoft Excel.
Functional analysis
Database for Annotation, Visualization and Integrated Discovery (DAVID v6.8, https://david.ncifcrf.gov/tools.jsp) [53] and The PANTHER database (Protein ANalysis THrough Evolutionary Relationships, http://pantherdb.org) [54] were used to annotate the functions of the differentially expressed genes. When comparing data groups, statistically significant differences were considered with the probability P < 0.05. To control the false discovery rate we used Benjamini–Hochberg procedure.
Additional files
Table S1. Characterization of ischemia at 4.5 after tMCAO using DWI mode of MRI. (XLSX 9 kb)
Table S2. DEGs that exhibited a reversed direction of expression in IR4.5 vs. SH4.5 and IR24 vs. SH24. (XLSX 13 kb)
Figure S1. RNA-Seq analysis of differentially expressed genes (DEGs) in tMCAO model conditions. (PPTX 2306 kb)
Table S4. Analysis of the functional categories of the proteins encoded by DEGs. (XLSX 21 kb)
Table S5. The RNA-Seq analysis of DEGs associated with the functional category “Adaptive immunity”. (XLSX 13 kb)
Table S6. The RNA-Seq analysis of DEGs associated with the functional category “Apoptosis”. (XLSX 23 kb)
Table S7. The RNA-Seq analysis of DEGs associated with the functional category “Cell adhesion”. (XLSX 23 kb)
Table S8. The RNA-Seq analysis of DEGs associated with the functional category “Differentiation”. (XLSX 28 kb)
Table S9. The RNA-Seq analysis of DEGs associated with the functional category “DNA replication”. (XLSX 15 kb)
Table S10. The RNA-Seq analysis of DEGs associated with the functional category “Inflammatory response”. (XLSX 14 kb)
Table S11. The RNA-Seq analysis of DEGs associated with the functional category “Innate immunity”. (XLSX 34 kb)
Table S12. The RNA-Seq analysis of DEGs associated with the functional category “Ion channel”. (XLSX 28 kb)
Table S13. The RNA-Seq analysis of DEGs associated with the functional category “Neurogenesis”. (XLSX 20 kb)
Table S14. The RNA-Seq analysis of DEGs associated with the functional category “Proteasome”. (XLSX 13 kb)
Table S15. The RNA-Seq analysis of DEGs associated with the functional category “Ribosomal protein”. (XLSX 23 kb)
Table S16. The RNA-Seq analysis of DEGs associated with the functional category “Stress response”. (XLSX 17 kb)
Table S17. The RNA-Seq analysis of DEGs associated with the functional category “Synapse”. (XLSX 29 kb)
Table S18. The RNA-Seq analysis of DEGs associated with the functional category “Transcription regulation”. (XLSX 49 kb)
Table S19. The RNA-Seq analysis of DEGs associated with the functional category “Translation regulation”. (XLSX 12 kb)
Table S20. Analysis of the signaling pathways associated with DEGs in the condition of the tMCAO model. (XLSX 19 kb)
Table S21. The characterization of the primers for real-time RT-PCR. (XLSX 12 kb)
Acknowledgments
The authors thank Dr. Vladimir P. Chekhonin, MD, PhD, Pirogov Russian National Research Medical University, Serbsky Federal Medical Research Centre of Psychiatry and Narcology, Moscow, Russia for help in organizing an MRI study. The authors thank Dr. Ekaterina V. Medvedeva from Institute for Regenerative Medicine, Sechenov First Moscow State Medical University for the help in conducting surgeries.
Funding
This work was supported by 16–14-00077 Grant of the Russian Science Foundation (RSF).
Availability of data and materials
All data supporting the conclusions of this article are included in this article and its Additional file (Additional file 1: Tables S1, Additional file 2: Tables S2, Additional file 4: Tables S4, Additional file 5: Tables S5, Additional file 6: Tables S6, Additional file 7: Tables S7, Additional file 8: Tables S8, Additional file 9: Tables S9, Additional file 10: Tables S10, Additional file 11: Tables S11, Additional file 12: Tables S12, Additional file 13: Tables S13, Additional file 14: Tables S14, Additional file 15: Tables S15, Additional file 16: Tables S16, Additional file 17: Tables S17, Additional file 18: Tables S18, Additional file 19: Tables S19, Additional file 20: Tables S20, Additional file 21: Tables S21; Additional file 3: Figure S1). RNA-sequencing data have been deposited in the Sequence Read Archive database under accession code SRP148632 (SAMN09235828-SAMN09235839), https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP148632.
Abbreviations
- 3D-TOF MRA
Magnetic resonance angiography
- cDNA
DNA Complementary to RNA
- Ct
Threshold cycle
- DEGs
Differentially expressed genes
- DNase
Deoxyribonuclease
- dNTP
Deoxyribonucleoside triphosphate
- DWI
Diffusion-weighted imaging
- Ef
The PCR efficiency
- HE
Haematoxylin and eosin
- IR
Ischemia–reperfusion conditions
- IR24
Ischemia–reperfusion at 24 h after tMCAO
- IR4.5
Ischemia–reperfusion at 4.5 h after tMCAO
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LQ
The lower quartiles for the 25 percentile interval
- Me
The median
- MRI
Magnetic resonance imaging
- nt
Nucleotide(s)
- Padj
p-values adjusted using the Benjamini–Hochberg procedure
- REST
Relative Expression Software Tool
- RNase
Ribonuclease
- RNA-Seq
High-throughput RNA sequencing
- RT–PCR
Reverse transcription polymerase chain reaction
- SH
Sham-operation conditions
- SH24
Sham-operation conditions at 24 h after surgical procedure
- SH4.5
Sham-operation conditions at 4.5 h after surgical procedure
- T2 WI
T2–weighted imaging
- tMCAO
Transient middle cerebral artery occlusion
- TTC
Triphenyl tetrazolium chloride
- UQ
The upper quartiles for the 75 percentile interval
Authors’ contributions
LVD and IBF analyzed the data and wrote the manuscript; LVD, SAL and LVG designed the experiments; IBF performed the bioinformatic analysis for transcriptomic data; VVS obtained the RNA from the tissue samples; IBF and VVS performed RT-PCR; AED performed the operations of experimental animals and MRI; VVY carried out the pathomorphological analysis of rat brain and conclusion on histological preparations; SAM carried out histological examination of rat brains; SAL coordinated the study and edited the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All manipulations with experimental animals were approved by the Animal Care Committee of the Pirogov Russian National Research Medical University (Approved ID: 15–2015, November 2, 2015) and were carried out in accordance with the Directive 2010/63/EU of the European Parliament and the Council of European Union on the protection of animals used for scientific purposes issued on September 22, 2010.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lyudmila V. Dergunova, Email: lvdergunova@mail.ru
Ivan B. Filippenkov, Email: filippenkov@img.ras.ru
Vasily V. Stavchansky, Email: bacbac@yandex.ru
Alina E. Denisova, Email: dalina543@gmail.com
Vadim V. Yuzhakov, Email: yuzh_vad@mail.ru
Sergey A. Mozerov, Email: mozerov@list.ru
Leonid V. Gubsky, Email: gubskii@mail.ru
Svetlana A. Limborska, Email: limbor@img.ras.ru
References
- 1.Kalaria RN, Ballard C. Stroke and cognition. Curr Atheroscler Rep. 2001;3:334–339. doi: 10.1007/s11883-001-0028-5. [DOI] [PubMed] [Google Scholar]
- 2.Seshadri S, Beiser A, Kelly-Hayes M, Kase CS, Au R, Kannel WB, et al. The lifetime risk of stroke: estimates from the Framingham study. Stroke. 2006;37:345–350. doi: 10.1161/01.STR.0000199613.38911.b2. [DOI] [PubMed] [Google Scholar]
- 3.Mukherjee D, Patil CG. Epidemiology and the global burden of stroke. World Neurosurg. 2011;76(6 Suppl):S85–S90. doi: 10.1016/j.wneu.2011.07.023. [DOI] [PubMed] [Google Scholar]
- 4.Hernandes MS, Lassègue B, Hilenski LL, Adams J, Gao N, Kuan C-Y, et al. Polymerase delta-interacting protein 2 deficiency protects against blood-brain barrier permeability in the ischemic brain. J Neuroinflammation. 2018;15:45. doi: 10.1186/s12974-017-1032-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, Liu H, Zhang L, Wang G, Zhang M, Yu Y. Neuroprotection of Dexmedetomidine against Cerebral Ischemia-Reperfusion Injury in Rats: Involved in Inhibition of NF-κB and Inflammation Response. Biomol Ther (Seoul) 2017;25:383–389. doi: 10.4062/biomolther.2015.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghazavi H, Hoseini SJ, Ebrahimzadeh-Bideskan A, Mashkani B, Mehri S, Ghorbani A, et al. Fibroblast growth factor type 1 (FGF1)-overexpressed adipose-derived Mesenchaymal stem cells (AD-MSCFGF1) induce neuroprotection and functional recovery in a rat stroke model. Stem Cell Rev. 2017;13:670–685. doi: 10.1007/s12015-017-9755-z. [DOI] [PubMed] [Google Scholar]
- 7.Kim Y, Kim YS, Kim HY, Noh M-Y, Kim JY, Lee Y-J, et al. Early treatment with poly(ADP-ribose) Polymerase-1 inhibitor (JPI-289) reduces infarct volume and improves long-term behavior in an animal model of ischemic stroke. Mol Neurobiol. 2018;55:7153–7163. doi: 10.1007/s12035-018-0910-6. [DOI] [PubMed] [Google Scholar]
- 8.Ismael S, Zhao L, Nasoohi S, Ishrat T. Inhibition of the NLRP3-inflammasome as a potential approach for neuroprotection after stroke. Sci Rep. 2018;8:5971. doi: 10.1038/s41598-018-24350-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berger C, Stauder A, Xia F, Sommer C, Schwab S. Neuroprotection and glutamate attenuation by acetylsalicylic acid in temporary but not in permanent cerebral ischemia. Exp Neurol. 2008;210:543–548. doi: 10.1016/j.expneurol.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 10.White BC, Sullivan JM, DeGracia DJ, O’Neil BJ, Neumar RW, Grossman LI, et al. Brain ischemia and reperfusion: molecular mechanisms of neuronal injury. J Neurol Sci. 2000;179(S 1–2):1–33. doi: 10.1016/S0022-510X(00)00386-5. [DOI] [PubMed] [Google Scholar]
- 11.Canazza A, Minati L, Boffano C, Parati E, Binks S. Experimental models of brain ischemia: a review of techniques, magnetic resonance imaging, and investigational cell-based therapies. Front Neurol. 2014:19. 10.3389/fneur.2014.00019. [DOI] [PMC free article] [PubMed]
- 12.Krafft PR, Bailey EL, Lekic T, Rolland WB, Altay O, Tang J, et al. Etiology of stroke and choice of models. Int J Stroke. 2012;7:398–406. doi: 10.1111/j.1747-4949.2012.00838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howells DW, Porritt MJ, Rewell SSJ, O’Collins V, Sena ES, van der Worp HB, et al. Different strokes for different folks: the rich diversity of animal models of focal cerebral ischemia. J Cereb Blood Flow Metab. 2010;30:1412–1431. doi: 10.1038/jcbfm.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nieswandt B, Kleinschnitz C, Stoll G. Ischaemic stroke: a thrombo-inflammatory disease? J Physiol. 2011;589:4115–4123. doi: 10.1113/jphysiol.2011.212886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryang Y-M, Dang J, Kipp M, Petersen K-U, Fahlenkamp AV, Gempt J, et al. Solulin reduces infarct volume and regulates gene-expression in transient middle cerebral artery occlusion in rats. BMC Neurosci. 2011;12:113. doi: 10.1186/1471-2202-12-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopes RD, Piccini JP, Hylek EM, Granger CB, Alexander JH. Antithrombotic therapy in atrial fibrillation: guidelines translated for the clinician. J Thromb Thrombolysis. 2008;26:167–174. doi: 10.1007/s11239-008-0272-4. [DOI] [PubMed] [Google Scholar]
- 17.Alexandrov AV, Hall CE, Labiche LA, Wojner AW, Grotta JC. Ischemic stunning of the brain: early recanalization without immediate clinical improvement in acute ischemic stroke. Stroke. 2004;35:449–452. doi: 10.1161/01.STR.0000113737.58014.B4. [DOI] [PubMed] [Google Scholar]
- 18.Nour M, Scalzo F, Liebeskind DS. Ischemia-reperfusion injury in stroke. Interv Neurol. 2013;1:185–199. doi: 10.1159/000353125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ford G, Xu Z, Gates A, Jiang J, Ford BD. Expression analysis systematic explorer (EASE) analysis reveals differential gene expression in permanent and transient focal stroke rat models. Brain Res. 2006;1071:226–236. doi: 10.1016/j.brainres.2005.11.090. [DOI] [PubMed] [Google Scholar]
- 20.Wang C, Liu M, Pan Y, Bai B, Chen J. Global gene expression profile of cerebral ischemia-reperfusion injury in rat MCAO model. Oncotarget. 2017;8:74607–74622. doi: 10.18632/oncotarget.20253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeGracia DJ. Regulation of mRNA following brain ischemia and reperfusion. Wiley Interdiscip Rev RNA. 2017;8 10.1002/wrna.1415. [DOI] [PubMed]
- 22.Xin Q, Cheng B, Pan Y, Liu H, Yang C, Chen J, et al. Neuroprotective effects of apelin-13 on experimental ischemic stroke through suppression of inflammation. Peptides. 2015;63:55–62. doi: 10.1016/j.peptides.2014.09.016. [DOI] [PubMed] [Google Scholar]
- 23.Lou Z, Wang A-P, Duan X-M, Hu G-H, Song G-L, Zuo M-L, et al. Upregulation of NOX2 and NOX4 mediated by TGF-β signaling pathway exacerbates cerebral ischemia/reperfusion oxidative stress injury. Cell Physiol Biochem. 2018;46:2103–2113. doi: 10.1159/000489450. [DOI] [PubMed] [Google Scholar]
- 24.Guo H, Adah D, James PB, Liu Q, Li G, Ahmadu P, et al. Xueshuantong injection (lyophilized) attenuates cerebral ischemia/reperfusion injury by the activation of Nrf2-VEGF pathway. Neurochem Res. 2018;43:1096–1103. doi: 10.1007/s11064-018-2523-x. [DOI] [PubMed] [Google Scholar]
- 25.McDonough A, Lee RV, Noor S, Lee C, Le T, Iorga M, et al. Ischemia/reperfusion induces interferon-stimulated gene expression in microglia. J Neurosci. 2017;37:8292–8308. doi: 10.1523/JNEUROSCI.0725-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fabian RH, Derry PJ, Rea HC, Dalmeida WV, Nilewski LG, Sikkema WKA, et al. Efficacy of novel carbon nanoparticle antioxidant therapy in a severe model of reversible middle cerebral artery stroke in acutely hyperglycemic rats. Front Neurol. 2018;9:199. doi: 10.3389/fneur.2018.00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krupka J, May F, Weimer T, Pragst I, Kleinschnitz C, Stoll G, et al. The coagulation factor XIIa inhibitor rHA-Infestin-4 improves outcome after cerebral ischemia/reperfusion injury in rats. PLoS One. 2016;11:e0146783. doi: 10.1371/journal.pone.0146783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu F, Schafer DP, McCullough LD. TTC, fluoro-jade B and NeuN staining confirm evolving phases of infarction induced by middle cerebral artery occlusion. J Neurosci Methods. 2009;179:1–8. doi: 10.1016/j.jneumeth.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Türeyen K, Vemuganti R, Sailor KA, Dempsey RJ. Infarct volume quantification in mouse focal cerebral ischemia: a comparison of triphenyltetrazolium chloride and cresyl violet staining techniques. J Neurosci Methods. 2004;139:203–207. doi: 10.1016/j.jneumeth.2004.04.029. [DOI] [PubMed] [Google Scholar]
- 30.Liu F, McCullough LD. Middle cerebral artery occlusion model in rodents: methods and potential pitfalls. J Biomed Biotechnol. 2011;2011:464701. doi: 10.1155/2011/464701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Z-W, Yang Q-D, Tang Q-P, Yang J, Guo R-J, Jiang W. Effect of willed movement training on neurorehabilitation after focal cerebral ischemia and on the neural plasticity-associated signaling pathway. Mol Med Rep. 2018;17:1173–1181. doi: 10.3892/mmr.2017.7964. [DOI] [PubMed] [Google Scholar]
- 32.Iadecola C, Ross ME. Molecular pathology of cerebral ischemia: delayed gene expression and strategies for neuroprotection. Ann N Y Acad Sci. 1997;835:203–217. doi: 10.1111/j.1749-6632.1997.tb48631.x. [DOI] [PubMed] [Google Scholar]
- 33.Takemoto O, Tomimoto H, Yanagihara T. Induction of c-fos and c-Jun gene products and heat shock protein after brief and prolonged cerebral ischemia in gerbils. Stroke. 1995;26:1639–1648. doi: 10.1161/01.STR.26.9.1639. [DOI] [PubMed] [Google Scholar]
- 34.Honkaniemi J, States BA, Weinstein PR, Espinoza J, Sharp FR. Expression of zinc finger immediate early genes in rat brain after permanent middle cerebral artery occlusion. J Cereb Blood Flow Metab. 1997;17:636–646. doi: 10.1097/00004647-199706000-00005. [DOI] [PubMed] [Google Scholar]
- 35.Abe K, Kawagoe J, Sato S, Sahara M, Kogure K. Induction of the “zinc finger” gene after transient focal ischemia in rat cerebral cortex. Neurosci Lett. 1991;123:248–250. doi: 10.1016/0304-3940(91)90942-M. [DOI] [PubMed] [Google Scholar]
- 36.Cox-Limpens KEM, Gavilanes AWD, Zimmermann LJI, Vles JSH. Endogenous brain protection: what the cerebral transcriptome teaches us. Brain Res. 2014;1564:85–100. doi: 10.1016/j.brainres.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 37.Rosenberg GA, Estrada EY, Dencoff JE. Matrix metalloproteinases and TIMPs are associated with blood-brain barrier opening after reperfusion in rat brain. Stroke. 1998;29:2189–2195. doi: 10.1161/01.STR.29.10.2189. [DOI] [PubMed] [Google Scholar]
- 38.Lochhead JJ, McCaffrey G, Quigley CE, Finch J, DeMarco KM, Nametz N, et al. Oxidative stress increases blood-brain barrier permeability and induces alterations in occludin during hypoxia-reoxygenation. J Cereb Blood Flow Metab. 2010;30:1625–1636. doi: 10.1038/jcbfm.2010.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ritter LS, Orozco JA, Coull BM, McDonagh PF, Rosenblum WI. Leukocyte accumulation and hemodynamic changes in the cerebral microcirculation during early reperfusion after stroke. Stroke. 2000;31:1153–1161. doi: 10.1161/01.STR.31.5.1153. [DOI] [PubMed] [Google Scholar]
- 40.van Lookeren Campagne M, Thomas GR, Thibodeaux H, Palmer JT, Williams SP, Lowe DG, et al. Secondary reduction in the apparent diffusion coefficient of water, increase in cerebral blood volume, and delayed neuronal death after middle cerebral artery occlusion and early reperfusion in the rat. J Cereb Blood Flow Metab. 1999;19:1354–1364. doi: 10.1097/00004647-199912000-00009. [DOI] [PubMed] [Google Scholar]
- 41.Hoehn M, Nicolay K, Franke C, van der Sanden B. Application of magnetic resonance to animal models of cerebral ischemia. J Magn Reson Imaging. 2001;14:491–509. doi: 10.1002/jmri.1213. [DOI] [PubMed] [Google Scholar]
- 42.Liu G, Song J, Guo Y, Wang T, Zhou Z. Astragalus injection protects cerebral ischemic injury by inhibiting neuronal apoptosis and the expression of JNK3 after cerebral ischemia reperfusion in rats. Behav Brain Funct. 2013;9:36. doi: 10.1186/1744-9081-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu K, Wu Y, Hu Y, Zhang Q, Xie H, Liu G, et al. Neuroprotective effects of prior exposure to enriched environment on cerebral ischemia/reperfusion injury in rats: the possible molecular mechanism. Brain Res. 2013;1538:93–103. doi: 10.1016/j.brainres.2013.09.036. [DOI] [PubMed] [Google Scholar]
- 44.Jover-Mengual T, Castelló-Ruiz M, Burguete MC, Jorques M, López-Morales MA, Aliena-Valero A, et al. Molecular mechanisms mediating the neuroprotective role of the selective estrogen receptor modulator, bazedoxifene, in acute ischemic stroke: a comparative study with 17β-estradiol. J Steroid Biochem Mol Biol. 2017;171:296–304. doi: 10.1016/j.jsbmb.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 45.Koizumi J, Yoshida Y, Nakazawa T, Ooneda G. Experimental studies of ischemic brain edema. Nosotchu. 1986;8:1–8. doi: 10.3995/jstroke.8.1. [DOI] [Google Scholar]
- 46.Li Y, Powers C, Jiang N, Chopp M. Intact, injured, necrotic and apoptotic cells after focal cerebral ischemia in the rat. J Neurol Sci. 1998;156:119–132. doi: 10.1016/S0022-510X(98)00036-7. [DOI] [PubMed] [Google Scholar]
- 47.Lipton P. Ischemic cell death in brain neurons. Physiol Rev. 1999;79:1431–1568. doi: 10.1152/physrev.1999.79.4.1431. [DOI] [PubMed] [Google Scholar]
- 48.Garcia JH, Liu KF, Ho KL. Neuronal necrosis after middle cerebral artery occlusion in Wistar rats progresses at different time intervals in the caudoputamen and the cortex. Stroke. 1995;26:636–642. doi: 10.1161/01.STR.26.4.636. [DOI] [PubMed] [Google Scholar]
- 49.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- 50.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 51.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 53.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 54.Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, et al. PANTHER version 11: expanded annotation data from gene ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Characterization of ischemia at 4.5 after tMCAO using DWI mode of MRI. (XLSX 9 kb)
Table S2. DEGs that exhibited a reversed direction of expression in IR4.5 vs. SH4.5 and IR24 vs. SH24. (XLSX 13 kb)
Figure S1. RNA-Seq analysis of differentially expressed genes (DEGs) in tMCAO model conditions. (PPTX 2306 kb)
Table S4. Analysis of the functional categories of the proteins encoded by DEGs. (XLSX 21 kb)
Table S5. The RNA-Seq analysis of DEGs associated with the functional category “Adaptive immunity”. (XLSX 13 kb)
Table S6. The RNA-Seq analysis of DEGs associated with the functional category “Apoptosis”. (XLSX 23 kb)
Table S7. The RNA-Seq analysis of DEGs associated with the functional category “Cell adhesion”. (XLSX 23 kb)
Table S8. The RNA-Seq analysis of DEGs associated with the functional category “Differentiation”. (XLSX 28 kb)
Table S9. The RNA-Seq analysis of DEGs associated with the functional category “DNA replication”. (XLSX 15 kb)
Table S10. The RNA-Seq analysis of DEGs associated with the functional category “Inflammatory response”. (XLSX 14 kb)
Table S11. The RNA-Seq analysis of DEGs associated with the functional category “Innate immunity”. (XLSX 34 kb)
Table S12. The RNA-Seq analysis of DEGs associated with the functional category “Ion channel”. (XLSX 28 kb)
Table S13. The RNA-Seq analysis of DEGs associated with the functional category “Neurogenesis”. (XLSX 20 kb)
Table S14. The RNA-Seq analysis of DEGs associated with the functional category “Proteasome”. (XLSX 13 kb)
Table S15. The RNA-Seq analysis of DEGs associated with the functional category “Ribosomal protein”. (XLSX 23 kb)
Table S16. The RNA-Seq analysis of DEGs associated with the functional category “Stress response”. (XLSX 17 kb)
Table S17. The RNA-Seq analysis of DEGs associated with the functional category “Synapse”. (XLSX 29 kb)
Table S18. The RNA-Seq analysis of DEGs associated with the functional category “Transcription regulation”. (XLSX 49 kb)
Table S19. The RNA-Seq analysis of DEGs associated with the functional category “Translation regulation”. (XLSX 12 kb)
Table S20. Analysis of the signaling pathways associated with DEGs in the condition of the tMCAO model. (XLSX 19 kb)
Table S21. The characterization of the primers for real-time RT-PCR. (XLSX 12 kb)
Data Availability Statement
All data supporting the conclusions of this article are included in this article and its Additional file (Additional file 1: Tables S1, Additional file 2: Tables S2, Additional file 4: Tables S4, Additional file 5: Tables S5, Additional file 6: Tables S6, Additional file 7: Tables S7, Additional file 8: Tables S8, Additional file 9: Tables S9, Additional file 10: Tables S10, Additional file 11: Tables S11, Additional file 12: Tables S12, Additional file 13: Tables S13, Additional file 14: Tables S14, Additional file 15: Tables S15, Additional file 16: Tables S16, Additional file 17: Tables S17, Additional file 18: Tables S18, Additional file 19: Tables S19, Additional file 20: Tables S20, Additional file 21: Tables S21; Additional file 3: Figure S1). RNA-sequencing data have been deposited in the Sequence Read Archive database under accession code SRP148632 (SAMN09235828-SAMN09235839), https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP148632.